Abstract
It has been recognized for 40 years that V(D)J recombination-mediated assembly of diverse B and T lymphocyte antigen receptor genes is essential for adaptive immunity, yet also a risk for autoimmunity and lymphoid malignancies. In the past few years, several studies have revealed that RAG endonuclease-induced DNA double strand breaks (DSBs) transcend hazardous intermediates during antigen receptor gene assembly. RAG cleavage within the genomes of lymphocyte progenitors and immature lymphocytes regulates the expression of ubiquitous and lymphocyte-specific gene transcripts to control the differentiation and function of both adaptive and innate immune cell lineages. These unexpected discoveries raise important new questions that have broad implications for basic immunology research and the screening, diagnosis, and treatment of human immunological disease.
Keywords: V(D)J Recombination, RAG endonuclease, DNA double strand breaks, DNA damage responses, lymphocyte differentiation, lymphocyte function
The Mechanism and Regulation of Lymphocyte Antigen Receptor Gene Assembly
In jawed vertebrates, the genes encoding B and T lymphocyte antigen receptor (AgR, see Glossary) variable regions are assembled through somatic recombination of variable (V), diversity (D), and joining (J) gene segments [1, 2]. The lymphocyte-specific RAG1/RAG2 (RAG) endonuclease initiates V(D)J recombination by introducing DNA double strand breaks (DSBs) adjacent to V, D, and J gene segments [3]. Ubiquitous DSB repair proteins process and join RAG-liberated DNA ends to form V(D)J rearrangements upstream of constant (C) region exons [4], thereby constructing immunoglobulin (Ig) genes in B lymphocytes and T cell receptor (TCR) genes in T lymphocytes. V(D)J recombination establishes AgR diversity through the number of possible V(D)J joining events within each AgR locus, imprecision in V(D)J joining, and pairing of distinct Ig (IgH/Igic or IgH/Igλ,) or TCR (TCRα/TCRβ or TCRy/TCR5) proteins to create B cell receptors (BCRs) or TCRs. The ability of V(D)J recombination in mice to generate about 1011 B [2] and 1015 T lymphocytes [1], that each expresses AgRs from a unique pair of Ig or TCR genes is the basis for adaptive immunity. Complete immune protection is provided by cooperation among B cells, T cells, and several distinct lineages of innate lymphocytes that use germ line-encoded antigen receptors to recognize foreign molecules.
AgR gene assembly is regulated to limit the risk of autoimmunity and lymphoid malignancies [5, 6]. V(D)J recombination creates both potentially hazardous self-reactive and potentially beneficial non-self-reactive AgRs [7]. The expression of a self-reactive AgR on immature lymphocytes can induce apoptosis, direct differentiation of anergic cells, or, for an IgH/Igκ B cell receptor (BCR), stimulate further Igκ recombination in an attempt to replace itself with a non-self-reactive BCR (receptor editing) [6, 8-10]. It remains unknown how an auto-reactive AgR triggers a particular cellular fate. Eukaryotic cells employ a conserved DSB response to protect themselves and their host organisms from hazards of DSBs, including apoptosis and oncogenic genomic lesions [11, 12]. This DSB response includes activation of DNA repair, cell cycle checkpoints, and apoptosis if DSBs are too many or cannot be repaired [11, 12]. RAG cleavage of AgR loci activates this DSB response and inactivation of DSB response factors causes lymphoid malignancies with RAG-dependent oncogenic AgR locus translocations [4, 5, 13]. The conserved cell cycle machinery promotes RAG2 degradation upon the G1/S phase transition and thereby cooperates with the DSB response to suppress oncogenic AgR translocations from RAG DSBs induced in S phase [14-16]. Moreover, the ability of RAG proteins to facilitate proper repair of RAG DSBs cooperates with the shared cellular DSB response to suppress oncogenic AgR translocations [17-19]. It is clear that ubiquitous and lymphocyte-specific mechanisms control the induction and repair of RAG DSBs to limit the frequency of fatal V(D)J recombination errors. However, it remains to be determined whether all V(D)J rearrangement events trigger responses to dangerous DSB intermediates or only a small subset in which RAG DSBs persist.
A study published in 2008 indicated that RAG DSBs transcend hazardous intermediates during V(D)J recombination [20]. It was shown that RAG cleavage of AgR loci activates several distinct signaling pathways to change the levels of transcripts from hundreds of genes, including many encoding lymphocyte-specific factors that regulate cellular processes important for lymphocyte differentiation and function [20]. This unexpected discovery prompted additional studies that have cemented the novel concept that RAG DSBs induced in lymphocyte progenitors and immature lymphocytes activate genetic programs that regulate the development and function of diverse lineages of adaptive and innate lymphocytes. This review summarizes these studies and how their findings raise important questions whose answers have broad implications for basic immunology research and clinical management of human immunological disease.
RAG DSBs Activate a Genetic Program that Transcends the Cellular DSB Response
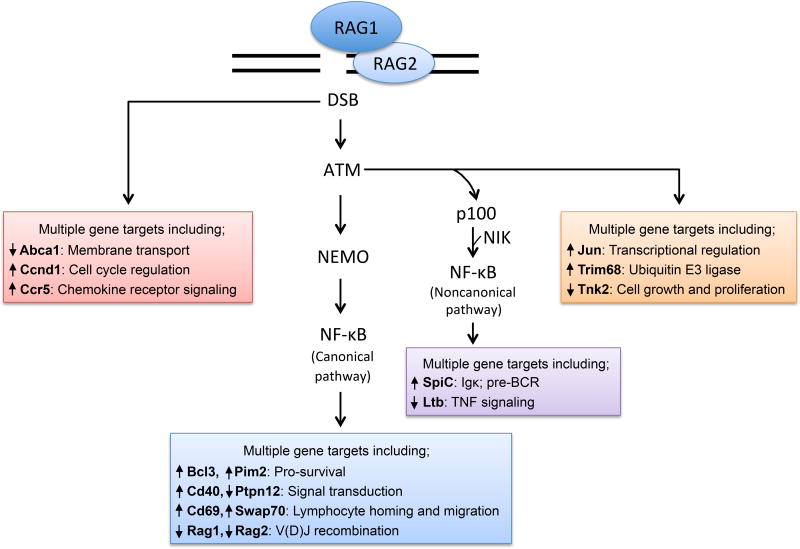
In 2008, an analysis of cellular DSB responses activated by RAG cleavage during V(D)J recombination was published [20]. The ATM kinase is a master regulator of the eukaryotic DSB response [21]. RAG DSBs activate ATM [22], which in turn stimulates phosphorylation of many proteins to promote DSB repair and activate cell cycle checkpoints or induce apoptosis [21]. Notably, ATM-mediated phosphorylation and activation of the Trp53 transcription factor drives expression of genes that trigger cell cycle checkpoints or apoptosis [21, 23]. It had been discovered that ATM-dependent phosphorylation of the NFκB essential upstream modifier (Nemo) protein activated canonical NFκB transcription factors upon induction of DSBs by ionizing radiation (IR) or genotoxic drugs [24]. Yet, neither the gene targets nor biological effects of NFκB activation in this setting were known. Thus, it was proposed that RAG DSBs may activate NFκB and its target genes. Indeed, it was found that RAG DSBs induced at Igκ loci in pre-B cells activate canonical NFκB transcription factors through ATM/Nemo-dependent mechanisms. In fact, RAG Igκ DSBs promote increases or decreases in the levels of transcripts from 364 genes, and corresponding changes in protein levels were assayed, through signaling pathways dependent on ATM and NFκB, only ATM, or neither (Figure 1, Key Figure). Notably, DSBs induced by genotoxic agents activate the same genetic program as RAG DSBs. The genes regulated by RAG DSBs encode DSB response proteins, antiapoptotic proteins, or generally expressed or lymphocyte-specific proteins that regulate cellular processes important for lymphocyte development and function. The lymphocyte-specific proteins up regulated by RAG DSBs include proteins that control lymphocyte homing and migration. Consistent with the expression and function of these proteins, co-injection into mice of pre-B cells harboring or not harboring RAG-cleaved Igκ loci leads to preferential localization of cells with RAG DSBs in the bone marrow. The migration of immature lymphocytes through distinct microenvironments within the bone marrow or thymus is important for their proper differentiation and selection. Accordingly, this study introduced the innovative concept that RAG DSBs transcend hazardous intermediates in that they can directly regulate lymphocyte differentiati on by signaling changes in gene transcription.
Figure 1. RAG DSBs Employ Distinct Signaling Pathways to Activate a Multifunctional Genetic Program that Transcends the Cellular DSB Response.
RAG DSBs induced at Igκ loci in pre-B cells signal through ATM-independent and ATM-dependent pathways, the latter involving canonical and non-canonical NFκB pathways, to control the levels of transcripts from numerous genes. Many of these genes encode proteins that regulate lymphocyte development and function. Representative genes regulated downstream of defined or undefined signaling pathways are indicated.
RAG DSBs Activate Pro-survival Signals to Direct Assembly of Broad AgR Repertoires
Within each stage of lymphocyte development where V(D)J recombination occurs, cells have a limited lifespan to assemble and express functional AgR genes, which then signal survival and further differentiation. Depending on their development stage, lymphocytes require at least one or two distinct V(D)J recombination events to construct an AgR gene. Since two thirds of V(D)J rearrangements form non-functional genes, most cells require one or two more recombination events. In pre-B cells and DP thymocytes that employ stringent mechanisms to select cells for developmental progression, multiple successive V-to-J rearrangements often are necessary for differentiation of B and T cells [6, 25]. Genetically altering the lifespan of DP thymocytes impacts the number of Tcra V-to-J rearrangements that occur in these cells, thereby regulating the TCRa repertoire of naïve T cells [26]. However, the mechanisms that control the survival of immature lymphocytes assembling AgR genes and the impact of these mechanisms on AgR repertoire are largely unknown. Two recent studies have shown that RAG DSBs activate anti-apoptotic factors to establish “feed-forward loops” that promote cellular survival to facilitate additional V(D)J recombination events.
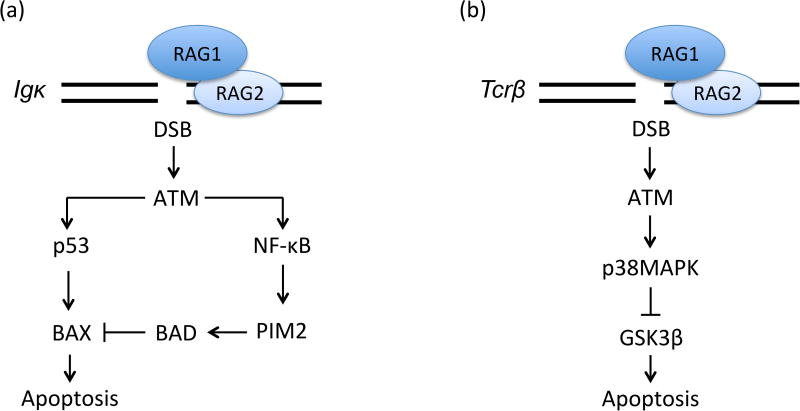
One study set out to determine how pre-B cells survive during V(D)J recombination to generate Igκ+ and Igλ+ B cells [27]. Upon the successful assembly of Igh genes in pro-B cells, IgH proteins complex with surrogate light chains to generate pre-BCRs [9]. Signals from pre-BCRs cooperate with signals from external IL7 cytokine to promote cellular survival, proliferation, and differentiation into large cycling pre-B cells [28]. Attenuation of IL7 signaling, likely by migration of cells away from an IL7-rich microenvironment [29, 30], results in G1 phase cell cycle arrest and loss of pro-survival signals, as well as initiation of Igκ recombination by unopposed pre-BCR signaling [28]. The expression of Igκ protein signals Igκ+ B cell differentiation, while unsuccessful Igκ gene assembly leads to Igλ recombination that, if successful, promotes Ig(&03BB)+ B cell development [31]. It was shown that RAG Igκ DSBs promote p53-mediated transcriptional activation of the pro-apoptotic BAX protein, yet paradoxically also prolong the survival of pre-B cells. Consistent with prior data that RAG DSBs signal ATM/NFicB-dependent up-regulation of transcripts encoding the Pim2 pro-survival and pro-proliferation kinase [20], it was demonstrated that RAG Igκ DSBs signal ATM-dependent induction of Pim2 protein expression. This increased Pim2 expression causes Pim2-dependent phosphorylation of the proapoptotic BAD protein, which blocks p53/BAX-mediated apoptosis (Figure 2A). Accordingly, Pim2-deficient pre-B cells attempting to assemble Igκ genes exhibit increased apoptosis. Considering that Pim2-deficient mice generate reduced numbers of Igλ+ B cells, these data helped to formulate a model wherein RAG Igκ DSBs signal via ATM, NFκB, and Pim2 to increase the lifespan of pre-B cells, thereby facilitating Igλ recombination and promoting Igλ+ B cell development. Subsequent data that Igλ+ B cell numbers are diminished in Nemo-deficient mice, but normal in ATM-deficient mice, raised questions about this model [32]. However, ATM has additional functions whose absence may have masked a role for RAG DSBs in stimulating production of Igλ+ B cells. The resolution of this issue may require creation and analysis of mice expressing a Nemo mutant that cannot be phosphorylated by ATM.
Figure 2. RAG DSBs Activate Signals that Prolong Survival of Immature Lymphocytes Assembling AgR Genes.
A. RAG Igκ DSBs induced in pre-B cells signal via ATM/p53 to transcriptionally activate the pro-apoptotic BAX protein and via ATM/NFκB to transcriptionally activate the Pim2 kinase. Pim2 promotes phosphorylation and inactivation of the pro-apoptotic BAD protein, which blocks BAX-induced cell death. B. RAG DSBs induced at Tcrb loci in DN thymocytes promote ATM-mediated phosphorylation and activation of p38MAPK, which in turn phosphorylates and inactivates the pro-apoptotic GSK3p kinase.
Another study sought to determine if the response to RAG DSBs includes pro-survival signals that protect immature lymphocytes [33]. GSK3β is a constitutively active kinase that induces cell death. Since GSK3p is ubiquitous, cellular survival requires mechanisms that limit its activity [34]. It had been shown that RAG DSBs induced in DN thymocytes activate p38MAPK [35], and this kinase can phosphorylate GSK3p to inhibit its kinase activity [36]. Thus, they measured phospho-GSK3p in thymocytes, detecting high levels within developmental stages where Tcrb (DN3 stage) and Tcra (DP stage) genes assemble [33]. The levels of phospho-GSK3p were reduced in RAG-deficient DN3 cells. Consistent with activation of p38MAPK by ATM [33], phospho-GSK3p levels were reduced in ATM-deficient DN3 thymocytes. To elucidate the role of ATM/p38MAPK-dependent phosphorylation of GSK3p, GSK3p-KI mice expressing a GSK3p mutant that cannot be modified by ATM were generated. The assembly and expression of functional Tcrb genes in G1 phase DN3 thymocytes drives rapid proliferation as cells differentiate into DN4 thymocytes [10]. GSK3p-KI mice had a lower frequency of DN3 thymocytes expressing TCRβ proteins and diminished numbers of DN4 cells. On each Tcrb allele, primary Vβ (Trbv) rearrangements can occur to DJp1 or DJp2 complexes, while secondary Trbv rearrangements only can occur to DJp2 complexes [37]. The frequencies of Trbv16 and Trbv19 rearrangements to DJp2 complexes are reduced in DN thymocytes of GSK3p-KI mice. Collectively, these data suggest that RAG Tcrb DSBs inactivate GSK3p to prolong the survival of DN3 thymocytes, thereby increasing the chance that cells with nonfunctional VpDJp1 rearrangements re-initiate Trbv rearrangements in another attempt to form TCRβ proteins (Figure 2B). Notably, the numbers of DP thymocytes and naive VaJa repertoires are normal in GSK3p-KI mice, implying that restraining GSK3p is not essential for promoting survival of all lymphocytes assembling AgR genes.
Neither of these studies assessed the role of RAG DSBs in promoting cellular survival of both immature B and T cells. It will be important to ascertain if RAG DSBs promote cellular survival by triggering mechanisms that are redundant (Pim2- and GSK3p-mediated), lineage-specific (Pim2 in B and GSK3β in T), and/or developmental stage specific (GSK3p in pro-B/T cells and Pim2 in pre-B/T cells).
RAG DSBs Signal Feedback Inhibition of V(D)J Recombination
In 1965, it was discovered that each rabbit B lymphocyte expresses IgH protein from only one allele, with both alleles represented equally within the B cell population [38]. Such allelic exclusion of Igh, Igk, and Tcrb loci appears conserved among organisms that assemble lymphocyte AgR genes through V(D)J recombination [39-41]. While this conservation suggests that allelic exclusion is important, the purpose for enforcing mono-allelic expression of Igh, Igk, and Tcrb loci remains enigmatic. Models include ensuring negative selection of self-reactive lymphocytes to suppress autoimmunity and facilitating antigen recognition and robust activation and clonal expansion of lymphocytes in response to antigens [10, 40, 41]. Since the 1980s, it has been appreciated that AgR allelic exclusion is achieved by asynchronous initiation of the V-to-(D)J recombination step between alleles and the ability of AgR proteins expressed from one allele to signal permanent feedback inhibition of V rearrangements on the other allele [39-43]. In 1980, it was hypothesized that V(D)J recombination events on one allele transiently feedback inhibit V rearrangements on the other allele [42]. Over the past several years, two studies have revealed that RAG DSBs signal ATM-dependent transient feedback inhibition of V recombination by modulating the expression of lymphocyte-specific proteins that regulate AgR gene assembly. These studies, along with three subsequent papers, have formulated a novel perspective for the relevance of AgR allelic exclusion.
To address this question one study sought to determine whether RAG DSBs induced on one Igκ allele signal via ATM to suppress RAG cleavage of the other allele, thereby helping regulate Igκ allelic exclusion [44]. It had previously been shown that immature B cells of ATM-deficient mice have increased frequencies of bi-allelic RAG cleavage at Igh and Igκ loci [45]. Also circumstantial evidence existed that RAG cleavage of one Igκ allele may trigger ATM-dependent inhibition of VK recombination on the other allele [45]. Thus, this study first monitored allele-specific Igκ protein expression in B cells of wildtype and ATM-deficient mice, finding that the loss of ATM resulted in a modest increase in the fraction of cells with bi-allelic IgK expression [44]. This less stringent Igκ allelic exclusion could arise from the absence of signals that regulate Igκ recombination between alleles and/or impaired DSB repair delaying IgK-signaled permanent feedback inhibition from one allele until after Igκ recombination initiates on the second allele. Upon the induction of Igκ recombination in pre-B cells incapable of repairing RAG DSBs, RAG cleavage at ∼50% of Igκ alleles within the cellular population was found. Notably, genetic or pharmacological inactivation of the ATM kinase leads to RAG cleavage on nearly 100% of Igκ alleles. These data provided firm evidence that RAG cleavage of one Igκ allele signals ATM-dependent feedback inhibition of Igκ recombination on the other allele. As DSB repair inactivates ATM, this feedback inhibition would be transient, terminating after completion of Igκ recombination on the first allele.
The ATM-dependent mechanism by which RAG cleavage at one Igκ allele prevents initiation of VK recombination at the other allele was then investigated. It was hypothesized that inactivation of RAG may be one mechanism. Indeed, it was found that Rag1, Rag2, and Gadd54α transcripts and Rag1 and Gadd45a protein each decrease in an ATM-dependent manner after induction of RAG Igκ DSBs [44]. Gadd45a had been demonstrated to induce Rag1 and Rag2 transcription in a single pre-B cell line [46]. Thus, this suggested that RAG Igκ DSBs signal ATM-dependent loss of Gadd45a, leading to transcriptional repression of RAG1 expression. However, it was shown that constitutive expression of Gadd45a in primary pre-B cells does not antagonize DSB-induced repression of RAG1 expression [47]. Moreover, it was also found that IR downregulates Rag1 and Rag2 transcripts via mechanisms dependent on Nemo [47], consistent with a role for repression of Rag1 and Rag2 transcription by the p50 NFκB protein [48]. Furthermore, another study showed that DSBs induced by IR or genotoxic drugs downregulate Rag1 and Rag2 transcripts concomitant with ATM-dependent loss of FOXO1 transcription factor binding to the Rag1/Rag2 locus Erag transcriptional enhancer [49]. Notably, the loss of RAG1 expression in response to DSBs induced by genotoxic drugs correlates with inhibition of V(D)J recombination within retroviral substrates and at Igκ loci [47, 49]. Collectively, these observations are consistent with RAG cleavage of one Igκ allele transiently blocking initiation of V(D)J recombination at both Igκ alleles via distinct ATM-dependent signaling pathways that repress Rag1 and Rag2 transcription (Figure 3). Additionally it has been shown that IR downregulates Rag1 and Rag2 transcripts in pro-B and pro-T cells and ATM-deficiency increases bi-allelic V recombination and allelic inclusion for Igh and Tcrb loci [47, 50]. These findings are consistent with a RAG DSB-induced feedback inhibition coordinating initiation of V(D)J recombination between alleles at Igh and Tcrb loci via ATM-dependent repression of RAG expression. However, an unequivocal conclusion rests on inactivating signals by which ATM inhibits Rag1 and Rag2 transcription without affecting mechanisms by which ATM repairs DSBs. Moreover, considering that IR had no effect upon Rag1 or Rag2 transcripts in pre-T cells, it will be important to determine whether DSBs repress RAG expression via lymphocyte developmental stage- and/or lineage- specific mechanisms.
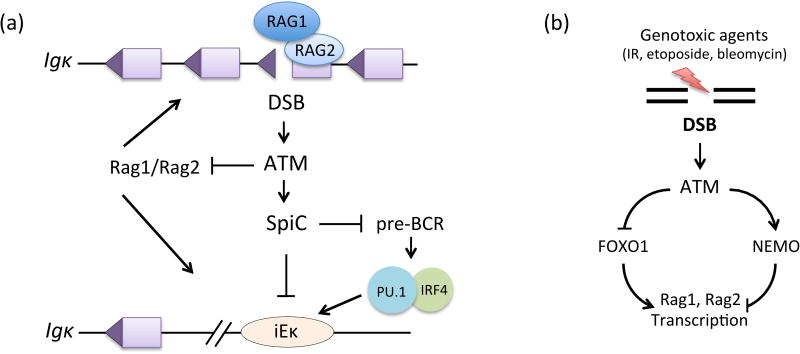
Figure 3. DSBs Signal to Inhibit V(D)J Recombination.
A. RAG DSBs induced at Igκ loci in pre-B cells signal via ATM to transiently feedback inhibit further initiation of Igκ recombination. ATM signals transcriptional repression of RAG expression and transcriptional up-regulation of the SpiC transcriptional repressor. SpiC antagonizes Igκ recombination potential by displacing PU.1 from the Igκ intronic enhancer (iEκ) and by inhibiting pre-BCR signals, which activate Igκ transcription via PU.1/IRF4 binding to iEκ. B. DSBs induced in pre-B cells by genotoxic agents signal via distinct ATM-dependent pathways to repress transcription of Rag1 and Rag2.
Additionally another study investigated whether RAG DSB-induced feedback inhibition might prevent V(D)J recombination through mechanisms in addition to down-regulation of RAG [51]. This study found that RAG Igκ DSBs induced in pre-B cells trigger ATM-dependent increases in the expression of RELB and NFκB2, which are non-canonical NFκB transcription factors. Gene expression profiling of NFκB2-deficient cells revealed that some genes whose transcripts are increased by RAG Igκ DSBs require both ATM and NFκB2 expression (Figure 1, Key Figure). One of these genes encodes the SpiC transcriptional repressor that recognizes the same DNA motif as the PU.1 transcription factor [52], which stimulates Igκ transcription and recombination [53]. It was shown that ectopic expression of SpiC interferes with binding of PU.1 at the Igκ intronic enhancer and inhibits Igκ transcription and recombination. SpiC over-expression had been shown to block differentiation of pre-B cells and to inhibit BCR signaling [51]. Since BCR and pre-BCR employ similar pathways, it seemed plausible that induction of SpiC also might inhibit Igκ recombination by inhibiting pre-BCR signals. Indeed, pre-B cells with RAG Igκ DSBs exhibit decreased pre-BCR signaling and lower expression of the SYK and BLNK proteins that transduce pre-BCR signals. Moreover, SpiC interferes with binding of PU.1 at Syk and Blnk transcriptional regulatory elements and represses expression of Syk and Blnk transcripts. These data indicate that, at least for IgκHoci, RAG DSB-induced feedback inhibition also might transiently block further V(D)J recombination at both Igκ alleles through distinct ATM-dependent pathways that suppress Igκ recombination potential (Figure 3A).
The discovery of ATM-dependent, DSB-induced feedback inhibition of V(D)J recombination led to a novel perspective for the importance of AgR allelic exclusion [41, 44, 50]. From this it was hypothesized that pressure to protect host organisms from fatal lymphoid malignancies prior to reproduction forced evolution of mechanisms that direct asynchronous V(D)J recombination between alleles of Igh and Tcrb loci. These loci recombine via D-to-J rearrangement and then V-to-DJ rearrangement in G1 phase pro-B/T cells. IgH/TCRp proteins expressed from in-frame VDJ rearrangements drive cells into S phase, where RAG is inactivated, and through many cell cycles as pro-B/T cells differentiate into pre-B/T cells. Asynchronous initiation and feedback inhibition regulates V-to-DJ rearrangements to enforce IgH and TCRβ allelic exclusion. While IgH/TCRβ-feedback inhibition does not prevent D-to-J recombination, at least in pre-B/T cells, DSB feedback inhibition could transiently block these rearrangements in any pro-B/T cells that do not simultaneously initiate D-to-J recombination on both alleles. As IgH/TCRβ proteins from one allele are driving pro-B/T cells through G1 phase, DSB feedback inhibition would suppress D-to-J and V-to-DJ recombination on the other allele to limit the frequency at which RAG DSBs evade the G1/S checkpoint and enter S phase where DSBs are prone to form translocations. Consistent with this notion, it was shown that ATM-deficiency increases the percentages of mature B and T cells harboring translocations of their non-selected Igh or Tcrb loci [50]. Moreover, these translocations depend on the Cyclin D3 protein, which IgH and TCRβ proteins activate to drive pro-lymphocytes through G1 phase [50]. It is important to note that this hypothesis does not exclude other hypotheses for the importance of allelic exclusion, such as preventing autoimmunity. As was previously proposed, RAG Igκ DSB-induced signals may suppress oncogenic Igκ translocations via feedback inhibition of Igκ recombination and activation of pre-B cell specific G1/S checkpoint mechanisms [27, 51]. In the latter context, it was demonstrated that RAG Igκ DSBs trigger G1 arrest by inhibiting pre-BCR proliferative signals and by transcriptional activation of the Pim2 kinase, which halts cell cycle through undefined mechanisms [27, 51]. Finally, the findings of that DSBs caused by genotoxic agents downregulate RAG expression and suppress initiation of V(D)J recombination imply that this lymphocyte-specific DSB response is important to maintain genomic integrity during AgR gene assembly [47, 49].
RAG DNA Cleavage Regulates Development and Function of Innate Immune Cells
A recent study has revealed that the ability of RAG to cleave DNA is vital for normal differentiation and function of natural killer (NK) cells [54]. NK cells are considered innate immune cells since they express germline-encoded antigen receptors and their development does not need RAG expression or V(D)J recombination [55-58]. Paradoxically, NK cells often have V(D)J rearrangements at AgR loci and RAG reporters showed that ∼40% of NK cells develop from RAG-expressing common lymphoid precursors [59-63]. By using reporters, it was found that NK cells lacking active or prior RAG expression are more activated, differentiated, and cytotoxic [54]. Relative to normal NK cells, RAG2-deficient NK cells are more activated, mature, and cytotoxic, yet impaired in ability to expand following viral infection [54, 64]. This study also showed that RAG2-deficient NK cells are more susceptible to apoptosis, exhibit a diminished DSB response, and express lower levels of transcripts encoding DSB repair/response proteins [54]. Consistent with these data, NK cells of mice that express an inactive DSB repair/response protein (DNA-PKcs) exhibit an activated phenotype and reduced survival in response to virus [54]. Moreover, NK cells from mice expressing a cleavage-incompetent RAG endonuclease share phenotypes of NK cells of mice lacking RAG2 or expressing functional DNA-PKcs [54]. Since RAG nicks DNA prior to cleaving DNA, it remains to be determined if RAG elicits these biological effects via single strand breaks, DSBs, or both. It was also shown that a history of RAG expression endows T cells with more transcripts from genes encoding DSB repair/response proteins, a more robust DSB response, and increased survival following activation by antigen. Finally, this study showed that RAG expression during ontogeny enhances DSB responses of mature innate lymphoid cells [54]. Based on their data, the authors proposed that RAG cleavage within the genomes of common lymphoid precursors endows adaptive and innate lymphocytes with a “fitness” that promotes their survival when proliferating in response to antigen [54]. Their findings introduce the innovative concept that developmentally programmed RAG cleavage of DNA induces heritable gene expression changes (independent of AgR gene assembly) that enhance the protective function of diverse lymphocytes [54].
Other Programmed DSBs Regulate Tissue-Specific Gene Expression and Differentiation
Additional studies over the past decade have revealed that programmed induction of DSBs by factors other than RAG can signal changes in expression of genes that regulate tissue-specific cellular function and/or differentiation. In response to antigen, mature B cells proliferate, induce expression of the AID protein to generate DSBs within Igh loci and thereby promote Igh class switch recombination, and differentiate into plasma cells that secrete antibodies [65]. One study showed that AID-dependent DSBs signal via ATM to repress a gene expression program that suppresses plasma cell differentiation [66]. Another study found that AID-mediated DSBs trigger an ATM-dependent positive feedback loop that stimulates AID activity, likely to enhance Igh class switch recombination [67]. In eukaryotes, the gamete-specific Spo11 endonuclease induces DSBs that stimulate pairing, recombination, and segregation of homologous chromosomes during meiosis [68]. Similar to RAG DSB feedback inhibition of V(D)J recombination, Spo11 DSBs signal via ATM to downregulate Spo11 expression and alter the genomic distribution of Spo11 DSBs [69, 70]. While it had been known for decades that cellular viability depends on the ability of topoisomerases to relieve DNA torsional stress from replication and transcription [71], several studies this past decade have revealed that regulated induction of topoisomerase DSBs at certain promoters stimulates transcription of genes that control tissue-specific functions [72-74]. The Caspase 3 protease promotes differentiation of many diverse cell types [75]. At least in skeletal muscle cells, Caspase 3 drives differentiation via the caspase-activated DNase (CAD), which creates DSBs throughout the nucleus and at the p21 promoter to increase p21 expression [76]. It remains to be determined whether DSBs induced by CAD, or topoisomerases, also regulate transcription of genes located away from sites of DNA cleavage. In response to infection, macrophages produce reactive oxygen species (ROS) that kill bacteria and induce DSBs to trigger gene expression changes that activates other immune cells [77]. Notably, unlike DSBs that promote differentiation of other cell types, these DSBs require signals from macrophage interferon receptors to drive macrophage-specific gene expression changes. Collectively, data from these studies and analyses of lymphocyte responses to RAG DNA cleavage have cemented the concept that DSBs function as signaling intermediates that control gene expression programs that regulate the differentiation and function of many, if not all, mammalian cell types.
Concluding Remarks
The unexpected discoveries that RAG DNA cleavage has important physiological roles beyond serving as an essential step in catalyzing AgR gene assembly raise important mechanistic questions (see Outstanding Questions) that have broad implications for basic research and the screening, diagnosis, and treatment of immunological disease. Undoubtedly, future immunology research will aim to answer these and related questions. Elucidating mechanisms by which RAG DSBs induced in distinct lymphocyte lineages and developmental stages signal transient and heritable gene expression changes should identify genes whose mutation causes lymphoid malignancies, autoimmunity, or immunodeficiency. In this context, it will be of interest to determine if non-endonuclease activities of the RAG proteins, such as the ubiquitin ligase of RAG1 [78, 79], coordinate AgR gene assembly with cell fate decisions to limit potential hazardous consequences of V(D)J recombination. It also will be important to evaluate if unique features of AgR loci, such as the distance between or chromatin environment over gene segments, serve to amplify or activate tissue-specific aspects of the cellular DSB response to signal changes in gene expression. Acquired knowledge of the precise mechanisms by which RAG DSBs signal has the potential to identify novel means to treat immunological diseases, such as genomic editing to fix mutations in lymphoid precursors or mature lymphocytes.
Outstanding Questions.
In addition to ATM, what proximal factor(s) do RAG DSBs engage to signal gene expression changes? Do RAG DSBs employ the intrinsic ubiquitin ligase of RAG1, the protein kinase that binds RAG1, and/or the ubiquitin ligase that associates with this kinase?
During V(D)J recombination of different AgR loci, do RAG DSBs stimulate cellular survival by mechanisms that are general, lymphocyte-specific, lymphocyte lineage-specific, lymphocyte developmental stage-specific? Do RAG DSBs induced at Igh, Tcrb, and/or Tcra loci activate Pim2 to increase cellular survival and modulate repertoires of V(D)J rearrangements?
Does activation of SpiC help control Igic receptor editing, suppress Igic translocations, and/or enforce Igtc allelic exclusion? Do RAG Igtc DSB-signaled activation of SpiC and inactivation of RAG cooperate to control Igκ receptor editing, suppress Igκ translocations, and/or enforce Igκ allelic exclusion
Do RAG DSBs induced at Igh and/or Tcrb loci utilize mechanisms in addition to repression of RAG expression to coordinate initiation of V(D)J recombination between alleles? Do RAG DSBs induced at Igh and/or Tcrb mediate transient feedback inhibition of V(D)J recombination through activation of the SpiC transcriptional repressor?
Is the increased AgR allelic exclusion observed in ATM-deficient mice due to loss of ATM functions in promoting DSB repair and/or in signaling RAG DSB-induced feedback inhibition?
Do nicks and/or DSBs induced by RAG in common lymphoid progenitors endow “fitness” on descendant lymphocytes? Does RAG cleavage during non-functional V(D)J rearrangements at AgR loci in common lymphoid progenitors impart this “fitness”? Is RAG directed to cleave at a specific non-AgR locus in common lymphoid progenitors? How does RAG cleavage within the genome of common lymphoid progenitors promote heritable changes in gene expression?
Does the ability of 2 RAG Igκ DSBs to activate DSB responses of similar magnitude as ∼100 DSBs induced throughout the genome by genotoxic agents reflect roles for the RAG proteins and/or features of AgR loci amplify DSB responses?
Trends.
RAG DSBs induced during AgR gene assembly signal through distinct pathways to activate a broad multifunctional gene transcription program that transcends the conserved cellular DSB response.
RAG DSBs generated during V(D)J recombination activate pro-survival signals that prolong the lifespan of immature lymphocytes and to ensure assembly of broad AgR gene repertoires.
RAG DSBs introduced during AgR gene assembly signal through distinct ATM-dependent mechanisms to transiently feedback inhibit further initiation of V(D)J recombination.
RAG cleavage within the genomes of common lymphoid progenitors promote heritable gene expression changes that increase the survival of innate and adaptive immune cells proliferating in response to antigen.
Acknowledgments
We acknowledge that space limitations prevented us from citing numerous important papers of our respected colleagues. The work on this review was supported by NIH R01 grant AI112621 (C.H.B).
Glossary
- AgR
Antigen Receptor refers to a surface protein on lymphocytes that recognizes and binds foreign molecules to initiate an immune response
- ATM
Ataxia Telangiectasia Mutated is a serine-threonine protein kinase that is the master regulator of the cellular response to DNA double strand breaks
- BCR
B Cell Receptor refers to the unique antigen receptor expressed on each B lymphocyte
- DN
Double Negative refers to the lack of two proteins (CD4 and CD8) expressed on immature T cells of later developmental stages
- DN3
Stage 4 Double Negative thymocytes in which Tcrb gene assembly is completed
- DN4
Stage 4 Double Negative thymocytes completing TCRβ selection and transitioning to DP0 stage
- DNA-PKcs
DNA-dependent Protein Kinase catalytic subunit is a serine-threonine protein that functions to repair DNA double strand breaks
- DP
Double Positive refers to the presence of two surface proteins (CD4 and CD8) expressed at later stages of T cell development
- DSBs
Double Strand Breaks are genomic lesions where both DNA strands are broken
- GSK3p
Glycogen Synthase Kinase-3 p isoform is a serine-threonine kinase that regulates cellular metabolism and survival
- Ig
Immunoglobulin refers to one of the protein subunits of B cell antigen receptors
- MAPK
Mitogen Activated Protein Kinase refers to a family of serine-threonine kinases that direct cellular responses to diverse stimuli
- Nemo
NF-κB Essential Modifier is a scaffolding protein that activates the kinases that activate NF-κB transcription factors
- NF-κB
Nuclear Factor kappa-light-chain-enhancer of activated B cells refers to a transcription factor complex that was identified by its role in regulating Igκ transcription
- NK
Natural Killer is a type of immune cell that helps protect from viral infections
- Pim2
Proviral Integrations of Moloney virus 2 is serine/threonine kinase that regulates cellular survival and proliferation
- Pre-B Cell
Precursor B Cell refers to cells of a late stage of B cell development where Igκ and IgA genes are assembled
- Pre-BCR
Pre B Cell Receptor refers to a complex of IgH proteins and surrogate light chains that drives early B cell development upon assembly and expression of a functional Igh gene
- Pre-T Cell
Precursor T Cell refers to cells of a late stage of T cell development where Tcrα genes are assembled
- Pro-B Cell
Progenitor B Cell refers to cells of an early stage of B cell development where Igh genes are assembled
- Pro-T Cell
Progenitor T Cell refers to cells of an early stage of T cell development where Tcrb genes are assembled
- RAG
Recombination Activating Gene refers to the RAG1/RAG2 endonuclease that catalyzes the assembly of lymphocyte antigen receptor genes
- TCR
T Cell Receptor refers to the unique antigen receptor expressed on each T lymphocyte
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 3.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 4.Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alt FW, et al. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemazee D. Receptor editing in lymphocyte development and central tolerance. Nat Rev Immunol. 2006;6:728–740. doi: 10.1038/nri1939. [DOI] [PubMed] [Google Scholar]
- 7.Grandien A, et al. Negative selection of multireactive B cell clones in normal adult mice. Eur J Immunol. 1994;24:1345–1352. doi: 10.1002/eji.1830240616. [DOI] [PubMed] [Google Scholar]
- 8.Pelanda R, Torres RM. Central B-cell tolerance: where selection begins. Cold Spring Harb Perspect Biol. 2012;4:a007146. doi: 10.1101/cshperspect.a007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melchers F. Checkpoints that control B cell development. J Clin Invest. 2015;125:2203–2210. doi: 10.1172/JCI78083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Boehmer H, Melchers F. Checkpoints in lymphocyte development and autoimmune disease. Nat Immunol. 2010;11:14–20. doi: 10.1038/ni.1794. [DOI] [PubMed] [Google Scholar]
- 11.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassing CH, Alt FW. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin WC, Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc Natl Acad Sci U S A. 1994;91:2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Desiderio S. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity. 1999;11:771–781. doi: 10.1016/s1074-7613(00)80151-x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, et al. Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis. Immunity. 2011;34:163–174. doi: 10.1016/j.immuni.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lescale C, et al. RAG2 and XLF/Cernunnos interplay reveals a novel role for the RAG complex in DNA repair. Nat Commun. 2016;7:10529. doi: 10.1038/ncomms10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee GS, et al. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 19.Deriano L, et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bredemeyer AL, et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 22.Perkins EJ, et al. Sensing of intermediates in V(D)J recombination by ATM. Genes Dev. 2002;16:159–164. doi: 10.1101/gad.956902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 24.Wu ZH, et al. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 25.Huang CY, et al. Revision of T cell receptor {alpha} chain genes is required for normal T lymphocyte development. Proc Natl Acad Sci U S A. 2005;102:14356–14361. doi: 10.1073/pnas.0505564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo J, et al. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 27.Bednarski JJ, et al. RAG-induced DNA double-strand breaks signal through Pim2 to promote pre-B cell survival and limit proliferation. J Exp Med. 2012;209:11–17. doi: 10.1084/jem.20112078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark MR, et al. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol. 2014;14:69–80. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson K, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Tokoyoda K, et al. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Gorman JR, Alt FW. Regulation of immunoglobulin light chain isotype expression. Adv Immunol. 1998;69:113–181. doi: 10.1016/s0065-2776(08)60607-0. [DOI] [PubMed] [Google Scholar]
- 32.Derudder E, et al. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nat Immunol. 2009;10:647–654. doi: 10.1038/ni.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thornton TM, et al. Inactivation of nuclear GSK3beta by Ser(389) phosphorylation promotes lymphocyte fitness during DNA double-strand break response. Nat Commun. 2016;7:10553. doi: 10.1038/ncomms10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs KM, et al. GSK-3beta: A Bifunctional Role in Cell Death Pathways. Int J Cell Biol. 2012;2012:930710. doi: 10.1155/2012/930710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedraza-Alva G, et al. Activation of p38 MAP kinase by DNA double-strand breaks in V(D)J recombination induces a G2/M cell cycle checkpoint. EMBO J. 2006;25:763–773. doi: 10.1038/sj.emboj.7600972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thornton TM, et al. Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science. 2008;320:667–670. doi: 10.1126/science.1156037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khor B, Sleckman BP. Intra- and inter-allelic ordering of T cell receptor beta chain gene assembly. Eur J Immunol. 2005;35:964–970. doi: 10.1002/eji.200425806. [DOI] [PubMed] [Google Scholar]
- 38.Pernis B, et al. Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J Exp Med. 1965;122:853–876. doi: 10.1084/jem.122.5.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mostoslavsky R, et al. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118:539–544. doi: 10.1016/j.cell.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 40.Vettermann C, Schlissel MS. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev. 2010;237:22–42. doi: 10.1111/j.1600-065X.2010.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brady BL, et al. Antigen receptor allelic exclusion: an update and reappraisal. J Immunol. 2010;185:3801–3808. doi: 10.4049/jimmunol.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alt FW, et al. Activity of multiple light chain genes in murine myeloma cells producing a single, functional light chain. Cell. 1980;21:1–12. doi: 10.1016/0092-8674(80)90109-9. [DOI] [PubMed] [Google Scholar]
- 43.Alt FW, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinel NC, et al. The ataxia telangiectasia mutated kinase controls Igkappa allelic exclusion by inhibiting secondary Vkappa-to-Jkappa rearrangements. J Exp Med. 2013;210:233–239. doi: 10.1084/jem.20121605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hewitt SL, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fisher MR, et al. Immature Lymphocytes Inhibit Rag1 and Rag2 Transcription and V(D)J Recombination in Response to DNA Double-Strand Breaks. J Immunol. 2017;198:2943–2956. doi: 10.4049/jimmunol.1601639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verkoczy L, et al. A role for nuclear factor kappa B/rel transcription factors in the regulation of the recombinase activator genes. Immunity. 2005;22:519–531. doi: 10.1016/j.immuni.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochodnicka-Mackovicova K, et al. The DNA Damage Response Regulates RAG1/2 Expression in Pre-B Cells through ATM-FOXO1 Signaling. J Immunol. 2016;197:2918–2929. doi: 10.4049/jimmunol.1501989. [DOI] [PubMed] [Google Scholar]
- 50.Steinel NC, et al. The ataxia telangiectasia mutated and cyclin D3 proteins cooperate to help enforce TCRbeta and IgH allelic exclusion. J Immunol. 2014;193:2881–2890. doi: 10.4049/jimmunol.1302201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bednarski JJ, et al. RAG-mediated DNA double-strand breaks activate a cell type-specific checkpoint to inhibit pre-B cell receptor signals. J Exp Med. 2016;213:209–223. doi: 10.1084/jem.20151048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batista CR, et al. PU.1 Regulates Ig Light Chain Transcription and Rearrangement in Pre-B Cells during B Cell Development. J Immunol. 2017;198:1565–1574. doi: 10.4049/jimmunol.1601709. [DOI] [PubMed] [Google Scholar]
- 53.Schwarzenbach H, et al. Involvement of the Ets family factor PU.1 in the activation of immunoglobulin promoters. J Biol Chem. 1995;270:898–907. doi: 10.1074/jbc.270.2.898. [DOI] [PubMed] [Google Scholar]
- 54.Karo JM, et al. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell. 2014;159:94–107. doi: 10.1016/j.cell.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goh W, Huntington ND. Regulation of Murine Natural Killer Cell Development. Front Immunol. 2017;8:130. doi: 10.3389/fimmu.2017.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondo M, et al. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 57.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 58.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 59.Borghesi L, et al. B lineage-specific regulation of V(D)J recombinase activity is established in common lymphoid progenitors. J Exp Med. 2004;199:491–502. doi: 10.1084/jem.20031800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Welner RS, et al. Asynchronous RAG-1 expression during B lymphopoiesis. J Immunol. 2009;183:7768–7777. doi: 10.4049/jimmunol.0902333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fronkova E, et al. Lymphoid differentiation pathways can be traced by TCR delta rearrangements. J Immunol. 2005;175:2495–2500. doi: 10.4049/jimmunol.175.4.2495. [DOI] [PubMed] [Google Scholar]
- 62.Lanier LL, et al. Expression of cytoplasmic CD3 epsilon proteins in activated human adult natural killer (NK) cells and CD3 gamma, delta, epsilon complexes in fetal NK cells. Implications for the relationship of NK and T lymphocytes. J Immunol. 1992;149:1876–1880. [PubMed] [Google Scholar]
- 63.Pilbeam K, et al. The ontogeny and fate of NK cells marked by permanent DNA rearrangements. J Immunol. 2008;180:1432–1441. doi: 10.4049/jimmunol.180.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrews DM, Smyth MJ. A potential role for RAG-1 in NK cell development revealed by analysis of NK cells during ontogeny. Immunol Cell Biol. 2010;88:107–116. doi: 10.1038/icb.2009.94. [DOI] [PubMed] [Google Scholar]
- 65.Methot SP, Di Noia JM. Molecular Mechanisms of Somatic Hypermutation and Class Switch Recombination. Adv Immunol. 2017;133:37–87. doi: 10.1016/bs.ai.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 66.Sherman MH, et al. AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling. Mol Cell. 2010;39:873–885. doi: 10.1016/j.molcel.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vuong BQ, et al. A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat Immunol. 2013;14:1183–1189. doi: 10.1038/ni.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 69.Lange J, et al. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell. 2016;167:695–708 e616. doi: 10.1016/j.cell.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lange J, et al. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pommier Y, et al. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 73.Abramson J, et al. Aire's partners in the molecular control of immunological tolerance. Cell. 2010;140:123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 74.Madabhushi R, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fernando P, Megeney LA. Is caspase-dependent apoptosis only cell differentiation taken to the extreme? FASEB J. 2007;21:8–17. doi: 10.1096/fj.06-5912hyp. [DOI] [PubMed] [Google Scholar]
- 76.Larsen BD, et al. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci U S A. 2010;107:4230–4235. doi: 10.1073/pnas.0913089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morales AJ, et al. A type I IFN-dependent DNA damage response regulates the genetic program and inflammasome activation in macrophages. Elife. 2017;6 doi: 10.7554/eLife.24655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jones JM, Gellert M. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc Natl Acad Sci U S A. 2003;100:15446–15451. doi: 10.1073/pnas.2637012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yurchenko V, et al. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 2003;17:581–585. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]