Abstract
Guanine-rich (G-rich) homopurine–homopyrimidine nucleotide sequences can block transcription with an efficiency that depends upon their orientation, composition and length, as well as the presence of negative supercoiling or breaks in the non-template DNA strand. We report that a G-rich sequence in the non-template strand reduces the yield of T7 RNA polymerase transcription by more than an order of magnitude when positioned close (9 bp) to the promoter, in comparison to that for a distal (∼250 bp) location of the same sequence. This transcription blockage is much less pronounced for a C-rich sequence, and is not significant for an A-rich sequence. Remarkably, the blockage is not pronounced if transcription is performed in the presence of RNase H, which specifically digests the RNA strands within RNA–DNA hybrids. The blockage also becomes less pronounced upon reduced RNA polymerase concentration. Based upon these observations and those from control experiments, we conclude that the blockage is primarily due to the formation of stable RNA–DNA hybrids (R-loops), which inhibit successive rounds of transcription. Our results could be relevant to transcription dynamics in vivo (e.g. transcription ‘bursting’) and may also have practical implications for the design of expression vectors.
INTRODUCTION
Certain nucleotide sequences in DNA that interfere with transcription can have important biological consequences (reviewed in (1)). Among these sequences are homopurine–homopyrimidine stretches, in which one DNA strand contains only purines and the complementary strand contains only pyrimidines. These sequences can cause partial transcription blockage when the homopyrimidine DNA strand is the template for transcription (2,3). The blockage increases with G-richness of the sequence, the sequence length, the presence of a break in the non-template strand near the sequence and with negative supercoiling (2,3).
All of these factors that contribute to the blockage correlate with the factors facilitating the formation of R-loops; these are structures in which an RNA strand ‘invades’ a longer DNA duplex, to generate an RNA–DNA duplex with a complementary region within one DNA strand, consequently displacing the homologous region within the other DNA strand. R-loops are widely distributed within the genome, and they produce multiple biological effects, both advantageous and deleterious (reviewed in (4–13)). They can form either co-transcriptionally, or by post-transcriptional RNA invasion ‘in trans’; the latter is usually mediated by specific proteins (reviewed in (8)).
R-loops preferably form within sequences for which the RNA–DNA duplex has superior stability over the corresponding DNA–DNA duplex; this is the case for G-rich sequences, provided that RNA strand is purine-rich, and that the complementary DNA strand is pyrimidine-rich ((14–16) and references therein). R-loop formation is also facilitated by any other factors that impair the propensity of the non-template DNA strand to hybridize with the template DNA strand in the wake of transcription, e.g. negative supercoiling (17,18) and breaks in the non-template DNA strand (17). The correlation between the factors causing transcription blockage and the factors facilitating R-loop formation has led to the model in which the transcription blockage is caused by R-loop formation, and several possible mechanisms for R-loop-mediated transcription blockage have been suggested ((2,3,19), reviewed in (1)). There are two major modes for R-loop interference with transcription: (i) R-loop formation in the wake of an RNA polymerase could affect transcription of that polymerase (2,3,19,20) and (ii) transcription by an RNA polymerase could be impacted by stable R-loops formed during preceding rounds of transcription (21).
It is important to note that homopurine–homopyrimidine sequences (or, more generally, sequences with strongly skewed purine-pyrimidine distributions between the DNA strands) could form a number of unusual DNA structures, e.g. H-DNA-type triplexes (reviewed in (22)) and (if they contain clusters of guanines in the purine-rich DNA strand) G-quadruplexes (reviewed in (23,24)). Both triplexes and quadruplexes could participate in formation of composite structures containing RNA–DNA hybrids ((25–27), reviewed in (1,28)); and they can facilitate R-loop formation and contribute to transcription blockage (29–33). However, the correlation between blockage patterns produced by the different factors that facilitate the R-loop formation implies some general ‘core’ mechanism for the R-loop-mediated transcription blockage (34).
An additional factor that enhances the R-loop formation is shortening of the distance between a sequence prone to R-loop formation and the transcription promoter (17). In that work (17), this effect was explained in terms of a thread-back model for R-loop formation: According to this model, the R-loop formation occurs after the nascent RNA is extruded from the transcription complex; thus, in order to form the R-loop the nascent RNA must ‘thread back’ into DNA duplex. The presence of a long RNA ‘tail’ behind the R-loop-forming sequence sterically interferes with this threading making the R-loop formation more difficult. Location of the R-loop-prone sequence closer to the promoter decreases the length of the RNA tail behind the R-loop forming sequence, thus facilitating R-loop formation.
If the transcription blockage by a G-rich homopurine–homopyrimidine sequence is caused by R-loop formation, we predict that placing that sequence closer to the promoter should increase the blockage. In the present study we have tested this prediction and we confirm that blockage is much stronger if the sequence of interest is localized closer to the promoter. We have also concluded that the predominant cause of blockage in this case is formation of stable R-loops during the initial round of transcription that inhibit following rounds of transcription.
MATERIALS AND METHODS
DNA substrates
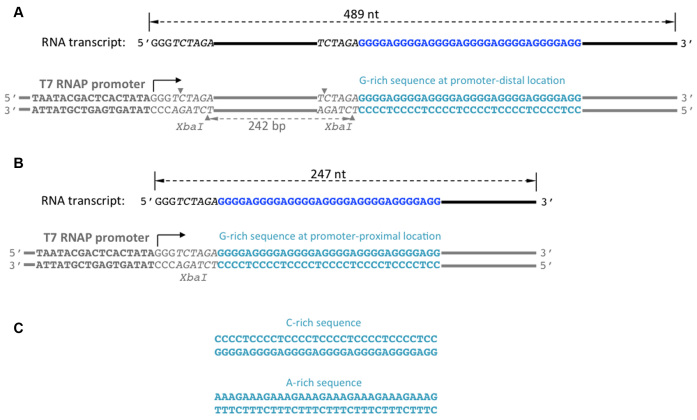
Plasmids containing homopurine/homopyrimidine sequences localized far from the T7 RNAP promoter (promoter–distal substrates) have been described in detail (3), and their important characteristics for the present work are shown in Figure 1A. Plasmids containing homopurine/homopyrimidine sequences localized close to the promoter (promoter–proximal substrates) were obtained by deletion of the fragment localized between two Xba I sites from the respective promoter–distal substrates (Figure 1B). All plasmids were purified using standard Qiagen maxiprep protocol, except that cell lysis time was reduced to several seconds. To obtained linearized substrates, the plasmids were digested by Hind III restriction enzyme and restriction products were purified from agarose gels, as described in (2,3). As a template for ‘spiking transcript’ (used to eliminate effects of loading errors and purification losses, see below), the plasmid pN-aga-hTel-C (3) linearized by Sca I was used. This substrate produces a run-off product of 1877 nt without any other detectable transcription products. This product is much longer than run-off products from the substrates of interest (489 nt and 247 nt for the promoter–distal and promoter–proximal substrates, respectively) and clearly separated from them during gel-electrophoresis.
Figure 1.
DNA substrates. The designation of homopurine/homopyrimidine sequences (i.e. G-rich, C-rich or A-rich) correspond to the non-template DNA strand. The bottom DNA strand is the template strand (i.e. the one that serves as a template for transcription), the top DNA strand is the non-template strand. DNA is shown in gray, except for the homopurine/homopyrimidine sequence insert shown in turquoise. T7 RNAP promoter is shown in bold, and the transcription start site is designated by a bent arrow. Xba I restriction sites are shown in italic, and the cleavage sites are shown by small gray triangles. DNA substrates are linear, obtained by restriction digestion of the respective supercoiled plasmid (see Materials and Methods). There are no specific transcription termination sites, so unobstructed transcription proceeds to the very end of the DNA template producing full-size (run-off) RNA products. Run-off RNA products are shown above the respective DNA templates in black, except for the homopurine sequence shown in dark-blue. The sizes of run-off transcription products are indicated by black dashed double-arrowed lines. (A) Substrate with promoter–distal location of the homopurine/homopyrimidine sequence (the G-rich sequence is shown). The distance between this sequence and the transcription start site is 252 bp. The substrate contains two Xba I restriction sites, one is localized 3 bp downstream from promoter, and the other is localized immediately upstream from the homopurine/homopyrimidine sequences. (B) Upon deletion of the fragment between these two sites, substrate with promoter–proximal location of the homopurine/homopyrimidine sequence (the G-rich sequence is shown) is obtained. In this substrate, the distance between the homopurine/homopyrimidine sequence and the transcription start site is only 9 bp. (C) C-rich and A-rich sequences.
Transcription
In the case of ‘high’ T7 RNAP concentration, the in vitro transcription reaction was performed for 30 min at 37°C in 12 μl of mixture containing 33 mM Tris–HCl (pH 7.9), 5 mM MgCl2, 8.3 mM NaCl, 1.7 mM spermidine, 4.2 mM DTT, 0.17 mM of each non-radioactive (‘cold’) NTP, 10 μCi of radioactive (α-32P) CTP (which corresponds to the final concentration about 0.0003 mM), 1.3 units/μl of RNasin, 1.7 units/μl of T7 RNAP (both from Promega corp, Madison, WI, USA) and 10 ng of DNA substrate. Below, we will refer to these concentrations of reagents as ‘standard concentrations’. For ‘low’ T7 RNAP concentration, all conditions were the same, except that the T7 RNAP concentration was 30-fold lower.
In the case of ‘pre-transcription’ experiments, the transcription was first performed for 30 min at 37°C with the standard concentrations of all reagents, except that radioactive CTP was omitted (and in the NTP-minus control ‘cold’ NTPs were omitted as well). Then, 1.2 μl of this mixture were mixed with 34.8 μl of solution containing the standard concentration of all transcription reagents, except that T7 RNAP and DNA substrate were omitted; and incubation was continued for another 30 min at 37°C.
In the case of transcription in the presence of RNase H, the reaction was supplemented with 0.42 units/μl of RNase H (NEB). Otherwise, conditions were standard, except that instead of 10 ng, 1/3 ng of DNA substrate were used.
Transcription reactions were stopped by adding EDTA up to 12.5 mM.
A spiking transcript was obtained in a separate transcription reaction running in parallel with the other samples; after all transcription reactions were stopped, identical amounts of spiking transcript were added to each sample. These amounts were usually adjusted so that the spiking signal would be comparable with that of the sample signal.
After stopping the transcription reaction and adding the spiking transcript, 1.5 μl of sample were mixed with 3 μl of formamide loading buffer, heated at 85°C for ∼2 min and analyzed by gel-electrophoresis in a 6% sequencing gel.
In some initial experiments, additional purification of the sample by SDS/Proteinase K treatment followed by ethanol precipitation was used, as previously described (32). However, this additional purification did not affect the results, so was later omitted.
Quantitation
Intensities of the transcript signals were measured by phosphorimaging and quantitated with Bio-Rad Image Lab software. Each signal was normalized upon the intensity of the spiking transcript in the same lane. This procedure would compensate random variations in the sample signals intensities due to purification losses and gel-loading errors, because purification losses and gel-loading errors would be the same for the sample and for the spiking transcript that was added to this sample immediately after the transcription reaction.
Since we were using radioactive (α-32P) CTP for the sample labeling, the radioactive labeling of each transcript is proportional to the number of cytosines within this transcript. Thus, to compare molar yields of transcripts with different lengths and compositions, their radioactive signals were normalized by the number of cytosines within a given transcript.
RESULTS
Experimental design
For in vitro transcription experiments, we used two types of DNA substrates: promoter–distal (Figure 1A) and promoter–proximal (Figure 1B). In promoter–distal substrates, a homopurine–homopyrimidine sequence 32 nt long (further referred as PuPy-insert) was localized around 250 bp from the starting point of transcription, while in the promoter–proximal substrates, most of the DNA sequence between the starting point of transcription and the PuPy-insert was deleted, and only 9 bp were left between the starting point of transcription and the PuPy-insert. The rest of the sequence for both types of substrates was the same.
In our experiments, we used linear DNA substrates without any specific termination signals. Within these substrates, unobstructed transcription proceeds from the promoter to the very end of the DNA template producing a well-defined complete (run-off) transcription product. The lengths of the run-off transcription product were 489 nt and 247 nt for the promoter–distal (Figure 1A), and promoter–proximal (Figure 1B) substrates, respectively. We used the amount of run-off transcription product (monitored by radioactive labeling) as a measure for the yield of transcription. To eliminate the effect of loading errors and losses during purification, after the termination of the transcription reaction, the equal amounts of radioactively-labeled spiking transcript were added to each sample, and each signal was normalized upon the intensity of the spiking transcript in the same lane. Note that in our experiments transcripts are ‘body-labeled’; consequently, the number of radioactive nucleotides within a transcript depends upon its length and composition. Thus, to compare the true (i.e. molar) yields of transcripts for different substrates, their radioactive signals were normalized by the number of radioactive nucleotides within the respective transcripts (see Materials and Methods section for details).
Effect of the sequence composition, location and RNAP concentration upon the yield of transcription
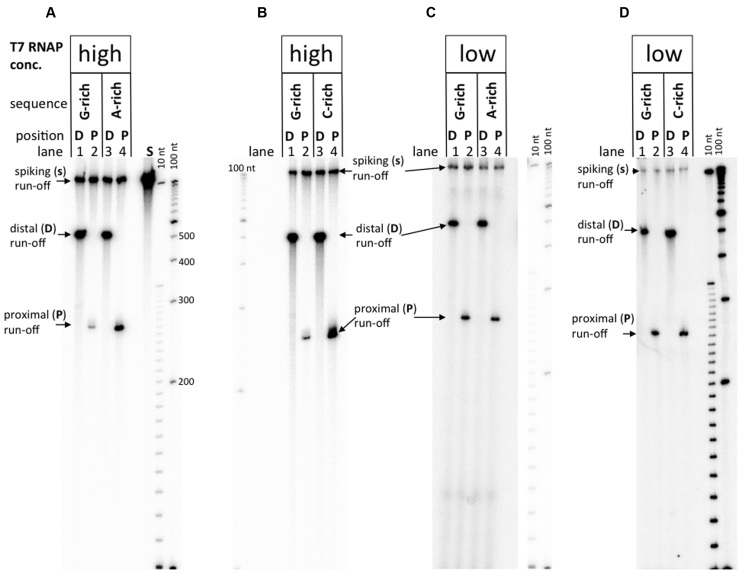
Transcription experiments for various promoter–distal and promoter–proximal substrates are shown in Figure 2, and their quantitation is shown in Figure 3.
Figure 2.
Transcription from DNA substrates with two different T7 RNAP concentrations. ‘High’ concentration of T7 RNAP corresponds to 1.7 units/μl; ‘low’ concentration is 30-fold less. Size standards are denatured radioactive labeled DNA fragment ‘ladders’ with step-sizes 100 nt and 10 nt. Panels A–D are representative gel images for various substrates and RNAP concentrations, as indicated in the lane headings.
Figure 3.
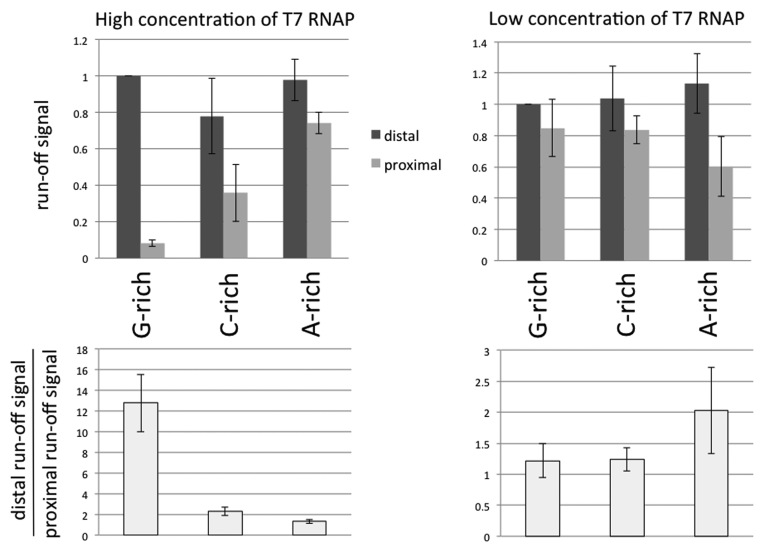
Comparison of transcriptional yields with two different T7 RNAP concentrations. The intensities of full-size (run-off) products (referred as ‘run-off’ signals) were used as a measure for the transcriptional yields. To obtain the molar amounts of the products, the signals were normalized to the number of radioactive nucleotides within the transcript; and to eliminate the effect of loading errors and losses during purification, each signal was normalized to the intensity of the spiking transcript in the same lane (see Materials and Methods). In addition, all run-off signals were normalized to the signal for substrate with promoter–distal G-rich insert; thus, the height of the column that corresponds to this signal is equivalent to 1, and it doesn't have error bars. All experiments were repeated at least twice.
We studied three PuPy-inserts: G-rich (Figure 1A and B), C-rich (Figure 1C, top), and A-rich (Figure 1C, bottom). (These designations correspond to the composition of the non-template DNA strand.). Note that the sequence motif of the non-template strand for the C-rich insert is the same as that of the template strand for the G-rich insert.
For all promoter–distal substrates, the yields of transcripts were similar (see dark-gray columns in top diagrams in Figure 3). However, a dramatically different result appears for the promoter–proximal substrates at high T7 RNAP concentration (see Figure 2A and B, and left diagrams in Figure 3).
For the G-rich PuPy insert, the yield of transcription for promoter–proximal substrate decreases ∼13-fold in comparison with that for the promoter–distal substrate. For the C-rich PuPy insert, this effect was much less pronounced (only 2-fold difference in yield between promoter–distal and promoter–proximal substrates), and for the A-rich insert, within the error of our experiments, this effect was not pronounced (see bottom-left diagram in Figure 3). Importantly, the G-rich sequence, for which this effect (further referred to as ‘transcription blockage’) was most pronounced, is the one that would form the most stable RNA–DNA hybrid ((3) and references therein), suggesting that R-loop formation is responsible for the blockage (see below).
Unexpectedly, the blockage effect also depended upon T7 RNAP concentration: the difference in transcription yields for the promoter–distal and promoter–proximal substrates for the G-insert became insignificant upon a 30-fold decrease in T7 RNAP concentration, i.e. the ratio of transcription yields for promoter–distal and promoter–proximal substrates drops from 13-fold to about unity (Figure 2C and D versus A and B; and the bottom-right versus bottom-left diagram in Figure 3; also see Supplementary Figure S1 for the intermediate RNAP concentrations). For the C-rich insert, this ratio also decreased upon decrease in RNAP concentration from two-fold to about unity. For the A-rich insert the blockage appears to increase from about unity to about two-fold upon RNAP dilution; however, this relatively minor effect is comparable with the errors of our experiments and validation of its significance would require further studies.
Since the magnitude of the blockage, as well as the effect of RNAP concentration was much more pronounced for the G-rich sequence than for other sequences, we used this sequence for further experiments to elucidate the mechanism of the blockage.
The blockage is caused by RNA–DNA hybrid formation during preceding rounds of transcription that inhibit further rounds of transcription
Dependence of the transcription blockage upon RNAP concentration suggests that different RNAP molecules are affecting each other during transcription, either directly (e.g. by active collisions between different RNAP molecules that could cause dissociation of one of RNAP from the template (35)), or indirectly (e.g. via alterations within the DNA substrate caused by transcription).
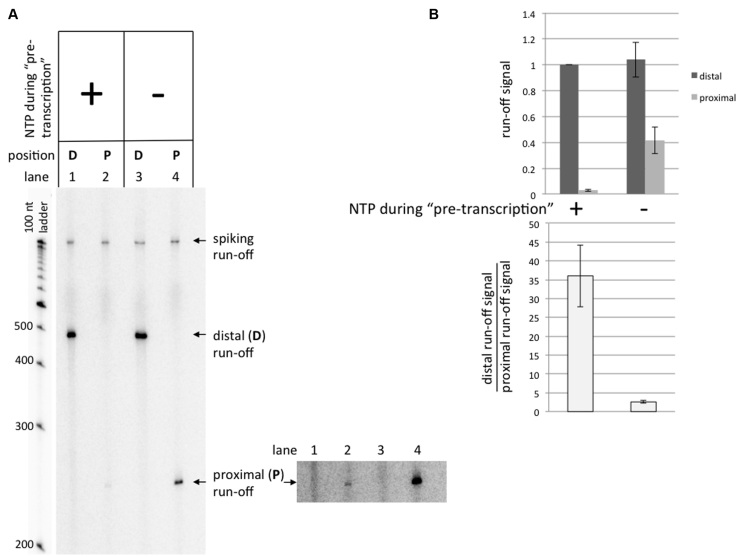
To distinguish between these two possibilities, we first asked whether preceding rounds of transcription could inhibit the following rounds. For that, we first ‘pre-transcribed’ the substrates with the G-rich PuPy insert with the high concentration of RNAP and non-radioactive NTPs (so that transcripts obtained at this stage would be ‘invisible’), and then added a small aliquot of the pre-transcribed mixture into the solution containing non-radioactive NTPs together with radioactive CTP in order to radioactively label the transcripts obtained at this stage. Control samples were treated exactly the same, except that they underwent ‘mock’ pre-transcription without NTPs (see Materials and Methods for details). It is seen (Figure 4) that transcription from the promoter–distal substrate is unaffected by pre-transcription with NTPs (Figure 4A, lane 1 versus lane 3; and dark-gray columns in the top diagram in Figure 4B), while for the promoter–proximal substrate the yield for pre-transcribed in the presence of NTPs samples is much smaller (∼14-fold) in comparison with the NTP-minus control (Figure 4A, lane 2 versus lane 4; and the bottom diagram in Figure 4B). Note that for pre-transcribed samples, the ratio of the transcription yields for the promoter–distal and promoter–proximal substrates was about 36 (Figure 4B, bottom diagram), which is even greater than under our standard conditions (∼13), and corresponds to ∼97% transcription blockage. Thus, transcription through the promoter–proximal G-rich PuPy insert strongly inhibits further rounds of transcription.
Figure 4.
‘Pre-transcription’ experiments. Substrates containing the G-rich sequence were used in these experiments. See Results section for description of the experiment. (A) Gel image. At the bottom-right, a higher exposure for the gel section containing run-offs from promoter–proximal substrate is shown. (B) Quantitation of the results. All run-off signals are normalized to the signal for the promoter–distal substrate pre-transcribed in the presence of NTPs.
To elucidate the mechanism of this inhibition, we performed the transcription reaction in the presence of RNase H to specifically degrade the RNA within RNA–DNA duplexes. It is seen (Figure 5A, lane 4 versus lane 2; and Figure 5B for quantitation) that the presence of RNase H during the transcription practically abolishes the blockage, indicating that the blockage is caused by RNA–DNA hybrids, i.e. R-loop formation.
Figure 5.
Effect of RNase H upon transcription. Substrates containing the G-rich sequence were used in these experiments. See the Results section for description of the experiment. (A) Gel image. (B) Quantitation of the results. All run-off signals are normalized to the signal for promoter–distal substrate transcribed without RNase H.
(Note that we mean RNA–DNA hybrids other than the short RNA–DNA hybrid (around 7–8 bp) that is present inside the transcription complex (reviewed in (36)). From the fact that the yield of the transcription product for the promoter–distal substrate is practically unaffected by RNase H (Figure 5B, dark-gray columns) we conclude that this short hybrid is well-protected from RNase by the transcription complex, and consequently, RNase H does not affect the process of normal transcription.) Note that the presence of RNase H also does not detectably affect the size of the run-off product in the major fraction of the sample (Figure 5A, left panel, lane 4 versus lane 2). However, in the higher exposure image of the same gel (Figure 5A, right panel, lane 4) it is seen that in the presence of RNase H, a minor fraction of shorter run-off products (shown by square bracket) appear, likely as a result of RNase H-mediated digestion of RNA within the R-loop (see Figure 6 for detailed explanation). Note that in the case of the promoter–distal substrate, a minor (∼1% relative to the run-off) band can be seen in the higher exposure image (Figure 5A, right panel, lane 1, white block arrow). This is the repeat-exiting blockage signal, described in detail in (3). We remark that this signal is not pronounced when transcription is performed in the presence of RNase H (Figure 5A, right panel, lane 3 versus lane 1), supporting our model that this signal is caused by R-loop formation (3).
Figure 6.
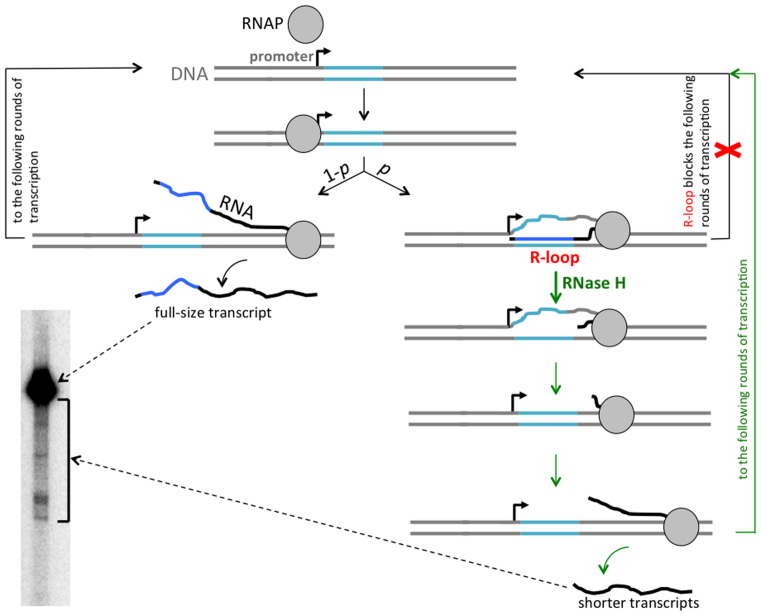
Model for transcription blockage by R-loop formation in the vicinity of the promoter. The R-loop-prone (G-rich) DNA sequence is shown in turquoise, the rest of DNA is shown in gray, transcript from the R-loop-prone sequence is shown in dark blue, the rest of RNA is shown in black, a bent arrow indicates the transcription start site. RNA polymerase (RNAP) is shown as a gray circle. During transcription, an R-loop is formed with a certain probability p, while transcription proceeds without R-loop formation with probability 1 – p. R-loop formation could be initiated somewhere within the R-loop-prone sequence, but then the nascent RNA tail is likely to invade the entire R-loop-prone sequence (probably, even further upstream to the very start of transcription) as shown. The RNAP that created the R-loop could continue transcription in the ‘R-loop mode’, and then stall, either within, or at some distance downstream from the R-loop-prone sequence. At least some of the stalled RNAPs may remain bound to the DNA template (as shown), or could dissociate (not shown). In any case, R-loop formation blocks further rounds of transcription (the blockage is symbolized by the red crisscross). Addition of RNase H during transcription (all arrows that symbolize transitions within RNase H-related pathway are shown in green) leads to R-loop removal and, consequently, eliminates the blockage (blockage elimination is symbolized by the green path parallel to the crisscrossed path). The substrate DNA molecules from which R-loop was removed, then become available for further rounds of transcription, and would produce some number of normal full-sized transcripts, before an R-loop would form again. In addition, an RNAP stalled within an R-loop could resume transcription upon R-loop removal, producing a shorter transcript. That accounts for the pattern of transcription products obtained in the presence of RNase H (lane 4 in Figure 5, the relevant part of it is placed in the present figure.).
RNAP sequestration does not detectably contribute to the transcription blockage in our system
The results described in previous sub-sections indicate that the decrease in transcription yield in the case of the promoter–proximal R-loop-prone sequence is caused by R-loop formation, primarily by inhibiting the following rounds of transcription. The most likely explanation for this is that the presence of an R-loop in close vicinity of the promoter within a given DNA template interferes with transcription initiation by new RNAP molecules. However, in principle, an alternative mechanism is possible, in which the R-loop sequesters RNAP molecules, preventing them from participation in following rounds of transcription. It has been shown that certain unusual DNA structures are capable of sequestering RNAP (30). Such a sequestration could occur within R-loops; and indeed, our data suggest that some RNAP molecules remain bound within the R-loop and can resume transcription after the R-loop removal (see Discussion and the legend to Figure 6). Thus, we examined whether RNAP sequestration contributes to the reduced yield of transcription in our system. Since RNAP sequestration reduces the concentration of active RNAP in solution, it would decrease the yield of transcription not only for the ‘causative’ substrates containing the RNAP-sequestering structure, but also for any other substrates if they are present in the transcription mixture together with the causative substrates (30). To check whether RNAP sequestration contributes to blockage in our system, we carried out the experiment with transcription promoter–distal and promoter–proximal substrates in the same transcription mixture under conditions in which the strong blockage for the promoter–proximal substrate is observed. It is seen (Supplementary Figure S2) that the yield of transcription for promoter–distal substrate is not detectably affected by the presence of the promoter–proximal substrate, indicating that RNAP sequestration does not contribute to the transcription blockage under our conditions.
The blockage is similar for transcription performed in high concentration of either potassium or lithium ions, suggesting that G-quadruplex formation does not contribute to the blockage
R-loop formation within G-rich insert could be accompanied by G-quadruplex formation within the displaced non-template DNA strand (27). To test whether G-quadruplex formation contributes to the blockage, we performed transcription at high concentration of either potassium ions that strongly stabilize G-quadruplex, or lithium ions, which do not stabilize G-quadruplexes (37). Transcription blockages were similar under these two conditions (see Supplementary Figure S3), suggesting that quadruplex formation does not detectably contribute to the blockage in our system. A possible explanation for this is that a very stable R-loop produced by a pure homopurine–homopyrimidine G-rich sequence in the vicinity of promoter causes practically complete transcription blockage by itself, thus an additional contribution of quadruplex cannot be detected.
Possibly, for quadruplex-forming sequences that produce less stable R-loops (i.e., in which G-stretches are interspersed by pyrimidine-rich or random sequences) the contribution of quadruplex to R-loop-induced transcription blockage would be significant.
DISCUSSION
The model
Based upon our results, we propose a model for transcription inhibition by the promoter–proximal G-rich PuPy sequence (further referred to as ‘R-loop-prone sequence’), which is consistent with all of our observations (Figure 6; for more detailed mathematical treatment see Supplementary Discussion).
According to this model, during transcription the R-loop is formed within the R-loop-prone sequence (shown in turquoise) with certain probability, p; and with probability 1 – p transcription proceeds without R-loop formation. When the R-loop does not form, transcription proceeds unobstructed, a full-size nascent RNA is released, and the DNA template can become involved in further rounds of transcription. However, after an R-loop is formed in close proximity to the promoter, it would interfere with transcription from this template by other RNAP molecules; thus, the following rounds of transcription from this DNA template would be inhibited (here we consider the inhibition of transcriptional rounds that occur after the one at which the R-loop is formed; the ‘fate’ of the RNAP that created R-loop will be discussed later in the context of experiments involving RNase H).
The assumption that R-loop formation at the start of transcription or in its close vicinity would very strongly inhibit further rounds of transcription is supported by the fact that before entering the stable elongation mode (which in the case of T7 RNAP occurs ∼10–14 bp from the start of transcription (see (38) and references therein), the transcription complex is unstable and, consequently, it would be very sensitive to obstacles. In our case the R-loop-prone sequence is localized 9 bp from the start of transcription, which is shorter than 10–14 bp required for transition to the stable elongation mode; moreover, due to the presence of three guanines in the non-template strand immediately after the promoter sequence (see Figure 1A and B), it seems probable that the upstream flank of the R-loop would be located immediately after the promoter (as it is shown in the Figure 6), which would likely to block further rounds of transcription at early initiation stages. (Here, we want to note that the position of the upstream flank of the R-loop does not necessarily coincide with the site at which the R-loop formation was initiated: R-loop formation is likely to be initiated somewhere within the R-loop-prone sequence, and then the nascent RNA ‘tail’ could invade the upstream DNA duplex as far as the sequence continues, for which RNA–DNA hybrid is more energetically favorable than DNA–DNA hybrid.) For the G-rich sequence motif used in this study, the RNA–DNA hybrid is much more stable, than the DNA–DNA hybrid (3); consequently, R-loop formation for this sequence is likely to be practically irreversible. Thus, according to our model, promoter–proximal R-loop formation gradually depletes DNA substrates available for transcription. This is also supported by the observation that in ‘pre-transcription’ experiments (Figure 4), almost complete transcription inhibition could be achieved.
The probabilistic nature of the R-loop formation postulated in our model predicts that the effect of R-loop formation upon the yield of transcription would increase upon increase in the number of transcriptional rounds that occur during the period of the transcription experiment: If the probability of the R-loop formation for the R-loop-prone substrate during one round of transcription is p, then the average number of the transcriptional rounds that occur before the R-loop formation in a given substrate molecule is 1/p (which includes the round of transcription at which the R-loop is formed; e.g. if P = 0.2, R-loop formation on average would occur at the fifth round of transcription within a given DNA molecule). If the number of transcriptional rounds during the period of the experiment is much less than 1/p, then the percentage of substrate molecules that can form the R-loop during this period would be small. Consequently, the impact of R-loop formation upon the yield of transcription would not be strongly pronounced, and the ratio of transcription yields for the non-R-loop-prone and R-loop-prone substrates would be close to unity (see Supplementary Discussion for the mathematical expression).
In contrast, if the number of transcriptional rounds is much greater than 1/p, then long before the end of the transcription experiment, practically all R-loop-prone substrate molecules would contain an R-loop that would block their transcription, while non-R-loop-prone substrate would continue to be efficiently transcribed. Consequently, at the end of the experiment the ratio of transcriptional yields for non-R-loop-prone and R-loop-prone substrates would be high (see Supplementary Discussion for the mathematical expression). More generally, the difference in the transcription yields for substrates with different propensity for R-loop formation would increase with the number of transcriptional rounds. The number of transcriptional rounds during a given time interval increases upon increase in RNAP concentration. That explains why the difference in yields of transcription products for substrates with different propensity for R-loop formation (e.g. promoter–proximal and promoter–distal substrates with the G-rich insert) increases upon increase in RNAP concentration (Figure 3).
As additional support for the R-loop-mediated transcription blockage model, the blockage is not pronounced in the presence of RNase H during transcription, which removes R-loops and thus ‘unblocks’ DNA templates for further rounds of transcription. Note that in the presence of RNase H, most of the run-off transcripts have the same (i.e. full-size) length, as in the absence of RNase H (Figure 5A, left panel, lane 4 versus lane 2), and the yields of full-size products in the presence of RNase H are similar for the promoter–proximal and promoter–distal substrates (Figure 5B); however, some minor fraction of shorter transcripts appears in the presence of RNase H (see the square bracket near the lane 4, Figure 5A, right panel). The following scenario explains all these features of the pattern of transcription products in the presence of of RNase H (Figure 6):
First, consider the round of transcription at which the R-loop is formed. Our previous results (2,3,34) suggest that R-loop formation behind the transcribing RNAP leads to RNAP stalling within or some distance downstream from the R-loop-prone (causative) sequence. Also, topological and energetic considerations (19) predict that once the R-loop is formed, transcription can continue only in the ‘R-loop mode’, i.e. newly synthesized RNA continues to hybridize to the DNA template strand during transcription, and the R-loop grows in size while transcription proceeds until the RNAP is stalled. If some of the stalled RNAPs remain bound to the DNA template after RNase H-mediated R-loop removal they could resume transcription, producing transcripts that are shorter than the full-size transcript by the length of RNA corresponding to the distance between the transcription start site and the downstream flank of the R-loop. According to this interpretation, the distribution of shorter transcription products within about 60 nt from the full-size product (see the square bracket near the lane 4, Figure 5A, right panel) means that the downstream flank of the R-loop typically extends not much farther than about 20 nt from the downstream flank of promoter–proximal G-rich PuPy sequence (41 nt).
In those R-loops, which did not retain the bound RNAP, RNA would be completely digested by RNase H. In any case, upon R-loop removal, the substrate DNA molecule becomes available for further rounds of transcription and would produce several (on average, (1/p) – 1) normal full-size transcripts before another R-loop is formed and the whole cycle is repeated. During this process, the fraction of transcripts which would be either shortened or completely digested by RNase H would be equivalent to the probability of the R-loop formation p, and the fraction of full-size transcripts would be 1 – p. The latter is equivalent to the ratio of the full-size transcript yields for R-loop-prone and non-R-loop-prone substrates in the case of transcription performed in the presence of RNase H. (More rigorously, 1 – p is an upper-bound for this ratio, which is achieved when RNase H-mediated R-loop removal occurs much faster than R-loop formation.) For example, if p = 0.2, the yield of full-size run-off transcript for R-loop-prone substrate in the presence of RNase H could reach up to 80% of the yield for non-R-loop-prone substrate. Thus, if the probability of R-loop formation during one round of transcription is sufficiently small, the yields of transcripts for R-loop-prone and non-R-loop-prone substrates in the presence of sufficiently high concentration of RNase H would be similar, which is observed in Figure 5B (though more precise experiments would be needed to reliably estimate probability p).
It is important to emphasize, that the probability of R-loop formation during one round of transcription could be quite small even for very R-loop-prone sequences, because R-loop formation within an intact DNA duplex is likely to be associated with overcoming of a large kinetic barrier. For example, within the framework of the thread-back model, nascent RNA invasion into DNA duplex would require an energetically unfavorable transient DNA unwinding. Thus, the kinetics for R-loop formation could be slow even when this formation is energetically favorable. As a result, the characteristic time for R-loop formation could be longer than the time required for the transcribing RNAP to pass through the R-loop-prone sequence, and, consequently, in most cases this passage would be completed without R-loop formation.
Potential biological implications
According to our model, the probability of an R-loop formation in the vicinity of a promoter defines an average number of transcriptional rounds performed from this promoter before transcription would be blocked (or strongly inhibited) by R-loop formation. Based upon this, one could speculate that in vivo, for certain promoters R-loop formation could serve as a negative feed-back regulator, and the probability of R-loop formation (which is defined by the sequence and the distance from promoter) would determine how many times the promoter would ‘fire’ before inhibition by R-loop formation, until the R-loop is removed by specific helicases or RNaseH activity. Thus, R-loop-mediated transcription inhibition could contribute to ‘transcriptional bursting’, that has been observed in many biological systems (reviewed in (39)). Since the R-loop-prone sequences have been found to localize in the vicinity of certain promoters (reviewed in (5)), the above-described mechanism could potentially contribute to gene regulation. Note that in the case, in which an R-loop initially forms some moderate distance (e.g. ≤100 bp) away from the promoter, RNA ‘tail’ upstream from the R-loop could branch-migrate into DNA duplex, displacing non-template DNA strand and expanding the R-loop up to the very start of transcription. This strand displacement would be facilitated by negative supercoiling, which is present (either transiently or permanently) in many genomes (reviewed in (40,41)). Thus, the above-described mechanism in principle could be also applicable to R-loops formed some distance away from the promoter; however, since the probability of the R-loop formation decreases upon decrease the distance from the promoter (17), so will the contribution of this mechanism to the promoter inhibition.
Our results also could have practical implications: since G-rich sequences in the vicinity of promoter strongly inhibit transcription, placing these sequences close to the promoter in the designed DNA templates (e.g. vectors used for in vivo gene expression, or in vitro RNA production) should be avoided.
In conclusion, R-loop mediated transcription blockage could be of significant importance, and its detailed mechanism deserves further investigations. In particularly, it would be important to study more detailed dependence of the transcription blockage upon the distance between the R-loop-prone sequence and promoter, and to design experiments to evaluate the probability of R-loop formation per one round of transcription.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute [CA077712 to P.C.H.]; Stanford Cancer Institute 2016 Fellowship Award [PTA 1164311-123-GHTDS to B.P.B.]; Undergraduate Research Grants at Stanford [to J.H.S.S.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Belotserkovskii B.P., Mirkin S.M., Hanawalt P.C.. DNA sequences that interfere with transcription: implications for genome function and stability. Chem. Rev. 2013; 113:8620–8637. [DOI] [PubMed] [Google Scholar]
- 2. Belotserkovskii B.P., Liu R., Tornaletti S., Krasilnikova M.M., Mirkin S.M., Hanawalt P.C.. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:12816–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belotserkovskii B.P., Neil A.J., Saleh S.S., Shin J.H., Mirkin S.M., Hanawalt P.C.. Transcription blockage by homopurine DNA sequences: role of sequence composition and single-strand breaks. Nucleic Acids Res. 2013; 41:1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 5. Chedin F. Nascent connections: R-loops and chromatin patterning. Trends Genet. 2016; 32:828–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sollier J., Cimprich K.A.. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015; 25:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richard P., Manley J.L.. Loops, R. and links to human disease. J. Mol. Biol. 2016; doi:10.1016/j.jmb.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costantino L., Koshland D.. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 2015; 34:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 10. Lin Y., Wilson J.H.. Transcription-induced DNA toxicity at trinucleotide repeats: double bubble is trouble. Cell Cycle. 2011; 10:611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Groh M., Gromak N.. Out of balance: R-loops in human disease, PLoS Genet. 2014; 10:e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Usdin K., Kumari D.. Repeat-mediated epigenetic dysregulation of the FMR1 gene in the fragile X-related disorders. Front Genet. 2015; 6:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim N., Jinks-Robertson S.. Transcription as a source of genome instability. Nat. Rev. Genet. 2012; 13:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roy D., Yu K., Lieber M.R.. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 2008; 28:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daniels G.A., Lieber M.R.. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995; 23:5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu K., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R.. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003; 4:442–451. [DOI] [PubMed] [Google Scholar]
- 17. Roy D., Zhang Z., Lu Z., Hsieh C.L., Lieber M.R.. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol. 2010; 30:146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Masse E., Drolet M.. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 1999; 274:16659–16664. [DOI] [PubMed] [Google Scholar]
- 19. Belotserkovskii B.P., Hanawalt P.C.. Anchoring nascent RNA to the DNA template could interfere with transcription. Biophys. J. 2011; 100:675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tomizawa J., Masukata H.. Factor-independent termination of transcription in a stretch of deoxyadenosine residues in the template DNA. Cell. 1987; 51:623–630. [DOI] [PubMed] [Google Scholar]
- 21. Tous C., Aguilera A.. Impairment of transcription elongation by R-loops in vitro. Biochem. Biophys. Res. Commun. 2007; 360:428–432. [DOI] [PubMed] [Google Scholar]
- 22. Mirkin S.M., Frank-Kamenetskii M.D.. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 1994; 23:541–576. [DOI] [PubMed] [Google Scholar]
- 23. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maizels N., Gray L.T.. The G4 genome. PLoS Genet. 2013; 9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reaban M.E., Griffin J.A.. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990; 348:342–344. [DOI] [PubMed] [Google Scholar]
- 26. Reaban M.E., Lebowitz J., Griffin J.A.. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J. Biol. Chem. 1994; 269:21850–21857. [PubMed] [Google Scholar]
- 27. Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N.. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004; 18:1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang G., Vasquez K.M.. Effects of replication and transcription on DNA structure-related genetic instability. Genes (Basel). 2017; 8:E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grabczyk E., Fishman M.C.. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J. Biol. Chem. 1995; 270:1791–1797. [DOI] [PubMed] [Google Scholar]
- 30. Grabczyk E., Usdin K.. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000; 28:2815–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tornaletti S., Park-Snyder S., Hanawalt P.C.. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 2008; 283:12756–12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belotserkovskii B.P., De Silva E., Tornaletti S., Wang G., Vasquez K.M., Hanawalt P.C.. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007; 282:32433–32441. [DOI] [PubMed] [Google Scholar]
- 33. Grabczyk E., Mancuso M., Sammarco M.C.. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007; 35:5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Belotserkovskii B.P., Hanawalt P.C.. PNA binding to the non-template DNA strand interferes with transcription, suggesting a blockage mechanism mediated by R-loop formation. Mol. Carcinog. 2015; 54:1508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou Y., Martin C.T.. Observed instability of T7 RNA polymerase elongation complexes can be dominated by collision-induced ‘bumping’. J. Biol. Chem. 2006; 281:24441–24448. [DOI] [PubMed] [Google Scholar]
- 36. Steitz T.A. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr. Opin. Struct. Biol. 2004; 14:4–9. [DOI] [PubMed] [Google Scholar]
- 37. Williamson J.R., Raghuraman M.K., Cech T.R.. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989; 59:871–880. [DOI] [PubMed] [Google Scholar]
- 38. Gong P., Martin C.T.. Mechanism of instability in abortive cycling by T7 RNA polymerase. J. Biol. Chem. 2006; 281:23533–23544. [DOI] [PubMed] [Google Scholar]
- 39. Sanchez A., Golding I.. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013; 342:1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lavelle C. Pack, unpack, bend, twist, pull, push: the physical side of gene expression. Curr. Opin. Genet. Dev. 2014; 25:74–84. [DOI] [PubMed] [Google Scholar]
- 41. Gilbert N., Allan J.. Supercoiling in DNA and chromatin. Curr. Opin. Genet. Dev. 2014; 25:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.