Abstract
The 7SK small nuclear ribonucleoprotein (snRNP) plays a central role in RNA polymerase II elongation control by regulating the availability of active P-TEFb. We optimized conditions for analyzing 7SK RNA by SHAPE and demonstrated a hysteretic effect of magnesium on 7SK folding dynamics including a 7SK GAUC motif switch. We also found evidence that the 5΄ end pairs alternatively with two different regions of 7SK giving rise to open and closed forms that dictate the state of the 7SK motif. We then used recombinant P-TEFb, HEXIM1, LARP7 and MEPCE to reconstruct a functional 7SK snRNP in vitro. Stably associated P-TEFb was highly inhibited, but could still be released and activated by HIV-1 Tat. Notably, P-TEFb association with both in vitro-reconstituted and cellular snRNPs led to similar changes in SHAPE reactivities, confirming that 7SK undergoes a P-TEFb-dependent structural change. We determined that the xRRM of LARP7 binds to the 3΄ stem loop of 7SK and inhibits the methyltransferase activity of MEPCE through a C-terminal MEPCE interaction domain (MID). Inhibition of MEPCE is dependent on the structure of the 3΄ stem loop and the closed form of 7SK RNA. This study provides important insights into intramolecular interactions within the 7SK snRNP.
INTRODUCTION
RNA polymerase II (Pol II) elongation control is the nexus of gene regulation. After initiation, Pol II becomes paused downstream of the transcription start site due to the influence of negative transcription factors that include DSIF and NELF (1,2). These promoter-proximal paused polymerases are present on essentially all expressed human genes (1). The transition of paused polymerases into productively elongating complexes is required to produce mature mRNA and is tightly regulated (3–5). This regulation is achieved, in part, by the control of the positive elongation factor P-TEFb. P-TEFb is a heterodimeric complex consisting of Cyclin T1 and cyclin dependent kinase Cdk9 (6,7). Targets for the kinase activity of P-TEFb include the carboxyl-terminal domain (CTD) of the large subunit of Pol II and NELF, but it is likely that the Spt5 subunit of DSIF is the main functional target (8–13). Phosphorylation of DSIF, which remains with the elongation complex, leads to loss of NELF and the subsequent incorporation of the PAF1 complex (14). Once the transition into productive elongation has occurred, Pol II transcribes the rest of the gene at a rate of about 4 kb/min (15,16).
Because so many genes contain Pol II in the paused state, the activity of P-TEFb must be properly regulated. Loss of control of P-TEFb leads to rapid productive elongation and aberrant gene expression (1,17,18). The activity of P-TEFb is restrained by the hexamethylene bisacetamide inducible protein HEXIM1 or HEXIM2 (19–22). HEXIM1 directly interacts with the active site in Cdk9 through the PYNT motif and inhibits the kinase activity of P-TEFb (23,24). Availability of the inhibitory motif in HEXIM1 is regulated by a conformational change in the protein upon binding to 7SK RNA (25). HEXIM1 exists as a homo-dimer and binds to 7SK through its KHRR repeats (21,26,27). The binding site of HEXIM1 on 7SK has been reported to a GAUC repeat in the 5΄ stem loop (25,28). HEXIM1 association with double stranded RNA exposes the PYNT motifs of each monomer and allows each to inhibit one molecule of P-TEFb (21,23). A core function of the 7SK small nuclear ribonucleoprotein (snRNP) complex is to mediate this HEXIM1-P-TEFb protein–protein interaction.
Human 7SK is a 332 nucleotide RNA transcribed by RNA polymerase III with a dynamic protein composition. The RNA is highly structured and forms the scaffold for protein interactions that ultimately regulate P-TEFb (29). After transcription by Pol III, 7SK is chaperoned by genuine La until it is transferred to the La related protein LARP7 (30–33). LARP7 contains an amino terminal La antigen motif (LAM) that binds and protects the 3΄ terminal U residues as well as a C-terminal xRRM that binds to the 3΄ stem loop structure of 7SK, required for 7SK snRNP stability (34–36). The 5΄ end of 7SK is protected by a γ-mono methyl cap added by the methyl-phosphate capping enzyme MEPCE (37–39). MEPCE remains associated with the 7SK snRNP complex after methylation, which may be due to the direct interaction with LARP7 (40,41). P-TEFb must be released by cellular or viral factors to facilitate productive elongation, and after release, there is a rearrangement of the 7SK snRNP (25,42). Loss of P-TEFb results in a loss of HEXIM1 binding. It has been hypothesized that the loss of HEXIM1 allows the association of heterogeneous ribonucleoproteins (hnRNPs) that in turn stabilize an alternative 7SK structure (32). LARP7 is always associated with 7SK RNA and MEPCE can be associated with complexes containing or lacking P-TEFb. Moreover, with the release of P-TEFb, 7SK also changes sensitivity to base modifying chemicals indicating a possible change in structure of the RNA (25). The cellular mechanism of release of P-TEFb is currently unknown, but the bromodomain protein Brd4 has been shown to be involved (43–45). The super elongation complex scaffolding proteins AFF4 and AFF1 are also involved in the regulation of P-TEFb at promoters and enhancers (46–49). The phosphatase PPM1G has been shown to bind to HEXIM1 and 7SK, thereby blocking the association of P-TEFb with the 7SK snRNP (50). In addition, the HIV-1 trans-activating protein Tat associates with P-TEFb (51) and leads to its release from the 7SK snRNP (52,53). Tat interacts intimately with P-TEFb and competes with 7SK RNA for P-TEFb binding (54). This interaction brings P-TEFb to the HIV genome through Tat interaction with HIV TAR RNA resulting in rapid viral gene expression. Through an unknown pathway, the 7SK snRNP is able to regain HEXIM1 and P-TEFb in the complex completing the 7SK snRNP cycle.
Because there are multiple forms of the 7SK snRNP in cells and due to limitations in some of the methods used in previous studies, a number of questions remain unanswered. The structure of the 7SK RNA that acts as a scaffold has not been determined for individual snRNP species. There are two published secondary structure models. The first was from Wassarman and Steitz who used chemical modifications of the RNA and RNP to predict structure (55). The second from Marz et al. added evolutionary conservation to the model (56). These models have very different predicted pairing of the 5΄ ends and both of these structures have been used as the basis for RNA experiments. The determination of the binding sites for each associated protein and how the proteins and RNA influence each other is needed. Some studies focused on the RNA using mutational analysis to examine protein interaction sites (30,42,57). Others have pulled out tagged proteins expressed in cells to examine association with 7SK and other proteins of the snRNP (33,41). While very informative, these studies in general did not analyze the entire 7SK snRNP. To examine the complex interactions within the 7SK snRNP we set out to assemble a functional 7SK snRNP in vitro. Using a combination of biochemical techniques, recombinant RNA and proteins, we were able to analyze these interactions in detail.
MATERIALS AND METHODS
Purification of RNA
7SK RNA constructs were cloned in pET21 vector with a DraI cut site at the 3΄ end of the template. Plasmids were digested with DraI for 6 hours before being cleaned up using Zymo DNA Clean and Concentrator columns. RNA was transcribed in vitro using life technologies MEGAscript T7 transcription kit. For radiolabeled 7SK in stoichiometric EMSAs, 1 μl of α-32P-CTP was added during the reaction. For radiolabeled 7SK in non-stoichiometric EMSAs MEGAscript conditions were modified in the following ways: 0.5 mM ATP, GTP and UPT, with 30 μM CTP and 2.5 μl of α-32P-CTP. Resulting RNAs were treated with DNase I before being cleaned up using a MEGAclear Transcription Clean-Up kit from Life Technologies. All purified RNAs were eluted in water and stored at –80°C.
Purification of LARP7, HEXIM1, P-TEFb, MEPCE, Brd4, Tat and DSIF
A LARP7 (PIP7S) construct was obtained from Qiang Zhou (Berkeley), and the coding sequence cloned in frame with an amino-terminal HIS-tag into pET47 vector. Truncations were made using site directed mutagenesis. All LARP7 plasmids were transformed into BL21 star cells, expressed by the addition of 100 μM IPTG, and purified over nickel NTA beads followed by a GE Mono S column. The following proteins were purified as described earlier: FL P-TEFb (Cdk9 and full length Cyclin T1) (6,54), HEXIM1 (58), MEPCE (37), Brd4 (25) and Tat (25). The plasmid containing the dual expression Spt4–Spt5 construct in pET14b vector was a gift from Brian Lewis (NIH) and was created as previously described (59). The protein purification was performed similarly to other proteins except transformation was into Agilent BL21 star RIPL expression cells.
SHAPE analysis
Recombinant RNAs were diluted in water, heated to 65°C for 5 min and snap cooled on ice before being folded. RNA folding was done in RNA folding buffer (25 mM HEPES pH 7.2, 100 mM potassium acetate, 0.1% Triton X-100 and magnesium acetate at indicated concentration) with SUPERase•In at 37°C for 10 min. 1 pmol RNA was reacted with 80 mM benzoyl cyanide (BzCN) in 10% DMSO in a 50 μl reaction. Background reactions were treated with only 10% DMSO. After 5 s of reaction time, RNA was isolated using a Zymo RNA Clean and Concentrator column. If proteins were in the SHAPE reaction, the RNA was separated using TRIzol LS and chloroform and the aqueous phase was applied to a Zymo Clean and Concentrator column. RNA was eluted and diluted to achieve a final concentration of 1 pmol in 10 μl of water.
RNA was reverse transcribed into cDNA fragments using Thermo Fisher SuperScript IV. The sequence of the SHAPE primer used was 5΄-CGCCTCATTTGGATGTGTCTG-3΄. For internal sequencing, a 5΄ 6FAM tag was added and for sample, reactions used a 5΄ HEX tag. Both primers were ordered from IDT. Primers were added to RNA and heated to 65°C for 5 min followed by an annealing step of 2 min at 42°C. Final RT conditions for sample reactions were: 1 pmol RNA, 1× SuperScript IV buffer, 0.5 mM dNTPs, 5 mM DTT, 20 U SUPERase•In, 3 U SuperScript IV enzyme, 250 nM primer in 20 μl. Sequencing RT reactions were similar except that 15 pmol of unmodified RNA was incubated with 10 U enzyme, 750 nM primer and 0.5 mM ddGTP. Reverse transcription reactions were incubated at 42°C for 2 min, followed by 65°C for 10 min, and finally raised to 70°C for 5 min. 1 μl of 4N NaOH was then added and the temperature raised to 95°C for 3 min to degrade the RNA. Individual reactions were EtOH precipitated and re-suspended in Hi-Di formamide (ABI) along with 0.25 pmol of the sequencing reaction product.
Fragmentation analysis of the resulting cDNAs was performed on an ABI 3730 by the Genomics Division of the Iowa Institute of Human Genetics at the University of Iowa. Raw sequencer (.abi) files were analyzed using QuShape V 1.0 software (60). The QuShape workflow was followed with the following adjustments: Smoothing was set to Gaussian with a window size of 3. During signal alignment, some peak matching was done manually to ensure proper sequence alignment. High signal to noise resulted in almost perfect sequence calling; however, minimal manual sequence alignment was required. Reactivity data was exported to Excel where conditional formatting was applied for visualization. A graded color scale with a minimum (black = 0 reactivity), a midpoint (goldenrod = reactivity of 1), and a maximum (yellow = reactivity of 7) was used. Reactivity values of less than –0.2 were colored dark blue. All of the SHAPE data described can be found in a supplementary Excel file. Secondary structure prediction was done using RNAstructure V 5.8.1. The standard protocol was followed and SHAPE data were used to assist the predication (61). The SHAPE slope was 2.6 and the intercept was –0.3. The predicated structure was exported as a dot bracket file and visualized with the forna webserver (62). The colors of the bases were matched to the colors of the excel images.
EMSAs
Optimization of EMSAs was carried out under a variety of conditions described in the Results section. Final optimized reactions contained 1 pmol of 32P-labeled, folded RNA with indicated amounts of proteins in a 12–20 μl reaction containing 20 mM HEPES 7.2, 100 mM potassium acetate, 100 μM magnesium acetate, 1 mM TCEP, 1% Ficoll 400 and 0.05 U/μl SUPERase•In. The samples were incubated at room temperature for 10 min before loading directly into a 4% 0.5× Tris/glycine native gel containing magnesium as specified. Running buffers were 0.5× Tris/glycine with magnesium matching that in the gel. The gels were then dried, exposed to phosphorimaging plates, and a Fuji FL7000 phosphorimager was used to detect incorporated label.
Kinase assays
After heating and snap cooling the RNA as for SHAPE, the RNA and purified proteins were diluted into buffer resulting in the condition of 20 mM HEPES pH 7.2, 100 mM potassium acetate, 5 mM magnesium acetate, 1 mM TCEP, 40 ng/μl BSA and 1 U SUPERase•In. 7SK RNA was added to the reaction first followed by the proteins. Full-length P-TEFb was kept constant at a concentration of 2 nM per reaction (reaction volume of 20 μl) and the concentration of the RNA and other proteins are indicated in the individual experiments. The RNA–protein mixtures were incubated at room temperature for 10 min to equilibrate. Kinase reactions were then started by the addition of 30 μM ATP with 2.5 μCi of γ-32P-ATP and 50–200 nM DSIF as indicated. Reactions were incubated at room temperature and stopped after 10 min with the addition of 5 μl of 5× SDS loading buffer containing EDTA. Samples were then heated at 95°C for 5 min and separated on a 9% SDS gel. Gels were silver stained, dried and exposed to phosphorimaging plates.
Optimized in vitro snRNP assembly
7SK RNA was diluted in water, heated to 65°C for 5 min and snap cooled on ice before being folded. RNA folding was done in RNA folding buffer (25 mM HEPES pH 7.2, 100 mM potassium acetate, 0.1% Triton X-100 and magnesium acetate at indicated concentration) with SUPERase•In at 37°C for 10 min. Stock proteins were diluted such that the final protein buffer conditions were: 25 mM HEPES pH 7.2, 100 mM potassium acetate, 100 μM magnesium acetate, 0.1% Triton X-100 and 1 mM TCEP. Equal molar amounts of proteins were added in the order of HEXIM1, P-TEFb, LARP7 and MEPCE and mixed by flicking. The reactions were incubated at 37°C for 10 min and then subjected to glycerol gradient purification.
Glycerol gradient analysis
5 ml gradients were poured using an FPLC from 45% to 5% glycerol and 20 mM HEPES (7.2), 100 mM potassium acetate, 100 μM magnesium acetate, 1 mM TCEP. 200 μl of in vitro assembled snRNP samples were carefully layered on top and centrifuged for 16 h at 50 000 rpm in an AH-650 swinging bucket rotor in a WX Ultra 80. After spinning, the gradients were fractionated by hand into 300 μl fractions. For western blotted fractions, 20 μl of samples were mixed with 5 μl of 5× SDS loading buffer and analyzed on a 9% SDS gel. Gel was transferred to nitrocellulose membrane using an Owl semi dry transfer system. Primary antibodies were AP anti-LARP7 (32), AP anti-MEPCE (37), Abcam ab28016 anti-HEXIM1, and Santa Cruz sc-484 anti-Cdk9. Secondary antibodies were Sigma A3415 anti-sheep HRP and Sigma A0545 anti-rabbit HRP.
Isolation of cellular 7SK snRNP
HeLa cells were grown in spinner flasks to a concentration of 1 × 106 cells/ml and harvested by centrifugation. The cells were washed once with ice cold PBS before being re-suspended in extraction buffer: 10 mM HEPES 7.6, 10 mM magnesium chloride, 10 mM potassium chloride, 0.5% IGEPAL CA-630, 1 mM EDTA, 1 mM DTT, 0.1% PMSF, 40 U/ml RNaseOUT and cOmplete protease inhibitor. Cells were added to a 7 ml Bellco dounce and extracted on ice for 10 min with intermittent douncing with a loose pestle. Samples were cleared by centrifugation at 45 000 RPM for 1 h in a T-1270 fixed angle rotor. The supernatant was used for the soluble snRNP isolation. The pelleted nuclear material was washed in the extraction buffer once and re-suspended in extraction buffer before being analyzed by SHAPE.
Immunoprecipitations were performed with Abcam ab27963 sheep anti-Cyclin T1 antibody on control lysates and AP anti-LARP7 antibodies on lysates from cells treated with 1 μM flavopiridol for 1 h. For each IP, 100 μl of Protein G Dynabeads were concentrated and washed twice with 200 μl IP wash buffer: 50 mM HEPES 7.2, 100 mM potassium acetate, 100 μM magnesium acetate, 0.02% Tween 20, 20 ng/μl BSA, 0.1 U/μl SUPERase•In and 1 mM TCEP. After the second wash, the beads were resuspended with 24 μg of antibody and incubated at room temperature for 2 h. The antibody-bound beads were then washed twice in 200 μl IP wash buffer and resuspended in 100 μl of cell lysate. After 2 h of incubation at 4°C the beads were washed three times with 200 μl IP wash buffer and resuspended in 90 μl IP SHAPE buffer: 20 mM HEPES 7.2, 100 mM potassium acetate, 100 μM magnesium acetate, 0.01% Tween 20, 20 ng/μl BSA, 0.1 U/μl SUPERase•In and 1 mM TCEP. Resuspended beads were added to new tubes containing 10 μl of 800 mM BzCN or DMSO and allowed to react for 5 s. Pelleted material was added directly to BzCN without any additional steps. After SHAPE reactions, the RNA was isolated with Trizol as previously described.
Methyltransferase assay
Purified proteins and RNA were diluted into buffers to a final reaction condition of: 20 mM HEPES 7.2, 100 mM potassium acetate, 100 μM magnesium acetate, 1 mM TCEP, 20 ng/μl BSA, 1% Ficoll 400 and 0.05 U/μl SUPERase•In. 250 nM 7SK and equal molar MEPCE were used in each reaction and LARP7 was titrated. The reactions were started by the addition of 14C-S-adenosylmethionine (Perkin Elmer) with a final concentration of 25 μM. Reactions were incubated at 37°C for 30 min and stopped with Trizol LS. RNA was cleaned up and concentrated as stated above, and run on a urea PAGE. The gels were dried, exposed to phosphorimaging plates and analyzed.
RESULTS
Optimization of 7SK snRNP assembly in vitro
We wanted to develop conditions for assembly of the 7SK snRNP in vitro that closely matched conditions in cells, but the EMSA and kinase assays needed for analysis of the snRNP have different requirements. Therefore, a large number of parameters were evaluated in the search for appropriate conditions. Before attempting snRNP assembly, P-TEFb kinase assays with DSIF as a substrate, were performed with or without DTT, at varying pH, and over a range of magnesium concentrations. Reducing conditions and pH 7.2 were optimal and increasing the magnesium to from 0.1 to 5 mM increased the activity of P-TEFb (Supplementary Figure S1A). Use of potassium acetate and potassium glutamate resulted in increased activity over potassium chloride and lower salt concentrations were optimal for the kinase assay (Supplementary Figure S1B). We then titrated LARP7, HEXIM1 and MEPCE onto labeled 7SK and examined binding by EMSA in the presence of 10, 100, 300 or 1000 μM MgCl2 in the reactions, gels and running buffers. Increasing magnesium reduced binding and 100 μM was chosen for its specificity (Supplementary Figure S1C). Addition of 5% glycerol to binding reactions increased both specific and non-specific binding of HEXIM1, LARP7 and MEPCE in non-stoichiometric EMSAs (Supplementary Figure S2). 1% Ficoll allowed well defined bands and binding affinities near 30 nM for HEXIM1, 0.3 nM for LARP7, and around 10 nM for MEPCE. Unexpectedly, increasing Ficoll to 3% or 10% was deleterious for binding for all three proteins. Based on these results, we selected 20 mM HEPES pH 7.2, 100 mM potassium acetate, 10 mM DTT and 1% Ficoll for all further work and adjusted magnesium as indicated.
7SK undergoes a magnesium-dependent conformational change in vitro
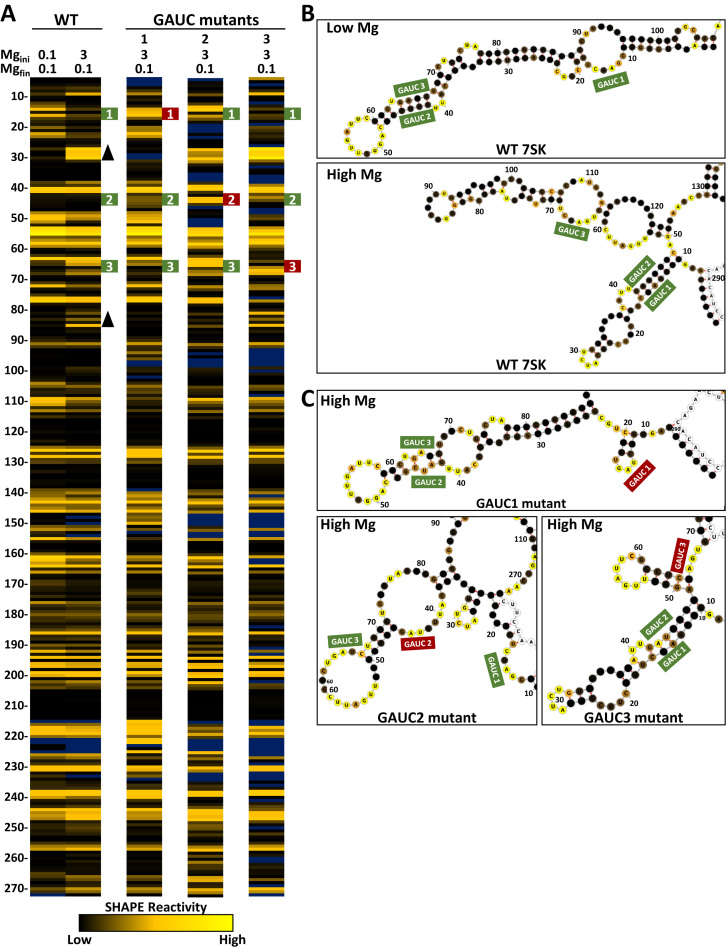
Because magnesium affects RNA folding, we sought to determine if there were any magnesium-dependent changes in the structure of 7SK RNA using Selective 2΄ Hydroxyl Acylation by Primer Extension (SHAPE). The method was optimized specifically for 7SK to give robust and highly reproducible raw signals. Because the 3΄ end of 7SK is critical for LARP7 binding, we tested internal primers instead of extending the 3΄ end for a primer-binding site. The best signals and best coverage of 7SK were obtained with a primer that is complementary to nucleotides 285–305 yielding accurate determination of base reactivities for nucleotide 4–272 (81% coverage). To enable interpretation of SHAPE data from this large region, vertical columns of colored cells that corresponded to normalized SHAPE reactivity of each base were used. Conditional formatting was used to color cells from black (zero to low reactivity), through goldenrod (moderate to high reactivity), and to yellow (very high reactivity). Dark blue cells represent bases with undetermined reactivities because of high background in the unreacted RNA. Low SHAPE reactivity normally corresponds to base paired regions with low flexibility. In contrast, high reactivity is associated with flexible nucleotides like those found in loops and bulges. The more flexible the nucleotide, the higher SHAPE reactivity. The overall reactivity profile of 7SK, with regions of low reactivity flanked by high reactivity, suggests a well-folded RNA (Figure 1, leftmost column).
Figure 1.
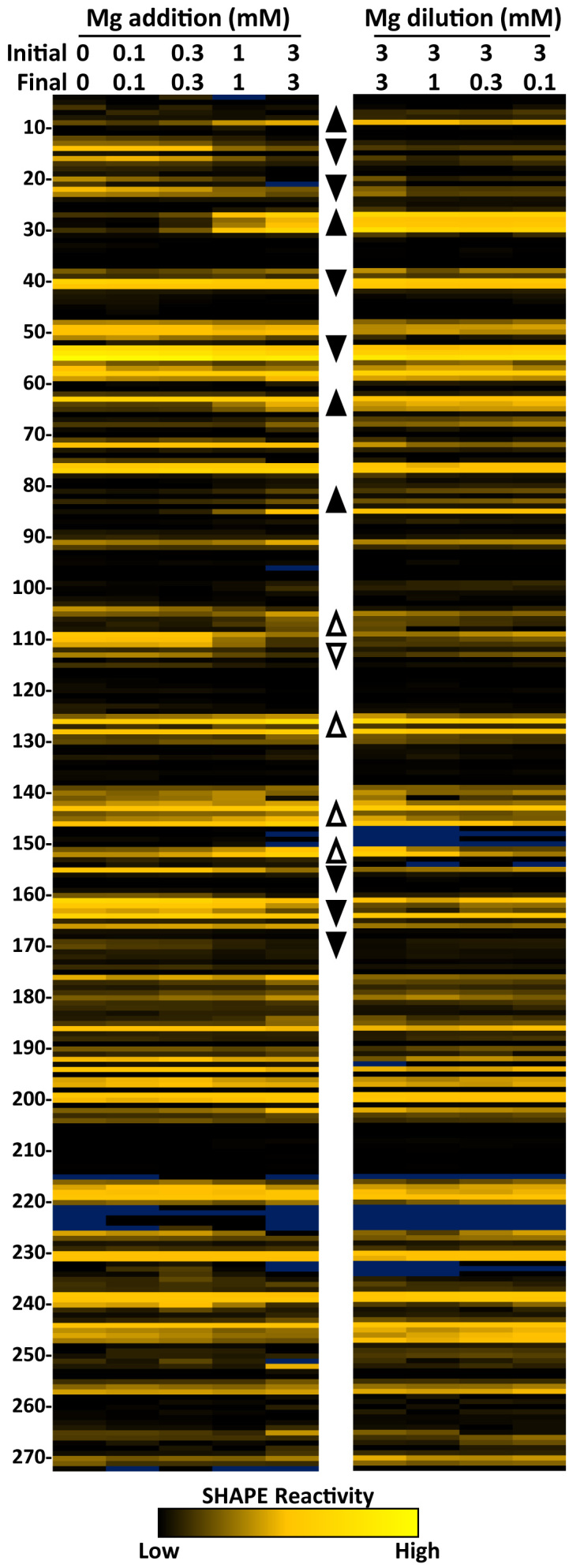
Magnesium induced structure of in vitro 7SK RNA. SHAPE reactivity heat maps showing flexibility of nucleotides 4–272. The color of each cell corresponds to the reactivity of that base from black (non-reactive), through goldenrod (reactive), to yellow (highly reactive). Bases with high background are colored blue. (Left) Magnesium acetate concentrations during the initial folding of 7SK RNA increase as indicated. Regions that change in reactivity are represented by up arrowheads for increases and down arrowheads for decreases. (Right) 7SK RNA was initially folded at 3 mM magnesium and diluted to the indicated concentrations for analysis. Regions that retain 3 mM reactivities are represented by filled arrowheads and open arrowheads represent regions that change after removal of magnesium.
Addition of increasing magnesium from 0.1 to 3 mM during folding caused a change in the reactivity of some bases but not others (Figure 1, left). There were both increases and decreases in reactivity as noted by up or down arrowheads, respectively. Areas of 7SK that started as non-reactive and became reactive were G9, A27–U30, G64–A65, G81, G83, G85 and C105–C108. Some flexible regions also increased in reactivity such as A125–C128, A143, G145–A146 and G151–A152. Some regions displayed decreased reactivity with increasing magnesium such as C12–C16, C20–C23, U40, G51–G55, A109–G113, A155, A160–A164 and A169–G170. Magnesium had little effect on the rest of 7SK. Interestingly, the most dramatic changes occurred within in the first 100 nucleotides. This suggests a magnesium induced structural rearrangement of this region of 7SK. These data reinforce the earlier hypothesis that 7SK has two different conformations that may be close energetically (25) and here we find that one may be favored by low and the other by high magnesium.
Because protein binding was optimal at 100 μM magnesium, we sought to determine what happened to 7SK when it was diluted after being folded at 3 mM magnesium. We were surprised that the majority of magnesium-induced changes to 7SK remained the same as the magnesium was diluted to 1 mM, 300 μM and finally 100 μM (Figure 1, right). Regions of 7SK that reverted back to low magnesium reactivities are indicated by open arrowheads and those that stayed the same as the high magnesium structure are indicated by filled arrowheads. Interestingly, the first 100 nucleotides all remained in the high magnesium conformation after removal of magnesium. Between nucleotides 101 and 150, the structure of 7SK depended on the final concentration of magnesium. There was a cluster of bases that retained the high magnesium structure between 155 and 170. SHAPE reactivities of nucleotides 170–270 were not impacted by magnesium during folding or during post-folding reduction of magnesium. As far as we know, the region of 7SK affected by magnesium during folding, but not during reduction of the magnesium is the first example of magnesium-induced hysteresis observed in an RNA.
One of the regions that became reactive at high magnesium and retained this reactivity as magnesium was reduced had an unusual alternating pattern of high and low reactivity with G81, G83, and G85 being reactive and the two bases in between completely unreactive. This region contains one of five 3G stretches in human 7SK (9-11, 50–52, 81–83, 188–190 and 262–264), all of which are conserved from human through frogs and four that are conserved through fish (Supplementary Figure S3). RNA G-quadruplexes form by the arrangement of the triplets so that there is a stack of three groups of four planer guanines (63–65). The G-quadruplex structure is stabilized by cations that are coordinated with the bases and between the planes. Potassium ions stabilize the formation of G-quadruplexes, while lithium ions do not (66). To test for the presence of a G-quadruplex in 7SK, folding was performed in a non-buffered solution (with 1 mM DTT instead of TCEP) containing lithium or potassium acetate or no monovalent salt, under three magnesium conditions (Supplementary Figure S4). As expected, in the presence of potassium, the effect of magnesium on folding of 7SK was similar in both non-buffered and buffered conditions. However, the 100 μM magnesium point in the non-buffered system had more reactivity in the regions that are the hallmarks of the high magnesium structure. Like the buffered system, addition of high magnesium resulted in a complete transition to the high magnesium structure. Surprisingly, folding in the presence of lithium resulted in the high magnesium structure regardless of magnesium concentration. This strongly suggests that the high magnesium form of 7SK is not stabilized by a traditional G-quadruplex. As expected, in the absence of monovalent cations, the pattern of reactivity reflects a mostly unstructured RNA. The fact that low magnesium and lithium caused the formation of the high magnesium structure may be evidence of metal stabilized tertiary structure such as kissing loops (67). In both potassium and lithium experiments, the reactivity of bases in the variable region (between 105 and 150) remained sensitive to magnesium levels.
In summary, we have found two types of responses to changes in magnesium during folding of 7SK. The most dramatic response occurred in the first 100 nt due to a stable conformational change in structure. A similar stabilization occurred from 155 to 170. High magnesium facilitated these changes but was not required to maintain them. The region between 100 and 150 responded to changes in magnesium but these were not stably maintained when magnesium was reduced. The rest of 7SK had little or no response to changing magnesium.
Changes in 7SK RNA are facilitated by a 7SK motif switch
There are three GAUC sequence elements within the 5΄ region of 7SK which are conserved (Supplementary Figure S3). The ‘7SK motif,’ a short helix that is flanked by unpaired uridines, is formed by pairing of GAUC2 (nt 42–45) and GAUC3 (nt 64–67) (28,68,69). GAUC1 (nt 13–16), however, has not been included in discussions of this motif. To test if alternating GAUC interactions contribute to the change in conformation in 7SK after folding in 3 mM magnesium, we mutated each of the elements individually to CUAG and performed SHAPE analysis (Figure 2A). Secondary structure prediction of wild type 7SK using SHAPE constraints suggest that the standard 7SK motif with GAUC2 paired with GAUC3 forms in low magnesium, but the data from high magnesium SHAPE predicts an alternative 7SK motif with GAUC1 paired with GAUC2 (Figure 2B). Mutation of GAUC1 led to an increase in reactivity in the mutated region, but not GAUC2 or GAUC3. Additionally, the GAUC1 mutant caused decreased reactivity in two regions where reactivities increased from low to high magnesium in wild type 7SK (Figure 2A, arrows).
Figure 2.
SHAPE analysis of GAUC mutants. (A) SHAPE results for wild type (WT, from Figure 1) and the three GAUC to CUAG mutations (green numbered boxes) folded under the indicated magnesium folding conditions. (B) Secondary structure predictions of low and high magnesium folded 7SK using RNAstructure and corresponding SHAPE constraints and modeled with forna. The GAUC regions are labeled and SHAPE value colors are mapped on the predicted structures. (C) Secondary structure predictions of GAUC mutants (red boxes) at high magnesium.
Mutation of GAUC2 resulted in increased reactivities of all three GAUC motifs. Besides a change reactivity around the mutation site, the GAUC3 mutation did not cause much change in the other GAUC motifs or in the high magnesium structure. Secondary structure analysis constrained by SHAPE data was performed on the GAUC mutants (Figure 2C). Mutation of GAUC1 eliminated the magnesium induced structural change and retained the standard 7SK motif. Mutation of GAUC2 resulted in reactivity that predicted disruption of both the standard and alternative 7SK motif structures without the formation of a GAUC1 and 3 pair. As expected, the mutation of GAUC3 led to the formation of the alternative 7SK motif. These data highlight the importance of the 7SK motif in the structure of 7SK RNA as well as introduce the idea of a ‘7SK motif switch’ that could play a key role in regulating P-TEFb activity through binding of HEXIM1.
Assembly and analysis of the 7SK snRNP in vitro
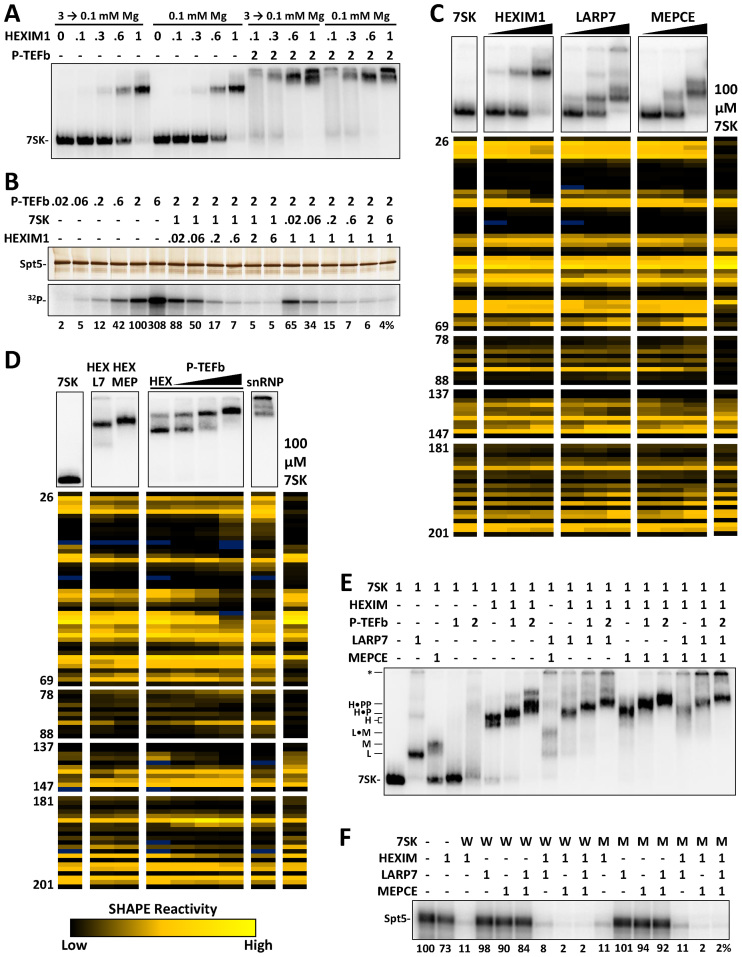
We next began to assemble the 7SK snRNP from individual components and characterize these complexes by EMSA, kinase assays, and SHAPE. We were interested to determine if the magnesium-induced 7SK motif switch would affect the binding of HEXIM1 and P-TEFb. Labeled 7SK RNA was folded at either 100 μM or 3 mM magnesium acetate and diluted down to 100 μM during protein binding. Using the optimized native EMSA conditions, HEXIM1 was titrated onto 7SK with and without a set amount of P-TEFb containing full length Cdk9 and Cyclin T1. Surprisingly, there was no effect on either HEXIM1 or subsequent P-TEFb binding (Figure 3A). Kinase assays were then used to examine the stoichiometry of HEXIM1 and P-TEFb binding to 7SK. A P-TEFb titration demonstrated that the assay was in a linear range of activity on excess Spt5 as visualized by silver stain (Figure 3B). Holding P-TEFb and 7SK constant at a 2:1 molar ratio and titrating in HEXIM1 demonstrated a saturation of inhibition at one molar equivalent of HEXIM1 dimer. Holding P-TEFb and HEXIM1 at a 2:1 molar ratio and titrating in 7SK demonstrated the 7SK dependence on inhibition.
Figure 3.
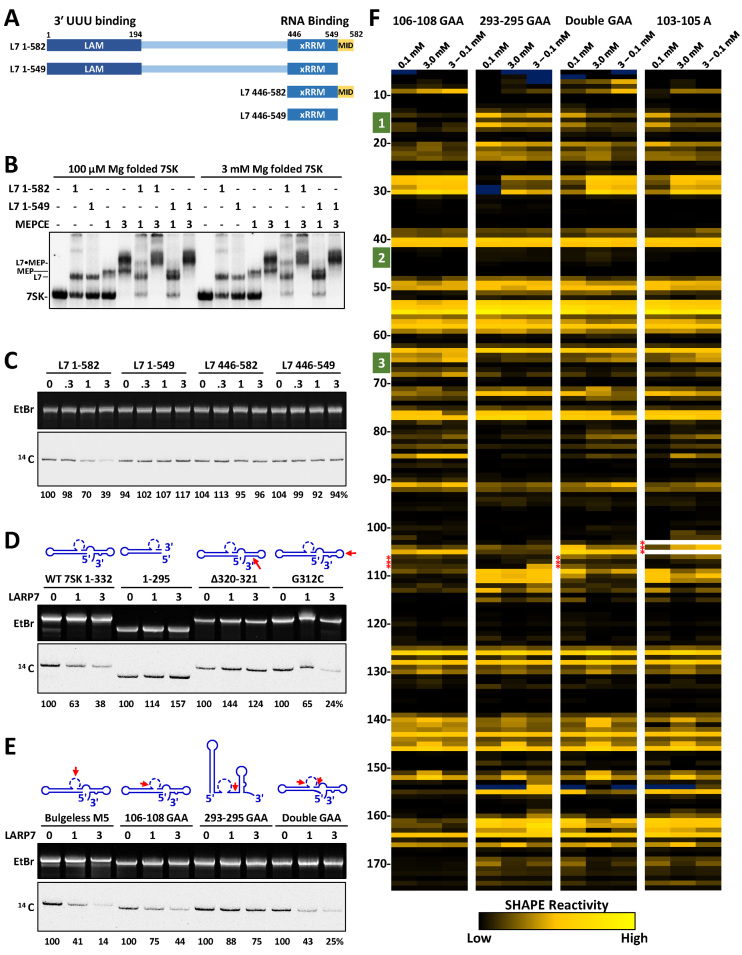
Stepwise in vitro assembly of the P-TEFb containing 7SK snRNP. Each reaction contained 1 pmol of 7SK folded at 3 mM magnesium acetate and diluted to 100 μM and the indicated amounts of proteins added, unless stated otherwise. EMSAs used 4% native PAGE and kinases assays 9% SDS PAGE. (A) Stoichiometric EMSA of HEXIM1 and P-TEFb binding to labeled 7SK RNA folded under the indicated magnesium conditions. (B) Kinase assay of P-TEFb activity using titrations of HEXIM1 and 7SK RNA using silver stain (above) and phosphorimaging (below). Amounts of proteins and RNA are in molar equivalents with 1 signifying 0.02 pmol. (C) Parallel EMSA and SHAPE analysis of individual HEXIM1, LARP7, and MEPCE proteins binding to 7SK RNA compared to low magnesium folded 7SK (from Figure 1). Triangles indicate increasing amounts of proteins (0.1, 0.3, 1 pmol). (D) Parallel EMSA and SHAPE of the indicated combination of proteins binding to 7SK. Proteins are at 1 pmol except for P-TEFb increasing from 0.5 to 1 to 2 pmol. 7SK–protein shifts are labeled with the first letter of the proteins where possible, asterisk denote complexes that did not enter the gel. (E) Stoichiometric EMSA of the assembly of snRNP complex in vitro. (F) Kinase assay as in B. Wild type (W) and GAUC1 mutant (M) RNA.
To determine if 7SK is structurally modified by the binding of proteins, parallel EMSA-SHAPE experiments were used (Figure 3C). After forming protein-RNA complexes under EMSA conditions, 10% of each reaction was analyzed by native gel and the rest by SHAPE. Because magnesium levels are between 2 and 4 mM in cells (70), RNA folding was standardized to 3 mM magnesium acetate followed by dilution to 100 μM to facilitate binding of proteins for these and all following EMSAs. Titration of HEXIM1 onto 7SK caused a reduction of reactivity at C38 though U40 and to a lesser extent 27–30, but overall the SHAPE reactivities indicated that the alternative 7SK motif (GAUC1/2) found in high magnesium was retained and suggest that HEXIM1 binding to this motif led to some protection of the adjacent flanking bases (Figure 3C). LARP7 has two RNA binding domains and mainly interacts with the 3΄ end of 7SK and an adjacent stem–loop. A number of complexes appeared as the protein was titrated onto 7SK and this complexity is likely explained by tethering multiple 7SK molecules as the two domains bind to different RNAs. The main binding sites for LARP7 are not covered by our SHAPE analysis and as expected, LARP7 appeared to maintain the alternative 7SK motif (Figure 3C). However, there was a slight decrease in reactivity C141 and G142 possibly due to an additional LARP7 interaction. Interestingly MEPCE caused a loss of reactivity at C38 and A39 similarly to HEXIM1. MEPCE also caused a slight loss of reactivity from 139 to 142. We conclude that HEXIM1, LARP7 and MEPCE individually do not lead to the switch from the alternative 7SK motif.
Parallel EMSA-SHAPE was then used to examine pairwise effects of LARP7, MEPCE and P-TEFb on HEXIM1. Except for the changes around the 137–140 region induced by LARP7 or MEPCE alone, neither protein had a significant effect on the HEXIM1 SHAPE pattern (Figure 3D). The most striking changes to 7SK structure occurred with the titration of P-TEFb to HEXIM1 which led to a dramatic loss of reactivity in the regions 48–51. This signal loss could be due to the direct interaction with P-TEFb (57). A loss of reactivity was also seen over 190–192 and an increase in the reactivity of A187 and G188. There were some changes in the regions characteristic of the 7SK motif switch, but the alternative 7SK motif was mostly unchanged. Together these data suggest P-TEFb association with HEXIM1 in the presence of 7SK RNA causes both local and long-range changes within the RNA. When all of the proteins were combined about half of the complexes formed entered the gel and contributions from each protein were found in SHAPE reactivities.
Our next step was to analyze the assembly of the complete 7SK snRNP in vitro by EMSA and kinase assay. To visualize the complex formation, native EMSAs with stoichiometric combinations of proteins were performed (Figure 3E). LARP7 caused a robust shift in the RNA and MEPCE shifted ∼50% of 7SK. The two concentrations of P-TEFb alone gave rise to an increasing amount of weak binding leading to a broad distribution of mobilities of the RNA. HEXIM1 led to a robust shift and now addition of P-TEFb gave two specific shifts as one and then two molecules of the kinase were added. A LARP7/MEPCE complex formed when both proteins were added, but MEPCE did not completely shift the remaining LARP7 complex. LARP7 and MEPCE, when individually added to HEXIM1, led to lower mobility bands that both interacted with one or two molecules of P-TEFb. Interestingly, in the presence of HEXIM1, regardless of the presence or amount of P-TEFb, MEPCE associated stoichiometrically. In the presence of all proteins at the correct stoichiometry more than half of the complexes formed a very tight band with most of the rest not being able to enter the gel due to higher order complexes with LARP7. These data suggest a complete, P-TEFb containing 7SK snRNP was formed in vitro. A kinase assay was performed to determine if MEPCE or LARP7 would influence the inhibition of P-TEFb by HEXIM1 with 7SK. In the absence of HEXIM1, neither LARP7 nor MEPCE inhibited P-TEFb (Figure 3F). However, MEPCE, but not LARP7 enhanced the inhibition by HEXIM1, again suggesting cooperation between the two factors. When the experiment was repeated with the GAUC1 mutant 7SK that forces the standard 7SK motif, essentially identical results were obtained. This suggests that the presence of all snRNA components drive the wild type RNA to the standard 7SK motif even in the presence of high magnesium.
In vitro assembly of the 7SK snRNP forms a stable and functional complex
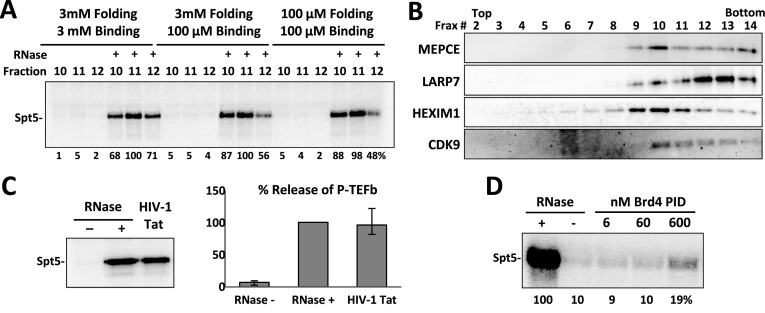
We next examined the stability of snRNPs assembled under the three magnesium conditions. The three reactions were scaled up to 20 pmol and glycerol gradient sedimentation was performed. The fractions that normally would contain the cellular snRNP were selected from each gradient and analyzed using kinase assays with and without RNase A. Regardless of the conditions, P-TEFb was inhibited until the RNA was degraded and all three conditions resulted in about the same amount of P-TEFb-containing snRNP (Figure 4A). Fractions across the gradient analyzing the snRNP formed from 7SK folded under the conditions used for all previous EMSAs were blotted for MEPCE, LARP7, HEXIM1 and Cdk9. All proteins co-sedimented in fractions 9–11 with higher order complexes containing LARP7 and multiple 7SK RNAs along with some of the other proteins further down the gradient (Figure 4B). Fraction 11 was incubated with HIV-1 Tat or the P-TEFb binding domain of Brd4 to determine if P-TEFb in the in vitro assembled snRNP could be released. A kinase assay revealed that all P-TEFb was released by Tat and a small fraction by Brd4 (Figure 4C and D) which matches the effects of these proteins on cellular snRNPs (25). These data suggest that the primary function of the 7SK snRNP, to sequester and release P-TEFb, is reconstituted under our in vitro assembly conditions.
Figure 4.
Functional analysis of the in vitro 7SK snRNP. Complete 7SK snRNP complexes were assembled under the indicated conditions and sedimented through a glycerol gradient as described in Materials and Methods. All gels are 9% SDS PAGE. (A) The indicated fractions were analyzed using a kinase assay with and without RNase A treatment. (B) Western blot analysis of the glycerol gradients for the indicated proteins. In vitro P-TEFb release assay by kinase assay using (C) HIV-1 Tat and (D) Brd4 P-TEFb interaction domain (PID).
Comparison of the in vitro 7SK snRNP with cellular snRNPs
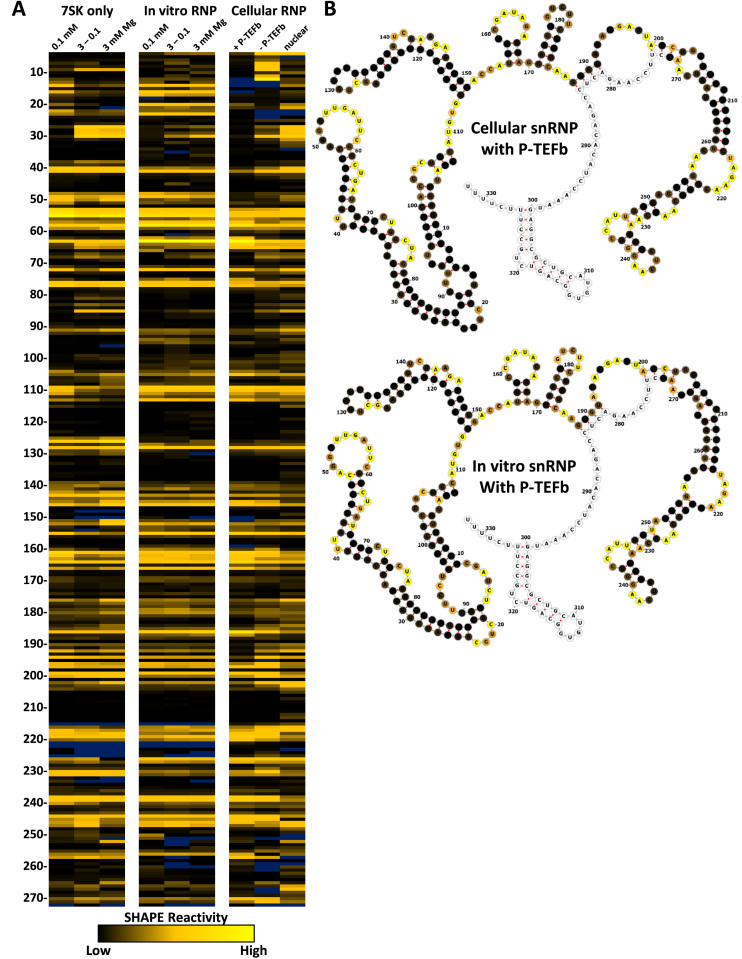
SHAPE analysis was carried out on glycerol gradient purified in vitro snRNPs (lacking contaminating higher order LARP7 complexes) formed under the three magnesium conditions. The RNA folded at 0.1 mM magnesium gave a pattern of SHAPE reactivity that was relatively unaffected by reconstitution of the RNP (Figure 5A). This strongly suggests that the standard 7SK motif is maintained during snRNP assembly. In contrast, when snRNPs were formed on RNAs folded at 3 mM magnesium, the patterns of SHAPE reactivity were significantly altered. In fact, after assembly, the patterns more closely resembled those of the low magnesium folded RNA. This suggests that association of the snRNP components force the standard 7SK motif.
Figure 5.
SHAPE analysis of cellular and in vitro 7SK snRNPs. (A) SHAPE results for (Left) 7SK folded under the indicated magnesium conditions (from Figure 1), (Center) in vitro 7SK snRNP complexes formed under the indicated magnesium conditions and purified by glycerol gradient, and (Right) the indicated cellular 7SK snRNP populations. (B) Secondary structure predications using RNAstructure and SHAPE constraints of cellular and in vitro P-TEFb containing snRNPs colored as in Figure 2.
To examine the 7SK motif switch in cells we isolated three populations of 7SK snRNPs and performed SHAPE analysis. HeLa cell lysates were fractionated on glycerol gradients and the P-TEFb containing snRNP was immunoprecipitated using antibodies to Cyclin T1. The snRNP lacking P-TEFb was immunoprecipitated using LARP7 antibodies from lysates of HeLa cells treated with flavopiridol, which causes a release of P-TEFb from the 7SK snRNP (71). We also found that a small fraction of LARP7 remained associated with the nuclear pellet from flavopiridol treated cells suggesting the existence of a third population of 7SK snRNP more tightly associated with chromatin. Loss of P-TEFb and HEXIM1 led to changes mainly in the first 100 nucleotides of 7SK where the 7SK motif switch occurs. In fact, SHAPE pattern of the P-TEFb containing snRNP was very similar to pattern from the low magnesium 7SK RNA (Figure 5A compare 27–30, 67–68 and 81–85). U63 which is the 5΄ flanking base of GAUC3, was ultra-reactive in the P-TEFb containing snRNP from cells and in all in vitro reconstituted snRNPs, but not in 7SK alone. There was a dramatic increase in the reactivity in the 4–12 region after the release of P-TEFb that could signify a change in 5΄ base pairing. Other changes were observed along the entire RNA suggesting a propagation of structure through 7SK after the release of P-TEFb. Some of these changes occurred and are the same as seen during the titration of P-TEFb on HEXIM1 in vitro (Figure 5A, 139–140, 181–194). The 7SK snRNP retained by the nucleus in flavopiridol treated cells had a very different pattern compared to the other cellular complexes. Interestingly, the pattern most closely resembled 7SK alone folded at high magnesium (Figure 5A). These data suggest the nuclear retained snRNP may be an alternate form of 7SK that is an intermediate between the soluble forms. The secondary structures of cellular 7SK snRNP with P-TEFb and the in vitro reconstituted snRNP were predicted using RNAstructure and visualized with forna (Figure 5B). Overall, there is good agreement between base paired regions with low SHAPE reactivity and bulges and loops with higher reactivity. The few changes could be due to additional proteins present in the cellular snRNP or post-translational modifications of some of the proteins. However, the two predicted structures are almost identical suggesting we have faithfully reconstituted the P-TEFb containing 7SK snRNP.
LARP7 C terminal domain interacts with MEPCE
To examine the function of LARP7 and MEPCE our approach was changed because their main sites of interaction occur outside of the region analyzed by SHAPE. LARP7 and MEPCE are known to interact (41), but the region of LARP7 responsible and the potential involvement of 7SK is not known. LARP7 has a highly conserved La antigen motif (LAM) that binds to the 3΄ terminal U repeat and an xRRM that is binds to the 3΄ stem loop (Figure 6A) (72). The LARP7 xRRM is most similar to the xRRM domain in the Tetrahymena telomerase accessory factor, p65 (34,73,74). However, LARP7 has a C-terminal extension predicted to be alpha helical. To determine if the helical extension affected binding of LARP7 or subsequent binding of MEPCE to 7SK, an EMSA with RNA folded at low and high magnesium was performed (Figure 6B). Both the full length LARP7 and LARP7 1–549 bound to 7SK and under the conditions used caused a similar mobility shift of about half of the 7SK. MEPCE shifted ∼25% of the RNA to the expected lower mobility position. An excess of MEPCE shifted all the RNA resulting in two complexes with the lower mobility complex likely a non-specific one. Mixing equal molar amounts of full length LARP7, but not truncated LARP7, and MEPCE gave rise to a new shift likely containing both proteins. Evidently, the MEPCE interaction domain (MID) of LARP7 helps tether MEPCE to 7SK. Identical results were obtained with 7SK folded at high magnesium indicating the 7SK switch is not involved in the interaction of LARP7 and MEPCE with 7SK.
Figure 6.
The role of the C-terminus of LARP7 and 7SK RNA in inhibition of MEPCE. Unless indicated, 6% TBE urea gels were used. (A) Structural domains of LARP7 and mutants indicating the two RNA binding domains of LARP7 and a predicted helical C-terminal region. (B) Stoichiometric EMSA of full length and truncated LARP7 with MEPCE on 7SK folded as indicated analyzed on a 4% native gel. Amounts of proteins and RNA are in pmol. (C) Methyltransferase assay using 5 pmol MEPCE to methylate recombinant 7SK with 14C-S-adenosylmethionine and the indicated molar equivalents of LARP7 or LARP7 mutants. (D and E) Methyltransferase assays with the indicated wild type or mutant 7SK RNAs and molar equivalents of LARP7. Gel was analyzed by ethidium bromide staining (EtBr) and phosphorimaging of 14C-labeled 7SK (7SK). 7SK RNAs are represented as cartoons with the mutated regions denoted by red arrows. (F) SHAPE analysis of the indicated mutant 7SK RNAs folded under indicated Mg conditions. GAUC motifs are shown as numbered green boxes and sites of mutations as ***.
7SK structure-dependent inhibition of MEPCE through the C-terminal domain of LARP7
Because previous experiments demonstrated that LARP7 can inhibit the methyltransferase activity of MEPCE (41) we hypothesized that the LARP7 MID was also responsible for the inhibition of MEPCE. A methyltransferase assay was set up with low magnesium folded 7SK substrate and equimolar MEPCE. LARP7 constructs were titrated into the reactions and after addition of 14C-S-adenosylmethionine for 30 min the RNA was analyzed by denaturing RNA gel and phosphorimaging. Full-length LARP7 inhibited methylation of 7SK by MEPCE (Figure 6C). As hypothesized, the LARP7 1–549 mutant that was unable to bind 7SK with MEPCE, was also unable to inhibit MEPCE. Interestingly, LARP7 constructs lacking the N-terminal La motif (446–582 and 446–549), which did not interact specifically with 7SK (Supplementary Figure S5), did not inhibit MEPCE even though one contained the MID. These results indicate that proper binding of LARP7 and an intact MID are needed for inhibition of MEPCE.
Because of conservation between Tetrahymena p65 and LARP7, we reasoned that they might interact with their RNA targets in a similar way and designed 7SK constructs to test this. There is an extension of the C-terminal α3 helix of the p65 xRRM upon interaction with the region of TER RNA near a 2-nucleotide bulge (73,74). The 3΄ stem loop of 7SK has a similar structure including a two-nucleotide bulge (72,75). We hypothesized that a similar interaction of 7SK with the α3 helix of LARP7 might expose the MID. As expected, in the context of full-length 7SK, LARP7 inhibited MEPCE, but that inhibition was lost when the final stem loop, the LARP7 binding region, was removed (Figure 6D, compare WT with 1–295). As predicted, deleting the bulge at 320–321 also caused a complete loss of LARP7 inhibition of MEPCE. Other studies demonstrated an interaction of LARP7 with G312 in the loop of the 3΄ stem–loop (30,72,76), however mutating G312 to C did not alter LARP7 inhibition of MEPCE (Figure 6D). SHAPE data described earlier suggested a possible interaction of LARP7 with 7SK in the region of 140–150. This region was mutated to remove bases 140, 141, 143 and 146 which were predicted to be bulges and had high reactivity (Bulgeless M5). Altering the RNA in this way, however, did not affect the inhibition of MEPCE by LARP7. The two published models of 7SK secondary structure differ in the proposed base pairing of the first 7 nt which is also substrate of MEPCE. Pairing with 289–295 as predicted by Marz et al. brings the 5΄ and 3΄ ends of 7SK together establishing a ‘closed’ structure. Pairing with 100–108 as predicted by Wassarman and Steitz creates an extended 5΄ stem loop and an ‘open’ RNA topology. The last 3 nt (UCC) in each potential pairing partner were mutated to GAA to block pairing. MEPCE alone was able to methylate both of these substrates but interestingly, only the RNA in the closed configuration allowed LARP7 inhibition of MEPCE, suggesting the proximity of the 5΄ and 3΄ are necessary for this inhibition. Interestingly, the double GAA mutant allowed inhibition suggesting that the remaining four complementary bases (289–292) favor the closed conformation of 7SK over the remaining three base complement (100–102) (see Figure 2B).
Open and closed conformations dictate the state of the 7SK motif
SHAPE analysis was carried out on mutants designed to manipulate the open or closed conformation of 7SK. The RNAs that were used were 106–108, 293–295 and Double GAA mutants as described in the preceding section as well as an additional mutant that deleted 103–CGC–105 and added a single A in their place (103–105 A) to favor the open conformation by pairing 1–11 with 96–106. RNAs were folded under the different magnesium conditions and subjected to SHAPE analysis (Figure 6F). The reactivity of bases in the region of 106–115 were used to determine the overall conformation. Low reactivity at nt 106–108 followed by a region of high reactivity reflects the open conformation while reactivity at 104–105 surrounded by low to moderate reactivity signifies the closed structure. As predicted, the 106–108 GAA mutant was forced into the closed conformation and the 293–295 GAA mutant was forced into the open conformation regardless of magnesium concentration. The Double GAA mutant and the 103–105 A mutant were in the open conformation at 100 μM magnesium, but at least part of each population switched to the closed conformation after exposure to 3 mM magnesium.
We next examined the effect of the mutants on the 7SK motif switch. The locked closed, 106–108 GAA mutant contained the alternative 7SK motif as indicated by low reactivity at GAUC1 and high reactivity at GAUC3 (Figure 6F, green boxes). The GAUC1 and three reactivities were reversed for the locked open 293–295 GAA mutant, indicating the presence of the standard 7SK motif. Importantly, the 7SK motifs in the locked mutants were impervious to changes in folding conditions indicating that the opening and closing of 7SK may control the 7SK motif switch. Both the Double GAA and 103–105 A RNAs that favored the open conformation contained the standard 7SK motif at low magnesium, but at least part of the populations switched to the alternative 7SK motif after high magnesium supporting the connection between open and closed conformation and the 7SK motif switch. The open conformation with the 5΄ end paired with nt 106–108 forces the standard 7SK motif (GAUC2–GAUC3), alternatively, the closed conformation with the 5΄ end paired with nt 293–295 forces the alternative 7SK motif (GAUC2–GAUC1). Interestingly, the region from 155 to 170 that responded to folding conditions like the 7SK motif switch (see Figure 1) responds to the mutations in a similar manner such that locked mutants are unaffected by magnesium. Overall, this data with free 7SK indicate that 5΄ end pairing directs the state of the 7SK motif.
DISCUSSION
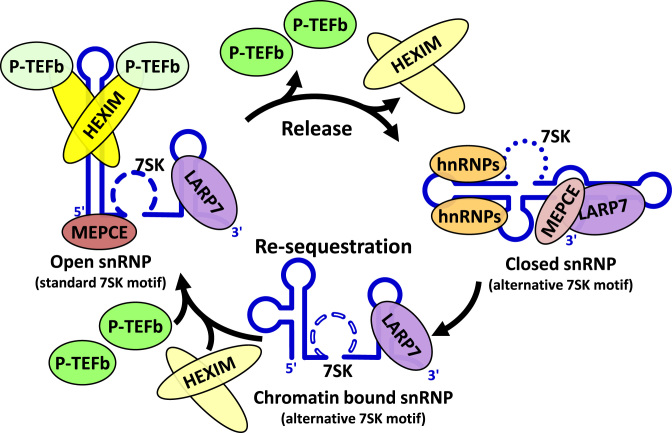
We successfully reconstituted a functional 7SK snRNP using optimized EMSAs, kinase assays, and SHAPE. A hysteretic effect of magnesium on the structure of 7SK and evidence for structural rearrangements that are not explained entirely by changes in secondary structure were found. During comparison of in vitro reconstituted and cellular snRNPs, we found 7SK snRNP strongly associated with chromatin. The domain of LARP7 (MID) and the requirement for a particular 7SK structure that are necessary for inhibition of MEPCE were determined. Our results support a cyclic model for the function of the 7SK snRNP in the release and re-sequestration of P-TEFb involving a 7SK motif switch (Figure 7). When P-TEFb and HEXIM1 are in the 7SK snRNP, the RNA is in an open conformation in which the 5΄ end forms an extended stem loop structure with a standard 7SK motif. When P-TEFb is released, HEXIM1 association is reduced and the alternative 7SK motif is stabilized by hnRNPs. Concomitantly, the RNA adopts a closed structure where the 5΄ and 3΄ ends are close to each other. The re-sequestration of P-TEFb may occur through a chromatin associated, LARP7-bound 7SK containing the alternative 7SK motif. Re-association of HEXIM1 and P-TEFb flicks the 7SK motif switch and converts 7SK back to the open snRNP.
Figure 7.
Model of 7SK snRNP cycle. 7SK in the P-TEFb-containing snRNP is in an ‘open’ conformation with a standard 7SK motif and an extended 5΄ stem loop, which allows HEXIM1 to bind and inhibit P-TEFb. MEPCE is associated with the 5΄ end and LARP7 is associated with the 3΄ stem loop. Upon release of P-TEFb, HEXIM1 is also released leading to an altered 7SK motif structure that is stabilized by hnRNPs. This structure has the 5΄ end of 7SK pairing near the 3΄ end resulting in a ‘closed’ conformation. In this conformation, LARP7 and MEPCE interact resulting in the inhibition of MEPCE. Re-sequestration of P-TEFb may be achieved through a chromatin bound 7SK intermediate containing the alternative 7SK motif. A 7SK motif switch allows HEXIM1 binding and subsequent recruitment of P-TEFb.
Using SHAPE, we demonstrated a magnesium-induced conformational change in the structure of 7SK RNA. The observed change involves a switch in base pairing of the standard 7SK motif (GAUC2 and GAUC3) to an alternative 7SK motif (GAUC2 and GAUC1) that was confirmed by SHAPE on 7SK RNAs containing GAUC mutants. Importantly, we also observed similar SHAPE reactivity changes in cellular 7SK after P-TEFb release. These extend our previous chemical modification studies that revealed a conformational change in the secondary structure of 7SK after the release of P-TEFb (25). During preparation of this manuscript, the crystal structure of 7SK 27–84 was published (68). We mapped SHAPE reactivities onto this structure and found that the low magnesium form fit the structure perfectly, unlike the high magnesium form, which had high reactivities for some paired bases (Supplementary Figure S6). Besides validating our SHAPE data, the packing of non-paired bases within the helix of the crystal structure explains why A34 was not reactive, even though it was predicted to be unpaired. We also found that secondary structure prediction was in good agreement with SHAPE data for the standard 7SK motif structure, but not for the alternative 7SK motif. Because of poor agreement with secondary structure prediction and because lithium caused similar effects as magnesium, we hypothesize that the alternative 7SK structure involves tertiary interactions that are stabilized by both monovalent and divalent cations. The 7SK motif switch provides a potential explanation for the loss of HEXIM1 from the snRNP after the release of P-TEFb (32). As discussed below there may be a competition between the formation of a more stable alternative 7SK structure and HEXIM1 binding. HEXIM1 by itself is quite promiscuous and can bind to both the standard and alternative 7SK motifs in vitro (77), however other proteins can affect the structure of 7SK and some could block HEXIM1 binding. We found a synergistic effect of P-TEFb, as well as MEPCE and LARP7, on stabilizing the standard 7SK motif. Relevantly, U63 became ultra-reactive only in the complete in vitro snRNPs and the P-TEFb containing cellular snRNP. Recently, the discovery of pseudouridylation at U250 was implicated in snRNP formation (78). We do notice a slight difference in reactivity of U250 between in vitro RNA and cellular P-TEFb containing complexes. However, the overall structure in this area of the RNA is unchanged between the two.
Our results clarify how LARP7 interacts with MEPCE and suggest additional constraints on the structure of 7SK. We found that the C-terminal helix of LARP7 is the MEPCE interaction domain (MID) which inhibits 7SK methylation. This inhibition required LARP7 interaction with 7SK and bringing the 5΄ and 3΄ ends of 7SK close to each other through pairing of 1–7 with 289–295. Recently published biochemical probing (72) and structural (76) studies of the LARP7 xRRM bound to the 3΄ stem loop of 7SK, report that the xRRM interacts with the apical loop of the RNA at G312 and does not require the two nucleotide bulge at 320–321. Those studies used only the xRRM domain and lacked the high affinity La motif and the MID. In contrast, using optimized conditions with full-length proteins and RNA, G312 was not required for LARP7 binding and inhibition of MEPCE, but the bulge was essential. Our results strongly support the idea that LARP7 interacts with 7SK in a highly analogous manner as p65 with TER RNA, which requires the two-nucleotide bulge in TER. Because binding of p65 across the bulge in TER stabilizes an extension of the C-terminal helix, we hypothesize a similar extension of the C-terminal helix of LARP7 makes the MID available for inhibition of MEPCE. Exposure of the LARP7 MID is not enough to achieve inhibition of MEPCE as the pairing of 1–7 with 289–295, forming the closed structure similar to that proposed by Marz et al. (56), is also required. Importantly, we also found that the closed structure of 7SK causes the alternative 7SK motif, and the open structure causes the standard 7SK motif.
The role of LARP7 inhibition of MEPCE is unclear. MEPCE interacts with 7SK before La is replaced by LARP7 (30) and could methylate the RNA before LARP7 is bound. Previous studies show that cellular 7SK is always methylated (41). We have also been unable to detect non-methylated 7SK (data not shown), however, the Jumonji domain-containing six protein has been shown to demethylate 7SK and may be involved in 7SK snRNP turnover (79). If a demethylase is required for regulation, MEPCE might be needed to re-methylate 7SK and this may be controlled by LARP7 when 7SK levels need to be reduced. It is also possible that LARP7 in the context of the 7SK snRNP regulates the activity of MEPCE on other targets such as U6. Alternatively, the interaction of MEPCE with LARP7 may play only a structural role in the 7SK snRNP, as we have demonstrated that the 7SK motif switch is intimately related to 5΄ end pairing. It is unclear which form of the 7SK motif is stabilized by MEPCE but is possible that MEPCE stabilizes the standard 7SK motif and the LARP7 disrupts this stabilization helping drive the RNA to the alternative motif. LARP7 may also regulate the state of the 7SK motif through hypothesized interactions between RRM1 and the 5΄ base paired region of 7SK (72).
Controlled transcription requires careful regulation of P-TEFb activity through release and re-sequestration of the kinase. P-TEFb is found either within the 7SK snRNP or associated with chromatin and uncontrolled release, such as after knockdown of 7SK, leads to the inappropriate transition into productive elongation from genes containing paused Pol II in a process we have called runaway transcription (18,80). Previous work has suggested release of P-TEFb from 7SK snRNP to chromatin is through Brd4 (43–45,81) or the super elongation complex by AFF4 or AFF1 (46,47,82). While these release factors are at promoters, it is not clear how P-TEFb is returned to the 7SK snRNP. During this study, we discovered a new form of the 7SK snRNP that lacks P-TEFb, is retained on chromatin, and has an alternative 7SK motif. Very recently, it was found that a 7SK snRNP containing LARP7 and MEPCE was associated with the little elongation complex (LEC) and that this complex was associated with Pol II transcribed snRNA and snoRNA genes (83). It is likely that this is the form of 7SK snRNP that we found associated with chromatin because both lack P-TEFb and HEXIM1 and both are just a few percent of the total 7SK. The P-TEFb-lacking snRNP associated with chromatin may also be involved in re-sequestration, as a P-TEFb lacking snRNP may be needed to recruit excess P-TEFb through association with HEXIM1 at the end of genes. In support of this idea, 7SK is recruited to genes only when they are shutting down (17). KAP1 may be involved because it has been shown to tether the 7SK snRNP to chromatin through an interaction with LARP7 (84). RNA helicases DDX6 (46), DDX21 (85) and RHA (42) can associate with 7SK and could provide the energy to remodel the RNA and remove the alternative snRNP factors such as hnRNPs.
The system we developed here has provided important new insights into 7SK RNA structure and the influence of the protein snRNP components. The new method of creating in vitro 7SK snRNP complexes should facilitate future structural work, especially by cryo-EM. Questions that remain mainly have to do with how P-TEFb is re-sequestered, although if and how P-TEFb is removed from the snRNP and delivered to genes also deserves further investigation.
Supplementary Material
ACKNOWLEDGEMENTS
Britt Griffin designed the primer used in all the SHAPE experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01 GM35500 to D.H.P.]. Funding for open access charge: NIH [GM35500].
Conflict of interest statement. None declared.
REFERENCES
- 1. Guo J., Price D.H.. RNA polymerase II transcription elongation control. Chem Rev. 2013; 113:8583–8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kwak H., Lis J.T.. Control of transcriptional elongation. Annu. Rev. Genet. 2013; 47:483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou Q., Li T., Price D.H.. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012; 81:119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J., Gilmour D.S.. Promoter proximal pausing and the control of gene expression. Curr. Opin. Genet. Dev. 2011; 21:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peterlin B.M., Price D.H.. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006; 23:297–305. [DOI] [PubMed] [Google Scholar]
- 6. Peng J., Zhu Y., Milton J.T., Price D.H.. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998; 12:755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marshall N.F., Price D.H.. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995; 270:12335–12338. [DOI] [PubMed] [Google Scholar]
- 8. Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H.. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006; 21:227–237. [DOI] [PubMed] [Google Scholar]
- 9. Marshall N.F., Peng J., Xie Z., Price D.H.. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996; 271:27176–27183. [DOI] [PubMed] [Google Scholar]
- 10. Narita T., Yamaguchi Y., Yano K., Sugimoto S., Chanarat S., Wada T., Kim D.K., Hasegawa J., Omori M., Inukai N. et al. . Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 2003; 23:1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wada T., Orphanides G., Hasegawa J., Kim D.K., Shima D., Yamaguchi Y., Fukuda A., Hisatake K., Oh S., Reinberg D. et al. . FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell. 2000; 5:1067–1072. [DOI] [PubMed] [Google Scholar]
- 12. Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H.. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999; 97:41–51. [DOI] [PubMed] [Google Scholar]
- 13. Wada T., Takagi T., Yamaguchi Y., Watanabe D., Handa H.. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998; 17:7395–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Y., Yamaguchi Y., Tsugeno Y., Yamamoto J., Yamada T., Nakamura M., Hisatake K., Handa H.. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009; 23:2765–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singh J., Padgett R.A.. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009; 16:1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Darzacq X., Shav-Tal Y., de Turris V., Brody Y., Shenoy S.M., Phair R.D., Singer R.H.. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007; 14:796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prasanth K.V., Camiolo M., Chan G., Tripathi V., Denis L., Nakamura T., Hubner M.R., Spector D.L.. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol. Biol. Cell. 2010; 21:4184–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castelo-Branco G., Amaral P.P., Engstrom P.G., Robson S.C., Marques S.C., Bertone P., Kouzarides T.. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013; 14:R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yik J.H., Chen R., Nishimura R., Jennings J.L., Link A.J., Zhou Q.. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003; 12:971–982. [DOI] [PubMed] [Google Scholar]
- 20. Chen R., Yang Z., Zhou Q.. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 2004; 279:4153–4160. [DOI] [PubMed] [Google Scholar]
- 21. Byers S.A., Price J.P., Cooper J.J., Li Q., Price D.H.. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J. Biol. Chem. 2005; 280:16360–16367. [DOI] [PubMed] [Google Scholar]
- 22. Yik J.H., Chen R., Pezda A.C., Zhou Q.. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J. Biol. Chem. 2005; 280:16368–16376. [DOI] [PubMed] [Google Scholar]
- 23. Michels A.A., Fraldi A., Li Q., Adamson T.E., Bonnet F., Nguyen V.T., Sedore S.C., Price J.P., Price D.H., Lania L. et al. . Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004; 23:2608–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kobbi L., Demey-Thomas E., Braye F., Proux F., Kolesnikova O., Vinh J., Poterszman A., Bensaude O.. An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:12721–12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krueger B.J., Varzavand K., Cooper J.J., Price D.H.. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One. 2010; 5:e12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dulac C., Michels A.A., Fraldi A., Bonnet F., Nguyen V.T., Napolitano G., Lania L., Bensaude O.. Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J. Biol. Chem. 2005; 280:30619–30629. [DOI] [PubMed] [Google Scholar]
- 27. Blazek D., Barboric M., Kohoutek J., Oven I., Peterlin B.M.. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res. 2005; 33:7000–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lebars I., Martinez-Zapien D., Durand A., Coutant J., Kieffer B., Dock-Bregeon A.C.. HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements. Nucleic Acids Res. 2010; 38:7749–7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peterlin B.M., Brogie J.E., Price D.H.. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA. 2011; 3:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muniz L., Egloff S., Kiss T.. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res. 2013; 41:4686–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Markert A., Grimm M., Martinez J., Wiesner J., Meyerhans A., Meyuhas O., Sickmann A., Fischer U.. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008; 9:569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krueger B.J., Jeronimo C., Roy B.B., Bouchard A., Barrandon C., Byers S.A., Searcey C.E., Cooper J.J., Bensaude O., Cohen E.A. et al. . LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008; 36:2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He N., Jahchan N.S., Hong E., Li Q., Bayfield M.A., Maraia R.J., Luo K., Zhou Q.. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008; 29:588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hussain R.H., Zawawi M., Bayfield M.A.. Conservation of RNA chaperone activity of the human La-related proteins 4, 6 and 7. Nucleic Acids Res. 2013; 41:8715–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bayfield M.A., Yang R., Maraia R.J.. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim. Biophys. Acta. 2010; 1799:365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barboric M., Lenasi T., Chen H., Johansen E.B., Guo S., Peterlin B.M.. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7798–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeronimo C., Forget D., Bouchard A., Li Q., Chua G., Poitras C., Therien C., Bergeron D., Bourassa S., Greenblatt J. et al. . Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007; 27:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shimba S., Reddy R.. Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for gamma-monomethyl-capped small RNAs. J. Biol. Chem. 1994; 269:12419–12423. [PubMed] [Google Scholar]
- 39. Gupta S., Busch R.K., Singh R., Reddy R.. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of gamma-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J. Biol. Chem. 1990; 265:19137–19142. [PubMed] [Google Scholar]
- 40. Singh N., Morlock H., Hanes S.D.. The Bin3 RNA methyltransferase is required for repression of caudal translation in the Drosophila embryo. Dev. Biol. 2011; 352:104–115. [DOI] [PubMed] [Google Scholar]
- 41. Xue Y., Yang Z., Chen R., Zhou Q.. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010; 38:360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van Herreweghe E., Egloff S., Goiffon I., Jady B.E., Froment C., Monsarrat B., Kiss T.. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007; 26:3570–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Z., He N., Zhou Q.. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 2008; 28:967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang Z., Yik J.H., Chen R., He N., Jang M.K., Ozato K., Zhou Q.. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005; 19:535–545. [DOI] [PubMed] [Google Scholar]
- 45. Chen R., Yik J.H., Lew Q.J., Chao S.H.. Brd4 and HEXIM1: multiple roles in P-TEFb regulation and cancer. Biomed. Res. Int. 2014; 232870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Muck F., Bracharz S., Marschalek R.. DDX6 transfers P-TEFb kinase to the AF4/AF4N (AFF1) super elongation complex. Am. J. Blood Res. 2016; 6:28–45. [PMC free article] [PubMed] [Google Scholar]
- 47. Scholz B., Kowarz E., Rossler T., Ahmad K., Steinhilber D., Marschalek R.. AF4 and AF4N protein complexes: recruitment of P-TEFb kinase, their interactome and potential functions. Am. J. Blood Res. 2015; 5:10–24. [PMC free article] [PubMed] [Google Scholar]
- 48. Liu P., Xiang Y., Fujinaga K., Bartholomeeusen K., Nilson K.A., Price D.H., Peterlin B.M.. Release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein (snRNP) activates hexamethylene bisacetamide-inducible protein (HEXIM1) transcription. J. Biol. Chem. 2014; 289:9918–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sobhian B., Laguette N., Yatim A., Nakamura M., Levy Y., Kiernan R., Benkirane M.. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 2010; 38:439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gudipaty S.A., McNamara R.P., Morton E.L., D’Orso I.. PPM1G binds 7SK RNA and Hexim1 to block P-TEFb assembly into the 7SK snRNP and sustain transcription elongation. Mol. Cell Biol. 2015; 35:3810–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu Y., Pe’ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M.B., Price D.H.. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997; 11:2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barboric M., Yik J.H., Czudnochowski N., Yang Z., Chen R., Contreras X., Geyer M., Matija Peterlin B., Zhou Q.. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007; 35:2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sedore S.C., Byers S.A., Biglione S., Price J.P., Maury W.J., Price D.H.. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007; 35:4347–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tahirov T.H., Babayeva N.D., Varzavand K., Cooper J.J., Sedore S.C., Price D.H.. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010; 465:747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wassarman D.A., Steitz J.A.. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 1991; 11:3432–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marz M., Donath A., Verstraete N., Nguyen V.T., Stadler P.F., Bensaude O.. Evolution of 7SK RNA and its protein partners in metazoa. Mol. Biol. Evol. 2009; 26:2821–2830. [DOI] [PubMed] [Google Scholar]
- 57. Egloff S., Van Herreweghe E., Kiss T.. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell. Biol. 2006; 26:630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Q., Price J.P., Byers S.A., Cheng D., Peng J., Price D.H.. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005; 280:28819–28826. [DOI] [PubMed] [Google Scholar]
- 59. Zhu W., Wada T., Okabe S., Taneda T., Yamaguchi Y., Handa H.. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 2007; 35:4064–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Karabiber F., McGinnis J.L., Favorov O.V., Weeks K.M.. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013; 19:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu Z.Z., Mathews D.H.. Experiment-Assisted Secondary Structure Prediction with RNAstructure. Methods Mol. Biol. 2016; 1490:163–176. [DOI] [PubMed] [Google Scholar]
- 62. Kerpedjiev P., Hammer S., Hofacker I.L.. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015; 31:3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Millevoi S., Moine H., Vagner S.. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA. 2012; 3:495–507. [DOI] [PubMed] [Google Scholar]
- 64. Wong H.M., Payet L., Huppert J.L.. Function and targeting of G-quadruplexes. Curr. Opin. Mol. Ther. 2009; 11:146–155. [PubMed] [Google Scholar]
- 65. Ji X., Sun H., Zhou H., Xiang J., Tang Y., Zhao C.. Research progress of RNA quadruplex. Nucleic Acid Ther. 2011; 21:185–200. [DOI] [PubMed] [Google Scholar]
- 66. Hagihara M., Yoneda K., Yabuuchi H., Okuno Y., Nakatani K.. A reverse transcriptase stop assay revealed diverse quadruplex formations in UTRs in mRNA. Bioorg. Med. Chem. Lett. 2010; 20:2350–2353. [DOI] [PubMed] [Google Scholar]
- 67. Lambert D., Leipply D., Shiman R., Draper D.E.. The influence of monovalent cation size on the stability of RNA tertiary structures. J. Mol. Biol. 2009; 390:791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Martinez-Zapien D., Legrand P., McEwen A.G., Proux F., Cragnolini T., Pasquali S., Dock-Bregeon A.C.. The crystal structure of the 5΄ functional domain of the transcription riboregulator 7SK. Nucleic Acids Res. 2017; doi:10.1093/nar/gkw1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martinez-Zapien D., Saliou J.M., Han X., Atmanene C., Proux F., Cianferani S., Dock-Bregeon A.C.. Intermolecular recognition of the non-coding RNA 7SK and HEXIM protein in perspective. Biochimie. 2015; 117:63–71. [DOI] [PubMed] [Google Scholar]
- 70. Romani A.M. Cellular magnesium homeostasis. Arch. Biochem. Biophys. 2011; 512:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Biglione S., Byers S.A., Price J.P., Nguyen V.T., Bensaude O., Price D.H., Maury W.. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007; 4:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Uchikawa E., Natchiar K.S., Han X., Proux F., Roblin P., Zhang E., Durand A., Klaholz B.P., Dock-Bregeon A.C.. Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7. Nucleic Acids Res. 2015; 43:3373–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Singh M., Choi C.P., Feigon J.. xRRM: a new class of RRM found in the telomerase La family protein p65. RNA Biol. 2013; 10:353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Singh M., Wang Z., Koo B.K., Patel A., Cascio D., Collins K., Feigon J.. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol. Cell. 2012; 47:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bourbigot S., Dock-Bregeon A.C., Eberling P., Coutant J., Kieffer B., Lebars I.. Solution structure of the 5΄-terminal hairpin of the 7SK small nuclear RNA. RNA. 2016; 22:1844–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Eichhorn C.D., Chug R., Feigon J.. hLARP7 C-terminal domain contains an xRRM that binds the 3΄ hairpin of 7SK RNA. Nucleic Acids Res. 2016; 44:9977–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li Q., Cooper J.J., Altwerger G.H., Feldkamp M.D., Shea M.A., Price D.H.. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 2007; 35:2503–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhao Y., Karijolich J., Glaunsinger B., Zhou Q.. Pseudouridylation of 7SK snRNA promotes 7SK snRNP formation to suppress HIV-1 transcription and escape from latency. EMBO Rep. 2016; 17:1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A.K., Rosenfeld M.G.. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013; 155:1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guo J., Li T., Price D.H.. Runaway transcription. Genome Biol. 2013; 14:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Patel M.C., Debrosse M., Smith M., Dey A., Huynh W., Sarai N., Heightman T.D., Tamura T., Ozato K.. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol. Cell. Biol. 2013; 33:2497–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lu H., Li Z., Xue Y., Schulze-Gahmen U., Johnson J.R., Krogan N.J., Alber T., Zhou Q.. AFF1 is a ubiquitous P-TEFb partner to enable Tat extraction of P-TEFb from 7SK snRNP and formation of SECs for HIV transactivation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E15–E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Egloff S., Vitali P., Tellier M., Raffel R., Murphy S., Kiss T.. The 7SK snRNP associates with the little elongation complex to promote snRNA gene expression. EMBO J. 2017; 36:934–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. McNamara R.P., Reeder J.E., McMillan E.A., Bacon C.W., McCann J.L., D’Orso I.. KAP1 recruitment of the 7SK snRNP complex to promoters enables transcription elongation by RNA polymerase II. Mol. Cell. 2016; 61:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Calo E., Flynn R.A., Martin L., Spitale R.C., Chang H.Y., Wysocka J.. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015; 518:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.