Abstract
In the late phase of the HIV virus cycle, the unspliced genomic RNA is exported to the cytoplasm for the necessary translation of the Gag and Gag-pol polyproteins. Three distinct translation initiation mechanisms ensuring Gag production have been described with little rationale for their multiplicity. The Gag-IRES has the singularity to be located within Gag ORF and to directly interact with ribosomal 40S. Aiming at elucidating the specificity and the relevance of this interaction, we probed HIV-1 Gag-IRES structure and developed an innovative integrative modelling strategy to take into account all the gathered information. We propose a novel Gag-IRES secondary structure strongly supported by all experimental data. We further demonstrate the presence of two regions within Gag-IRES that independently and directly interact with the ribosome. Importantly, these binding sites are functionally relevant to Gag translation both in vitro and ex vivo. This work provides insight into the Gag-IRES molecular mechanism and gives compelling evidence for its physiological importance. It allows us to propose original hypotheses about the IRES physiological role and conservation among primate lentiviruses.
INTRODUCTION
Translation in eukaryotic cells largely relies on a strictly cap-dependent initiation mechanism. During this process the ribosome is recruited at the 5΄ terminus of the mRNA through a series of interactions that indirectly attaches the 40S subunit of the ribosome to the cap structure. A minimal set of at least 10 cellular proteins, known as eukaryotic Initiation Factors (eIF), is involved. The cap binding protein eIF4E interacts with both the 5΄ terminus of the mRNA and eIF4G, a central protein complexed to the RNA helicase eIF4A and its activator eIF4B. The preformed 43S pre-initiation complex (PIC) comprising eIF3, eIF2, the Met-tRNAi, eIF1, eIF1A and eIF5 bound onto the 40S small ribosomal subunit is then recruited on the mRNA owing to the eIF4G–eIF3 interaction. The 40S ribosomal subunit does not have any specific affinity for the mRNA, and eIF4E–eIF4G–eIF3 can be seen as a ‘molecular bridge’ linking the small ribosomal subunit to the 5΄-cap structure of the RNA. Part or all of this ribonucleoprotein complex then scans the 5΄UTR until it reaches the first AUG triplet, where the large ribosomal subunit joins the complex to form a ribosome competent for translation (1). Viruses are obligatory parasites, and evolution has selected many different mechanisms allowing the recruitment of the cellular translation machinery onto the viral mRNAs. Of peculiar interest, RNA viruses that are generally replicated by viral RNA-dependent RNA polymerase are not capped. Some of these mRNAs, such as picoRNAviruses, are translated through a cap-independent initiation mechanism known as the ‘internal entry of the ribosome’ (2,3). In this phenomenon, the translation machinery is recruited directly on the initiation codon, or at immediate vicinity. This process has been shown to rely on the secondary and tertiary structure of the 5΄UTR which is then called the Internal Ribosome Entry Site (IRES) (1). IRESes have been identified in many different viruses and have been classified in four major classes. Type I and type II IRESes, epitomized by the Poliovirus and the Encephalomyocarditis virus (EMCV) IRESes respectively, require all the initiation factors but eIF4E. These IRESes have a specific affinity for eIF4G/4A, and the IRES/eIF4G/4A ribonucleoprotein then recruits the 43S PIC through the interaction between eIF4G and eIF3 (4–8). Type III IRES which archetype is found in the Hepatitis C Virus (HCV) directly binds the 40S subunit of the ribosome placing the initiation codon in the P site (9–12). In terms of initiation factors, it requires only eIF2 to deliver the Met-tRNAi, eIF5 and eIF5B to promote the 60S ribosomal subunit joining (10). Type III IRESes also directly bind eIF3, but do not necessitate this factor to initiate translation per se. The suggested role of this interaction is to displace eIF3 from the 40S ribosomal subunit, and to titrate it away from the translation machinery (13). Finally, type IV IRES which was first characterized in the Cricket Paralysis Virus (CrPV), also directly binds the 40S ribosomal subunit of the ribosome and does not require the Met-tRNAi. A region of the IRES that mimics the codon–anticodon interaction within the P site promotes the first translocation event leaving the P site available for initiation at a non-AUG triplet (14–17). Since this classification was first issued, exceptions, and additional IRESes that do not readily fall into these classes have been described (18–25). Interestingly, type III and IV IRESes have the characteristic to directly interact with the 40S ribosomal subunit without any initiation factors. In addition, our lab has recently shown that type II IRESes molecular mechanisms could be slightly amended knowing that the recruitment of the PIC on EMCV IRES has been found to be independent of the eIF3/eIF4G interaction (5) but rather relies on direct binding of the 40S ribosomal RNA (26).
This ability to directly bind the 40S ribosome has also been reported to RNA sequences located in the human immunodeficiency virus (HIV) genomic mRNA (gRNA) (27). The HIV unspliced genomic RNA is exported to the cytoplasm in the late phase of the virus cycle and allows expression of both Gag and Gag–Pol polyproteins. As retroviruses, lentivirus mRNAs are produced by the cellular RNA polymerase II and are therefore capped and polyadenylated. Remarkably, their long 5΄ UTRs (335 and 545 nucleotides for HIV-1 and HIV-2, respectively) conceals some non-cognate initiation codons (CUG, UUG, GUG) but are deprived of AUG triplets (28). The counter selection of initiation codons within the 5΄UTR is a good indication for a cap-dependent initiation pathway involving ribosome scanning of this region. These two characteristics favor a canonical cap-dependent translation initiation pathway; however additional elements could impede cap-dependent initiation and could therefore be indicative of alternative pathways. First, lentiviral 5΄UTRs contain many stable structures that have distinct roles along the virus cycle (29–33). Particularly the 5΄UTR begins immediately with the long and very stable TAR stem-loop which is very detrimental for translation (34–37). Second, the viral Vpr protein induces the cell cycle arrest of infected cells in phase G2/M where the hypo-phosphorylation of eIF4E is known to severely downregulate cap-dependent translation (38). In this context, several mechanisms allowing cap-dependent translation of HIV-1 gRNA to overcome these detrimental factors have been proposed. In particular, Soto-Rifo et al. demonstrated that the cellular helicase DDX3 is required for translating mRNA which 5΄ terminus is involved in a stable stem loop, and specifically allows translation of transcripts beginning with TAR (36). RNA helicase A (also known as DHX9) has also been shown to stimulate cap-dependent translation of HIV-1 mRNA (39). Independently, it has been hypothesized that HIV-1 gRNA could be translated during G2/M phase, using the cap-binding complex, composed of CBP80/CBP20, instead of eIF4E (40). In parallel, internal ribosome entry within HIV-1 5΄UTR has been described and extensively documented (41–45), it is stimulated by an oxidative stress (46), in specific cell lines (47) and in G2/M phase of the cell cycle (42,48). In contrast, HIV-2 gRNA does not contain an IRES in the 5΄UTR but within the 350 first nucleotides of Gag ORF. It has been shown to direct translation from the bona fide Gag initiation codon, but also from two in-frame AUG triplets yielding N-terminally truncated isoforms of Gag (49). This occurs in infected Jurkat cells and shorter isoforms can be incorporated in virions (49). This IRES known as HIV Gag-IRES has been identified in other primate lentiviruses such as HIV-1 and SIVMAC (27,28,43,50–52). The HIV1 Gag-IRES can direct initiation at 336AUG yielding the full length Gag polyprotein (Gag p55) and at 759AUG to translate an N-truncated isoform named Gag p40. The role of this short isoform is unknown but 759AUG mutation decreases the mutant virus infectivity ((43) and C. Swanson, personal communication). Interestingly, the presence of 759AUG and its nucleotide context are very conserved in clinical isolates derived from HIV-1 positive patients. Furthermore, the expression of a p40 truncated Gag isoform has been observed for these clinical isolates in both reticulocytes lysate and Jurkat T-cells (52,53). The exact initiation mechanisms involved for both IRESes remain elusive, but the Gag-IRES was shown to have the conserved ability to directly and independently bind the 40S subunit of the ribosome and eIF3 (27). Analysis of the initiation complexes paused on HIV-2 Gag-IRES revealed that in addition to the 40S, they contain eIF2, eIF3, eIF4A, eIF4G and eIF5 but neither eIF1 nor eIF4E (27).
In summary, translation initiation on the unspliced gRNA can clearly be achieved through at least three different mechanisms (34,54,55). However, a role for each of them is still lacking to date. Are they alternatively used at different stages of the virus cycle? Are they cell-type specific? What is the role of the different isoforms? Are they all used indifferently to maximise Gag production? Because the three mechanisms are intricate, it is difficult to assay them separately. For this reason, IRES-dependent Gag translation remains controversial (56–58). Here, following a ‘reverse’ strategy, we further characterize the Gag-IRES structure and properties focusing on its ability to bind the 40S ribosome to gain insight into its function. This led us to propose an original mechanism for ribosome entry on this IRES and to suggest a potential physiological role for this non-conventional initiation mode.
MATERIALS AND METHODS
DNA constructs
The HIV-1 sequence used in this study corresponds to the 5΄UTR followed by the gag coding region from the NL-4.3 strain (Genbank: AF324493.2). Deletion of the site 1 (nt. 363 to nt. 485), the site 2 (nt. 632 to nt. 736) or both was performed by site-directed mutagenesis with corresponding primers (supplementary Table) on the pHIV-1 UTR-AUG (761)-Renilla plasmid (52) and on the NL-4.3 strain. Fragments of the IRES Gag used for filter-binding assays were generated by PCR using the primers described in the supplementary table. The bicistronic construct pT7-Puromycin/Renilla was built by inserting into the pRenilla vector digested by PvuII and BamHI (59), a PCR fragment, flanking with the Pvu II restriction site in 5΄ and a multiple restriction site (Pst I/Sal I/Bam HI) in 3΄, and coding for the T7 promoter followed by the globin 5΄UTR and the puromycin coding region. The IRES gag coding region (nt. 331 to nt. 851, WT) and associated deletions (named ΔP1, ΔP1–P3, ΔP1–P4, ΔN1, ΔP1–N6, ΔP8–P10, ΔN5–P10, ΔN1–P10, ΔP4–P10, ΔP1–P4/P8–P10 and ΔN1/P8–P10) were amplified by PCR with corresponding primers (supplementary Table) to be inserted into the intergenic region (IGR) of the pT7-Puromycin/Renilla plasmid digested by Sal I and Bam HI restriction enzymes. DNA template production for in vitro transcription was performed either by digestion of pT7-Renilla and pT7-Puromycin/Renilla plasmids with the Eco RI restriction enzyme, either by PCR using the T7 5΄UTR HIV-1 and the Hiv1-3΄UTR primer (the sequences of all the oligonucleotides used in this study are provided in the Supplementary table).
In vitro transcription, toeprinting and filter binding assays were essentially as in Chamond et al. (26), ribosome purification, sucrose density gradients, IRES probing and footprinting by SHAPE, and in vitro translation were as in Angulo et al. (9), these methods are detailed in the supplementary Materials and Methods.
T1 or V1 RNAse footprinting assays
Twelve picomoles of RNA were resuspended in 96 μl of water (for a final concentration of 100 nM), and were denatured and renatured under the same conditions in the same buffer as described for the SHAPE analysis (supplementary Materials and Methods). After a 10 min incubation at 37°C, three molar equivalent of the purified 40S ribosomal subunit (300 nM final of 40S), or the same volume of buffer (sample—40S) were added, and the mixture was incubated for 10 min at 37°C. For each condition, the mixture was split in two tubes. In one, 1 μl of RNAse V1 (Ambion) diluted at 1/1000 from the commercial stock tube or RNAse T1 (Ambion) diluted at 1/10 000 was added. In the other tube, 1 μl of water was added (negative control). The mixture was incubated for 10 min at 37°C. The reaction was blocked by the addition of SDS 0.2%, EDTA 5 mM, AcONH4 0.5 M and 20 ng of glycogen. After phenol chloroform extraction, the RNA was precipitated with 2.5 volumes of ethanol. The cleavage sites were revealed by elongating fluorescently labeled primers, exactly as described for the SHAPE experiment. All experiments were repeated three times and the mean and the standard error to the mean values for each nucleotide were reported and analysed. The thresholds of moderate and strong hits were 1 and 1.5 respectively. Were considered as footprint site, nucleotides which reactivity varies by 2-fold, by at least 0.2 (in absolute value) between the two conditions, and that reach a minimal value of 0.7 in at least one of the conditions.
RNA secondary structure modeling flowchart
Reactivity profiles were used as constraints for a secondary structure modeling based on the RNAsubopt software (60,61). SHAPE data (62) were integrated as ‘soft constraints’, meaning that the reactivity values were translated into pseudo-energies as described (63).
Susceptibility to RNAses cleavage was used as hard constraints, by converting reactivity scores into ‘hard’ structural constraints (64). For the V1 and T1 enzymes, positions whose reactivity exceeded a given cutoff were respectively forced to be paired and unpaired. For each enzyme, we derived two sets of constraints associated with two cutoffs, identifying moderate to highly reactive nucleotides, having a score >1, or >1.5, respectively. This led to six sets of parameters for RNAsubopt: 1M7, NMIA, V1++, V1+/–, T1++ and T1+/–.
We outline our integrative modeling approach, which will be further detailed in a forthcoming manuscript (A.S et al., in preparation). At the thermodynamic equilibrium, a given RNA can theoretically adopt multiple stable alternative structures, where each structure is associated with a probability within the space of all the possible conformations (Boltzmann ensemble). We reasoned that the correct structure(s) should be, at the same time, energetically stable and supported by several experimental conditions. For this reason, we coupled a stochastic sampling from the Boltzmann ensembles associated with the experimentally derived constraints, with a clustering across experimental conditions, to generate a structural models that are well-supported by available data.
We first used Boltzmann sampling (65) to generate a predefined number of stable structures, compatible with the constraints derived for each condition. We used the stochastic sampling mode of RNAsubopt (-p option) to generate energetically stable structures that are either fully compliant with constraints derived from enzymatic data (hard constraints (64)), or constitute reasonable trade-offs between thermodynamic stability and compatibility with SHAPE data (soft constraints, using the pseudo-potentials of Deigan et al. (66), see (63) for details). We assumed the folding process to be modular, and our functional folding to be mainly composed of short structural elements. For this reason, we limited the maximal base pair span to 240 nts, thereby penalizing - frequently spurious—long-range interactions. We sampled 2000 structures per condition (12 000 structures in total), and merged the structures while keeping labels to retain the origin of each structure. In order to detect recurrent RNA architectures, the merged sets of models were clustered, using the classic base-pair distance as a measure of dissimilarity. Two clustering algorithms (affinity propagation (67) and Mini Batch K-means (68)), both implemented in the scikit-learn Python package (69), were tested to agglomerate and identify recurrent structures. One of the advantages of affinity propagation resides in its low computational requirements, and its capacity to determine an optimal number of clusters according to coherence criteria. Unfortunately, we observed that the inherently noisy nature of Boltzmann sampling induced too many clusters, leading to reproducibility issues. For this reason, we ultimately adopted the Mini Batch K-means clustering, explicitly setting the number of clusters to 6. We then sought to identify clusters that are homogeneous, stable and well supported by experimental evidences, leading to the identification of the following objective criteria:
Represented conditions (A): Our primary target is to favor clusters that are compatible with multiple experimental conditions. However, the larger sampled sets required for reproducibility tend to populate each cluster with structures from all conditions. We thus associated with each cluster the number of represented conditions, defined as the number of conditions for which the accumulated Boltzmann probability in the cluster exceeds a predefined threshold;
Boltzmann weight (B): Structures that are found in a given cluster may be unstable, and should be treated as outliers. For this reason, we computed the cumulated normalized Boltzmann probabilities within the cluster, to favor stable clusters consisting of stable structures;
Average cluster distance (C): We observed a general tendency of clustering algorithms to create heterogeneous clusters when faced with noisy data. We thus associated with each cluster the mean distance between pairs of structures, estimated as the average distance to the MEA (70) for the sake of efficiency, in order to neglect clusters that were too diverse.
We then restricted our analysis to clusters that were found on the 3D Pareto Frontier (71) with respect to these criteria. In other words, we only retained the subset of clusters that were not strictly dominated by another cluster with respect to all of these criteria. This strategy resulted in two optimal Pareto clusters. After detecting the optimal Pareto cluster(s), we need to identify representative structure for each cluster. We chose the maximum expected accuracy (MEA) structure (70) as the representative structure for each cluster, which is defined as the secondary structure whose structural elements have highest accumulated Boltzmann probability within the cluster. This resulted in two structures which we narrowed down to a single candidate using compatibility with the 1M7 SHAPE data as our final discriminatory criterion. Specifically, we built a set of highly reactive positions in the SHAPE profile by using an arbitrary threshold (0.5) on the reactivity. For each structure, we considered the overlap between the SHAPE-derived set, and the set of positions predicted to be unpaired. Interestingly, the most compatible structure turned out to be quite robust, and was impervious to the choice of alternative thresholds (0.4–0.7), or reagent type (NMIA). Finally, the selected structure was evaluated for covariation using R-Chie (72) and 67 sequences from the ‘los Alamos compendium’ representing the variability of HIV-1 (see Supplementary Figure S6), the results were used as a basis for a marginal manual refinement.
Stoichiometry assay
The stoichiometry of 40S ribosomal subunit binding to the entire Gag-IRES or the first ribosome binding site was determined by filter binding assay, using conditions previously described (73). Briefly, radiolabeled RNA was denatured in water for 1 min at 80°C and cooled down to at 37°C in FB buffer (20 mM Tris pH 7.5, 100 mM AcOK, 200 mM KCl, 2.5 mM MgCl2 and 1 mM DTT) and the solution was equilibrated for 15 min at 37°C. Then, different concentrations of the purified 40S ribosomal subunit were added (for a final 40S concentration of 750, 375, 187.5, 93.8, 46.9, 23.4 or 0 nM) to 250 nM final of radiolabeled RNA. After a 15 min incubation at 37°C, the mixture was filtered through a nitrocellulose and a nylon membrane. For each condition, the radioactivity was quantified on both membranes to determine the proportion of free and 40S bound RNA. Results were treated with Prism5 (Graphpad software).
Cell culture—RNA transfection and western blot analysis
Jurkat T-cell line, obtained from ATCC, was cultured in RPMI-1640 medium supplemented with 10% fetal calf serum, penicillin, streptomycin, Hepes pH 7.5, l-glutamine and sodium pyruvate.
For RNA transfection, 5 × 105 cells were resuspended in 10 μl of buffer R in presence of capped mRNAs, and immediately transfected using the neon transfection kit (Invitrogen). Cells were incubated in RPMI medium for 90 min at 37°C and then resuspended in RIPA buffer (150 mM NaCl, 1% Na-deoxycholate, 1% Triton X-100, 0.1% SDS, 10 mM Tris pH 7.8) supplemented with protease inhibitor cocktail (Roche). Cellular extracts were sonicated; equal amount of protein was resolved on a 12% SDS-PAGE and transferred to a PVDF membrane. Proteins of interest were detected by western blot with antibody targeting the HA tag or the HIV-1 p24 (183-H12-5C) (74–76) and revealed with the Supersignal West Dura extended Duration signal kit (Pierce).
RESULTS
Footprinting the Gag-IRES structure with the 40S ribosome
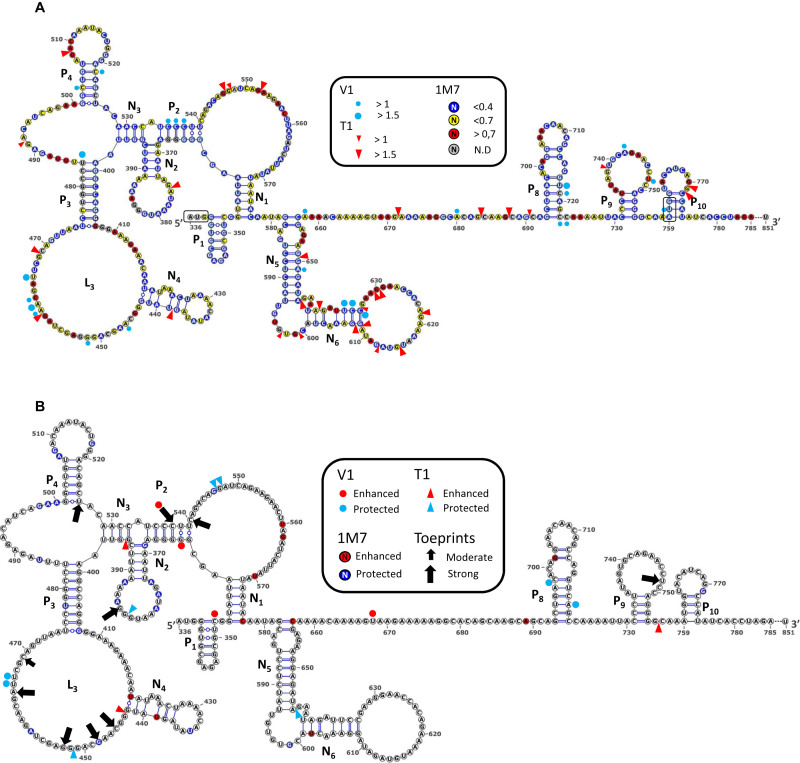
HIV-1, HIV-2 and SIVMac Gag IRESes have the ability to directly recruit 40S ribosomal subunit (27). In order to identify the determinants involved in HIV-1 Gag IRES ribosome binding, we undertook to footprint the 40S subunit on the IRES using chemical and enzymatic probes. First, the isolated HIV-1 Gag-IRES (nucleotides 336–851) was incubated with T1 or V1 RNAse, N-Methyl Isatoic Anhydre (NMIA) or 1 methyl-7 isatoic anhydre (1M7). T1 RNase specifically cleaves RNA 3΄ of single stranded guanosines, whereas V1 RNase cleaves double-stranded positions essentially independently of the nucleotide identity (77,78). NMIA and 1M7 are SHAPE reagents that react with flexible riboses, thus detecting unpaired nucleotides (79,80). The main difference between the two SHAPE reagents is their half-life, resulting in essentially compatible but slightly different modification profiles (81). Experiments with each probe were carried out in triplicate, and the results obtained were reported on the secondary structure model proposed by Weill et al. (28). The reactivity patterns obtained (Supplementary Figures S1, S2, S3 and S4A and ‘probing data’ provided in the supplementary material) are essentially in agreement with the model proposed that had been obtained with other probes (28). In order to further improve the quality of the RNA structure prediction, we developed an original integrative modelling strategy (see Materials and Methods) in which the secondary structure was independently modeled with RNAsubopt (60) using RNAse T1, RNAse V1, NMIA or 1M7 reactivity maps as constraints. In brief, two thousand models issued from each modelling batch were sampled and the (12 000) models gathered were clustered according to their base pair distance. Six families of structural models were obtained and ranked according to their representativeness, their homogeneity and their Boltzmann weight. Each cluster was represented by its MEA structure (maximum expected accuracy) (70), the fit of which with the SHAPE data was calculated (see material and methods). To refine our model, we further analysed its base pairs in a comparative setting, using R-Chie (72) on sequences from the Los Alamos compendium (82) to produce covariation scores (Supplementary Figure S6). Most of our predicted model was confirmed by the results of the covariation analysis, but we marginally eliminated a few base pairs which appeared not to be supported by most isolates, and somewhat reactive to one or another probe. Figure 1A shows the structural model that best fitted with all our data. Our novel model features most of the pairings originally proposed but P5, P6 and P7 for which a new arrangement that takes better the data into account was identified (pairings N5 and N6). In addition, we propose new pairings (N2 and N3) forming a three-way junction between P2 and P3. This new model is compatible with most of the experimental data, meaning that nucleotides in double strand are poorly reactive to 1M7 and are in some occurrences substrate for RNAse V1; while unpaired positions are reactive to 1M7 among which some of the Gs are cleaved by RNAse T1. This observation suffers some exceptions, for example nucleotides in the N2 loop are very poorly reactive to 1M7. This may reflect the existence of a pseudoknot yet to be identified.
Figure 1.
RNA secondary structure model of the HIV1 Gag-IRES and 40S ribosome subunit footprints and toeprints. (A) Schematic representation of the secondary structure model of Gag-IRES. Nucleotides are colored according to their reactivity toward 1M7 as indicated in the box. Red triangles and blue dots represent RNAse T1 and V1 cleavages. Pairings numbering (Pn and Nn) are as described in the text. Nucleotide numbering is from the +1 of transcription (First nucleotide of TAR). Experimental values result from the mean of three independent experiments (see material and methods and supplementary material). (B) Footprints and toeprints of the 40S ribosomal subunit on Gag-IRES. As indicated in the box, the colors and signs compare the reactivities without and with saturating concentration of 40S ribosomal subunit. Nucleotides in red are more reactive to 1M7, while nucleotides in blue are less. Red and Blue triangles indicate nucleotides exposed or protected to RNAse T1 respectively. Red and Blue dots indicate nucleotides exposed or protected to RNAse V1 respectively. Black Arrows indicate premature RT stops observed in the presence of 40S ribosomal subunits. Small and big arrows are respectively moderate and strong toeprints. Experimental values result from the mean of three independent experiments (see material and methods and supplementary material).
The same experiments were repeated in presence of saturating purified 40S ribosomal subunit. Cleavages or reactivities obtained were compared with those observed in absence of 40S ribosomal subunit. Both experiments were carried in triplicate, positions considered to be more or less reactive toward each of the probes in presence of the ribosome were defined following a statistical approach detailed in the material and methods section. Modification of the reactivity patterns, known as footprints, may have several significations. First the ribosome bound to the RNA may hinder the access of the reagent to the nucleotide, in which case a protection reveals the actual binding site. In this case, steric hindrance from large probes such as RNAses map the global binding site, while small SHAPE reagents reveal short distance interactions. Alternatively, a modification of the reactivity pattern may reveal a structural rearrangement of the RNA upon ribosome binding. This is clearly the case for reactivity enhancement. Note that positions more reactive toward the SHAPE reagent may be close to the ribosome if not involved in an atomic interaction (see examples in (9)). Reactivity alterations in presence of the ribosome were reported on the structure and are shown in Figure 1B (Supplementary Figures S1–S3 and ‘probing data’ provided in the supplementary material). Although they appear more concentrated in some regions, the footprints are scattered all over the secondary structure and do not allow to readily define one binding site. Dispersed footprints could reflect a three dimensional determinant or else, the effect of local structural rearrangements in addition to the ribosome protection.
Toeprinting the 40S ribosome on Gag-IRES
To further characterize the interaction between the 40S and the IRES, we undertook to localize the ribosome on the IRES by toeprinting assay. In this assay, the HIV-1 Gag-IRES RNA is reverse transcribed in the presence or in the absence of the small ribosomal subunit. The reverse transcription premature stops observed only in the presence of the 40S ribosomal subunit are known as ‘toeprints’ and are assumed to reflect the reverse transcriptase bumping against the ribosome. When 48S or 80S complexes are paused with the initiation codon properly accommodated within the P site, toeprints are usually observed 15–18 nucleotides downstream the A of the AUG triplet (83,84). In addition, as mentioned above for the footprinting assay, ribosome binding may also modify the secondary structure of the RNA stabilizing some structures and thus inducing premature reverse transcription stops which do not directly reflect the 40S position. Ten unambiguous toeprints of moderate and strong intensity (see material and methods) were observed (Figure 1B and Supplementary Figure S5). Surprisingly, we did not detect any toeprint within the twenty first nucleotides downstream any of the two initiation codons. Consistent with what was observed for the footprinting assay, a toeprint is observed in the N2 loop while five of them cluster in the P3 loop (L3). This suggests that these regions are in close contact with the 40S subunit. Additional three premature RT stops were detected at U526, C537 and C540, and a very strong toeprint was consistently observed few nucleotides upstream of the second AUG, at U748.
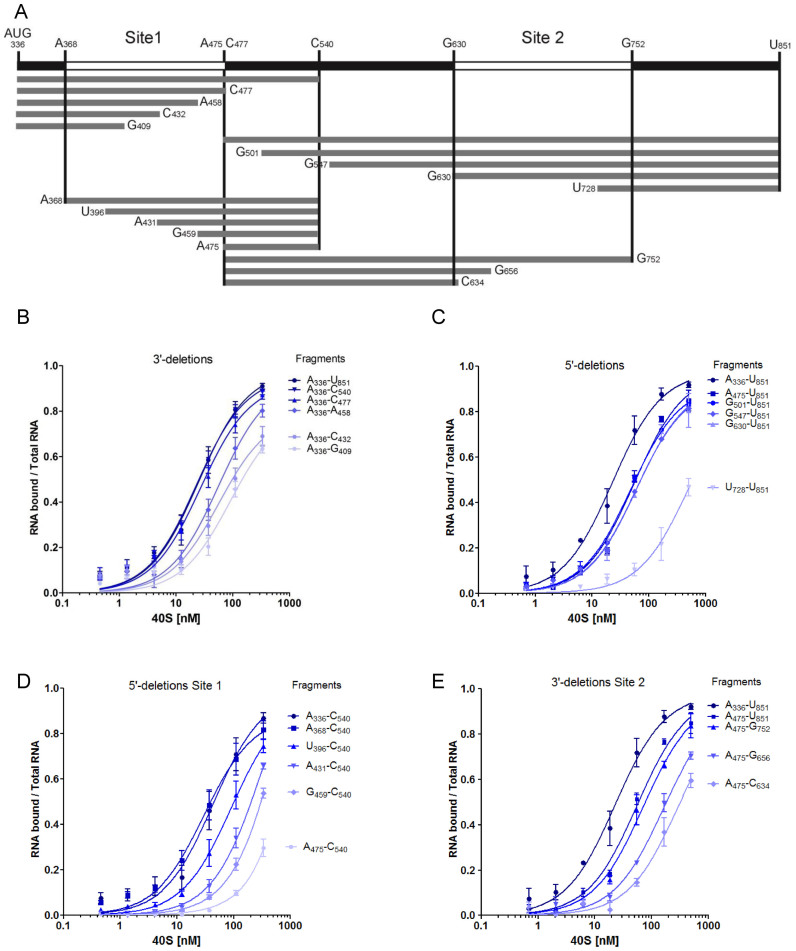
Minimal ribosome binding sites as defined by progressive deletion
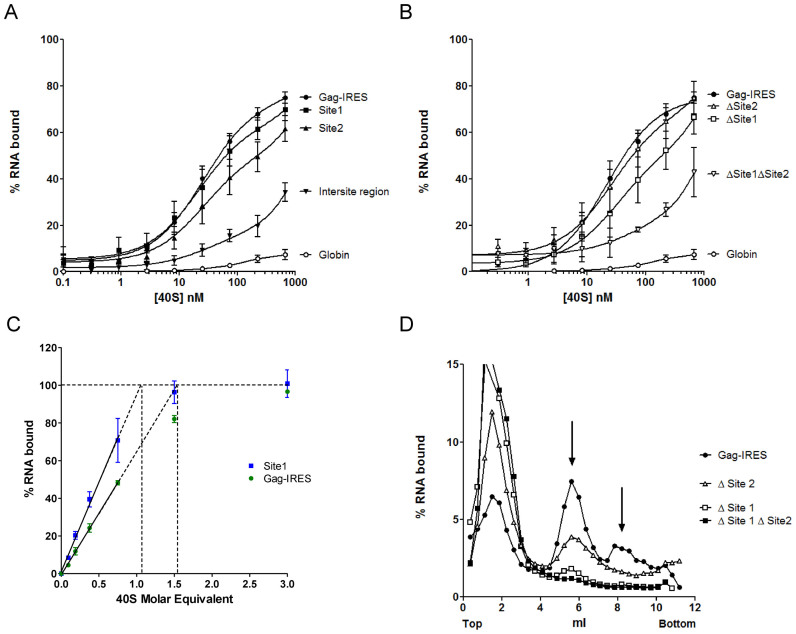
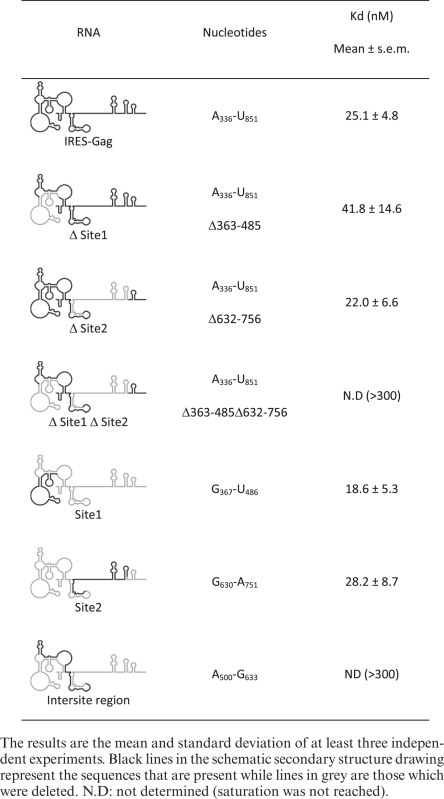
Toeprinting and footprinting assays suggest that N2 and P3 loops and probably some sequences downstream are important determinants for ribosome recruitment. However, they do not permit to define the minimal region necessary for binding. We undertook to map the minimal binding site by progressive deletions from both the 5΄ and the 3΄ terminus (Figure 2A). In a previous report, we had shown that a fragment of the Gag-IRES ending at nucleotide 572 efficiently binds the ribosome (27). To extend these results, we made several truncations ending at C540 (3΄ of P2), C477 (3΄end of L3), A458, C432 and G409. The affinity of these RNA fragments for purified 40S subunit was assayed by filter binding assay. Transcripts terminating at C540 and C477 bind the 40S subunit as well as a fragment encompassing the full length IRES (336–851) (Figure 2B). Further elimination of twenty nucleotides within L3 (fragment A336–A458) increases the Kd by ∼2-fold (Table 1). For transcripts further truncated within L3, saturation is not reached at 300 nM of 40S impeding the determination of a precise Kd, but it is evaluated to be superior to 100 nM. The same strategy was carried out from the 5΄ terminus, and transcripts beginning at A475, G501, G547, G630 and U728 were assayed for their ability to bind the 40S ribosome. Surprisingly, transcripts starting at the end of L3 (C475–U851) which lacks the above defined binding site (A336–C477) still recruit the 40S ribosome with only a 2-fold drop of the affinity (equivalent to a 2-fold Kd increase) (Figure 2C and Table 2). Even more strikingly, further 5΄ deletion up to G630 does not have any notable effect on ribosome recruitment. Further deletion in the 3΄ direction yields an RNA fragment (U728–U851) that does not bind the ribosome over the background. These two series of experiment defined two non-overlapping RNA fragments A336–C477 and G630–U851 that specifically bind the 40S ribosomal subunit with a significant affinity and specificity (Kd = 27 ± 3 and 61 ± 8 nM respectively). This suggests that the Gag open reading frame actually harbors two 40S binding sites. In order to precisely map the first binding site, we constructed a series of transcripts ending at C540 while starting 3΄ to A336 which were assayed for 40S subunit recruitment. Ribosome binding is not affected for transcripts preserving N2 (A368–C540), but significantly drops upon N2 deletion (U396–C540), and is completely lost upon further deletion within L3 (Figure 2D and Table 1). This circumscribes a first minimal binding region between A368 and C477 (Site 1). In parallel 3΄ truncations of transcripts starting with A475 were assayed for purified 40S subunit binding. While deletion of all nucleotides up to G752 does not significantly alter the Kd, further truncation within P9 rapidly leads to the loss of significant binding activity (Figure 2E and Table 2). Together with the first series of deletion, these results define a second 40S binding site laying between G630 to G752 (Site 2). To confirm these conclusions, the two putative binding sites were synthetized as independent fragments (‘Site 1’ and ‘Site 2’). The fragment lying between the two sites (intersite region: A500–G633) and the globin gene were used as negative controls (Figure 3A and Table 3). As expected, the transcript corresponding to Site 1 (G367–U486) binds the 40S ribosome subunit with an affinity similar to the full length Gag IRES (Figure 3A), while the fragment corresponding to Site 2 (G630–A751) also independently recruits the ribosome but with a slightly reduced affinity as compared to the full length Gag-IRES. The intersite fragment (A500–G633) does not show a significant affinity for the 40S subunit, although we observed some more RNA-ribosome complex than when using the globin mRNA (Figure 3A and Table 3). In parallel, we followed the complementary approach which consists in deleting the nucleotides corresponding to Site 1 and/or Site 2 from the full length IRES, and evaluate the ability of these fragments to recruit the ribosome. As can be observed in Figure 3B, Site 2 deletion (Δ Site 2: A336 to U851Δ632–736) does not significantly influences 40S binding, while a transcript devoid of the region corresponding to Site 1 (Δ Site 1: A336 to U851 Δ363–485) is still able to significantly bind the ribosome although with a slightly reduced affinity. Finally, a transcript from which both sites have been deleted (Δ Site 1 Δ Site 2) is no longer able to recruit the 40S subunit (Figure 3B).
Figure 2.
Mapping the 40S ribosomal subunit on HIV1 Gag-IRES using truncated fragments of the IRES. (A) Schematic representation of the fragments used in this study. Nucleotides numbering is from the +1 of transcription (first nucleotide of Tar). (B–E) Binding curves of 32P-labeled fragments to purified 40S ribosomal subunits as measured by filter binding assays (see Materials and Methods). Fragments were obtained by successive deletions from the 3΄ (B) and the 5΄ (C) end of Gag-IRES, or from the 5΄ of A336–C540 fragment (D) or by the 3΄ of the C477–U851 fragment (E). The results are the mean of at least three independent experiments ± s.e.m.
Table 1. 40S binding affinities of Gag-IRES Site1 deletion mutants as evaluated by filter binding assay.

|
Table 2. 40S binding affinities of Gag-IRES Site2 deletion mutants as evaluated by filter binding assay.

|
Figure 3.
Two sites within the Gag IRES can independently recruit the 40S ribosomal subunit. (A) Binding curves of 32P-labeled Site 1 (▪, G367–U486), Site 2 (▴, G630–A751), full length HIV-1 Gag-IRES (•, A336–U851), the intersite region (▾), and the globin transcript (○) with purified 40S ribosomal subunits. (B) Binding curves for HIV-1 Gag-IRES deleted of site 1 (Δ Site 1: A336–U851Δ363–485), of site 2 (Δ Site 2: A336-U851Δ632–756), or of both sites (Δ Site 1 Δ Site 2: A336–U851Δ363–485Δ632–756) as compared to the full length HIV-1 Gag-IRES (A336–U851), and to the globin transcript. (C) Determination of the stoichiometry of the 40S ribosomal subunit and the Gag-IRES RNA. A constant concentration of RNA corresponding to the whole IRES (▪) or to the isolated site 1 (G367–U486 •), tenfold over the Kd of the Gag-IRES/40S complex (250 nM) was incubated with increasing concentration of 40S ribosome. The proportion of RNA in complex in the mix was determined by filter binding assay. The % of complex was plotted as a function of [RNA] equivalent of 40S, the linear part of the curve was calculated drawn, and extrapolated to 100% binding. The values are the mean of at least three independent experiments ± standard deviation. The two curves are statistically different (P < 10-4). (D) Analysis of the complexes formed by the full length HIV-1 Gag-IRES deleted of site 1 (Δ Site 1: A336–U851Δ363–485), of site 2 (Δ Site 2: A336–U851Δ632–756), or of both sites (Δ Site 1 Δ Site 2: A336–U851Δ363–485Δ632–756) and the full length HIV-1 Gag-IRES (A336–U851) with the 40S ribosomal subunit. 32P-labeled RNA transcripts were separated on 10–30% sucrose gradients. Peaks were identified by comparison with UV profiles obtained with purified 40S ribosomal subunits.
Table 3. 40S binding affinities of the two sites as evaluated by filter binding assay.
|
Two regions can recruit the 40S subunit independently
The identification of two distinct binding sites raises the question of whether two 40S subunits can be recruited on the same RNA, or else if these two sites are mutually exclusive. In order to answer this question, we determined the maximal ratio of 40S subunit over the full length RNA in a 40S/IRES complex (85). An RNA concentration 10-fold higher than the dissociation constant for the 40S subunit (250 nM) was incubated in the presence of increasing amount of 40S subunit. The fraction of RNA bound to the 40S was determined by filter binding assay (Figure 3C). The experiment was carried out with an RNA containing the minimal site one and with the full length RNA. Using the isolated Site 1, 100% of binding would be reached with 1.05 molar equivalent of 40S ribosome which is consistent with the presence of only one site. In contrast, when using the full length IRES, 1.5 molar equivalent would be required to reach saturation. This result is compatible with the presence of two 40S binding sites on this RNA and could suggest that binding of the ribosome on one site disfavor the simultaneous recruitment on the second site.
The 40S subunit–RNA complexes were then analysed on sucrose gradients using the full length IRES, transcripts in which the sequences corresponding to one or the other site have been deleted (Δ Site 1 or Δ Site 2) and transcripts in which both sites have been removed (Δ Site 1 Δ Site 2) (Figure 3D). In addition to the peak corresponding to the free RNA, the full length IRES sediments essentially as one 40S subunit, but we also observed one peak in a heavier fraction that could correspond to two 40S ribosomal subunits bound on the same RNA. Deletion of Site 2 yields a RNA that sediments as a complex with a single 40S subunit although with a lower efficiency than the WT construct. In contrast, Δ Site 1 RNA essentially runs as a free RNA and only a small amount of complex was repeatedly observed. Finally, no high molecular weight complex is observed with the Δ Site 1 Δ Site 2 RNA. Altogether these results show that there are multiple determinants of 40S subunit recruitment along the 400 first nucleotides of Gag open reading frame. Site 1 and Site 2 can recruit the ribosome independently, although Site 2 presents a lower affinity and yields complexes that appear unstable on a sucrose gradient.
Impact of the ribosome binding site on Gag translation in vitro and ex vivo
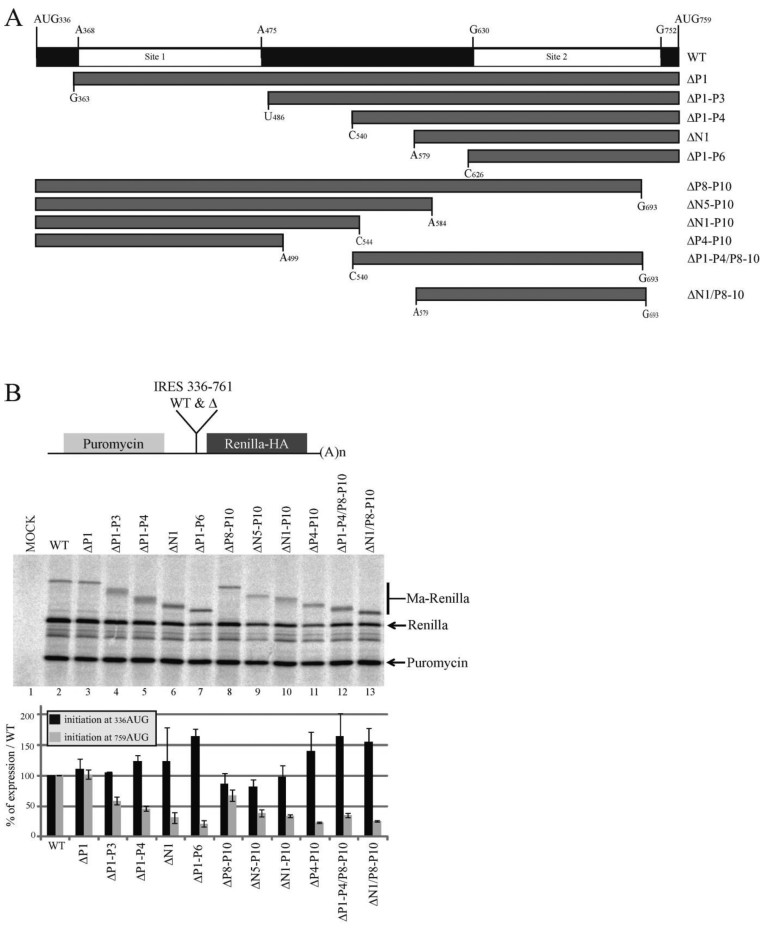
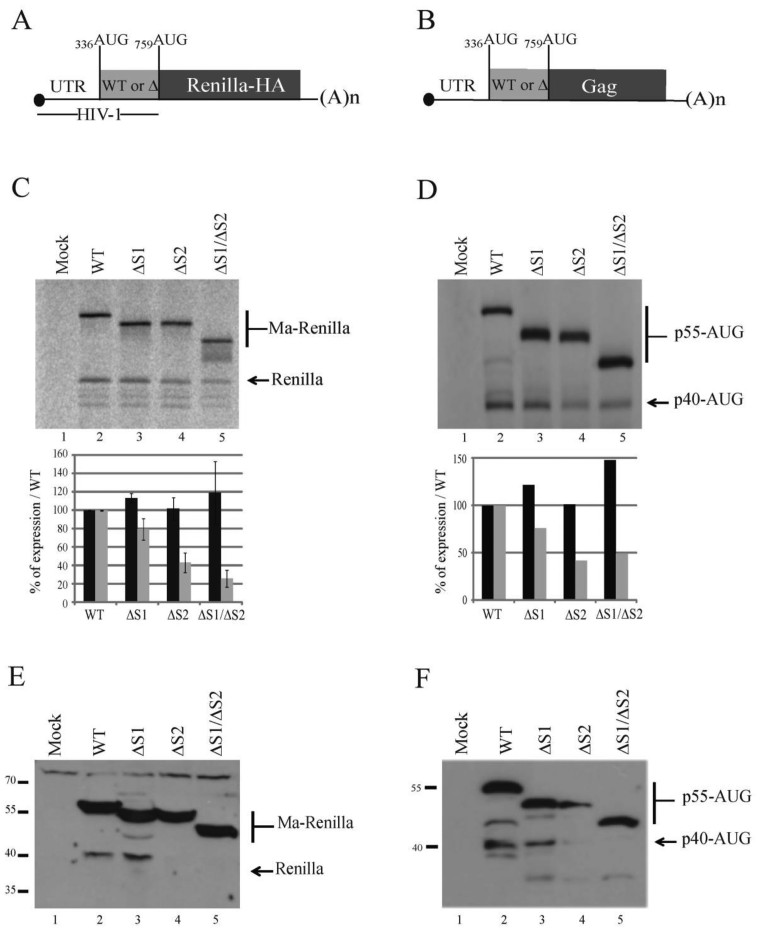
We then assayed the impact of Site 1 and Site 2 ribosome binding sites on Gag translation. First we intended to monitor only the IRES activity in vitro, to this end the whole IRES, from 336AUG to 759AUG was inserted in the intergenic region of a bicistronic reporter. In this construct 759AUG is fused to the HA-tagged Renilla luciferase ORF (Figure 4). When this mRNA was used to program a Rabbit Reticulocyte Lysate containing 35S methionine, we observed the production of the two isoforms (MA-Renilla and Renilla) confirming once again that the Gag-IRES can direct initiation to the upstream AUG (Figure 4B, lane 2). The intergenic fragment was then progressively truncated from the 5΄ or the 3΄ end eliminating ribosome binding site 1 or 2 (Figure 4A). All the constructs were designed to preserve an open reading frame encoding a shortened full length protein, but an intact isoform translated from 759AUG. As observed on Figure 4B, progressive deletion in Site 1 stimulates translation at 336AUG. This is specifically obvious after the elimination of the 5΄ highly structured domain comprising N1, P2, N2, N3, P3 and P4 (see ΔP1–N6, Figure 4B, lane 7). In contrast, Site 2 deletion does not strongly influence initiation at 336AUG. Translation from 759AUG was gradually impaired concomitantly with the progressive deletion of both Site 1 and Site 2 (Figure 4B). The impact of the two ribosome binding sites was then evaluated in monocistronic constructs encompassing the 5΄UTR and the Gag IRES followed by either the HA-tagged Renilla luciferase ORF (Figure 5A) or the Gag ORF (Figure 5B). In this context, in vitro translation from 336AUG remains unaffected by the deletion of both sites independently or together (Figure 5C and D), confirming that under such conditions the translation of p55 Gag is mainly driven by the 5΄UTR in RRL. In contrast translation of the p40 isoform is slightly reduced upon Site 1 deletion, strongly affected after Site 2 elimination and almost abolished when both ribosome binding sites were removed (Figure 5C and D). Translation efficiency of the same constructs ex vivo was then addressed by directly transfecting the different capped and poly-adenylated RNAs in Jurkat T cells and analyzing Renilla (Figure 5E) or Gag (Figure 5F) production by western blot analysis. Results were similar to those observed in vitro, in summary, Site 1 deletion does not affect translation, while deletion of Site 2 abrogates the shortest isoform translation without affecting Gag p55 translation. Finally, only the Full length Gag protein is produced when using an artificial construct deleted for both sites (Δ Site 1 Δ Site 2) (Figure 5E and F). In conclusion, Site 2 is strictly required for translation from 759AUG which is expected to be essentially IRES-dependent. Although the translation of the full length Gag protein can undoubtedly be promoted by the sequences downstream 336AUG most of the translation observed using 5’UTR-containing constructs rather reflect cap-dependent initiation preventing the observation of an IRES-dependent translation.
Figure 4.
The presence of the ribosome binding sites influence IRES driven HIV-1 Gag translation in vitro. (A) Schematic representation of the fragments of the Gag coding region IRES used as intergenic region in the in vitro bicistronic translation assay (nucleotides numbering are from the +1 of transcription). (B) Schematic representation of the bicistronic construct. The first gene encodes for the puromycin N-acetyl transferase. In the intergenic region are cloned the Gag IRES fragments with 759AUG fused to the HA-tagged Renilla luciferase open reading frame (upper panel). Translation of the second gene from 336AUG yields a protein which is a fusion of the Gag Matrix sequence with the Renilla Luciferase (Ma-Renilla), while initiation from 759AUG yields the Renilla luciferase from. RRL was programmed for 45 min at 30°C with bicistronic mRNAs that contain in the intergenic region the full length Gag IRES (nt.331 to nt.761) or the different truncated fragments described in panel A (lanes 2–13), or with water (lane 1). Protein products were resolved by 12% SDS-PAGE and quantified using with a PhosphorImager. Expression initiated at both 336AUG and 759AUG codons was normalized to wild type expression and to the puromycin; Black and grey squares of the histograms represent the quantification of translation initiated at 336AUG (p55) and 759AUG (p40) codons respectively. Data are representative of three independent experiments, error bars are ± the S.E.M.
Figure 5.
The presence of the ribosome binding sites influences the translation of HIV1 Gag short isoform both in vitro and ex vivo. Schematic representation of the monocitronic mRNA encoding for (A) the HA-tagged renilla luciferase reporter gene under controlled of the HIV-1 sequence from the +1 of transcription up to the 759AUG codon, or (B) the HIV-1 Gag mRNA starting from the +1 of transcription and ending at the stop codon of Gag open reading frame. Both initiation codons are indicated. HIV-1-Renilla (C) or UTR-GAG (D) mRNAs (200 fmol) containing either the Full length Gag IRES (lane 2), or fragments of the Gag IRES (Δsite 1 (Δ363–485), Δsite 2 (Δ632–756) and Δsite 1/Δ site 2 (Δ363–485Δ632–756); lanes 3–5) were translated in RRL during 45 min at 30°C. Protein products were analyzed and quantified as previously (Figure 4). Jurkat T-cells were transfected with mRNAs coding with the HA-tagged renilla luciferase constructs (lanes 2–5) (E) or the constructs carrying HIV-1 5’UTR and Gag ORF (lanes 2–5) (F), or water (lane 1) (E and F). Cells were then incubated for 90 min at 37°C. The cellular extracts were sonicated and equal amount of proteins were loaded on 12% SDS-PAGE. Proteins of interest were revealed by western blotting. Data are representative of three independent experiments, error bars are ± the S.E.M.
DISCUSSION
A novel secondary structure model obtained by an integrative approach
In this study, the structure of an RNA corresponding to the 515 first nucleotides of HIV-1 Gag open reading frame has been probed with two different chemical reagents and two specific RNAses. Although these data were originally produced to evidence and characterize 40S ribosomal subunit binding to this RNA, we took advantage of their availability to refine the Gag-IRES secondary structure model. Software, such as mfold (86), RNAStructure (87) or RNAfold (88) which are based on thermodynamical data generate secondary structure models using experimental structure probing data as constraints. These softwares generate many different models, rank them according to their predicted free energy and yield one or several models for the experimenter to choose. Most of the time, the resulting models are mostly, but not completely in agreement with the probing data. This can be due to model inaccuracy or to experimental artefacts, but in many cases most probably reflects the inherent property of RNA to fold into several conformations. In addition, each probe has its own specificity and can reflect slightly different things. For example some ‘single strand’ specific reagents react with nucleotides involved in non-canonical base-pairs while others do not (81). As a result, the modelling process often leaves the experimenter with different RNA structure models among which it is often difficult to choose. In addition, when modelling the secondary structure with multiple constraints sets, one faces the necessity to compare many different structures (20 more stable for each data set) in order to identify structures (ideally one) that can hold most of the probing information. This step is extremely time-consuming when manually performed and necessitates a rationalization that cannot be obtained with available structure prediction softwares. Here, we have developed an integrative approach which identifies the model that fits the best with the reactivity map obtained with different structural probes. The structure probing and modelling of this portion of Gag ORF has been reported in two previous studies (89,90). Both rely on the SHAPE technology and have been conducted in the context of the full length genomic RNA. In the first report, positions reactive to 1M7 were resolved through electrophoresis as in this study, while the second used the recently developed SHAPE-Map technology which relies on Next Generation Sequencing (90). The 1M7 reactivity maps presented in both reports are in good agreement although not identical. Both are also globally comparable to the 1M7 reactivity map reported in this study, but for two noticeable exceptions. The nucleotides between U559 and U570 and those involved in N2 stem were found to be mostly reactive in both studies while we found them poorly reactive. We have no explanation for such discrepancy although it could reflect an alternative folding of these regions in the isolated IRES as compared to the whole gRNA. Although grounded on similar reactivity maps, the three studies proposed three different models. Four pairings are identical (P3 and N5) or similar (N6) in the three models. The model proposed by Watts et al. which is more similar to ours also predicted P4 and P8. In both models proposed by the Weeks laboratory, the sequence 363GGGGG368 is base paired 746CCUCC750 instead of forming P2 with 535CCCUU539 as in our model. This roots the difference between their models and ours, but it is interesting to note that both possibilities are essentially compatible with the three reactivity maps. In summary, the main difference is the modelling strategy and not the experimental results. Here we propose a novel integrative strategy that reflects the results obtained with various probes, and smooths the effect of potential artifacts that may reside in one particular experimental set up. As the model obtained does not give a rationale to all the probing data, it should be considered as a working model to be further refined.
40S ribosomal subunit recruitment
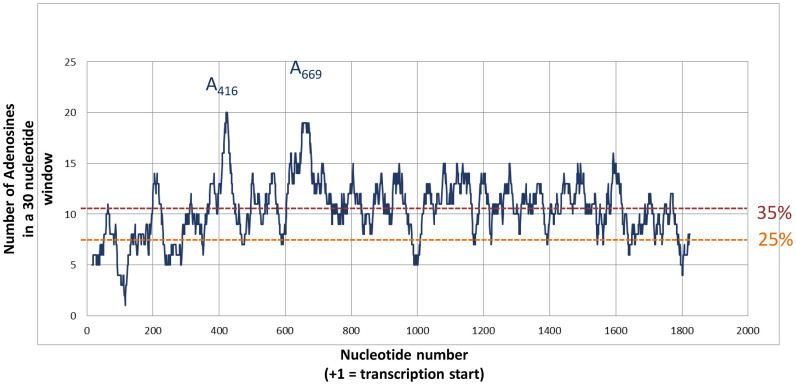
We had previously shown that 40S recruitment by the Gag open reading frame is a conserved property among primate lentiviruses. Here, we set out to better define the minimal binding site and its structure. Using chemical and enzymatic probes, we observed ribosome footprints dispatched all over the sequence although they are more abundant in some regions. In addition to reactivity protection that could reveal sites of direct contact with the ribosome, we also observed reactivity enhancement to 1M7 and T1 nuclease which rather indicate the destabilisation of the structure upon ribosome binding. The dispersion of the footprints could suggest the existence of a large contact surface between the ribosome and a compactly folded IRES as observed for type III and IV IRESes. Therefore, we were surprised to observe that non-overlapping fragments of the Gag-IRES can independently recruit the 40S ribosomal subunit. These two fragments appear to have a slightly different affinity for the small ribosomal subunit, but are not cooperative as expected if these were two fragments of a same site. Finally, stoichiometry determination and sucrose gradients showed that the full length IRES is able to recruit two 40S subunits while Site 1 or Site 2 deleted fragments are not. The two independent sites thus defined are Site 1 lying from A368 to C477, and Site 2 encompassing nucleotides from G634 to G752. Consistent with this finding, N2 and P3 loops gather six toeprints, while site two comprises U748 where we observed the strongest toeprint. Somehow confusing, we noticed that except if they fold in alternative conformations when isolated, the two sites are mostly unstructured. The isolated Site 1 retains N2, while Site 2 features P8 and P9. This suggests that binding determinants may rather lie in the sequence than in the RNA structure. We therefore looked for sequences that could base pair with the 18S rRNA. Among others, we found a twelve nucleotide long sequence in N2 370AAUUAGAUAAAU381 complementary to 1368UUGGUCUGUUUUAG1356 of the 18S rRNA. Arguing for a potential base-pairing A374, A376 and A378 are protected from 1M7 modification upon 40S subunit binding. Interestingly, a similar sequence that could base pair with the 18S rRNA is also found in an equivalent region of HIV-2 and SIVMac. The eight first positions (370 to 377) are over ninety percent conserved in amongst the HIV-1 isolates collected at the ‘HIV sequence database’ of Los Alamos. However, this region also encodes a conserved sequence of the Gag polyprotein which could account for the nucleotide conservation. The possibility of the involvement of such base-pairing in the 40S recruitment is being investigated. Independently, adenosine rich sequences have been shown to recruit the 40S subunit and to be involved in atypical IRES activity (18,91–93). This led us to investigate adenosine distribution in the HIV-1 genome (sequence pNL4.3 Accession Number AF324493.2). As previously described (94), there is a bias in the nucleotide distribution of both the Gag open reading frame and the HIV full genome which are about 34% A-rich. This trend is accentuated in the Gag-IRES (from 336AUG to 759AUG) which is 39% rich. This is in contrast with the isolated 5’ and 3΄ UTR which are only 24% A-rich. We then used a 30 nucleotide sliding window corresponding to the occupancy of a ribosome on an RNA, to examine the local abundance in adenosine residues. As can be observed in Figure 6, two regions appear to be particularly A-rich with up to 20 adenosine residues out of thirty. These A-rich sequences culminate in A416–A428 and G650–A674 which are the center of Site 1 and Site 2, respectively. In contrast, in the region in between the two sites, the average number of adenosine in a 30 nucleotide window is 11, and the structured region between U476 and U539 is ‘only’ 30% A-rich. This is nevertheless over 25% and could explain why this sequence shows some residual affinity for the 40S subunit. More precisely, nucleotides 600–630 are A-rich which may account for the residual binding of the intersite, this region could be included in Site2. We therefore make the assumption that its high adenosine content endows the Gag-IRES with the capacity of recruiting the ribosome in multiple sites, and especially in two hot spots that we have defined as Site 1 and Site 2. This may be modulated by sequences complementary to the 18S rRNA, and independently by the IRES-Gag structure hindering or exposing A-rich sequences, or else hindering ribosome progression. Obviously, we do not define here a structured compact binding site as can be observed in type III and IV IRESes, but sequences on which the 40S ribosomal subunits are loaded with an unusually high efficiency.
Figure 6.
Adenosine distribution in HIV-1 Gag open reading frame. For each nucleotide within the 5’ UTR and the whole Gag ORF of the PNL4.3, the number of Adenosine within the 20 surrounding positions (9 in 5΄ and 10 in 3΄) was plotted. Two dotted lines indicate the level of 25% adenosine (as expected in a random distribution) and 35% (as the mean observed in Gag ORF).The nucleotides corresponding to the two major peaks (A416 and A669) are indicated.
IRES driven translation and physiological consequences
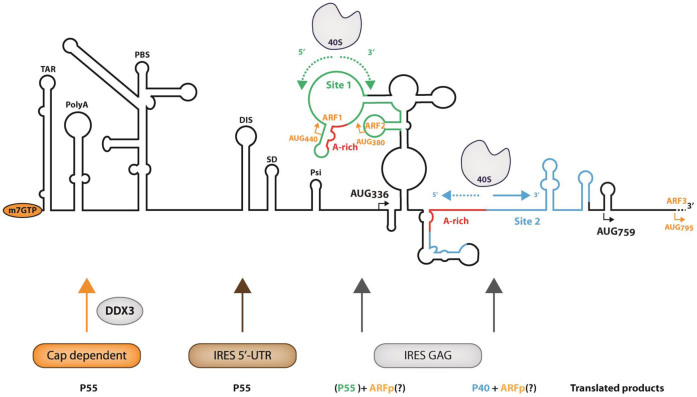
The molecular mechanism underlying translation initiation driven by A-rich IRES has been recently reported by Abeava et al. (18). In summary, the 40S subunit is recruited in one or several A-rich regions within the IRES, then the ribosome subunit scans the surrounding region for an initiation triplet, not only in the canonical 5’ to 3΄ direction, but also and even preferentially in the 3΄ to 5΄5΄ direction. The ‘backward’ scanning itself may only require the 40S subunit, but initiation on such IRES also requires eIF3, eIF2 and the Met-tRNAi. In addition, eIF4G/eIF4A favors 5΄ to 3΄ scanning, and eIF1 helps to discriminate AUG triplets (18). Such a mechanism offers a rationale for most of the finding on HIV-1 and HIV-2 gag-IRES reported in this study and in earlier works (27,28,43,49,50,52,95–97). In such a hypothesis, ribosomal small subunit would be recruited within L3 and/or N2 loop for Site 1, and in the single strand A-rich sequence between 658A and 696A. Ribosomes would then scan either backward or forward until they encounter an initiation triplet. Note that in the absence of the Met-tRNA, 40S progression may be arrested only by stable structures. As a consequence, 40S ribosomal subunit may be trapped within L3 of Site 1 while those recruited on Site 2 are relatively free to diffuse until the 5΄5΄ or 3΄ terminus in the Δ Site 1 construct. This could explain the relative instability on sucrose gradients of complexes recruited on Δ Site 1 construct and is reminiscent of what was observed for the Halastavi Arva Virus (18). This also gives a rationale for the presence of footprints and toeprints outside of the recruiting region, and for the efficient translation of transcripts lacking a large part of the IRES ((97) and this work). Such mechanism is compatible with the originally proposed provocative idea of an IRES located downstream the initiation codon (49). In this respect, it is interesting to note that Site 1 deletion brings 336AUG close to Site 2 and eliminates most of the secondary structure between the recruitment site and the initiation codon. This probably accounts for the stimulation of p55 Gag translation observed upon P1–P4 deletion in a bicistronic context where only IRES-dependent initiation is observed. Such observation suggests that RNA structure would rather be involved in constraining ribosome movement and in the initiation triplet choice than in the ribosome recruitment per se.
The presence of the Gag-IRES and the translation of N-terminal truncated Gag isoforms are conserved amongst both primate lentiviruses and clinical isolates derived from HIV-1 infected patients (28,52,53). Here by a combination of biochemical and functional analyses, we have characterized two redundant ribosome binding regions driving p40 expression and modulating p55 in some contexts. These results converge with the conservation of p40 initiation sites (53) and its requirement for optimal infectivity ((43) and C. Swanson, personal communication) providing compelling evidence of the importance of the Gag IRES. However, translation of the full length Gag polyprotein can clearly be driven by a canonical cap mechanism stimulated by a cellular helicase (36,98), or by another IRES located within the 5΄UTR (42,45–48). In this context, full length Gag translation through the Gag-IRES may be required at a very specific stage of the viral life cycle, or in specific cell type. Alternatively, the observation of the full length Gag as a result of ribosome entry within the Gag ORF could be a side effect of a physiological phenomenon selected for other reasons. Highly structured viral IRESes such as HCV (type III) or CrPV (type IV) IRES strongly constrain the position of the ribosome on the initiation site, although to a lesser extent, this holds true for type I and type II IRESes. In the case of the ‘poly-A’ type IRES where the ribosome is recruited in many locations and scans both forward and backward, virtually any AUG triplet can be used as an initiation codon (18). Indeed, translation from illegitimate initiation codons within the Gag-IRES has been observed in several occasions. For instance, translation can initiate from triplets that were introduced by site-directed mutagenesis (49), or that are uniquely present in patients isolates (52), or which are non-AUG initiation codons (28,97), or are located downstream 749AUG (see Figures 4B and 5C). All these initiation events could be observed because they result from initiation at triplets in phase with the Gag ORF thus yielding relatively long proteins. Importantly, this raises the possibility that numerous significant translation events happen from non-in frame initiation codons yielding peptides unrelated to Gag. For example, a 14 and a 12 amino acid long peptide could be translated from 380AUG and 440AUG respectively (see Figure 7). Interestingly enough, one such peptide which translation is initiated at 795AUG has been shown to be immunogenic and activated T-cells against this peptide have been found in patients’ blood (99,100). Initiation at 795AUG can undoubtedly be driven from the Gag-IRES, since we recurrently observe the translation of shorter isoforms initiated downstream from 759AUG. In addition to the production of p40, translation of such peptide could be the main role of HIV Gag-IRES, and explain the conservation of this mechanism amongst primate lentiviruses.
Figure 7.
Summary of the translation initiation events on HIV-1 genomic RNA. The secondary structure of HIV-1 gRNA is schematically represented with the names of the structural elements in the 5΄UTR: TAR (TARget of Tat), PolyA stem loop, PBS (Primer Binding Site), DIS (Dimerization Initiation Site), SD (Splicing Donor), Psi (Encapsidation). AUG336 and AUG759 responsible for the production of Gag p55 and p40 are represented. AUG440, AUG380 and AUG795 represented in orange are non in frame potential initiation triplets on which could be initiated the translation of ARFp (Alternative Reading Frame peptide). The ribosome binding sites 1 and 2 as defined in this work are represented in green and blue respectively, they both include an ‘A-rich region (in red). The arrows under the representation of the 40S ribosomal subunits represent the potential ribosome scanning, dotted arrows suggest that the progression of the ribosome might be difficult due to the presence of stable structures. At the bottom of the figure are schematized the different translation initiation pathways and the proteins yielded from each individual pathway. Under the IRES gag label p55 is in green and between parentheses because its translation from site 1 is probably only marginal.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank C. Swanson for sharing unpublished data. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 Monoclonal Antibody (183-H12-5C) from Dr Bruce Chesebro and Kathy Wehrly. Many thanks to G. De Bisschop for help with Figure 6 and A. Morris for fruitful discussions.
Footnotes
Present address: Jules Deforges, Université de Lausanne, Département de biologie moléculaire végétale Bâtiment Biophor, CH-1015 Lausanne, Suisse.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research in B.S. and T.0 laboratory was funded by ANRS [AO 2012-2 – AO2014-2]; J.D. and M.A. were recipient of a PhD fellowship from the French Ministry for Research and Education (Allocation MRE); Research in Y.P. and B.S. laboratories and A.S. PhD fellowship are funded by the French ‘Fondation pour la Recherche Medicale’ [FRM DBI20141423337]. Funding for open access charge: Fondation pour la recherche Medicale (FRM).
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson R.J., Hellen C.U., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2009; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jang S.K., Krausslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E.. A segment of the 5΄ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988; 62:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pelletier J., Sonenberg N.. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988; 334:320–325. [DOI] [PubMed] [Google Scholar]
- 4. Kolupaeva V.G., Lomakin I.B., Pestova T.V., Hellen C.U.. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 2003; 23:687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lomakin I.B., Hellen C.U., Pestova T.V.. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000; 20:6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pestova T.V., Shatsky I.N., Hellen C.U.. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996; 16:6870–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sweeney T.R., Abaeva I.S., Pestova T.V., Hellen C.U.. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014; 33:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Breyne S., Yu Y., Unbehaun A., Pestova T.V., Hellen C.U.. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9197–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Angulo J., Ulryck N., Deforges J., Chamond N., Lopez-Lastra M., Masquida B., Sargueil B.. LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res. 2016; 44:1309–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U.. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998; 12:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Quade N., Boehringer D., Leibundgut M., van den Heuvel J., Ban N.. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-A resolution. Nat. Commun. 2015; 6:7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamamoto H., Unbehaun A., Loerke J., Behrmann E., Collier M., Burger J., Mielke T., Spahn C.M.. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 2014; 21:721–727. [DOI] [PubMed] [Google Scholar]
- 13. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Pestova T.V., Hellen C.U., Frank J.. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013; 503:539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Costantino D.A., Pfingsten J.S., Rambo R.P., Kieft J.S.. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 2008; 15:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernandez I.S., Bai X.C., Murshudov G., Scheres S.H., Ramakrishnan V.. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014; 157:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koh C.S., Brilot A.F., Grigorieff N., Korostelev A.A.. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9139–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilson J.E., Pestova T.V., Hellen C.U., Sarnow P.. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000; 102:511–520. [DOI] [PubMed] [Google Scholar]
- 18. Abaeva I.S., Pestova T.V., Hellen C.U.. Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi arva virus IRES. Nucleic Acids Res. 2016; 44:2362–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ali I.K., McKendrick L., Morley S.J., Jackson R.J.. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 2001; 75:7854–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borman A.M., Michel Y.M., Kean K.M.. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J. Virol. 2001; 75:7864–7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caceres C.J., Contreras N., Angulo J., Vera-Otarola J., Pino-Ajenjo C., Llorian M., Ameur M., Lisboa F., Pino K., Lowy F. et al. Polypyrimidine tract-binding protein binds to the 5΄untranslated region of the mouse mammary tumor virus mRNA and stimulates cap-independent translation initiation. FEBS J. 2016; 283:1880–1901. [DOI] [PubMed] [Google Scholar]
- 22. Olivares E., Landry D.M., Caceres C.J., Pino K., Rossi F., Navarrete C., Huidobro-Toro J.P., Thompson S.R., Lopez-Lastra M.. The 5΄ untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. J. Virol. 2014; 88:5936–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Othman Z., Sulaiman M.K., Willcocks M.M., Ulryck N., Blackbourn D.J., Sargueil B., Roberts L.O., Locker N.. Functional analysis of Kaposi's sarcoma-associated herpesvirus vFLIP expression reveals a new mode of IRES-mediated translation. RNA. 2014; 20:1803–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Terenin I.M., Dmitriev S.E., Andreev D.E., Royall E., Belsham G.J., Roberts L.O., Shatsky I.N.. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol. Cell. Biol. 2005; 25:7879–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vallejos M., Ramdohr P., Valiente-Echeverria F., Tapia K., Rodriguez F.E., Lowy F., Huidobro-Toro J.P., Dangerfield J.A., Lopez-Lastra M.. The 5΄-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation. Nucleic Acids Res. 2010; 38:618–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chamond N., Deforges J., Ulryck N., Sargueil B.. 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of Type II IRESs. Nucleic Acids Res. 2014; 42:10373–10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Locker N., Chamond N., Sargueil B.. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011; 39:2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weill L., James L., Ulryck N., Chamond N., Herbreteau C.H., Ohlmann T., Sargueil B.. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010; 38:1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baudin F., Marquet R., Isel C., Darlix J.L., Ehresmann B., Ehresmann C.. Functional sites in the 5΄ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993; 229:382–397. [DOI] [PubMed] [Google Scholar]
- 30. Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996; 54:1–34. [DOI] [PubMed] [Google Scholar]
- 31. Dirac A.M., Huthoff H., Kjems J., Berkhout B.. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 2002; 30:2647–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. James L., Sargueil B.. RNA secondary structure of the feline immunodeficiency virus 5΄UTR and Gag coding region. Nucleic Acids Res. 2008; 36:4653–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kenyon J.C., Ghazawi A., Cheung W.K., Phillip P.S., Rizvi T.A., Lever A.M.. The secondary structure of the 5΄ end of the FIV genome reveals a long-range interaction between R/U5 and gag sequences, and a large, stable stem-loop. RNA. 2008; 14:2597–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guerrero S., Batisse J., Libre C., Bernacchi S., Marquet R., Paillart J.C.. HIV-1 replication and the cellular eukaryotic translation apparatus. Viruses. 2015; 7:199–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parkin N.T., Cohen E.A., Darveau A., Rosen C., Haseltine W., Sonenberg N.. Mutational analysis of the 5΄ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988; 7:2831–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soto-Rifo R., Rubilar P.S., Limousin T., de Breyne S., Decimo D., Ohlmann T.. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012; 31:3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Svitkin Y.V., Pause A., Sonenberg N.. La autoantigen alleviates translational repression by the 5΄ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994; 68:7001–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pyronnet S., Sonenberg N.. Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 2001; 11:13–18. [DOI] [PubMed] [Google Scholar]
- 39. Bolinger C., Sharma A., Singh D., Yu L., Boris-Lawrie K.. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010; 38:1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharma A., Yilmaz A., Marsh K., Cochrane A., Boris-Lawrie K.. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathogens. 2012; 8:e1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Amorim R., Costa S.M., Cavaleiro N.P., da Silva E.E., da Costa L.J.. HIV-1 transcripts use IRES-initiation under conditions where Cap-dependent translation is restricted by poliovirus 2A protease. PLoS One. 2014; 9:e88619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brasey A., Lopez-Lastra M., Ohlmann T., Beerens N., Berkhout B., Darlix J.L., Sonenberg N.. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003; 77:3939–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Buck C.B., Shen X., Egan M.A., Pierson T.C., Walker C.M., Siliciano R.F.. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001; 75:181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carvajal F., Vallejos M., Walters B., Contreras N., Hertz M.I., Olivares E., Caceres C.J., Pino K., Letelier A., Thompson S.R. et al. Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation. FEBS J. 2016; 283:2508–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Monette A., Valiente-Echeverria F., Rivero M., Cohen E.A., Lopez-Lastra M., Mouland A.J.. Dual mechanisms of translation initiation of the full-length HIV-1 mRNA contribute to gag synthesis. PLoS One. 2013; 8:e68108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gendron K., Ferbeyre G., Heveker N., Brakier-Gingras L.. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 2011; 39:902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Plank T.D., Whitehurst J.T., Kieft J.S.. Cell type specificity and structural determinants of IRES activity from the 5΄ leaders of different HIV-1 transcripts. Nucleic Acids Res. 2013; 41:6698–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vallejos M., Deforges J., Plank T.D., Letelier A., Ramdohr P., Abraham C.G., Valiente-Echeverria F., Kieft J.S., Sargueil B., Lopez-Lastra M.. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 2011; 39:6186–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Herbreteau C.H., Weill L., Decimo D., Prevot D., Darlix J.L., Sargueil B., Ohlmann T.. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005; 12:1001–1007. [DOI] [PubMed] [Google Scholar]
- 50. Chamond N., Locker N., Sargueil B.. The different pathways of HIV genomic RNA translation. Biochem. Soc. Trans. 2010; 38:1548–1552. [DOI] [PubMed] [Google Scholar]
- 51. Nicholson M.G., Rue S.M., Clements J.E., Barber S.A.. An internal ribosome entry site promotes translation of a novel SIV Pr55(Gag) isoform. Virology. 2006; 349:325–334. [DOI] [PubMed] [Google Scholar]
- 52. de Breyne S., Chamond N., Decimo D., Trabaud M.A., Andre P., Sargueil B., Ohlmann T.. In vitro studies reveal that different modes of initiation on HIV-1 mRNA have different levels of requirement for eIF4F. FEBS J. 2012; 279:3098–3111. [DOI] [PubMed] [Google Scholar]
- 53. Daude C., Decimo D., Trabaud M.A., Andre P., Ohlmann T., de Breyne S.. HIV-1 sequences isolated from patients promote expression of shorter isoforms of the Gag polyprotein. Arch. Virol. 2016; 161:3495–3507. [DOI] [PubMed] [Google Scholar]
- 54. de Breyne S., Soto-Rifo R., Lopez-Lastra M., Ohlmann T.. Translation initiation is driven by different mechanisms on the HIV-1 and HIV-2 genomic RNAs. Virus Res. 2013; 171:366–381. [DOI] [PubMed] [Google Scholar]
- 55. Rojas-Araya B., Ohlmann T., Soto-Rifo R.. Translational control of the HIV unspliced genomic RNA. Viruses. 2015; 7:4326–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miele G., Mouland A., Harrison G.P., Cohen E., Lever A.M.. The human immunodeficiency virus type 1 5΄ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 1996; 70:944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smirnova V.V., Terenin I.M., Khutornenko A.A., Andreev D.E., Dmitriev S.E., Shatsky I.N.. Does HIV-1 mRNA 5΄-untranslated region bear an internal ribosome entry site. Biochimie. 2016; 121:228–237. [DOI] [PubMed] [Google Scholar]
- 58. Yilmaz A., Bolinger C., Boris-Lawrie K.. Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr. HIV Res. 2006; 4:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soto Rifo R., Ricci E.P., Decimo D., Moncorge O., Ohlmann T.. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007; 35:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorith. Mol. Biol.: AMB. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wuchty S., Fontana W., Hofacker I.L., Schuster P.. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999; 49:145–165. [DOI] [PubMed] [Google Scholar]
- 62. Merino E.J., Wilkinson K.A., Coughlan J.L., Weeks K.M.. RNA structure analysis at single nucleotide resolution by selective 2΄-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005; 127:4223–4231. [DOI] [PubMed] [Google Scholar]
- 63. Lorenz R., Luntzer D., Hofacker I.L., Stadler P.F., Wolfinger M.T.. SHAPE directed RNA folding. Bioinformatics. 2016; 32:145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H.. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:7287–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ding Y., Chan C.Y., Lawrence C.E.. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005; 11:1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Deigan K.E., Li T.W., Mathews D.H., Weeks K.M.. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang K., Zhang J., Li D., Zhang X., Guo T.. Adaptative affinity propagation clustering. Automatica. 2007; 33:85–115. [Google Scholar]
- 68. Sculley D. Web scale k-means clustering. Proceedings of the 19th international conference on World wide web WWW. 2010; 10:1177. [Google Scholar]
- 69. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. et al. Scikit-learn: machine learning in python. JMLR. 2011; 12:2825–2830. [Google Scholar]
- 70. Lu Z.J., Gloor J.W., Mathews D.H.. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA. 2009; 15:1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mattson M., Messac A.. Pareto frontier based concept selection under uncertainty, with visualization. Optimiz. Eng. 2005; 6:85–115. [Google Scholar]
- 72. Lai D., Proctor J.R., Zhu J.Y., Meyer I.M.. R-CHIE: a web server and R package for visualizing RNA secondary structures. Nucleic Acids Res. 2012; 40:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hellman L.M., Fried M.G.. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007; 2:1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chesebro B., Wehrly K., Nishio J., Perryman S.. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 1992; 66:6547–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Toohey K., Wehrly K., Nishio J., Perryman S., Chesebro B.. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995; 213:70–79. [DOI] [PubMed] [Google Scholar]
- 76. Wehrly K., Chesebro B.. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997; 12:288–293. [DOI] [PubMed] [Google Scholar]
- 77. Lowman H.B., Draper D.E.. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 1986; 261:5396–5403. [PubMed] [Google Scholar]
- 78. Sobczak K., Michlewski G., de Mezer M., Krol J., Krzyzosiak W.J.. Trinucleotide repeat system for sequence specificity analysis of RNA structure probing reagents. Anal. Biochem. 2010; 402:40–46. [DOI] [PubMed] [Google Scholar]
- 79. Mortimer S.A., Weeks K.M.. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007; 129:4144–4145. [DOI] [PubMed] [Google Scholar]
- 80. Wilkinson K.A., Gorelick R.J., Vasa S.M., Guex N., Rein A., Mathews D.H., Giddings M.C., Weeks K.M.. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008; 6:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Steen K.A., Rice G.M., Weeks K.M.. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J. Am. Chem. Soc. 2012; 134:13160–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Foley B., Leitner T., Apetrei C., Hahn B., Mizrachi I., Mullins J., Rambaut A., Wolinsky S., Korber B.. Los Alamos National Laboratory, theoretical biology and biophysics, Los alamos, New mexico. 2016; LA-UR-16-25625.
- 83. Dmitriev S.E., Pisarev A.V., Rubtsova M.P., Dunaevsky Y.E., Shatsky I.N.. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 2003; 533:99–104. [DOI] [PubMed] [Google Scholar]
- 84. Kozak M. Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res. 1998; 26:4853–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ryder S.P., Recht M.I., Williamson J.R.. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008; 488:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xu Z.Z., Mathews D.H.. Secondary structure prediction of single sequences using RNAstructure. Methods Mol. Biol. 2016; 1490:15–34. [DOI] [PubMed] [Google Scholar]
- 88. Hofacker I.L. RNA secondary structure analysis using the Vienna RNA package. Curr. Protoc. Bioinformatics. 2009; doi:10.1002/0471250953.bi1202s04. [DOI] [PubMed] [Google Scholar]
- 89. Watts J.M., Dang K.K., Gorelick R.J., Leonard C.W., Bess J.W. Jr, Swanstrom R., Burch C.L., Weeks K.M.. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009; 460:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Siegfried N.A., Busan S., Rice G.M., Nelson J.A., Weeks K.M.. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods. 2014; 11:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dorokhov Y.L., Skulachev M.V., Ivanov P.A., Zvereva S.D., Tjulkina L.G., Merits A., Gleba Y.Y., Hohn T., Atabekov J.G.. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5301–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gilbert W.V., Zhou K., Butler T.K., Doudna J.A.. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007; 317:1224–1227. [DOI] [PubMed] [Google Scholar]
- 93. Shirokikh N.E., Spirin A.S.. Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10738–10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Berkhout B., van Hemert F.J.. The unusual nucleotide content of the HIV RNA genome results in a biased amino acid composition of HIV proteins. Nucleic Acids Res. 1994; 22:1705–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Balvay L., Lastra M.L., Sargueil B., Darlix J.L., Ohlmann T.. Translational control of retroviruses. Nat. Rev. Microbiol. 2007; 5:128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ricci E.P., Soto Rifo R., Herbreteau C.H., Decimo D., Ohlmann T.. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem. Soc. Trans. 2008; 36:690–693. [DOI] [PubMed] [Google Scholar]
- 97. Ricci E.P., Herbreteau C.H., Decimo D., Schaupp A., Datta S.A., Rein A., Darlix J.L., Ohlmann T.. In vitro expression of the HIV-2 genomic RNA is controlled by three distinct internal ribosome entry segments that are regulated by the HIV protease and the Gag polyprotein. RNA. 2008; 14:1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Soto-Rifo R., Rubilar P.S., Ohlmann T.. The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic Acids Res. 2013; 41:6286–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cardinaud S., Consiglieri G., Bouziat R., Urrutia A., Graff-Dubois S., Fourati S., Malet I., Guergnon J., Guihot A., Katlama C. et al. CTL escape mediated by proteasomal destruction of an HIV-1 cryptic epitope. PLoS Pathogens. 2011; 7:e1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cardinaud S., Moris A., Fevrier M., Rohrlich P.S., Weiss L., Langlade-Demoyen P., Lemonnier F.A., Schwartz O., Habel A.. Identification of cryptic MHC I-restricted epitopes encoded by HIV-1 alternative reading frames. J. Exp. Med. 2004; 199:1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.