Abstract
Riboswitches are regulatory elements that control gene expression by altering RNA structure upon the binding of specific metabolites. Although Bacillus subtilis riboswitches have been shown to control premature transcription termination, less is known about regulatory mechanisms employed by Escherichia coli riboswitches, which are predicted to regulate mostly at the level of translation initiation. Here, we present experimental evidence suggesting that the majority of known E. coli riboswitches control transcription termination by using the Rho transcription factor. In the case of the thiamin pyrophosphate-dependent thiM riboswitch, we find that Rho-dependent transcription termination is triggered as a consequence of translation repression. Using in vitro and in vivo assays, we show that the Rho-mediated regulation relies on RNA target elements located at the beginning of thiM coding region. Gene reporter assays indicate that relocating Rho target elements to a different gene induces transcription termination, demonstrating that such elements are modular domains controlling Rho. Our work provides strong evidence that translationally regulating riboswitches also regulate mRNA levels through an indirect control mechanism ensuring tight control of gene expression.
INTRODUCTION
Bacteria must adapt to a wide range of environmental conditions such as changes in temperature, pH, ions and metabolite levels. The bacterial adaptive response is exemplified by riboswitches, which are structured RNA elements specifically recognizing cellular metabolites (1–3). Riboswitches are located in the 5΄ untranslated regions (UTR) of several bacterial mRNAs and control the expression of downstream genes upon metabolite binding. These RNA elements exert their regulatory effects by controlling transcription termination, translation initiation or mRNA decay (1). Such RNA switches are composed of two modular domains: an aptamer involved in metabolite recognition and an expression platform that undergoes structural change to regulate gene expression (1). Riboswitch aptamer domains have been shown to respond to a variety of small cellular metabolites including amino acids, carbohydrates, coenzymes and nucleobases (1,2), and were recently shown to have a trans-acting activity (4–6), clearly indicating that riboswitches are involved in a wide range of cellular processes to regulate bacterial biochemical pathways.
In Bacillus subtilis, the majority of characterized riboswitches have been shown to control premature transcription termination by modulating the formation of Rho-independent terminators (1,2). In contrast, close inspection of Escherichia coli riboswitch sequences (7) suggests that they control gene expression by modulating ribosome access to the ribosome-binding site (RBS) and/or AUG start codon. However, in addition to controlling translation initiation, recent studies have reported that E. coli riboswitches make use of other mechanisms to regulate mRNA levels. For example, in agreement with chromatin immunoprecipitation (ChIP) and microarray data (ChIP-chip) (8), Hollands et al. have shown that the E. coli ribB riboswitch controls Rho-dependent transcription termination (9). Moreover, Caron et al. discovered a different mechanism for the E. coli lysC lysine-sensing riboswitch that directly modulates RNase E-dependent degradation of the lysC mRNA (10). At last, it was also shown that while the btuB riboswitch only represses translation initiation, regulatory elements located in its open reading frame (ORF) negatively modulate btuB mRNA levels in the presence of adenosylcobalamin (AdoCbl) (11). Together, these studies suggest that riboswitches can efficiently regulate gene expression by combining translational and mRNA level controls, the latter ensuring irreversible gene repression.
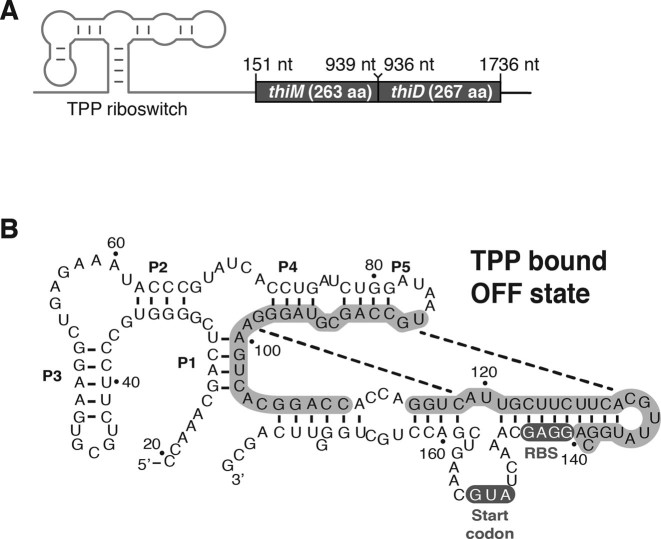
The thiM thiamin pyrophosphate (TPP)-sensing riboswitch is one of the most studied E. coli riboswitches (12–17). The thiM riboswitch is found upstream of the thiMD operon, which encodes enzymes involved in TPP biosynthesis, an essential cofactor for carbohydrate metabolism (see Figure 1A for schematic description of thiMD operon). Although this riboswitch was previously reported to strictly regulate translation initiation (12,13), we recently have obtained experimental evidence suggesting that thiM mRNA levels are modulated in TPP-rich growth conditions (10). According to the current thiM riboswitch regulation model (13,16,18), TPP binding stabilizes the aptamer domain, leading to the formation of a RBS/AUG sequestering stem, thereby inhibiting translation initiation (Figure 1B). Since the riboswitch secondary structure does not show any obvious Rho-independent transcription terminator, it suggests that alternative mechanisms are in place to regulate mRNA levels. However, because Rho- and RNase E-dependent regulatory processes do not rely on specific RNA target sequences, it is difficult to predict whether such protein cofactors are involved in the TPP-dependent mRNA regulatory mechanisms.
Figure 1.
Genetic regulation of the Escherichia coli thiM riboswitch. (A) Schematic representing the thiM riboswitch and the thiMD operon. The secondary structure of the thiM riboswitch aptamer is shown upstream of the thiM and thiD genes. Genomic locations and sizes of both proteins are indicated. Note that there is an overlap of three nucleotides between thiM and thiD. (B) Predicted secondary structure of the thiM riboswitch in the presence of thiamin pyrophosphate (TPP) (OFF state). Nucleotides involved in the formation of the anti-sequestering stem (ON state) are shown in gray and indicated by dotted lines. The ribosome-binding site (RBS) and AUG start codons are highlighted. The nomenclature indicates the nucleotide positions and the paired regions (P1–P5).
Herein, we characterized the importance of mRNA regulation among E. coli riboswitches. Strikingly, we found that most known E. coli riboswitches modulate Rho-dependent transcription termination, suggesting that mRNA control is widespread among translationally acting riboswitches. By investigating the thiM riboswitch regulation mechanism, we observed that TPP binding to the riboswitch initiates a series of regulatory events involving the Rho transcription terminator. Our data indicate that the TPP-induced translation inhibition promotes Rho-dependent transcription termination in a region located between codons 20 and 34 of thiM. The Rho-dependent modulation can be obtained in an unrelated gene by introducing the Rho-regulating thiM region, emphasizing the modular nature of this regulation mechanism. Furthermore, we found that TPP binding to the riboswitch occurs both co- and post-transcriptionally, indicating that the riboswitch controls the outcome of nascent transcripts through Rho-dependent transcription termination and the stability of pre-existing thiMD mRNAs. Together, our results provide a comprehensive view showing that the thiM riboswitch regulates at both translational and mRNA levels, a feature that is most likely shared by the majority of known E. coli riboswitches.
MATERIALS AND METHODS
DNA oligonucleotides, bacterial strains and plasmids
DNA oligonucleotides were purchased from Integrated DNA Technologies. Strains used in this study are derived from E. coli MG1665. Strain genotypes are listed in Supplementary Table S1. Strain DH5α was used for routine cloning procedures (19), and BL21 (DE3) was used for overproduction of Rho and NusG proteins. The thiM transcriptional and translational fusions were constructed with the PM1205 strain (Supplementary Table S2) (10). The polymerase chain reaction (PCR) strategy to obtain thiM riboswitch translational constructs is listed in Supplementary Table S3. Transcriptional fusions used an additional PCR step (LB1–LB47) to insert a stop codon. Mutations performed in thiM were made using three PCR steps (Supplementary Table S3). PCR1 and PCR2 were performed with the genomic DNA, and PCR3 was performed using products of the PCR1 and PCR2 reactions. DNA oligonucleotides used in this study are listed in the Supplementary Table S4. The R66S rho mutant strains were constructed by P1 transduction, selected for tetracycline resistance and sequenced.
Enzymes and chemical
T7 polymerase, RNA degradosome, Rho and NusG protein were purified as described previously (20). The E. coli RNA polymerase (RNAP) was purchased from Epicentre. Bicyclomycin (BCM) was obtained from Santa Cruz Biotech. TPP and coenzyme B12 were purchased from Sigma Aldrich.
ChIP-qPCR
Wild-type MG1655 and MG1655 rhoR66S mutant strains were grown in Luria broth (LB) at 37°C to mid-exponential phase. ChIP of RNAP was performed as described previously (21) using anti-β mouse monoclonal antibody (NeoClone). After purification of ChIP and input (starting material) DNA samples, real-time PCR was performed using an ABI 7500 instrument with the primers listed in Supplementary Table S4. In each reaction, forward and reverse primers were used to amplify either the UTR or ORF region of rho or selected riboswitch genes. The percentage of immunoprecipitation (IP) efficiency was calculated for each region of interest as the ratio of DNA in the ChIP sample compared to the input sample.
Northern blot analysis and mRNA half-life determination
Bacterial cultures were grown at 30 or 37°C in M63 0.2% glucose minimal medium to mid log-phase (OD600 of 0.35). Cells were then centrifuged and resuspended with sterile water, and total RNA was extracted immediately using the hot phenol method (10). TPP (500 μg/ml) or BCM (25 μg/ml) was added at indicated times. RNA transcription was blocked by adding rifampicin (250 μg/ml) to determine mRNA half-life. Northern blot experiments were performed as described previously (10). Probes were generated by PCR (see Supplementary Table S3).
β-Galactosidase assays
Kinetic assays for β-galactosidase experiments were performed as described previously (10). Briefly, an overnight bacterial culture grown in M63 0.2% glycerol minimal medium was diluted to an OD600 of 0.02 in 50 ml of fresh medium. The culture was incubated at 37°C until an OD600 of 0.1 was obtained. Arabinose (0.1%) was then added to induce the expression of lacZ constructs. TPP (500 μg/ml) or AdoCbl (80 μg/ml) was added where indicated. β-Galactosidase experiments determining Rho factor involvement were performed in 3 ml of culture media as described above. BCM (25 μg/ml) was added where indicated. As for the Rho R66S mutant strains, the β-galactosidase experiments were performed in 50 ml of LB medium, following the same method described above.
Rho-dependent transcription termination assays
In vitro transcriptions were performed using either the first 400 nt of thiM mRNA or the first 336 nt of the 34-codon thiM–lacZ transcriptional fusion. The transcription buffer contained 40 mM Tris–HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2 and 1.5 mM dithiothreitol (DTT). In a reacting vessel, 300 fmol DNA template, 10 μM CUGC tetranucleotide and 400 nM ATP/GTP were incubated in the transcription buffer at 37°C for 5 min. Rho (50 nM) and/or NusG were added where indicated. A mixture containing 0.2 U E. coli RNAP and 10 μCi [α-32P] UTP were then added to the transcription reaction and incubated at 37°C for 15 min. A solution containing 50 μM of nucleoside triphosphates (NTPs) and 1.5 μg/ml rifampicin was added to complete the transcription reaction. Resulting reactions were phenol/chloroform treated and mixed 1:1 with a stop solution containing 95% formamide, 10 mM ethylenediaminetetraacetic acid (EDTA) and 0.1% sodium dodecyl sulphate (SDS).
TPP binding kinetics monitored using RNase H assays
In vitro transcription assays were performed using the first 300 nt of thiM mRNA. The transcription buffer contained 20 mM Tris–HCl (pH 8.0), 20 mM NaCl, 20 mM MgCl2, 100 μM EDTA and 14 mM β-mercaptoethanol. Radiolabeled transcripts were mixed 1:1 with a solution containing 0.12 U/μl RNase H and 200 μM DNA oligonucleotide (AC3, see Supplementary Table S4) in 5 mM Tris–HCl (pH 8.0), 20 mM MgCl2, 100 mM KCl, 50 μM EDTA and 10 mM β-mercaptoethanol at 37°C for 15 s. Reactions were stopped with denaturing buffer containing 95% formamide, 10 mM EDTA and 0.1% SDS.
Cotranscriptional and post-transcriptional TPP binding analysis was performed by adding TPP either at the beginning (cotranscriptionally) or after the transcription reaction (post-transcriptionally), which was stopped through the addition of heparin. In both cases, samples were taken at various time points and treated with RNase H for 15 s and stopped by adding the denaturing buffer. Ligand binding analysis assumed a 1:1 stoichiometry between the aptamer and TPP, consistent with in-line probing data (13) and crystal structure (18,22,23). The fitting was done using a single exponential decay to yield the pseudo-first order rate of ligand binding.
Rho binding monitored using RNase H assays
Using a DNA template corresponding to the first 400 nt of thiM mRNA, transcripts were prepared in the absence of Rho, as described in the section ‘Rho-dependent transcription termination assays.’ After transcription was complete, the mixture was filtered twice through a G50 column to remove free nucleotides. Purified transcripts were then incubated in presence of 70 nM Rho for 5 min at 37°C. The resulting mixture was incubated with a solution containing 10 μM of a DNA oligonucleotide (130 or 201, see Supplementary Table S4) and 0.12 U/μl RNase H in 5 mM Tris–HCl (pH 8.0), 20 mM MgCl2, 100 mM KCl, 50 μM EDTA and 10 mM β-mercaptoethanol at 37°C for 5 min. Reactions were stopped using 95% formamide, 10 mM EDTA and 0.1% SDS.
Bioinformatic analyses of riboswitch sequences for rut determination
A bash shell script was written to calculate the proportion of cytosines and guanines using riboswitch sequences obtained from public databases. A scanning window of 25 nt was used with a stepping value of 1 nt. The validity of the technique was verified using pgaA, which was shown to be a rut site recognized by Rho (24).
RESULTS
Monitoring Rho-dependent transcription termination using ChIP assays
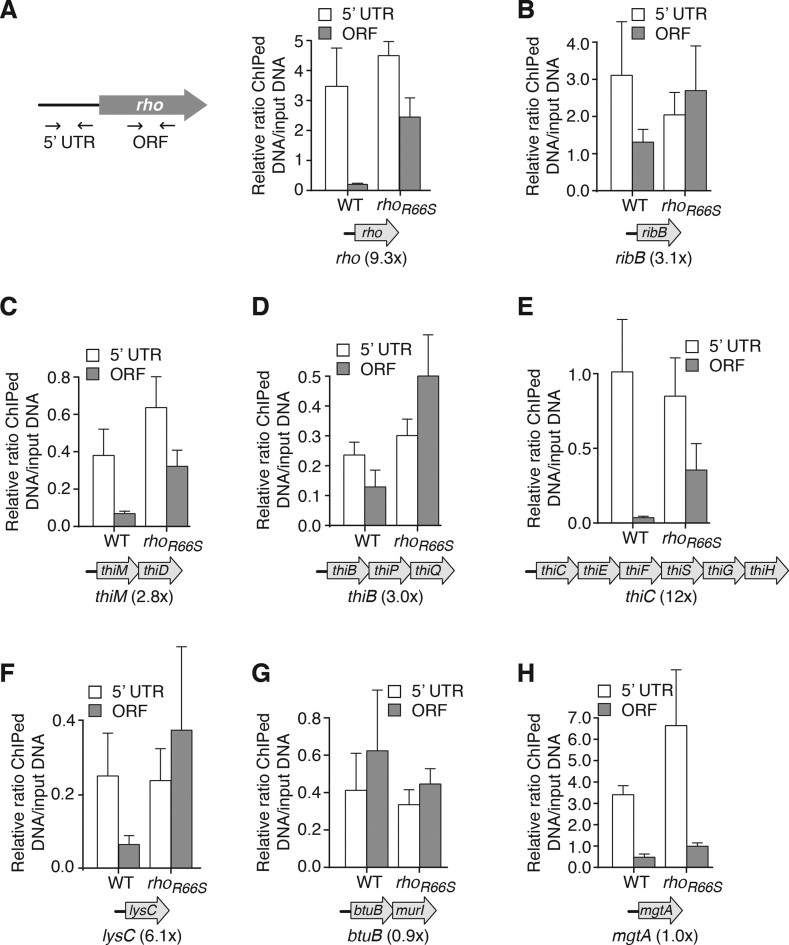
In addition to modulating gene expression at the translational level, recent studies have shown that specific E. coli riboswitches can regulate mRNA levels by controlling either ribonuclease access or Rho-dependent transcription termination (9,10). To determine how widespread mRNA regulation is among E. coli riboswitches, we first monitored Rho-dependent transcription termination by performing ChIP of the RNAP (β subunit) with quantitative PCR (ChIP-qPCR). Because our approach relies on a sonication step generating small DNA fragment sizes (∼200–300 bp), it limits the possibility of ChIP enrichment being due to RNAP binding to upstream or downstream regions, thus reducing contamination from non-overlapping transcripts. Also, to ensure that riboswitches would be bound to their respective metabolite, these experiments were performed on cells that were grown in rich media. We used a wild-type and a R66S Rho mutant strain that is defective in the termination of many RNAs, presumably due to a defect in its ability to bind RNA (25). RNAP local occupancy was first assessed by measuring the amount of DNA from the rho gene co-immunoprecipitated with RNAP, using two pairs of primers targeting specific regions of the 5΄ UTR and ORF domains (see schematic, Figure 2A). In the wild-type strain, we found lower RNAP occupancy within rho ORF compared to the 5΄ UTR, suggesting transcriptional termination within the early coding sequence (Figure 2A). However, a smaller difference in RNAP occupancy between 5΄ UTR and ORF domains was detected in the rho mutant cells (Figure 2A), consistent with Rho inactivation leading to increased transcription efficiency across rho ORF. To summarize the effect of the R66S Rho mutant on RNAP association across a gene, we calculated a ‘Rho termination score’, defined as the ratio of RNAP association in the ORF versus the transcript 5΄ UTR in the rho mutant, normalized to the equivalent ratio in the wild-type. Hence, Rho termination scores >1.0 suggest Rho-dependent termination within the gene. We determined a Rho termination score of 9.3 for rho, indicating Rho-dependent termination within rho (Figure 2A), which is consistent with a previously reported auto-regulation mechanism (26). We next investigated the flavin mononucleotide (FMN)-binding ribB riboswitch that was previously shown to modulate Rho-dependent transcription termination (9). In contrast to the wild-type strain, we observed a complete lack of transcriptional control in the rho mutant (Figure 2B), resulting in a Rho termination score of 3.1 for ribB. These results are consistent with ribB using Rho-dependent transcription termination to modulate ribB mRNA levels (9).
Figure 2.
Monitoring the RNA polymerase (RNAP) occupancy of Escherichia coli riboswitches using chromatin immunoprecipitation (ChIP). (A) Left, schematic representing ChIP-qPCR targeting 5΄ untranslated regions (UTR) and ORF regions for rho. Right, ChIP-qPCR data showing RNAP (β subunit) occupancy at indicated locations of rho. Values indicated represent the relative enrichment of ChIP DNA to the input control in wild-type cells or cells expressing the R66S Rho mutant from its native locus. The Rho termination score is represented below the plot by the ratio of RNAP association in the ORF versus the transcript 5΄ UTR in the rho mutant, normalized to the equivalent in the wild-type (ORFR66S/UTRR66S)/(ORFWT/UTRWT). (B–H) ChIP-qPCR data showing RNAP occupancy for ribB (B), thiM (C), thiB (D), thiC (E), lysC (F), btuB (G) and mgtA (H). Values represent the relative enrichment of ChIP DNA to the input control in wild-type cells or cells expressing the R66S Rho mutant from its native locus. The Rho termination score is shown as ‘(#x)’ below each plot.
To establish whether Rho-dependent transcription termination is widespread among all other known E. coli riboswitches, we used ChIP-qPCR for riboswitch-controlled genes thiM, thiB, thiC, lysC, btuB and mgtA. When monitoring TPP-responsive riboswitches, we obtained Rho termination scores of 2.8 for thiM, 3.0 for thiB and 12 for thiC (Figure 2C–E), indicative of Rho termination within these genes. We determined a Rho termination score of 6.1 for lysC (Figure 2F), also indicating significant Rho-dependent transcription termination. In contrast, we calculated the Rho termination score for btuB to be 0.9 (Figure 2G), suggesting that Rho is not involved in btuB regulation. The similar level of RNAP occupancy for both 5΄ UTR and ORF domains in the wild-type strain suggests either that no transcriptional control occurs or that a low AdoCbl concentration is present in the growth medium (27). At last, we obtained a score of 1.0 for mgtA (Figure 2H), suggesting that Rho-dependent transcription termination is not involved for this riboswitch. Thus, our data strongly suggest that, in addition to their translational regulation, most known E. coli riboswitches use Rho-dependent termination to modulate gene expression.
The presence of TPP leads to a decrease in thiM mRNA levels
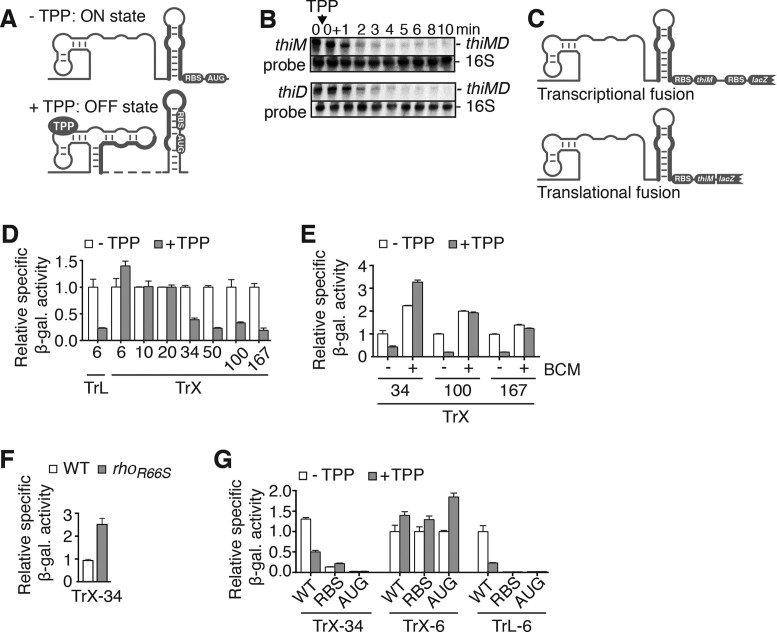
We have previously observed that thiM mRNA levels are decreased in TPP-rich conditions (10). However, no mechanism was proposed to explain this ligand-dependent regulation. According to available data (10,12,13), TPP binding to the thiM riboswitch induces a structural change leading to translation inhibition through the formation of a RBS/AUG sequestering stem (Figure 3A). We first investigated thiM mRNA regulation by northern blot experiments using a probe targeting the thiM ORF (Figure 3B, see top panel). A large decrease in mRNA was observed when TPP was added to the culture medium, consistent with the modulation of thiM mRNA levels in presence of TPP. A similar ligand-induced mRNA decrease was also observed with a probe directed against thiD (Figure 3B, bottom panel), indicating that the riboswitch downregulates mRNA levels of the complete thiMD operon. Together, our results indicate that thiMD mRNA levels are significantly reduced in the presence of TPP, consistent with a Rho-mediated regulation mechanism.
Figure 3.
The Escherichia coli thiM riboswitch uses Rho-dependent transcription termination to modulate mRNA levels. (A) Schematic representing thiM riboswitch translational control. In the absence of TPP, the anti-sequestering stem (bold) exposes the RBS and allows translation initiation. TPP binding sequesters both the RBS and the AUG start codon. The dotted line represents the immediate connectivity between linked regions. (B) Northern blot analysis of thiMD mRNA levels. Total RNA was extracted at the indicated times immediately before (0−) and after (0+) addition of TPP (0.5 mg/ml). Probes were designed to detect thiM (top panel) and thiD (bottom panel). RNA species are indicated on the right of the gels. 16S rRNA was used as a loading control. (C) Transcriptional (thiM–lacZ) and translational (ThiM–LacZ) fusions of the thiM riboswitch. The transcriptional fusion contains RBS sequences for both thiM and lacZ, thus allowing to monitor mRNA levels. In contrast, the translational fusion comprises a single RBS sequence and reports on both mRNA and protein levels. (D) β-Galactosidase assays of translational ThiM–LacZ (TrL) and transcriptional thiM–lacZ (TrX) fusions performed in the absence or presence of 500 μg/ml TPP. The number of thiM codons is indicated below the histograms. Values were normalized to the activity obtained in the absence of TPP. The average values of three independent experiments with standard deviations (SDs) are shown. (E) β-Galactosidase assays of the wild-type strain performed in the absence or presence of 500 μg/ml TPP or 25 μg/ml bicyclomycin (BCM). The number of thiM codons is indicated below histograms. Values were normalized to enzymatic activity obtained for wild-type constructs without ligand. The average values of three independent experiments with SDs are shown. (F) β-Galactosidase assays of the wild-type and R66S strains performed in LB media using a transcriptional thiM–lacZ fusion (TrX-34). Values were normalized to enzymatic activity obtained for the wild-type construct. The average values of three independent experiments with SDs are shown. (G) β-Galactosidase assays of transcriptional thiM–lacZ (TrX) or translational ThiM–LacZ (TrL) fusions. The numbers of thiM codons are indicated below each data set. Assays were performed in the absence or presence of 500 μg/ml TPP using the wild-type, and RBS (G142C) and AUG start codon (U152A) mutants. Values were normalized to enzymatic activity obtained for the WT construct in the absence of TPP. The average values of three independent experiments with SDs are shown.
The thiM coding region is important for TPP-dependent mRNA control
Given that our northern blot data do not indicate which region of the transcript is important for mRNA regulation, we used transcriptional lacZ chromosomal fusions (Figure 3C) containing various thiM ORF sizes. Constructs containing 34 codons or more resulted in a substantial reduction in β-galactosidase activity in the presence of TPP (Figure 3D). In contrast, shorter fusions containing 6, 10 or 20 codons did not result in repression, indicating that the minimum sequence for mRNA modulation is between 20 and 34 codons. A noticeable increase in gene expression was observed with the 6-codon transcriptional construct (Figure 3D), suggesting that TPP binding stabilizes mRNA levels in this context. As expected, a translational lacZ fusion containing the riboswitch fused to only the first 6 thiM codons demonstrated TPP-dependent repression (Figure 3D). These results suggest that while the thiM riboswitch regulates translation initiation in the presence of only 6 codons, the mRNA regulation requires specific elements located between codons 20 and 34.
To assess whether such regulatory elements rely on Rho, we measured β-galactosidase activity using a thiM–lacZ transcriptional fusion in the absence or presence of the Rho inhibitor BCM (28). These experiments were performed using a lacZ transcriptional construct containing 34 codons, which is the shortest fusion producing TPP-dependent mRNA regulation (Figure 3D). As shown in Figure 3E, addition of BCM led to complete loss of TPP-induced repression, indicating that Rho-dependent transcription termination is involved in the first 34 thiM codons. An increased activity was observed in the presence of TPP, suggesting that the transcript corresponding to the transcriptional fusion is more stable when bound to TPP. An absence of regulation was also obtained using longer fusions of 100 and 167 codons (Figure 3E), consistent with the prevalence of Rho regulation. In addition, when performing β-galactosidase assays using the 34-codon thiM–lacZ transcriptional construct in a R66S Rho mutant strain, we observed an increased expression compared to the wild-type (Figure 3F), consistent with Rho being involved in the regulation. The implication of Rho in thiM regulation is corroborated with previously obtained RNA-seq data showing that the thiM transcript is present at higher levels following BCM treatment (29).
Given that our results suggest an underlying model in which Rho-dependent transcription termination occurs upon TPP-induced translation inhibition (Figure 3D), we next investigated the role of translation repression on thiM mRNA decrease. As shown in Figure 3G, RBS (G142C) and AUG (U152A) mutations resulted in low β-galactosidase activities both in the absence and presence of TPP, suggesting that translation elongation is crucial for maintaining mRNA levels. Nonetheless, β-galactosidase activity for the RBS mutant was still detectable, and showed no decrease in the presence of TPP. Reporter gene assays performed with transcriptional fusions containing RBS or AUG mutations in the presence of only the first six thiM codons yielded high β-galactosidase activities (Figure 3G), demonstrating the absence of any mRNA regulatory element. As expected, no β-galactosidase expression was detected when altering the RBS and AUG start codon in the context of translational fusions (Figure 3G). Together, these results suggest that thiM codons 20–34 are required to modulate mRNA levels upon the TPP-dependent inhibition of translation.
Rho-dependent transcription termination encompasses the 20–34 thiM codon region
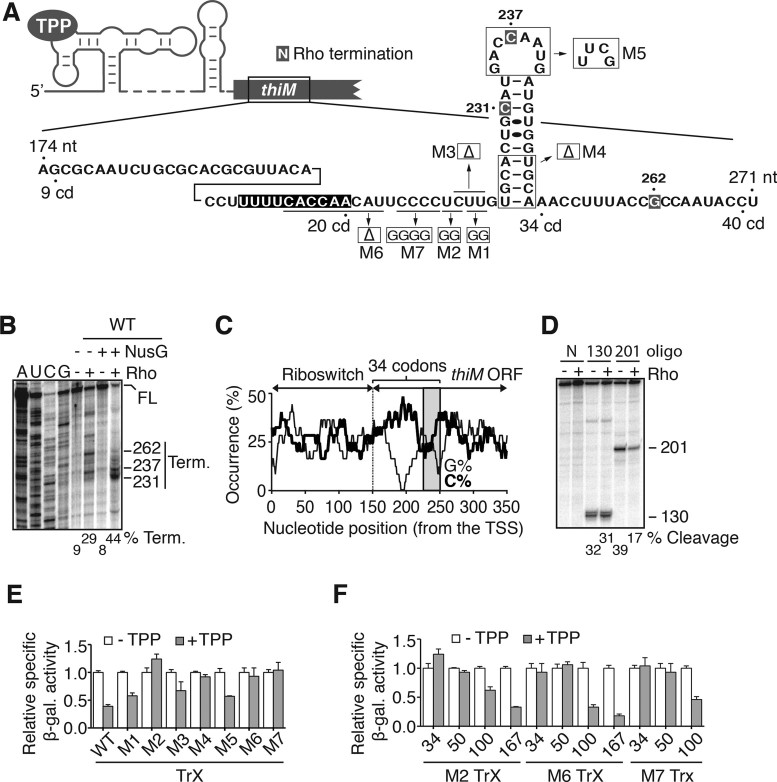
We next carried out single-round in vitro assays to determine the location of Rho-dependent transcription termination (Figure 4A). In the presence of Rho, we observed specific thiM mRNA transcription termination at several sites, including positions 237 and 262 (Figure 4A and B). An additional strong termination site was observed at position 231 when Rho and the transcription elongation factor NusG (30,31) were simultaneously included in the transcription reaction, leading to a transcription termination efficiency of 44% (Figure 4B). As expected, Rho termination occurred in the context of a transcriptional 34-codon lacZ–thiM fusion (Supplementary Figure S1), consistent with β-galactosidase assays (Figure 3E and F).
Figure 4.
A defined mRNA region is used for the control of Rho-dependent transcription termination. (A) Schematic representing thiM ORF comprising codons 9–40. Nucleotides highlighted in gray and in black represent Rho termination sites and the 201–210 nt region, respectively. Mutations and deletions (Δ) are shown in the structure. (B) In vitro transcriptions performed using wild-type thiM mRNA. Transcriptions were done in the absence (−) or presence (+) of 50 nM NusG or 50 nM Rho. Read-through and termination transcripts are indicated at the right. Termination efficiencies are indicated below. Note that a read-through product of 400 nt is expected. (C) Sequence analysis of cytosine (C%) and guanine (G%) distribution in the thiM sequence. A scanning window of 25 nt was used to determine C and G occurrences as a function of the transcription start site (TSS). The shaded region represents the position of the stem-loop. (D) RNase H probing of Rho binding on thiM mRNA. RNase H assays were performed in the absence (−) or presence (+) of 50 nM Rho. Cleavage assays were done using DNA oligonucleotides targeting regions 130–139 (130) or 201–210 (201). Cleavage products and cleavage efficiencies are indicated on the right and below, respectively. Non-cleaved transcripts (N) are shown as controls. (E) β-Galactosidase assays of transcriptional thiM–lacZ fusions performed in the absence or presence of 500 μg/ml TPP. Measurements were performed for mutants targeting the C-rich region (M1–M3, M6 and M7) and the stem-loop (M4 and M5). Values were normalized to the activity obtained in the absence of TPP. The average values of three independent experiments with SDs are shown. (F) β-Galactosidase assays of transcriptional thiM–lacZ fusions of M2, M6 and M7 mutants in the context of various sizes of thiM open reading frame. Enzymatic activities were conducted in the absence or presence of 500 μg/ml TPP. Values were normalized to the enzymatic activity obtained in the absence of TPP. The average values of three independent experiments with SDs are shown.
Close examination of the first 34-codon thiM sequence shows high and low proportions of cytosines and guanines, respectively, often associated with Rho utilization sites (rut) (Figure 4C) (9,24). To establish the location of the Rho loading site, we employed a well-established RNase H assay that cleaves at RNA:DNA hybrid regions (32). In these experiments, 5΄-radiolabeled thiM mRNA transcripts produced by using the E. coli RNAP were incubated in presence of Rho, but without ATP, to avoid Rho translocation activity. We used a DNA oligonucleotide to target residues 201–210 (oligonucleotide 201, Figure 4D). We expect these residues to be part of the rut site, given their high and low composition in cytosines and guanines respectively (Figure 4C). In the absence of Rho, we observed efficient cleavage of the thiM transcript by RNase H (39%), indicating that this region is accessible for the binding of oligonucleotide 201 (Figure 4D). However, the presence of Rho resulted in much decreased RNase H cleavage (17%), suggesting that Rho binding protects the 201–210 nt region from oligonucleotide binding. No such Rho-dependent protection was observed when using an oligonucleotide targeting residues 130–139 (oligonucleotide 130, Figure 4D). Thus, our data are consistent with Rho loading at the predicted rut site in the first 34 codons of thiM mRNA (Figure 4C).
Importance of RNA regulatory elements for mRNA control
To investigate RNA elements important for Rho-dependent thiM regulation, we next carried out a mutagenesis study by using a portion of the thiM mRNA sequence (174–271 nt) encompassing codons 9–40 (Figure 4A). Mutations were designed to investigate the importance of the C-rich region (M1–M3, M6 and M7) and the importance of the downstream stem-loop (M4 and M5). In addition to disrupting the identity of the C-rich region by introducing purine residues into specific sites (M1, M2 and M7), deletions were also performed (M3 and M6) to determine if the length of the C-rich region is important for the regulation. Of the mutants we tested, only M2, M4, M6 and M7 significantly abolished TPP-dependent gene regulation in thiM–lacZ transcriptional fusions (Figure 4E). These results suggest that the integrity of the C-rich region and the stem-loop is crucial for thiM mRNA regulation. We next used mutants disrupting TPP-dependent mRNA regulation in the context of various ORF lengths to determine whether thiMD mRNA levels were controlled solely through the identified Rho targeted region. When employing the M2 mutant that completely inhibits mRNA regulation (Figure 4E), increasing the ORF size to >100 codons recovered TPP-dependent mRNA regulation (Figure 4F). Similar results were obtained in a background of M6 and M7 mutants (Figure 4F). Together, our data indicate the presence of additional downstream sequences that control the level of thiMD mRNA in the presence of TPP.
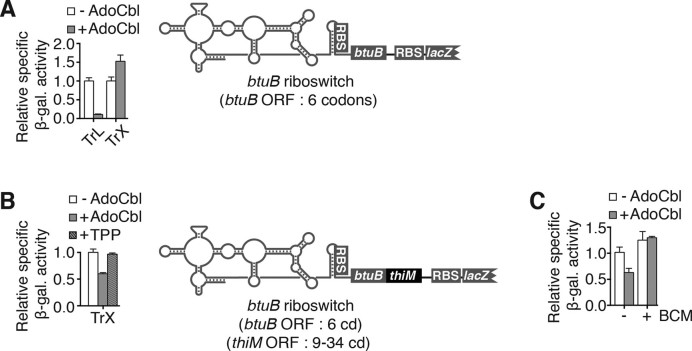
Since the first 23 nt of thiM ORF mRNA are part of the riboswitch expression platform (Figure 1B, residues 151–172), we determined whether riboswitch sequence element(s) were involved in mRNA control. To do so, we used the E. coli AdoCbl-sensing btuB riboswitch that was shown to strictly control translation initiation (33). We first assayed a translational BtuB–LacZ fusion containing only the first six codons of btuB in the absence or presence of AdoCbl, and observed ligand-dependent repression (Figure 5A). Importantly, no ligand-dependent repression was observed using a lacZ transcriptional fusion (Figure 5A), consistent with btuB translation inhibition not modulating mRNA levels. However, adding a sequence corresponding to codons 9–34 of thiM, downstream of btuB codons, significantly reduced β-galactosidase activity only in the presence of AdoCbl (Figure 5B). As expected, the presence of BCM abolished AdoCbl-dependent repression (Figure 5C), equivalent to the effect of BCM on TPP-dependent repression in the thiMD mRNA context (Figure 3E). Hence, thiM mRNA regulation does not rely on specific riboswitch elements but, rather, on inhibiting translation initiation.
Figure 5.
The control of mRNA levels is not dependent on riboswitch identity. (A and B) β-Galactosidase assays using translational BtuB–LacZ and transcriptional btuB–lacZ fusions (A) and transcriptional btuB–thiM–lacZ fusions (B). Enzymatic activities were measured in the absence or presence of 5 μM adenosylcobalamin (AdoCbl) or 500 μg/ml TPP. Values were normalized to enzymatic activity obtained without ligand. Schematics representing transcriptional constructs are shown on the right. The average values of three independent experiments with SDs are shown. (C) β-Galactosidase assays of the wild-type strain performed in the absence or presence of 5 μM AdoCbl or 25 μg/ml BCM. Values were normalized to enzymatic activity obtained for wild-type constructs without ligand. The average values of three independent experiments with SDs are shown.
TPP binding kinetics is consistent with both co- and post-transcriptional controls
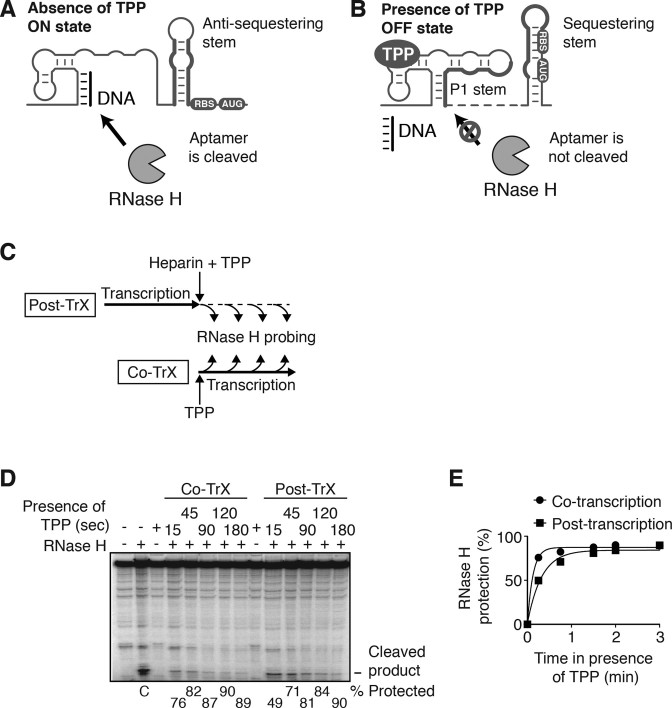
Our results suggest that TPP binding to the riboswitch occurs cotranscriptionally, thus leading to translation inhibition and Rho-dependent transcription termination. To investigate the efficiency of cotranscriptional TPP recognition by the riboswitch, we used RNase H cleavage assays to monitor TPP binding kinetics to nascent thiM transcripts. In this assay, a DNA oligonucleotide is added after transcription completion to probe TPP-dependent formation of the P1 stem (Figure 6A and B). Thus, by monitoring P1 formation as a function of time (Figure 6C), it is possible to estimate the apparent rate of TPP binding to the riboswitch. We first performed transcription of the thiM riboswitch without TPP and observed efficient RNase H cleavage of nascent transcripts (Figure 6D, lane C). However, transcription reactions with TPP (cotranscriptional binding) yielded cleavage protection for all transcription time points, consistent with efficient TPP binding (Figure 6D). Post-transcriptional TPP addition also resulted in RNase H protection, although to a lesser degree (Figure 6D). Fitting the data with a single-exponential model yielded apparent binding rates of 0.13 ± 0.02 and 0.05 ± 0.01 s−1 for co- and post-transcriptional TPP addition, respectively (Figure 6E). No TPP-dependent protection was observed when using a G40A mutant preventing ligand binding (Supplementary Figure S2). These apparent binding rates indicate that TPP recognition is performed only 2.5-fold slower post-transcriptionally, suggesting that the riboswitch can sense TPP both co- and post-transcriptionally. Accordingly, we observed a TPP-dependent decrease of thiMD mRNA half-life in vivo (Supplementary Figure S3A–C), consistent with the thiMD transcript being targeted by cellular ribonucleases upon post-transcriptional TPP binding. Together, our in vitro and in vivo results indicate that the thiM riboswitch performs both co- and post-transcriptional TPP sensing to regulate the thiMD operon.
Figure 6.
RNase H cleavage assays monitoring co- and post-transcriptional TPP binding to the thiM riboswitch. (A) Representation of the thiM riboswitch in the absence of TPP. Because the riboswitch is in the ON state, the incubation of a DNA oligonucleotide specifically hybridizing to the aptamer domain results in RNase H cleavage activity of the riboswitch. (B) Representation of the thiM riboswitch in presence of TPP. TPP binding leads to the adoption of the OFF state in which the P1 stem is formed. In this condition, DNA oligonucleotide binding to the riboswitch is not possible and RNase H cleavage of the riboswitch is not observed. (C) Experimental setup assessing TPP binding to the thiM riboswitch. Addition of TPP is performed either at the beginning or at the end of the transcription reaction, thereby allowing TPP binding to occur either cotranscriptionally or post-transcriptionally, respectively. Under these conditions, full-length transcription is achieved in <15 s and the presence of heparin ensures that no transcription re-initiation occurs. (D) RNase H assays performed on nascent thiM mRNAs. Transcription was performed using Escherichia coli RNAP and RNase H reactions were performed for 15 s in all cases. A control (lane C) shows the cleaved product in the absence of TPP, consistent with accessibility of the P1 stem to oligonucleotide binding and RNase H cleavage. Cotranscriptional studies were carried out by adding TPP (10 μM) at the beginning of the transcription and by performing RNase H assays at the indicated times. Post-transcriptional TPP binding studies were conducted similarly, but TPP was added at the end of transcription. The DNA probe was designed to target the P1 stem. (E) Quantification analysis of RNase H experiments shown in panel A. Data were fitted with a single-exponential model and yielded apparent TPP binding rates of 0.13 ± 0.02 and 0.05 ± 0.01 s−1 for co- and post-transcriptional binding, respectively. The model assumes that no cleavage protection is obtained in the absence of TPP.
DISCUSSION
In the course of this study, we investigated the importance of Rho-mediated regulation for several known E. coli riboswitches. Strikingly, all riboswitches we tested regulate gene expression at the mRNA level, and at least five of seven riboswitches employ Rho-dependent transcription termination (Figure 2). Indeed, our ChIP data strongly suggest that riboswitches sensing TPP (thiB, thiC and thiM), FMN (ribB) and lysine (lysC) control Rho-dependent transcription termination. Accordingly, a previous study indicated that the ribB riboswitch directly controls Rho-dependent termination (9), and RNA sequencing analysis revealed an increase in mRNA levels for thiM and thiC (∼8- and ∼12-fold respectively) in the presence of BCM (29). In contrast, while the Salmonella enterica mgtA riboswitch is known to control Rho-dependent transcription termination, our results indicate that its E. coli homolog may use a different mechanism. Indeed, despite our ChIP data clearly showing a decrease in RNAP occupancy at the 3΄ end of the mgtA ORF, consistent with transcription termination, no obvious regulation appears to be performed in a Rho-dependent manner (Figure 2H). These data suggest either that the E. coli mgtA controls transcription termination independently of Rho or, alternatively, that the R66S Rho mutant may not prevent mgtA Rho-dependent termination. Consistent with this notion, in our experimental conditions, the R66S mutant grows almost as well as the wild-type, despite Rho being essential for viability. Moreover, another RNA-binding domain mutant of Rho, F62S, is only defective in Rho termination for a subset of RNAs (30). In the case of the AdoCbl-sensing riboswitch (btuB), additional experiments will need to be performed to decipher the regulation mechanism. Nonetheless, it is expected that mRNA degradation would be the main mechanism employed by this translationally acting riboswitch to control its mRNA level (11).
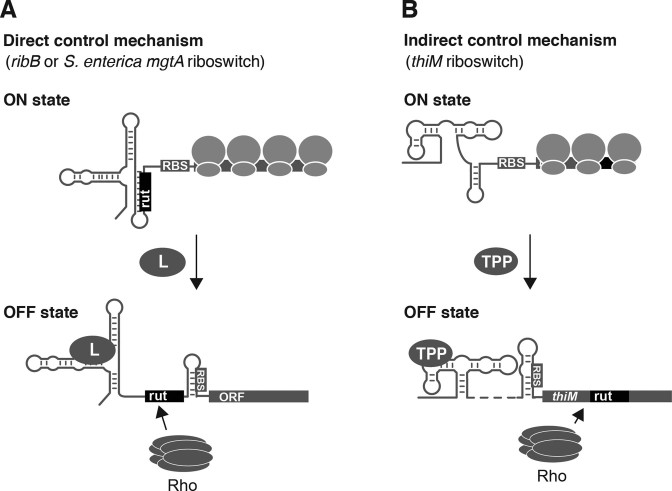
Rho was previously shown to be involved in the regulation of the E. coli ribB and S. enterica mgtA riboswitches (9). It was suggested that the conformational state of the riboswitch directly dictates the access of the Rho factor to the rut site located in the riboswitch expression platform (Figure 7A). In contrast, we describe here a riboswitch-based mechanism in which Rho is modulated as a consequence of translation inhibition. Based on our study, low intracellular TPP concentrations allow efficient transcription and translation of the thiMD operon (Figure 7B). However, upon cotranscriptional TPP binding, the riboswitch adopts the OFF state where ribosome access to the RBS is precluded, thereby inhibiting translation initiation (Figure 7B). As a result, the rut sequence becomes accessible for Rho binding, and premature transcription termination is established to reduce thiMD mRNA levels. The Rho termination mechanism is consistent with both the presence of a rut site, determined by sequence analysis and RNase H assays (Figure 4C and D), and Rho-dependent transcription termination sites occurring at positions 231, 237 and 262 (Figure 4B). Interestingly, our results are in agreement with a recent study suggesting that Rho is involved in the control mediated by the thiM riboswitch (34). Taken as a whole, these results indicate that Rho recognizes target sequences located within thiM coding sequence, which are only accessible upon the inhibition of translation (Figure 7B). The dependence of Rho transcription termination on the efficiency of translation initiation ensures that thiMD mRNA levels are efficiently modulated in a riboswitch-dependent manner at high intracellular TPP concentrations. Given that such a Rho-dependent regulating mechanism can be readily introduced in a distinct gene such as btuB (Figure 5B), it suggests that other translationally controlling riboswitches may rely on a similar strategy to modulate mRNA levels upon translation inhibition.
Figure 7.
Proposed model describing how the thiM riboswitch modulates Rho-dependent transcription termination upon the inhibition of translation. (A) Direct control mechanism of Rho-dependent transcription termination. The Escherichia coli ribB and Salmonella enterica mgtA riboswitches allow efficient genetic expression at low ligand (L) concentrations (ON state). However, the recognition of their respective metabolite leads to the selective exposition of a rut site, thus resulting in Rho-dependent transcription termination (OFF state). (B) Indirect control mechanism of Rho-dependent transcription termination. The thiM riboswitch adopts the ON state in which the initiation of thiM translation is efficiently performed. However, upon TPP binding, the riboswitch folds into the OFF state whereby translation initiation is inhibited. As a result, a rut sequence located in the coding region is available for Rho binding, therefore promoting premature transcription termination of the thiMD operon.
β-Galactosidase experiments using a series of thiM–lacZ transcriptional fusions indicate that Rho-dependent transcription termination elements are localized within the first 34 thiM codons (Figure 3D). Interestingly, although no TPP-dependent regulation was observed for the M2, M6 and M7 mutants, the use of longer thiM fusions allowed to recover riboswitch regulation (Figure 4F). These results suggest that additional mechanisms are in place to regulate thiMD mRNA levels. One possibility is that there are additional rut sequences downstream of codon 34. Another possibility is that thiMD mRNAs are actively degraded by cellular RNases, as suggested by the ∼2-fold decrease of thiMD mRNA half-life in the presence of ligand (Supplementary Figure S3C). It is worth noting that half-life measurements strictly report the effect of TPP on already transcribed mRNAs, suggesting that control of thiM mRNA decay must occur through post-transcriptional TPP binding to the riboswitch. Recently obtained biochemical data showed that the E. coli lysC riboswitch contains regulatory elements within the riboswitch domain to initiate mRNA decay through RNase E cleavage activity (10). The location of RNase E cleavage sites presumably allows the lysC riboswitch to rapidly downregulate mRNA levels upon lysine binding using a nucleolytic mechanism (35). Interestingly, in agreement with others (34), our current analysis reveals that Rho is also involved in lysC regulation (Figure 2F), suggesting that lysC is tightly regulated through multiple biological mechanisms. A precedent for a dual acting mechanism implying Rho and mRNA degradation was recently described in Corynebacterium glutamicum where both Rho and RNase G are involved in the control mechanism of the FMN riboswitch (36).
Rho-dependent transcription termination is involved in a variety of processes such as maintaining translation–transcription coupling through polarity (37,38) and suppressing antisense transcription (29). Although no specific consensus sequence is known, Rho-binding rut sites typically exhibit C-rich/G-low content and are ∼80 nt in length (37). This is mostly explained by the fact that bioinformatic prediction of rut sites is not straightforward as few conserved elements are observed across regulatory sites (37,39). In the case of the thiM riboswitch, bioinformatic analysis of C/G content is consistent with the composition of typical rut sites (Figure 4C). Our predictions agree well with RNase H data indicating that the rut site is approximately located at position ∼200 of thiM (Figure 4D), thus making a 30–60 nt distance between rut and transcription termination sites. Although 30–60 nt is shorter than the distance between Rho loading and termination at other described terminators, the presence of a stem-loop nearby Rho termination sites could be involved in making a transcriptional pause important for Rho termination (40). Strikingly, a sequence analysis of thiM in other bacterial species revealed a similar sequence and secondary structure pattern in the same narrow region (Supplementary Figure S4). Specifically, a C-rich/G-low content located immediately upstream of a stem-loop was observed in each species, suggesting that a rut site is present and that a Rho-dependent transcription termination mechanism is used to modulate thiM mRNA levels. It is plausible that the evolution of Rho-dependent transcription termination mechanism is facilitated within ORF regions compared to riboswitch domains, given that the latter are under evolutionary pressure to maintain an active metabolite-binding sequence.
Our work provides the first detailed mechanism describing how ligand binding to translation-controlling riboswitches orchestrates the simultaneous regulation of both translation initiation and premature transcription termination. The strategic location of such regulatory sequences at the beginning of regulated mRNAs ensures that they are recognized early in the expression pathway, thus providing efficient coordination of translation and mRNA regulation. Importantly, our study suggests that this mechanism is widespread among riboswitches and most likely among diverse bacterial mRNAs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Alain Lavigueur for discussion and critical reading of the manuscript. We thank the laboratory of James Berger and Irina Artsimovitch for providing plasmids for the purification of Rho and NusG protein, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CIHR) [MOP82877 to D.A.L., MOP69005 to E.M.]; National Institutes of Health through the NIH Director's New Innovator Award Program [1DP2OD007188 to J.T.W.]; Fonds de Recherche Santé Québec Senior Scholarship (to E.M., D.A.L.). Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Serganov A., Nudler E.. A decade of riboswitches. Cell. 2013; 152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breaker R.R. Riboswitches and the RNA World. Cold Spring Harb. Perspect. Biol. 2012; 4:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haller A., Souliere M.F., Micura R.. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc. Chem. Res. 2011; 44:1339–1348. [DOI] [PubMed] [Google Scholar]
- 4. Loh E., Dussurget O., Gripenland J., Vaitkevicius K., Tiensuu T., Mandin P., Repoila F., Buchrieser C., Cossart P., Johansson J.. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009; 139:770–779. [DOI] [PubMed] [Google Scholar]
- 5. Mellin J.R., Koutero M., Dar D., Nahori M.-A., Sorek R., Cossart P.. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science. 2014; 345:940–943. [DOI] [PubMed] [Google Scholar]
- 6. DebRoy S., Gebbie M., Ramesh A., Goodson J.R., Cruz M.R., van Hoof A., Winkler W.C., Garsin D.A.. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science. 2014; 345:937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrick J.E., Breaker R.R.. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007; 8:R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peters J.M., Mooney R.A., Kuan P.F., Rowland J.L., Keles S., Landick R.. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:15406–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hollands K., Proshkin S., Sklyarova S., Epshtein V., Mironov A., Nudler E., Groisman E.A.. Riboswitch control of Rho-dependent transcription termination. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:5376–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caron M.P., Bastet L., Lussier A., Simoneau-Roy M., Masse E., Lafontaine D.A.. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E3444–E3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nou X., Kadner R.J.. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J. Bacteriol. 1998; 180:6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ontiveros-Palacios N., Smith A.M., Grundy F.J., Soberon M., Henkin T.M., Miranda-Rios J.. Molecular basis of gene regulation by the THI-box riboswitch. Mol. Microbiol. 2008; 67:793–803. [DOI] [PubMed] [Google Scholar]
- 13. Winkler W., Nahvi A., Breaker R.R.. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002; 419:952–956. [DOI] [PubMed] [Google Scholar]
- 14. Lang K., Rieder R., Micura R.. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007; 35:5370–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miranda-Rios J. The THI-box riboswitch, or how RNA binds thiamin pyrophosphate. Structure. 2007; 15:259–265. [DOI] [PubMed] [Google Scholar]
- 16. Rentmeister A., Mayer G., Kuhn N., Famulok M.. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007; 35:3713–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L., Cressina E., Leeper F.J., Smith A.G., Abell C.. A fragment-based approach to identifying ligands for riboswitches. ACS Chem. Biol. 2010; 5:355–358. [DOI] [PubMed] [Google Scholar]
- 18. Edwards T.E., Ferre-D’Amare A.R.. Crystal structures of the Thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006; 14:1459–1468. [DOI] [PubMed] [Google Scholar]
- 19. Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., Court D.L.. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rabhi M., Espeli O., Schwartz A., Cayrol B., Rahmouni A.R., Arluison V., Boudvillain M.. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 2011; 30:2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stringer A.M., Currenti S., Bonocora R.P., Baranowski C., Petrone B.L., Palumbo M.J., Reilly A.A., Zhang Z., Erill I., Wade J.T.. Genome-scale analyses of Escherichia coli and Salmonella enterica AraC reveal noncanonical targets and an expanded core regulon. J. Bacteriol. 2014; 196:660–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Serganov A., Polonskaia A., Phan A.T., Breaker R.R., Patel D.J.. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006; 441:1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thore S., Leibundgut M., Ban N.. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006; 312:1208–1211. [DOI] [PubMed] [Google Scholar]
- 24. Figueroa-Bossi N., Schwartz A., Guillemardet B., D’Heygere F., Bossi L., Boudvillain M.. RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 2014; 28:1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinez A., Burns C.M., Richardson J.P.. Residues in the RNP1-like sequence motif of Rho protein are involved in RNA-binding affinity and discrimination. J. Mol. Biol. 1996; 257:909–918. [DOI] [PubMed] [Google Scholar]
- 26. Matsumoto Y., Shigesada K., Hirano M., Imai M.. Autogenous regulation of the gene for transcription termination factor rho in Escherichia coli: localization and function of its attenuators. J. Bacteriol. 1986; 166:945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lawrence J.G., Roth J.R.. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J. Bacteriol. 1995; 177:6371–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohn H., Widger W.. The molecular basis for the mode of action of bicyclomycin. Curr. Drug Targets Infect. Disord. 2005; 5:273–295. [DOI] [PubMed] [Google Scholar]
- 29. Peters J.M., Mooney R.A., Grass J.A., Jessen E.D., Tran F., Landick R.. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012; 26:2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shashni R., Qayyum M.Z., Vishalini V., Dey D., Sen R.. Redundancy of primary RNA-binding functions of the bacterial transcription terminator Rho. Nucleic Acids Res. 2014; 42:9677–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J., Mason S.W., Greenblatt J.. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993; 7:161–172. [DOI] [PubMed] [Google Scholar]
- 32. Treiber D.K., Williamson J.R.. Kinetic oligonucleotide hybridization for monitoring kinetic folding of large RNAs. Methods Enzymol. 2000; 317:330–353. [DOI] [PubMed] [Google Scholar]
- 33. Nou X., Kadner R.J.. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7190–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sedlyarova N., Shamovsky I., Bharati B.K., Epshtein V., Chen J., Gottesman S., Schroeder R., Nudler E.. sRNA-mediated control of transcription termination in E. coli. Cell. 2016; 167:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dreyfus M. Killer and protective ribosomes. Prog. Mol. Biol. Transl. Sci. 2009; 85:423–466. [DOI] [PubMed] [Google Scholar]
- 36. Takemoto N., Tanaka Y., Inui M.. Rho and RNase play a central role in FMN riboswitch regulation in Corynebacterium glutamicum. Nucleic Acids Res. 2015; 43:520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peters J.M., Vangeloff A.D., Landick R.. Bacterial transcription terminators: The RNA 3΄-end chronicles. J. Mol. Biol. 2011; 412:793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Crombrugghe B., Adhya S., Gottesman M., Pastan I.. Effect of Rho on transcription of bacterial operons. Nat. New Biol. 1973; 241:260–264. [DOI] [PubMed] [Google Scholar]
- 39. Ciampi M.S. Rho-dependent terminators and transcription termination. Microbiology. 2006; 152:2515–2528. [DOI] [PubMed] [Google Scholar]
- 40. Chan C.L., Landick R.. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J. Mol. Biol. 1993; 233:25–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.