Abstract
One third of tumor suppressors are haploinsufficient transcriptional regulators, yet it remains unknown how a 50% reduction of a transcription factor is translated at the cis-regulatory level into a malignant transcriptional program. We studied CUX1, a haploinsufficient transcription factor that is recurrently mutated in hematopoietic and solid tumors. We determined CUX1 DNA-binding and target gene regulation in the wildtype and haploinsufficient states. CUX1 binds with transcriptional activators and cohesin at distal enhancers across three different human cell types. Haploinsufficiency of CUX1 altered the expression of a large number of genes, including cell cycle regulators, with concomitant increased cellular proliferation. Surprisingly, CUX1 occupancy decreased genome-wide in the haploinsufficient state, and binding site affinity did not correlate with differential gene expression. Instead, differentially expressed genes had multiple, low-affinity CUX1 binding sites, features of analog gene regulation. A machine-learning algorithm determined that chromatin accessibility, enhancer activity, and distance to the transcription start site are features of dose-sensitive CUX1 transcriptional regulation. Moreover, CUX1 is enriched at sites of DNA looping, as determined by Hi-C analysis, and these loops connect CUX1 to the promoters of regulated genes. We propose an analog model for haploinsufficient transcriptional deregulation mediated by higher order genome architecture.
INTRODUCTION
Tumor suppressor genes outnumber oncogenes, and one third of tumor suppressor genes encode transcriptional regulators (1,2). Many tumor suppressors are thought to be haploinsufficient, i.e. inactivation of one allele contributes to tumorigenesis (1,2). Tumor suppressors present a therapeutic challenge, as it is relatively easier to develop a drug that inhibits the gain of function of an oncogenic protein, compared to restoring the normal function of an inactivated tumor suppressor. An alternative approach is to target aberrant downstream pathways of haploinsufficient transcriptional regulators. Yet, it remains unknown how a 50% reduction of a transcription factor (TF) is deciphered at the molecular level into a malignant transcriptional program. Uncovering this mechanism, and identifying ‘dose-sensitive’ target genes, is essential to determine new therapeutic strategies.
CUX1 encodes a haploinsufficient TF that is recurrently mutated in cancer (3,4). CUX1 (named cut in Drosophila) is highly conserved, ubiquitous, and essential in mice and Drosophila (5–8). We previously demonstrated that deletion or inactivation of a single allele of CUX1 leads to a 50% reduction in CUX1 protein and a tumorigenic phenotype (3). Indeed, CUX1/Cut is exquisitely dosage-sensitive; under or over-expression alters the fate of numerous cell types (8–11). In Drosophila, Cut abundance is tightly controlled, and specific levels of Cut alternately direct lineage specification, proliferation, or survival (12,13).
The full-length p200 CUX1 protein has one homeodomain and three CUT repeat DNA-binding domains. Post-translation cleavage of p200 removes one CUT repeat, generating the p110 isoform. An alternative start site generates the p75 isoform, which contains one CUT repeat and the homeodomain (14). While p200 has a similar DNA binding affinity compared to the DNA binding domains of the short isoforms, full-length CUX1 has a faster on/off rate (15). These rates were determined by electromobility shift assays measuring the formation and stability of CUX1 binding to an oligonucleotide probe over time. Thus p200 CUX1 is thought to bind more transiently to DNA compared to the other isoforms. Reporter assays indicate that the p200 isoform represses gene expression, while the other isoforms can both activate or repress gene expression (14). Little is know about the tissue-specificity or genome-wide function of these isoforms. To date, there has been scant analysis of CUX1 transcriptional activity (4,16), and no study of endogenous, genome-wide CUX1/Cut DNA binding activity in any species.
The cis-regulatory logic of TF concentration and gene expression is well studied in TF morphogen gradients (17). Some TF morphogens fit an ‘affinity-threshold model,’ wherein target genes with high-affinity TF DNA binding motifs respond at low TF concentration. Genes with low-affinity binding motifs require the TF to reach a higher concentration threshold for activation. In the affinity-threshold model, target gene expression is ‘on’ or ‘off’ in a binary fashion. Other TFs ascribe to a linear or ‘analog’ gene regulation model, wherein target genes have graded transcriptional responses proportional to TF abundance (18–20). Linearity is associated with multiple TF binding sites per target gene, low-affinity TF binding, and an excess number of competing binding sites (18–20).
To test if CUX1 fits one of these models, and to characterize CUX1 genomic functions, we performed RNA-seq and CUX1 ChIP-seq in the wildtype and haploinsufficient states. We intersected these data to determine dose-sensitive CUX1 target genes. To identify genomic properties of CUX1 DNA binding, we took advantage of the extensive functional genomics datasets of the ENCODE consortium (21). We demonstrate that CUX1 binds distal enhancers, along with the transcriptional co-activator EP300 and cohesin components in three human cell types. Long-distance CUX1 binding sites loop to the promoter of dose-sensitive genes. We identified additional genomic features associated with the transcriptional response to CUX1 haploinsufficiency. Finally, we provide evidence that p200 CUX1 best fits the analog model of gene regulation. In summary, we provide the first comprehensive analysis of CUX1 functional DNA binding genome-wide and a model for haploinsufficient CUX1 target gene deregulation.
MATERIALS AND METHODS
Cell culture and transfections
K562 cells stably expressing shCUX1-A, shCUX1-B and control shRNA were maintained as described (3). shCUX1.338, shCUX1.775, shCUX1.810 and control shRNA targeting renilla-luciferase were provided by Mirimus in the LT3GEPIR vector (22). shRNA sequences are provided in Supplemental Table S1. K562 cells were transfected by Lipofectamine (Thermo Fisher), selected with 3 μg/ml puromycin (Invitrogen), and shRNAs induced with 2 μg/ml of doxycycline (Thermo Fisher). Cell viability was measured by CellTiter-Glo® (Promega).
Western blot
Ten microgram total protein was electrophoresed on a 4–15% Mini-PROTEAN TGX Gradient gel (Bio-Rad), transferred to nitrocellulose, and probed with anti-β-actin (C4, sc-47778), anti-HSC-70 (sc-1059), or anti-CUX1 (B-10, sc-514008) from Santa Cruz. Secondary antibodies were anti-mouse-HRP (A9044, Sigma) or anti-goat HRP (sc-2020, Santa Cruz), detected with Chemiluminescence Supersignal West Pico kit (Thermo Scientific), and quantified with ImageJ (23).
ChIP-seq
We followed the Myers Lab ChIP-seq Protocol v042211.2 (www.encodeproject.org). Two biological replicates of ChIP-seq were performed on 100 E6 K562 cells stably expressing shCUX1-A, shCUX1-B, or non-specific control shRNA with 10 μg of anti-CUX1 (sc-6327 Santa Cruz) (3). Libraries were made with Ovation Ultralow Library kit (NuGEN) and size selected with SPRIselect beads (Beckman Coulter). 50 bp single-end sequencing was obtained by Illumina HiSeq. Alignment statistics are provided in Supplemental Table S2. Sequencing data are available in the GEO Database (accession no.: GSE92882).
Peak calling and occupancy analysis
We aligned reads to hg19 using bwa (version 0.7.5) (24) and called peaks using Q (25) with input control. We combined replicates with the Irreproducibility Discovery Rate (IDR) (26) and removed peaks in poor mapability regions (ENCODE Data Coordination Center, DCC, ENCSR365LFZ). ENCODE (27) data was retrieved from the UCSC genome browser (28) or www.encodeproject.org. We assigned peaks to the single nearest transcription start site (TSS) within 1 Mb using GREAT (29). We used the same pipeline to call peaks with ENCODE CUX1 ChIP-seq data (DCC ENCSR000DYR, ENCSR000EFO and ENCSR049KIZ) with input controls (DCC ENCSR000FAK, ENCSR000EYX and ENCSR000EVT).
We used DiffBind (30) for occupancy analysis. Within the control peaks (IDR 0.10), we counted the number of reads per peak in each replicate, normalized by input control and library size. We compared the mean occupancy from the two controls to the mean occupancy across the four knockdowns. Occupancy data are provided in Supplemental Table S3.
ChIP-seq comparison with other datasets
Chromatin state predictions were derived from hidden Markov model analysis of eight chromatin marks and CTCF ChIP-seq data (31). TF binding sites were obtained from Uniform Peak calls (wgEncodeRegTfbsClusteredWithCellsV3hg19.bed). To obtain a background bed file of open chromatin, we merged all sites bound by any factor. To establish the enrichment between different genomic regions, we used permutation tests (32). We shuffled regions throughout the genome or open chromatin with Bedtools (v2) (33).
DNase-seq were from ENCODE (DCC ENCSR000EPC, ENCSR000EMT, and ENCFF000SQM). We counted the number of DNase cuts in a region and generated average footprint profiles centered on the ATCRAT motif (34). To control for sequence bias of DNase-I (35), the same analysis was performed on regions of open chromatin containing the ATCRAT motif not bound by CUX1.
RNA-seq analysis
K562 expressing shCUX1.338, shCUX1.810, or shRen control were treated with 2 μg/mL of doxycycline (Thermo Fisher) to induce shRNA expression for four days. We performed three biological replicates as we performed previously (3). We used the generalized linear model option in EdgeR (36) to derive P-values, creating effects for batch and shRNA, and corrected for multiple testing (37). Supplemental Table S4 contains RNA-seq analysis results. Sequencing data are available in the GEO Database (accession GSE92882).
Machine learning model
We used support vector machines (38) with leave-one-out cross validation. We calculated the area under the curve (AUC), statistical significance, and confidence intervals using bootstrapping and pROC (39). To determine the significance of individual features, we used a resampling-based approach. For each variable, we reshuffled the values of that variable and tested whether the model was as accurate. If the reshuffled values were equally accurate to the unshuffled values, the variable was deemed not useful to the model. Separately, we performed a regression-based analysis to predict the log-fold change of gene expression based on the characteristics of the associated site.
For enhancer RNA quantification, we downloaded poly-A depleted RNA-seq data from ENCODE experiments (DCC ENCSR000COS, ENCSR000CPG, and ENCSR000CPD). We counted the number of RNA-seq reads in a CUX1 peak.
Hi-C data
We obtained Hi-C data from (40). Rather than processing it independently, we relied on the analysis from (41), which produced a list of 96 137 interacting loci between genomic locations. For each putative contact, we built a window of 5 kb surrounding the contact point. Any sites or gene transcription start sites falling within this window were considered to be participating in Hi–C loops.
RESULTS
CUX1 binds distal enhancers
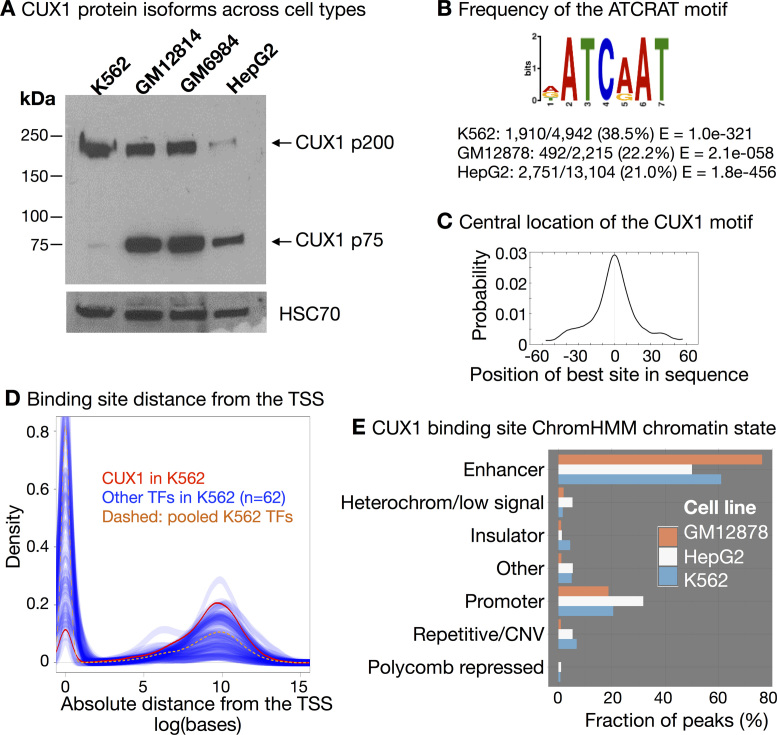
To identify CUX1 genomic targets, we studied human ENCODE cell lines, which are richly annotated with functional genomic datasets (21). ENCODE-generated CUX1 ChIP-seq data were available for three cell types: K562 blast phase chronic myelogenous leukemia cells (42), GM12878 B lymphoblastoid cells (43) and HepG2 hepatoblastoma cells (44). We identified which CUX1 isoforms these cell types express by Western blot. K562 predominantly express the p200 isoform, HepG2 largely express the p75 isoform, while lymphoblastoid cells express both (Figure 1A). We did not detect the p110 isoform.
Figure 1.
CUX1 binds to distal enhancers across three cell types. (A) Western blot of CUX1 in human cell lines from three cell types: myeloid leukemia (K562), lymphoblastoid (GM12814 and GM6984) and liver (HepG2). (B) The ATCRAT DNA binding motif is enriched in CUX1 binding sites. The position weight matrix from MEME-ChIP (46) analysis of K562 peaks (IDR < 0.05) is shown. The position weight matrix was similar in the other cell types. The frequency of the ATCRAT DNA binding motif within CUX1 peaks across cell types is indicated. (C) The ATCRAT motif is enriched in the center of CUX1 binding sites. The graph shows the probability of an ATCRAT motif for each base pair upstream and downstream of the center of K562 CUX1 peaks (P < 4.1e–38). The graph was similar in the other cell types. (D) Graph shows the density of CUX1 sites (in red) in relationship to the absolute distance from the single nearest protein-coding genes’ transcription start site (TSS). Blue lines represent the same densities calculated for 62 other ENCODE transcription factor (TF) datasets in K562. The yellow dashed line represents all 62 TFs pooled together. TSSs were defined by Gencode V.24 annotation. Data are representative of other cell types (shown in Supplemental Figure S1). (E) CUX1 is enriched in predicted enhancer chromatin states. Chromatin states are derived from hidden Markov analysis of eight chromatin marks and CTCF ChIP-seq data in each cell type (31). The height of each bar indicates the proportion of peaks in each cell type that fall within the predicted chromatin state for the respective cell type.
We analyzed the two available CUX1 ChIP-seq replicates for each cell line (21). We called peaks for each replicate and combined replicates according to ENCODE standards using an Irreproducible Discovery Rate (IDR) cut-off of 5% (25,26,45). This analysis yielded 4942 peaks in K562, 2215 in GM12878 and 13 104 in HepG2.
To identify endogenous, in vivo CUX1 DNA-binding motif preferences, we performed de novo motif analysis (46). In agreement with prior studies (16,47) the ATCRAT motif (where R represents A or G) was the most significantly enriched, centrally located motif in each cell type, and was present in 21.0–38.5% of peaks (Figure 1B and C). Despite this association, the vast majority of ATCRAT motifs within open chromatin lacked a CUX1 peak. Of the ∼30 000 ATCRAT motifs present in DNase-I hypersensitivity sites in each cell type, <4% contained a CUX1 peak. Thus, there is a large reservoir of available CUX1 binding sites that are unoccupied. This suggests that the number of potential CUX1 binding sites outnumbers the available CUX1 protein in the cell, and CUX1 genomic occupancy remains unsaturated, a feature of analog TFs (18,19).
CUX1 binding shared certain similarities across cell types. For example, CUX1 peaks were most often distant from the transcription start site (TSS) of the nearest gene, even compared to 62 other TFs assayed by ENCODE (Figure 1D). While all TFs showed some enrichment at the TSS compared to randomly permuted intervals (P < 0.01), CUX1 showed one of the lowest fractions of binding sites at the TSS, compared to other TFs. This tendency towards distal binding was also present in GM12878 and HepG2 (Supplemental Figure S1). Thus in comparison to other TFs, CUX1 binds distal to the promoter, rather than at the promoter. This is of interest because distal cis-regulatory elements are associated with cell type–specific gene expression (48).
CUX1 binding in all cell types also showed stable preferences for particular chromatin states. We used chromatin state predictions derived from published hidden Markov model analysis of eight chromatin marks and CTCF ChIP-seq data for each cell type (31). CUX1 was most enriched at predicted ‘enhancer’ chromatin states, comprising 50–76% of CUX1 peaks, and enriched for H3K4me1, H3K4me2, and H3K27ac (Figure 1E). Promoter binding was second-most common, with repressive and repetitive chromatin binding less common. Thus, across cell types and protein isoforms, CUX1 binds distal cis-regulatory elements associated with gene activation, i.e. enhancers.
CUX1 p75 has a stronger DNase-I footprint than p200
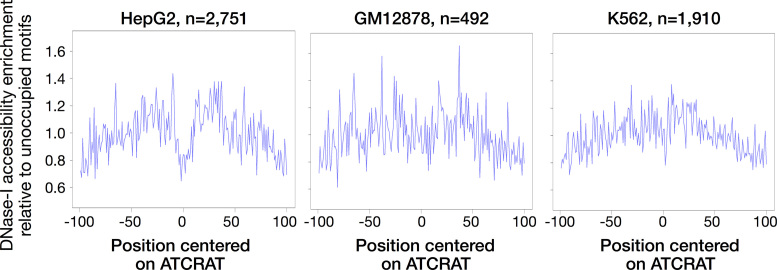
p200 CUX1 binds weakly to DNA, with a faster off-rate, compared to p75 in vitro (15). We tested this finding in vivo by examining the patterns of DNase-I protection afforded by CUX1 binding. Strong, direct binding of a protein to DNA protects the DNA from DNase-I digestion, producing a characteristic signature of read depletion within the bound region (the footprint). It has been suggested that TFs with short DNA residence times (such as nuclear hormone receptors) lack such footprints, while factors with longer residency times (such as CTCF and AP1) have deeper footprints (49,50). To search for a CUX1 footprint, we examined DNase-seq cuts surrounding ATCRAT motifs within CUX1 peaks. We observed a strong depletion of DNase-seq cuts in the central region of HepG2 CUX1 peaks (Figure 2, P < 0.005, permutation test), but no similar signature of DNase protection in K562 (P > 0.10). GM12878, which contains both isoforms, showed an intermediate depletion of cuts in the central six base pairs, suggesting a potentially weak (albeit nonsignificant, P > 0.10) footprint present only in a subset of sites. These observations are consistent with higher affinity binding by the p75 isoform in HepG2 compared to p200 in K562.
Figure 2.
CUX1 p75 has a DNase footprint in HepG2, while CUX1 p200 in K562 does not. Each panel shows the DNase-seq cuts per base pair, as a function of the position within each peak centered on the ATCRAT motif. To control for sequence bias in DNase-I cleavage (35), the data are normalized to the DNase cuts centered at ATCRAT motifs in open chromatin not bound by CUX1. While HepG2 shows a significant (P < 0.01) depletion of cuts in the center of CUX1 sites, indicative of a footprint, DNase data from other cell types showed no significant depletion in the central seven base pairs of sites. The number of ATCRAT-containing CUX1 peaks is indicated.
CUX1 co-occupies sites with transcriptional activators and cohesin
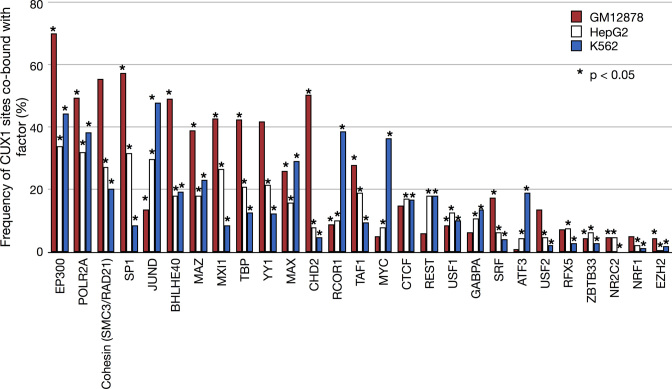
An alternative explanation for differences in DNase-I sensitivity of p75 in HepG2 and p200 in K562 is that CUX1 could potentially form a complex with another TF in HepG2 that contributes to DNase footprinting. To determine the frequency of CUX1 co-occupancy with other proteins, we compared CUX1 peaks to 140 different factors with publically available ChIP-seq data in K562, GM12878, or HepG2. Of the factors, 28 had data in all three lines. The frequency of CUX1 sites co-bound with each of the 28 factors is shown in Figure 3 (Supplemental Table S5 provides data for all 140 factors). We discovered some consistent relationships that replicated across cell types. For example, RNA polymerase II (POLR2A) frequently co-occurred with CUX1, occupying to 32–49% of CUX1 sites. The histone acetyltransferase and transcriptional co-activator, EP300, bound 34–70% of CUX1 sites. We did not observe increased co-occupancy of any TF with CUX1 in HepG2 that could explain the DNase-seq footprints observed in Figure 2. Overall, across cell types and CUX1 isoforms, CUX1 most frequently associated with transcriptional activators.
Figure 3.
CUX1 sites are co-bound with the p300 transcriptional co-activator, RNA polymerase II, and cohesin components. The graph shows the fraction of CUX1 sites that overlap with 28 other factors. The factors are ranked by the mean overlap with CUX1 across the three cell types. The significance of the overlap was determined by obtaining a random expectation (P = 0.05) via permutation of the peaks for each factor over open chromatin, which was defined as any region of the genome bound by any factor from the Uniform Peak calls for each respective cell type. * Indicates P < 0.05. Supplemental Table S5 contains the complete list of factors with overlap frequencies and enrichment.
There was also significant CUX1 co-occupancy with components of the cohesin complex, which is involved in DNA looping (51). Of the cohesin complex members with available data, RAD21 was found at 11–37% of CUX1 peaks and SMC3 at 9–29%. Overall 20–55% of CUX1 sites contained a RAD21 and/or SMC3 protein. This result implies that CUX1 overlaps sites involved in DNA loops and may participate in higher-order chromatin architecture. In summary, CUX1 co-occurs most often with transcriptional activators and cohesin across cell types and isoforms.
CUX1 regulates cell viability pathways
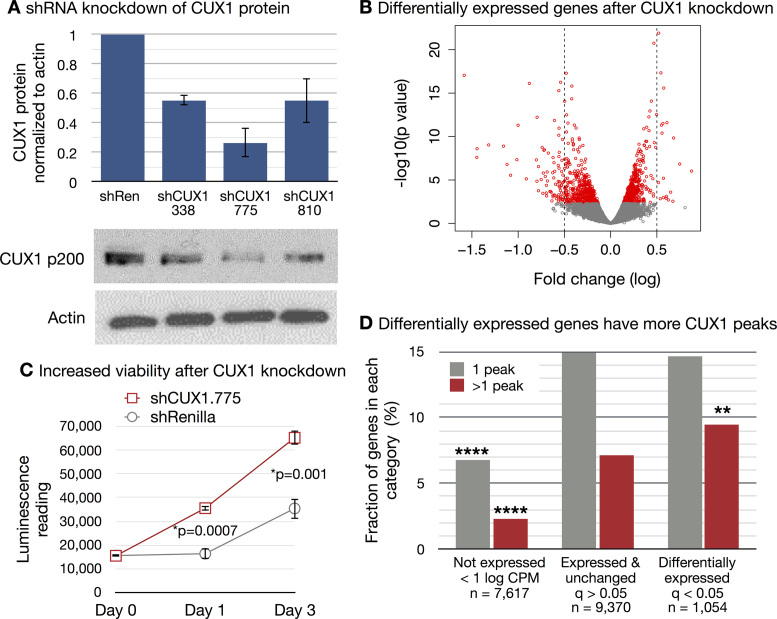
We sought to identify those genes that change expression in response to CUX1 haploinsufficiency. We focused on the K562 myeloid leukemia cells, as we identified CUX1 inactivation in half of high-risk myeloid leukemias (3), whereas CUX1 mutations are uncommon in liver and lymphoid cancers (52). To this end, we generated K562 cells stably expressing inducible shRNA targeting CUX1. Of three shRNAs tested, shCUX1.338 and shCUX1.810 best modeled haploinsufficiency, with 55% residual CUX1 protein (Figure 4A). To identify differentially expressed genes (DEGs) after CUX1 knockdown, we performed RNA-seq on control and two independent shRNAs targeting CUX1. There was a significant concordance between gene expression changes after knockdown with shCUX1.810 compared to shCUX1.338 (Spearman's r = 0.68, P = 2.2e–16). So we identified DEGs from a combined analysis of shCUX1.338 and shCUX1.810 using a generalized linear model (36).
Figure 4.
CUX1 tends towards transcriptional activation and regulates cellular viability. (A) K562 cells were transfected with doxycycline-inducible vectors expressing three independent shRNAs targeting CUX1. A vector expressing shRNA targeting Renilla luciferase (shRen) was used as a control. CUX1 protein was measured by western blot after 7 days of doxycycline treatment. One representative blot of three independent experiments is shown. The graph depicts the quantification of CUX1 protein levels across the three independent experiments, with standard deviations. (B) RNA-seq was performed after four days of doxycycline treatment of K562 cells expressing shCUX1.810, shCUX1.338 or shRen control. Three independent biological replicates were performed. Volcano plot of differentially expressed genes from combined analysis of sh.338 and sh.810 by a generalized linear model. Red indicates differentially expressed genes with FDR < 0.05. (C) Doxycycline-treated K562 cells expressing shCUX1.775 or shRenilla control were cultured in 0.1% FBS containing media. ATP-lite luminescence was measured starting on day 0. One representative experiment of three independent experiments is shown. Three technical replicates were performed within each experiment. (D) Genes were categorized as not-expressed if they had a mean of <1 log counts per million reads (CPM), expressed but unchanged after CUX1 knockdown, or differentially expressed after CUX1 knockdown (FDR < 0.05). The number of genes within each category is indicated. Within each category, the fraction of genes targeted by one or more than one CUX1 peak is plotted. CUX1 peaks from ENCODE were assigned to the single nearest gene within 1 Mb. **** indicates P< 0.0001 chi-squared test compared to expressed genes. ** indicates P = 0.0069 chi-squared test compared to expressed, unchanged genes.
CUX1 knockdown led to 1175 DEGs at a 5% false discovery rate (FDR). The effect size was modest across this large number of DEGs (Figure 4B). 45.6% of DEGs went down after knockdown. Restricting the analysis to genes bound by CUX1 (n = 254), 42.1% went down after knockdown, indicating that in this cell type, CUX1 can both activate or repress target genes.
We looked for pathways altered by CUX1 haploinsufficiency and identified ‘GO:0000278∼mitotic cell cycle’ as the most significant pathway of up-regulated genes (FDR = 2.60E–15), and ‘GO:0042981∼regulation of apoptosis’ as the most significant pathway for down-regulated genes (FDR = 5.74E–04, Supplemental Table S6) (53). In accordance with alteration of these pathways, CUX1 knockdown led to increased K562 cell viability when grown in reduced serum conditions (Figure 4C). Thus, CUX1 regulates viability pathways in K562 cells.
CUX1 fits an analog model of dose-sensitive gene regulation
We then identified the DEGs that are direct targets of CUX1 binding. Genes were categorized as DEG, unchanged, or not expressed in K562 cells. The fraction of genes with one, or more than one, CUX1 binding site was quantified (Figure 4D). CUX1-bound genes were more likely to be expressed, but were not significantly associated with differential expression upon a 50% reduction of CUX1 (P = 0.14). Notably, the presence of multiple CUX1 binding sites was correlated with DEGs. Specifically, 9.49% of DEGs had two or more CUX1 binding sites, significantly more than other expressed genes (Figure 4D, P = 0.0069). Overall, these data implicate the analog model for CUX1 target gene regulation, in which target genes have multiple TF binding sites and modest effect sizes on expression (Figure 4B).
We next tested the affinity-threshold model. We defined CUX1 binding sites as ‘dose-sensitive’ when they were associated with DEGs, and all other binding sites as ‘dose-resistant.’ Per the affinity-threshold model, dose-sensitive sites should be lower affinity. Assuming that CUX1 has higher affinity for the ATCRAT motif (16,47), dose-sensitive sites would be predicted to lack the ATCRAT motif. This result was not evident, as dose-sensitive sites had a similar frequency of the ATCRAT motif compared to dose-resistant sites (P > 0.05). De novo motif analysis (46) did not reveal underlying sequence differences at dose-resistant or dose-sensitive sites. Indeed, Jensen–Shannon divergence analysis (54), of all possible k-mers from 1–10 bp in size, failed to uncover significant differences of the DNA sequence at dose-sensitive and dose-resistant sites. In summary, these results conflict with the affinity-threshold model.
As a second approach to test the affinity-threshold model, we directly measured CUX1 genomic occupancy in the wildtype and haploinsufficient states by ChIP-seq. If CUX1 adheres to the affinity-threshold model, we would expect to observe low- and high-affinity CUX1 binding sites. To test this, we used K562 cells stably expressing CUX1-targeting shRNA-A or shRNA-B, with 24% and 55% residual CUX1 protein, respectively (3). ChIP-seq was performed on two biological replicates for each shRNA and a control shRNA. Of the 2591 CUX1 peaks identified in our control shRNA line (IDR 0.10), 1497 (57.8%) of these were also called in the ENCODE K562 dataset (IDR 0.05).
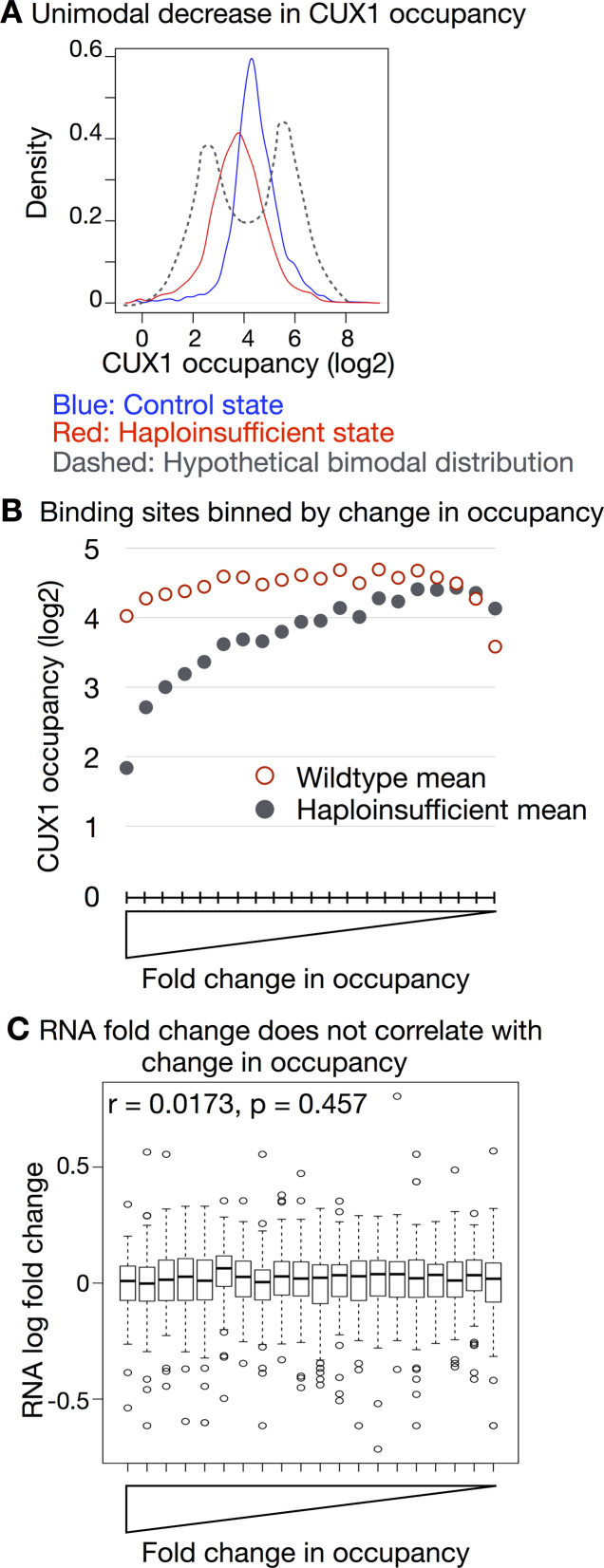
We predicted that low-affinity binding sites would exhibit less occupancy in the haploinsufficient state compared to high-affinity sites. As such, a density plot of CUX1 occupancy would be bimodal, reflecting two populations of sites with either high- or low-occupancy (Figure 5A, dashed line). To test this, we quantified CUX1 occupancy in control peaks before and after CUX1 knockdown. As shown in Figure 5A, CUX1 occupancy showed a unimodal distribution in the control cells. After knockdown, CUX1 occupancy unimodally decreased. The finding that the vast majority of CUX1 peaks decrease after CUX1 knockdown confirms the specificity of the CUX1 peaks identified. This unimodal distribution has also been reported for MYC, which is more analog than binary (55). We did not observe a class of lower-occupancy sites that were particularly dose-sensitive. Indeed, we did not identify any significantly differentially occupied peaks after knockdown after multiple testing correction (30). These results argue against the affinity-threshold model.
Figure 5.
Binding site affinity does not correlate with dose-sensitive gene regulation. ChIP-seq was performed in K562 cells expressing shRNA targeting CUX1 (shCUX1-A or shCUX1-B) or control shRNA. Two biological replicates were performed for each shRNA. (A) CUX1 occupancy after CUX1 knockdown shows a unimodal reduction. We identified peaks in the control shRNA cells (IDR 0.10), and then counted the number of ChIP-seq reads within those regions in the control and the haploinsufficient conditions. Occupancy was defined as the log2 read counts per peak, input subtracted and normalized to library size. The mean occupancy for control (blue) and knockdown (red) conditions is plotted. The dashed line indicates a hypothetical bimodal density plot. (B) Peaks were binned by fold change in occupancy. The bins are ranked from largest fold change to least. The mean CUX1 ChIP-seq occupancy in the control and haploinsufficient conditions is plotted per bin. (C) Peaks were assigned to genes. The boxplots depict the fold change in RNA expression after CUX1 knockdown for genes within the same bins as (B).
Further refuting the affinity-threshold model, CUX1 change in occupancy did not correlate with differential gene expression. First we binned CUX1 peaks according to their fold change in occupancy. This produced an affinity-rank of binding sites with lower affinity binding sites exhibiting the largest change in occupancy (Figure 5B). However, those genes targeted by low-affinity CUX1 binding did not have greater gene expression changes compared to other binding sites (Figure 5C). We also determined that peaks at DEGs did not have significantly different occupancy in the control or haploinsufficient state, nor did they have significantly different change in occupancy (P > 0.05). To remove the potential effects of using different thresholds for occupancy or expression changes, we also tested RNA fold change in expression versus ChIP-seq fold change in occupancy across all peaks and did not observe a correlation (Spearman r = -0.004, P = 0.840). Overall, these data disagree with the affinity-threshold model for CUX1 transcriptional regulation.
In contrast, several features of CUX1 invoke the analog model (19). p200 CUX1 has low-affinity DNA binding (Figure 2 and ref.(15)), there is a large excess of unbound CUX1 binding sites, and there are multiple CUX1 binding sites at dose-sensitive genes (Figure 4C) (18–20). Collectively, these data argue against the affinity-threshold model, and in favor of the analog model of CUX1 gene regulation.
Cis-regulatory features of dose-sensitive CUX1 binding sites
Similar to other TFs (56), most genes associated with CUX1-binding sites genes do not change in expression after CUX1 knockdown. CUX1 directly binds 2314 genes by ChIP-seq, of which 254 are DEG after CUX1 knockdown. To better understand the differences between dose-sensitive and dose-resistant binding sites, we adopted a machine learning approach. We sought to predict whether a given binding site affected the nearest gene's expression, contingent on characteristics of the site.
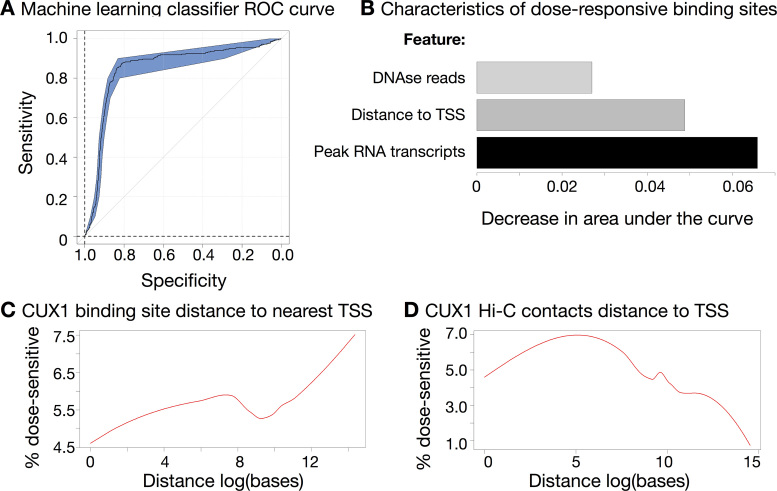
We annotated the binding sites with functional genomic features generated by ENCODE, such as measures of open chromatin, other TF ChIP-seq peaks, and chromatin marks. We found that dose-sensitivity was readily predictable using only three genomic characteristics. A support-vector machine-based model was able to achieve extremely accurate performance with leave-one-out cross validation (Figure 6A; area under the curve (AUC) = 0.89; P < 0.001). We replicated the AUC with multiple methods (39,57). The most informative features of dose-sensitive binding sites were: distance between each binding site and the nearest TSS; the number of DNase-seq reads mapping to each peak; and the number of RNA reads mapping to each peak (enhancer RNA) (58). We found that the three characteristics differed in the extent to which they improved classifier performance (Figure 6B), but worked much better in combination than individually, suggesting interactions between the features. No single feature showed a statistically significant difference between dose-sensitive and -resistant sites (P > 0.05). To control for a possible threshold effect of using a 5% FDR cut-off for DEGs, we also tested the prediction accuracy when using fold change in gene expression instead. This result was also significant (P = 0.045). We conclude that enhancer activity (enhancer RNA and DNase-seq reads) and distance to the TSS are important features of CUX1 function.
Figure 6.
Dose-sensitive CUX1 binding sites loop to the promoter. (A) A Support-Vector Machine approach can accurately classify dose-sensitive and non-sensitive sites. A receiver-operator curve shows an area-under-the-curve (AUC) value of 0.89, significantly above 0.5 (P < 0.01), indicating correct classification of dose-sensitive sites. (B) The support vector machine makes use of three characteristics for each site. Length of each bar indicates the decrease in AUC value when that variable is removed from the classifier. (C) CUX1 peaks more distal from the TSS are more likely to be dose-sensitive. Graph shows the proportion of dose-sensitive CUX1 sites (on the y-axis) as a function of the absolute distance from the TSS (plotted as the natural log of the base pair number on the x-axis). (D) Dose-sensitive CUX1 binding sites loop to the promoter. As in (C), the graph shows the proportion of dose-sensitive CUX1 sites as a function of the distance from the TSS distance, but after incorporating Hi-C contact information. For CUX1 binding sites within 5 kb of one end of a Hi-C contact, the distance of the other end of the Hi-C contact to the nearest TSS is plotted. CUX1 binding sites not assigned to Hi-C contacts are also included.
CUX1 is enriched at sites of DNA looping
We inferred that CUX1 regulates genes via looping to target promoters for the following reasons: (i) CUX1 binds to distal enhancers (Figure 1D and E); (ii) CUX1 cobinds with cohesin (Figure 3) and (iii) Enhancer RNAs are associated with promoter looping (Figure 6B) (58). To test the relationship between CUX1 and DNA looping, we used Hi–C data from K562 cells (41). For each Hi–C contact, we built a window of 5 kb surrounding the contact loci, a distance based on the resolution limits of Hi–C data (41). Any CUX1 peak or gene TSS within this window was considered to be participating in DNA loops.
We found that 54.2% of CUX1 binding sites fell within a Hi–C contact. This was a significant enrichment of 2679 overlaps compared to 1374 expected by chance (P < 0.01, permutation test over open chromatin). We next assessed how looping relates to differential expression of CUX1 target genes. First, we plotted the proportion of dose-sensitive CUX1 binding sites as a function of the distance from the nearest gene (Figure 6C). This analysis revealed distal CUX1 peaks to be enriched for dose-sensitive targets. The fraction of dose-sensitive peaks did not plateau even at a distance as far as 1.7 Mb (14.3 on the natural log scale, Figure 6C). To determine if we could improve upon the peak to gene association by accounting for distal DNA looping, we repeated the analysis after incorporating Hi–C contact information. For CUX1 binding sites within 5 kb of one end of a Hi–C contact, the distance of the other end of the Hi–C contact to the nearest TSS is plotted. With this analysis, a greater proportion of dose-sensitive CUX1 sites is proximal to the TSS, and a maximum dose-sensitive fraction is apparent close to the TSS (Figure 6D). Therefore dose-sensitive CUX1 binding sites loop to promoters.
DISCUSSION
The CUX1 transcription factor is a conserved, essential, and ubiquitous protein recurrently mutated across cancer types. The genome-scale properties and targets of endogenous CUX1 DNA-binding have remained unknown, in any species, creating a substantial gap in our knowledge of CUX1 function. In this study, we provide the first identification of CUX1 genome-wide binding across multiple human cell types and the cis-regulatory features of haploinsufficient gene targets.
We showed that CUX1 predominantly binds distal enhancers, which is significant because distal cis-regulatory elements are associated with cell type–specific gene expression (48). CUX1 is exceptional in this regard when compared to other TFs, which are more likely to bind promoter-proximal regions (Figure 1D). Distal enhancer binding may be an integral property of CUX1, which dictates unique pathways across a breadth of cell types (59).
A previous study performed chromatin affinity purification of an exogenously expressed, tagged, p110 isoform of CUX1 (16). Binding sites were identified by microarrays, which were limited to probes for promoter and coding regions. Based on our results, those microarrays likely missed the majority of CUX1 binding sites. Despite that limitation, the authors observed the ATCRAT motif in just under half of binding sites, and noted that most ATCRAT motifs on the array remained unbound, similar to our findings. After knockdown of CUX1, they determined that some DEG were bound distally from the TSS, indicating that p110 may function similarly to p200 in long-distance gene regulation.
We found that the p75 isoform of CUX1 is associated with greater DNase-I protection, consistent with higher affinity binding. In contrast, the full-length p200 isoform afforded less DNase-I protection, in agreement with prior EMSAs demonstrating faster on-off rates of p200 DNA binding. Despite weaker DNA binding in K562 cells, 1,175 DEGs were identified after 50% CUX1 knockdown, revealing that CUX1 is transcriptionally active in this cell line. Reporter assays previously showed that p200 CUX1 is a transcriptional repressor (14). We observed that CUX1 in K562 cells, which predominantly express the p200 isoform, had both activating and repressive transcriptional activities, in agreement with prior microarray data (4). These apparently dual activities may result from activating functions of the low level of p75 in K562. However, prior reporter assays lacked a native chromatin context and architecture, thus it remains possible that p200 CUX1 may have a role in gene activation in a chromosomal context.
p200 CUX1 gene regulation is more analog than binary. Supporting this, p200 CUX1 had low-affinity binding and a large reservoir of unoccupied binding sites. DEGs contained more than one CUX1 binding site and exhibited modest changes in expression. In contrast to a binary model, analog regulation may provide a more nuanced means for CUX1 to regulate a broad range of pathways across a large concentration range. Stewart-Ornstein et al. proposed that gene expression in proportion to TF dosage might also enable stoichiometric activation of a group of genes (19). This property may explain how changes in CUX1/Cut abundance can to lead to proliferation, lineage fate, or apoptosis, within a single cell type (12,13).
Directly or indirectly, CUX1 is associated with DNA looping. CUX1 binding sites were enriched for cohesin components and Hi-C contact points that linked binding sites to the promoters of dose-sensitive genes. In line with this finding, Pollard and colleagues recently queried hundreds of functional genomic datasets to identify features of interacting enhancer-promoter pairs (60). They independently identified CUX1 binding to be highly predictive of looping (60). It remains to be determined if either: (i) CUX1 promotes DNA looping; or (ii) CUX1 is recruited to pre-existing DNA loops. Intriguingly, SATB1, a member of the CUT homeobox superclass, regulates gene expression by fostering DNA looping and TF recruitment (61). In future work, it will be key to test if CUX1 directly facilitates DNA looping and TF recruitment in a manner similar to SATB1. Most factors show a significant enrichment for binding with CUX1 (Figure 3 and Supplemental Table S5), implying that CUX1 may promote TF recruitment.
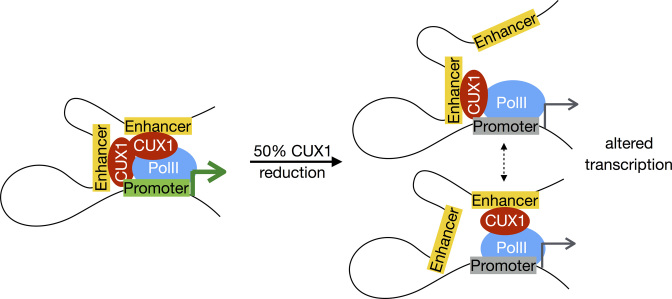
In conclusion, we propose a model for CUX1 haploinsufficient target gene deregulation (Figure 7). At a normal concentration, CUX1 binds distal enhancers and regulates target genes via DNA loops. When CUX1 concentration is halved, CUX1 binding decreases across binding sites. At the single cell level, this may manifest as reduced probability of CUX1 binding at any specific locus; in a population of cells, this is seen as decreased occupancy at all loci. Binding sites characterized by looping to the promoter and the interaction of other cis-regulatory features (Figure 6B) have a quantitative impact on gene expression. We speculate that given the distinct features of CUX1 genomic binding properties compared to other TFs, this model may be unique to CUX1 and not generalizable to most other haploinsufficient transcriptional regulators.
Figure 7.
Model for haploinsufficient CUX1 target gene deregulation. At a normal concentration, CUX1 binds at sites of chromatin contacts between distal enhancers and gene promoters, promoting their expression. When CUX1 dosage is halved, CUX1 binding decreases across binding sites. At the single cell level, this may manifest as reduced probability of CUX1 binding at any specific locus; in a population of cells, this is seen as decreased occupancy at all loci. Binding sites characterized by looping to the promoter and the interaction of other genomic characteristics (Figure 6B) will have a quantitative impact on gene expression. It remains to be determined if either: (i) CUX1 promotes DNA looping; or (ii) CUX1 is recruited to pre-existing DNA loops.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Yoav Gilad, Barbara Kee, and Angela Stoddart for critical reading of the manuscript. RNA-seq library preparation and RNA and ChIP next-generation sequencing was performed at The University of Chicago High-Throughput Genome Analysis Core facility and the Genomics Core Facility.
Footnotes
Present address: Megan E. McNerney, Department of Pathology, The University of Chicago, Knapp Center for Biological Discovery Room 5128, 900 E. 57th Street, Chicago, IL 60637, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [5K08CA181254 to M.E.M.]; The V Foundation for Cancer Research V Foundation Scholar Award; The University of Chicago Medicine Comprehensive Cancer Center Support Grant [P30CA14599]; American Cancer Society Institutional Research Grant [IRG-58-004-53], Riviera Country Club United-4 A Cure; The University of Chicago Cancer Research Foundation Auxiliary Board. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Davoli T., Xu A.W., Mengwasser K.E., Sack L.M., Yoon J.C., Park P.J., Elledge S.J.. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013; 155:948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A. Jr, Kinzler K.W.. Cancer genome landscapes. Science. 2013; 339:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McNerney M.E., Brown C.D., Wang X., Bartom E.T., Karmakar S., Bandlamudi C., Yu S., Ko J., Sandall B.P., Stricker T. et al. . CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. 2013; 121:975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong C.C., Martincorena I., Rust A.G., Rashid M., Alifrangis C., Alexandrov L.B., Tiffen J.C., Kober C., Green A.R. Chronic Myeloid Disorders Working Group of the International Cancer Genome, C. et al. . Inactivating CUX1 mutations promote tumorigenesis. Nat. Genet. 2014; 46:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ludlow C., Choy R., Blochlinger K.. Functional analysis of Drosophila and mammalian cut proteins in files. Dev. Biol. 1996; 178:149–159. [DOI] [PubMed] [Google Scholar]
- 6. Rong Zeng W., Soucie E., Sung Moon N., Martin-Soudant N., Berube G., Leduy L., Nepveu A.. Exon/intron structure and alternative transcripts of the CUTL1 gene. Gene. 2000; 241:75–85. [DOI] [PubMed] [Google Scholar]
- 7. Johnson T.K., Judd B.H.. Analysis of the cut locus of Drosophila melanogaster. Genetics. 1979; 92:485–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sinclair A.M., Lee J.A., Goldstein A., Xing D., Liu S., Ju R., Tucker P.W., Neufeld E.J., Scheuermann R.H.. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 2001; 98:3658–3667. [DOI] [PubMed] [Google Scholar]
- 9. Ledford A.W., Brantley J.G., Kemeny G., Foreman T.L., Quaggin S.E., Igarashi P., Oberhaus S.M., Rodova M., Calvet J.P., Vanden Heuvel G.B.. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev. Biol. 2002; 245:157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ellis T., Gambardella L., Horcher M., Tschanz S., Capol J., Bertram P., Jochum W., Barrandon Y., Busslinger M.. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001; 15:2307–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001; 270:1–15. [DOI] [PubMed] [Google Scholar]
- 12. Pitsouli C., Perrimon N.. The homeobox transcription factor cut coordinates patterning and growth during Drosophila airway remodeling. Sci. Signal. 2013; 6:ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grueber W.B., Jan L.Y., Jan Y.N.. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003; 112:805–818. [DOI] [PubMed] [Google Scholar]
- 14. Ramdzan Z.M., Nepveu A.. CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat. Rev. Cancer. 2014; 14:673–682. [DOI] [PubMed] [Google Scholar]
- 15. Moon N.S., Berube G., Nepveu A.. CCAAT displacement activity involves CUT repeats 1 and 2, not the CUT homeodomain. J. Biol. Chem. 2000; 275:31325–31334. [DOI] [PubMed] [Google Scholar]
- 16. Vadnais C., Awan A.A., Harada R., Clermont P.L., Leduy L., Berube G., Nepveu A.. Long-range transcriptional regulation by the p110 CUX1 homeodomain protein on the ENCODE array. BMC Genomics. 2013; 14:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Briscoe J., Small S.. Morphogen rules: design principles of gradient-mediated embryo patterning. Development. 2015; 142:3996–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lorberbaum D.S., Barolo S.. Gene regulation: when analog beats digital. Curr. Biol. 2013; 23:R1054–R1056. [DOI] [PubMed] [Google Scholar]
- 19. Stewart-Ornstein J., Nelson C., DeRisi J., Weissman J.S., El-Samad H.. Msn2 coordinates a stoichiometric gene expression program. Curr. Biol. 2013; 23:2336–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giorgetti L., Siggers T., Tiana G., Caprara G., Notarbartolo S., Corona T., Pasparakis M., Milani P., Bulyk M.L., Natoli G.. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol. Cell. 2010; 37:418–428. [DOI] [PubMed] [Google Scholar]
- 21. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y. et al. . An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013; 5:1704–1713. [DOI] [PubMed] [Google Scholar]
- 23. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hansen P., Hecht J., Ibrahim D.M., Krannich A., Truss M., Robinson P.N.. Saturation analysis of ChIP-seq data for reproducible identification of binding peaks. Genome Res. 2015; 25:1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Q., Brown J.B., Huang H., Bickel P.J.. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 2011; 5:1752–1779. [Google Scholar]
- 27. Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R. et al. . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. et al. . Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. et al. . Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011; 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De S., Pedersen B.S., Kechris K.. The dilemma of choosing the ideal permutation strategy while estimating statistical significance of genome-wide enrichment. Brief Bioinform. 2014; 15:919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piper J., Elze M.C., Cauchy P., Cockerill P.N., Bonifer C., Ott S.. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013; 41:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He H.H., Meyer C.A., Hu S.S., Chen M.W., Zang C., Liu Y., Rao P.K., Fei T., Xu H., Long H. et al. . Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods. 2014; 11:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Storey J.D. A direct approach to false discovery rates. J. Roy. Stat. Soc.: Ser. B (Statistical Methodology). 2002; 64:479–498. [Google Scholar]
- 38. Karatzoglou A., Meyer D., Hornik K.. Support vector machines in R. 2006; 15:28. [Google Scholar]
- 39. Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Muller M.. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011; 12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. et al. . Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lan X., Witt H., Katsumura K., Ye Z., Wang Q., Bresnick E.H., Farnham P.J., Jin V.X.. Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic Acids Res. 2012; 40:7690–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lozzio C.B., Lozzio B.B.. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975; 45:321–334. [PubMed] [Google Scholar]
- 43. International HapMap, C. A haplotype map of the human genome. Nature. 2005; 437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aden D.P., Fogel A., Plotkin S., Damjanov I., Knowles B.B.. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979; 282:615–616. [DOI] [PubMed] [Google Scholar]
- 45. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. et al. . ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G. et al. . DNA-binding specificities of human transcription factors. Cell. 2013; 152:327–339. [DOI] [PubMed] [Google Scholar]
- 48. Natarajan A., Yardimci G.G., Sheffield N.C., Crawford G.E., Ohler U.. Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res. 2012; 22:1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sung M.H., Guertin M.J., Baek S., Hager G.L.. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol. Cell. 2014; 56:275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gusmao E.G., Allhoff M., Zenke M., Costa I.G.. Analysis of computational footprinting methods for DNase sequencing experiments. Nat. Methods. 2016; 13:303–309. [DOI] [PubMed] [Google Scholar]
- 51. Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S. et al. . Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010; 467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. . The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 54. Lin J. Divergence measures based on the Shannon entropy. IEEE Trans. Infor. Theory. 1991; 37:145–151. [Google Scholar]
- 55. Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R. et al. . c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012; 151:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cusanovich D.A., Pavlovic B., Pritchard J.K., Gilad Y.. The functional consequences of variation in transcription factor binding. PLoS Genet. 2014; 10:e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sing T., Sander O., Beerenwinkel N., Lengauer T.. ROCR: visualizing classifier performance in R. Bioinformatics. 2005; 21:3940–3941. [DOI] [PubMed] [Google Scholar]
- 58. Li W., Notani D., Rosenfeld M.G.. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016; 17:207–223. [DOI] [PubMed] [Google Scholar]
- 59. Sansregret L., Nepveu A.. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. 2008; 412:84–94. [DOI] [PubMed] [Google Scholar]
- 60. Whalen S., Truty R.M., Pollard K.S.. Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat. Genet. 2016; 48:488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cai S., Lee C.C., Kohwi-Shigematsu T.. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006; 38:1278–1288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.