Abstract
Polintons (polintoviruses), polinton-like viruses (PLVs) and virophages belong to a recently described major class of eukaryotic viruses that is characterized by a distinct virion morphogenetic protein module and, in many members, a protein-primed family B DNA polymerase (pDNAP). All Polintons, by definition, encode a pDNAP and a retrovirus-like integrase. Most of the PLV lack these genes and instead encode a large protein containing a superfamily 1 (SF1) helicase domain. We show here that the SF1 helicase domain-containing proteins of the PLV also contain an inactivated pDNAP domain. This unique helicase-pDNAP fusion is also encoded by transpovirons, enigmatic plasmid-like genetic elements that are associated with giant viruses of the family Mimiviridae. These findings indicate the directionality of evolution of different groups of viruses and mobile elements in the Polinton-centered class. We propose that the PLV evolved from a polinton via fusion of the pDNAP gene with a helicase gene that was accompanied by mutations in the pDNAP active site, likely resulting in inactivation of the polymerase activity. The transpovirons could have evolved from PLV via the loss of several genes including those encoding the morphogenetic module proteins. These findings reaffirm the central evolutionary position of the Polintons in the evolution of eukaryotic viruses and other mobile genetic elements.
1. Introduction
Eukaryotic viruses with double-stranded (ds) DNA genomes are classified into 19 viral families, with several additional groups (e.g. pandoraviruses and pithoviruses) still awaiting formal classification (Koonin et al. 2015). The largest, most diverse and widely distributed is the monophyletic viral assemblage known as the Nucleo-cytoplasmic large DNA viruses or the proposed order ‘Megavirales’ (Iyer et al. 2006; Colson et al. 2013). Members of the ‘Megavirales’ fall into seven families and propagate in organisms from three of the five eukaryotic supergroups, namely Archaeplastida, Chromalveolata, and Uniconta (Colson et al. 2013; Krupovic and Koonin, 2015). The diversity of other eukaryotic dsDNA viruses is far more modest, with the vast majority, in particular the expansive order Herpesvirales, being restricted to animals (Metazoa) (Davison et al. 2009; Koonin et al. 2015). However, the newly discovered major group of dsDNA viruses, referred to as polintoviruses, appears to match and possibly even surpass ‘Megavirales’ in terms of the distribution and abundance in Eukarya (Krupovic and Koonin 2015).
Polintons (also known as Mavericks) have been originally described as the largest eukaryotic DNA transposons (Feschotte and Pritham 2005; Kapitonov and Jurka 2006; Pritham et al. 2007). These elements are found in a wide range of eukaryotes and universally encode two signature genes, namely protein-primed DNA polymerase (pDNAP) and a retrovirus-like integrase (RVE) (hence the name of these elements: POLINTons). Phylogenetic analysis of these two genes suggests antiquity of polintons and their long-term co-evolution with eukaryotes (Haapa-Paananen et al. 2014). The polintons are known as self-synthesizing transposons given that they encode the key enzyme of their own replication (Kapitonov and Jurka 2006). Indeed, biochemical characterization of the pDNAP from Entamoeba histolytica polintons has demonstrated intrinsic strand displacement, processivity and lesion bypass by this enzyme (Pastor-Palacios et al. 2012). However, recent evidence suggests that Polintons are viruses in disguise. Indeed, most of the Polintons encode the viral genome-packaging ATPase and cysteine protease homologous to viral capsid maturation proteases (Kapitonov and Jurka 2006; Pritham et al. 2007). Furthermore, it has been recently shown that majority of Polintons encode conserved homologs of the two capsid proteins that are required for the formation of icosahedral virions, namely the major capsid protein (MCP) with the double jelly roll (DJR) fold and the minor capsid protein (mCP) with the single jelly roll fold (Krupovic and Bamford 2008; Krupovic et al. 2014). These observations strongly suggest that most of the Polintons are actually polintoviruses, i.e. can form bona fide virions that, however, remain to be discovered experimentally (Krupovic and Koonin 2015).
Comparative genomic and phylogenetic analyses suggest that Polintons played a key role in the evolution of several groups of eukaryotic DNA viruses and plasmids (Krupovic and Koonin 2016). Polintons are related to adenoviruses and virophages (recently classified as the family LavidaviridaeKrupovic et al. 2016)) with which they share the four genes for virion morphogenesis as well as the pDNAPs (present only in a subset of the virophages (Fischer and Suttle 2011; Yutin et al. 2015)). Polintons could have also contributed to the evolution of single-stranded DNA viruses of the Bidnaviridae family by supplying the pDNAP gene (Krupovic and Koonin 2014). Recent metagenome mining has led to the identification of another putative group of viruses that resemble Polintons in several respects and have been denoted Polinton-like viruses (PLVs) (Yutin et al. 2015). Notably, most PLV genomes were assembled from sequence reads derived from pre-filtered fractions enriched in viral particles. Furthermore, virions of one PLV representative, namely Tetraselmis viridis virus S1 (TVS1), have been isolated from green algae and represent an actual virus (Sizov and Polischuk 2006). The PLVs encode the MCP, (in most cases) the mCP and the genome packaging ATPase but not the maturation protease (Yutin et al. 2015). Another major difference between Polintons and most PLVs is that the latter lack pDNAP and RVE integrase genes. Instead, many PLVs encode a predicted tyrosine recombinase of a distinct family and also often possess helicases of superfamilies 1 or 3 (SF1 and SF3, respectively) that are implicated in viral genome replication (Yutin et al. 2015). Transposable elements of another group, named Tlr1, are integrated into the genome of the ciliate Tetrahymena thermophila and are also tightly linked to Polintons (Wuitschick et al. 2002). The Tlr1 elements are present in ∼30 copies per genome and encode MCP, mCP, packaging ATPase and the RVE integrase, suggesting that they also form virions (Krupovic et al. 2014). Similar to the PLVs, Tlr1 elements lack genes for the protease and pDNAP but instead encode a distinct variety of SF1 helicase. In addition to the clear evolutionary relationships between all these (predicted) viruses and mobile elements, Polintons also appear to have contributed the virion morphogenesis module (encompassing all four genes) and possibly some other genes, such as that for a SF3 helicase, to the evolution of the much larger viruses of the ‘Megavirales’ (Krupovic and Koonin 2015).
Virophages are obligate parasites of the giant mimiviruses and negatively affect the reproduction of the latter (La Scola et al. 2008; Fischer and Suttle 2011; Desnues et al. 2012). Some virophages can integrate into the mimivirus genome (Desnues et al. 2012), whereas others are capable of integration into the genome of their cellular hosts, as in the case of the green alga Bigelowiella natans (Blanc et al. 2015; Fischer 2015). Unlike Polintons, virophages show a much more restricted host range and are currently exclusive to protists. Although it has been suggested that virophages, in particular Mavirus, gave rise to Polintons (Fischer and Suttle 2011; Katzourakis and Aswad 2014), given the much broader taxonomic distribution of Polintons (Krupovic and Koonin 2015), their long-lasting co-evolution with eukaryotes (Haapa-Paananen et al. 2014)] as well as their broad genetic diversity (Krupovic and Koonin 2016), the reverse evolutionary scenario appears more compelling. However, it cannot be ruled out that Polintons, similar to virophages, lead a lifestyle dependent on helper viruses and, accordingly, adhere to the definition of virophages. Thus, the debate will be put to rest only when the life cycle of Polintons and PLVs is characterized experimentally.
Mimiviruses are also parasitized by another group of mobile elements, named transpovirons (Desnues et al. 2012). The transpovirons are small (∼7 kb), linear dsDNA plasmid-like molecules with terminal inverted repeats. Transpovirons are incorporated into mimivirus particles in high copy numbers and can also be integrated into the viral genome (Desnues et al. 2012). Recent analysis of the B. natans genome has shown that, besides the integrated virophages, this alga contains several copies of integrated elements which were identified as transpovirons (Blanc et al. 2015). Although transpovirons do not encode viral structural proteins, they carry genes for a SF1 helicase related to those of Tlr1 elements and PLVs (Desnues et al. 2012; Yutin et al. 2013; Blanc et al. 2015). However, the origin of transpovirons remains enigmatic.
Polintons, PLVs, virophages and Tlr1 are connected into a network by the shared gene content (Fig. 1A) and represent an extremely diverse, widely distributed, recently identified supergroup of eukaryotic DNA viruses (Krupovic and Koonin 2015; Yutin et al. 2013, 2015). However, the exact scenario of their evolution and relationship to other groups of mobile elements, such as transpovirons, remain obscure. Here we present the results of sequence analysis of the Tlr1-like helicases from transpovirons, Tlr1 transposons and PLVs which clarify the relationships between these different types of elements and indicate their origin from Polintons.
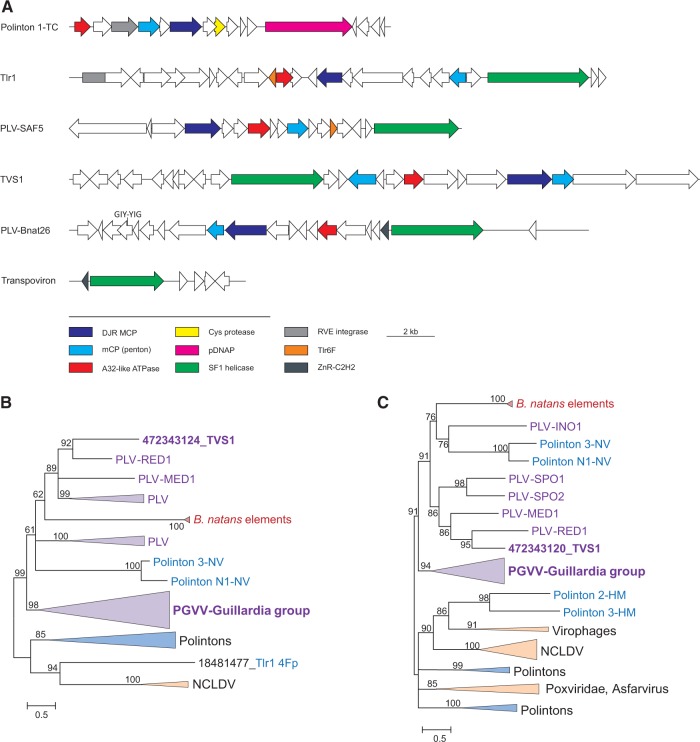
Figure 1.
PLVs of B. natans. (A) Genome maps of various evolutionarily related mobile genetic elements. Homologous genes are indicated by arrows of the same color. The color key is provided at the bottom of the panel. (B) ML phylogenetic tree of the MCPs. (C) ML phylogenetic tree of the genome packaging ATPases. Numbers at the branch points represent the Bayesian-like transformation of aLRT (aBayes) local support values. Branches with support values <50% were collapsed. Tlr1 4Fp and Tlr 6F denote the MCP (Krupovic et al. 2014) and a conserved hypothetical protein (Yutin et al. 2015) of T. thermophyla element Tlr1, respectively. PGVV-Guillardia group represents a previously described group of PLVs (Yutin et al. 2015). Abbreviations: TVS1, T. viridis virus S1; PGVV, Phaeocystis globosa virus virophages; PLV, Polinton-like viruses; NCLDV, Nucleo-cytoplasmic large DNA viruses; HM, H. magnipapillata; NV, Nematostella vectensis.
Methods
The non-redundant database of protein sequences at the NCBI was searched using the PSI-BLAST (Altschul et al. 1997). Profile-against-profile searches were performed using HHpred (Söding 2005) against different protein databases, including PFAM (Database of Protein Families), PDB (Protein Data Bank), CDD (Conserved Domains Database), and COG (Clusters of Orthologous Groups), which are available via the HHpred website. For phylogenetic analyses protein sequences were aligned with Promals3D (Pei and Grishin 2014) (MCP, SF1 helicase, and pDNAP) or Muscle (Edgar 2004) (ATPase). The alignment was visualized using Jalview (Waterhouse et al. 2009). The quality of the MCP and mCP alignments was evaluated using the Transitive Consistency Score (TCS), a sequence alignment reliability measure which helps to estimate alignment accuracy (Chang et al. 2014, 2015). Poorly aligned (low information content) positions were removed using the Gappyout function of Trimal (Capella-Gutierrez et al. 2009). Representative MCP and ATPase sequences were acquired from Yutin et al. (2015). Representative sequences from different families of SF1B helicases were collected from PFAM database with following accession numbers PF05970 (PIF1), PF05127 (RecD), PF02689 (Herpesvirus UL5). Viral and plasmid pDNAP sequences were collected from GenBank, whereas those from Polintons were obtained from the Repase Update database (Jurka et al. 2005). Phylogenetic trees were constructed using the PhyML program (Guindon et al. 2010) the latest version of which (http://www.atgc-montpellier.fr/phyml-sms/) includes automatic selection of the best-fit substitution model for a given alignment. The best models identified by PhyML for ATPase, SF1 helicase and pDNAP domains were LG +G6 +I +F. LG, Le-Gascuel matrix; G6 +I +F, Gamma shape parameter: fixed; number of categories: 6; Proportion of Invariable sites: fixed; Equilibrium frequencies: empirical. The best model identified by PhyML for MCP was the same as above but with estimated Proportion of invariable sites. All the alignments used for the tree construction are available from the authors upon request. A Bayesian-like transformation of aLRT (aBayes) (Anisimova et al. 2011), as implemented in PhyML (Guindon et al. 2010), and non-parametric bootstrapping (1,000 replicates) were used to estimate branch support. Alternative topologies for the SF1 helicase tree were tested using the CONSEL software (Shimodaira and Hasegawa 2001). The likelihoods for the initial tree and the alternative trees, used by CONSEL, were calculated by PhyML (Guindon et al. 2010).
Results and discussion
Putative integrated transpovirons in B.Natans are PLVs
The original analysis of the putative transpovirons of B. natans (Blanc et al. 2015) has shown that these elements are considerably larger than the transpovirons associated with the mimiviruses (Desnues et al. 2012) and encompass several viral genes. To better understand the provenance of the putative transpovirons of B. natans and to investigate their link to viruses, we reanalyzed their gene content. BLASTp searches against the NCBI viral database seeded with the protein sequences of the putative transpoviron BnTV26 resulted in a match between protein BnTV26_9 and TVSG_00021 of TVS1(Supplementary Table S1). The latter protein has been recently identified as the putative MCP of TVS1, related to the DJR MCPs of the other PLVs (Yutin et al. 2015). The hit encompassed only 16% of BnTV26_9. However, considering that viral capsid proteins are notoriously divergent (Krupovic and Bamford 2011), sometimes to the extent that, even within the same viral family, homologous capsid proteins are recognizable only at the structural level (Laurinmaki et al. 2005), we further explored the significance of the match between BnTV26_9 and TVSG_00021. Additional sequence analysis has shown that the two proteins display not only sequence but also clear secondary structure similarity (Supplementary Figure S1A). Finally, we evaluated the quality of the alignment of the two proteins using TCS (Chang et al. 2014, 2015). With the TCS score of 706, the alignment was found to be reliable (Supplementary Figure S2A; scores below 500 are considered poor (Chang et al. 2014, 2015)). Collectively, these results suggest that BnTV26_9 is the MCP of BnTV26 (Figure 1A). Considering that viruses with DJR MCPs typically encode an additional mCP with a single jelly-roll fold, the protein set of BnTV26 was searched against a custom database of PLV proteins (Yutin et al. 2015). A significant hit was obtained between the mCP of PLV-MED1 and BnTV26_10 (Supplementary Table S1) which is encoded immediately downstream of the MCP (Fig. 1A). The same arrangement of the two genes is also found in TVS1. As in the case of the MCP, multiple sequence alignment (TCS score 645; Supplementary Figure S2B) confirmed the conservation of both sequence and secondary structure in the mCPs of PLV and BnTV26 (Supplementary Figure S1B). Other significant matches were identified between proteins BnTV26_5 and A32-like genome packaging ATPase of PLV-MED1, BnTV26_13 and GIY-YIG nuclease of PLV-ACE1, BnTV26_18 and Tlr1-like SF1 helicase of PLV-SAF5 as well as between BnTV26_14 and a hypothetical protein of PLV-RED1 (Supplementary Table S1).
Conservation of the PLV-like MCP, mCP, and A32-like ATPase, the three proteins that constitute the morphogenetic module of PLVs and other viruses with DJR MCPs (Fig. 1A), in the putative B. natans transpovirons strongly suggests that these elements are bona fide PLVs (hereinafter denoted PLV-Bnat) rather than transpovirons. Maximum likelihood (ML) phylogenetic trees of the MCP, A32-like ATPase, and Tlr1-like helicase further support the inclusion of B. natans elements into a distinct family of PLVs (Figs. 1B and 2).
Transpovirons associated with mimiviruses evolved from PLVs
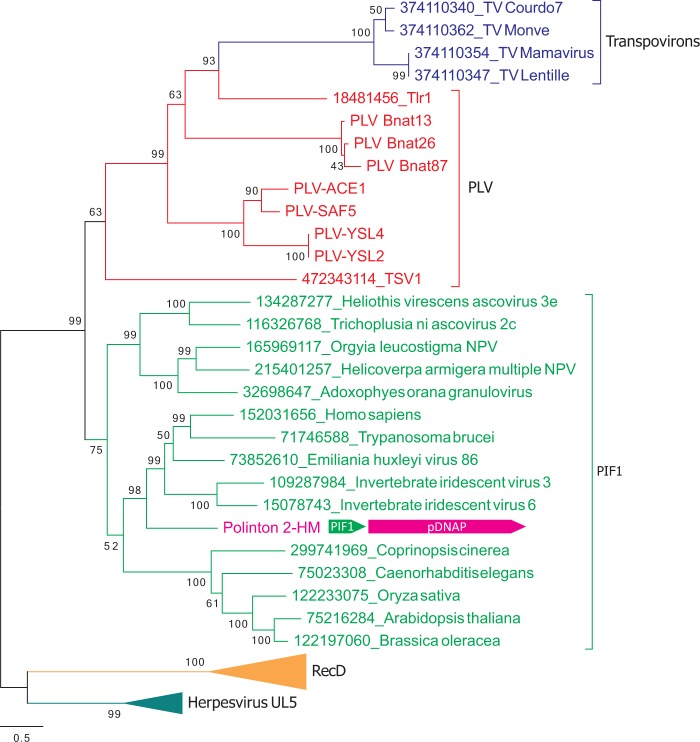
Transpovirons and some PLVs, including PLV-Bnat, share a gene for the SF1 helicase which is most closely related to the helicase first described in Tlr1 (Wuitschick et al. 2002) (Fig. 1), indicating that all these elements might be evolutionarily related. Previous analysis has shown that transpoviron helicases form a sister clade to proteins of the PIF1 family which falls into the SF1B group (Desnues et al. 2012), a division of SF1 helicases that translocate in the 5′-3′ direction (Singleton et al. 2007). However, at the time of the analysis, several groups of elements encoding Tlr1-like helicases have not been yet identified. To clarify the relationships between transpovirons and other groups of mobile elements, we performed phylogenetic analysis of the Tlr1-like helicases jointly with other SF1B helicase families, namely PIF1 (PF05970), RecD (PF05127), and Herpesvirus UL5 (PF02689). In the resulting phylogenetic tree, the transpoviron helicases formed a clade with the helicases of Tlr1 and PLVs (Fig. 2). Consistent with the previous observations (Desnues et al. 2012), the transpoviron-PLV clade was a sister group to PIF1 helicases, with the transpovirons emerging from within the PLV diversity (Fig. 2 and Supplementary Figure S3). To further scrutinize the topology of the phylogenetic tree, we performed several statistical tests, including the approximately unbiased test (Shimodaira 2002). All tests rejected all tree topologies with transpovirons placed outside the PLV branch (Supplementary Table S2).
Figure 2.
ML phylogenetic tree of SF1 helicases. Clades corresponding to different families within SF1 are shown with different colors. The arrangement of the genes encoding PIF1 helicase and pDNAP in Polinton 2-HM is shown next to the corresponding branch. Numbers at the branch points represent the Bayesian-like transformation of aLRT (aBayes) local support values (Supplementary Figure S3 shows the SF1 helicase tree with the bootstrap support values). The scale bar represents the number of substitutions per site. Abbreviations: PLV, polinton-like viruses (ACE, from ACE Lake; RED, Red Sea; MED, Mediterranean Sea; INO, Indian Ocean; SPO, South-Pacific Ocean; YSL, Yellowstone Lake); HM, H. magnipapillata; TV, transpoviron.
Besides the helicase gene, transpovirons and PLV-Bnat share a gene for a Zn-ribbon protein. The helicase and Zn-ribbon genes are adjacent and divergently oriented in both types of elements (Fig. 1A). Furthermore, transpovirons display the same genome organization as PLV-Bnat and some other PLVs as well as Tlr1: all these elements are linear dsDNA molecules terminating with inverted repeats (Wuitschick et al. 2002; Desnues et al. 2012; Blanc et al. 2015; Yutin et al. 2015). Collectively, these similarities strongly suggest that transpovirons have evolved from PLVs, conceivably by losing the virion morphogenesis module and other genes.
Tlr1-like helicases contain a family B DNA polymerase domain
Tlr1-like helicases are large proteins (1,000–1,400 aa) in which the actual SF1 helicase domain (∼350 aa) is located in the C-terminal portion of the polypeptide. To gain insight into the origins and possible functions of the remaining regions of these proteins and to further explore the possibility of a common origin of the helicases encoded by transpovirons, Tlr1 and PLVs, we performed a detailed domain organization analysis using sensitive profile-profile searches with HHpred. The previous study has detected a GIY-YIG endonuclease domain at the C-terminus of the Tlr1 protein, following the SF1 helicase domain (Dunin-Horkawicz et al. 2006), and the present analysis confirmed this identification (Fig. 3A, Supplementary Figure S4). Notably, GIY-YIG nucleases are encoded as stand-alone proteins in Bnat26 (Fig. 1A, Supplementary Table S1) and some other PLVs (Yutin et al. 2015). However, homologous domains could not be identified in the helicases of PLVs or transpovirons. Instead, the PLVs, including those from B. natans, contain homing endonuclease domains of the HNH superfamily at the corresponding location of the SF1 helicase domain-containing protein (Fig. 3A, Supplementary Figure S4). Notably, three insertion elements encoding HNH homing endonucleases have been found to be specifically associated with Tlr1 elements (Yutin et al. 2015). Thus, it appears that PLVs and Tlr1 elements are parasitized by smaller insertion elements and that GIY-YIG and HNH homing endonuclease domains were independently appended to the helicases of Tlr1 and PLVs, respectively.
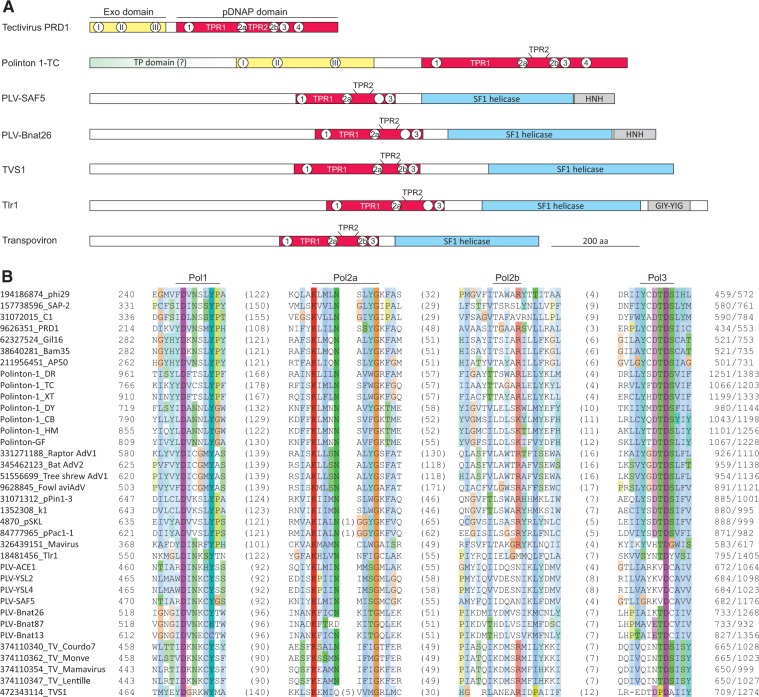
Figure 3.
DNA polymerase domain of Tlr1-like helicases. (A) Domain organizations of Tlr1-like helicases from various elements and their comparison to pDNAPs of tectivirus PRD1 and Polinton 1-TC. Conserved motifs of the exonuclease and polymerization domains are indicated with Roman and Arabic numbers, respectively. Open circles indicate that the particular motif is missing. TPR1, TPR2 and TP (terminal protein) domains are also shown. (B) Multiple sequence alignment of the conserved motifs from pDNAP domains of Tlr1-like helicases and pDNAPs encoded by various viruses and plasmids. Sequences are identified with their GI identifiers. The conserved motifs are indicated above the alignment. The alignment is colored according to sequence conservation using the standard Clustal color scheme.
Strikingly, the HHpred searches seeded with the sequences of transpoviron Lentille and PLV-SAF5 returned moderately significant matches (HHpred Probabilities of 90 and 83, respectively) to protein-primed family B DNA polymerases (pDNAP) (Supplementary Figure S5). To further validate this assignment, the corresponding regions of Tlr1-like helicases were aligned with pDNAPs from various mobile genetic elements, including bacterial phi29-like podoviruses (subfamily Picovirinae) and tectiviruses (Tectiviridae) as well as eukaryotic adenoviruses (Adenoviridae), the virophage Mavirus (Lavidaviridae), cytoplasmic plasmids and Polintons. The region of homology was found to be restricted to the pDNAP polymerization domain and encompassed motifs 1 (also referred to as motif A (Blanco and Salas 1996)), 2a (B), 2b, and 3 (C), whereas motif 4 and the N-terminal exonuclease proofreading domain were missing (Fig. 3B). Notably, the SF1 helicase of TVS1, although originally not considered to be closely related to Tlr1-like helicases (Yutin et al. 2015), also contains the unique pDNAP-like domain (Fig. 3). This shared domain architecture is consistent with the results of the phylogenetic analysis which places TVS1 protein in the transpoviron-PLV clade albeit as the deepest branch (Fig. 2 and Supplementary Figure S3). The pDNAP motifs 1 and 2 are well conserved in all Tlr1-like helicases, whereas motifs 3 and 4 contain non-conservative substitutions in some members of the group (Fig. 3B). The regions corresponding to the TPR1 and TPR2 subdomains, which are specific to protein-primed DNAPs and are responsible for the interaction with the terminal proteins as well as processivity and strand displacement capacity (Redrejo-Rodriguez and Salas 2014), are also present in the helicase domain-containing proteins of PLVs, Tlr1 and transpovirons (Fig. 3A). The elimination of motif 4 and mutations of some other active site residues (particularly in the key motif 3) of the pDNAP domain strongly suggest that the pDNAP homologous domains of the Tlr1-like helicases have lost the DNA polymerization capacity. However, in the absence of biochemical data, a remote possibility remains that the substitutions in the catalytic motifs of the transpoviron pDNAP domain are compensated by other mutations, in less strongly conserved parts of the domain such that some enzymatic activity is retained. This possibility and the role of the pDNAP domain in the Tlr1-like helicases await experimental investigation.
Inactivated family B DNAP domains have been detected previously, in particular in the eukaryotic DNA polymerase ε (Tahirov et al. 2009), in a distinct family of archaeal DNAP homologs (Rogozin et al. 2008; Makarova et al. 2014), in a poxvirus virion protein (Yutin et al. 2014) and in one of the subunits of the herpesvirus helicase-primase complex (Kazlauskas and Venclovas 2014). The biological functions of these inactivated DNAP homologs have not been studied in detail but they are predicted to play structural roles in replisomes and beyond (Tahirov et al. 2009; Kazlauskas and Venclovas 2014; Yutin et al. 2014).
The BLASTp and HHpred searches seeded with the N-terminal ∼450 aa regions of the Tlr1-like helicases did not result in informative hits. In pDNAPs, the polymerization domain is typically preceded by the exonuclease domain (Fig. 3A), and in Polintons and cytoplasmic plasmids also by an N-terminal extension which corresponds to the terminal protein involved in priming of the DNA replication (Klassen and Meinhardt 2007; Krupovic and Koonin 2014). The unannotated regions in Tlr1-like helicases might be derived from the ancestral exonuclease and/or terminal protein domains which have diverged beyond recognition.
The domain organization of Tlr1-like helicases clarifies the evolutionary relationships between Polintons, PLVs, Tlr1 elements and transpovirons
As indicated earlier, PLVs and Tlr1 appear to be evolutionarily related to Polintons. All the Polintons, by definition, encode RVE integrases and pDNAPs (Krupovic and Koonin 2015; Yutin et al. 2015) but in the PLVs these enzymes are represented sporadically, with many members carrying helicase genes instead of the pDNAPs (Yutin et al. 2015). Thus, the exact evolutionary relationship between these two types of elements remained unclear. Our finding that PLVs encode remnants of pDNAPs fused to SF1 helicases (Fig. 3) implies that pDNAP is an ancestral gene in PLVs. Furthermore, the unique fusion of an inactivated pDNAP with SF1 helicase is shared by PLV and transpovirons suggesting that the latter elements, the provenance of which so far remained enigmatic, are highly derived forms of PLV.
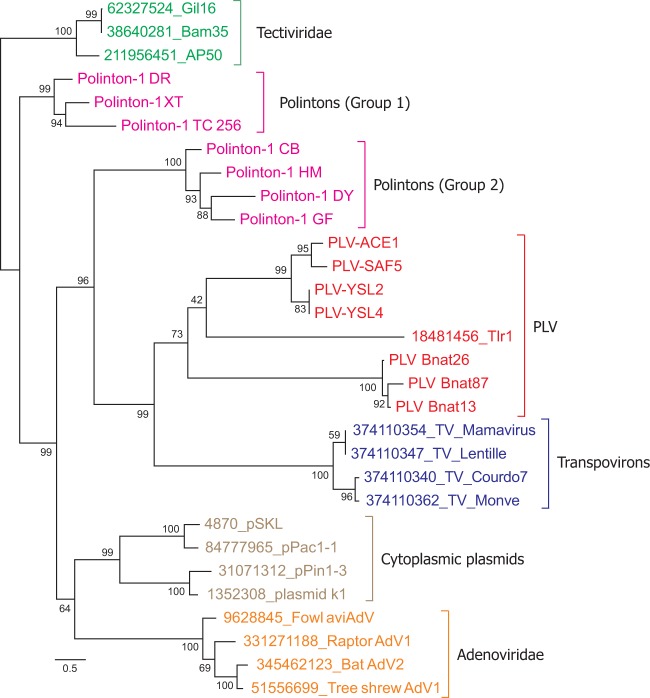
To further investigate these evolutionary relationships, we constructed an ML tree that included pDNAP sequences from various mobile genetic elements (Fig. 4 and Supplementary Figure S6). The general branching of different pDNAP groups was fully consistent with the previous results which show that pDNAPs of adenoviruses and cytoplasmic plasmids emerge from amidst the Polinton diversity (Kapitonov and Jurka 2006; Krupovic and Koonin 2014; Krupovic and Koonin 2015). The clade including all PLVs, Tlr1 and transpovirons formed a sister group to one of the two subdivisions of Polintons, Group 2 (Fig. 4). Taken together, the results of our analysis suggest that all elements encoding Tlr1-like helicases shown here to be fused to inactivated derivatives of pDNAPs arise from within the Polintons, strongly suggesting that Polintons (polintoviruses) are the ancestral forms, whereas PLVs are a derived group of viruses.
Figure 4.
ML phylogenetic tree of pDNAPs. Clades corresponding to different groups of mobile genetic elements are shown with different colors. Numbers at the branch points represent the Bayesian-like transformation of aLRT (aBayes) local support values (Supplementary Figure S6 shows the pDNAP tree with the bootstrap support values). The pDNAP domain of TVS1 was omitted from the analysis due to its high divergence. The tree is rooted with bacterial tectiviruses. The scale bar represents the number of substitutions per site. Abbreviations: AdV, adenovirus; DR, Danio rerio; DY, Drosophila yakuba; CB, Caenorhabditis briggsae; HM, H. magnipapillata; TC, Tribolium castaneum; GF, Glyptapanteles flavicoxis; TV, transpoviron; XT, Xenopus tropicalis.
Concluding remarks
The present results indicate that the purported transpovirons of B. natans are actually PLVs and encode a characteristic set of virion morphogenesis proteins which are not present in the bona fide transpovirons (Fig. 1A). Notably, some of the PLV genes are expressed in B. natans (Blanc et al. 2015). Whether these integrated PLVs elicit any effect on giant viruses of the proposed order ‘Megavirales’ infecting the host organism, as has been proposed for the provirophages (Blanc et al. 2015; Fischer 2015), remains an interesting open question.
The PLVs and Polintons share the virion morphogenetic module including the MCP, mCP, and packaging ATPase indicating that the two groups of elements are evolutionarily related. However, unlike Polintons, the PLVs generally lack genes for pDNAPs and instead contain helicase-encoding genes (Yutin et al. 2015). In principle, Polintons and PLVs could have evolved from two distinct types of mobile elements by (independent) acquisition of the morphogenetic modules. Alternatively, one group could be the ancestral with the other group being a derived one, evolving through replacement of the pDNAP or helicase genes, respectively. The detailed sequence and phylogenetic analysis of the Tlr1-like helicases allows distinguishing between the two possibilities and provides insights into the origin of Tlr1 elements and the PLV group of viruses. The conservation of the Polinton-like pDNAP domain in SF1 helicases of Tlr1, PLVs, and transpovirons is consistent with the monophyly of all these elements and their evolution from bona fide Polintons. Moreover, the inactivation of the pDNAP polymerization domain and the apparent loss of the exonuclease and domain clearly indicate that Polintons are the ancestral forms in this supergroup of viruses and mobile elements, whereas all other groups are derived. Such directionality of evolution, from Polintons to PLVs, is also supported by phylogenetic analysis of the pDNAP domains from various mobile elements (Fig. 4). The present findings further suggest that the Tlr1 elements, although typically viewed as a separate group of large DNA transposons or sometimes as non-autonomous polintons (Wuitschick et al. 2002; Kapitonov and Jurka 2006; Pritham et al. 2007), based on the gene content, should be considered a genuine, even if distinct, family of PLVs. Conceivably, the PLV ancestor encoding a Tlr1-like helicase evolved from a typical Polinton in which the SF1 helicase gene was fused to the pDNAP gene, likely leading to elimination of Motif 4 of the polymerization domain and abrogation of the polymerase activity. This sequence of events appears more parsimonious than horizontal acquisition of the inactivated polymerase domain by the ancestor of PLVs from Polintons, especially given that co-localization of the intact helicase and pDNAP genes is indeed observed in some bona fide Polintons. For example, in Polinton 2 from Hydra magnipapillata, the SF1 helicase gene is located immediately upstream of the pDNAP, although the helicase does not appear to be closely related to the corresponding domains of Tlr1-like helicases and is instead embedded within the PIF1 clade (Fig. 2). We note that although phylogenetic analyses indicate that transpovirons, PLVs and Tlr1 elements form a monophyletic assemblage (Figs. 2 and 4), the internal relationship between these elements could not be fully resolved. Phylogenetic analysis of the corresponding helicase domains suggests that PLVs occupy a basal position in this assemblage, whereas in the pDNAP phylogeny, PLVs form a sister group to transpovirons associated with mimiviruses. One possible explanation for this discrepancy is the short size (∼230 aa) and high divergence of the pDNAP domain in the PLVs/transpovirons which might affect the results of corresponding phylogenetic analysis. Indeed, when comparing highly divergent sequences there is unavoidable uncertainty in the alignment and substitution models might not fully represent complex biological reality. Furthermore, phylogenetic results can be dependent on current sampling, particularly if a diverse group is hugely undersampled compared with other groups.
Given that both transpovirons and PLVs are derived from Polintons (Fig. 4) and taking into account the apparent inactivation of pDNAP in the transpovirons, the most parsimonious scenario includes evolution of the mimivirus-associated transpovirons from PLVs through the reductive evolution route which involved the loss of several genes including those of the morphogenetic module. The alternative, convoluted evolutionary scenario involving emergence of PLVs from transpovirons via re-acquisition of the virion morphogenetic module, although not formally excluded, appears much less likely. Collectively, the results of the present analysis are consistent with the central role of Polintons (polintoviruses) in the evolution of diverse groups of eukaryotic DNA viruses and plasmids.
Supplementary data
Supplementary data are available at Virus Evolution online.
Supplementary Material
Acknowledgements
N.Y. and E.V.K. are supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
Conflict of interest: None declared.
References
- Altschul S. F., Madden T. L., Schäffer A. A.. et al. (1997) ‘Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs’, Nucleic Acids Research, 25: 3389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Gil M., Dufayard J. F.. et al. (2011) ‘Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-Based Approximation Schemes’, System Biology, 60: 685–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Gallot-Lavallee L., Maumus F. (2015) ‘Provirophages in the Bigelowiella Genome Bear Testimony to Past Encounters with Giant Viruses’, Proceedings of the National Academy of Sciences of the United States of America, 112: E5318–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L., Salas M. (1996) ‘Relating Structure to Function in phi29 DNA Polymerase’, The Journal of Biological Chemistry, 271: 8509–12 [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M., Gabaldon T. (2009) ‘trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses’, Bioinformatics, 25: 1972–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. M., Di Tommaso P., Notredame C. (2014) ‘TCS: A New Multiple Sequence Alignment Reliability Measure to Estimate Alignment Accuracy and Improve Phylogenetic Tree Reconstruction’, Molecular Biology and Evolution, 31: 1625–37 [DOI] [PubMed] [Google Scholar]
- Chang J. M., Di Tommaso P., Lefort V.. et al. (2015) ‘TCS: A Web Server for Multiple Sequence Alignment Evaluation and Phylogenetic Reconstruction’, Nucleic Acids Research, 43: W3–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., De Lamballerie X., Yutin N.. et al. (2013) ‘Megavirales”, A Proposed New Order for Eukaryotic Nucleocytoplasmic Large DNA Viruses’, Archives in Virology, 158: 2517–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Eberle R., Ehlers B.. et al. (2009) ‘The Order Herpesvirales’, Archives in Virology, 154: 171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnues C., Boyer M., Raoult D. (2012) ‘Sputnik, a Virophage Infecting the Viral Domain of Life’, Advance in Virus Research, 82: 63–89 [DOI] [PubMed] [Google Scholar]
- Desnues C., La Scola B., Yutin N.. et al. (2012) ‘Provirophages and Transpovirons as the Diverse Mobilome of Giant Viruses’, Proceedings of the National Academy of Sciences of the United States of America, 109: 18078–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunin-Horkawicz S., Feder M., Bujnicki J. M. (2006) ‘Phylogenomic Analysis of the GIY-YIG Nuclease Superfamily’, BMC Genomics, 7: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32/5: 1792–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Pritham E. J. (2005) ‘Non-Mammalian C-Integrases are Encoded by Giant Transposable Elements’, Trends in Genetics, 21: 551–2 [DOI] [PubMed] [Google Scholar]
- Fischer M. G. (2015) ‘Virophages go Nuclear in the Marine Alga Bigelowiella natans’, Proceedings of the National Academy of Sciences of the United States of America, 112: 11750–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. G., Suttle C. A. (2011) ‘A Virophage at the Origin of Large DNA Transposons’, Science, 332: 231–4 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V.. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, System Biology, 59: 307–21 [DOI] [PubMed] [Google Scholar]
- Haapa-Paananen S., Wahlberg N., Savilahti H. (2014) ‘Phylogenetic Analysis of Maverick/Polinton Giant Transposons Across Organisms’, Molecular Phylogenetics and Evolution, 78: 271–4 [DOI] [PubMed] [Google Scholar]
- Iyer L. M., Balaji S., Koonin E. V., Aravind L. (2006) ‘Evolutionary Genomics of Nucleo-Cytoplasmic Large DNA VIRUSES’, Virus Research, 117: 156–84 [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V. V., Pavlicek A.. et al. (2005) ‘Repbase Update, A Database of Eukaryotic Repetitive Elements’, Cytogenetic Genome Research, 110-4: 462–7 [DOI] [PubMed] [Google Scholar]
- Kapitonov V. V., Jurka J. (2006) ‘Self-Synthesizing DNA Transposons in Eukaryotes’, Proceedings of the National Academy of Sciences of the United States of America, 103: 4540–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., Aswad A. (2014) ‘The Origins of Giant Viruses, Virophages and Their Relatives in Host Genomes’, BMC Biology, 12: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas D., Venclovas C. (2014) ‘Herpesviral Helicase-Primase Subunit UL8 is Inactivated B-Family Polymerase’, Bioinformatics, 30: 2093–7 [DOI] [PubMed] [Google Scholar]
- Klassen R., Meinhardt F. (2007) ‘Linear Protein-Primed Replicating Plasmids in Eukaryotic Microbes’, Microbiology Monographs, 7: 188–216. [Google Scholar]
- Koonin E. V., Dolja V. V., Krupovic M. (2015) ‘Origins and Evolution of Viruses of Eukaryotes: The Ultimate Modularity’, Virology, 479–480: 2–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Bamford D. H. (2008) ‘Virus Evolution: How Far Does the Double Beta-Barrel Viral Lineage Extend?’, Nature Reviews Microbiology, 6: 941–8 [DOI] [PubMed] [Google Scholar]
- Krupovic M.,, Bamford D. H. (2011) ‘Double-Stranded DNA Viruses: 20 Families and Only Five Different Architectural Principles for Virion Assembly’, Current Opinion in Virology, 1: 118–24 [DOI] [PubMed] [Google Scholar]
- Krupovic M., Koonin E. V. (2014) ‘Evolution of Eukaryotic Single-Stranded DNA Viruses of the Bidnaviridae Family from Genes of Four Other Groups of Widely Different Viruses’, Sciencetific Reports, 4: 5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Koonin E. V. (2015) ‘Polintons: A Hotbed of Eukaryotic Virus, Transposon and Plasmid Evolution’, Nature Reviews Microbiology, 13: 105–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Koonin E. V. (2016) ‘Self-Synthesizing Transposons: Unexpected Key Players in the Evolution of Viruses and Defense Systems’, Current Opinion in Microbiology, 31: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Bamford D. H., Koonin E. V. (2014) ‘Conservation of Major and Minor Jelly-Roll Capsid Proteins in Polinton (Maverick) Transposons Suggests that they are Bona Fide Viruses’, Biology Direct, 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Kuhn J. H., Fischer M. G. (2016) ‘A Classification System for Virophages and Satellite Viruses’, Archives in Virology, 161: 233–47 [DOI] [PubMed] [Google Scholar]
- La Scola B., Desnues C., Pagnier I.. et al. (2008) ‘The Virophage as a Unique Parasite of the Giant Mimivirus’, Nature, 455: 100–4 [DOI] [PubMed] [Google Scholar]
- Laurinmaki P. A., Huiskonen J. T., Bamford D. H., Butcher S. J. (2005) ‘Membrane Proteins Modulate the Bilayer Curvature in the Bacterial Virus Bam35’, Structure, 13: 1819–28 [DOI] [PubMed] [Google Scholar]
- Makarova K. S., Krupovic M., Koonin E. V. (2014) ‘Evolution of Replicative DNA Polymerases in Archaea and Their Contributions to the Eukaryotic Replication Machinery’, Frontiers in Microbiology, 5: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Palacios G. López-Ramírez V. Cardona-Felix C. S., Brieba L. G.(2012) ‘A Transposon-Derived DNA Polymerase from Entamoeba Histolytica Displays Intrinsic Strand Displacement, Processivity and Lesion Bypass’, PLoS One, 7: e49964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Grishin N. V. (2014) ‘PROMALS3D: Multiple Protein Sequence Alignment Enhanced with Evolutionary and Three-Dimensional Structural Information’, Methods in Molecular Biology, 1079: 263–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham E. J., Putliwala T., Feschotte C. (2007) ‘Mavericks, a Novel Class of Giant transposable elements widespread in eukaryotes and related to DNA viruses’, Gene, 390: 3–17 [DOI] [PubMed] [Google Scholar]
- Redrejo-Rodriguez M., Salas M. (2014) ‘Multiple Roles of Genome-Attached Bacteriophage Terminal Proteins’, Virology, 468–470: 322–9 [DOI] [PubMed] [Google Scholar]
- Rogozin I. B., Makarova K. S., Pavlov Y. I., Koonin E. V. (2008) ‘A Highly Conserved Family of Inactivated Archaeal B Family DNA Polymerases’, Biology Direct, 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. (2002) ‘An Approximately Unbiased Test of Phylogenetic Tree Selection’, System Biology, 51: 492–508 [DOI] [PubMed] [Google Scholar]
- Shimodaira H., Hasegawa M. (2001) ‘CONSEL: for assessing the confidence of phylogenetic tree selection’, Bioinformatics, 17: 1246–7 [DOI] [PubMed] [Google Scholar]
- Singleton M. R., Dillingham M. S., Wigley D. B. (2007) ‘Structure and Mechanism of Helicases and Nucleic Acid Translocases’, Annual Review of Biochemistry, 76: 23–50 [DOI] [PubMed] [Google Scholar]
- Sizov D. V., Polischuk V. P. (2006) ‘Cultivation, Purification and Crystallization of Virus of Green Algae Tetraselmis viridis’, Biopolym Cell, 22: 243–5 [Google Scholar]
- Söding J. (2005) ‘Protein Homology Detection by HMM-HMM Comparison’, Bioinformatics, 21: 951–60 [DOI] [PubMed] [Google Scholar]
- Tahirov T. H., Makarova K. S., Rogozin I. B.. et al. (2009) ‘Evolution of DNA Polymerases: An Inactivated Polymerase-Exonuclease Module in Pol Epsilon and A Chimeric Origin of Eukaryotic Polymerases from Two Classes of Archaeal Ancestors’, Biology Direct, 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M.. et al. (2009) ‘Jalview Version 2–A Multiple Sequence Alignment Editor and Analysis Workbench’, Bioinformatics, 25: 1189–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuitschick J. D., Gershan J. A., Lochowicz A. J., et al. (2002) ‘A Novel Family of Mobile Genetic Elements is Limited to the Germline Genome in Tetrahymena thermophila’, Nucleic Acids Research, 30: 2524–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuitschick J. D., Lindstrom P. R., Meyer A. E., Karer K. M. (2004) ‘Homing Endonucleases Encoded by Germ Line-Limited Genes in Tetrahymena Thermophila Have APETELA2 DNA Binding Domains’, Eukaryotic Cell, 3: 685–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Faure G., Koonin E. V., Mushegian A. R. (2014) ‘Chordopoxvirus Protein F12 Implicated in Enveloped Virion Morphogenesis is an Inactivated DNA Polymerase’, Biology Direct, 9: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Kapitonov V. V., Koonin E. V. (2015) ‘A New Family of Hybrid Virophages from an Animal Gut Metagenome’, Biology Direct, 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Raoult D., Koonin E. V. (2013) ‘Virophages, polintons, and Transpovirons: A Complex Evolutionary Network of Diverse Selfish Genetic Elements with Different Reproduction Strategies’, Virology Journal, 10: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Shevchenko S., Kapitonov V.. et al. (2015) ‘A Novel Group of Diverse Polinton-Like Viruses Discovered by Metagenome Analysis’, BMC Biology, 13: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.