Abstract
Mutations in SOX10 cause neurocristopathies which display varying degrees of hypopigmentation. Using a sensitized mutagenesis screen, we identified Smarca4 as a modifier gene that exacerbates the phenotypic severity of Sox10 haplo-insufficient mice. Conditional deletion of Smarca4 in SOX10 expressing cells resulted in reduced numbers of cranial and ventral trunk melanoblasts. To define the requirement for the Smarca4 -encoded BRG1 subunit of the SWI/SNF chromatin remodeling complex, we employed in vitro models of melanocyte differentiation in which induction of melanocyte-specific gene expression is closely linked to chromatin alterations. We found that BRG1 was required for expression of Dct, Tyrp1 and Tyr, genes that are regulated by SOX10 and MITF and for chromatin remodeling at distal and proximal regulatory sites. SOX10 was found to physically interact with BRG1 in differentiating melanocytes and binding of SOX10 to the Tyrp1 distal enhancer temporally coincided with recruitment of BRG1. Our data show that SOX10 cooperates with MITF to facilitate BRG1 binding to distal enhancers of melanocyte-specific genes. Thus, BRG1 is a SOX10 co-activator, required to establish the melanocyte lineage and promote expression of genes important for melanocyte function.
INTRODUCTION
Neurocristopathies are a diverse set of genetic disorders that involve defects in cells derived from the embryonic neural crest (1). Waardenburg syndrome (WS) is a type of neurocristopathy primarily caused by melanocyte deficiencies and manifested by varying degrees of skin and hair hypopigmentation, pigmentation defects of the choroid and iris, and deafness from loss of inner ear melanocytes. There are four clinically distinct WS types that are due to mutations in different genes (2–6): WS1 and WS3 result from mutations in the PAX3 gene, some cases of WS2 are due to mutations in the Microphthalmia-associated transcription factor (MITF) gene and some cases of WS2 and WS4 are due to haploinsufficiency of SOX10, which can also result in aganglionosis of the distal colon, and peripheral neuropathies. Individuals with SOX10 haploinsufficiency display phenotypic variability, suggesting the existence of additional modifier genes.
SOX10 is a member of the SOX (Sry-related high-mobility-group box) family of transcription factors that is essential for neural crest stem cell survival and for differentiation into melanocytes and Schwann cells (7). Members of this family generally cooperate with other transcriptional regulators to synergistically activate transcription, binding to AT-rich motifs in the minor groove of DNA (8). During Schwann cell differentiation, SOX10 interacts with a different set of transcription factors to activate genes required for myelination (9–13). SOX10 has been shown to interact with the SWI/SNF chromatin remodeling complex and to recruit the complex to Schwann cell-specific loci (14,15).
SWI/SNF chromatin remodeling enzymes utilize energy from ATP hydrolysis to physically remodel chromatin and regulate gene expression (16). At the core of the complex is either of two ATPases, BRG1 or BRM, which are encoded by the Smarca4 and Smarca2 genes, respectively. BRG1 and BRM each have a conserved ATPase domain that utilizes the energy from ATP hydrolysis to alter DNA-histone interactions, rendering sites accessible to factor binding and in some cases promoting nucleosome repositioning or eviction (17–19). Distinct SWI/SNF complexes containing BRG1 or BRM are formed through associations with 9–11 additional subunits known as BRG1/BRM-associated factors (BAFs) (20). SWI/SNF enzymes lack sequence specificity and are thought to be recruited to target regions primarily by interactions with major transcriptional regulators such as C/EBP factors in myeloid cells and adipocytes (21–23), MYOD in muscle (24,25), NEUROD in neurons (26), SOX10 in Schwann cells (14,15) and MITF in melanocytes (27–29). SWI/SNF enzymes play essential roles in maintaining embryonic pluripotency as well as promoting lineage specification and differentiation (30). Disruption of Smarca4 causes early embryonic lethality (31). Mice with melanocyte-specific disruption of Smarca4 fail to develop melanocytes and thus lack pigmentation (29). Schwann cell-specific disruption of Smarca4 severely compromises Schwann cell differentiation and causes peripheral neuropathy (14). Thus, Smarca4-encoded BRG1 is critically required for the development of two neural crest cell-derived lineages, which are both also highly dependent on SOX10. It has been well established that BRG1 interacts with MITF and co-activates MITF target genes in melanocytes and melanoma cells. BRG1 is a SOX10 co-activator in Schwann cells, however it remains to be determined if BRG1 also interacts with SOX10 to regulate melanocyte development and function.
In this study, we employed a previously established N-ethyl-N-nitrosourea (ENU) mutagenesis screen and identified Smarca4 as a modifier gene that exacerbates the phenotypic severity of SOX10LacZ/+ mice (32). We confirmed that the Smarca4 mutation was responsible for the pigmentation phenotype by deleting Smarca4 in SOX10-expressing cells. To mechanistically define the functional interaction between SOX10 and BRG1 in the regulation of genes that promote melanocyte function, we utilized in vitro models of melanocyte differentiation and characterized chromatin structural changes at the regulatory regions of melanocyte-specific genes that are induced during differentiation. We found that BRG1 is required for melanocyte differentiation, expression of melanocyte-specific genes, and chromatin remodeling at regulatory sites on SOX10 and MITF target genes. SOX10 was found to interact with BRG1 and to cooperate with MITF in recruiting BRG1 to melanocyte-specific regulatory regions.
MATERIALS AND METHODS
Mouse genetics and husbandry
Maintenance of the Sox10LacZ line (SOX10tm1Weg) was performed as previously described (32). SOX10LacZ and Smarca4Mos6 mice were bred and housed in an NHGRI animal facility according to NIH guidelines. Animal care was done in accordance with NHGRI institutional standards and was approved by the Institutional Animal Care and Use Committee, NHGRI, and NIH Institutional Review Board. Mice carrying the Tg(SOX10-cre)1Wdr transgene (33) and a floxed BRG1 allele, Smarca4tm1.2Pcn (34) were bred in the Erlangen animal facility in accord with EU animal welfare laws with approval of the responsible local committees and government bodies.
Mapping and exome sequencing of the Smarca4 Mos6 mutation
A low density genome scan was completed with seven affected mice using 61 SSLP markers (a subset of MIT markers described in (35) polymorphic between the founder strains C57BL/6J and BALBcJ). Because of a mixed genetic background of the founder mouse, a portion of the genome was not informative in this initial mapping. Following this preliminary genome scan, 11 regions were identified as ‘potentially linked’ (two regions of linkage plus an additional nine uninformative regions). One sample from an affected mouse was used for exome sequencing. Sequence variants were filtered to remove low quality variants, variants not predicted to alter coding sequences, variants present in dbSNP, and variants previously identified in other exome sequencing projects of independent mutants on the same genetic background within our laboratory. After filtering, 69 variants remained within the 11 regions of ‘potential linkage’. These variants included two on Chr 9, both within Smarca4. Subsequent breeding confirmed segregation of the Smarca4 variants with the Mos6 phenotype in 42 mice across three generations.
Genotyping
The Taqman assay or RT-PCR was used for genotyping.
In situ hybridization, immunohistochemistry and β-galactosidase staining
Whole mount in situ hybridization was performed essentially as described (36). Embryos underwent overnight fixation in 4% paraformaldehyde at 4°C. After dehydration, bleaching and rehydration, in situ hybridization was performed with DIG-labeled antisense riboprobes for Kit, Dct and MITF. All steps except probe hybridization and final colorimetric detection were performed on a Biolane HTI (Hölle & Hüttner AG, Tübingen, Germany).
Immunohistochemistry was performed on 10-μm-thick sections from the trunk of embryos after fixation, embedding, freezing and cryotome sectioning using the following primary antibodies: anti-Islet1 mouse monoclonal (1:500 dilution, Developmental Studies Hybridoma Bank), anti-MITF rabbit antiserum (1:1000 dilution, gift of H. Arnheiter, NIH, Bethesda, MD, USA), anti-FABP7 rabbit antiserum (1:300 dilution; Millipore), anti-SOX10 guinea pig antiserum [1:1000 dilution (37)]. Secondary antibodies conjugated to Cy2, Cy3 (1:200 dilution, Dianova) or Alexa488 (1:500 dilution, Molecular Probes/Invitrogen) immunofluorescent dyes were used for detection. Nuclei were counterstained with 4΄,6-diamidin-2-phenylindole (DAPI).
β-Galactosidase (β-gal) staining was performed on whole embryos as previously described (32).
Cell culture and expression plasmids
MITF-M and SOX10 retroviral plasmids were described in (15,27). B22 cells were cultured as described in (38). Melb-a cells were obtained from the Wellcome Trust Functional Genomics Cell Bank (St. George's, University of London, UK) and cultured in growth medium (RPMI 1640 with 10% fetal bovine serum, 40 pM fibroblast growth factor and 10 ng/ml stem cell factor) at 37°C under 5% CO2. Differentiation was induced when cultures were 70% confluent by replacing growth medium with differentiation medium (DMEM with 10% fetal bovine serum, 2 nM alpha melanocyte stimulating hormone (NDP-alpha MSH) and 200 nM phorbol–myristate–acetate (39). C2C12 and 3T3L1 cells were from the American Type Culture Collection (ATCC) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum.
RNA isolation and quantitative PCR (qPCR)
Total RNA was isolated using Trizol (Life Technologies, Carlsbad, CA, USA) and cDNA was prepared using the Quantitect Reverse Transcription kit (Qiagen, Valencia, CA, USA). Quantitative PCR (qPCR) was performed in SYBR Green master mix (Qiagen) and analyzed as described (28). Primers used for mouse Dct: 5΄-GGACCGGCCCCGACTGTAATC-3΄ and 5΄-GGGCAACGCAAAGGACTCAT-3΄, mouse Tyrp1: 5΄-GCCCCAACTCTGTCTTTTCTCAAT-3΄ and 5΄-GATCGGCGTTATACCTCCTTAGC-3΄, mouse Tyrosinase (Tyr: 5΄-CGGCCCAAATTGTACAGAGAAGC-3΄ and 5΄-CTGCCAGGAGAAGAAGGATTG-3΄, mouse Trpm1: 5΄-CCTACGACACCAAGCCAGAT-3΄, and 5΄-GACGACACCAGTGCTCACAC-3΄, and mouse Rab27a: 5΄-CGACCTGACAAATGAGCAAA-3΄ and 5΄-GGCAGCACTGGTTTCAAAAT-3΄. Mouse Mpz, Mbp, and Rpl7 primers were described in (15).
Cell extracts and immunoblot analysis
Total cell extracts were prepared and Western blots were performed as described (28). Antiserum to BRG1 was previously described (38). The BRM and MITF antibodies were from Abcam (Cambridge, MA, USA), the SOX10 antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA), the FLAG antibody was from Sigma (St. Louis, MO, USA), BAF60A and BAF60B antibodies were from Bethyl Laboratories (Montgomery, TX, USA) and the Tubulin antibody was from Cell Signaling Technology (Boston, MA, USA).
Melanin assays
Cells were analyzed for melanin content as described in (40). Briefly, cells were trypsinized and counted using the Scepter 2.0 handheld automated cell counter (Millipore) Cells were lysed in 0.1 M NaOH and vortexed for 20 min. Melanin content was calculated based on the absorbance at 475 nm as compared to the standard curve obtained using synthetic melanin (Sigma, St. Louis, MO, USA) and normalized to cell number.
siRNA knockdown
A non-targeting siRNA and siRNA targeting MITF as reported in (41–44), siRNA targeting rodent SOX10 as reported in (15,45), siBRG1/BRM as reported in (28,46) and a SMART pool of siRNAs targeting BRG1 (siBRG1-D) were obtained from Dharmacon Inc. (Lafayette, CO, USA). Two additional siRNAs that target mouse BRG1 (siBRG1-II3.1: 5΄-GUCAGACAGUAAUAAAUUAAAGCAA-3΄ and siBRG1-I13.3:5΄-GAUGAUGAGACCGUCAACCAGAUGA-3΄) and siRNAs that target mouse BRM (siBRM-I13.2: 5΄-AUCAUUGAUACUGUGAUAAACUACA-3΄ and siBRM-I13.3: 5΄-GAUGUGGGUGGAUAGUAUAUUUCTA-3΄) were purchased from Integrated DNA Technologies (Coralville, IA, USA). siRNas were transfected with Dharmafect 1 according to manufacturer's instructions. Undifferentiated Melb-a cells were transfected in growth medium for 48 h and harvested or the medium was then replaced and the cells were cultured in differentiation medium for an additional 48 h.
Formaldehyde-assisted Isolation of regulatory elements (FAIRE)
FAIRE was performed as essentially as described in (47). Cells were crosslinked in 1% formaldehyde for 6 min at room temperature and quenched with 125 mM glycine. Cells were then lysed using the buffers listed in the alternative lysis method as described in (48) and sonicated as described in (28). Sonicated chromatin was subjected to two rounds of phenol/chloroform extractions, back extracted once with TE and then once with chloroform. The aqueous phase was ethanol precipitated and digested with 0.2 mg/ml Proteinase K for 1 h at 55°C. Cross-links were then reversed by heating overnight at 65°C. DNA was then purified by an additional phenol chloroform extraction and ethanol precipitation. Control Inputs were 10% of each sample that was heated at 65°C overnight to reverse crosslinking prior to purification. The primers used were: mouse Dct distal region: 5΄-ACGGTTCCACGCAAATTAAA-3΄ and 5΄-TACTTCTCCCCCATGACTGC-3΄, mouse Dct proximal region: 5΄-GGCGAGCCAGAGAGAATAAA-3΄ A and 5΄-TTCTCGCCAGTCTTCCTTGT, mouse Tyrp1 distal region: 5΄-TGACAGTGAGGGCACATTTC-3΄ and 5΄-ACCCATGTTCCCAGACTGAA-3΄, mouse Tyrp1 proximal region: 5΄-GCAAAATCTCTTCAGCGTCTC-3΄ and 5΄-AGCCAGATTCCTCACACTGG-3΄, mouse tyrosinase (Tyr) distal region 5΄-TGCCAGCTGACTTTGTCAAG-3΄ and 5΄-AATATTGTGGTTTGCCAGGA-3΄, mouse Tyr proximal: 5΄-AGTCATGTGCTTTGCAGAAGAT-3΄ and 5΄-CAGCCAAGAACATTTTCTCCTT-3΄ mouse Scn2a1: 5΄-AAGCAGCTGCCTTTGGGAAG-3΄ and CACAGGACACTGAGCATCAAG-3΄ and mouse MyoD CER as reported in (49).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described (28) using the same antibodies as in Westerns as well as antibodies to histone H3, H3K4me3 and H3K27ac that were obtained from Abcam. Primers were the same as used in FAIRE.
Statistical analysis
Statistical significance was calculated by the Student's t test. One way ANOVA followed by Tukey's post-hoc test was used for comparing multiple groups.
RESULTS
ENU alleles of Smarca4
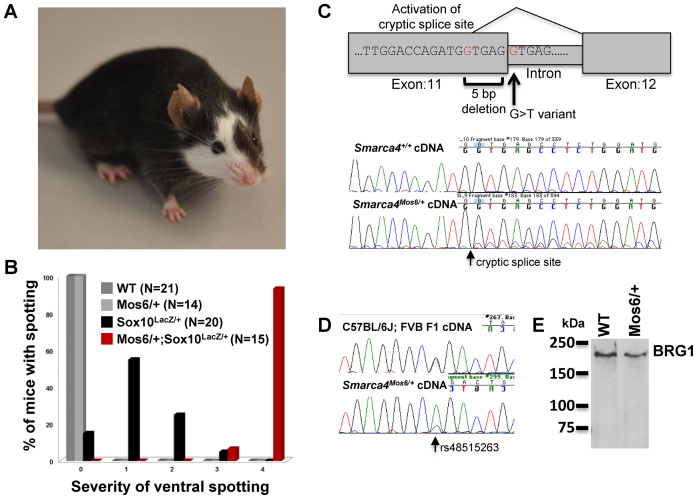
We used a previously established N-ethyl-N-nitrosourea (ENU) mutagenesis screen to identify novel modifiers of SOX10 neurocristopathies (Mos) (32). Briefly, BALB/cJ male mice were given weekly ENU injections for 3 weeks, and after regaining fertility, mated with C57BL/6J females to generate first generation (G1) offspring. G1 males were mated with Sox10LacZ/+ females and second generation (G2) offspring were then scored for hypopigmentation. One of the founder mutants, Mos6, had more severe white spotting than is typically observed in the Sox10LacZ/+ haploinsufficient mice (Figure 1A). Further breeding of this founder and quantitation of ventral spotting in the pedigree showed that Mos6 synergistically increased the severity of white spotting observed in SOX10 haploinsufficient mice (Figure 1B). To map the Mos6 locus, seven affected Mos6/+; SOX10LacZ/+ double heterozygotes were genotyped in a low density genome scan (61 simple sequence length polymorphism markers), and in parallel with this linkage mapping, one Mos6/+; SOX10LacZ/+ double heterozygote sample was used for exome sequencing. Candidate sequence variants within regions of potential linkage were further refined by subsequent breeding that confirmed linkage of Mos6 to two sequence variants within Smarca4, the gene that encodes BRG1.
Figure 1.
Identification of the Smarca4Mos6 allele. (A) SOX10LacZ/+; Mos6/+ double heterozygous mice exhibited a head spot and extensive belly spotting, demonstrating that the presence of the Mos6 allele increased the spotting phenotype of SOX10LacZ/+. (B) Hypopigmentation in the trunk of adult mice was quantitated using a standardized scale of 0–4, in which 0 indicates no spotting, and 1–4 indicates progressively greater hypopigmentation (32). Mos6 synergistically increased the ventral white spotting observed in SOX10 haploinsufficient mice. (C) The Smarca4 mutation splice site variant (mm9 Chr9:21447196;G>T) that alters a consensus 5΄ donor splice site at the junction of exon 11 and intron 11 (c.1812+1G>T, NM_001174078.1). Low levels of a mutant transcript were detected (15% of total transcript), suggesting the mutant causes activation of a cryptic splice site within exon 11, as illustrated. (D) In cDNA from an outcross C57BL/6J and FVB/N F1 hybrid embryo, the two strain-specific alleles at rs48515263, a SNP located within exon 15 of Smarca4, were present in equal proportion. In cDNA from an embryo carrying the Mos6 ENU-induced C57BL/6J mutation, the C57BL/6J SNP allele detected in the mutant transcript was reduced relative to the FVB/N wild-type allele, consistent with instability of the Mos6 mutant allele. (E) Western blots detected only wild-type BRG1 expressed in Mos6/+ heterozygous embryos. Similar results were observed using four embryos of each genotype run in triplicate Western blots with no detection of a truncated protein even with overloading or overexposure.
The two Smarca4 ENU mutations include a splice site variant at the consensus 5΄ donor splice site of exon 11 (mm9 Chr9:21447196;G>T) as well as a downstream variant in exon 22 (mm9 Chr9:21474974;G>A). Subsequent genotyping of both Smarca4 ENU mutations confirmed strong linkage with the Mos6 phenotype throughout the colony (0/42 recombinants over three generations; x2 = 42; P < 0.0001). Because of the proximity of the two ENU mutations within Smarca4, they could not be segregated in the colony and thus we could not determine the relative contribution of the two mutations individually to the phenotype. However, the splice site variant at the junction of exon 11 and intron 11 (c.1812+1G>T, NM_001174078.1) is predicted to introduce a frameshift resulting in protein truncation upstream of the variant in exon 22, thus rendering the exon 22 variant incapable of contributing to the in vivo phenotype. To confirm the consequence of the splice site variant, RT-PCR products from Mos6 heterozygotes were sequenced. There was no evidence of exon skipping or intron retention resulting from the splice site variant, however, a mutant transcript was detected that results from activation of a cryptic splice site 5 bp upstream of the Exon 11 splice site disrupted by the mutation (Figure 1C). The ratio of chromatogram peaks detected by sequencing of the RT-PCR product suggested that the mutant transcript was present at low levels (∼15% of the total transcript (Figure 1C). To further characterize the stability of this Mos6 mutant transcript, the relative ratio of wild type and mutant transcript was measured in an outcross to FVB/N, where a strain-specific SNP (rs48515263) within exon 15 of Smarca4 could be used to assess relative expression of the two alleles. In cDNA from a wild-type F1 hybrid embryo, the two SNP alleles (C57BL/6J and FVB/N) were detected in equal proportion. In contrast, cDNA from a Mos6 heterozygous F1 hybrid embryo confirmed that the C57BL/6J SNP allele carrying the linked Mos6 mutations was barely detectable relative to the FVB/N wild-type allele, thus indicating that the mutant transcript was unstable (Figure 1D). Consistent with this, Western blotting failed to detect any evidence of the truncated protein (605 aa for Smarca4 mutant versus 1617 aa for wild-type) that might result from any residual Mos6 mutant transcript (Figure 1E). In summary, the low levels of mutant transcript along with the negative Western data indicate that the splice site mutation is likely responsible for the phenotype observed in Mos6 mice and that the Mos6 allele is functionally null. We subsequently use the allele name Smarca4Mos6 to refer to the allele that contains both point mutations.
Because of the visible white spotting in Smarca4Mos6/+;Sox10LacZ/+ double heterozygote adult mice, we sought to further characterize the effect of Smarca4 mutation on melanocyte development. A reduction in melanoblasts was first evident at E13.5 when the number of cranial melanoblasts marked by whole mount β-gal staining was significantly reduced in Smarca4Mos6/+;Sox10LacZ/+ double heterozygotes compared to Sox10LacZ/+ heterozygotes (Supplementary Figure S1). The reduction in melanoblast number was Sox10-dependent, as a similar reduction was not observed in Smarca4Mos6/+ heterozygote embryos where melanoblasts were marked with a Dct-LacZ transgene (data not shown). Further characterization of melanoblast development was limited as we were not able to recover Smarca4 Mos6/Mos6 homozygous embryos at E9.5 or later time points, consistent with published observations of early embryonic lethality resulting from a Smarca4 null allele (31).
Conditional deletion of Smarca4 in melanoblasts
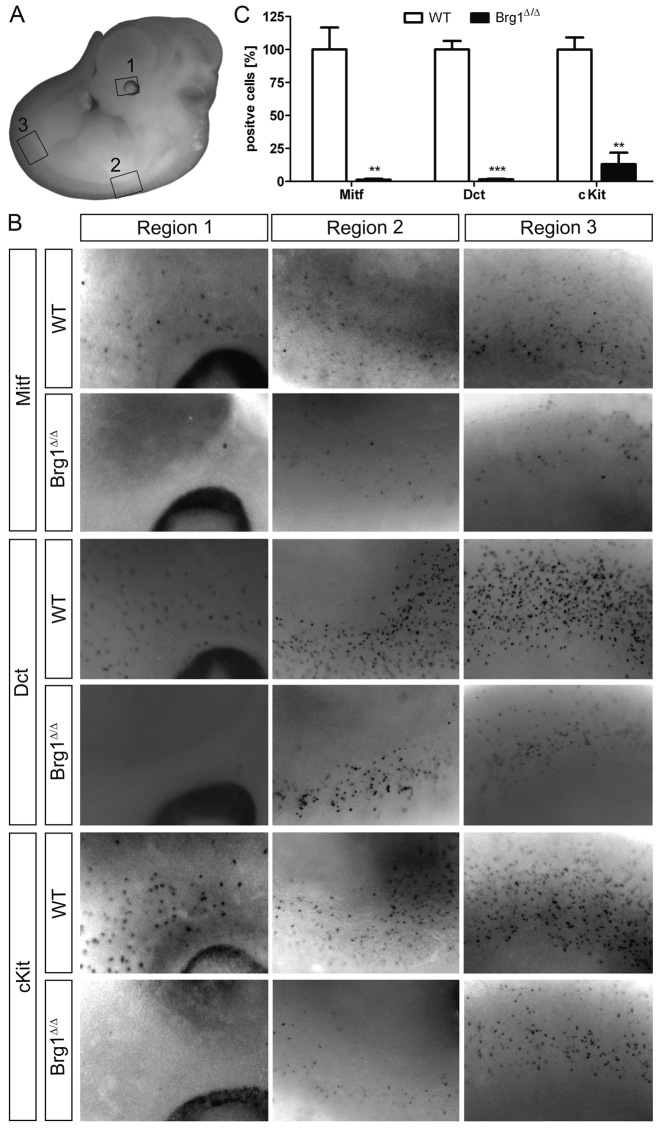
To further characterize the impact of Smarca4-deficiency on melanocyte development and confirm Smarca4Mos6 as the causative mutation for the Mos6 phenotype, we characterized a second allele of Smarca4. Melanocyte development was examined when Smarca4 was specifically deleted in SOX10-expressing cells using a SOX10-Cre construct (33,34). This conditional deletion of Smarca4 and subsequent loss of BRG1 expression resulted in a striking reduction in the numbers of cranial and ventral trunk melanoblasts at E12.5 as visualized by Mitf, Dct and Kit in situ hybridization (Figure 2A and B). Quantification of cells in the eye region that were positive for Mitf, Dct and Kit revealed a drastic reduction of melanocytes (Figure 2B, region 1). Mitf-positive cells were reduced to 1.3 ± 0.7%, Dct-posititve cells to 1.6 ± 0.4% and Kit-positive cells to 13.0 ± 8.8% (Figure 2C). Immunohistochemistry of trunk sections indicated a less pronounced, but qualitatively similar reduction of Mitf-positive cells to 43.6 ± 10.6% (Supplementary Figure S2). This correlates with a Sox10-Cre mediated deletion rate of 57 ± 5% (n = 3) that we determined for a Rosa26-stopflox-YFP reporter in trunk melanocytes (data not shown). We also investigated the consequences of the Smarca4 deletion on other neural crest lineages as well as the possibility of a fate switch in melanoblasts. However, FABP7-expressing Schwann cell precursors along peripheral nerves (Supplementary Figure S3A, left) and ISLET1-positive sensory neurons in dorsal root ganglia (Supplementary Figure S2B) are still present in the Smarca4-deleted mice despite near complete deletion of the Rosa26–stopflox–YFP reporter. We also have no evidence that the remaining melanocytes convert to FABP-expressing glia (Supplementary Figure S3A, right) or islet1-expressing neurons (data not shown). This conditional Smarca4 mutant verifies the causative mutation in Mos6 mice and confirms an essential role for the Smarca4-encoded protein, BRG1, in establishing the SOX10-expressing melanoblast lineage.
Figure 2.
Conditional deletion of Smarca4 resulted in a striking reduction in the numbers of cranial and ventral trunk melanoblasts at E12.5. (A) The melanoblast-containing regions labeled 1 (cranial melanoblasts near optic cup), 2 (anterior trunk at forelimb) and 3 (posterior trunk at hindlimb) are shown at greater magnification in B, as indicated. (B) Comparison of melanoblast number, as measured by whole mount in situ hybridization for MITF (top), Dct (center) and Kit (bottom) shows fewer melanoblasts in embryos where Smarca4 was conditionally deleted in melanoblasts using a SOX10-Cre construct (BRG1Δ/Δ) as compared to normal littermates (WT). (C) Quantification of cells in the eye region from the ISH whole mounts that were positive for the indicated melanocyte markers (***P < 0.001, **P < 0.01, Student's t test).
The requirement for BRG1 in the activation of melanocyte-specific genes by SOX10
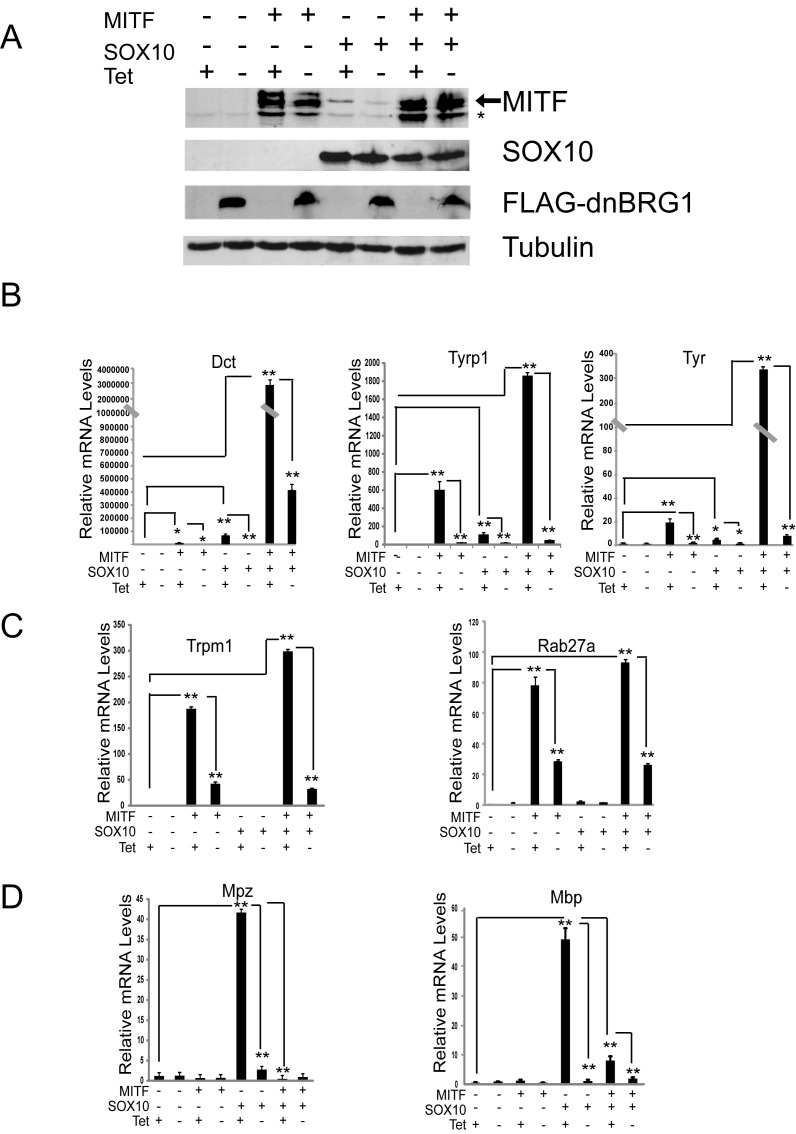
To elucidate the transcriptional mechanisms by which BRG1 cooperates with SOX10 to establish the melanocyte lineage, we turned to a previously described fibroblast cell line that can be induced to express a dominant negative version of BRG1 under the control of the tetVP16 activator, hereafter referred to as dnBRG1 (38). dnBRG1 lacks chromatin remodeling activity due to a mutation in the ATPase domain and acts as a dominant negative because it suppresses gene activation events that normally require SWI/SNF function. By ectopically expressing either SOX10 or MITF in B22 cells, we previously found that SOX10 interacts with BRG1 to promote myelin gene expression while MITF interacts with BRG1 to promote melanocyte-specific gene expression (15,27). To determine the requirement for BRG1 in SOX10-mediated melanocyte-specific gene expression, we ectopically expressed MITF, SOX10, or a combination of MITF and SOX10 in B22 cells and cultured them in the presence or absence of tetracycline. Western blotting confirmed that MITF, SOX10, or both MITF and SOX10 were expressed regardless of the presence or absence of tetracycline, while expression of dnBRG1 was detected only in cells that had been cultured in the absence of tetracycline (Figure 3A).
Figure 3.
Dominant negative BRG1 inhibits activation of melanocyte-specific genes by MITF and SOX10. (A) B22 cells were infected with a pBABE control vector, pBABE-MITF, pBABE-SOX10 or pBABE-MITF with pBABE-SOX10 in the presence (dominant negative BRG1 off) or absence of tetracycline (dominant negative BRG1 on) and then cultured in low serum medium to promote differentiation. Western Blot analysis showing expression of MITF and SOX10 in cells that were cultured in the presence and absence of tetracycline, and the expression of FLAG-tagged dominant BRG1 when cells were cultured in the absence of tetracycline. Protein expression was detected from cell extracts and tubulin was used as a loading control. (B–D) Quantitative RT-PCR (qRT-PCR) of MITF and/or SOX10 target genes from cells transfected with pBABE, pBABE-MITF, pBABE-SOX10 or pBABE-MITF together with pBABE-SOX10 in the presence or absence of tetracycline demonstrated that dominant negative BRG1 blocked the (B) synergistic activation of Dct, Tyrp1 and Tyr gene expression by SOX10 and MITF, (C) activation of Trpm1 and Rab27a by MITF and (D) activation of Mpz and Mbp by SOX10. Expression of each gene was normalized to expression of Rpl7. The data are the average of at least two independent experiments performed in triplicate. Standard error bars and statistical significance are shown (**P < 0.01, *P < 0.05, ANOVA).
Consistent with our previous semi-quantitative data (27), we found that ectopic expression of MITF in B22 cells that were cultured in the presence of tetracycline activated expression of tyrosinase family members Dct (110-fold), Tyrp1 (600-fold) and Tyr (20-fold) and removal of tetracycline and expression of dominant negative BRG1 dramatically inhibited expression of these three genes (Figure 3B). SOX10 is known to induce Mitf expression when partnered with PAX3 (50). We found that in the absence of Pax3, ectopic expression of SOX10 increased Mitf expression <2-fold compared to a control empty vector (data not shown). Interestingly, SOX10 induced expression of MITF target genes to a greater extent (Figure 3B). Dct was activated by SOX10 (57 000-fold) which was greater than Dct activation by MITF. However, SOX10 was less effective than MITF in activating Tyrp1 (100 fold) and Tyr (5-fold). Co-expression of MITF and SOX10 resulted in a synergistic increase in Dct expression (300 000-fold), Tyrp1 (2000-fold) and Tyr (350-fold). Activation of these three genes by MITF, SOX10 or MITF in combination with SOX10 was dramatically inhibited by expression of dnBRG1. These data suggest that BRG1 functionally interacts with SOX10 and that its catalytic activity is required for expression of the MITF- and SOX10-regulated genes that encode enzymes involved in melanin synthesis.
We also investigated whether SOX10 could activate the expression of two other MITF target genes that are important for melanocyte function. The transient receptor potential cation channel member 1 (Trpm1), associated with terminal melanocyte differentiation, and the small GTPase, Rab27a, which regulates melanosome transport are known MITF target genes (51–53). We found that MITF induced Trpm1 (200-fold) and Rab27a (80-fold) in a BRG1-dependent manner (Figure 3C). However, the expression of these genes did not change significantly in SOX10 expressing cells relative to control. The combination of MITF and SOX10 increased the mRNA levels of Trpm1 and Rab27a to a small extent (1.6- and 1.25-fold respectively) compared to the levels induced by MITF alone. Thus, expression of Trpm1 and Rab27a is highly dependent on MITF and BRG1 but less so on SOX10 in these cells.
Our previous work had indicated that ectopic expression of SOX10 in B22 cells can induce expression of Schwann cell genes that encode myelin protein zero (Mpz) and myelin basic protein (Mbp) (15). These genes were recently found to be regulated by MITF in melanoma cells (54,55). Therefore, we queried whether SOX10 synergizes with MITF to increase expression of Mpz and Mbp in a BRG1-dependent manner. We found that MITF alone did not significantly induce either Mpz or Mbp (Figure 3D). As we determined previously, SOX10 activated both Mpz and Mbp when cells were cultured in the presence of tetracycline but not when expression of dnBRG1 was induced by the removal of tetracycline. Interestingly, co-expression of MITF with SOX10 prevented SOX10 from inducing these myelin genes. Thus, in this cellular context, SOX10 cooperates with MITF to activate a subset of melanocyte-specific genes in a BRG1-dependent manner but works antagonistically with MITF to regulate genes important for myelination.
Temporal analysis of melanin synthesis and gene expression in differentiating melanoblasts
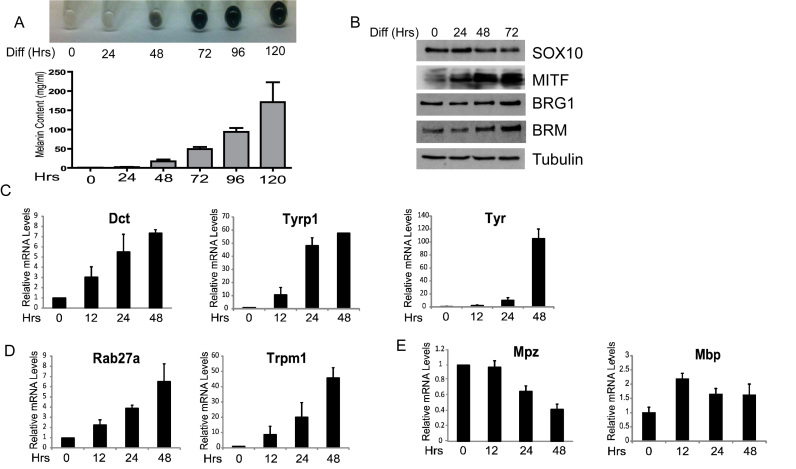
In vitro models of muscle and adipocyte differentiation, such as C2C12 myoblasts and 3T3L1 pre-adipocytes, have provided a wealth of knowledge regarding the mechanisms by which previously silent genes embedded in repressive chromatin structure are activated in a lineage-specific manner (25,56–58). However, in vitro models of melanocyte differentiation have been rarely used for such purposes. Therefore, we characterized mouse Melb-a cells, which are immortalized mouse melanoblasts derived from black nonagouti mice (39,59), to elucidate the mechanisms by which BRG1 co-activates SOX10 target genes in melanocytes. These cells can be maintained in an undifferentiated proliferative state when cultured in growth medium containing stem cell factor (SCF) and fibroblast growth factor 2 (FGF2) and then differentiated into pigmented melanocytes by shifting sub-confluent cells to differentiation medium containing a stable analog of alpha-MSH (NDP-α-MSH). We observed a time-dependent increase in melanin accumulation when Melb-a cells were shifted from growth to differentiation medium (Figure 4A). MITF expression was strongly induced concomitantly with the observed increase in melanin synthesis while SOX10 levels and BRG1 levels remained constant (Figure 4B). Interestingly, there was a small increase in the expression of the alternative ATPase BRM after 24 h of differentiation, suggesting that BRM may have a role in melanocyte differentiation (Figure 4B).
Figure 4.
Melanin synthesis and changes in gene expression during a time course of melanoblast differentiation (A) Melb-a cells were cultured in growth medium containing SCF and FGF until 70% confluent (time 0), then growth medium was replaced with differentiation medium containing NDP-α-MSH and the cells were cultured for the indicated periods of time. Cells were harvested at each time point and photographed. Cells were counted and an equal number were subjected to the melanin assay. The data are the average of at least two independent experiments performed in triplicate. Standard error bars and statistical significance compared to siC are shown (**P < 0.01, *P < 0.05, Student's t test). (B) Protein extracts were prepared from differentiating Melb-a cells and subjected to Western blotting with the indicated antibodies. Tubulin was used as a loading control. (C–E) RNA was isolated at the indicated time points, reversed transcribed and the specific transcript of interest quantified by qRT-PCR. The CT value for each gene was normalized to Rpl7. The data are the average of at least two independent experiments performed in triplicate. (C) Melanogenic enzyme gene expression increased as the cells differentiated. (D) Expression of genes associated with melanocyte differentiation increased as the cells differentiated. (E) Expression of myelin genes either decreased (Mpz) or exhibited a transient modest increase (Mbp). Standard error bars and statistical significance compared to siC are shown (**P < 0.01, *P < 0.05, Student's t test).
We performed a temporal analysis of gene expression to determine if genes required for melanin synthesis are induced during the differentiation process. Our data indicate that expression of these genes increases over a 48-h time period (Figure 4C and D). This long time-course of gene induction is likely due to asynchronous differentiation of individual cells as was previously demonstrated for B16 melanoma cells (60). Induction of Dct and Tyrp1 expression occurred 12 h after shifting to differentiation medium and peaked after 24 h with a 7–8-fold induction of Dct and 50–60-fold induction of Tyrp1, while Tyr expression was first detected at 24 h but increased substantially after 48 h with an ∼120-fold induction (Figure 4C). Rab27a and Trpm1 were induced with similar kinetics as Dct and Tyrp1, with a maximum induction of 8-fold and 50-fold respectively (Figure 4D). Whereas these MITF-dependent genes were consistently induced during the time course of Melb-a differentiation, the expression of Mpz, a gene which was activated by SOX10 but repressed by MITF in B22 cells, decreased during the time course of Melb-a differentiation while Mbp levels fluctuated and exhibited a slight increase of 1.5-fold after 48 h (Figure 4E).
Temporal analysis of gene expression and chromatin accessibility in differentiating melanoblasts
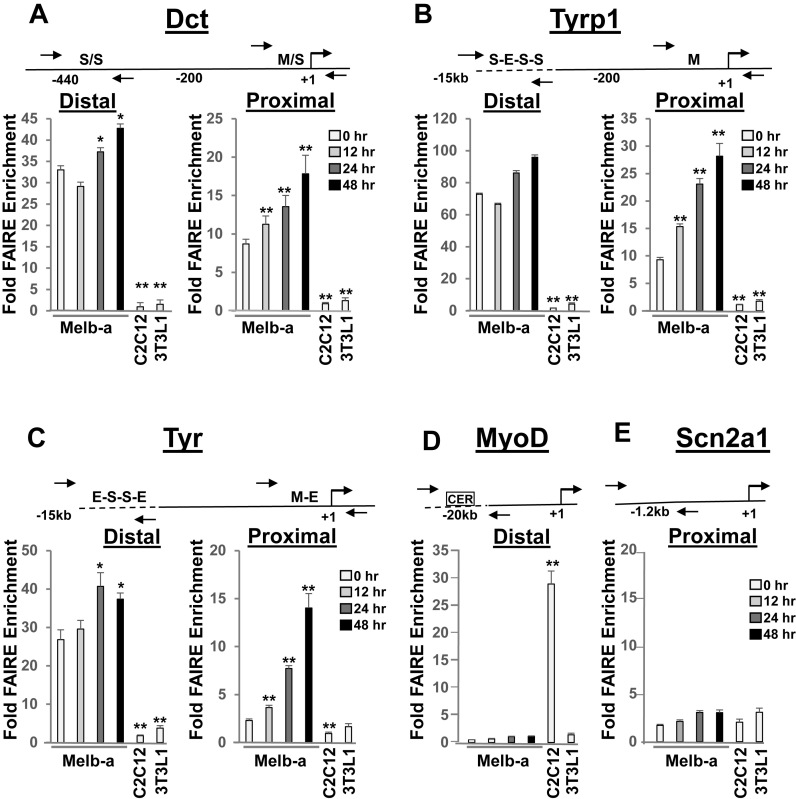
We performed a temporal analysis of chromatin accessibility in differentiating Melb-a cells to determine if the observed changes in melanocyte-specific gene expression were associated with alterations in chromatin structure. The FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) assay enriches for accessible chromatin sites that correspond closely with DNase I hypersensitive sites, occurring at transcriptional start sites, promoters, and enhancers (61). We assayed the regions of Dct, Tyrp1 and Tyr that contain SOX10 and MITF binding sites: SOX10 binding sites are located in distal enhancers, while M boxes which bind MITF are located in proximal promoter regions and E boxes that also bind MITF are also contained within distal enhancers (50,62–65). We detected significant FAIRE enrichment of the Dct distal enhancer and proximal promoter both in undifferentiated and in differentiated Melb-a cells but not in myoblasts (C2C12) nor pre-adipocytes (3T3L1). (Figure 5A). There was only a small increase in the FAIRE enrichment of the distal enhancer after 48 h of differentiation while enrichment of the Dct proximal promoter increased ∼2-fold. The enrichment level of the Tyrp1 distal enhancer was also high in undifferentiated Melb-a cells but not in C2C12 or 3T3L1 cells and increased slightly during the time course of differentiation, whereas there was a significant increase in the enrichment level of the Tyrp1 proximal promoter during the time course of differentiation (3-fold) (Figure 5B). We digested nuclei with MNase I and confirmed that FAIRE enrichment levels of the Tyrp1 distal and proximal regions reflect changes in chromatin accessibility (data not shown). Similarly, enrichment of the Tyr distal enhancer was high in undifferentiated Melb-a cells compared to C2C12 and 3T3LI cells and increased to a small extent during differentiation, while enrichment of the Tyr proximal promoter was only slightly higher in undifferentiated Melb-a cells than in C2C12 and 3T3L1 cells and increased greatly (7-fold) during the time course of differentiation (Figure 5C). Thus, the distal enhancers of Dct, Tyrp1 and Tyr were observed to be in a constitutively open chromatin state prior to differentiation while the chromatin at the proximal promoters of the highly induced Tyrp1 and Tyr became increasingly accessible upon differentiation. As a control for FAIRE specificity, we probed a distal regulatory region (CER) of MyoD, a muscle-specific gene that is expressed in proliferating C2C12 myoblasts (49). As expected, we found high FAIRE enrichment of this region in C2C12 cells compared to Melb-a and 3T3L1 cells (Figure 5D) while the level of FAIRE enrichment of the control Scn2a1 upstream region was similarly low in all cells (Figure 5E).
Figure 5.
Time course of chromatin accessibility at distal and proximal control regions of melanogenic enzyme genes during Melb-a differentiation. (A–C) Melb-a cells were cultured in growth medium containing SCF and FGF2 until 70% confluent (time 0). Growth medium was replaced with differentiation medium containing NDP-α-MSH and the cells were cultured for the indicated periods of time. C2C12 and 3T3L1 cells were cultured in growth media and harvested at 70% confluency. Cells were cross-linked and subjected to FAIRE analysis. Enrichment was quantified by qPCR by normalizing to an input (UnFAIRE) control for each primer set. A schematic of the distal enhancer and proximal promoter elements for each locus is shown above each graph. (A) Dct, (B) Tyrp1, (C) Tyr. (D) MyoD, (E) Scn2a1 (M: Mbox, S: SOX10 binding site, E: E box). The data are the average of at least two independent experiments performed in triplicate. Standard error bars and statistically significant differences compared to undifferentiated (0hr) Melb-a cells are shown (**P < 0.01, *P < 0.05, Student's t test).
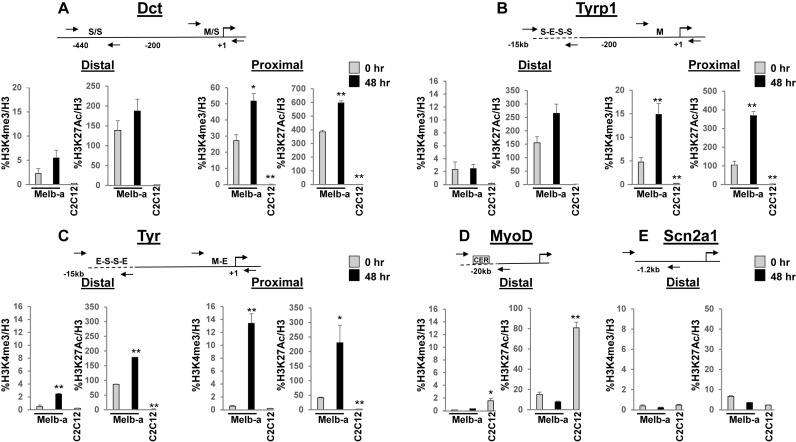
We confirmed FAIRE analysis with chromatin immunoprecipitations (ChIPs) to detect histone modifications that are associated with active gene expression. Active enhancers are marked by high levels of histone H3 acetylated on lysine 27 (H3K27ac) and low levels of histone H3 trimethylated on lysine 4 (H3K4me3), while active promoters are marked by high levels of H3K4me3 and variable levels of H3K27ac (66). We found that the distal enhancers of Dct, Tyrp1 and Tyr had low levels of H3K4me3 and high levels of H3K27ac which did not significantly increase (Dct, Figure 6A, left) or increased to a small extent upon differentiation (Tyrp1, Figure 6B, left, Tyr, Figure 6C, left). The proximal promoters of Dct, Tyrp1 and, Tyr displayed variable increases in both H3K27ac and H3K4me3 enrichment upon differentiation, with Tyr characterized by the lowest relative levels of these histone modifications in undifferentiated Melb-a cells and the highest relative increase upon differentiation (Figure 6A, B, C, right). ChIP specificity was demonstrated by low enrichment of melanocyte-specific regulatory regions and high H3K27ac enrichment of the MyoD CER in C2C12 cells (Figure 6D) and low enrichment of the Scn2a1 locus in all cells (Figure 6E). These experiments confirm FAIRE analysis which suggests that the distal regulatory regions of melanocyte-specific genes are primed by open chromatin structure in undifferentiated melanoblasts, while proximal promoters become increasingly accessible upon differentiation.
Figure 6.
Changes in histone modifications at distal and proximal control regions of melanogenic enzyme genes during Melb-a differentiation. Chromatin immunoprecipitations (ChIPs) were performed with an antibodies to histone H3, histone H3 acetylated on lysine 27 (H3K27ac), histone H3 trimethylated at lysine 4 (H3K4me3). or a control IgG antibody. Enrichment was quantified by qPCR by normalizing to H3 for each primer set. ChIP with IgG resulted in <1% of the enrichment obtained with histone antibodies (data not shown). (A) Dct, (B) Tyrp1, (C)Tyr, (D) MyoD CER, (E) Upstream region of Scn2a1. The data are the average of at least two independent experiments performed in triplicate. Standard error bars and statistical significance compared to undifferentiated (0hr) Melb-a cells are shown (**P < 0.01, *P < 0.05, Student's t test).
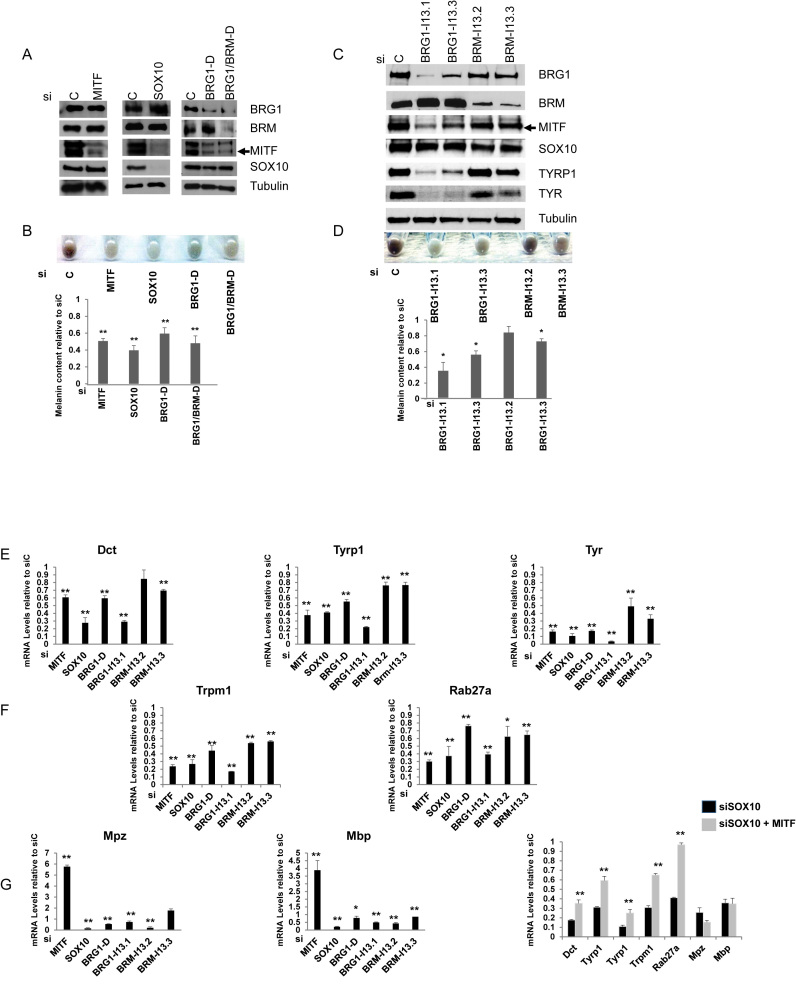
Depletion of SOX10 and BRG1 inhibits melanocyte differentiation and melanocyte-specific gene expression
To investigate the requirement for SOX10 and BRG1 in Melb-a differentiation, we depleted expression of these proteins by transiently transfecting short interfering RNAs (siRNA). Because SOX10 regulates MITF expression, we also knocked down MITF to compare the effects of concurrent depletion of SOX10 and MITF with depletion of MITF alone. Furthermore, because our previous data in melanoma cells suggested that the alternative ATPase, BRM, can partially compensate for BRG1 loss (28), we utilized an siRNA sequence that targets only BRG1 and an siRNA sequence that targets both BRG1 and BRM. These siRNAs or a non-targeting siRNA (siC) were transfected into undifferentiated Melb-a cells and then cells were shifted to differentiation medium 48 h later. The extent of knockdown was evaluated by Western blotting (Figure 7A and B). Knockdown of SOX10, BRG1 or BRG1/BRM inhibited MITF expression in Melb-a cells and knockdown of BRG1 or BRG1/BRM also caused a small decrease in SOX10 expression (Figure 7A). These results are consistent with other reports that SOX10 transcriptionally activates MITF expression (67) and that BRG1 can also contribute to the regulation of MITF and SOX10 in melanoma (68) and Schwann cells (69) respectively. However, it is important to note that the effect of BRG1 or BRG1/BRM depletion on the expression of either MITF or SOX10 was small compared to the effects seen by depletion with siMITF or siSOX10.
Figure 7.
SOX10 and BRG1 are required for melanocyte differentiation. Undifferentiated Melb-a cells were transfected with the indicated siRNAs for 48 hours. The medium was then replaced by differentiation medium and cells were cultured for an additional 48 h. (A) Melb-a cells were subjected to Western blotting with antibodies to BRG1, BRM, MITF and SOX10. Tubulin was used as a loading control. (B) Melb-a cells transfected with the indicated siRNAs were pelleted and photographed. Cells were counted, and an equal number were subjected to the melanin assay. Each of the siRNAs resulted in a significant reduction in melanin relative to the siC. (C) Melb-a cells transfected with siRNAs that uniquely target BRG1 or BRM were subjected to Western blotting as in (A). Protein extracts were also evaluated for TYRP1 and TYR expression. (D) Melb-a cells transfected with siRNAs that uniquely target BRG1 or BRM were pelleted, photographed and subjected to the melanin assay as in (B). (E–G) RNA was isolated from siC, siMITF, siSOX10, siBRG1-D, siBRG1-I13.1, siBRM-I13.2 and siBRM-I13.3 transfected Melb-a cells, reversed transcribed and quantified by qRT-PCR. The CT value for each gene was normalized to Rpl7. (E) Melanogenic enzyme gene expression. (F) Expression of genes associated with melanocyte differentiation. (G) Expression of myelin genes. The data are the average of at least two independent experiments performed in triplicate. Standard error bars and statistical significance compared to siC are shown (**P < 0.01, *P < 0.05, Student's t test). (H) Melb-a cells were co-transfected with siSOX10 and CMV-MITF. RNA was isolated from siC, siSOX10, and siSOX10/CMV-MITF transfected Melb-a cells, reversed transcribed and quantified by qRT-PCR. The CT values for each gene were normalized to Rpl7 and are presented relative to values obtained with siC transfected cells. The data are the average of at least two independent experiments performed in triplicate. Standard error bars are shown. Stars indicate statistical difference between the MITF rescued siSOX10 cells compared to siSOX10 (**P < 0.01, *P < 0.05).
We then assayed the effects of the knockdowns on melanin content (Figure 7B). Transfection with siSOX10, which not only depleted SOX10 but also severely inhibited MITF expression, had a slightly greater effect on observable pigmentation than transfection of siMITF which depleted MITF but did not affect SOX10 expression. Depletion of both BRG1 and BRM had a greater effect on observable pigmentation compared to depletion of BRG1 alone, suggesting that BRM may contribute to activation of genes required for melanin synthesis.
To more precisely determine the relative contributions of BRG1 and BRM in promoting pigmentation and melanin synthesis, we utilized additional siRNAs that uniquely target each of the ATPases. Two siRNAs against BRG1 (siBRG1-II3.1 and siBRG1-II3.3) and two siRNAs directed against BRM (siBRM-I13.2) and siBRM-I13.3) depleted the respective proteins with varying efficiencies (Figure 7C). Depletion of BRG1 by either siBRG1-I13.1 or siBRG1-I13.3 reduced MITF, TYRP1 and TYR at the protein level, but had only a slight effect on SOX10 expression, while depletion of BRM by either siRNA had minimal effects on MITF and SOX10. Only the more efficient siBRM-I13.3 substantially reduced TYRP1 and TYR expression at the protein level. Depletion of BRG1 by either siRNA reduced observable pigmentation and melanin content while only the more efficient siRNA targeting BRM had an inhibitory effect on observable pigmentation and on melanin content (Figure 7D).
We next investigated the effects of the knockdowns on expression of SOX10 and MITF target genes. As expected, knockdown of either SOX10 or MITF inhibited expression of genes that regulate melanin synthesis and melanocyte function (Figure 7E and F). Knockdown of BRG1 by either siBRG1-D (60% knockdown by qRT-PCR, data not shown) or siBRG1-I13.1 (80% knockdown by qRT-PCR, data not shown) significantly reduced expression of these genes such that the more efficient knockdown of BRG1 resulted in a greater effect on melanocyte-specific gene expression (Figure 7E and F). Knockdown of BRM alone by either siBRM-I13.2 (65% knockdown by qRT-PCR, data not shown) or siBRM-I13.3 (80% knockdown by qRT-PCR) resulted in small but significant effects on most melanocyte-specific genes. Therefore, both BRG1 and BRM likely contribute to the regulation of these melanocyte-specific genes, with BRG1 depletion having a more severe effect on expression of DCT, TYRP1 and TYR at both the protein and gene levels as well as on observable pigmentation.
Consistent with the inhibitory effect of MITF expression on Mpz and Mbp expression in NIH 3T3 cells (Figure 3D), we found that knockdown of MITF in Melb-a cells led to an increase in the expression of these genes, while knockdown of SOX10, BRG1 and BRM decreased their expression (Figure 7G). Ectopic expression of MITF in SOX10-depleted cells partially restored Dct, Tyrp1, Tyr and Trpm1 expression and fully restored Rab27a levels to those in cells expressing control siRNA (siC), but did not have a significant effect on the expression of Mpz and Mbp (Figure 7H). These data show that MITF cannot fully compensate for SOX10 loss in the regulation of a subset of melanocyte-specific genes that require BRG1, including Dct, Tyrp1 and Tyr. This suggests that the requirement for SOX10 in the transcriptional regulation of these genes extends beyond activation of MITF expression.
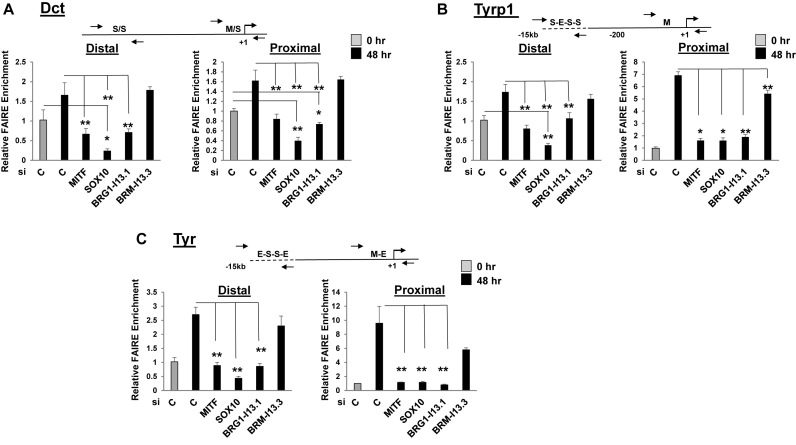
SOX10 and BRG1 promote chromatin accessibility at common sites in the regulatory regions of co-regulated genes
To determine whether SOX10 and BRG1 are both required to promote chromatin accessibility at SOX10 binding sites in the distal enhancers of Dct, Tyrp1 and Tyr, we transfected undifferentiated Melb-a cells with the respective siRNAs for 48 h, then shifted the cells to differentiation medium and performed FAIRE (Figure 8A–C). SOX10 depletion reduced chromatin accessibility at both the distal and proximal regulatory regions of Dct and the distal enhancer of Tyrp1 to levels that were significantly below what was observed in undifferentiated Melb-a cells and reduced chromatin accessibility at proximal promoters as well as the Tyr distal enhancer to levels significantly lower than in control differentiated cells. Since SOX10 regulates MITF expression and the proximal promoters have M boxes while the distal regions have E boxes that potentially bind MITF, we knocked down MITF to determine if the changes in accessibility that were observed in SOX10 knockdown cells might be indirectly due to loss of MITF. Depletion of MITF reduced chromatin accessibility at both the distal and proximal regulatory regions to levels that were significantly below those in control differentiated cells, bringing levels down to the levels in undifferentiated cells. Importantly, chromatin accessibility at the Dct and Tyrp1 distal enhancers was significantly lower in SOX10 depleted cells than in MITF depleted cells. Knockdown of BRG1also decreased accessibility at both the distal and proximal regions of Dct, Tyrp1 and Tyr while knockdown of BRM slightly reduced chromatin accessibility at the distal and proximal promoters of Tyrp1 and Tyr but had no effect on either region of Dct. Importantly, depletion of BRG1 resulted in reduced chromatin accessibility at all regulatory regions examined compared to depletion of BRM. These data suggest that SOX10 likely has a role in regulating chromatin structure at distal enhancers of some melanocyte-specific genes that extends beyond its role in regulating MITF expression and that BRG1 is the primary SWI/SNF ATPase that regulates chromatin accessibility at these sites.
Figure 8.
Knockdown of SOX10, MITF, or BRG1/BRM reduces chromatin accessibility at distal and proximal control regions of the melanogenic enzyme genes. (A–C). Undifferentiated Melb-a cells were transfected with siC, siMITF, siSOX10, siBRG1-I3.1 or siBRMI3.3 for 48 h. The medium was then replaced by differentiation medium and cells were cultured for an additional 48 h. One set of cells was processed for FAIRE prior to differentiation (siC-undifferentiated). Cells were cross-linked and subjected to FAIRE analysis. Enrichment was quantified by qPCR by normalizing to an input (UnFAIRE) control for each primer set and to the Scn2a1 region as a negative control. The data are the average of at least two independent experiments performed in triplicate. Standard error bars and statistical significance compared to siC undifferentiated (0hr) and differentiated (48 h) cells are shown (**P < 0.01, *P < 0.05, ANOVA).
SOX10 physically interacts with BRG1 and recruits BRG1 to the Tyrp1 enhancer in Melb-a cells
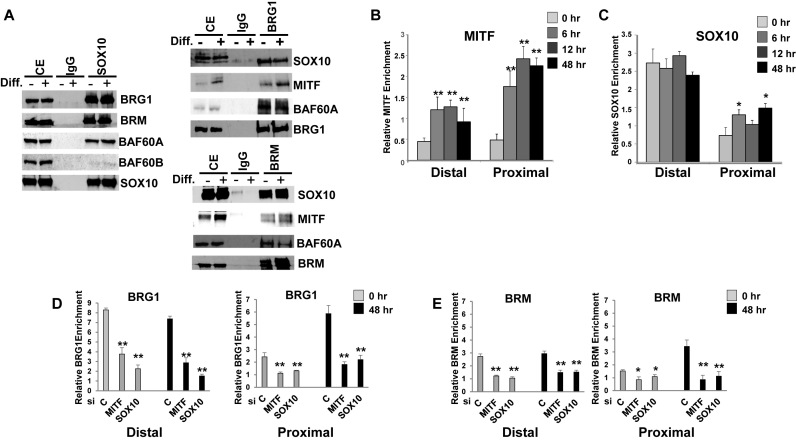
We performed co-immunoprecipitations to detect physical interactions between SOX10 nd BRG1 in Melb-a cells. Using an antibody to pull down SOX10, we detected SOX10-BRG1 as well as SOX10-BRM complexes in both undifferentiated and differentiated Melb-a cells (Figure 9A, left). We also probed for the BAF60A subunit of the SWI/SNF complex which was previously shown to interact directly with SOX10 and promote BRG1recruitment to Schwann cell-specific loci (14). As in Schwann cells, we detected a stronger interaction between SOX10 and BAF60A compared to a different subunit, BAF60B. This is consistent with the previous study that BAF60A mediates SOX10-SWI/SNF interactions and suggests that BRG1/BRM interact indirectly with SOX10 in melanocytes. Reciprocal co-immunopreciptations using antibodies to BRG1 or BRM confirmed that SOX10 interacts with both BRG1 and BRM. We also probed for MITF and found that although BRG1-MITF as well as BRM-MITF complexes can be detected in undifferentiated cells, there is an increase in these complexes upon differentiation when MITF levels are higher (Figure 9A, right). These results show that SOX10 interacts with SWI/SNF complexes in both undifferentiated melanoblasts and in differentiated melanocytes and may facilitate SWI/SNF recruitment to distal regulatory regions of melanogenic enzyme genes where SOX10 has a more pronounced role in chromatin remodeling.
Figure 9.
SOX10 physically interacts with BRG1 and recruits BRG1 to a melanocyte-specific enhancer in Melb-a cells. (A) Growing Melb-a cells or cells that had been differentiated for 18 h and either immunoprecipitated with an irrelevant antibody (IgG) or with an antibody to SOX10 (left), antiserum to BRG1 (top, right), or BRM (bottom, right). Cell extract (CE) or the immunoprecipitated material was run on an SDS-polyacrylamide gel and blotted with the indicated antibodies. (B) Chromatin immunoprecipitations (ChIPs) were performed with an antibody to MITF or control IgG. Enrichment of the Tyrp1 distal and proximal regions was quantified by qPCR by normalizing to the IgG control for each primer set and to the Scn2a1 region as a negative control region. The data are the average of at least two independent experiments performed in triplicate. Standard error bars and statistical significance compared to undifferentiated cells (0 h) are shown (**P < 0.01, *P < 0.05, Student's t test). (C) ChIP was performed and analyzed as in B using an antibody to SOX10 or control IgG. (D–E) Undifferentiated Melb-a cells were transfected with a control siRNA or siRNAs targeting MITF or SOX10 for 48 h. The medium was then replaced by differentiation medium and cells were cultured for an additional 48 h. (D) ChIP was performed and analyzed as in B using an antibody to BRG1 or as a control to IgG. (E) ChIP was performed and analyzed as in B using an antibody to BRM or as a control to IgG. The data are the average of at least two independent experiments performed in triplicate. Standard error bars and statistical significance compared to siC undifferentiated (0 h) and differentiated (48 h) cells are shown (**P < 0.01, *P < 0.05, ANOVA).
Since chromatin at distal enhancers was found to be accessible in both undifferentiated and differentiated Melb-a cells and to be highly affected by knockdown of SOX10 but less so by MITF knockdown, we hypothesized that SOX10 recruits BRG1 to this site in order to remodel chromatin. The Tyrp1 proximal region contains an M box which is the consensus binding site for MITF, and the distal region contains E boxes that potentially bind MITF. We found that there was little enrichment of MITF at either of these sites in undifferentiated Melb-a cells and that MITF became associated with both these sites 12 h after differentiation but that enrichment of MITF at the proximal region was higher than at the distal region (Figure 9B). SOX10 was constitutively enriched at the Tyrp1 distal region in both undifferentiated and differentiated cells while a small but significant level of SOX10 enrichment occurred at the proximal region beginning 12 h after differentiation (Figure 9C). The significance of SOX10 enrichment at the proximal promoter regions is not clear since this region does not contain SOX10 binding sites. It may reflect non-specific binding which has been demonstrated by many transcription factors, including the SOX family member, SOX2, or it may reflect long range promoter-enhancer interactions, brought about by chromatin looping (70–72). Like SOX10, BRG1 was found to be constitutively associated with the Tyrp1 distal enhancer in both undifferentiated and differentiated Melb-a cells while recruitment to the Tyrp1 proximal promoter increased during differentiation (Figure 9D). BRG1 recruitment to both these regions was decreased by depletion of either MITF or SOX10, with SOX10 depletion having a slightly greater effect on BRG1 recruitment to the distal enhancer than MITF depletion (Figure 9D). BRM also constitutively occupied the Tyrp1 distal enhancer in both undifferentiated and differentiated cells, while recruitment to the proximal promoter increased upon differentiation and in an MITF- and SOX10-dependent manner. However, enrichment of BRM at the distal enhancer was substantially less than that of BRG1 (Figure 9E). In combination, these data suggest SOX10 facilitates recruitment of BRG1 to distal enhancers. Thus, BRG1 is a SOX10 co-activator, required to establish the melanocyte lineage and promote expression of genes important for melanocyte function.
DISCUSSION
SOX10 interacts with a diverse set of transcriptional regulators to promote development of multiple cell lineages and is essential for the survival and differentiation of neural crest-derived melanocytes and Schwann cells (7,8). As ‘architectural transcription factors,’ SOX proteins were originally thought exert their effects on transcription by virtue of their ability to bend DNA (73). However, activation of transcription by SOX10 has recently been associated with recruitment of chromatin remodeling enzymes to regulatory regions (14,15). SOX10 was found to genetically and functionally interact with Smarca4, the gene encoding the BRG1 component of the SWI/SNF chromatin remodeling complex, to activate Schwann cell-specific gene expression at multiple steps in Schwann cell differentiation (14,15). BRG1-deficient mice exhibit Schwann cell defects strikingly similar to those seen in SOX10-deficient mice and these defects are exacerbated by additional heterozygous loss of one SOX10 allele in BRG1-deficient mice. Our studies reveal that SOX10 also interacts genetically and functionally with BRG1 to promote melanocyte development. Utilizing an ENU-based mutagenesis screen, we found that heterozygous mutations in Smarca4 exacerbate the spotting phenotype exhibited by SOX10 haploinsufficient mice, resulting in severe hypopigmentation. This is strong evidence that SOX10 and BRG1 act together in the melanocyte lineage. In contrast, a genetic interaction between SOX10 and BRG1 was not observed during oligodendrocyte differentiation, despite a requirement for both SOX10 and BRG1 in this process (74). Instead, the requirement for BRG1 in oligodendrocyte differentiation may occur at a stage prior to when SOX10 is required. Thus, the observed interactions between SOX10 and BRG1 in melanocytes and Schwann cells may not be essential for differentiation of all cell types that express these proteins.
To confirm that disruption of BRG1 function was responsible for the observed hypopigmentation phenotype, we deleted Smarca4 using a SOX10-Cre construct (33,34). Conditional deletion of Smarca4 resulted in a striking loss of cranial and trunk melanoblasts (Figure 2). The observed loss of melanocytes is consistent with a previous study that utilized a Tyr-Cre construct to delete Smarca4 (29), and confirms that BRG1 is required to establish the melanocyte lineage in vivo. It was previously unclear what happens to the melanocyte lineage when BRG1 is deleted. We now show that loss of BRG1 does not result in trans-differentiation to other neural crest lineages. Therefore, it is likely that loss of BRG1, like SOX10 and MITF, compromises melanoblast survival during embryonic development.
These results suggest that the alternative SWI/SNF ATPase, BRM, cannot compensate for the loss of BRG1 in the embryonic development of the melanocyte lineage. An absolute requirement for BRG1 and lack of compensation by BRM has been demonstrated in several other lineages including muscle (14,75–77). Indeed, our in vitro studies confirm that knockdown of BRG1 has a more profound effect on pigmentation, melanocyte-specific gene expression, and chromatin remodeling at several melanocyte-specific loci, than knockdown of the alternative ATPase, BRM. However, our data also show that BRM can interact with both SOX10 and MITF and be recruited to the regulatory regions of melanocyte-specific genes. It was previously shown that although BRM cannot compensate for BRG1 loss during embryonic development of vascular endothelial cells, BRM can compensate for BRG1 in maintaining these cells in the adult mouse heart (78). BRM null mice also display defects in muscle regeneration and BRM was found to be distinctly required for cell cycle arrest and late muscle specific gene expression (79). Our observations as well as other reports that associated BRM with highly differentiated cells (80) as well as melanocyte senescence (81) suggest that although BRM cannot compensate for BRG1 in early stages of melanocyte development, BRM may be important at later stages of melanocyte development or for melanocyte maintenance.
We found that SOX10 and BRG1 physically and functionally interact to promote melanocyte differentiation in cultured cells. The requirement for BRG1 in the melanocyte lineage has previously been attributed primarily to physical and functional interactions with MITF, which serve to recruit BRG1 to target genes involved in melanocyte differentiation, proliferation, and survival (28–29,41). However, BRG1 had also been reported to regulate MITF expression, to promote melanoma survival through MITF independent mechanisms, and to co-occupy SOX10 binding sites in melanoma cells (29,68,82). Establishing a functional interaction between SOX10 and BRG1 in melanocytes and melanoma cells has been complicated because melanocyte genes are co-regulated by SOX10 and MITF, including MITF itself. Therefore, knockdown of SOX10 results in depletion of MITF, which then abrogates BRG1 recruitment. Our findings suggests that SOX10 facilitates recruitment of BRG1 to the Tyrp1 distal enhancer. We found that SOX10 physically interacts with BRG1 in undifferentiated melanoblasts that express very low levels of MITF and that exhibit constitutively open chromatin structure at distal enhancers containing SOX10 binding sites. Furthermore, the timing of SOX10 and BRG1 binding to the Tyrp1 distal enhancer coincide in undifferentiated melanoblasts, occurring when MITF binding to this region is very low. However, both SOX10 and MITF depletion compromised BRG1 recruitment to the Tyrp1 distal enhancer, suggesting that SOX10 and MITF cooperate to recruit BRG1. In combination, these data suggest that in melanocyte precursors, SOX10 marks some melanocyte genes for later activation by MITF in part by recruiting BRG1 to distal enhancers.
In summary, we provide evidence that BRG1 is required for establishment of the melanocyte lineage in vivo and closely correlate this function with the role of SOX10 during melanocyte development. Our in vitro studies show that BRG1 is also required for melanin synthesis during melanocyte differentiation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Laura Baxter for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) [R01(ARO59379) to I.L.D.]; Deutsche Forschungsgemeinschaft (DFG) [We1326/8 to M.W.]; Wellcome Trust [078327 to D.C.B.]. Funding for open access charge: University of Toledo.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bolande R.P. Neurocristopathy: its growth and development in 20 years. Pediatr. Pathol. Lab. Med. 1997; 17:1–25. [PubMed] [Google Scholar]
- 2. Bondurand N., Dastot-Le Moal F., Stanchina L., Collot N., Baral V., Marlin S., Attie-Bitach T., Giurgea I., Skopinski L., Reardon W. et al. Deletions at the SOX10 gene locus cause Waardenburg syndrome types 2 and 4. Am. J. Hum. Genet. 2007; 81:1169–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pingault V., Bondurand N., Kuhlbrodt K., Goerich D.E., Prehu M.O., Puliti A., Herbarth B., Hermans-Borgmeyer I., Legius E., Matthijs G. et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998; 18:171–173. [DOI] [PubMed] [Google Scholar]
- 4. Baldwin C.T., Hoth C.F., Amos J.A., da-Silva E.O., Milunsky A.. An exonic mutation in the HuP2 paired domain gene causes Waardenburg's syndrome. Nature. 1992; 355:637–638. [DOI] [PubMed] [Google Scholar]
- 5. Tassabehji M., Read A.P., Newton V.E., Harris R., Balling R., Gruss P., Strachan T.. Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature. 1992; 355:635–636. [DOI] [PubMed] [Google Scholar]
- 6. Tassabehji M., Newton V.E., Read A.P.. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat. Genet. 1994; 8:251–255. [DOI] [PubMed] [Google Scholar]
- 7. Mollaaghababa R., Pavan W.J.. The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene. 2003; 22:3024–3034. [DOI] [PubMed] [Google Scholar]
- 8. Wegner M. Secrets to a healthy Sox life: lessons for melanocytes. Pigment Cell Res. 2005; 18:74–85. [DOI] [PubMed] [Google Scholar]
- 9. Jones E.A., Jang S.W., Mager G.M., Chang L.W., Srinivasan R., Gokey N.G., Ward R.M., Nagarajan R., Svaren J.. Interactions of Sox10 and Egr2 in myelin gene regulation. Neuron Glia Biol. 2007; 3:377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. LeBlanc S.E., Ward R.M., Svaren J.. Neuropathy-associated Egr2 mutants disrupt cooperative activation of myelin protein zero by Egr2 and Sox10. Mol. Cell. Biol. 2007; 27:3521–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei Q., Miskimins W.K., Miskimins R.. Sox10 acts as a tissue-specific transcription factor enhancing activation of the myelin basic protein gene promoter by p27Kip1 and Sp1. J. Neurosci. Res. 2004; 78:796–802. [DOI] [PubMed] [Google Scholar]
- 12. Jagalur N.B., Ghazvini M., Mandemakers W., Driegen S., Maas A., Jones E.A., Jaegle M., Grosveld F., Svaren J., Meijer D.. Functional dissection of the Oct6 Schwann cell enhancer reveals an essential role for dimeric Sox10 binding. J. Neurosci. 2011; 31:8585–8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghislain J., Charnay P.. Control of myelination in Schwann cells: a Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006; 7:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weider M., Kuspert M., Bischof M., Vogl M.R., Hornig J., Loy K., Kosian T., Muller J., Hillgartner S., Tamm E.R. et al. Chromatin-remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev. Cell. 2012; 23:193–201. [DOI] [PubMed] [Google Scholar]
- 15. Marathe H.G., Mehta G., Zhang X., Datar I., Mehrotra A., Yeung K.C., de la Serna I.L.. SWI/SNF Enzymes Promote SOX10- Mediated Activation of Myelin Gene Expression. PLoS ONE. 2013; 8:e69037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imbalzano A.N. Energy-dependent chromatin remodelers: complex complexes and their components. Crit. Rev. Eukaryot. Gene Expr. 1998; 8:225–255. [DOI] [PubMed] [Google Scholar]
- 17. Zhou C.Y., Johnson S.L., Gamarra N.I., Narlikar G.J.. Mechanisms of ATP-dependent chromatin remodeling motors. Annu. Rev. Biophys. 2016; 45:153–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imbalzano A.N., Kwon H., Green M.R., Kingston R.E.. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994; 370:481–485. [DOI] [PubMed] [Google Scholar]
- 19. Kwon H., Imbalzano A.N., Khavari P.A., Kingston R.E., Green M.R.. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994; 370:477–481. [DOI] [PubMed] [Google Scholar]
- 20. Vinod Saladi S., Marathe H., de la Serna I.L.. SWItching on the transcriptional circuitry in melanoma. Epigenetics. 2010; 5:469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kowenz-Leutz E., Leutz A.. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell. 1999; 4:735–743. [DOI] [PubMed] [Google Scholar]
- 22. Pedersen T.A., Kowenz-Leutz E., Leutz A., Nerlov C.. Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001; 15:3208–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Salma N., Xiao H., Mueller E., Imbalzano A.N.. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol. Cell. Biol. 2004; 24:4651–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de la Serna I.L., Ohkawa Y., Berkes C.A., Bergstrom D.A., Dacwag C.S., Tapscott S.J., Imbalzano A.N. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 2005; 25:3997–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simone C., Forcales S.V., Hill D.A., Imbalzano A.N., Latella L., Puri P.L.. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 2004; 36:738–743. [DOI] [PubMed] [Google Scholar]
- 26. Seo S., Richardson G.A., Kroll K.L.. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005; 132:105–115. [DOI] [PubMed] [Google Scholar]
- 27. de la Serna I.L., Ohkawa Y., Higashi C., Dutta C., Osias J., Kommajosyula N., Tachibana T., Imbalzano A.N.. The microphthalmia-associated transcription factor requires SWI/SNF enzymes to activate melanocyte-specific genes. J. Biol. Chem. 2006; 281:20233–20241. [DOI] [PubMed] [Google Scholar]
- 28. Keenen B., Qi H., Saladi S.V., Yeung M., de la Serna I.L.. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene. 2010; 29:81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laurette P., Strub T., Koludrovic D., Keime C., Le Gras S., Seberg H., Van Otterloo E., Imrichova H., Siddaway R., Aerts S. et al. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. eLife. 2015; 4: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saladi S.V., de la Serna I.L.. ATP dependent chromatin remodeling enzymes in embryonic stem cells. Stem Cell Rev. 2010; 6:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G. et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000; 6:1287–1295. [DOI] [PubMed] [Google Scholar]
- 32. Matera I., Watkins-Chow D.E., Loftus S.K., Hou L., Incao A., Silver D.L., Rivas C., Elliott E.C., Baxter L.L., Pavan W.J.. A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum. Mol. Genet. 2008; 17:2118–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsuoka T., Ahlberg P.E., Kessaris N., Iannarelli P., Dennehy U., Richardson W.D., McMahon A.P., Koentges G.. Neural crest origins of the neck and shoulder. Nature. 2005; 436:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sumi-Ichinose C., Ichinose H., Metzger D., Chambon P.. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol. Cell. Biol. 1997; 17:5976–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Witmer P.D., Doheny K.F., Adams M.K., Boehm C.D., Dizon J.S., Goldstein J.L., Templeton T.M., Wheaton A.M., Dong P.N., Pugh E.W. et al. The development of a highly informative mouse Simple Sequence Length Polymorphism (SSLP) marker set and construction of a mouse family tree using parsimony analysis. Genome Res. 2003; 13:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stolt C.C., Lommes P., Hillgartner S., Wegner M.. The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic Acids Res. 2008; 36:5427–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maka M., Stolt C.C., Wegner M.. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005; 277:155–169. [DOI] [PubMed] [Google Scholar]
- 38. de La Serna I.L., Carlson K.A., Hill D.A., Guidi C.J., Stephenson R.O., Sif S., Kingston R.E., Imbalzano A.N. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 2000; 20:2839–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sviderskaya E.V., Hill S.P., Balachandar D., Barsh G.S., Bennett D.C.. Agouti signaling protein and other factors modulating differentiation and proliferation of immortal melanoblasts. Dev. Dyn. 2001; 221:373–379. [DOI] [PubMed] [Google Scholar]
- 40. Eller M.S., Ostrom K., Gilchrest B.A.. DNA damage enhances melanogenesis. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saladi S.V., Wong P.G., Trivedi A.R., Marathe H.G., Keenen B., Aras S., Liew Z.Q., Setaluri V., de la Serna I.L.. BRG1 promotes survival of UV-irradiated melanoma cells by cooperating with MITF to activate the melanoma inhibitor of apoptosis gene. Pigment Cell Melanoma Res. 2013; 26:377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carreira S., Goodall J., Aksan I., La Rocca S.A., Galibert M.D., Denat L., Larue L., Goding C.R.. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005; 433:764–769. [DOI] [PubMed] [Google Scholar]
- 43. Carreira S., Goodall J., Denat L., Rodriguez M., Nuciforo P., Hoek K.S., Testori A., Larue L., Goding C.R.. Mitf regulation of Dia 1 controls melanoma proliferation and invasiveness. Genes Dev. 2006; 20:3426–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bourseguin J., Bonet C., Renaud E., Pandiani C., Boncompagni M., Giuliano S., Pawlikowska P., Karmous-Benailly H., Ballotti R., Rosselli F. et al. FANCD2 functions as a critical factor downstream of MiTF to maintain the proliferation and survival of melanoma cells. Sci. Rep. 2016; 6:36539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuspert M., Weider M., Muller J., Hermans-Borgmeyer I., Meijer D., Wegner M.. Desert hedgehog links transcription factor Sox10 to perineurial development. J. Neurosci. 2012; 32:5472–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramirez-Carrozzi V.R., Nazarian A.A., Li C.C., Gore S.L., Sridharan R., Imbalzano A.N., Smale S.T.. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006; 20:282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simon J.M., Giresi P.G., Davis I.J., Lieb J.D.. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012; 7:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements). Methods. 2009; 48:233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Law C., Cheung P.. Expression of non-acetylatable H2A.Z in myoblast cells blocks myoblast differentiation through disruption of MyoD expression. J. Biol. Chem. 2015; 290:13234–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lang D., Lu M.M., Huang L., Engleka K.A., Zhang M., Chu E.Y., Lipner S., Skoultchi A., Millar S.E., Epstein J.A.. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005; 433:884–887. [DOI] [PubMed] [Google Scholar]
- 51. Miller A.J., Du J., Rowan S., Hershey C.L., Widlund H.R., Fisher D.E.. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res. 2004; 64:509–516. [DOI] [PubMed] [Google Scholar]
- 52. Zhiqi S., Soltani M.H., Bhat K.M., Sangha N., Fang D., Hunter J.J., Setaluri V.. Human melastatin 1 (TRPM1) is regulated by MITF and produces multiple polypeptide isoforms in melanocytes and melanoma. Melanoma Res. 2004; 14:509–516. [DOI] [PubMed] [Google Scholar]
- 53. Chiaverini C., Beuret L., Flori E., Busca R., Abbe P., Bille K., Bahadoran P., Ortonne J.P., Bertolotto C., Ballotti R.. Microphthalmia-associated transcription factor regulates RAB27A gene expression and controls melanosome transport. J. Biol. Chem. 2008; 283:12635–12642. [DOI] [PubMed] [Google Scholar]
- 54. Hoek K.S., Schlegel N.C., Eichhoff O.M., Widmer D.S., Praetorius C., Einarsson S.O., Valgeirsdottir S., Bergsteinsdottir K., Schepsky A., Dummer R. et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008; 21:665–676. [DOI] [PubMed] [Google Scholar]
- 55. Strub T., Giuliano S., Ye T., Bonet C., Keime C., Kobi D., Le Gras S., Cormont M., Ballotti R., Bertolotto C. et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011; 30:2319–2332. [DOI] [PubMed] [Google Scholar]
- 56. Serra C., Palacios D., Mozzetta C., Forcales S.V., Morantte I., Ripani M., Jones D.R., Du K., Jhala U.S., Simone C. et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell. 2007; 28:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Salma N., Xiao H., Imbalzano A.N.. Temporal recruitment of CCAAT/enhancer-binding proteins to early and late adipogenic promoters in vivo. J. Mol. Endocrinol. 2006; 36:139–151. [DOI] [PubMed] [Google Scholar]
- 58. Xiao H., Leblanc S.E., Wu Q., Konda S., Salma N., Marfella C.G., Ohkawa Y., Imbalzano A.N.. Chromatin accessibility and transcription factor binding at the PPARgamma2 promoter during adipogenesis is protein kinase A-dependent. J. Cell Physiol. 2011; 226:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sviderskaya E.V., Wakeling W.F., Bennett D.C.. A cloned, immortal line of murine melanoblasts inducible to differentiate to melanocytes. Development. 1995; 121:1547–1557. [DOI] [PubMed] [Google Scholar]
- 60. Bennett D.C. Differentiation in mouse melanoma cells: initial reversibility and an on-off stochastic model. Cell. 1983; 34:445–453. [DOI] [PubMed] [Google Scholar]
- 61. Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D.. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007; 17:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lowings P., Yavuzer U., Goding C.R.. Positive and negative elements regulate a melanocyte-specific promoter. Mol. Cell. Biol. 1992; 12:3653–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ganss R., Schutz G., Beermann F.. The mouse tyrosinase gene. Promoter modulation by positive and negative regulatory elements. J. Biol. Chem. 1994; 269:29808–29816. [PubMed] [Google Scholar]
- 64. Murisier F., Guichard S., Beermann F.. A conserved transcriptional enhancer that specifies Tyrp1 expression to melanocytes. Dev. Biol. 2006; 298:644–655. [DOI] [PubMed] [Google Scholar]
- 65. Murisier F., Guichard S., Beermann F.. The tyrosinase enhancer is activated by Sox10 and Mitf in mouse melanocytes. Pigment Cell Res. 2007; 20:173–184. [DOI] [PubMed] [Google Scholar]
- 66. Yao L., Berman B.P., Farnham P.J.. Demystifying the secret mission of enhancers: linking distal regulatory elements to target genes. Crit. Rev. Biochem. Mol. Biol. 2015; 50:550–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Potterf S.B., Furumura M., Dunn K.J., Arnheiter H., Pavan W.J.. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum. Genet. 2000; 107:1–6. [DOI] [PubMed] [Google Scholar]
- 68. Vachtenheim J., Ondrusova L., Borovansky J.. SWI/SNF chromatin remodeling complex is critical for the expression of microphthalmia-associated transcription factor in melanoma cells. Biochem. Biophys. Res. Commun. 2010; 392:454–459. [DOI] [PubMed] [Google Scholar]
- 69. Limpert A.S., Bai S., Narayan M., Wu J., Yoon S.O., Carter B.D., Lu Q.R.. NF-kappaB forms a complex with the chromatin remodeler BRG1 to regulate Schwann cell differentiation. J. Neurosci. 2013; 33:2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Biggin M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011; 21:611–626. [DOI] [PubMed] [Google Scholar]
- 71. Soufi A., Garcia M.F., Jaroszewicz A., Osman N., Pellegrini M., Zaret K.S.. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015; 161:555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. LeBlanc S.E., Wu Q., Lamba P., Sif S., Imbalzano A.N.. Promoter-enhancer looping at the PPARgamma2 locus during adipogenic differentiation requires the Prmt5 methyltransferase. Nucleic Acids Res. 2016; 44:5133–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Werner M.H., Burley S.K.. Architectural transcription factors: proteins that remodel DNA. Cell. 1997; 88:733–736. [DOI] [PubMed] [Google Scholar]
- 74. Bischof M., Weider M., Kuspert M., Nave K.A., Wegner M.. Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation. J. Neurosci. 2015; 35:21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lickert H., Takeuchi J.K., Von Both I., Walls J.R., McAuliffe F., Adamson S.L., Henkelman R.M., Wrana J.L., Rossant J., Bruneau B.G.. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004; 432:107–112. [DOI] [PubMed] [Google Scholar]
- 76. Griffin C.T., Brennan J., Magnuson T.. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008; 135:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Griffin C.T., Curtis C.D., Davis R.B., Muthukumar V., Magnuson T.. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:2282–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Willis M.S., Homeister J.W., Rosson G.B., Annayev Y., Holley D., Holly S.P., Madden V.J., Godfrey V., Parise L.V., Bultman S.J.. Functional redundancy of SWI/SNF catalytic subunits in maintaining vascular endothelial cells in the adult heart. Circ. Res. 2012; 111:e111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Albini S., Coutinho Toto P., Dall’Agnese A., Malecova B., Cenciarelli C., Felsani A., Caruso M., Bultman S.J., Puri P.L.. Brahma is required for cell cycle arrest and late muscle gene expression during skeletal myogenesis. EMBO Rep. 2015; 16:1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Reisman D.N., Sciarrotta J., Bouldin T.W., Weissman B.E., Funkhouser W.K.. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl. Immunohistochem. Mol. Morphol. 2005; 13:66–74. [DOI] [PubMed] [Google Scholar]
- 81. Bandyopadhyay D., Curry J.L., Lin Q., Richards H.W., Chen D., Hornsby P.J., Timchenko N.A., Medrano E.E.. Dynamic assembly of chromatin complexes during cellular senescence: implications for the growth arrest of human melanocytic nevi. Aging Cell. 2007; 6:577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ondrusova L., Vachtenheim J., Reda J., Zakova P., Benkova K.. MITF-independent pro-survival role of BRG1-containing SWI/SNF complex in melanoma cells. PLoS ONE. 2013; 8:e54110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.