Abstract
Many tumors, including cervical carcinoma, show dysregulated expression of the microRNA processing machinery, specifically DROSHA and DICER. Some cervical cancers exhibit chromosome 5p amplifications and DROSHA is the most significantly upregulated transcript and is observed in all tumors with 5p gain. DROSHA and DICER mRNA levels, however, are higher in HPV positive cancer lines than in an HPV negative cervical carcinoma line. We show that high-risk HPV E6/E7 expression in HPV negative C33A cervical carcinoma cells and primary human epithelial cell causes increased expression of DROSHA and DICER mRNA and protein. Most importantly, many DROSHA regulated microRNAs are dysregulated in HPV16 E6/E7 expressing cells. These results suggest that increased DROSHA levels contribute to HPV16 E6/E7 dysregulation of cellular microRNA expression.
Keywords: microRNA, cervical cancer, human papillomavirus
INTRODUCTION
DROSHA is a double-stranded RNA-specific ribonuclease (RNAse III) and the catalytic component of the microprocessor protein complex, which is rate limiting for the initial processing of primary microRNA transcripts (pri-miRs) into precursor microRNAs (pre-miRs) in the nucleus (Denli et al., 2004; Han et al., 2004; Lee et al., 2003; Xie and Steitz, 2014). After export to the cytoplasm, pre-miRs are processed by another RNAse III enzyme, DICER, into the mature miR duplex (Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001; Knight and Bass, 2001). The miR duplex is then incorporated into the RNA-induced silencing complex (RISC) containing DICER, an AGO (Argonaute) protein (Liu et al., 2004; Meister et al., 2004), and the RNA binding proteins TRBP (TAR RNA binding protein) (Chendrimada et al., 2005; Haase et al., 2005) and PACT (protein activator of PKR) (Lee et al., 2006). After RISC binding, one strand of the miR duplex is selected as the guide strand (miR), based on the strength of base-pairing at its 5′ end, to form the functional miR-RISC complex (miRISC) (Khvorova et al., 2003; Schwarz et al., 2003). The other strand, known as the passenger strand (miR*), is displaced and degraded. The miRISC, the final product of this carefully orchestrated maturation process, can then function to silence miR targets via mRNA degradation/destabilization, translational repression, or a combination of both mechanisms (Djuranovic et al., 2012).
The miR biogenesis pathway, including DROSHA and DICER expression, is frequently dysregulated in human cancers (reviewed in (Adams et al., 2014; Hata and Lieberman, 2015). DROSHA is frequently amplified or mutated whereas DICER is most often mutated (Cerami et al., 2012; Gao et al., 2013). Altered DROSHA and DICER expression has also been noted in cervical carcinoma. Specifically, a gain of chromosome 5p has been observed in most advanced cervical squamous cell carcinomas. DROSHA is the most significantly upregulated transcript associated with this chromosomal gain and DROSHA overexpression has been observed in all tumors with 5p gain, suggesting that DROSHA may be a critical 5p target with a potential role in cervical cancer progression (Muralidhar et al., 2007; Scotto et al., 2008). DROSHA mRNA levels were also shown to be increased in the HPV positive SiHa and HeLa cervical cancer cell lines and to a lesser extent in the C33A HPV negative cervical cancer cell line (Muralidhar et al., 2007(Zhou et al., 2013).
Interestingly, modulation of DROSHA expression in cervical carcinoma lines did not result in global alterations of miR levels and only a small subset of 45 out of 319 miRs examined showed changes in expression upon modulation of DROSHA levels (Muralidhar et al., 2011). Modulation of these miRs was linked to alterations in cell migration and invasiveness (Muralidhar et al., 2011), two important hallmarks of cancer progression.
Reports on DICER mRNA levels in cervical cancers are not consistent. A small study reported lower DICER mRNA levels in cervical cancer tissues (Zhao et al., 2014), whereas another study reported varied DICER mRNA expression among cervical cancer specimens, with low DICER mRNA expression in 36.7% of samples and higher DICER mRNA expression in 63.3% of samples (He et al., 2014).
We have previously investigated modulation of cellular miR levels in response to HPV16 E6/E7 expression in primary human epithelial cells (Harden et al., 2017). Given that the high-risk HPV E6 and E7 proteins are consistently expressed in cervical carcinoma lines, that HPV positive cervical carcinoma lines expressed DROSHA at higher levels than the HPV negative C33A line and the fact that DROSHA expression was shown to only affect expression of a small subset of miRs in cervical cancer lines, we set out to determine whether HPV E6 and/or E7 expression may cause altered DROSHA and/or DICER levels. Additionally, we aimed to investigate whether some of the alterations in cellular miR expression that we previously observed may potentially be caused by altered DROSHA expression.
MATERIALS AND METHODS
Cell Culture
Primary human foreskin keratinocytes (HFKs) were isolated from a pool of de-identified newborn foreskins and cultured as previously described (Harden et al., 2017). CaSki, C33A, HeLa and SiHa cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin and 50 mg/ml streptomycin. HFKs were transduced with LXSN based recombinant retroviruses encoding HPV16 E6 and/or E7 or a control LXSN vector (Halbert et al., 1991) as previously described (Harden et al., 2017). C33As were transiently transfected with pCMV HPV16 E6 and/or pCMV HPV16 E7 plasmids, a control pCMV plasmid as a control utilizing FuGENE 6 Transfection Reagent (Promega) according to the manufacturer’s protocol. HFKs were grown to 80% confluence prior to passaging and passaged up to 8 times. In all experiments, donor and passage matched HFK populations were used.
Reverse Transcription quantitative PCR (RT-qPCR)
Total RNA was isolated using the miRNeasy Mini Kit (Qiagen) per the manufacturer’s instructions. For RT-qPCR of DROSHA and DICER, following RNA isolation, total RNA was DNAase-treated with the TURBO DNA-free kit (Ambion/Thermo Fisher Scientific). DNAse-treated total RNA was then reverse transcribed utilizing TaqMan Reverse Transcription Reagents (Applied Biosystems/Thermo Fisher Scientific). TaqMan Assay IDs Hs00203008_m1 and Hs00229023_m1 (Applied Biosystems/Thermo Fisher Scientific) were employed to detect DROSHA and DICER, respectively, using the comparative Ct method with the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). RT-qPCR assays were performed in triplicate and 18S ribosomal RNA was utilized as an internal control.
Western Blotting
Protein lysates were prepared by incubating the cells in ML buffer (300 mM NaCl, 0.5% Nonidet P-40 [NP-40], 20 mM Tris-HCl [pH 8.0], 1 mM EDTA) supplemented with one Complete EDTA-free Protease Inhibitor Cocktail tablet (Roche) per 50 ml lysis buffer. The cells were incubated on ice for 20 minutes, scraped and rotated at 4°C for 20 minutes and then cleared by centrifugation at 16,000 × g for 20 min. Protein concentrations were determined via the Bradford method (Bradford, 1976). Samples containing 200 μg of protein were boiled in NuPAGE® LDS Sample Buffer (4X) (Invitrogen), separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore). The membranes were blocked for 2 h in 5% nonfat dry milk in TBST buffer (137 mM NaCl, 2.7 mM KCl, 25 mM Tris [pH 7.4], 0.1% Tween 20). Primary antibodies were used as follows: DROSHA (ab12286; Abcam) at 1:500, DICER (3363; Cell Signaling Technology) at 1:1,000 and β-actin (MAB1501; Millipore) at 1:1,000. Secondary anti-mouse IgG and anti-rabbit IgG horseradish peroxidase-conjugated antibodies (Amersham) were used at 1:10,000 dilutions. Proteins were visualized by enhanced chemiluminescence (Luminata™ Crescendo Western HRP Substrate; Millipore) and electronically acquired with a Syngene G:BOX image station (Syngene) equipped with GeneSys software, v1.5.6.0. Loading was assessed using β-actin immunoblots.
RESULTS
Increased DROSHA and DICER mRNA levels in HPV positive cervical cancer cell lines
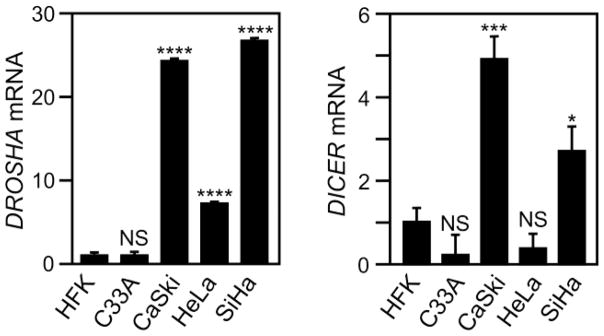
DROSHA is frequently expressed at high levels in cervical carcinomas. Therefore, we examined DROSHA mRNA levels by RT-qPCR in the HPV16 positive CaSki and SiHa, the HPV18 positive HeLa and the HPV-negative C33A cervical cancer cell lines. Compared to primary human keratinocytes (HFKs), DROSHA mRNA was significantly upregulated in CaSki (24.4 fold; P<0.0001), SiHa (26.8 fold; P<0.0001), HeLa (7.2 fold; P<0.0001) but was not significantly modulated in C33A cells (1.1 fold; P=0.72) (Fig. 1A). These results partially confirm and extend a previous report that documented increased DROSHA mRNA expression in HeLa, SiHa and C33A cervical cancer cell lines (Zhou et al., 2013).
Fig. 1.
RNA levels of DROSHA and DICER mRNA in cervical cancer cell lines. Total RNA was harvested from HFKs and C33A (HPV negative), CaSki (HPV16), SiHa (HPV16) and HeLa (HPV18). (A) DROSHA and (B) DICER mRNA levels were determined by RT-qPCR. Expression of 18S ribosomal RNA (18S) was utilized as a control and results were normalized to DROSHA and DICER expression in HFKs. Results show averages of three independent experiments and error bars depict standard deviation. Unpaired, two-tailed t-tests with a 95% confidence level were performed to determine statistical significance. *, **, *** and **** indicate statistical significance at a P<0.05, P<0.01, P<0.001 and P<0.0001 respectively. “NS”: Not Statistically Significant.
Reports in the literature on DICER mRNA expression in cervical cancers are inconsistent (He et al., 2014; Zhao et al., 2014). Therefore, we next determined DICER mRNA expression in cervical carcinoma lines. Our analysis revealed that compared to HFKs, DICER mRNA levels were significantly higher in the HPV16 positive CaSki (4.9 fold; P=0.0005) and SiHa (2.7 fold; P=0.0136) but not significantly altered in HPV18 positive HeLa (−2.7 fold; P=0.0978) and the HPV negative C33A (−4.9 fold; P=0.0946) lines (Fig. 1B). In summary, our data are partially consistent with a previous report (Zhou et al., 2013), and show that DROSHA mRNA levels are significantly increased in all HPV positive cervical cancer lines, whereas DICER mRNA levels were only significantly increased in the HPV16 positive CaSki and SiHa but not in the HPV18 positive HeLa or the HPV negative C33A cervical carcinoma lines.
HPV16 E6/E7 expression in HPV negative C33A cells increases DROSHA mRNA
Our results suggested that DROSHA mRNA levels in cervical cancer lines may correlate with HPV status (Fig. 1). Hence, we investigated whether HPV16 E6 and/or E7 expression in C33A cells may increase DROSHA mRNA levels. We transiently transfected C33A cells with CMV based E6 or E7 expressing vectors either alone or in combination and determined DROSHA mRNA levels by RT-qPCR. DROSHA mRNA levels were most upregulated upon HPV16 E6/E7 co-transfection (2.3 fold; P=0.0335), followed by transfection with HPV16 E7 alone (2.0 fold; P=0.0073) but were not significantly altered by HPV16 E6 expression alone (1.3 fold; P=0.2372) (Fig. 2). These results suggest that HPV16 E6/E7 expression causes increased DROSHA mRNA expression in C33A cells. DROSHA mRNA levels in HPV16 E6/E7 expressing C33A cells were lower than in HPV16 positive cervical cancer cell lines however, it is difficult to directly compare the various cell lines and the effect of HPV16 E6/E7 on DROSHA mRNA levels in C33A cells depends on several experimental parameters, including E6/E7 expression levels and the cellular background of C33A cells.
Fig. 2.
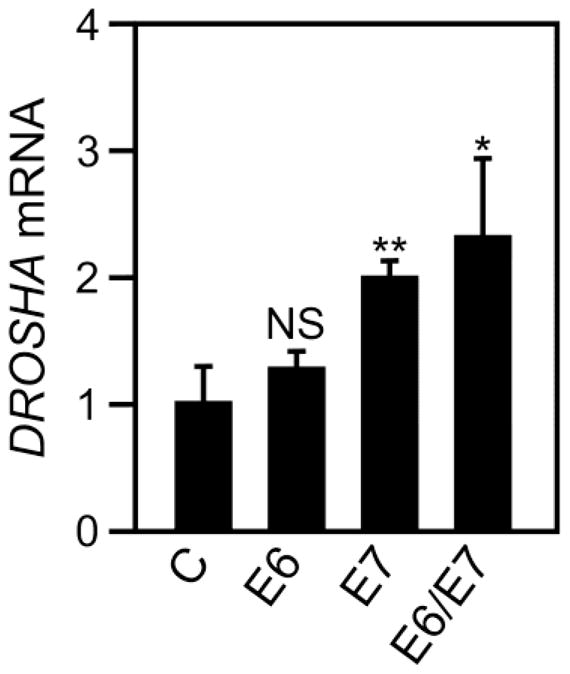
DROSHA mRNA expression in HPV negative C33A cervical cancer cells with transient expression of HPV16 E6/E7. C33A cells were transiently transfected with empty vector (C), HPV16 E6 alone (E6), HPV16 E7 alone (E7) or both HPV16 E6/E7 (E6/E7). DROSHA mRNA levels were analyzed by RT-qPCR. Expression of 18S ribosomal RNA (18S) was utilized as a control and values were normalized to control vector transduced C33A cells. Results represent averages of three independent experiments and error bars show standard deviation. * and ** indicate statistical significance at a P<0.05 and P<0.01, respectively. “NS”: Not Statistically Significant.
HPV16 E6/E7 expression in primary human epithelial cells increases DROSHA and DICER mRNA levels
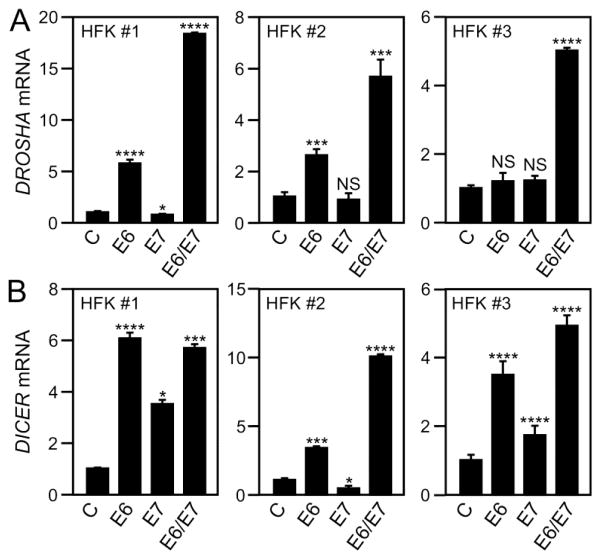
Since our results from cervical cancer cell lines suggested a possible correlation between DROSHA and DICER mRNA levels and HPV16 status, we hypothesized that expression of the HPV E6 and/or E7 oncoproteins in primary human epithelial cells may be sufficient to alter DROSHA and DICER mRNA levels. To test this hypothesis, we analyzed DROSHA and DICER mRNA levels by RT-qPCR in three independent, donor and passage matched HFK populations each with stable expression of HPV16 E6 and/or E7 or a control vector. In two of the three HFK populations tested, HPV16 E6 expression caused a significant increase in DROSHA mRNA levels compared to control HFKs. In contrast HPV16 E7 expressing HFKs had DROSHA mRNA levels that were similar to or slightly lower than control HFKs. HPV16 E6/E7 expressing HFKs, however, consistently exhibited significantly increased DROSHA mRNA levels that exceeded those in HPV16 E6 expressing HFKs (Fig. 3A). Similarly, DICER mRNA levels were higher in HPV16 E6 expressing HFKs. Except in population 2, HPV16 E7 expressing HFKs also had significantly higher DICER mRNA levels and DICER mRNA levels were consistently and significantly increased in HPV16 E6/E7 co-expressing cells (Fig. 3B).
Fig. 3.
DROSHA and DICER mRNA levels in HPV16 E6 and/or E7 expressing primary human foreskin keratinocytes. Three donor and passage matched HFK populations with stable expression of HPV16 E6 (E6), HPV16 E7 (E7), HPV16 E6/E7 (E6/E7) or an empty vector (C) were analyzed for (A) DROSHA and (B) DICER mRNA expression by RT-qPCR. Expression of 18S ribosomal RNA (18S) was utilized as a control and values were normalized to control vector transduced HFKs. Similar results were obtained with three additional HFK populations. Error bars show standard deviation. Unpaired, two-tailed t-tests with a 95% confidence level were performed to determine statistical significance. *, **, *** and **** indicate statistical significance at a P<0.05, P<0.01, P<0.001 and P<0.0001 respectively. “NS”: Not Statistically Significant. Please note that these HFK populations are distinct from those shown in Figure 4.
HPV16 E6/E7 expression in primary epithelial cells increases DROSHA and DICER protein levels
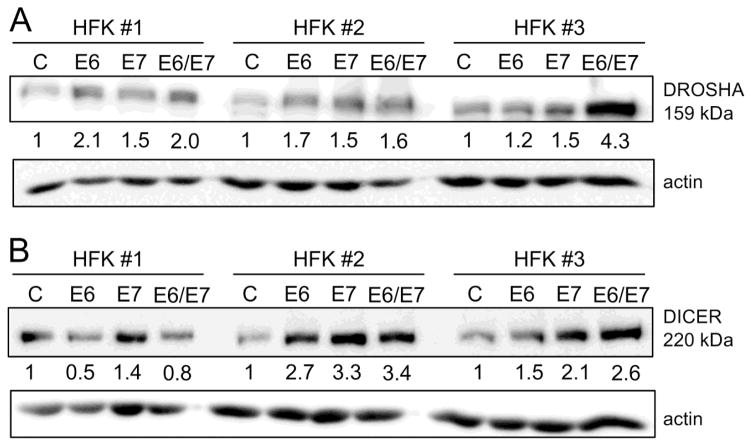
We next examined by immunoblot experiments, whether the observed modulation of DROSHA and DICER mRNA levels resulted in similar alterations in DROSHA and DICER protein levels. DROSHA protein levels were consistently increased in HPV16 E6 and/or E7 expressing HFKs although the magnitude of the effect varies between the different HFK populations (Fig. 4A). DICER protein levels are also increased in HPV16 E6 and/or E7 expressing HFKs although only the HPV16 E7 expressing HFKs expressed higher DICER protein in population 1 (Fig. 4B).
Fig. 4.
Protein levels of DROSHA and DICER protein levels in HPV16 E6 and/or E7 expressing primary human foreskin keratinocytes. Three donor and passage matched HFK populations with stable expression of HPV16 E6 (E6), HPV16 E7 (E7), HPV16 E6/E7 (E6/E7) or an empty vector (C) were analyzed for (A) DROSHA and (B) DICER protein expression by immunoblot analysis. Steady state levels of actin served as a loading control and quantifications normalized to actin levels are indicated underneath the blots. Please note that these HFK populations are distinct from those shown in Figure 3.
In combination with the data shown in Figure 3, these results show that HPV16 E6/E7 expression in primary human epithelial cells can cause increased DROSHA and DICER levels and these increases are, at least in part, due to increased mRNA levels.
Expression of DROSHA-regulated miRs is altered in HPV16 E6/E7 expressing primary human epithelial cells
A previous study showed that modulation of DROSHA expression in cervical carcinoma cells levels significantly altered expression of only 45 out of 319 miRs tested (Muralidhar et al., 2011). Since we observed increased DROSHA levels in HPV16 E6/E7 expressing HFKs, we wanted to determine if the levels of these DROSHA-regulated miR were correspondingly altered in HPV16 E6/E7 expressing HFKs. We previously performed a comprehensive analysis of HPV16 E6/E7 induced alterations of cellular miR expression (Harden et al., 2017). Hence, we assessed expression of the 45 DROSHA-regulated miRs (Muralidhar et al., 2011) in our data set (Table 1). A total of 37 out of 45 (82%) of DROSHA-regulated miRs are regulated in HPV16 E6/E7 expressing HFKs consistent with increased DROSHA expression. This analysis suggests that at least some of the HPV16 E6/E7 induced alterations in cellular miR expression that we observed may be a consequence of the increased DROSHA levels in HFKs expressing HPV16 E6/E7.
Table 1.
Expression of DROSHA regulated miRs in cervical squamous cell carcinoma lines.
| DROSHA-regulated miR | Regulation (Observed)* | Regulation (Literature)** | Overlap |
|---|---|---|---|
| miR-193a-3p | down-regulated | down-regulated | |
| miR-138-5p | down-regulated | down-regulated | |
| miR-370 | up-regulated | down-regulated | |
| miR-380-3p | down-regulated | down-regulated | |
| miR-520e | down-regulated | down-regulated | |
| miR-7-5p | up-regulated | up-regulated | |
| miR-148b-3p | up-regulated | up-regulated | |
| miR-98-5p | up-regulated | up-regulated | |
| miR-378a-3p | up-regulated | up-regulated | |
| miR-141-3p | up-regulated | up-regulated | |
| miR-107 | down-regulated | up-regulated | |
| let-7c | up-regulated | up-regulated | |
| let-7a-5p | up-regulated | up-regulated | |
| miR-30c-5p | up-regulated | up-regulated | |
| let-7f-5p | up-regulated | up-regulated | |
| miR-125b-5p | up-regulated | up-regulated | |
| miR-23a-3p | up-regulated | up-regulated | |
| miR-125a-5p | up-regulated | up-regulated | |
| let-7d-5p | up-regulated | up-regulated | |
| miR-23b-3p | down-regulated | up-regulated | |
| miR-100-5p | up-regulated | up-regulated | |
| miR-185-5p | up-regulated | up-regulated | |
| let-7e-5p | up-regulated | up-regulated | |
| let-7b-5p | up-regulated | up-regulated | |
| miR-200c-3p | down-regulated | up-regulated | |
| miR-7i-5p | up-regulated | up-regulated | |
| miR-15b-5p | up-regulated | up-regulated | |
| miR-26a-5p | up-regulated | up-regulated | |
| miR-106b-5p | up-regulated | up-regulated | |
| miR-22-3p | down-regulated | up-regulated | |
| let-7g-5p | up-regulated | up-regulated | |
| miR-24-3p | up-regulated | up-regulated | |
| miR-16-5p | up-regulated | up-regulated | |
| miR-130a-3p | up-regulated | up-regulated | |
| miR-151a-3p | up-regulated | up-regulated | |
| miR-27b-3p | up-regulated | up-regulated | |
| miR-27a-3p | up-regulated | up-regulated | |
| miR-203a-3p | down-regulated | up-regulated | |
| miR-338-3p | up-regulated | up-regulated | |
| miR-99b-5p | up-regulated | up-regulated | |
| miR-330-3p | down-regulated | up-regulated | |
| miR-31-5p | down-regulated | up-regulated | |
| miR-342-3p | up-regulated | up-regulated | |
| miR-191-5p | up-regulated | up-regulated | |
| miR-10b-5p | up-regulated | up-regulated |
Data from (Harden et al., 2017).
Data from (Muralidhar et al., 2011).
We also compared the DROSHA-regulated miRs to our list of the 15 most up- or down regulated miRs identified by a comprehensive target pairing analysis in HPV16 E6/E7 expressing HFKs (Harden et al., 2017). Only one DROSHA-regulated miR, miR-203a-3p, was on that list. We, and others (Harden et al., 2017; McKenna et al., 2010; Melar-New and Laimins, 2010), however, had shown that this miR is downregulated in HPV16 E6/E7 expressing HFKs, the opposite of what would have been expected based on increased DROSHA expression (Muralidhar et al., 2011). Hence, DROSHA-regulated miRs are not amongst the most highly modulated miRs in HPV16 E6/E7 expressing HFKs and DROSHA-regulated miRs must also be modulated by HPV16 E6/E7 through other pathways.
DROSHA-regulated microRNAs and microRNAs modulated by HPV16 E6/E7 are involved in modulating similar cellular pathways
To examine pathways relevant to the DROSHA-regulated miRs, we utilized the core analysis function of Ingenuity Pathway Analysis (IPA). This type of analysis identifies relationships, mechanisms, functions and pathways of relevance to a particular dataset. Out of the 45 DROSHA-responsive miRs, 30 were associated with “cancer”, 26 were associated with “organismal injury and abnormalities” and 25 were associated with “reproductive system disease.” Top molecular and cellular functions affiliated with these miRs were “cell movement, development, growth and proliferation” as well as “cell cycle” and “DNA replication, recombination and repair.” Interestingly, the top five molecular and cellular functions associated with the DROSHA responsive miRs were the same as those associated with all miRs altered by HPV16 E6/E7 expression. However, the order of significance of association, based on p-value, is different. For miRs associated with DROSHA levels “cell movement” was the top molecular and cellular function followed by “cell development, growth and proliferation” and lastly “cell cycle.” For all miRs perturbed by HPV16 E6/E7 expression “cell cycle” was the top molecular and cellular function followed by “cell movement”, and “cell development, growth and proliferation.” These results suggest that DROSHA regulated miRs participate in similar, cellular processes than miRs previously identified to be modulated by HPV16 E6/E7 expression.
DISCUSSION
Similar to other cancers, expression of the miR biosynthesis machinery is dysregulated in cervical carcinomas. DROSHA expression is frequently upregulated and 5p gene amplification involving the DROSHA gene has also been reported in cervical cancers. Our experiments partially confirm and extend a previous publication (Zhou et al., 2013) and document higher DROSHA mRNA levels in HPV16 positive SiHa and CaSki, as well as HPV18 positive Hela, but not in the HPV negative C33A cervical cancer line. We show that HPV16 E6/E7 expression in C33A cells causes an increase in DROSHA expression although not to the levels observed in HPV positive cervical cancer lines.
In our experimental system, we introduce expression of HPV16 E6/E7 into low passage primary HFKs that have not yet acquired the amplification of chromosome 5p, carrying the DROSHA gene, which is frequently observed in cervical cancers. Given the relatively high frequency of chromosome 5p amplification, on may hypothesize that high level DROSHA expression is important for HPV positive cervical cancers. Therefore, we hypothesize that increased DROSHA expression by HPV16 E6/E7 may be a mechanism to establish higher DROSHA expression at very early stages of HPV carcinogenesis until the gain of chromosome 5p can occur.
Our results with HFKs suggest that HPV16 E6 is the main driver of DROSHA expression. While E6 expression also causes increased DROSHA expression in HPV-negative C33A cervical carcinoma cells HPV16 E7 is the major driver of DROSHA mRNA upregulation. Given that C33A cells express mutant p53, this E6-mediated increase in C33A cells is p53 independent, but given that E6 generally caused a more marked increase in DROSHA expression in HFKs that express wild type p53, it is conceivable that p53 inactivation by E6 may contribute to increased DROSHA expression. Co-expression of HPV16 E6/E7 in primary human epithelial cells resulted in the most dramatic increases in DROSHA and DICER mRNA and protein expression. This may be the result of the well-known functional cooperativity of the HPV16 E6 and E7 oncoproteins (Moody and Laimins, 2010).
We also noted differences in the HPV16 E6/E7-dependent regulation of DROSHA/DICER at the mRNA and protein levels. This suggests that DROSHA/DICER expression by HPV16 E6/E7 may also be regulated post-transcriptionally, which has been previously observed for both DROSHA (Han et al., 2009) and DICER (Wiesen and Tomasi, 2009).
It was previously noted that modulation of DROSHA levels in cervical cancer lines altered expression of only 45 of the 315 miRs that were tested (Muralidhar et al., 2011). Since not all miRs were evaluated in this study, there are likely additional DROSHA-regulated miRs to be discovered. While most (40/45) of the “DROSHA-regulated” miRs were expressed at higher levels, some (5/45) were expressed at lower levels. We showed that 37 out of 45 (82%) of the DROSHA-regulated miRs are expressed in HPV16 E6/E7 expressing primary human epithelial cells consistent with DROSHA regulation. Hence, at least some of the HPV16 E6/E7 mediated changes in cellular miR expression might be through a mechanism involving increased DROSHA expression. Nonetheless, DROSHA was not the major driver of expression of some other DROSHA-regulated miRs, including miR-203-3p, in HPV16 E6/E7 expressing cells.
To directly test whether increased DROSHA expression in E6 and E6/E7 expressing cells contributes to increased expression of these miRs, we attempted to silence DROSHA expression in our engineered HFK populations. We used both lentiviral transduction with multiple shRNAs and transfections with siRNAs, however, we were unable to decrease DROSHA levels sufficiently to examine the effects of DROSHA silencing on HPV16 E6/E7 expressing HFKs. We were only able to decrease DROSHA levels by ~50% in HFKs using transient transfection of several different siRNAs. HPV16 E6/E7 expressing HFKs transduced with DROSHA shRNAs did not survive selection post-transduction and perhaps this is due to other non-miR related functions of DROSHA (Johanson et al., 2013) that may be important in primary human epithelial cells. This, unfortunately precluded us from directly and conclusively determining whether and how increased DROSHA expression by E6 and E7 contributes to modulation of cellular miR levels.
To determine whether DROSHA-regulated miRs affect specific cellular signaling pathways, we compared a core analysis of these miRs to the entire miRseq dataset (−1 ≥ FC ≥ 1). We utilized less stringent fold change cutoffs since the reported fold changes for the DROSHA-regulated miRs ranged from 1.8–4.2 fold (Muralidhar et al., 2011). This comparative analysis revealed that many of the same molecular and cellular functions are associated with both DROSHA regulated and HPV16 E6/E7 modulated miRs.
In summary, our study shows that HPV16 E6/E7 oncoprotein expression alters RNA and protein levels of DROSHA and DICER, two critical enzymes in the canonical miR biogenesis pathway. Examination of known DROSHA-regulated miRs suggests that HPV16 E6/E7 perturbation of DROSHA levels may be one mechanism by which HPV16 E6/E7 expression perturbs cellular miR expression. In addition, given the miR biogenesis independent activities of DROSHA and DICER, their increased expression may also contribute to cervical carcinogenesis through miR independent mechanisms. Further studies will be necessary to arrive at a mechanistic understanding of DICER and DROSHA modulation by HPV16 E6/E7.
Highlights.
DROSHA is frequently overexpressed due to gene amplification in human papillomavirus associated cancers.
We showed that HPV16 E6 and E7 cause increased DROSHA and DICER expression.
This is correlated to modulation of DROSHA regulated microRNAs in HPV16 E6/E7 expressing cells.
Acknowledgments
Supported by PHS grants F31CA180516 (MEH) and R01CA066980 (KM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden ME, Prasad N, Griffiths A, Munger K. Modulation of microRNA-mRNA Target Pairs by Human Papillomavirus 16 Oncoproteins. MBio. 2017;8 doi: 10.1128/mBio.02170-16. pii: e02170–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal. 2015;8:re3. doi: 10.1126/scisignal.2005825. [DOI] [PubMed] [Google Scholar]
- He L, Wang HY, Zhang L, Huang L, Li JD, Xiong Y, Zhang MY, Jia WH, Yun JP, Luo RZ, Zheng M. Prognostic significance of low DICER expression regulated by miR-130a in cervical cancer. Cell Death Dis. 2014;5:e1205. doi: 10.1038/cddis.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Johanson TM, Lew AM, Chong MM. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 2013;3:130144. doi: 10.1098/rsob.130144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- McKenna DJ, McDade SS, Patel D, McCance DJ. MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J Virol. 2010;84:10644–10652. doi: 10.1128/JVI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212–5221. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–60. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, Pett MR, Coleman N. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- Muralidhar B, Winder D, Murray M, Palmer R, Barbosa-Morais N, Saini H, Roberts I, Pett M, Coleman N. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol. 2011;224:496–507. doi: 10.1002/path.2898. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Subramaniyam S, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Schneider A, Arias-Pulido H, Murty VV. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer. 2008;7:58. doi: 10.1186/1476-4598-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Steitz JA. Versatile microRNA biogenesis in animals and their viruses. RNA Biol. 2014;11:673–681. doi: 10.4161/rna.28985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Jin X, Su H, Deng X, Fang Y, Shen L, Xie C. Down-regulation of Dicer expression in cervical cancer tissues. Med Oncol. 2014;31:937. doi: 10.1007/s12032-014-0937-0. [DOI] [PubMed] [Google Scholar]
- Zhou J, Cai J, Huang Z, Ding H, Wang J, Jia J, Zhao Y, Huang D, Wang Z. Proteomic identification of target proteins following Drosha knockdown in cervical cancer. Oncol Rep. 2013;30:2229–2237. doi: 10.3892/or.2013.2672. [DOI] [PubMed] [Google Scholar]