Abstract
The significance of islet antigen-reactive T cells found in peripheral blood of type 1 diabetes (T1D) subjects is unclear, partly because similar cells are also found in healthy control (HC) subjects. We hypothesized that key disease-associated cells would show evidence of prior antigen exposure, inferred from expanded T cell receptor (TCR) clonotypes, and essential phenotypic properties in their transcriptomes. To test this, we developed single-cell RNA sequencing (RNA-seq) procedures for identifying TCR clonotypes and transcript phenotypes in individual T cells. We applied these procedures to analysis of islet- antigen reactive CD4+ memory T cells from the blood of T1D and HC individuals following activation with pooled immunodominant islet peptides. We found extensive TCR clonotype sharing in antigen-activated cells, especially from individual T1D subjects, consistent with in vivo T cell expansion during disease progression. The expanded clonotype from one T1D subject was detected at repeat visits spanning more than 15 months, demonstrating clonotype stability. Notably, we found no clonotype sharing between subjects, indicating a predominance of “private” TCR specificities. Expanded clones from two T1D subjects recognized distinct IGRP peptides, implicating this molecule as a trigger for CD4+ T cell expansion. While overall transcript profiles of cells from HC and T1D subjects were similar, profiles from the most expanded clones were distinctive. Our findings demonstrate that islet- antigen reactive CD4+ memory T cells with unique antigen specificities and phenotypes are expanded during disease progression and can be detected by single-cell analysis of peripheral blood.
Introduction
Accumulating evidence for a role of islet- antigen reactive CD4+ T cells in development of T1D has spurred efforts to utilize them to investigate disease mechanisms and as therapeutic targets and biomarkers for beta cell destruction (1–6). While levels of islet- antigen reactive cells may be increased in the pancreas (2, 3), biopsy of this organ is not tenable in humans. Instead, most efforts in humans have focused on peripheral blood, which is readily available for testing. Numerous studies have reported detection of islet- antigen reactive CD4+ T cells in blood of at-risk and T1D subjects, but these cells are often detected in healthy control subjects as well (7–9). Distinctive phenotypic properties of islet- antigen reactive CD4+ T cells in T1D subjects (8–11) suggest their relationship to disease. Early findings suggested that T1D was a Th1 disease (12), whereas subsequent studies suggest involvement of additional T cell subsets (13).
Another consideration in identifying CD4+ T cells important for disease progression is their proliferation in response to an antigenic peptide. This results in clonal expansion (14) of a population of cells with identical antigen specificity and unique, identically rearranged TCR α– and β– chains. Characterization of rearranged TCR sequence variation thus provides a measure of T cell diversity, and antigen specificity, which can then be used to interrogate the role of those cells in disease.
Transcript profiling is a widely utilized tool for unbiased identification of phenotypic characteristics of cell populations. Increasingly, genome-wide transcriptome analysis by RNA-seq has been extended to the single-cell level (15, 16), revealing heterogeneity that is masked in bulk profiling studies. Combining flow cytometry-based assays and single-cell RNA sequencing, we have developed methods to identify TCR sequences in parallel with full transcriptome phenotypes from individual islet antigen-reactive CD4+ memory T cells. We have used this approach to perform an exploratory study of TCR clonotype expansion among islet T cells from HC and T1D subjects. We detected CD4+ memory T cells with expanded clonotypes in peripheral blood and identified their targets and transcript phenotypes.
Materials and Methods
Human subjects
Samples were obtained from HLA DRB1*0401 (DRB1*0401) healthy control and T1D subjects under informed consent (Table I). Healthy controls were matched for age and gender to T1D patients, and had no personal or family history of T1D. All protocols were approved by the Institutional Review Board at Benaroya Research Institute.
Table I.
Subject characteristics.
| Subject | Visit | Gender | Age (years) | Disease duration (months) | HLA class II1 | C-peptide (ng/ml)2 | Auto-Abs3 |
|---|---|---|---|---|---|---|---|
| T1D2 | 1 | F | 38 | 28 |
DRB1*0401/unknown DQB1*0302/unknown (DQ8) |
1.29 | GAD, IA2, Ins |
| T1D2 | 2 | F | 38 | 31 |
DRB1*0401/unknown DQB1*0302/unknown (DQ8) |
0.95 | NT4 |
| T1D2 | 3 | F | 39 | 43 |
DRB1*0401/unknown DQB1*0302/unknown (DQ8) |
0.41 | NT |
| T1D4 | 1 | M | 32 | 18 |
DRB1*0401/unknown DQB1*0302/unknown (DQ8) |
0.27 | GAD, IA2, Ins, ZNT8 |
| T1D5 | 1 | M | 27 | 23 |
DRB1*0401/*13 DQB1*06/unknown |
0.37 | GAD, IA2, Ins, ZNT8 |
| HC2 | 1 | F | 38 | NA5 |
DRB1*0401/*03 DQB1*02/unknown |
NA | NT |
| HC3 | 1 | F | 30 | NA |
DRB1*0401/*03 DQB1*02/unknown |
NA | NT |
| HC5 | 1 | M | 30 | NA |
DRB1*0401/1502 DQB1 unknown |
NA | NT |
DRB1 unknown, not DRB1*01, *04, *03, *13, *1501, *1502; DQB1 unknown, not DQB1*02, *0302, *0303; DQB1*06, not *0602 or *0603
C-peptide limit of detection, 0.05 ng/ml; subjects were not all fasting at C-peptide determination
Auto-Abs, autoantibodies;. Ins autoantibodies may be due to insulin therapy.
NT, not tested
NA, not applicable
Isolation of islet antigen-reactive T cells
Peripheral blood (100cc) was drawn by venous puncture using heparin as anti-coagulant. Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque centrifugation and cultured in RPMI media supplemented with 10% commercial human serum (Gemini Bio Products, West Sacramento, CA), penicillin/streptomycin (100 U/ml, 100 μg/ml), sodium pyruvate (1 mM), and L-glutamine (2 mM). Immediately following isolation, PBMC (10e6/ml) were stimulated with a pool of 28 islet peptides (Table II, 1.7μg/ml each, Mimotopes, San Diego CA) and 1 μg/ml of anti-CD40 blocking mAb (Miltenyi Biotec, San Diego CA) for 12–14 hr at 37°C. As controls, PBMC were cultured with an equal volume of DMSO (vehicle, negative control) or two influenza peptides as a positive control (MP p8 57–76 KGILGFVFTLTVPSERGLQR, MP p54 97–116 VKLYRKLKREITFHGAKEIS). Cells were harvested, labeled with PE-conjugated anti-CD154 mAb followed by anti-PE magnetic beads, and enriched using a magnetic column (Miltenyi Biotec). Enriched cells were stained with a live/dead dye (BD Via-Probe, BD Biosciences, Franklin Lakes, NJ) and antibodies targeting surface markers: CD14-PerCP Cy5.5 (61D3, eBiosciences, San Diego CA), CD19-PerCP Cy5.5 (HIB19, eBiosciences), CD4-Alexa Fluor 700 (OKT4, BioLegend, San Diego CA), CD69-APC (FN50, Biolegend), CD45RA-AmCyan (HI100, BD Biosciences, San Jose CA), CD45RO-APC Cy7 (UCHL1, Biolegend), CCR6-FITC (11A9, BD Biosciences), CXCR3-PE Cy7 (1C6/CXCR3, BD Biosciences), and CD38-Pacific Blue (HB7, eBiosciences). Cells were gated as shown in Figure S2: lymphocytes, singlets, CD4+Via-Probe-CD14-CD19-, CD154+CD69+, CD45RA-RO+. CD4+CD154+CD69+CD45RA-RO+ T cells were sorted on a BD FACSAria™ II flow cytometer directly into a 96 well C1 microfluidic chip (Fluidigm, San Francisco CA) for single cell capture. For each sample, the CD154+CD69+ gate in islet stimulated cultures was set based on the DMSO treated culture. Each experiment was performed with PBMC from a single blood draw.
Table II. Islet antigen peptides used for stimulation.
Shown are peptides from islet antigens used in this study:
| Peptide | Protein | Pool | Start | End | Length | Sequence | Reference |
|---|---|---|---|---|---|---|---|
| p1 | GAD651 | GAD1 | 1 | 20 | 20 | MASPGSGFWSFGSEDGSGDS | (44) |
| p10 | GAD65 | GAD1 | 73 | 92 | 20 | CACDQKPCSCSKVDVNYAFL | (44) |
| p14 | GAD65 | GAD1 | 105 | 124 | 20 | RPTLAFLQDVMNILLQYVVK | (44) |
| p15 | GAD65 | GAD1 | 113 | 132 | 20 | DVMNILLQYVVKSFDRSTKV | (44) |
| p34 | GAD65 | GAD1 | 265 | 284 | 20 | KGMAALPRLIAFTSEHSHFS | (44) |
| p35 | GAD65 | GAD2 | 273 | 292 | 20 | LIAFTSEHSHFSLKKGAAAL | (44) |
| p36 | GAD65 | GAD2 | 281 | 300 | 20 | SHFSLKKGAAALGIGTDSVI | (44) |
| p37 | GAD65 | GAD2 | 289 | 308 | 20 | AAALGIGTDSVILIKCDERG | unpublished |
| p38 | GAD65 | GAD2 | 297 | 316 | 20 | DSVILIKCDERGKMIPSDLE | (44) |
| p39 | GAD65 | GAD2 | 305 | 324 | 20 | DERGKMIPSDLERRILEAKQ | (44) |
| p41 | GAD65 | GAD3 | 321 | 340 | 20 | EAKQKGFVPFLVSATAGTTV | (44) |
| p45 | GAD65 | GAD3 | 353 | 372 | 20 | ICKKYKIWMHVDAAWGGGLL | (44) |
| p47 | GAD65 | GAD3 | 369 | 388 | 20 | GGLLMSRKHKWKLSGVERAN | (44) |
| p48 | GAD65 | GAD3 | 377 | 396 | 20 | HKWKLSGVERANSVTWNPHK | (44) |
| p55 | GAD65 | GAD4 | 433 | 452 | 20 | YDLSYDTGDKALQCGRHVDV | (44) |
| p60 | GAD65 | GAD4 | 473 | 492 | 20 | KCLELAEYLYNIIKNREGYE | (44) |
| p69 | GAD65 | GAD4 | 545 | 564 | 20 | VSYQPLGDKVNFFRMVISNP | (44) |
| p70 | GAD65 | GAD4 | 553 | 572 | 20 | KVNFFRMVISNPAATHQDID | (44) |
| p3 | IGRP2 | IGRP | 17 | 36 | 20 | KDYRAYYTFLNFMSNVGDPR | (45) |
| p31 | IGRP | IGRP | 241 | 260 | 20 | KWCANPDWIHIDTTPFAGLV | (45) |
| p39 | IGRP | IGRP | 305 | 324 | 20 | QLYHFLQIPTHEEHLFYVLS | unpublished |
| np1 | ZNT83 | ZNT8 | 1 | 20 | 20 | MEFLERTYLVNDKAAKMYAF | unpublished |
| np2 | ZNT8 | ZNT8 | 9 | 28 | 20 | LVNDKAAKMYAFTLESVELQ | unpublished |
| np3 | ZNT8 | ZNT8 | 17 | 36 | 20 | MYAFTLESVELQQKPVNKDQ | unpublished |
| p20 | ZNT8 | ZNT8 | 202 | 221 | 20 | GHNHKEVQANASVRAAFVHA | unpublished |
| p28 | ZNT8 | ZNT8 | 266 | 285 | 20 | ILKDFSILLMEGVPKSLNYS | unpublished |
| p36 | ZNT8 | ZNT8 | 330 | 349 | 20 | VRREIAKALSKSFTMHSLTI | unpublished |
| p28 | PPI4 | GAD4 | 76 | 90 | 15 | SLQPLALEGSLQKRG | (43) |
GAD65, GAD, glutamate decarboxylase 2, NP_000809.1
IGRP, G6PC2, glucose-6-phosphatase 2 isoform 1, NP_066999.1
ZNT8, SLC30A8, zinc transporter 8 isoform a, NP_776250.2
PPI, INS, insulin preproprotein, NP_000198.1
Class II tetramers
DRB1*0401 MHC class II tetramers (class II Tmr) labeled with PE were produced at Benaroya Research Institute Tetramer Core Laboratory and loaded with exogenous islet peptides (Table II) as described (17). As an irrelevant control, tetramers were loaded with an influenza hemagglutinin peptide (HA 306–318 PRYVKQNTLKLAT). CD4+ T cell clones or transduced 5KC murine hybridoma cells (18) expressing human CD4+ (provided by Maki Nakayama) were incubated with Tmr at 37° C for 1–2 hrs, then surface stained with anti-CD4 and analyzed by flow cytometry. Tmr staining was assessed in gated CD4+ T cells.
T cell clones
The GAD65-specific DRB4–restricted T-cell clone BRI4.13 was described previously (19). Cells were used directly (unstimulated) or were stimulated prior to use. Polyclonal stimulation by mAbs was achieved by incubation with immobilized anti-human CD3 plus soluble anti-human CD28 mAbs (eBioscience). Stimulation by Tmr (antigen-specific stimulation) was achieved by incubation in 96 well flat-bottom plates coated with class II Tmr loaded with GAD 555–567 (20) at 20 μg/ml. Cells were stimulated at 37°C for 12 hrs prior to use.
T cell clones were established from islet- antigen reactive CD4+ memory T cells from visit 3 of subject T1D2 using successive rounds of non-specific activation with PHA and irradiated PBMC in the presence of IL-2 (10 U/ml, Roche Applied Sciences, Mannheim, Germany). Clones were screened for expression of TRBV6-6 using a specific mAb (JU74.3, Beckman Coulter, Brea CA), by flow cytometry and clones testing positive were sequenced to confirm expression of the expanded TCR pair from T1D2.
Retroviral TCR expression
Oligonucleotides (Genscript, Piscataway, NJ) encoding codon-optimized rearranged TRAV and TRBV sequences from expanded clonotypes were cloned into the modified ‘TCR flex’ pMP71 retroviral backbone upstream of the murine Trac and Trbc genes (21). Recombinant retroviruses were packaged using Phoenix-AMPHO (CRL-3213, ATCC, Rockville MD) by transfection of 5 μg retroviral vector DNA with Lipofectamine 3000 transfection reagent (ThermoFisher Scientific, Waltham, MA). Viral supernatants were collected at 48 and 72 hrs post transfection. Purified human CD4+ T cells from peripheral blood (106) were cultured in ImmunoCult™-XF T Cell Medium (Stem Cell Technologies, Cambridge, MA) and activated with CD3/CD28 T cell activator (Stem Cell Technologies) in the presence of 100 IU/ml IL-2 and 5ng/ml of recombinant human IL-15 (BD Biosciences, San Jose, CA) for 48 hrs. Activated CD4+ T cells (0.2–0.5x106) were suspended in 1 ml of retroviral supernatant diluted 1:2 and polybrene (final concentration 10 μg/ml), and transduced by spin inoculation (2000 rpm, 90 min). To maximize transduction efficiency, spin inoculation was repeated after 24 hrs. After 3 days, transduction efficiency was determined flow cytometry using a murine TCRβ constant region mAb (H57-597, BD Biosciences). 5KC murine T cell hybridoma cells were transduced with recombinant TCR retroviruses as described above and sorted by flow cytometry to yield homogenous populations of human TCR-expressing cells for Tmr binding experiments.
Antigen specific proliferation assays
Peptide-induced proliferation was detected by 3H-thymidine incorporation for T cell clones; CellTrace Violet (ThermoFisher Scientific, Grand Island, NY, plus Ki57 staining (clone B56, BD Pharmingen) for PBMC; and CFSE dye dilution for transduced CD4+ T cells (22). T cell clones were screened in triplicate for proliferation to the original islet peptide pool in the presence of irradiated DRB1*0401 PBMC for 96 hr, followed by deconvolution to smaller pools and individual peptides. Peptide vehicle (DMSO) and anti-CD3/anti-CD28 mAbs were used as negative and positive controls, respectively. T cell clones with a stimulation index >3-fold over the DMSO control were considered to have proliferated. For CFSE proliferation assays, CFSE labeled CD4+ T cells (104) were mixed with 2x104 irradiated antigen presenting cells (Priess lymphoblastoid cells, ATCC), previously loaded with antigenic peptides (5 μg/ml, 1 hr), and were cultured for up to 5 days. Priess cells (DRB1*0401, *0401) have been used to present T1D antigens in the context of that class II molecule (23). Cells were stained with mAbs specific for CD4 and the murine TCR β chain. CFSE intensity was measured by flow cytometry, and quantified by gating on the murine TCR+ or TCR- populations in the CD4+ population.
Cell capture and RNA-seq library construction
Since islet- antigen reactive CD4+ memory T cells are rare (a median of ~700 cells recovered in our experiments), it was important to optimize recovery of single cells. A major advance in our procedure was to sort islet- antigen reactive T cells directly into microfluidic chips (Fluidigm C1), which decreased the input cell number required, increased the cell capture yield, and resulted in better quality libraries (https://github.com/linsleyp/Cerosaletti_Linsley). This direct sorting procedure resulted in a median of ~33 high quality libraries per 96 well chip (N=3 samples each for T1D and HC subjects). Neither the number of cells captured nor the number of high quality libraries recovered differed between T1D and HC subjects (p-value >0.4 by two-sided t-test). After capture, cells were lysed, followed by reverse transcription (SMART-Seq v1 Ultra Low Input RNA Kit, Takara, Mountain View, CA) and cDNA amplification according to the manufacturers protocols (Fluidigm, San Jose, CA). Sequencing libraries were prepared using Nextera XT DNA kits (Illumina, San Diego, CA).
RNA-seq
Single cell libraries were sequenced on an HiScanSQ or HiSeq2500 sequencers (Illumina, San Diego, CA) using single read 100 bp dual indexed reads (T cell clones) or 58 bp single read dual indexed reads (antigen reactive T cells) to an average read depth of ~1.8 million raw reads, a value that yields saturating numbers of genes detected in single cell assays (24). Bulk RNA-seq libraries were sequenced to target depths of ~10 million reads. RNA-seq pipeline analysis methods have previously been described (25). Quality metrics for aligned reads were obtained using the Picard (v.1.56) suite of tools (https://broadinstitute.github.io/picard/). For transcriptome analysis, reads were processed to remove reads with identical genome coordinates, which likely resulted from PCR amplification during library construction and comprised a large fraction of the raw reads (mean ~ 42% of T cell clone reads).
To ensure the highest data quality for islet antigen-reactive CD4+ T cells, we sequentially examined the distribution of values for several un-related quality control (https://broadinstitute.github.io/picard/) and eliminated outlier libraries (cutoff values for elimination in parentheses): PF_ALIGNED_BASES (≤3.5e6); MEDIAN_CV_COVERAGE (<0.4 or >2.0); PCT_USABLE_BASES (<0.25); and MEDIAN_3PRIME_BIAS (log10 value+1 <0.1 and > 0.4). The combined use of these quality control metric filters eliminated ~24% of initial libraries (89/364 initial libraries eliminated). We also eliminated libraries that did not yield at least one in-frame rearranged TCR junction (~10% of libraries that had passed quality control metric filters). Altogether, the use of these conservative metrics led us to consider ~68% of initial profiles (246/364) as having sufficient quality for subsequent analyses. Data from retained profiles were deposited in the GEO repository (GEO accession number: GSE96569; https://www.ncbi.nlm.nih.gov/geo/). We analyzed a total of 93, 35 and 25 cells from subjects, T1D2, T1D4 and T1D5; and 37, 31 and 22 from subjects, HC2, HC3 and HC5, respectively). The higher number of cells for subject T1D2 resulted from pooling of profiles from three different visits. Prior to analysis, counts were normalized for read counts using the TMM method (26) and transformed to reads per kilobase of transcript per million reads mapped (RPKM) values.
TCR clonotype identification
To determine the sequence of rearranged TCR sequences, which include non-templated nucleotides in the CDR3 junction not present in the reference genome, we utilized methods for genome-independent (de novo) assembly to construct a set of overlapping DNA segments (or contigs) (27). In initial experiments, we pre-filtered RNA-seq reads from the BRI-4.13 T cell clone to identify reads aligning (28) to TCR genes (29), and assembled them de novo into contigs (27). We found that each cell yielded ~1, 000–3,000 reads matching TCR genes which could be assembled (27) into TCR contigs of ~100–1,000 base pairs in length. Submission of these contigs to IMGT/V-QUEST (30) identified productive TCR chain rearrangements. In subsequent experiments, we found that performing de novo assembly on total reads without TCR gene pre-filtering gave very similar results; we consequently omitted the TCR gene pre-filtering step in later experiments. Unique TCR chains for all cells were sequentially filtered for: TRAV/TRBV gene usage (i.e., no TRDV or TRGV); productive rearrangements (i.e., no in frame stop codons); and length (7–25 amino acids).
Gene expression analysis
Differential gene expression was performed using the MAST R package (31).. Linear models for gene expression contained terms for cellular detection rate (31), group (T1D or HC) or frequency of TCR sharing. X and Y chromosome genes were removed prior to differential gene expression comparisons. Protein-protein interaction were obtained from STRING (32) (http://string-db.org/) or Genemania (33) (http://genemania.org/) and visualized using Cytoscape (34).
Statistics
Statistical tests were performed using the R programming language and software environment. For continuous, normally distributed variables, we utilized t-tests; for non-normally distributed variables, Wilcoxon tests; and for categorical variables, Fisher’s exact test. One-sided tests were performed when testing whether a given parameter was larger than the value given by the null hypothesis, a. A two-sided test was used when the test was that a parameter was simply not equal to the value given by the null hypothesis (i.e., that the direction did not matter). A false-discovery rate (FDR) of <0.1 was used to define differential gene expression. The specific test used to derive each p-value is listed in the text.
For comparing fractions of cells sharing clonotypes, we devised a permutation testing procedure. We originally utilized a down-sampling approach for comparing TCR frequencies (35), but found it difficult to run statistical tests on these down-sampled data, as sample sizes were insufficient for standard non-parametric tests, and the distributions violate normality assumptions. Because of these problems, we devised an alternative permutation approach which estimated the probability of recovering differences as large as those actually observed if all patients had the same distribution of TCRs. We generated a single distribution of TCRs by pooling all the TCR sequences recovered from all patients. For each replicate, we drew simulated sets of TCRs from that pooled distribution that were equal in size to the actual samples we obtained from each patient. We then quantified the percentage of shared TCRs within each patient at each sharing threshold, calculated the mean percent across patients within each group (T1D or HC), and determined the between-group difference in mean percent shared. We repeated this process 1000 times to generate a distribution of expected between-group differences if all patients have the same TCR repertoire. If the observed differences between HC and T1D were due to unequal sample sizes, sampling error, or a combination of the two, the observed values should fall within this distribution. Significance values were calculated as the proportion of replicates for which the permuted difference was greater than or equal to the observed difference.
Results
Detecting rearranged TCR chains and transcript profiles of individual T cells
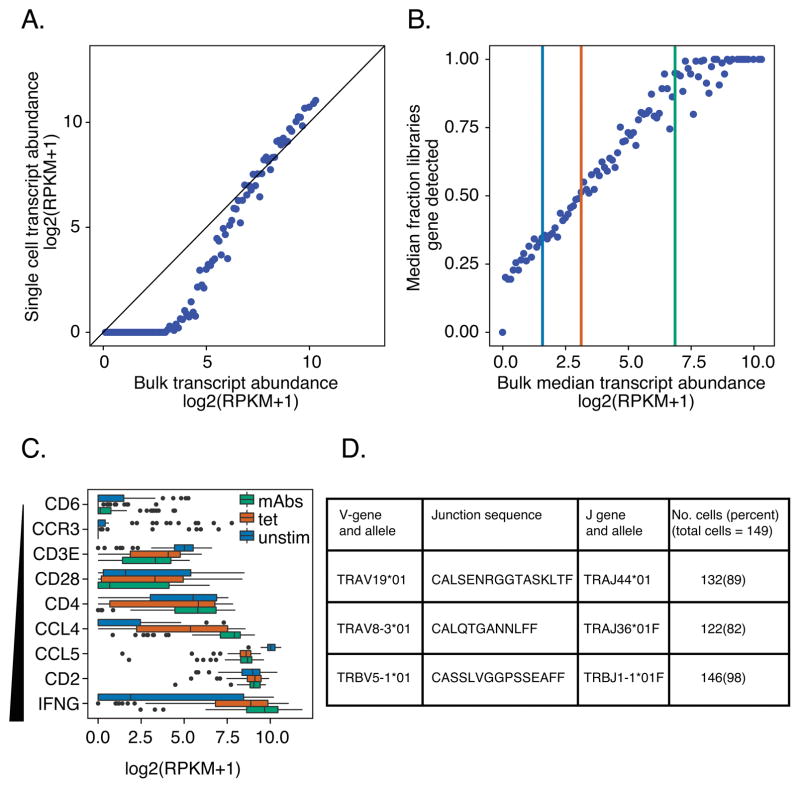
We hypothesized that single cell RNA-seq profiling would allow parallel determination of both the rearranged TCR chains and transcriptome phenotypes of individual T cells. We tested this hypothesis by comparing individual cell and bulk profiles from BRI-4.13 (19), an islet antigen-reactive CD4+ T cell clone from an individual with T1D (Materials and Methods). Although single cell transcript profiling has been successfully used with several cell types (36), less is known (37) about the performance of single cell techniques with antigen-specific T cells, which contain very limited amounts of RNA. In our experiments, we detected non-linearity between individual cell and bulk profiles in expression of low abundance genes, indicating that genes expressed at low levels are less likely to be detected at the single cell level than in a bulk measurement (Figure 1A). Median expression in single cells was ~0 for genes expressed at less than the top quartile of expression in bulk samples (log2(RPKM + 1) ~3.4 or ~5 RPKM). We calibrated our ability to detect expression of genes of different abundance (Figure 1B): single copy genes (median expression ~2 RPKM (38)) were detected in ~35% of cells; genes expressed at ~8 RPKM (~4 copies/cell) were detected in ~50% of cells; and genes expressed at ~115 RPKM (~60 copies/cell) detected in ~90% of cells. Finally, we tested the consistency of expression in single cell profiles for genes of different abundance in bulk samples (Figure 1C). This revealed that low abundance genes tended to show bimodal gene expression (39, 40) in single cell profiles. For these genes, one mode was near zero, indicating a population of cells in which the gene was either not expressed or the transcript failed to be amplified during library construction (Figure 1C).
Figure 1. Calibrating transcript detection and TCR recovery in single cell profiles from CD4+ T cells.
Clone BRI4.13 cells were left unstimulated or were stimulated with anti-CD3/anti-CD28 mAbs or with GAD peptide-loaded Tmr. Single cell profiles were collected from unstimulated, mAb- and Tmr-stimulated cells; bulk profiles, from mAb-stimulated cells only. A) Non-linearity between bulk and single cell profiles. Transcript counts from mAb-stimulated cells were grouped into 100 bins by their median expression levels (RPKM) in three bulk sample replicates. Shown is a comparison of median expression levels of genes in bins from bulk samples (X axis) versus the median expression levels in bins of genes in single cells (Y axis). The diagonal line represents perfect concordance. B) Calibrating the frequency of transcript detection in single cell profiles of islet- antigen reactive T cells. Shown is a comparison of median expression levels of genes in bins from bulk samples (X axis) versus the median fraction of libraries in which the genes in bins were detected in single cells (>0 RPKM). Vertical lines correspond to 2, 8 and 115 RPKM, which were detected in 35%, 50% and 90% of libraries, respectively. C) T cell genes show skewed expression patterns. Box plots show expression of T cell marker and cytokine genes, selected for increasing expression in bulk libraries (range: 0.5–8.5 RPKM). Tops, centerlines, and bottoms of the boxes represent the 25th, 50th percentiles, 75th percentiles, respectively. The “dots” at the ends of the boxplots represent outliers. D) The efficiency of TCR chain recovery in individual cells of an autoreactive T cell clone, BRI4.13. Shown are TRAV and TRBV sequences identified, together with the numbers and percentages of cells yielding each chain. Sequencing was performed on 149 single cells and 9 bulk replicates.
We next tested our ability to recover rearranged TCR chains from individual cells of the BRI-4.13 T cell clone, where rearranged TRAV-CDR3-TRAJ and TRBV-CDR3-TRBJ sequences that bind antigenic peptide were known (41). Using our RNA-seq and TCR clonotype identification pipeline (Materials and Methods), we identified the expected rearranged TCR chains as well as a previously undescribed productively rearranged TRAV chain in 82–98% of cells (Figure 1D). Most cells (~ 74%) yielded all three sequences (https://github.com/linsleyp/Cerosaletti_Linsley).
To extend our approach to analyze primary antigen-specific T cells in peripheral blood, we isolated influenza tetramer (Tmr)-reactive CD8 T cells from a healthy subject and subjected them to single cell profiling (Materials and Methods, Figure S1A). We recovered rearranged TCRs from ~76% of cells (34/45), of which TRAV and TRBV chains were found in 33% (15/45) and 42% (19/45) of cells, respectively, and both TRAV and TRBV chains were found in in 18% (8/45) of cells (https://github.com/linsleyp/Cerosaletti_Linsley). The recovery of both TRAV and TRBV chains from the same cells was lower with influenza-reactive cells than with the BRI-4.13 T cell clone or islet- antigen reactive cells (see below), likely because we used frozen PBMC for identification of influenza-reactive cells. Most of the recovered TCR sequences (~88% or 30/34) were expanded (Figure S1B). These expanded rearranged TCRs shared sequence identity with previously identified immunodominant TCR chains for influenza (https://github.com/linsleyp/Cerosaletti_Linsley, Figure S1C). Using flow cytometry of influenza tetramer-reactive cells, we confirmed the expansion of the TRBV19 gene segment predicted by single cell RNA-seq (Figure S1D). Together, these results validate the sensitivity and specificity of our procedures for determining transcript profiles and TCR sequences from RNA-seq profiles of individual antigen-specific T cells.
Isolation of islet- antigen reactive CD4+ memory T cells in blood
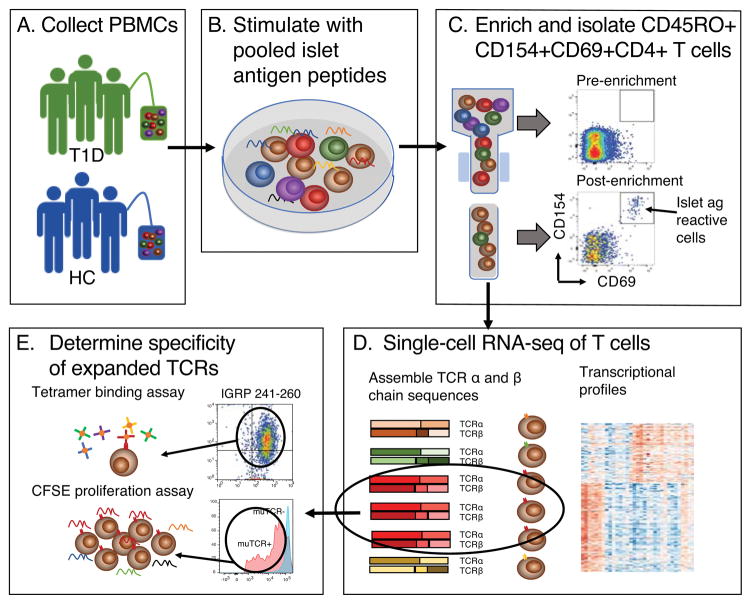
To investigate the diversity of islet specific CD4+ T cells in disease and health, we extended our methods include comparisons of islet antigen-specific T cells in blood from HC and T1D individuals (Figure 2). We relied on CD154 up-regulation (42) to identify CD4+ T cells that became activated when pooled islet antigen peptides were added to PBMC. We then isolated and sorted these activated cells into microfluidic chips using flow cytometry, and subjected them to single-cell RNA-seq. We then processed RNA-seq reads along two parallel paths to identify rearranged TCR chains and elucidate transcript phenotypes. From these results, we identified paired TCR chains that were found in multiple individual cells (expanded), expressed them in recombinant form, and deconvoluted the islet antigen peptide pool to identify the specific antigenic peptides recognized (Materials and Methods and Figure 2).
Figure 2. Determining TCR clonotypes and transcript phenotypes of antigen specific T cells.
Shown is a schematic view of the experimental process for determining expanded TCR clonotypes and transcript phenotypes from single islet- antigen reactive CD4 memory T cells.
We recruited a group of well-characterized high-risk DRB1*0401 T1D subjects and matched HC subjects (Table I). All T1D subjects were adult, within two years of diagnosis at their initial visit, and had detectable levels of C-peptide and multiple islet autoantibodies (Table I). HC subjects had no personal or family history of T1D. To identify islet- antigen reactive CD4+ memory T cells, we stimulated fresh PBMC by exposure to a pool of DRB1*0401-restricted islet antigen peptides (Table II) and enriched for cells with up-regulated CD154 using magnetic bead separation (Figure S2A). We then sorted enriched cells for those that had upregulated expression of the CD154 (42) and CD69 activation markers (Materials and Methods, Figure S2B). We included two activation markers to increase the specificity of the CD154 assay. We chose stimulation times of 12–14 hrs because we found that this condition yielded improved downstream library quality. CFSE labeling and Ki67 staining experiments showed that relative to DMSO-treated (negative control) cells, < 1% of islet antigen- or influenza- reactive cells proliferated under these conditions (Figure S2C). The peptides we used (Table II) represent consensus immunodominant epitopes recognized by CD4+ T cells in DRB1*0401 T1D subjects over many functional and epitope mapping studies (43–45). We also tested additional unpublished peptides identified using similar procedures, including several derived from ZNT8, a major islet autoantigen (46, 47). From these previous studies, we expected that most DRB1*0401 T1D subjects would have cells reactive with some, but not all, of the peptides used. After flow cytometry, the isolated cells (Figure S2B) were predominantly effector T cells, as regulatory T cells do not strongly up-regulate CD154 under these experimental conditions (48) and we did not detect genes clearly related to regulatory T cells in subsequent transcript analyses.
Rearranged TCRs of islet- antigen reactive CD4+ memory T cells in blood
If islet- antigen reactive T cells expand after encountering islet antigen(s), we expected to detect expanded TCR clonotypes of shared TRAV and TRBV chains. To test this prediction, we isolated CD4+ memory T cells from T1D and HC subjects following stimulation with islet antigen peptides and performed single-cell RNA-seq. We applied quality metrics to restrict RNA-seq analysis to 246 high quality libraries (Materials and Methods).
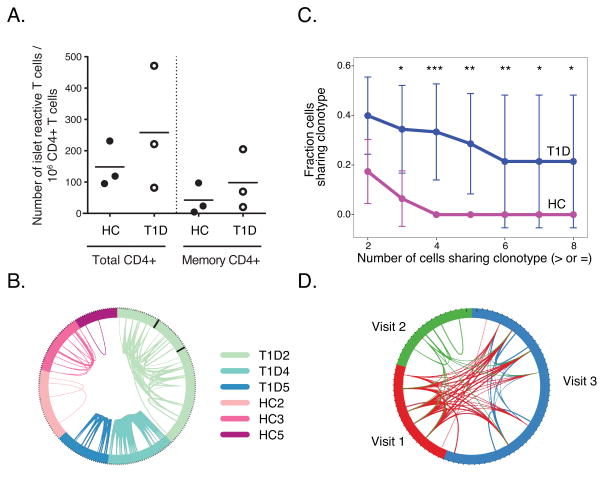
The frequencies of islet- antigen reactive CD4+ T cells detected in total or CD4+ memory T cell populations were similar between T1D and HC subjects, (p-value ≥0.35, one-sided Wilcoxon test) (Figure 3A). Likewise, similar numbers of cells were captured and passed quality filters between T1D and HC subjects at each visit (Materials and Methods) (p-value = >0.48, two-sided t-test). We then examined the distribution of rearranged CDR3 junctions, TRAV and/or TRBV, detected in individual islet- antigen reactive CD4+ memory T cells from T1D and HC subjects (https://github.com/linsleyp/Cerosaletti_Linsley). From N = 246 total cells, N = 165 (67%) contained both TRAV and TRBV chains; N = 30 (12%) contained TRAV chains only; and N=51 (21%) contained TRBV chains only (https://github.com/linsleyp/Cerosaletti_Linsley). Some cells (N=21, 8.5%) contained two TRAV chains; fewer contained two TRBV chains (N = 9, 3.7%). Many rearranged CDR3 junctions, particularly from T1D subjects, were shared between cells from the same subject (Figure 3B). The fraction of shared junctions within cells from the same participant was higher than sharing between subjects, where we detected no shared CDR3 junctions (p-value =2.5e-9, Fishers exact test). This indicates that the islet CD4+ T cell responses were subject-specific, or “private”. Despite similarities in frequencies of islet- antigen reactive CD4+ T cells between T1D and HC subjects (Figure 3A), fractions of cells sharing CDR3 junctions were higher in T1D than HC subjects (Figure 3C). These differences were significant (by permutation testing) over a range of thresholds (2–8 cells) (Figure 3C). To calculate these p-values, we utilized permutation testing (Materials and Methods). In accord with the observation of increased CDR3 junction sharing, we also found that values for Shannon entropy, a measure of clonal diversity (35), were higher for HC than T1D subjects (means of 4.3 versus 3.4, respectively; p-value = 0.04, one-sided Wilcoxon test). Shannon entropy values were calculated using down-sampling (35).
Figure 3. Sharing of rearranged TCRs from islet- antigen reactive CD4+ memory T cells.
A) Levels of islet- antigen reactive CD4+ memory T cells in T1D and HC subjects studied. Cell frequency per million CD4+ T cells was calculated as E/(Tx50) where E is the number of CD4+CD154+CD69+ T cells (Total CD4+) or CD4+CD154+CD69+CD45RA-RO+ (memory CD4+) following enrichment, and T is the total number of CD4+ T cells in 1/50th of the sample pre-enrichment as determined by flow cytometry (p-values ≥0.35, one-sided Wilcoxon test. Symbols represent individual subjects and the bars indicate the mean islet T cell frequency for the subjects in a column. B) TCR sharing in individual islet- antigen reactive T cells. Shown is a circos plot where segments in the circle represent individual cells yielding a rearranged TCR sequence. Black lines for subject T1D2 separate cells from different visits. Arcs connect cells sharing identically rearranged TCR genes. Line thickness is proportional to the number of junctions shared between cells, generally indicating that both TRAV and TRBV junctions were identified. Libraries from three different visits for subject T1D2 were combined for this and subsequent analyses (N = 22, 19 and 52 libraries for visits 1–3, respectively). C) Fraction of cells with expanded clonotypes is higher in T1D than HC cells. Shown are mean fractions of cells ± SD (Y axis) sharing clonotypes with different numbers of cells (X axis). Significance of mean differences between groups was calculated by permutation testing (Materials and Methods) (*, p-value <0.05 and ≥0.01; **, p-value <0.01 and ≥0.001; ***, p-value <0.001). D) Sharing of rearranged TCR junctions over time in subject T1D2. The circos plot depicts each visit in a different color and segments represent individual cells yielding a rearranged TCR sequence at a given visit. Lines connect cells sharing clonotypes at the same or different visits. Experiments were performed on cells from 3 healthy individuals and 3 T1D patients. 92, 35, and 28 cells were analyzed from T1D2, T1D4, and T1D5, respectively. 37, 31, and 22 cells were analyzed from HC2, HC3, and HC4 respectively.
Within T1D subjects, we found identical rearranged TRAV and TRBV protein sequences for the most highly shared junctions (Sequences 1–8, Table III) in 4–18 individual cells per subject (frequencies ~0.10–0.47). From each subject, nearly all cells with one of these rearranged chains also contained the other rearranged chain, thereby permitting unambiguous determination of TRAV/TRBV pairing (Table III). Expanded TCR sequences in individual cells were identical at the amino acid (Table III) and nucleotide levels (not shown) through the V, J and D genes (for TRBV chains), and CDR3 regions. This indicates clonal rather than convergent origin of the expanded clonotype sequences. Subjects T1D2 and T1D4 yielded a single expanded clone, whereas T1D5 yielded two. In one T1D subject (T1D2), who was sampled three times, we observed extensive sharing of expanded rearranged TCR chains (Sequences 1 and 2, Table III) over a period spanning >15 months (Figure 3D). Approximately 13% of junctions (12/93) were shared between visits 1 and 3, and 1 expanded junction was shared between all three visits by subject T1D2. The reduced TCR sharing at visit 2 may be due to in part lower cell yield at this visit. In comparison, there was no sharing of junctions (0/355) between any two T1D or HC subjects (p-value = 1.8e-6, Fisher’s exact test).
Table III.
Shared rearranged productive TCR sequences found in at least four individual cells from a T1D subject.
| TRAV chain | TRBV chain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Subject | Sequence | V gene | J gene | Junction | No. cells | Frequency | Sequence | V gene | J gene | Junction | No. cells | Frequency |
| T1D2 | 1 | TRAV29 | TRAJ40 | CAATRTSGTYKYIF | 10 | 0.11 | 2 | TRBV6-6 | TRBJ2-3 | CASSPWGAGGTDTQYF | 9 | 0.10 |
| T1D4 | 3 | TRAV2 | TRAJ15 | CAVEDLNQAGTALIF | 17 | 0.45 | 4 | TRBV5-1 | TRBJ2-1 | CASSLALGQGNQQFF | 18 | 0.47 |
| T1D5 | 5 | TRAV25 | TRAJ36 | CAGQTGANNLFF | 5 | 0.18 | 6 | TRBV4-3 | TRBJ1-5 | CASSQEVGTVPNQPQHF | 5 | 0.18 |
| T1D5 | 7 | TRAV26-2 | TRAJ48 | CILRDTISNFGNEKLTF | 4 | 0.14 | 8 | TRBV7-9 | TRBJ1-2 | CASSFGSSYYGYTF | 4 | 0.14 |
This demonstrates stability of a shared clonotype over time, a feature expected in cells relevant for disease progression. Taken together, these findings illustrate more extensive clonotype sharing among islet- antigen reactive CD4+ memory T cells in T1D than HC subjects. This suggests the possibility of in vivo clonal expansion of T cells with certain clonotypes, as the result of repeated encounters with antigen. The higher clonotype expansion in T1D subjects may indicate that such encounters are more frequent in T1D than HC subjects.
Identification of antigens recognized by islet- antigen reactive CD4+ memory T cells
We then identified specific antigenic peptide(s) from the pool used to trigger T cell activation to clarify whether expanded TCRs from different individuals recognize the same or different islet antigens and/or epitopes. Our procedures involved isolation and characterization of islet- antigen reactive T cell clones; and retroviral expression of recombinant TCR sequences.
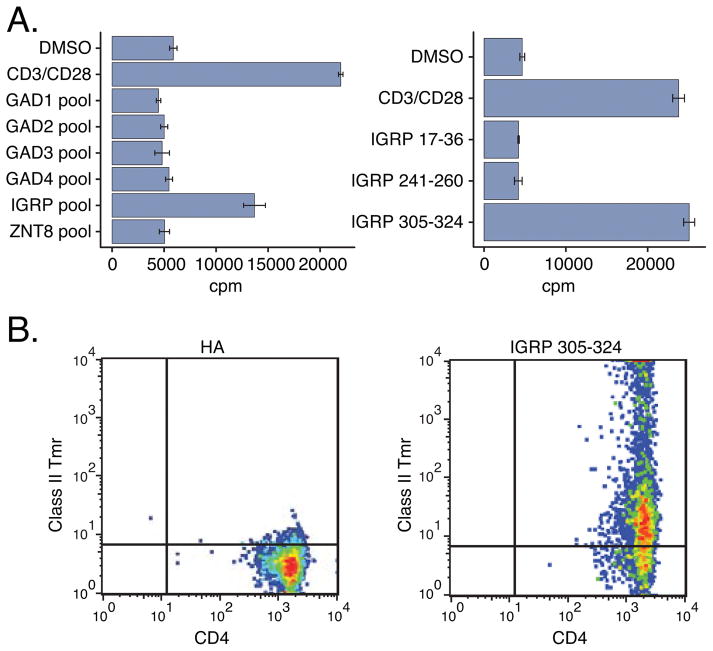
We first identified the antigen recognized by the TCR clonotype expanded in subject T1D2 (Sequences 1 and 2, Table III) (Figure 4). We generated T cell clones from islet- antigen reactive CD4+ memory T cells from this subject and screened them by flow cytometry for expression of different TRBV genes. We found that ~23% (11/47) of clones expressed TRBV6-6, the TRBV segment expanded in T1D2 (Sequence 2). We selected five of these TRBV6-6+ clones for RNA-seq analysis and confirmed that they yielded rearranged TCR chains identical to Sequences 1 and 2. We then tested these TRBV6-6+ clones for proliferation in response to pooled and individual peptides. All five TRBV6-6+ T cell clones proliferated in response to a pool of peptides from IGRP, but not to other pooled peptides (Figure 4A, Table 2). Testing against individual peptides showed that only IGRP 305–324 (QLYHFLQIPTHEEHLFYVLS, Table II) triggered specific proliferation (Figure 4A). Consistent with this finding, the T cell clones expressing the expanded clonotype bound class II Tmr loaded with IGRP 305–324, but not Tmrs loaded with an influenza HA peptide (49) or other IGRP peptides (Figure 4B). These experiments demonstrate that the expanded TCR pair comprising Sequences 1 and 2 (subject T1D2) recognizes IGRP 305–324 in the context of DRB1*0401 MHC class II molecules.
Figure 4. Demonstration of islet specificity of an expanded TCR clonotype from subject T1D2 using T cell clones.
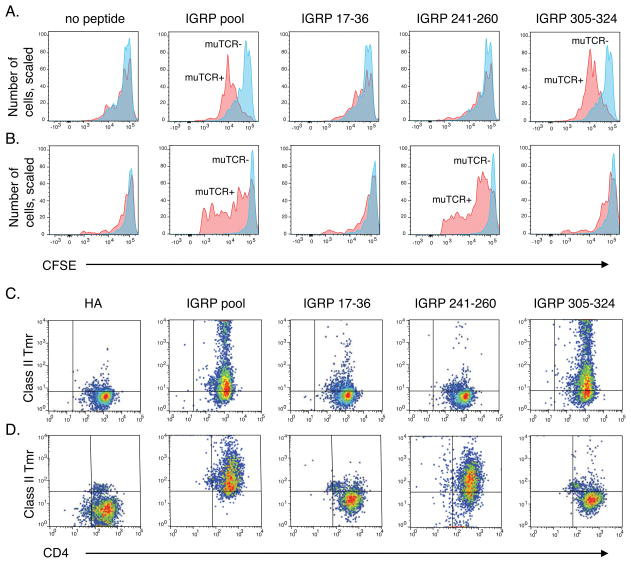
A) A representative T cell clone expressing the expanded clonotype from subject T1D2 (Sequences 1 and 2, Table III) was tested for proliferation by 3H-thymidine incorporation (Materials and Methods) after incubation with pooled (left panel) or individual (right panel) islet peptides. Values represent mean ± standard deviation cpm of triplicate wells. DMSO, negative (vehicle) control; CD3/CD28, anti-CD3/antiCD28 mAbs, positive control. B) T cell clone from (A) was tested by flow cytometry for binding of DRB1*0401 class II Tmrs loaded with HA 306–318 as a negative control (left panel), or the individual IGRP peptide IGRP 305–324 (right panel).
Although we successfully used T cell clones to elucidate the specificity of the expanded TCR clonotype from subject T1D2, there are clear drawbacks to this approach, including difficulties in access to patients and isolating T cell clones of some specificities. We therefore developed recombinant and ectopic retroviral expression methods to demonstrate the specificity of expanded TCR clonotypes (Figure 5). We transduced primary human CD4+ T cells with retroviruses expressing recombinant rearranged TRAV and TRBV sequences expanded in subjects T1D2 and T1D4 (Sequences 1 and 2, and Sequences 3 and 4, respectively). We then tested transduced T cells for proliferation in response to islet peptides by flow cytometry using CFSE dye dilution. Since autologous antigen presenting cells were not available for these experiments, we instead used Priess lymphoblastoid cells, which have been used to present T1D antigens in the context of DRB1*0401 class II molecules (23). Although these cells are DRB1*0401, *0401, they may have other HLA mismatches with patient cells (MHC class-1 etc.). To control for alloreactivity and other potential background issues, we compared proliferation in T cells expressing the recombinant TCR clonotype (17–29% of total cells), with un-transduced T cells in the same culture, as a negative control. As shown in Figure 5A, we observed minimal proliferation in either transduced or un-transduced T cells in the absence of peptide, indicating that alloreactivity was not a major concern under these conditions. As expected from Figure 4, we found that transduced T cells expressing the expanded TCR clonotype from subject T1D2 proliferated in response to the IGRP pool and IGRP 305–324 peptide but not with other peptides (Figure 5A). These results confirm that the expanded TCR clonotype from subject T1D2 specifically recognizes the IGRP 305–324 peptide. Surprisingly, however, transduced T cells expressing the expanded clonotype in subject T1D4 (Sequences 3 and 4) also proliferated in response to the IGRP peptide pool, but not other peptide pools (Figure 5B and data not shown). In contrast to subject T1D2, however, proliferation with the expanded clone from subject T1D4 was specific for peptide IGRP 241–260 (KWCANPDWIHIDTTPFAGLV, Table 2), a different IGRP peptide than recognized by the clonotype from subject T1D2. 5KC cells expressing these same recombinant TCRs from subjects T1D2 and T1D4 specifically bound class II Tmrs loaded with IGRP 305–324 and IGRP 241–260, respectively, but not an influenza HA peptide (Figure 5C–D). Thus, the expanded clones from two subjects with T1D, comprising Sequences 1 and 2, and Sequences 3 and 4, respectively, recognize distinct epitopes of the IGRP protein in the context of DRB1*0401.
Figure 5. Islet specificity of expanded TCR clonotypes from subjects T1D2 and T1D4 using recombinant and ectopic retroviral methods.
Antigen specificities of the expanded TCR pairs from subject T1D2, Sequences 1 and 2, and T1D4, Sequences 3 and 4, were determined by ectopically expressing the TCRs in primary human CD4+ T cells (A, B) or the TCR deficient murine hybridoma cell line 5KC (C, D) by retroviral transduction. A–B) Proliferation of primary CD4+ T cells transduced with the TCR sequences from T1D2 (A) or T1D4 (B) was measured by CFSE dye dilution at day 5 after co-culture with DRB1*0401 APC loaded with the indicated IGRP peptides using flow cytometry. A sample to which no peptide was added served as a negative control. muTCR+, cells expressing the murine TCR constant region encoded by the recombinant TCR (magenta); muTCR-, non-transduced cells. (cyan); C–D) 5KC hybridoma cells transduced with the TCR sequences from T1D2 (C) or T1D4 (D) were tested for binding to DRB1*0401 class II Tmrs loaded with the indicated IGRP peptides by flow cytometry. DRB1*0401 Tmr loaded with the irrelevant HA peptide served as a negative control. Data shown are representative of ≥ 3 experiments for each of the 2 individuals.
Gene expression differences between islet- antigen reactive CD4+ memory T cells in T1D and HC subjects
To examine whether transcript profiles of individual cells differed by disease status, we performed global comparisons of transcript profiles from T1D and HC subjects using an unsupervised approach (Principal Component Analysis or PCA). Comparison across the top three principal components showed small group differences between T1D and HC cells (Figure 6 A–B). Moreover, there were no robustly differentially expressed genes between the groups (False Discovery Rate (50) (FDR) <0.1, with an estimable log fold change (i.e., not NA), https://github.com/linsleyp/Cerosaletti_Linsley).
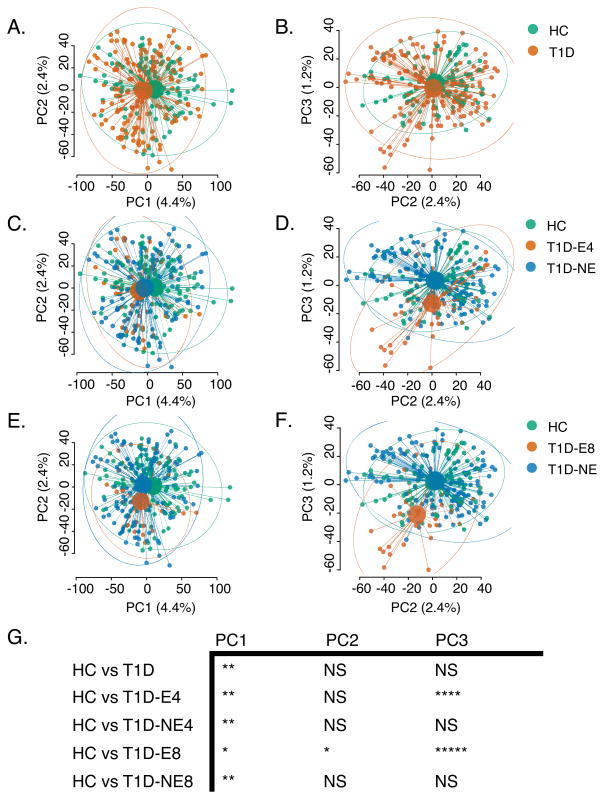
Figure 6. Differential gene expression by subject of islet- antigen reactive CD4+ cells.
PCA plots for single-cell transcript profiles from islet- antigen reactive CD4+ memory T cells from T1D and HC subjects. Small dots represent individual cells; large dots, centroids for each group; lines, connections between individual cells and centroids; and ellipses, 95% confidence intervals for each group. A–B) coloring by disease status (HC or T1D); C–D) coloring of cells designated as: HC cells; T1D-NE cells, T1D cells with non-expanded TCRs (TCR junction shared with <4 cells); or T1D-E4 cells, T1D cells with expanded TCRs (TCR junction shared with ≥4 cells); E–F) coloring of cells designated as: HC cells; T1D-NE cells, T1D cells with non-expanded TCRs (TCR junction shared with <8 cells); or T1D-E4 cells, T1D cells with expanded TCRs (TCR junction shared with ≥8 cells). Significance of different between specified groups with PC1-PC3 was calculated by multivariate linear models (NS, p-value >0.05; *, p-value <0.05 and ≥1e-2; **, p-value <1e-2 and ≥1e-3; ***, p-value <1e-3 and ≥1e-4; ****, p-value <1e-4 and ≥1e-6; ****, p-value <1e-6). PCA plots show cells from 3 healthy individuals and 3 T1D patients. 92, 35, and 28 cells were analyzed from T1D2, T1D4, and T1D5, respectively. 37, 31, and 22 cells were analyzed from HC2, HC3, and HC4, respectively.
We reasoned that group differences between T1D and HC might be obscured by heterogeneity at the cellular level within subjects and groups. To examine whether clonal expansion was associated with transcriptional heterogeneity within the T1D group, we focused on comparing T1D cells having expanded TCR sequences (T1D-E cells) with cells having non-expanded TCR sequences (T1D-NE). We selected T1D cells with TCRs shared in ≥4 cells (T1D-E4), i.e., cells expressing expanded clonotypes Sequences 1–8 (Table III). We compared the distribution of T1D-E4 cells, T1D cells with TCRs shared in <4 cells (T1D-NE), and HC cells (Figure 6 C–D) by PCA. In principal component space, T1D-E4 profiles were shifted towards lower PC3 values than T1D-NE or HC profiles (Figure 6 C–D). We found even more pronounced differences (Figure 6 E–F) when repeating this analysis using T1D cells with TCRs shared in ≥8 cells (T1D-E8 cells), indicating that the greatest differences in gene expression were among cells with the most expanded TCRs. These shifts in T1D-E4 and T1DE-8 versus HC profiles were highly significant (Figure 6G).
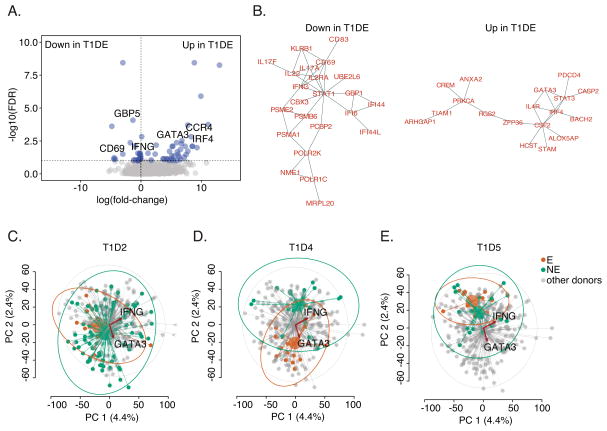
We found more genes showing significant differences in expression (N=62, FDR<0.1, https://github.com/linsleyp/Cerosaletti_Linsley) when including frequency of TCR sharing as a term in a linear model for gene expression (31), again demonstrating that degree of expansion is correlated with variation in gene expression. Genes positively related to degree of clonotype expansion (up in T1D-E cells) were enriched in T cell activation and leukocyte differentiation genes (http://genemania.org/; FDR = 6.5e-3 for both terms). Genes negatively associated with clonotype expansion (down in T1D-E cells) were enriched in type I interferon signaling genes (FDR = 4.0e-10). Volcano plots showed that expression of selected Th2-related genes (e.g., GATA3, CCR4, IRF4) and genes involved in interferon responses (e.g., IFNG, CD69, GBP5) was higher and lower, respectively, in T1D-E cells (Figure 7A). Projection of differentially expressed genes onto protein-protein interaction (PPI) networks showed significant interconnectedness of genes down and up in T1D-E cells (Figure 7B). Other differentially expressed genes that were not represented in the PPI networks were also noted (Figure 7B, https://github.com/linsleyp/Cerosaletti_Linsley), but these have not been investigated further.
Figure 7. Differential gene expression by expanded clones of islet- antigen reactive CD4+ cells.
A) Genes differentially expressed in islet- antigen reactive CD4+ memory T cells with the most expanded TCRs was determined using a model comprising terms for TCR clonotype frequency and cellular detection rate (31) (https://github.com/linsleyp/Cerosaletti_Linsley). Blue dots, genes differentially expressed with FDR <0.10 (N=62); grey dots, all other genes (N = 5,259 total). Selected immune genes are labeled. Up in T1D-E, genes higher in cells having the most expanded TCRs (including GATA3, CCR4, and IRF4); Down in T1D-E, genes lower in cells having the most expanded TCRs (including IFNG, CD69, and GBP5). B) Significantly interconnected (FDR ≤1.8e-3) PPI networks (32) were found in differentially expressed genes that are preferentially associated with TCR clonotype frequency as defined in (A) (31). Similar interactions were seen in other PPI networks (33). C–E) PCA plots showing PC1 versus PC2 for single cell transcript profiles from islet- antigen reactive CD4+ memory T cells from individual T1D subjects. In each panel, cells from a specified subject are highlighted. Small dots represent individual cells; large dots, centroids for each group; lines, connections between individual cells and centroids; and ellipses, 95% confidence intervals for each group. T1D-E, cells from the specified subject having a rearranged TCR shared with ≥4 cells (orange); T1D-NE, cells from the specified subject having a rearranged TCR shared with <4 cells (green); or others, cells from all other subjects (grey). Biplot vectors show information on expression of individual genes, IFNG and GATA3. Plots summarize the results from 3 T1D patients. 92, 35, and 28 cells were analyzed from T1D2, T1D4, and T1D5, respectively.
To explore the consistency of gene expression within and between subjects, we used PCA plots to examine transcript profiles from expanded and non-expanded cells from each of the three T1D subjects individually, compared to all other cells in the data set (Figure 7C–E). To these PCA plots we added vectors (biplots) for expression of individual differentially expressed genes from https://github.com/linsleyp/Cerosaletti_Linsley, including GATA3, a Th2 cell marker, and IFNG, a Th1 cell marker, (Figure 4A–C). These projections showed that T1D-E4 cells were shifted relative to T1D-NE cells in principal component space for each subject, especially for subject T1D4, and had different relationships with the GATA3 and IFNG gene expression biplots. T1D-E cells from subject T1D4 were more shifted in the dimension of the Th2 marker, suggesting that these cells were the primary source of the Th2-like genes differentially expressed at the group level. Taken together, these findings demonstrate that cells expressing expanded TCR clonotypes differ from cells with non-expanded clonotypes, but that there is heterogeneity between donors for the extent of differences.
Discussion
The presence of antigen specific T cells in peripheral blood of T1D as well as HC subjects, even cells with a memory phenotype, has been a surprising finding (7–9, 51). The widespread detection of these islet antigen-reactive T cells may result from their expression of TCRs that cross react with pathogen derived antigens, as has been reported for the islet antigens insulin and IGRP (53, 54). Attempts to identify disease-related differences in islet T cells have yielded inconsistent results, confounding efforts to use them to elucidate mechanisms of pathophysiology or as biomarkers of disease progression and therapeutic targets.
In this study, we show that novel features of islet- antigen reactive T cells from the peripheral blood of T1D subjects can be uncovered by utilizing the power of single-cell RNA-seq profiling to identify their TCR clonotypes in parallel with full transcript profiles. One of our key findings was the demonstration of expanded clones of islet- antigen reactive T cells, particularly in T1D subjects. Since we utilized 12–14 hr activation times our CD154 assays in order to increase RNA yield, we were concerned that some of the TCR sharing, or expansion, we observed resulted from proliferation in culture rather than in vivo. However, direct measurements showed negligible cell proliferation occurred under our culture conditions (Figure S2). Even if some cells did proliferate during culture, resulting in daughter cells sharing the same clonotypes, the length of the cell cycle in human lymphocytes (10–15 hrs) (52) would have precluded more than a single cell division during the culture period. At most, in vitro proliferation could account for no more than 2 daughter cells sharing the same clonotype in an experiment. To further mitigate this concern, we focused on more highly expanded clones (>4 cells) which were most likely to have resulted from in vivo expansion. The presence of more expanded clonotypes in cells from T1D compared with HC subjects links expansion of these cells to disease progression, possibly occurring during immune destruction of the pancreas. Although we have been able to analyze longitudinal visits from only a single subject thus far, our finding of the same rearranged TCR sequences in multiple visits from subject T1D2 suggests that expanded clonotypes can be stable over time. It is important to note that we have examined only adult T1D subjects in this study, primarily because of the blood volumes needed for our technology in its current form. It will be important to determine in future studies how the present results compare with results in children with T1D.
Another of our key findings was the demonstration of the specificity of expanded clones of islet- antigen reactive CD4+ T cells from different individuals for distinct peptides from the immunodominant islet protein IGRP. While numerous T cell islet antigens and epitopes have been described in T1D (Table 2), it remains unclear which of these are most important in disease progression. The CD154 assay (42) provides an opportunity to compare the relative frequencies of CD4+ T cells recognizing different published and un-published antigens and epitopes in side-by-side testing in the same assays. In our studies, this “competitive” approach, has highlighted the importance of IGRP as a target for expanded clones of islet antigen-reactive CD4+ T cells, including IGRP 305–324, a previously unpublished epitope.
IGRP is a metabolic enzyme (glucose-6-phosphatase 2) which is recognized as a major CD8+ T cell autoantigen for T1D in the NOD mouse model (56–59). IGRP also was recognized by T1D-related CD4+ T cells in mouse (57) and human studies (45). Our present studies showed the presence of expanded clonotypes of IGRP-specific CD4+T cells in two T1D subjects where TCR specificity was established, suggesting a pathogenic role for IGRP-specific CD4+T cells. One reason for the immunodominance of this islet protein may stem from activation of IGRP-specific T cells by molecular mimicry with microbial antigens (58, 59).
Intriguingly, we observed heterogeneity of transcriptional responses in islet- antigen reactive CD4+ T cells. While group differences between T1D and HC cells were small, differences related to clonotype frequency were larger. We do not currently know how the in vitro stimulation in our assay affects transcriptome differences between T1D and HC cells. In fact, our activation conditions may have obscured subtle differences in cells from T1D and HC subjects (9). While, to date, our attempts to used less stimulated cells for single cell analysis have yielded poor results, future iterations of our technology, perhaps using Tmr staining under non-activating conditions, may be useful in addressing this limitation. However, between individual subjects, transcript phenotypes of individual cells differed both qualitatively and quantitatively, demonstrating phenotypic heterogeneity. IGRP-reactive T1D-E4 cells from one subject (T1D4) that recognized peptide IGRP 241–260, had a more Th2-like phenotype. In contrast, T1D-E4 cells from other subjects, including cells from T1D2 that recognized IGRP 305–324, did not show this phenotype as clearly. It is also worth considering whether there is intraclonal heterogeneity in gene expression profiles. Consistent with this possibility, the PCA plots in Figure 7C-E show cells that seem to cluster separately from other cells having the same clonotypes. Although the present studies were not powered sufficiently to conclusively demonstrate whether these outlier cells represent intraclonal heterogeneity, the possibility should be considered in future studies. Importantly, neither inter- nor intraclonal heterogeneity would be apparent in bulk analyses of islet- antigen reactive T cells.
While early studies on autoreactive T cell responses suggested Th1-type pro-inflammatory polarization in CD4+ T cells in T1D (12), other studies have indicated a more complex scenario (13). More recent studies showed different ratios of Th1, Th2 and T regulatory type 1 (Tr1) cells in IGRP-reactive CD4+ T cells from adult- and juvenile-onset T1D subjects (60). A role for Th2 cells in T1D has been suggested by increased levels of type 2 cytokines in the serum of T1D subjects (61) and by genetic and epigenetic fine mapping studies of causal autoimmune disease genetic variants (62). Taken together with previous studies, our findings challenge a simple classification of T1D as a Th1-mediated pathology. Our results suggest either the existence of different disease subtypes (60) or changes in disease over time that were not resolved in our study, which mostly were taken from single visits.
Our results demonstrate the power of single-cell RNA-seq profiling for simultaneously determining T cell clonotypes, and linking these with expression profiles. When utilized in conjunction with technologies for isolating antigen-specific T cells, our methods allow an unprecedented view of specific T cells likely to be involved in pathogenic responses. In contrast with other methods (63, 64), our procedures for clonotype determination do not require the use of multiple sets of PCR primers for determining TCR sequences, and couple the power of unbiased, full transcriptome analysis with TCR clonotype determination. Similar procedures for linking TCR clonotypes and single-cell RNA-seq transcriptomes were recently published (65, 66), as was a computational method to infer the CDR3 sequences of tumor-infiltrating T cells in RNA-seq profiles from tumors profiles (67). Our procedures extend these previous studies by using short single end reads, rather than longer paired end reads, which reduces the cost of sequencing; and by eliminating initial TCR gene filtering, which reduces the number of steps in data processing. We also confirmed the accuracy of our approach by defining the specificity of two of the TCR clonotypes for individual islet peptides. One limitation of single cell RNA-seq is that transcripts present at low to moderate abundance (i.e. ≤ ~4–8 copies per cell in our studies), were not uniformly detected in transcriptomes of individual cells. Bulk RNA-seq is likely to be more useful for detection of genes with low expression, albeit at the price of averaging expression over all cells in the sample.
A key feature of single cell RNA-seq data sets is that they are typically more powered for detecting gene expression differences between individual cells than between individual subjects. For example, our exploratory studies utilized 246 cells, but only six subjects, three T1D and HC subjects each. The limited number of subjects in our studies means that our findings, while significant (i.e., unlikely to have happened by chance), should be confirmed in more highly powered and/or differently designed studies. These studies should include expanded cross-sectional studies to verify the extent and specificity of expanded islet- antigen reactive CD4+ T cell clones in T1D and HC subjects, and longitudinal studies to verify the extent and stability of expanded clonotypes, and to determine their relationship to disease progression. Further studies also will be needed to compare the extent of clonotype expansion in T1D versus other autoimmune conditions.
Our findings could have implications for the treatment of T1D. CD4+ T cells with expanded clonotypes may provide new biomarkers for disease progression, and potential targets for antigen-specific therapies. Biomarkers and therapeutics involving islet- antigen reactive T cells will likely need to be individualized, since we observed that they had mostly unique or “private” sequences with distinctive specificities. T cells with expanded clonotypes may also provide new and better targets for immunotherapy than islet antigen-reactive T cells that have not expanded and are therefore less likely to be involved in the disease process. Extending our approaches to include more subjects and/or longitudinal studies may reveal how levels of T cells with expanded clonotypes change during disease progression, how their levels are modified during therapeutic intervention, and which specificities will provide the best therapeutic targets.
Supplementary Material
Acknowledgments
We acknowledge the NIH Tetramer Core Facility for provision of the influenza tetramer (contract HHSN272201300006C); Jane Buckner for comments on the manuscript; Chester Ni, Mark Robinson, Masanao Yajima and Greg Finak for bioinformatics assistance; Janice Chen, John P. McNevin, Kimberly O’Brien and Quynh-Anh Nguyen for technical assistance; and Anne Hocking for assistance preparing the manuscript. Author contributions: GTN, WK and EW conceived the basic experimental approach; KC, JY, FW, EI, CS, SAM and MP performed or directed laboratory experiments; VG directed RNA-seq profiling; JAE performed pipeline analysis of the RNAseq data; PSL and HD analyzed the RNA-seq profiling data; FW and KC analyzed flow cytometry data; HR, MP and CJG provided materials; KH, ME and JN aided in data interpretation; RG provided statistical advice; PSL and KC conceived the experiments, analyzed the data and wrote the manuscript. All authors made contributions to the final manuscript.
Footnotes
Grant Support: This work was funded by NIH grants DP3DK110867, DP3DK106909, DP2 DE023321 and 5UM1AI109565, awarded to PSL, WWK, MP and GTN, respectively; and by JDRF grant 1-PNF-2014-97-Q-R to KC and JY.
Data and materials availability. Data from retained RNA-seq profiles have been deposited in the GEO repository (GSE96569). Flow cytometry data are available at http://flowrepository.org/. Data files and R code used to generate figures were deposited in the GitHub Repository, (https://github.com/linsleyp/Cerosaletti_Linsley).
References
- 1.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 2.Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature. 2005;435:224–228. doi: 10.1038/nature03625. [DOI] [PubMed] [Google Scholar]
- 3.Pathiraja V, Kuehlich JP, Campbell PD, Krishnamurthy B, Loudovaris T, Coates PTH, Brodnicki TC, O’Connell PJ, Kedzierska K, Rodda C, Bergman P, Hill E, Purcell AW, Dudek NL, Thomas HE, Kay TWH, Mannering SI. Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes. 2015;64:172–182. doi: 10.2337/db14-0858. [DOI] [PubMed] [Google Scholar]
- 4.Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol. 2009;155:173–181. doi: 10.1111/j.1365-2249.2008.03860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lennon GP, Bettini M, Burton AR, Vincent E, Arnold PY, Santamaria P, Vignali DAA. T cell islet accumulation in type 1 diabetes is a tightly regulated, cell-autonomous event. Immunity. 2009;31:643–653. doi: 10.1016/j.immuni.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rigby MR, Ehlers MR. Targeted immune interventions for type 1 diabetes: not as easy as it looks! Curr. Opin Endocrinol Diabetes Obes. 2014;21:271–278. doi: 10.1097/MED.0000000000000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danke NA, Koelle DM, Yee C, Beheray S, Kwok WW. Autoreactive T cells in healthy individuals. J Immunol Baltim Md 1950. 2004;172:5967–5972. doi: 10.4049/jimmunol.172.10.5967. [DOI] [PubMed] [Google Scholar]
- 8.Danke NA, Yang J, Greenbaum C, Kwok WW. Comparative study of GAD65-specific CD4+ T cells in healthy and type 1 diabetic subjects. J Autoimmun. 2005;25:303–311. doi: 10.1016/j.jaut.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Skowera A, Ladell K, McLaren JE, Dolton G, Matthews KK, Gostick E, Kronenberg-Versteeg D, Eichmann M, Knight RR, Heck S, Powrie J, Bingley PJ, Dayan CM, Miles JJ, Sewell AK, Price DA, Peakman M. β-cell-specific CD8 T cell phenotype in type 1 diabetes reflects chronic autoantigen exposure. Diabetes. 2015;64:916–925. doi: 10.2337/db14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alleva DG, Crowe PD, Jin L, Kwok WW, Ling N, Gottschalk M, Conlon PJ, Gottlieb PA, Putnam AL, Gaur A. A disease-associated cellular immune response in type 1 diabetics to an immunodominant epitope of insulin. J Clin Invest. 2001;107:173–180. doi: 10.1172/JCI8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monti P, Scirpoli M, Rigamonti A, Mayr A, Jaeger A, Bonfanti R, Chiumello G, Ziegler AG, Bonifacio E. Evidence for in vivo primed and expanded autoreactive T cells as a specific feature of patients with type 1 diabetes. J Immunol Baltim Md 1950. 2007;179:5785–5792. doi: 10.4049/jimmunol.179.9.5785. [DOI] [PubMed] [Google Scholar]
- 12.Arif S, Tree TI, Astill TP, Tremble JM, Bishop AJ, Dayan CM, Roep BO, Peakman M. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J Clin Invest. 2004;113:451–463. doi: 10.1172/JCI19585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker LSK, von Herrath M. CD4 T cell differentiation in type 1 diabetes. Clin Exp Immunol. 2016;183:16–29. doi: 10.1111/cei.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janeway CA, Jr, Travers P, Walpert M, Shlomchik MJ. Immunobiology: The Immune System in Health and Disease. 5. Garland Science; New York: 2001. [Google Scholar]
- 15.Baslan T, Hicks J. Single cell sequencing approaches for complex biological systems. Curr Opin Genet Dev. 2014;26C:59–65. doi: 10.1016/j.gde.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 17.James EA, LaFond R, Durinovic-Bello I, Kwok W. Visualizing antigen specific CD4+ T cells using MHC class II tetramers. J Vis Exp JoVE. 2009;25:1–5. doi: 10.3791/1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White J, Pullen A, Choi K, Marrack P, Kappler JW. Antigen recognition properties of mutant V beta 3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J Exp Med. 1993;177:119–125. doi: 10.1084/jem.177.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reijonen H, Mallone R, Heninger A-K, Laughlin EM, Kochik SA, Falk B, Kwok WW, Greenbaum C, Nepom GT. GAD65-specific CD4+ T-cells with high antigen avidity are prevalent in peripheral blood of patients with type 1 diabetes. Diabetes. 2004;53:1987–1994. doi: 10.2337/diabetes.53.8.1987. [DOI] [PubMed] [Google Scholar]
- 20.Mallone R, Kochik SA, Laughlin EM, Gersuk VH, Reijonen H, Kwok WW, Nepom GT. Differential recognition and activation thresholds in human autoreactive GAD-specific T-cells. Diabetes. 2004;53:971–977. doi: 10.2337/diabetes.53.4.971. [DOI] [PubMed] [Google Scholar]
- 21.Linnemann C, Heemskerk B, Kvistborg P, Kluin RJC, Bolotin DA, Chen X, Bresser K, Nieuwland M, Schotte R, Michels S, Gomez-Eerland R, Jahn L, Hombrink P, Legrand N, Shu CJ, Mamedov IZ, Velds A, Blank CU, Haanen JBAG, Turchaninova MA, Kerkhoven RM, Spits H, Hadrup SR, Heemskerk MHM, Blankenstein T, Chudakov DM, Bendle GM, Schumacher TNM. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat Med. 2013;19:1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 22.Lyons AB, Parish CR. Determination of lymphocyte division by flow cytometry. J Immunol Methods. 1994;171:131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 23.Nepom GT, Lippolis JD, White FM, Masewicz S, Marto JA, Herman A, Luckey CJ, Falk B, Shabanowitz J, Hunt DF, Engelhard VH, Nepom BS. Identification and modulation of a naturally processed T cell epitope from the diabetes-associated autoantigen human glutamic acid decarboxylase 65 (hGAD65) Proc Natl Acad Sci U S A. 2001;98:1763–1768. doi: 10.1073/pnas.98.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, Quake SR. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long SA, et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci Immunol. 2016;1:1–9. doi: 10.1126/sciimmunol.aai7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brochet X, Lefranc M-P, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321:182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 36.Marr C, Zhou JX, Huang S. Single-cell gene expression profiling and cell state dynamics: collecting data, correlating data points and connecting the dots. Curr Opin Biotechnol. 2016;39:207–214. doi: 10.1016/j.copbio.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eltahla AA, Rizzetto S, Pirozyan MR, Betz-Stablein BD, Venturi V, Kedzierska K, Lloyd AR, Bull RA, Luciani F. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunol Cell Biol. 2016;94:604–611. doi: 10.1038/icb.2016.16. [DOI] [PubMed] [Google Scholar]
- 38.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 39.McDavid A, Dennis L, Danaher P, Finak G, Krouse M, Wang A, Webster P, Beechem J, Gottardo R. Modeling bi-modality improves characterization of cell cycle on gene expression in single cells. PLoS Comput Biol. 2014;10:e1003696. doi: 10.1371/journal.pcbi.1003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinforma Oxf Engl. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gebe JA, Yue BB, Unrath KA, Falk BA, Nepom GT. Restricted Autoantigen Recognition Associated with Deletional and Adaptive Regulatory Mechanisms. J Immunol. 2009;183:59–65. doi: 10.4049/jimmunol.0804046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bacher P, Schink C, Teutschbein J, Kniemeyer O, Assenmacher M, Brakhage AA, Scheffold A. Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naive and memory Th cell repertoire. J Immunol Baltim Md 1950. 2013;190:3967–3976. doi: 10.4049/jimmunol.1202221. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Danke N, Roti M, Huston L, Greenbaum C, Pihoker C, James E, Kwok WW. CD4+ T cells from type 1 diabetic and healthy subjects exhibit different thresholds of activation to a naturally processed proinsulin epitope. J Autoimmun. 2008;31:30–41. doi: 10.1016/j.jaut.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Yang J, James EA, Sanda S, Greenbaum C, Kwok WW. CD4+ T cells recognize diverse epitopes within GAD65: implications for repertoire development and diabetes monitoring. Immunology. 2013;138:269–279. doi: 10.1111/imm.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Danke NA, Berger D, Reichstetter S, Reijonen H, Greenbaum C, Pihoker C, James EA, Kwok WW. Islet-specific glucose-6-phosphatase catalytic subunit-related protein-reactive CD4+ T cells in human subjects. J Immunol Baltim Md 1950. 2006;176:2781–2789. doi: 10.4049/jimmunol.176.5.2781. [DOI] [PubMed] [Google Scholar]
- 46.Wenzlau JM, Hutton JC, Davidson HW. New antigenic targets in type 1 diabetes. Curr Opin Endocrinol Diabetes Obes. 2008;15:315–320. doi: 10.1097/MED.0b013e328308192b. [DOI] [PubMed] [Google Scholar]
- 47.Dang M, Rockell J, Wagner R, Wenzlau JM, Yu L, Hutton JC, Gottlieb PA, Davidson HW. Human type 1 diabetes is associated with T cell autoimmunity to zinc transporter 8. J Immunol Baltim Md 1950. 2011;186:6056–6063. doi: 10.4049/jimmunol.1003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoenbrunn A, Frentsch M, Kohler S, Keye J, Dooms H, Moewes B, Dong J, Loddenkemper C, Sieper J, Wu P, Romagnani C, Matzmohr N, Thiel A. A converse 4-1BB and CD40 ligand expression pattern delineates activated regulatory T cells (Treg) and conventional T cells enabling direct isolation of alloantigen-reactive natural Foxp3+ Treg. J Immunol Baltim Md 1950. 2012;189:5985–5994. doi: 10.4049/jimmunol.1201090. [DOI] [PubMed] [Google Scholar]
- 49.Hennecke J, Wiley DC. Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity. J Exp Med. 2002;195:571–581. doi: 10.1084/jem.20011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 51.Chow I-T, Yang J, Gates TJ, James EA, Mai DT, Greenbaum C, Kwok WW. Assessment of CD4+ T cell responses to glutamic acid decarboxylase 65 using DQ8 tetramers reveals a pathogenic role of GAD65 121–140 and GAD65 250–266 in T1D development. PloS One. 2014;9:e112882. doi: 10.1371/journal.pone.0112882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auf der Maur P, Berlincourt-Böhni K. Human lymphocyte cell cycle: studies with the use of BrUdR. Hum Genet. 1979;49:209–215. doi: 10.1007/BF00277644. [DOI] [PubMed] [Google Scholar]
- 53.Han B, Serra P, Amrani A, Yamanouchi J, Marée AFM, Edelstein-Keshet L, Santamaria P. Prevention of diabetes by manipulation of anti-IGRP autoimmunity: high efficiency of a low-affinity peptide. Nat Med. 2005;11:645–652. doi: 10.1038/nm1250. [DOI] [PubMed] [Google Scholar]
- 54.Han B, Serra P, Yamanouchi J, Amrani A, Elliott JF, Dickie P, Dilorenzo TP, Santamaria P. Developmental control of CD8 T cell-avidity maturation in autoimmune diabetes. J Clin Invest. 2005;115:1879–1887. doi: 10.1172/JCI24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieberman SM, Evans AM, Han B, Takaki T, Vinnitskaya Y, Caldwell JA, Serreze DV, Shabanowitz J, Hunt DF, Nathenson SG, Santamaria P, DiLorenzo TP. Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc Natl Acad Sci U S A. 2003;100:8384–8388. doi: 10.1073/pnas.0932778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vincent BG, Young EF, Buntzman AS, Stevens R, Kepler TB, Tisch RM, Frelinger JA, Hess PR. Toxin-Coupled MHC Class I Tetramers Can Specifically Ablate Autoreactive CD8+ T Cells and Delay Diabetes in Nonobese Diabetic Mice. J Immunol. 2010;184:4196–4204. doi: 10.4049/jimmunol.0903931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee R, Wagar D, Stephens TA, Lee-Chan E, Singh B. Identification of CD4+ T cell-specific epitopes of islet-specific glucose-6-phosphatase catalytic subunit-related protein: a novel beta cell autoantigen in type 1 diabetes. J Immunol Baltim Md 1950. 2005;174:5306–5315. doi: 10.4049/jimmunol.174.9.5306. [DOI] [PubMed] [Google Scholar]
- 58.Tai N, Peng J, Liu F, Gulden E, Hu Y, Zhang X, Chen L, Wong FS, Wen L. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J Exp Med. 2016;213:2129–2146. doi: 10.1084/jem.20160526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cole DK, Bulek AM, Dolton G, Schauenberg AJ, Szomolay B, Rittase W, Trimby A, Jothikumar P, Fuller A, Skowera A, Rossjohn J, Zhu C, Miles JJ, Peakman M, Wooldridge L, Rizkallah PJ, Sewell AK. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J Clin Invest. 2016;126:2191–2204. doi: 10.1172/JCI85679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chujo D, Nguyen T-S, Foucat E, Blankenship D, Banchereau J, Nepom GT, Chaussabel D, Ueno H. Adult-onset type 1 diabetes patients display decreased IGRP-specific Tr1 cells in blood. Clin Immunol Orlando Fla. 2015;161:270–277. doi: 10.1016/j.clim.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 61.Hundhausen C, Roth A, Whalen E, Chen J, Schneider A, Long SA, Wei S, Rawlings R, Kinsman M, Evanko SP, Wight TN, Greenbaum CJ, Cerosaletti K, Buckner JH. Enhanced T cell responses to IL-6 in type 1 diabetes are associated with early clinical disease and increased IL-6 receptor expression. Sci Transl Med. 2016;8:356ra119. doi: 10.1126/scitranslmed.aad9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farh KK-H, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJH, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eugster A, Lindner A, Heninger A-K, Wilhelm C, Dietz S, Catani M, Ziegler A-G, Bonifacio E. Measuring T cell receptor and T cell gene expression diversity in antigen-responsive human CD4+ T cells. J Immunol Methods. 2013;400–401:13–22. doi: 10.1016/j.jim.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 64.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stubbington MJT, Lönnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eltahla AA, Rizzetto S, Pirozyan MR, Betz-Stablein BD, Venturi V, Kedzierska K, Lloyd AR, Bull RA, Luciani F. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunol Cell Biol. 2016 doi: 10.1038/icb.2016.16. [DOI] [PubMed] [Google Scholar]
- 67.Li B, Li T, Pignon JC, Wang B, Wang J, Shukla SA, Dou R, Chen Q, Hodi FS, Choueiri TK, Wu C, Hacohen N, Signoretti S, Liu JS, Liu XS. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat Genet. 2016;48:725–732. doi: 10.1038/ng.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.