Abstract
Unlike stable RNAs that require processing for maturation, prokaryotic cellular mRNAs generally follow an ‘all-or-none’ pattern. Herein, we used a 5΄ monophosphate transcript sequencing (5΄P-seq) that specifically captured the 5΄-end of processed transcripts and mapped the genome-wide RNA processing sites (PSSs) in a methanogenic archaeon. Following statistical analysis and stringent filtration, we identified 1429 PSSs, among which 23.5% and 5.4% were located in 5΄ untranslated region (uPSS) and intergenic region (iPSS), respectively. A predominant uridine downstream PSSs served as a processing signature. Remarkably, 5΄P-seq detected overrepresented uPSS and iPSS in the polycistronic operons encoding ribosomal proteins, and the majority upstream and proximal ribosome binding sites, suggesting a regulatory role of processing on translation initiation. The processed transcripts showed increased stability and translation efficiency. Particularly, processing within the tricistronic transcript of rplA-rplJ-rplL enhanced the translation of rplL, which can provide a driving force for the 1:4 stoichiometry of L10 to L12 in the ribosome. Growth-associated mRNA processing intensities were also correlated with the cellular ribosomal protein levels, thereby suggesting that mRNA processing is involved in tuning growth-dependent ribosome synthesis. In conclusion, our findings suggest that mRNA processing-mediated post-transcriptional regulation is a potential mechanism of ribosomal protein synthesis and stoichiometry.
INTRODUCTION
Messenger RNA (mRNA) decay and sequence- or structure-dependent translation efficiency are well known mechanisms involved in post-transcriptional regulation in prokaryotes (1–5). However, unlike the extensive processing and maturation of stable RNAs (e.g. ribosomal RNA and transfer RNA) (6,7), prokaryotic cellular mRNAs are generally considered as following an ‘all-or-none’ pattern (i.e. no processing and maturation but decay) except for the intergenic regions (IGRs) of polycistronic transcripts that encode proteins/enzymes of the same metabolic pathway (8,9). The IGRs of such polycistronic transcripts could be processed by an endonucleolysis-based selective RNA processing and stabilization (SRPS) mechanism to allow differential expression of individual genes in the same operon to achieve correct stoichiometry for function (10). SRPS has been reported for a number of bacterial operons such as rpsU-danG-rpoD and glmU-glmS in Escherichia coli; and cggR-gapA, ilvBHC-leuABCD and infC-rpml-rplT in Bacillus subtilis (11–15). However, unlike IGR processing, SRPS occurring in the 5΄ untranslated region (UTR) has been sporadically reported in some biologically important prokaryotic transcripts, including E. coli rpoS, tisAB, dinQ, B. subtilis thrS, Clostridium perfringens colA, Streptococcus pyogenes ska and Staphylococcus aureus cspA (16–22). An earlier study in a methanogenic archaeon Methanocaldococcus jannaschii also showed evidence of mRNA processing upstream of the corresponding translation start codon (23). Hence, SRPS may be an underappreciated mechanism in prokaryotic post-transcriptional regulation, and requires further analysis via genome-wide studies on its prevalence and function.
Genome-wide identification of RNA processing sites has been made possible by the development of the next-generation sequencing technology (24). Using this technology as platform, global studies on RNA processing have been reported in a few bacterial species and one archaeon (25–29). In these studies, RNA processing sites in the maturation step of stable RNA were experimentally validated. However, the regulatory roles of mRNA processing were rarely studied. Using differential RNA-seq (dRNA-seq), Xu et al. reported that the IGR processing within the cip-cel operon can be involved in tuning the stoichiometry of the hetero-multiprotein complex—cellulosome (28). These studies, although limited in scope, suggested that SRPS is widespread in both bacteria and archaea, and may play an important role in post-transcriptional regulation.
In a previous study, we obtained a genome-wide transcription start site (TSS) map for a psychrophilic archaeon Methanolobus psychrophilus R15 (30). Furthermore, we found that 51% of the mRNAs harbored long 5΄ UTRs (>50 nt), a high percentage of which also contained processing site-like sequences. However, whether these putative processing sites are actually utilized, and the biological function of these processing sites remain unknown. In the present study, we used 5΄ monophosphate transcript sequencing (5΄P-seq), modified from the selective strategy of dRNA-seq (31,32), to specifically determine the 5΄ ends of processed transcripts. By applying statistical analysis and stringent filtration, we obtained a genome-wide processing site (PSS) map of M. psychrophilus R15. Additional biochemical analyses with a selected set of ribosomal gene operons suggested that the 5΄ UTR PSS (uPSS) may play an important role in ribosomal protein translation, and the IGR PSS (iPSS) can be involved in fine-tuning stoichiometry of ribosomal proteins.
MATERIALS AND METHODS
Strain growth and RNA extraction
Methanolobus psychrophilus R15 was grown at 18°C or 8°C in a defined mineral medium containing 20 mM trimethylamine and under a gas phase of 80:20 N2:CO2 as described elsewhere (30). Methanococcus maripaludis S2 was grown in a rich medium reduced with 3 mM cysteine (33). Escherichia coli strains (DH5α, JM109, and BL21) were grown at 37°C in Luria-Bertani (LB) medium supplemented with 100 μg/ml of ampicillin. For RNA extraction, R15 cells at mid-log phase (an OD600 of 0.4) were harvested, and the total RNA was extracted and purified using TRIzol™ reagent (Invitrogen) for 5΄P-seq, primer extension, and Northern blot analysis. RNA purity and integrity were assessed using a NanoPhotometer spectrophotometer (Implen) and a RNA 6000 Nano Assay Kit on an Agilent Bioanalyzer 2100 system (Agilent Technologies).
Construction of 5΄-end cDNA library for 5΄ monophosphate transcript sequencing (5΄P-seq)
The prokaryotic cellular RNA pools consist of primary transcripts of carrying a 5΄ triphosphate (5΄PPP), and processed transcripts with 5΄ monophosphate (5΄P). To identify genome-wide RNA processing sites (PSSs) in M. psychrophilus R15, we constructed an mRNA library that specifically enriched 5΄P RNA based on the principle of dRNA-seq library construction (30–32) (Supplementary Figure S1A). Approximately 5 μg of total RNA was used for construction of 5΄P-seq library. Briefly, after rRNA depletion using a Ribo-Zero™ rRNA Removal Kit (Epicentre), RNAs were directly ligated to the 5΄ RNA adapter without any enzymatic pretreatment. Next, a cDNA library was constructed using an RNA-seq Library Preparation Kit for Transcriptome Discovery (Gnomegen). The amplified DNA was size-fractioned to a size range of 300–600 nt on agarose gels, and purified using a QIAquick Gel Extraction Kit (Qiagen). Sequencing was performed on an Illumina HiSeq 2000 system with 100-bp paired-end reads. After quality control processing of raw data, filtered reads were aligned to the M. psychrophilus R15 reference genome using Bowtie (34). Sequence alignment was performed with default parameters except for allowing three mismatches.
RNA processing site calling and statistics
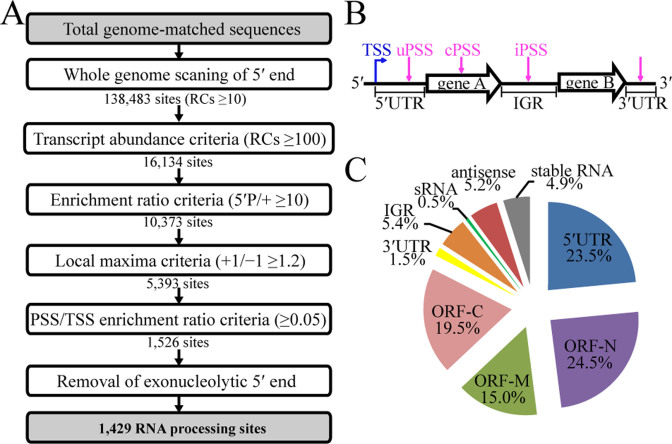
Stringent criteria were used to ensure a high level of confidence for the identification of 5΄P sites. As shown in Figure 1, read counts (RCs) of the sequencing reads whose first base aligned to the position in the R15 genome were calculated. A total of 138 483 sites showed RCs of ≥10 in the 5΄P-seq library. Next, this criterion was raised to RCs of ≥100 in consideration of transcript abundance. Based on the principle of dRNA-seq method, 5΄P sites would not be enriched in the (+) library because of enzymatic digestion. Therefore, the third criterion was ≥10 enrichment ratio of a site in 5΄P-seq to dRNA-seq (+) library. To ascertain the enrichment of a 5΄P site, the fourth criterion was that a position represents the local maxima within a 2-nt window, and the RC of +1/−1 ≥ 1.2 will define a processing site by its higher RCs than the upstream site. Through these four filtration criteria, a total of 5393 5΄P sites were identified. To remove possible degradative 5΄P transcripts, RCs of a given site were compared with the corresponding TSS in the dRNA-seq (−) library, and the processing site/transcription start site (PSS/TSS) ratio criterion was set. Using a 0.05 threshold of PSS/TSS, 3,937 5΄P sites were eliminated, including 97 exonucleolytic 5΄P sites in the 5΄ UTR that were located at ≤6 nt to the corresponding TSSs (Supplementary Figure S2). The final dataset with high confidence contained a total of 1429 5΄P sites including 1359 mRNA and 70 stable RNA processing sites.
Figure 1.
Determination of the processing sites in the transcriptome of M. psychrophilus R15. (A) Flowchart for the filtration steps in identification of the authentic RNA processing sites (PSSs). On the basis of statistics described in Materials and Methods, the criterion for each filtration is indicated. RC, read count; 5΄P/+, count ratio of a site in (5΄P) to (+) library; +1/−1, count ratio of PSS to that of 1 nt upstream; PSS/TSS, count ratio of a PSS to the corresponding TSS in (−) library. (B) A diagram for PSS definition. uPSS, cPSS and iPSS represent the processing sites in the 5΄ UTR, coding region and IGR, respectively. Blue and magenta arrows indicate TSS and PSS, respectively. (C) A pie chart showing distribution of the identified PSS in the transcriptome. ORF-N, -M and -C specify the PSS location at the N-terminal, middle, and C-terminal region, respectively, of the encoding sequence.
Primer extension
Primer extension was used to validate the 5΄P-seq determined processing sites and to determine RNA stabilities as previously described (30). Briefly, a DNA fragment located downstream and proximal to the start codon of a gene was used as a specific primer (Supplementary Table S1) for reverse transcription and in sequencing the PCR product. Primers were radiolabeled at the 5΄ end using T4 polynucleotide kinase (Thermo Scientific) and [γ-32P]ATP (PerkinElmer) and incubated at 37°C for 1 h. cDNA was synthesized at 55°C for 1 h using SuperScript III reverse transcriptase (Invitrogen). Sequencing ladders were generated by using a Thermo Sequenase™ Cycle Sequencing Kit (USB). The products of reverse transcription and DNA sequencing reactions were separated on a 6% polyacrylamide sequencing gel with 7 M urea, and visualized by autoradiography on an X-ray film.
Northern blot
Northern blot was employed to assay the integrity and stability of the processed transcripts. For small RNA molecules, RNA was denatured for 10 min at 65°C in loading buffer containing 95% (v/v) formamide, and 2–5 μg was loaded to each lane of a 4% or 6% polyacrylamide gel with 7.6 M urea. The gel was run in 0.5 × TBE buffer. For large RNA molecules, RNA was denatured and 10 μg was loaded in each lane of a 1.2% agarose gel with formamide and MOPS buffer. The agarose gel was run in 1 × MOPS buffer. A low range and regular single-stranded RNA (ssRNA) ladder (New England Biolabs) served as a size marker on denaturing polyacrylamide-urea gels and on denaturing agarose gels, respectively. After separation, RNAs were transferred onto Hybond-N+ membranes (GE Healthcare) by electroblotting or capillary blotting and crosslinked to the membrane using UV. Membranes were prehybridized at 42°C, followed by hybridization with 2–10 pmol of biotin-labeled DNA probes for 12 h. After three rounds of washing for 10 min each in 1 ×, 0.2 × and 0.1 × SSC–0.1% SDS solutions, signals were visualized using a chemiluminescent nucleic acid detection module (Thermo Scientific) according to the manufacturer's protocol.
In vivo RNA-decay assay
Actinomycin D (MP Biomedicals) at a final concentration of 100 μg/ml was added to the mid-exponential culture to inhibit transcription. Cells were then collected after 0, 5, 10, 15 and 30 min post-treatment, and total RNA was extracted. For mono- and poly-cistronic operons, Northern blot and primer extension assays, respectively, were used to determine RNA stability as described above. The quantity of transcript at each time point was calculated based on band intensity using the Quantity One software (Bio-Rad). RNA decay rate was determined by the residual RNA at a given time point over that at time 0. RNA half-life was calculated based on the exponential regression curve as described (35), where the abundance of the initial transcript decreased to 50%.
lacZ reporter gene construction
To assess the effect of 5΄ UTR or IGR on translation efficiency of the downstream genes, a lacZ reporter was translationally fused to the entire downstream open reading frame (ORF). For large genes like the thermosome (Mpsy_2247), the N-terminal 30 amino acids were fused with the lacZ reporter. The reporter fusions were constructed by overlapping PCR or Gibson assembly method using a NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). All DNA fragments were amplified individually by high-fidelity PCR with KOD DNA polymerase (Toyobo). The resulting fusion DNA fragment was cloned into pET-23b at the BglII and XhoI sites and transformed into E. coli JM109 (DE3). lacZ expression of each fusion was visualized by streaking the reporter strain on an agarose plate containing X-gal/IPTG.
β-Galactosidase assay
A β-galactosidase assay kit (Beyotime) was used to quantify β-galactosidase activity in each of lacZ reporter strain of E. coli. Briefly, an overnight culture was 100-fold diluted with fresh LB liquid and grown at 37°C. IPTG was added to a final concentration of 0.5 mM when the cultures reach an OD600 of 0.4–0.6. After 1- or 3-h induction, 500 μl of culture was used for β-galactosidase activity assay using the standard method following manufacturer's recommended protocol. The same amount of culture sampled before induction was used as control. β-Galactosidase activity was expressed in Miller units (36).
Protein overexpression and purification
For purification of ribosomal proteins RP-L10, RP-L12 and RP-S15, C-terminal 6 × His-Tag fusions were constructed. The genes of rplJ, rplL and rpsO were PCR amplified and cloned via NdeI/HindIII or NdeI/XhoI sites into pET-23b. Escherichia coli BL21(DE3)pLysS cells carrying the corresponding plasmid were cultured until an OD600 of 0.6 was reached. Expression was induced with 1 mM IPTG, and cells were harvested by centrifugation (10 min, 5000g) after 3 h induction at 37°C, or after 16 h induction at 22°C. The pellet was resuspended in binding buffer [20 mM Tris–HCl (pH 8.0), 500 mM NaCl, 20 mM imidazole. and 5% (w/v) glycerol] and lysed by sonication. The ribosomal proteins were purified from the cell lysates by immobilized metal ion affinity chromatography using a HisTrap HP column (GE Healthcare). Bound proteins were eluted with a linear gradient of 20 mM to 500 mM imidazole. The proteins were then loaded onto a HiPrep 26/10 desalting column (GE Healthcare) to remove imidazole, and eluted with a desalting buffer [20 mM Tris–HCl (pH 8.0), 500 mM NaCl and 5% (w/v) glycerol]. Purified proteins were analyzed on 12% SDS-PAGE, and protein concentration was determined by using a Pierce™ BCA protein assay kit (Thermo Scientific).
Western blot
After resuspending in lysis buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 10 (w/v) glycerol, 0.1% (w/v) SDS, 0.1% (v/v) Triton X-100], cells of M. psychrophilus were lysed by sonication. Lysates were precipitated at 10 000g for 1 min at 4°C. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Western blot was performed using polyclonal rabbit antiserum raised against purified archaeal RP-L10, RP-L12 and RP-S15 (MBL). The RP-L10 and L12 antibodies were used at a 1:20 000 dilution, and the RP-S15 antibody was at a 1:50 000 dilution. For detection, a horseradish peroxidase-linked secondary conjugate was used at a 1:5000 dilution, and reactive bands were visualized by using an Amersham ECL Prime Western blotting detection reagent (GE Healthcare). Quantitation was performed using Quantity One (Bio-Rad).
In silico analysis of RNA motif and structure
RNA sequences around the processing sites were used for the motif search. Putative processing motifs were identified by using the MEME suite (37). A sequence logo was generated by WebLogo (38). RNA secondary structures were predicted using the Mfold software under 18°C or 37°C (39). Secondary structure with minimum free energy for folding (ΔG) was used in further statistical analysis of the location of the processing sites. The secondary structure of the RNA molecules was created by Rnaviz2.0.3 (40) based on the Mfold ct files.
RESULTS
Genome-wide analysis of RNA processing sites in a methanogenic archaeon
To gain insight into RNA processing in the methanogenic archaeon M. psychrophilus R15, we adapted a well-established differential RNA sequencing (dRNA-seq) technology to specifically enrich RNAs with a 5΄ monophosphate, named 5΄P-seq (Supplementary Figure S1A, see Materials and Methods). We first examined the utility of this technology by accurately detecting the processing sites of all 53 known tRNA species in R15 (Supplementary Table S2). The 5΄P-seq captured each mature 5΄-end of the 53 tRNAs and the splice sites of four intron-containing tRNAs. For example, the 5΄P library highly enriched the 5΄ splice site of pre-tRNATrp intron (Supplementary Figure S1B), and this 121-nt intron contained a sR3 C/D box sRNA (41). Using Northern blot analysis, we detected the excised intron, sR3 C/D box sRNA, which was a full-length intron and not subject to further processing (Supplementary Figure S1B). Taken together, these results confirmed that the 5΄P-seq method efficiently captured the processing sites of pre-tRNA maturation, thus can be used for genome-wide mRNA processing sites analysis.
To distinguish processing from decay of RNA, as both will generate 5΄ monophosphate in a RNA molecule, we devised stringent exclusion criteria for the raw 5΄P-seq data based on statistical analysis, which is shown in a flowchart shown in Figure 1A and described in detail in Materials and Methods. Particularly, 97 5΄P sites located within 6-nt downstream of the TSSs had ≤0.35 PSS/TSS ratios (Supplementary Figure S2); these were likely the exonucleolytic decay intermediates, therefore were excluded. Finally, a total of 1429 5΄P sites, upon removal of exonucleolytic 5΄P sites, were defined as the processing sites in the transcriptome of M. psychrophilus R15 (Supplementary Table S3). Among which, 23.5%, 5.4% and 59% occurred within the 5΄ UTR (uPSS), IGR (iPSS) and CDS (cPSS), respectively (Figure 1B and C).
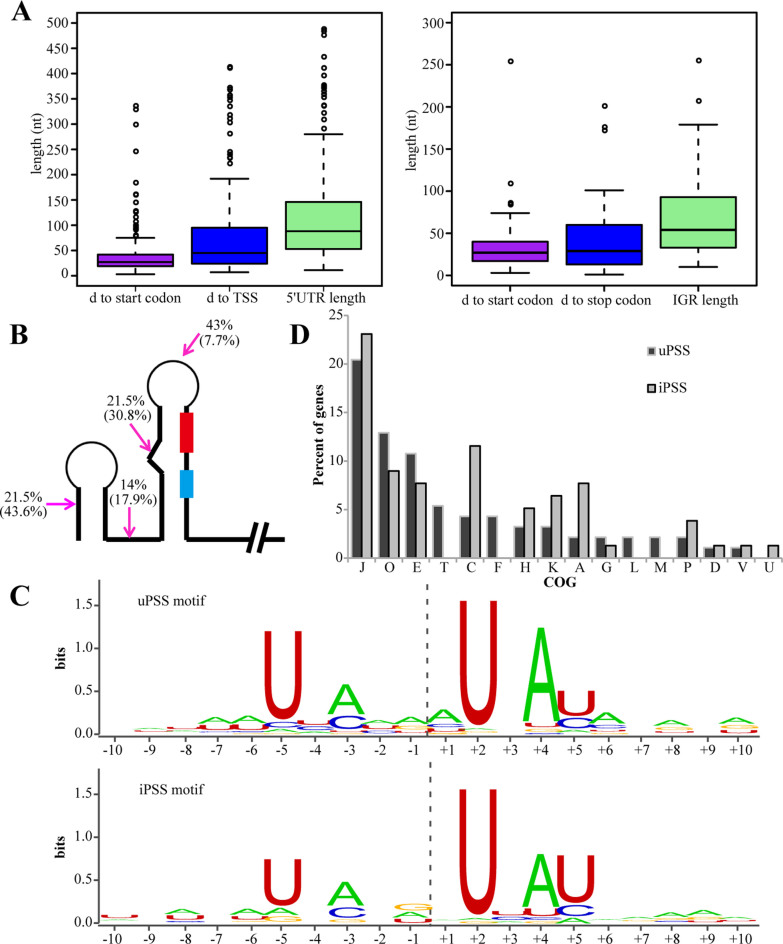
As RNA processing at the 5΄ UTR and IGR is more likely to exert biological effects on the resulting transcript, we conducted detailed analysis of those uPSS and iPSS. As shown in Figure 2A, uPSSs (total: 197) and iPSSs (total: 50) were overrepresented in relatively longer 5΄ UTRs (median length: 88 nt) and IGRs (median length: 54 nt), and most were located at 20–30 nucleotides upstream of the start codon. This indicated that RNA processing frequently occurred proximal and upstream of the ribosome binding site (RBS) and start codon, suggesting a regulatory function in translation initiation. To gain further insight into these PSSs, we chose the most abundant PSS from each transcript for in silico structure and sequence motif analysis. Of the 93 uPSSs and 39 iPSSs analyzed, most (78.5%) of uPSSs were located in single-strand regions, whereas approximately half (43.6%) of the iPSSs were located within RNA duplexes (Figure 2B), suggesting that they were likely the cleavage products of different families of ribonucleases. MEME program searching identified an A/U-rich sequence motif around both uPSSs and iPSSs, with uridine (U) occurring at the highest frequencies at nucleotides +2, −5 and +5 relative to the PSS (Figure 2C).
Figure 2.
Characteristic patterns of the processing sites and functional enrichment of PSS-associated genes in R15. (A) Boxplots show distance (d) distribution of uPSSs to their TSS/start codon and the 5΄ UTR lengths (left), and iPSSs to the downstream start codon/upstream stop codon and the IGR length (right). (B) A diagram shows the location distribution percentage of uPSS in 5΄ UTR and iPSS within IGR shown inside the parentheses. PSS (magenta arrow), RBS (red box) and translation start codon (light blue box) are shown. (C) The processing motif of uPSS and iPSS based on alignment of sequence around processing sites. Positions on the x-axis are relative to the PSS. (D) Distribution of uPSS- and iPSS-associated genes amongst different COGs. Note that the majority of uPSS- and iPSS-associated genes belong to four COGs: J, O, E and C.
To understand the biological significance of mRNA processing, genes that were associated with the above mentioned 93 uPSSs and 39 iPSSs were examined for functional categories as defined by Cluster of Orthologous Groups (COG). As shown in Figure 2D and Supplementary Table S4, most of the genes associated with uPSS (26.4%) and iPSS (29%) belonged to COG J (translation and ribosome biogenesis), followed by COG O (protein turnover and chaperones), COG E (amino acid transport and metabolism), and COG C (energy production and conservation). Interestingly, genes in COG T (signal transduction) appeared to contain only uPSS, while genes belonging to COG A (RNA processing and modification) predominantly contained iPSS. A similar COG distribution was found for 197 uPSS- and 50 iPSS-associated genes (Supplementary Figure S3). Such a non-random distribution of uPSS and iPSS among different functional groups of genes suggests a regulatory role of these uPSSs and iPSSs in gene expression.
Confirmation of the 5΄P-seq data by biochemical analyses
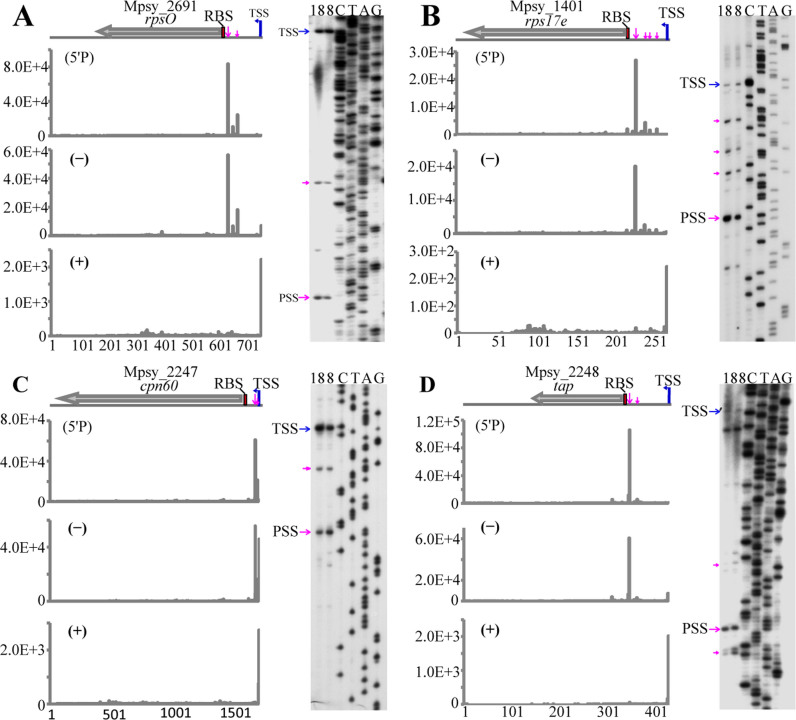
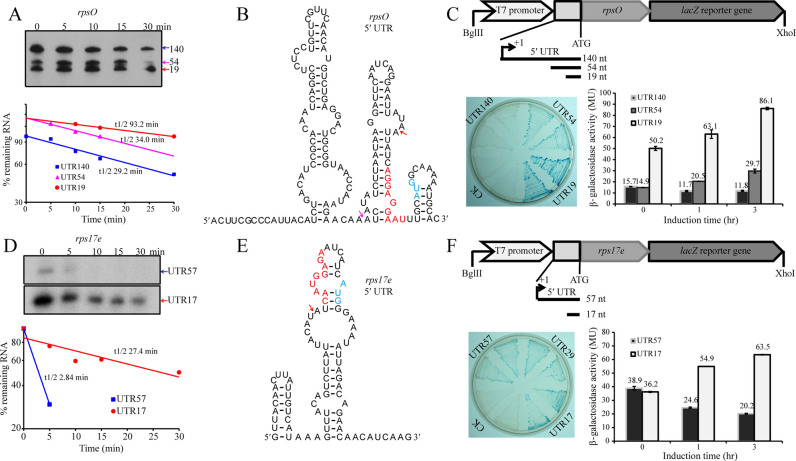
To ascertain that the uPSSs and iPSSs identified by our 5΄P-seq approach were accurate, primer extension was performed on monocistronic and polycistronic mRNAs with highly abundant PSSs (Figures 3 and 4). To assess whether PSS occurred under different growth conditions, RNA was isolated from cells grown at 18°C (optimal growth) and 8°C (cold adapted). As shown in Figure 3, primer extension revealed all transcripts processed from uPSS that were identified in our 5΄P-seq in operons encoding ribosomal proteins [rpsO (Mpsy_2691, Figure 3A) and rps17e (Mpsy_1401, Figure 3B)] and thermosome protein [cpn60 (Mpsy_2247, Figure 3C) and tap (thermosome-associated protein, Mpsy_2248, Figure 3D)]. For example, two PSSs were captured in the 5΄ UTR of rpsO in both 5΄P-seq library and dRNA-seq (−) library (Figure 3A, left panel). Primer extension detected the primary transcript (140-nt 5΄ UTR) and two processed transcripts (54-nt and 19-nt 5΄ UTR) with the same 5΄-ends as identified in the 5΄P-seq (Figure 3A, right panel). Likewise, one major and three minor processed transcripts identified by 5΄P-seq in the rps17e operon were also demonstrated by primer extension. It is worth noting that signals for the processed transcripts of rps17e and tap were much stronger than their corresponding primary transcripts (compare TSS with PSS in Figure 3B and D) under both conditions, indicating that processed RNA was the dominant species for those two transcripts. Primer extension also verified uPSSs in the transcripts of ribosomal protein genes rps8e, rps24e, and rplK (Supplementary Figure S4).
Figure 3.
Primer extension analysis of uPSS-mediated transcripts of ribosomal protein genes rpsO (A) and rps17e (B), and thermosome-related genes cpn60 (C) and tap (D). Blue and magenta arrows indicate TSSs and PSSs, respectively. RBSs are shadowed by red box. Sequencing reads of the TSS in (+) library, both TSS and PSS in (−) library and PSS only in (5΄P) library are shown in the left panels. The y-axis scale represents the read count of a given site. Primer extension gels are shown in the right panels. The numbers 18 and 8 on the gel top indicate RNA isolated from R15 cultures grown at 18°C and 8°C, respectively.
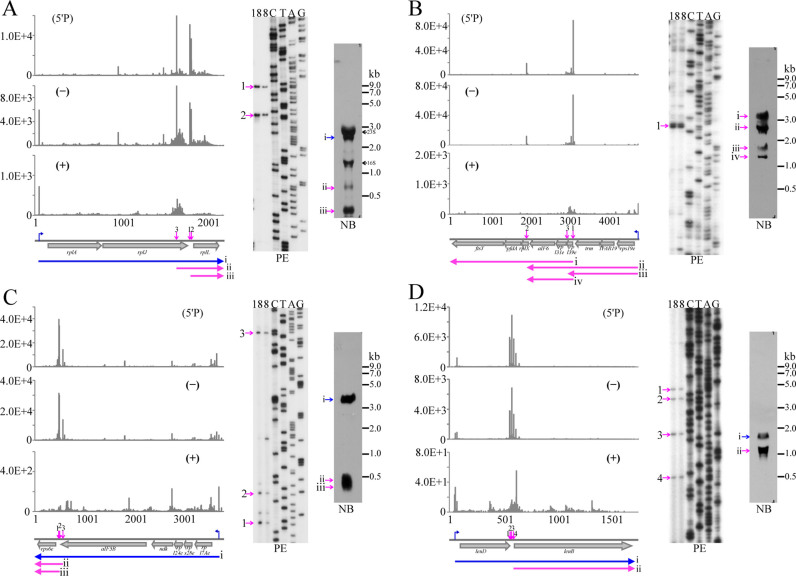
Figure 4.
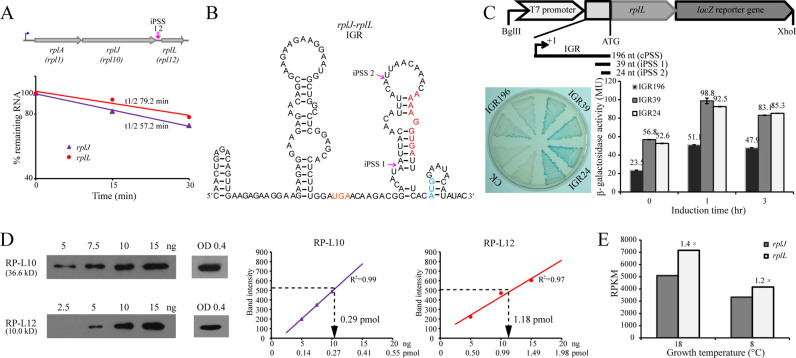
Primer extension and Northern blot analyses of iPSS in the rplA (A), rps19e (B), rpl7Ae (C) and leuD (D) operons. Sequencing reads of the TSS in (+) library, both TSS and PSS in (−) library and PSS only in (5΄P) library are shown in the left panels, primer extension gels are shown in the middle panels, and Northern blot gels are shown in the right panels. Magenta arrows indicate PSSs, and blue arrows indicate TSSs. iPSSs on primer extension gels are indicated as the same Arabic numerals as those in the operon diagram. Roman numerals on Northern blot gels indicate the transcripts with the expected length according to TSS and PSS. The numbers 18 and 8 on the gel top indicate RNA isolated from R15 cultures grown at 18°C and 8°C, respectively. The 23S and 16S rRNA bands in (A) were due to cross hybridization with the probe used.
Using the same approach, iPSSs within three polycistronic operons that encode ribosomal protein and amino acid biosynthesis genes were also verified in R15 cells (Figure 4). In addition, Northern blot analyses using downstream genes as probes were employed to further confirm the existence and relative abundance of the primary and processed transcripts. As predicted from the operon sequence and revealed by 5΄P-seq, both primer extension and Northern blot analyses showed the presence of two iPSS products in rplA operon (Figure 4A). The relatively long distance from the primer binding site to the 5΄ end of the mRNA precluded the detection of the primary transcript by primer extension; however, Northern blot was capable of detecting this transcript (Figure 4A, right panel). Nonetheless, primer extension mapped the processing sites in the other operons as well (Figure 4C and D), except for rps19e operon where the location of the primer (within the rpl39e gene) allowed mapping only processing site 1 (Figure 4B, left and middle panels). It is also important to note that due to low resolution of the agarose gels used in Northern blot analysis, products from processing sites 1–3 in Figure 4C and 1–4 in Figure 4D could not be separated.
Processing at the 5΄ UTRs stabilizes transcripts and activates translation
The methanogen R15 genome contains 59 ribosomal protein genes, which are located on four monocistronic and 16 polycistronic operons (42). Our previous work revealed that most of the ribosomal operons in R15 possess large 5΄ UTRs with an average length of 154 nt (30), a feature shared by some other prokaryotic ribosomal operons. Our primer extension data shown in Figure 3 demonstrated higher signal intensity for the processed RNA at uPSS than the primary transcript, especially for transcripts of the rps17e operon (Figure 3B). These results suggested that the processed RNAs were more stable than the full length transcript, and/or they were translated more efficiently, which may contribute to the stabilizing effect.
To test these two possibilities, we first measured RNA stability of transcripts from the two ribosomal operons rpsO and rps17e. By using 100 μg/ml of actinomycin D to inhibit new transcription in the exponential phase culture of R15, steady-state RNA was isolated and measured by Northern blot (Figure 5A and D, upper panel) at 5 min intervals up to 30 min post actinomycin D addition. A linear curve was constructed by plotting the percentage of residual RNA over time (Figure 5A and D, lower panel), and the half-life of each transcript was then calculated. Our results demonstrated a significant increase in half-life for the processed transcripts, especially for the rps17e UTR17 transcript, which showed a nearly 10-fold increase in half-life than the primary UTR57 transcript (27.4 min versus 2.8 min, Figure 5D), while the half-life of the processed rpsO transcript was >3-fold longer than the primary transcript (93.2 min versus 29.2 min, Figure 5A). The half-life of the transcripts from the thermosome gene cpn60 and tap was also measured, and similar results were obtained (Supplementary Figure S5A and D).
Figure 5.
5΄ UTR processing stabilizes the transcript of the ribosomal protein gene rpsO and activates its translation. (A, D) Northern blot or primer extension probes the stabilities of PSS-mediated mRNA isoforms of rpsO and rps17e in R15 cells. The 5΄ UTR lengths for primary and processed mRNA isoforms are shown to the right of the gel (upper panel). Half-lives are calculated from the regression curve of mRNA remnant (lower panel). (B, E) 5΄ UTR secondary structures of rpsO and rps17e transcripts predicted by Mfold. Red and light blue letters show RBS and translation start codon, respectively. Magenta and red arrows indicate the PSSs. (C, F) Translation efficiencies of mRNA isoforms of rpsO and rps17e with different lengths of 5΄ UTR by processing. A schematic diagram shows the construction of lacZ translational fusion (upper panel). A LB plate containing X-gal visualizes the expression of lacZ (left), and a histogram shows the β-galactosidase activities upon IPTG induction (right). The β-galactosidase activities are the mean determination of three reporter strains, and standard deviations are shown.
To determine whether RNA processing also affected translation of the downstream gene because of the vicinity of PSS to RBS (Figure 5B and E), we used lacZ translational fusions to test the translation efficiency of rpsO and rps17e in an E. coli host. The lacZ gene was fused in-frame to the entire ORF of rpsO or rps17e with upstream fragments reflecting the primary and processed transcripts of the two operons (Figure 5C and F, upper panels). Expressions of lacZ in these constructs were visualized on plates and quantified by standard β-galactosidase assays (Figure 5C and F, lower panels). In general, lacZ fusions containing upstream regions equivalent to processed transcripts expressed higher levels of β-galactosidase activity than those containing the equivalent of a full-length transcript. The highest increase was observed in the fusion to the equivalent of the UTR19 transcript of rpsO (Figure 5C). lacZ translational fusion was also performed with the thermosome gene cpn60 and tap, and similar results were obtained (Supplementary Figure S5). It is noteworthy that IPTG even did not induce translation of the primary transcript of rpsO (Figure 5C, UTR140), implying that RBS embedding prevents ribosome access so translation (Figure 5B). This was also supported by that 5΄ UTR processing did not increase the translation of processed transcripts of rps24e and rplK due to innate RBS exposure in their primary transcripts (Supplementary Figure S6). Taken together, our results suggest that RNA processing at the 5΄ UTR of ribosomal and thermosome genes can increase RNA stability and/or translation efficiency.
IGR processing fine-tunes the stoichiometric ratio of the ribosome subunits
A typical bacterial ribosome is composed of ∼60 protein subunits with a single copy of each except L12, which usually exists as four copies (43). Since the gene encoding L12 (rplL) is co-transcribed with genes encoding L1 (rplA) and L10 (rplJ), it is important to address how the bacterium ensures 4-fold more L12 than L1 and L10. Our results described above demonstrated that RNA processing at the 5΄ UTR stabilized the processed transcripts and/or enhanced translation by unmasking the RBS (Figure 5B and C). Based on these results we hypothesized that RNA processing at the polycistronic RNA transcript may play an important role in stoichiometry among ribosomal proteins by differentially affecting the stability or translatability of different resulting transcripts. To test this, we measured the RNA stability and translation efficiency of genes in the rplA operon, as well as the abundance of L10 and L12.
We have shown in Figure 4A that processing at the IGR between rplJ (encodes L10) and rplL (encodes L12) produced a monocistronic rplL and a dicistronic rplA-rplJ. Using genome-wide mRNA stability sequencing (unpublished data), we first determined that the RNA half-life of the two transcripts was 79.2 min for rplL and 57.2 min for rplA-rplJ (Figure 6A). Using Mfold, the putative secondary structure of the primary transcript was predicted (Figure 6B), which suggested that processing at the two PSSs could unmask the RBS for the rplL transcript, thus increasing its translatability. To test this, lacZ translational fusion was done with the primary and the two processed rplL transcripts (39- and 24-nt IGR) (Figure 6C), and LacZ activity was measured. lacZ fused to the two processed transcripts displayed about 2-fold higher β-galactosidase activity than fusion with the primary transcript (Figure 6C), suggesting that processing at the IGR also increased the translation efficiency of rplL. Because the lacZ fusions were done in E. coli, which may not accurately reflect the situation in M. psychrophilus R15, we used Western blot analysis to determine the expression level of L10 and L12 in exponential culture of R15. Using known amounts of L10 and L12 proteins and specific antibodies to generate standard curves, the expression of the two proteins was estimated to be 0.29 and 1.18 pmol, respectively. This ratio, L12/L10 of 4:1, was similar as reported in the bacterial ribosome (Figure 6D). It is important to note that the in vivo mRNA abundance of rplL is slightly higher than rplJ (Figure 6E), which could not be the cause of proportional protein synthesis. These findings indicate that transcript processing may be one of the driving forces for the proportional translation of ribosomal proteins, thereby fine-tuning the stoichiometry.
Figure 6.
IGR processing within the tricistronic operon rplA-rplJ-rplL enhances rplL translation and tunes the stoichiometric ratio of L10 and L12. (A) A diagram shows gene organization of the tricistronic operon rplA-rplJ-rplL with two iPSSs within the IGR of rplJ-rplL (upper panel), and the half-lives of the two transcripts calculated from an unpublished transcriptomic data (lower panel). (B) Mfold predicts the secondary structures within the IGR of rplJ-rplL dicistronic transcript. Red and light blue letters show RBS and translation start codon, respectively. Magenta arrows indicate the PSSs. (C) Translation efficiencies of iPSSs-mediated mRNA isoforms of rplL. Interpretations for the diagrams and X-gal plate are the same as for Figure 5C. (D) Western blot analyses determine the protein ratio of L12 to L10 to be 4:1. Purified proteins L12 and L10 are loaded with the indicated contents on SDS-PAGE gel, and by using the antibodies protein content calibrations are determined for L12 and L10, respectively (left). The cellular L12 and L10 levels (ng) are determined at an OD600 of 0.4, and the protein molecules are calculated based on the calibrations (right). (E) mRNA abundance of rplJ and rplL in R15 at different temperatures. The fold changes of rplL to rplJ are shown above the histogram of rplL.
5΄UTR and IGR processing of the ribosomal transcripts are associated with growth-related ribosome biosynthesis
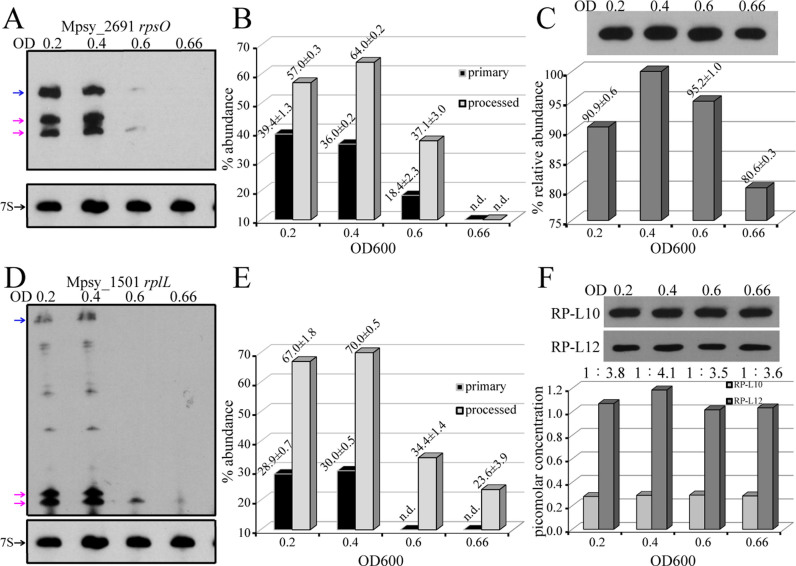
Given that bacterial ribosome synthesis is correlated with growth rates and phases (44,45), we investigated the correlation between growth-associated RNA processing and ribosomal protein synthesis in consideration of translational activation by processing. The monocistronic rpsO and the tricistronic operon rplA-rplJ-rplL were selected as the models for the 5΄ UTR and IGR processing, respectively. Total RNA and cellular protein were collected during the growth of methanogen R15 (Supplementary Figure S7). For rpsO (encodes S15), Northern blot probed the maximal processed transcript, particularly the rpsO UTR54 and UTR19 transcripts, at the mid-exponential phase (OD600: 0.4), and both the primary and processed mRNAs showed rapid decreases in abundance at the late-exponential stage (OD600: 0.6) (Figure 7A and B). Specifically, none of the primary and processed transcripts of rpsO were detectable at the stationary phase (OD600: 0.66, Figure 7A). Consistently, Western blot detected a relatively higher abundance of the S15 protein at the mid-exponential phase, but a reduced level at the stationary phase (Figure 7C).
Figure 7.
Growth-associated mRNA processing and abundance of ribosomal proteins. (A) Northern blot detects the primary and processed transcripts of rpsO during growth. OD600 0.2, 0.4, 0.6, and 0.66 correspond respectively to the early, middle, late exponential, and stationary growth phases referenced in Supplementary Figure S7. Primary (blue arrow) and processed (magenta arrows) transcripts are indicated and 7S RNA serves as internal standard. (B) Histogram of transcripts level of rpsO primary and processed isoforms. (C) The cellular protein level of S15 shown as Western blot bands (upper panel) and quantitative analysis displayed by histogram (lower panel). (D) Northern blot probe of the iPSS-mediated processed transcript of rplL and the tricistronic primary transcript (left). (E) Histogram of transcripts level of rplL primary and processed isoforms. (F) Western blot displaying the protein expression level of L10 and L12 during growth (upper panel) and quantitative analysis of the stoichiometric ratio of L10:L12 (lower panel).
For rplA-rplJ-rplL transcripts, Northern blot detected a higher level of the processed rplL transcript started in the earlier exponential culture (OD600: 0.2), whereas the level of the primary tricistronic transcript was constantly low (Figure 7D and E), indicating that IGR processing in the ribosomal polycistronic operon occurred when growth was initiated. At the late exponential and stationary phases, only the processed rplL transcripts were traceable (Figure 7D). On the other hand, the two processed rplL transcripts were the active translational form with 2-fold increase in translation efficiency than the primary transcript (Figure 6C). Therefore, the processed rplL transcripts acted as the main translation template for protein synthesis. Western blot determined a similar protein content profile for L12 as the processing intensity, and reached the maxima at OD600 of 0.4 (Figure 7F). To investigate whether the L12:L10 stoichiometry changed along the growth phase, we further detected the protein expression level of L10. Unlike L12, L10 appeared to be constitutively translated during growth (Figure 7F). Based on the calibration curve in Figure 6D, we then calculated the molar concentration of L12 and L10. Remarkably, although the RNA processing intensity was compatible with the growth rate, a 4:1 molecular ratio for L12 and L10 was maintained during the growth (Figure 7F, lower panel).
It is worth noting that 5΄ UTR or IGR processing of the ribosomal transcripts was onset as long as growth was initiated (OD600: 0.2), and reached the maxima at the mid-exponential growth. The ribosomal protein abundance followed a similar pattern in terms of growth, which was retained until the stationary phase mostly because proteins are more stable than mRNAs. Altogether, these findings revealed a RNA processing-mediated post-transcriptional regulatory mode in coordinated and proportional archaeal ribosomal protein synthesis.
DISCUSSION
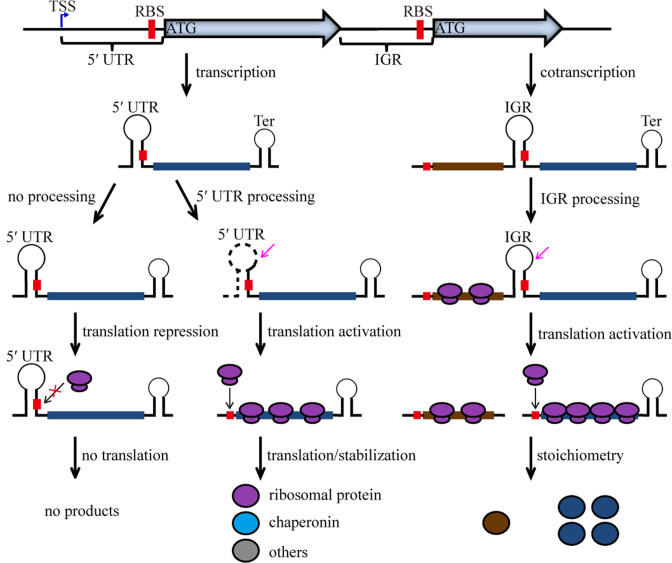
Although genome-wide RNA processing was recently found in few bacterial species, its prevalence and biological significance remain unclear. In the present work, through 5΄P-seq and in vivo and in vitro molecular approaches, we revealed the genome-wide mRNA processing in a methanogenic archaeon, and further determined that mRNA processing is involved in the regulation of ribosomal protein translation and stoichiometry. A proposed working model is shown in Figure 8, which indicates that genes or operons encoding methanogenic archaeal ribosomal and housekeeping proteins generally possess long 5΄ UTRs or IGRs; thus, complex RNA secondary structures would be formed at the 5΄ end of the transcripts. Meanwhile, RNA processing removes the bulk structures and liberates ribosome binding sites. Processing at either the 5΄ UTRs or the IGRs enhances translation initiation of the downstream gene, and the IGR processing also harnesses the stoichiometry of the hetero-multiprotein complex such as ribosome. More intensive translation also stabilizes the processed transcripts. Growth demand on ribosomes and other housekeeping proteins initiates transcript processing-modulated translation. Through data collection of genome-wide TSS and processing site of some bacteria, we predict that RNA processing occurs widely in prokaryotes and could be significantly used in post-transcriptional regulation.
Figure 8.
A schematic model of RNA processing involved in the post-transcriptional regulation of the methanogenic archaeal ribosomal protein synthesis and stoichiometry. The methanogenic archaeal genes or operons that encode ribosomal and other housekeeping proteins frequently possess a long 5΄ UTR or IGR that form complex RNA structures. RNA processing removes the bulk structure and liberates the RBS. Processing of the 5΄ UTRs enhances translation initiation, while processing of the IGRs would tune the proportional translation of the co-transcribed transcripts, thus harness the stoichiometry of the hetero-multiprotein complex like ribosome. Efficient translation also stabilizes the processed transcripts. The blue and magenta arrows specify TSS and PSS, respectively. Red box indicates RBS and Ter represents transcription terminator.
5΄UTR/IGR processing activates coordinated gene expression in Archaea
It is generally accepted that gene expression is regulated at three levels: transcription, translation and protein turnover. Although the effects of transcription and protein degradation indeed help to tune gene expression, it is important to note that the product yield for a given mRNA is strictly governed by the rate of translation initiation (46). The sequence surrounding translation start codon, in particular the stronger structure, strongly affects the translation efficiency of an mRNA, thus the protein level (47,48). Similarly, Sharp and Bechhofer found that a 5΄-terminal secondary structure and the RBS strength and localization were related to mRNA stability (49). Similarly, we found that in Archaea, both 5΄-terminal secondary structure and exposure of RBS control the translation efficiency, while this is controlled by mRNA processing event. For instance, processing site 1 at the 5΄ UTR of rpsO removes a stem-loop structure which elevates about 3-fold translation efficiency but without mRNA stability fluctuation; while processing site 2 liberates RBS and increases about 5–7-fold translation efficiency concomitant with 3-fold increase in mRNA stability (Figure 5). Albeit an arbitrary estimation of the processing-mediated translation efficiency may be obtained from a bacterial system used in the present work, the protein level of S15 in the M. psychrophilus cells was consistent with the processing intensity (Figure 7). It is most likely that the bacterial ribosomes recognize the archaeal RBS as the high sequence similarity of the 3΄ termini of 16S rRNA that bind to RBS in the two types of prokaryotes.
RNA processing within the IGR of polycistronic operons has been reported for physically uncoupling of transcripts, which in turn may result in variation in the transcript lifespans and translation efficiencies (10), and further to tune the proportional expression of the multiprotein cellulosome in C. cellulolyticum (28). Through in silico analysis, differential translation initiation is predicted as the main determinant for the observed uneven stoichiometry among proteins in various bacterial complexes such as L12 occurs as 4–6 copies, but the other subunits present as a single copy for ribosome biogenesis (50). Furthermore, ribosomal profiling confirmed that translation initiation controls hierarchical expression, which regulates the stoichiometric ratio of the subunits in multiprotein complexes, such as four copies of L12 per ribosome (51). However, the determinants that control the translation initiation rates remain unclear. This work discloses that IGR processing mediates the uncoupling of rplL from the tricistronic transcript of rplA-rplJ-rplL, facilitates rplL translation, and reveals RNA processing being one of the possible determinants for the proportional synthesis of the ribosomal proteins. Comparably, different stoichiometry of subunits in the atp operon (encoding F0F1 ATP synthase complex) of E. coli was attributable to differential translational control, and to a lesser extent, to control of segmental difference in mRNA stability (52). In contrast to the translation uncoupling of rplJ-rplL, translation coupling seems to play an important role in permitting the subunit stoichiometry of ATP synthase. Ribosomal proteins, transcription/translation factors, and chaperone genes are all among the predicted highly expressed (PHX) genes in prokaryotes through biased codon usages (53). Consistently, the processing event in M. psychrophilus is predominantly associated with genes encoding ribosomal proteins and thermosome (Figure 2D and Supplementary Table S4). Given that the processing level is positively correlated with growth rate and the cellular protein level, we conclude that mRNA processing plays a vital role for translation, especially in PHX genes.
RNA processing is a novel regulatory mechanism for ribosomal protein synthesis in archaea and bacteria
Autoregulation of ribosomal protein synthesis has been reported in bacteria. Dean and Nomura first reported that some ribosomal proteins selectively inhibit the synthesis of ribosomal protein genes located in the same operon (54). Recently, Fu et al. indicated that 72% of the ribosomal proteins of E. coli were controlled by 12 distinct RNA intrinsic regulatory elements within the ribosomal protein transcripts, thus set a RNA-based regulatory mechanism of ribosomal protein biosynthesis in bacteria (55). Through 5΄P-seq and molecular methods, the present study has revealed a different post-transcriptional regulatory mechanism for methanogenic archaeal ribosomal protein synthesis. This mechanism might be used by another methanogen, M. maripaludis, wherein the processed transcripts of rpsO (MMP1579) and nusG-rplK (MMP1434-MMP1433) were also probed (data not shown). It is highly possible that other archaea whose mRNAs possess long 5΄ UTRs or IGRs utilize a similar processing pattern, like Methanosarcina mazei, whose mRNAs consist of long 5΄ UTRs with an average length of 150–200 nt (32). Uniquely, the 5΄ UTR length of ribosomal protein genes in these species are exceptionally long (Supplementary Figure S8), making them the suitable RNA processing targets. RNA processing can happen and be involved in the modulation of bacterial ribosomal protein synthesis as well. Lioliou et al. reported that the majority 5΄ UTRs of ribosomal protein mRNAs in S. aureus were among the transcripts bound by RNase III, an endonuclease that specifically cleaves double-stranded RNA (22). A more recent study also showed that RNase E-mediated processing sites enriched at 5΄ UTR of ribosomal protein genes in Salmonella enterica (56). Further quantitative analysis for the cleavage sites and TSS would clarify the nature of cleavage (i.e. processing or decay).
An evolutionary conserved uridine ruler-and-cut mechanism may sustain the regulatory RNA processing
Ribonucleases that initiate archaeal transcript processing have yet to be elucidated. This is mainly attributed to the rare archaeal ribonucleases that are valid for biological function, especially in their cellular mRNA targets and precise processing sites (57,58). We hypothesized that processing at 5΄ UTR/IGR is an endoribonucleolytic action based on two observations. First, >50% processing events occurred at single-strand RNA region (Figure 2B). Second, the consensus motif of processing sites in M. psychrophilus is ‘UNMND↓NUNAY’ (Figure 2C). In contrast, we could not enrich an exonucleolytic motif (Supplementary Figure S2C). Besides, the only structural trait of exonucleolysis was the stem-loop after the processing sites (Supplementary Figure S2D). Our unpublished work indicates that the archaeal cleavage and polyadenylation specificity factor 1 (aCPSF1), which belongs to the conserved archaeal β-CASP ribonucleases (59), exhibits dual in vitro endo- and exonucleolytic activities and most likely implements this particular function, as knockdown of the gene increased the total RNA half-life in M. maripaludis.
Most recently, Chao et al. reported the processing sites of RNase E in S. enteric and also determined that uridine was the most conserved +2 base in the 5 nt core motif of ‘RN↓WUU’ (56). Furthermore, mutating U+2 to a disfavored G strongly diminished both the in vitro and in vivo processing intensity of RNase E, indicating that U+2 is crucial for RNase E cleavage. Strikingly, the processing motif in M. psychrophilus of ‘UNMND↓NUNAY’ also has a preference for uridine at position +2 (Figure 2C). This suggests that the predicted endoribonuclease in M. psychrophilus also uses a U+2 ruler-and-cut mechanism to mediate specific cleavage. Moreover, the human kinase homology endoribonuclease RNase L also recognizes an identity nucleotide (U) at the −2 position (60). This establishes the uridine ruler-and-cut mechanism as an evolutionarily conserved hallmark for different types of endoribonucleases.
In summary, this work has revealed the genome-wide mRNA processing sites in a methanogenic archaeon using 5΄P-seq, thereby paving a new avenue for exploiting the significance of post-transcriptional regulation of biological processes and also the cleavage mechanism of sequence-specific endoribonucleases. Remarkably, mRNA processing at the 5΄ UTRs and IGRs of the polycistronic operons that encode ribosomal proteins stabilizes transcripts and enhances translation. Therefore, mRNA processing at the noncoding regions exerts a regulatory role in ribosomal protein synthesis and in establishment of the stoichiometric ratio of the different subunits. This work provides a new post-transcriptional regulatory mode in ribosome biogenesis in Archaea, and possibly Bacteria.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tong Sun and Huanmin Liu for experimental support. We also thank Beijing Novogene Bioinformatics Technology Co., Ltd. for their work in sequencing. All members of the Anaerobes Research Group are acknowledged for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31430001, 31670049, 31100085]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kaberdin V.R., Blasi U.. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 2006; 30:967–979. [DOI] [PubMed] [Google Scholar]
- 2. Geissmann T., Marzi S., Romby P.. The role of mRNA structure in translational control in bacteria. RNA Biol. 2009; 6:153–160. [DOI] [PubMed] [Google Scholar]
- 3. Picard F., Dressaire C., Girbal L., Cocaign-Bousquet M.. Examination of post-transcriptional regulations in prokaryotes by integrative biology. C. R. Biol. 2009; 332:958–973. [DOI] [PubMed] [Google Scholar]
- 4. Condon C., Bechhofer D.H.. Regulated RNA stability in the Gram positives. Curr. Opin. Microbiol. 2011; 14:148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Evguenieva-Hackenberg E., Klug G.. New aspects of RNA processing in prokaryotes. Curr. Opin. Microbiol. 2011; 14:587–592. [DOI] [PubMed] [Google Scholar]
- 6. Deutscher M.P. Maturation and degradation of ribosomal RNA in bacteria. Prog. Mol. Biol. Transl. Sci. 2009; 85:369–391. [DOI] [PubMed] [Google Scholar]
- 7. Redko Y., de la Sierra-Gallay I.L., Condon C.. When all's zed and done: the structure and function of RNase Z in prokaryotes. Nat. Rev. Microbiol. 2007; 5:278–286. [DOI] [PubMed] [Google Scholar]
- 8. Bechhofer D.H. Messenger RNA decay and maturation in Bacillus subtilis. Prog. Mol. Biol. Transl. Sci. 2009; 85:231–273. [DOI] [PubMed] [Google Scholar]
- 9. Laalami S., Zig L., Putzer H.. Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 2014; 71:1799–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rochat T., Bouloc P., Repoila F.. Gene expression control by selective RNA processing and stabilization in bacteria. FEMS Microbiol. Lett. 2013; 344:104–113. [DOI] [PubMed] [Google Scholar]
- 11. Yajnik V., Godson G.N.. Selective decay of Escherichia coli dnaG messenger RNA is initiated by RNase E. J. Biol. Chem. 1993; 268:13253–13260. [PubMed] [Google Scholar]
- 12. Urban J.H., Vogel J.. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008; 6:631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ludwig H., Homuth G., Schmalisch M., Dyka F.M., Hecker M., Stulke J.. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 2001; 41:409–422. [DOI] [PubMed] [Google Scholar]
- 14. Mader U., Hennig S., Hecker M., Homuth G.. Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes. J. Bacteriol. 2004; 186:2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bruscella P., Shahbabian K., Laalami S., Putzer H.. RNase Y is responsible for uncoupling the expression of translation factor IF3 from that of the ribosomal proteins L35 and L20 in Bacillus subtilis. Mol. Microbiol. 2011; 81:1526–1541. [DOI] [PubMed] [Google Scholar]
- 16. Resch A., Afonyushkin T., Lombo T.B., McDowall K.J., Blasi U., Kaberdin V.R.. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5΄-leader. RNA. 2008; 14:454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vogel J., Argaman L., Wagner E.G.H., Altuvia S.. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004; 14:2271–2276. [DOI] [PubMed] [Google Scholar]
- 18. Kristiansen K.I., Weel-Sneve R., Booth J.A., Bjoras M.. Mutually exclusive RNA secondary structures regulate translation initiation of DinQ in Escherichia coli. RNA. 2016; 22:1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Condon C., Putzer H., Grunberg-Manago M.. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:6992–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Obana N., Shirahama Y., Abe K., Nakamura K.. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5΄ leader sequence. Mol. Microbiol. 2010; 77:1416–1428. [DOI] [PubMed] [Google Scholar]
- 21. Ramirez-Pena E., Trevino J., Liu Z.Y., Perez N., Sumby P.. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol. Microbiol. 2010; 78:1332–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lioliou E., Sharma C.M., Caldelari I., Helfer A.C., Fechter P., Vandenesch F., Vogel J., Romby P.. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012; 8:e1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J., Olsen G.J.. Messenger RNA processing in Methanocaldococcus (Methanococcus) jannaschii. RNA. 2009; 15:1909–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Güell M., Yus E., Lluch-Senar M., Serrano L.. Bacterial transcriptomics: what is beyond the RNA horiz-ome. Nat. Rev. Microbiol. 2011; 9:658–669. [DOI] [PubMed] [Google Scholar]
- 25. Linder P., Lemeille S., Redder P.. Transcriptome-wide analyses of 5΄-ends in RNase J mutants of a gram-positive pathogen reveal a role in RNA maturation, regulation and degradation. PLoS Genet. 2014; 10:e1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romero A.D., Hasan A.H., Lin Y.F., Kime L., Ruiz-Larrabeiti O., Urem M., Bucca G., Mamanova L., Laing E.E., van Wezel G.P. et al. A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing. Mol. Microbiol. 2014; 94:963–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Innocenti N., Golumbeanu M., D’Herouel A.F., Lacoux C., Bonnin R.A., Kennedy S.P., Wessner F., Serror P., Bouloc P., Repoila F. et al. Whole-genome mapping of 5΄ RNA ends in bacteria by tagged sequencing: a comprehensive view in Enterococcus faecalis. RNA. 2015; 21:1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu C.G., Huang R.R., Teng L., Jing X.Y., Hu J.Q., Cui G.Z., Wang Y.L., Cui Q., Xu J.. Cellulosome stoichiometry in Clostridium cellulolyticum is regulated by selective RNA processing and stabilization. Nat. Commun. 2015; 6:6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wurtzel O., Sapra R., Chen F., Zhu Y.W., Simmons B.A., Sorek R.. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010; 20:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J., Qi L., Guo Y., Yue L., Li Y.P., Ge W.Z., Wu J., Shi W.Y., Dong X.Z.. Global mapping transcriptional start sites revealed both transcriptional and post-transcriptional regulation of cold adaptation in the methanogenic archaeon Methanolobus psychrophilus. Sci. Rep.-UK. 2015; 5:9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., Chabas S., Reiche K., Hackermuller J., Reinhardt R. et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010; 464:250–255. [DOI] [PubMed] [Google Scholar]
- 32. Jäger D., Sharma C.M., Thomsen J., Ehlers C., Vogel J., Schmitz R.A.. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:21878–21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sarmiento B.F., Leigh J.A., Whitman W.B.. Genetic systems for hydrogenotrophic methanogens. Methods Enzymol. 2011; 494:43–73. [DOI] [PubMed] [Google Scholar]
- 34. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Redder P., Linder P.. DEAD-box RNA helicases in gram-positive RNA decay. Methods Enzymol. 2012; 511:369–383. [DOI] [PubMed] [Google Scholar]
- 36. Miller J.H. Experiments in Molecular Genetics. 1972; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 37. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J.Y., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Rijk P., Wuyts J., De Wachter R.. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003; 19:299–300. [DOI] [PubMed] [Google Scholar]
- 41. Clouet-d’Orval B., Bortolin M.L., Gaspin C., Bachellerie J.P.. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 2001; 29:4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen Z.J., Yu H.Y., Li L.Y., Hu S.N., Dong X.Z.. The genome and transcriptome of a newly described psychrophilic archaeon, Methanolobus psychrophilus R15, reveal its cold adaptive characteristics. Environ. Microbiol. Rep. 2012; 4:633–641. [DOI] [PubMed] [Google Scholar]
- 43. Davydov I.I., Wohlgemuth I., Artamonova I.I., Urlaub H., Tonevitsky A.G., Rodnina M.V.. Evolution of the protein stoichiometry in the L12 stalk of bacterial and organellar ribosomes. Nat. Commun. 2013; 4:1387. [DOI] [PubMed] [Google Scholar]
- 44. Scott M., Klumpp S., Mateescu E.M., Hwa T.. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 2014; 10:747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bosdriesz E., Molenaar D., Teusink B., Bruggeman F.J.. How fast-growing bacteria robustly tune their ribosome concentration to approximate growth-rate maximization. FEBS J. 2015; 282:2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fredrick K., Ibba M.. How the sequence of a gene can tune Its translation. Cell. 2010; 141:227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kudla G., Murray A.W., Tollervey D., Plotkin J.B.. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009; 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burkhardt D.H., Rouskin S., Zhang Y., Li G.W., Weissman J.S., Gross C.A.. Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. Elife. 2017; 6:e22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sharp J.S., Bechhofer D.H.. Effect of 5΄-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol. Microbiol. 2005; 57:484–495. [DOI] [PubMed] [Google Scholar]
- 50. Quax T.E.F., Wolf Y.I., Koehorst J.J., Wurtzel O., van der Oost R., Ran W.Q., Blombach F., Makarova K.S., Brouns S.J.J., Forster A.C. et al. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 2013; 4:938–944. [DOI] [PubMed] [Google Scholar]
- 51. Li G.W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCarthy J.E. Post-transcriptional control in the polycistronic operon environment: studies of the atp operon of Escherichia coli. Mol. Microbiol. 1990; 4:1233–1240. [DOI] [PubMed] [Google Scholar]
- 53. Karlin S., Mrazek J., Ma J., Brocchieri L.. Predicted highly expressed genes in archaeal genomes. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:7303–7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dean D., Nomura M.. Feedback regulation of ribosomal protein gene expression in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fu Y., Deiorio-Haggar K., Anthony J., Meyer M.M.. Most RNAs regulating ribosomal protein biosynthesis in Escherichia coli are narrowly distributed to Gammaproteobacteria. Nucleic Acids Res. 2013; 41:3491–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chao Y., Li L., Girodat D., Forstner K.U., Said N., Corcoran C., Smiga M., Papenfort K., Reinhardt R., Wieden H.J. et al. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell. 2017; 65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dominski Z., Carpousis A.J., Clouet-d’Orval B.. Emergence of the beta-CASP ribonucleases: highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta. 2013; 1829:532–551. [DOI] [PubMed] [Google Scholar]
- 58. Clouet-d’Orval B., Phung D.K., Langendijk-Genevaux P.S., Quentin Y.. Universal RNA-degrading enzymes in Archaea: prevalence, activities and functions of beta-CASP ribonucleases. Biochimie. 2015; 118:278–285. [DOI] [PubMed] [Google Scholar]
- 59. Phung D.K., Rinaldi D., Langendijk-Genevaux P.S., Quentin Y., Carpousis A.J., Clouet-d’Orval B.. Archaeal beta-CASP ribonucleases of the aCPSF1 family are orthologs of the eukaryal CPSF-73 factor. Nucleic Acids Res. 2013; 41:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Han Y., Donovan J., Rath S., Whitney G., Chitrakar A., Korennykh A.. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014; 343:1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.