Abstract
DNA assembly forms the cornerstone of modern synthetic biology. Despite the numerous available methods, scarless multi-fragment assembly of large plasmids remains challenging. Furthermore, the upcoming wave in molecular biological automation demands a rethinking of how we perform DNA assembly. To streamline automation workflow and minimize operator intervention, a non-enzymatic assembly method is highly desirable. Here, we report the optimization and operationalization of a process called Twin-Primer Assembly (TPA), which is a method to assemble polymerase chain reaction-amplified fragments into a plasmid without the use of enzymes. TPA is capable of assembling a 7 kb plasmid from 10 fragments at ∼80% fidelity and a 31 kb plasmid from five fragments at ∼50% fidelity. TPA cloning is scarless and sequence independent. Even without the use of enzymes, the performance of TPA is on par with some of the best in vitro assembly methods currently available. TPA should be an invaluable addition to a synthetic biologist's toolbox.
INTRODUCTION
DNA assembly, which is the precise and ordered arrangement of functional DNA parts, plays a pivotal role in the implementation of synthetic biology designs (1–3). The holy grail of DNA assembly will be a method that allows scarless, sequence independent, multi-fragment assembly of large constructs at high efficiency and high fidelity (4,5). Despite the numerous assembly techniques now in the molecular biology toolbox, to our knowledge, none has been able to fulfill the ideal and each still has its limitations. As such, a complex construction often requires the use of more than one technique to complement each other's strengths and weaknesses (6), and is frequently carried out through a multi-step hierarchal assembly scheme (7).
Some of the most widely adopted techniques for scarless, sequence independent, multi-fragment assembly are Gibson assembly (GA) (8), Golden Gate assembly (9), uracil-specific excision reagent cloning (USER) (10), ligase cycling reaction (LCR) (11) and DNA assembler (yeast homologous recombination) (12). Very large constructs can be built using DNA assembler. However, because it is an in vivo cloning method, the process is slow and complex. It is therefore usually used only when all in vitro methods have failed or are expected to fail.
Modern in vitro DNA assembly methods can be generally classified into either ligase-dependent or ligase-independent methods. Ligase-dependent methods require a DNA ligase and often one or more other enzymes to perform the reaction, whereas ligase-independent methods generally require fewer enzymes (13). Among the ligase-dependent methods, GA is fast and versatile, but its efficiency and fidelity drops precipitously when the number of fragments goes beyond four (14). Furthermore, promoters, ribosomal binding sites (RBS) and terminators, which are common components in genetic pathways, are notoriously difficult for GA due to their secondary structures (15). Golden Gate assembly is very robust and is capable of assembling more than 15 fragments at high efficiency and fidelity (16). However, due to the limited number of commercial Type IIs endonucleases, it is often difficult to find an appropriate enzyme to avoid naturally occurring Type IIs sites within DNA parts, especially for large gene fragments. Otherwise, additional effort will be needed to recode DNA parts to eliminate the unwanted cut sites. Lastly, while LCR can assemble 12 small fragments with over 60% fidelity, the fidelity drops sharply for constructs above 12 kb (6). Furthermore, the long (60–90 nt) bridging oligonucleotides that are required by LCR add significant cost to the method (11).
Ligation-independent cloning (LIC) methods include circular polymerase extension cloning (CPEC) (17), In-fusion (18), Hot fusion (19), USER (10), sequence and LIC (SLIC) (20), seamless ligation cloning extract (SLiCE) (21) and multiple overlap extension polymerase chain reaction (MOE-PCR) (22) (Table 1). Among these, SLIC offers the best capability, allowing the assembly of 10 fragments with 40 bp overlaps with around 20% fidelity (20). In general, the current LIC methods lag behind the ligation-dependent methods in most performance parameters (13). Furthermore, since LIC methods still require at least one enzyme, they are not fundamentally different from the ligase-dependent methods.
Table 1. Summary of common ligation- and enzyme-independent assembly methods.
| Number of fragments (fidelity) | Enzyme required | Reference | |
|---|---|---|---|
| Ligase-independent assembly methods | |||
| CPEC | 5 fragments for a 9 kb plasmid (90%) | DNA polymerase | (17) |
| In-fusion | 5 fragments for a 5 kb plasmid (90%) | In-fusion HD enzyme mix | (18) |
| Hot fusion | 8 fragments for a 8 kb plasmid (90%) | T5 Exonuclease, DNA polymerase | (19) |
| MOE-PCR | 7 fragments for a 7 kb plasmid (25%) | DNA polymerase | (22) |
| SLICa | 10 fragments for a 8 kb plasmid (20%) | T4 DNA polymerase, RecA | (20) |
| SLiCEa | 7 fragments for a 5 kb plasmid (90%) | Escherichia coli extract | (21) |
| USER | 8 fragments for a 11 kb plasmid (60%) | USER enzyme mix | (10) |
| Enzyme-independent assembly methods | |||
| EFC | 2 fragments for a 3 kb plasmid (90%) | (23) | |
| PIPE | 2 fragments for a 5 kb plasmid (90%) | (13) | |
| SLICb | 2 fragments for a 15 kb plasmid | (20) | |
| TPAa | 10 fragments for a 7 kb plasmid (80%) | This study | |
The largest demonstrated number of fragments is shown for each method with the corresponding fidelity in bracket.
aThe largest demonstrated assembly sizes are as indicated except for SLIC (12 kb from 2 fragments), SLiCE (24 kb from 2 fragments) and TPA (31 kb from 5 fragments).
bSLIC has an enzyme independent protocol.
On the other hand, enzyme-independent cloning methods are able to assemble DNA fragments without any enzyme at all, for example, enzyme-free cloning (EFC) (23) and polymerase incomplete primer extension (PIPE) (13). Although non-enzymatic assembly methods may not show much advantage for day-to-day bench top DNA assembly, they become significant in a high-throughput setting. In the quest to accelerate the design-build-test cycle, many synthetic biologists are turning to automation as a way to test multiple designs in parallel (3). Due to the lack of integrated refrigerated storage space on most automation platforms, enzymatic steps are problematic. Operator intervention is usually required to provide the temperature-sensitive reagents just before the reaction. A non-enzymatic DNA assembly method can potentially remove such bottleneck. However, existing non-enzymatic DNA assembly methods have low efficiency, in most cases even for a two-fragment assembly (Table 1). Hence, automation and high-throughput cloning cannot be achieved with these methods.
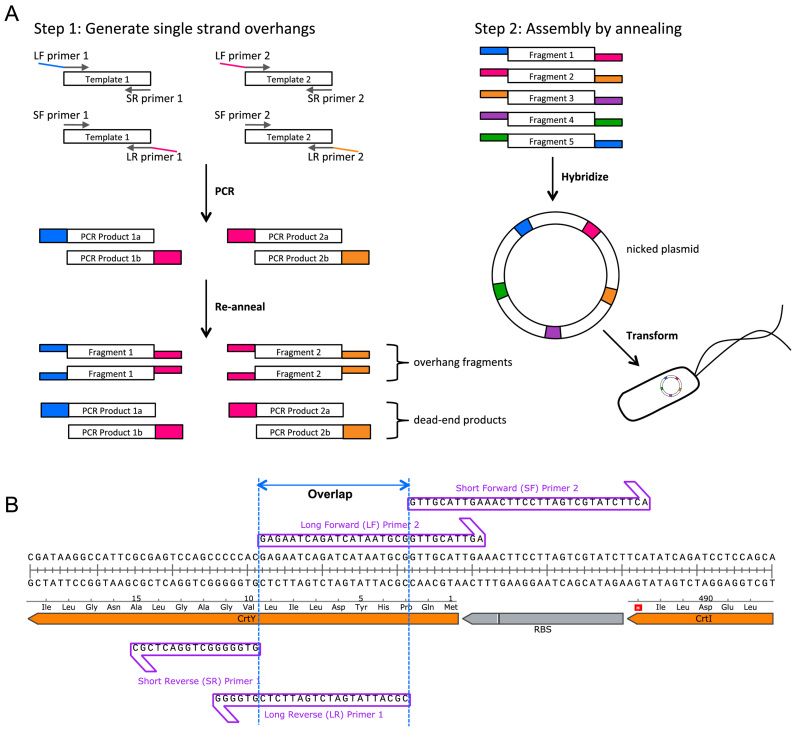
Here we introduce a scarless high efficiency non-enzymatic assembly method with performance that rivals even the best of the ligation dependent methods. Named ‘Twin-Primer Assembly (TPA)’, it can assemble PCR-amplified fragments without the use of enzymes after the initial PCR, without phosphorylated or long oligonucleotides, and without sequence limitation. Each fragment used in TPA requires two PCR products, amplified separately using two sets of primers (Figure 1A). One product has an overlap to the fragment that comes before and the other has an overlap to the fragment that comes after. The length of the overlap region is determined by the melting temperature (TM) of the overlap region, and all overlaps used in an assembly are designed to have the same TM. For TM = 50°C, the overlap will usually be between 16–20 bp, given moderate GC content. The two PCR products are mixed and fully denatured at high temperature before they are allowed to re-anneal slowly, generating intermediates that have overhangs on both sides. These intermediates are then allowed to hybridize at an elevated temperature to form a circular nicked plasmid. Upon Escherichia coli transformation, the cells phosphorylate and ligate the nicks to create the final product plasmid. TPA cloning is capable of assembling a 7 kb plasmid from 10 fragments at around 80% fidelity and a 31 kb plasmid with repetitive sequences from five fragments at around 50% fidelity.
Figure 1.
TPA design and workflow. (A) TPA workflow showing a five-fragment assembly. For step 1, only two of five fragment preparations are represented. After re-annealing, half of the DNA is expected to contain hybridizable overhangs, while the other half is expected to be blunt-end products that will not participate in step 2. For step 2, only one set of compatible fragments with 5΄ overhangs is shown. The set of fragments with 3΄ overhangs should work in a similar fashion. (B) A typical TPA junction showing the long and short primers of two adjacent fragments.
MATERIALS AND METHODS
Strains, reagents and cell cultivation
Escherichia coli OmniMAX [F΄ {proAB lacIq lacZΔM15 Tn10(TetR) Δ(ccdAB)} mcrA Δ(mrr hsdRMS-mcrBC) Φ 80(lacZ)ΔM15 Δ(lacZYA-argF)U169 endA1 recA1 supE44 thi-1 gyrA96 relA1 tonA panD] (Thermo Fisher Scientific, US) was used for cloning. E. coli cultures were grown at 37°C in Lysogeny Broth (LB) selection medium (100 μg/ml carbenicillin, 25 g/l LB Broth Miller, Axil Scientific, Singapore) for plasmid amplification and verification; LB selection plates (20 g/l LB Broth Miller, 100 μg/ml carbenicillin) were used for E. coli transformation. For better yellow/white visualization, 5 g/l peptone, 1 g/l beef extract could also be added to the LB plate formulation to assist zeaxanthin production. All restriction enzymes were obtained from New England Biolabs (NEB) (Ipswich, MA, USA). PCR reactions were carried out using KOD Xtreme Hotstart (Merck, Darmstadt, Germany) or Q5 (NEB, Ipswich, MA, USA), and primers were ordered from Integrated DNA Technologies (IDT) (Coralville, IA, USA).
Design of TPA oligonucleotides
For each DNA fragment to be assembled, four oligonucleotides were ordered: long forward (LF), short forward (SF), long reverse (LR) and short reverse (SR). The long primers (LF and LR) were analogous to that used in GA, and they defined the overlap region, whereas the short primers stopped exactly before the overlap. The TPA oligonucleotides design algorithm was as follows:
The melting temperature (TM) of the overlap region (Figure 1B) was set at 50°C as predicted by IDT's OligoAnalyzer using the following modified conditions: Oligo concentration = 0.015 μM, Na+ = 50 mM, Mg2+ = 10 mM. All junctions used in an assembly should have the same TM.
As much as possible, G/C locks (i.e. the nucleotide closest to the nick being a G or C) were used at both sides of the nicks when choosing junction sequences. In situations where such locks were not possible, consecutive T/A close to the nicks should be avoided.
The overlap regions of the long primers were defined by their TM rather than by length. Long primers with TM = 50°C junctions were usually GA compatible.
The short primers (SF or SR) were ordinary PCR primers adjacent to the overlap sequences.
Two-step assembly: preparation of fragments
For each DNA fragment to be assembled, two PCR products were generated: one with primers LF and SR, the other with primers SF and LR. PCR reactions were performed using either KOD Xtreme Hotstart (Merck, Darmstadt, Germany) or Q5 (NEB, Ipswich, MA, USA) DNA polymerase according to the respective manufacturer's recommended protocol. Depending on PCR purity, either gel purification (QIAquick Gel Extraction Kit, Qiagen, Hilden, Germany) or PCR purification (QIAquick PCR Purification Kit, Qiagen, Hilden, Germany) were used to clean up the products. The two PCR products for each fragment were mixed in 1:1 molar ratio, to a final concentration of 40 fmol/μl DNA (40 fmol/μl ≈ 24 ng/kb/μl DNA) in 1× CutSmart buffer (NEB, Ipswich, MA, USA). A lower final concentration (down to 10 fmol/μl) was used for some fragments when the higher final concentration could not be achieved due to low PCR yield. An excel worksheet was used to facilitate the calculations (see Supplementary Protocol for details). If plasmid template was used in the PCR, 0.5 μl DpnI was added to eliminate the plasmid template so as to reduce the transformation background. The re-annealing step that generates overhangs was carried out using the following temperature profile: 37°C for 30 min (for DpnI digest), 98°C for 2 min, 85°C for 2 min (at a ramping rate of 0.1°C/s), 75°C for 2 min (0.1°C/s), 65°C for 2 min (0.1°C/s), 55°C for 2 min (0.1°C/s) and 8°C hold (0.1°C /s). The annealed DNA fragments were stored at 4°C until the assembly step.
Two-step assembly: assembly of fragments
The assembly was carried out by hybridizing the re-annealed fragments. All the re-annealed DNA fragments were mixed in 1:1 molar ratio, to a final concentration of 4 fmol/μl/fragment (4 fmol/μl ≈ 2.4 ng/kb/μl) in 1× CutSmart buffer. The hybridization step that generates nicked circular plasmid was carried out using the following temperature profile: 65°C (TM + 15) for 10 s, 55.5°C (TM + 5.5) for 1–2 h and 8°C hold (0.1°C/s). The hybridized nicked plasmid was stored at 4°C until E. coli transformation.
One-step assembly
In one-step assembly, all PCR products for the construct were put together in one-pot at a final concentration of 2 fmol/μl/product in 1× CutSmart buffer, 0.5 μl DpnI was added if necessary. The one-step assembly was carried out using the following temperature profile: 37°C for 30 min (DpnI digest), 98°C for 2 min, 85°C for 2 min (0.1°C/s), 75°C for 2 min (0.1°C /s), 65°C for 2 min (0.1°C/s), 55.5°C (TM + 5.5) for 1–2 h (0.1°C/s) and 8°C hold (0.1°C/s). The hybridized nicked plasmid was stored at 4°C until E. coli transformation.
For a detailed example of the TPA protocol, please refer to Supplementary Protocol.
E. coli transformation
Mix & Go OmniMAX competent cells were prepared using the Mix & Go E. coli Transformation Kit (Zymo Research, Orange, CA, USA). For each Mix & Go transformation, 4 μl of the TPA assembly mix was transformed into 50 μl of competent cells, held on ice for 30 min and spread onto selection plate. Our Mix & Go OmniMax cells had displayed a typical transformation efficiency of around 5 × 107 CFU/μg pUC19. Chemical heat-shock transformation was found to be compatible with TPA (Supplementary Figure S1). However, electroporation had not been successful even with desalting. In our hands, all electroporation experiments had drastically reduced TPA fidelity.
Gibson assembly
Gibson assemblies were carried out using Gibson Assembly Cloning Kit (NEB, Ipswich, MA, USA). A total of 30 ng backbone was used for each GA and all other fragments with ∼30 bp overlap regions were mixed together with the backbone fragment in 1:1 molar ratio together with an equal volume of Gibson master mix, incubated at 50°C for 3 h and immediately transformed into E. coli.
Analysis of the assembled plasmids
Assemblies were evaluated if the negative controls (either lacking the backbone fragment or one of the insert fragments) had less than a tenth of the number of colonies on the assembly plate. The efficiency of the DNA assembly was calculated by counting the total colony number. The fidelity of assembly for the zeaxanthin pathway plasmid was calculated by dividing the number of yellow colonies by the total colony number. The fidelity of non-zeaxanthin pathway assemblies was assessed by restriction digestion followed by gel electrophoresis, and by DNA sequencing. Plasmid DNA was isolated from resistant clones (Plasmid SV mini, GeneAll Biotechnology, Seoul, Korea), digested with respective enzymes at 37°C for 1 h, electrophoresed in 1% agarose gel at 120V for 20 min and visualized using SYBR Safe DNA Gel Stain (Invitrogen, Carlsbad, CA, USA) on a BioRad GEL Doc (BioRad, Hercules, CA, USA) with a XcitaBlue Conversion Screen (BioRad, Hercules, CA, USA). All junction regions of correctly digested constructs were sequenced for junction errors by Sanger sequencing (first BASE, Singapore). For all DNA assemblies performed in this work, the plasmid backbone, which included the selection marker and replication origin in E. coli, was counted as one DNA fragment.
RESULTS
Optimization of TPA
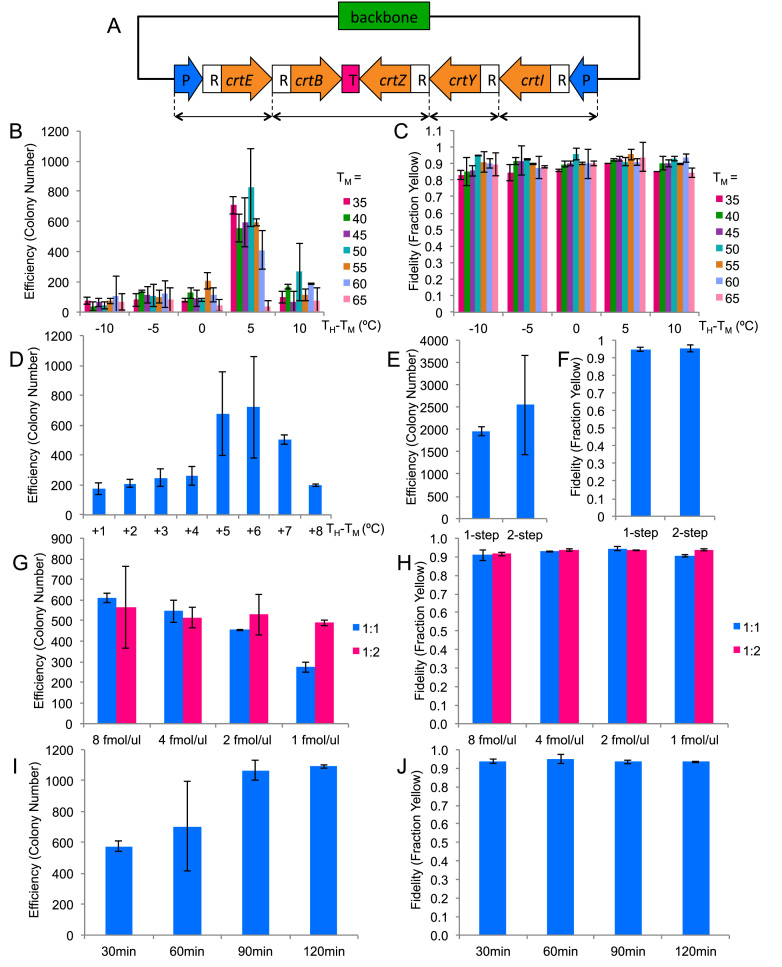
To find the optimal reaction condition for TPA, we performed a serial optimization of six factors that we had hypothesized to be important for TPA (Table 2). In each step, we tested a small subset of factor(s) and selected the conditions that had showed the best assembly efficiency and fidelity for the optimization of more factors.
Table 2. Summary of optimal TPA conditions.
| Factor | Unit | Tested conditions | Optimal condition |
|---|---|---|---|
| Junction TM | °C | 35, 40, 45, 50, 55, 60, 65 | 50 |
| Annealing temperature | °C | T M - 10, TM - 5, TM, TM + 5, TM + 10 | T M + 5 |
| Protocol | one-step, two-step | ||
| DNA inputs | fmol/μl/frag | 0.5, 1, 2, 4 | 2 |
| DNA ratios | insert:backbone | 1:1, 1:2 | 1:1 |
| Incubation time | min | 30, 60, 90, 120 | 120 |
In order to quickly gauge assembly fidelity, we designed a five-fragment assembly of a plasmid (pAmp-EC-Zeax) harboring the zeaxanthin pathway (Figure 2A and Supplementary Figure S3) which made a colony expressing the functional pathway yellow. To show that this coloration test was a good gauge of assembly fidelity, we randomly picked 14 yellow colonies from one of the assembly plates for restriction digestion. All of them showed the correct digestion pattern (Supplementary Figure S2).
Figure 2.
TPA optimization based on five-fragments assembly. The columns represent the averages of two independent experiments, and the error bars represent the standard deviations. (A) The zeaxanthin pathway plasmid used for the optimization experiments. CrtE, B, Z, Y, I are the coding regions for the enzymes in the pathway. P represents promoter, R represents RBS and T represents terminator. The double-headed arrows at the bottom denote how the pathway has been broken up. (B and C) Efficiency and fidelity as a function of junction TM and hybridization TH. Horizontal axis represents the difference between TH and TM. (D) Efficiency as a function of TH for TM = 50°C. (E & F) Efficiency and fidelity as a function of annealing protocol. (G & H) Efficiency and fidelity as a function of DNA input amount and backbone:insert ratio. (I & J) Efficiency and fidelity as a function of hybridization time.
Optimization of overlap TM and hybridization temperature (TH)
We started the optimization with a full combinatorial testing of the effect of overlap TM and TH (Figure 2B). The tested TM ranged from 35 to 65°C in 5°C intervals, and the tested TH ranged from −10°C to +10°C from each TM in 5°C intervals. The optimization was done using the two-step assembly protocol. Our results showed that although there was a relatively large range of optimal TM, the range of the corresponding optimal TH was markedly narrower. Good assembly efficiency was achieved for TM between 35 and 60°C, and the optimal TH was determined to be a consistent +5°C offset from their respective TM. The parameters tested did not influence the assembly fidelity and assembly fidelity remained at ∼90% (Figure 2C). We picked a moderate TM of 50°C and tested a finer graduation of TH in 1°C intervals (Figure 2D). The optimal TH turned out to be just a 1°C window between 55 and 56°C. For further optimization, we picked a TM of 50°C and a TH of TM + 5.5°C.
Optimization of the DNA assembly program
Since we could achieve high assembly efficiency using the optimal TM and TH through the two-step process, we proceeded to simplify the protocol to a one-step process by combining the re-annealing and hybridization into a one-pot reaction. As shown in Figure 2E and F, the efficiency and fidelity were not significantly different when we switched to the one-step protocol. We therefore chose the one-step assembly process for the rest of study unless stated otherwise.
Optimization of the input DNA concentration and ratio
Next we tested the effect of the total DNA concentration as well as the molar ratio between the backbone and inserts. For 1:1 ratio, we tested four different DNA concentrations by adding each PCR product (before re-annealing) to 0.5, 1, 2 and 4 fmol/μl/product, which is equivalent to 1, 2, 4 and 8 fmol/μl/fragment respectively. For 1:2 ratio, we added insert PCR products to the above concentrations and halved the backbone PCR products. As shown in Figure 2G, while the assembly efficiency for 1:2 ratio did not change significantly within the concentration range tested, it did decline for 1:1 ratio for concentrations lower than 2 fmol/μl/product. Assembly fidelity did not vary significantly for all conditions tested (Figure 2H). We decided to use 2 fmol/μl/product for further experiments because, at this concentration, the reaction was less sensitive to fragment ratio, which would allow more experimental variance. We chose 1:1 ratio for easier pipetting.
Optimization of the hybridization time
Up to this point, we had used a 60 min hybridization step for all our tests. To find the optimal hybridization time, we tested the effect of the hybridization time on the assembly efficiency. Four different time points—30, 60, 90, 120 min—were tested, as shown in Figure 2I. The assembly efficiency did increase with longer incubation time and plateaued after 90 min. Despite the observed plateau, we chose 120 min for future experiments, because we conjectured that a longer hybridization time would be beneficial for the more challenging assemblies to be attempted later. Likewise, less challenging assembly would require less time. In fact, 15 min hybridization was sufficient for a two-fragment reaction (data not shown). In all conditions, the fidelity was around 90% (Figure 2J).
Characterization of capability and limitations
Having identified the optimal reaction conditions for TPA, we then probed the limits of its capabilities. We tested the two dimensions that are the most important when constructing synthetic pathways—number of fragments and size of construct. We would first find the limits of the one-step protocol, and then see if we could get further with the two-step protocol.
Number of fragments
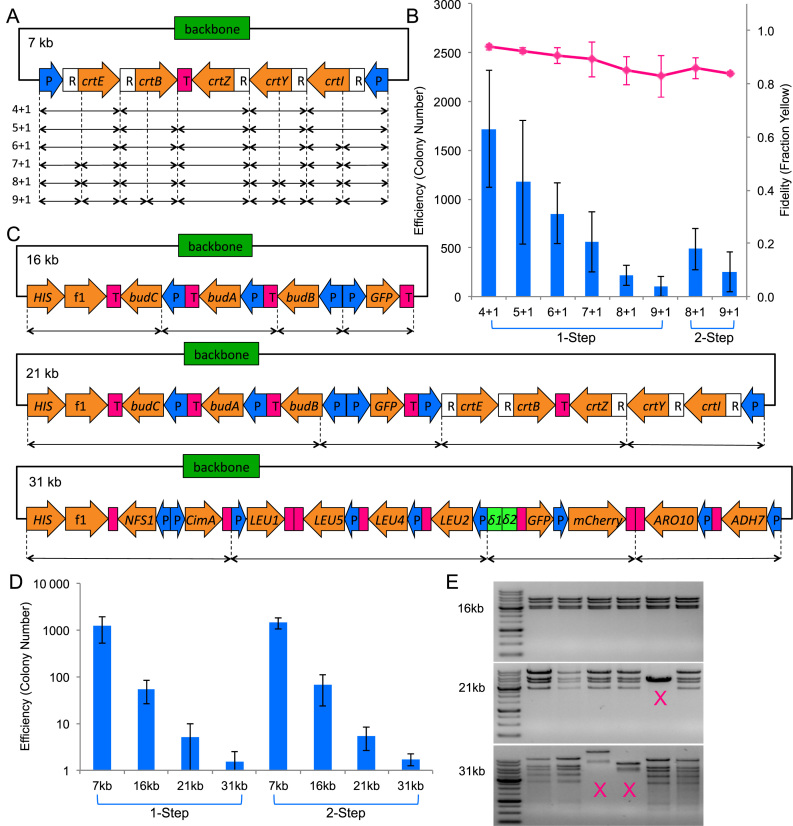
To evaluate the effect of fragment number on efficiency, we assembled the zeaxanthin pathway plasmid (pAmp-EC-Zeax) from varying number of DNA fragments (Figure 3A and Supplementary Figure S3). By keeping the resultant construct constant, the effect due to construct size would not confound the effect due to varying fragment number. In all cases, we defined the backbone of the plasmid, including the selection marker and replication origin in E. coli, as one fragment. As was expected for any DNA assembly method, the assembly efficiency decreased when the number of fragments increased. Using the one-step protocol, the number of colony on the plate dipped to around 100 for assemblies of 10 fragments (Figure 3B). We reattempted these more challenging assemblies using the two-step protocol, and obtained over 200 colonies for a 10-fragment assembly. The fidelity dropped slightly but remained >80% even for a 10-fragment assembly.
Figure 3.
TPA characterization. The columns represent the averages of 4 independent experiments, and the error bars represent the standard deviations. The specific TPA protocol used (one-step or two-step) is indicated at the bottom of the data points. (A) The zeaxanthin pathway plasmid used for number of fragments characterization. CrtE, B, Z, Y, I are the coding regions for the enzymes in the pathway. P represents promoter, R represents RBS, and T represents terminator. The double-headed arrows at the bottom denote how the pathway has been broken up to generate varying numbers of fragment. The total number of fragments for each assembly is indicated on the left of the double-headed arrows, in the following format: number of insert fragment + number of backbone fragment. (B) Efficiency and fidelity as a function of fragment number. Pink line chart represents the fidelity and is plotted on the secondary vertical axis. (C) The 16 kb BDO-GFP (24), 21 kb BDO-GFP-zeaxanthin (24), and 31 kb n-butanol pathway (25) used for the size of construct characterization. (D) Efficiency as a function of construct size. (E) Restriction digestion of 16 kb, 21 kb, and 31 kb clones. Incorrect patterns are marked with an ‘X’. For the full set of restriction digest verification data, refer to Supplementary Figure S6.
Size of construct
To evaluate the effect of the final plasmid size on efficiency, we assembled four different plasmids of increasing sizes while keeping the number of fragments at five (Figure 3C and Supplementary Figure S3). By keeping the fragment number constant, the effect due to varying fragment number would not confound the effect due to varying construct size. However, we did not account for the decrease in transformation efficiency due to increase in plasmid size. The four chosen plasmids were a 7 kb plasmid pAmp-EC-Zeax harboring the zeaxanthin pathway (also used during optimization and fragment number test), a 16 kb plasmid pSSB100 harboring the (R,R)-2,3-butanediol (BDO) pathway and green fluorescent protein (GFP) (24), a 21 kb plasmid pSSB100-Zeax harboring the zeaxanthin pathway as well as the BDO pathway and GFP (24), and a 31 kb plasmid pSSB-144 harboring an n-butanol pathway (25). As shown in Figure 3D, one-step assembly performed admirably for up to 16 kb assembly, beyond which, two-step assembly showed a slight edge. However, due to the large experimental variation typical of transformation experiments, the difference was not statistically significant. The sharp non-linear downward trend in efficiency suggested that TPA was highly sensitive to the plasmid size. The efficiency dropped by a factor of ∼1000 when we increased the plasmid size from 7 to 31 kb, decreasing from over a thousand colonies to just a few. Although higher efficiency E. coli transformation methods such as electroporation should, in theory, improve the colony number, we found that electroporation drastically decreased the assembly fidelity (data not shown). On the other hand, heat-shock transformation was found to be compatible with TPA (Supplementary Figure S1).
In order to test the fidelity of TPA on large DNA construct, we picked between 15 and 20 colonies for restriction digestion analysis. As shown in Table 3, all 20 clones of the 16 kb plasmid, 13 of 19 clones of the 21 kb plasmid, and 7 of 15 clones of the 31 kb plasmid, were assembled correctly. For those plasmids that digested correctly, up to eight were sent for DNA sequencing to detect any junction error. Gross rearrangements that were detectable by restriction digest became more common with increasing plasmid size. Junction errors that were detectable only by sequencing were above PCR error background, but they appeared to be randomly distributed. Assuming that the chance of error at each junction was independent, we estimated the probability of obtaining a sequence correct clone (Table 3). For five-fragment assembly of large plasmids, the probability was about 60% on average. Detailed sequencing results can be found in Supplementary Figures S4, 5 and all restriction digestion gel pictures can be found in Supplementary Figure S6.
Table 3. Summary of restriction digest and DNA sequencing verification for 16, 21 and 31 kb assemblies.
| Assembly | RE digest | P(Dig correct) | Seq J1 | Seq J2 | Seq J3 | Seq J4 | Seq J5 | P(Seq correct) |
|---|---|---|---|---|---|---|---|---|
| 16kb TPA | 20/20 | 100% | 8/8 | 6/8 | 7/8 | 8/8 | 6/8 | 49% |
| 21kb TPA | 13/19 | 68% | 8/8 | 8/8 | 6/7 | 8/8 | 8/8 | 86% |
| 31kb TPA | 7/15 | 47% | 6/7 | 6/7 | 6/6 | 7/7 | 6/7 | 63% |
| GC TPA | 14/14 | 100% | 7/8 | 8/8 | 8/8 | 8/8 | - | 88% |
| 16kb GA | 11/13 | 85% | 7/7 | 0/8 | 6/8 | 7/8 | 8/8 | 0% |
| GC GA | 11/13 | 85% | 4/8 | 8/8 | 5/8 | 7/8 | - | 27% |
RE (Restriction Endonuclease) digest column shows the fraction of clones that were digested correctly. Seq J1-5 columns show the fraction of clones that were sequenced correctly at each junction. P (Dig correct) is the probability of a random clone having the correct digestion pattern, and P(Seq Correct) is the probability of a random digestion correct clone having all correct junctions as determined by DNA sequencing.
Error analysis
While TPA efficiency varied depending on the number of fragments and the size of construct, its fidelity remained generally high. To find out what happened when TPA failed, we analyzed four white and two pink colonies from our 10-fragment zeaxanthin pathway assembly. The lack of yellow coloration meant that the zeaxanthin pathway was non-functional or incomplete. Surprisingly, all the colonies showed the correct digestion pattern (data not shown). We proceeded to sequence the six clones and found point deletion or substitution in all of them (Supplementary Table S1). Out of the 10 mutations detected, 5 resided within junction regions. Since 697 out of the 7130 bases read were in the junction regions, there was an enrichment of errors within junction regions. The total number of junction analyzed was 60, giving a junction error probability of around 5/60 per junction. The other half of the detected mutations was likely to be PCR-induced. Indeed, when we switched our polymerase from KOD Xtreme to Q5 DNA polymerase, which had much higher fidelity, we observed a five-fragment zeaxanthin pathway assembly fidelity of more than 99.5% (Supplementary Figure S7). However, in our hands, Q5 failed to amplify several fragments needed for our large plasmid and high GC content plasmid assemblies, we therefore continued with KOD Xtreme despite its lower fidelity.
TPA versus Gibson assembly
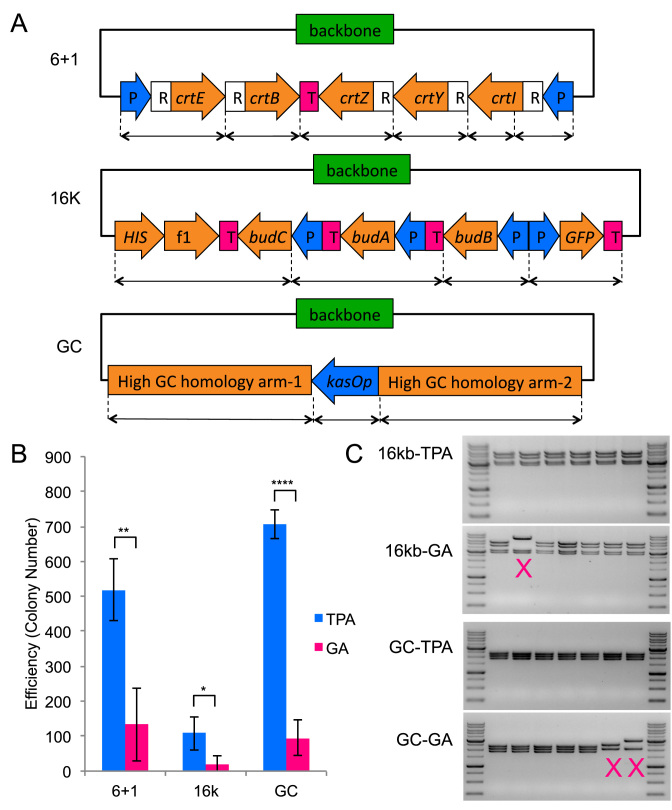
Next, we compared the two-step TPA with the widely used GA method in three aspects: number of fragments, plasmid size and the assembly of high GC content fragments. For high GC assembly comparison, we built a 6-kb plasmid pAMP-BLUE3 that had two high GC homology arms (66 and 71% respectively) for genome integration in Streptomyces flanking a kasOp promoter in a four-fragment assembly (Figure 4A and Supplementary Figure S3). Even though two of the four assembly fragments were high GC, the junctions to assemble them were designed to have moderate GC content for both TPA and GA. Assembly using higher GC junctions did not work for both methods (data not shown). As shown in Figure 4B, TPA resulted in a much high number of colonies than GA did (P = 1.5E-6). A total of 14 of 14 TPA colonies showed the expected digestion pattern while 11 of 13 GA colonies did (Table 3). DNA sequencing of assembly junctions found significantly more errors in the GA assembled plasmids than in TPA assembled plasmids. The probability of obtaining a sequence correct TPA clone was around 88%, compared to 27% for a GA clone.
Figure 4.
TPA versus Gibson Assembly (GA). The columns represent the averages of four independent experiments, and the error bars represent the standard deviation. All TPA data points have been obtained using the two-step protocol. (A) Schematic of the three constructs used for this comparison. kasOp is a Streptomyces promoter. (B) Efficiency comparison between TPA and GA for the three different assemblies. (*P < 0.05, **P < 0.01, ****P < 0.0001) (C) Restriction digestion of TPA and GA clones. Incorrect patterns are marked with an ‘X’. For the full set of restriction digest verification data, refer to Supplementary Figure S6.
For fragment number comparison, we assembled the 7-kb zeaxanthin pathway plasmid from 7 (6 + 1) fragments using either TPA or GA (Figure 3A). As shown in Figure 4B, TPA resulted in approximately four times the number of colonies as GA (P = 0.0086), and both methods achieved similar assembly fidelity. For plasmid size comparison, we assembled the 16 kb BDO-GFP plasmid from five fragments using either TPA or GA. TPA showed significant advantage over GA for assembly of the 16 kb plasmid (P = 0.016), with 5-fold higher efficiency and also higher fidelity. A total of 20 of 20 colonies from TPA showed the correct digestion pattern, whereas only 11 of 13 colonies from GA did. For comparison of capability of assembly high GC content plasmid, DNA sequencing of the assembly junctions found a particularly problematic junction for GA where none of the eight clones was correct (Table 3). The problematic junction did not appear to affect TPA assembled clones. Detailed sequencing results can be found in Supplementary Figures S5 and 8.
DISCUSSION
Here, we report the development of a new method called TPA for rapid in vitro DNA assembly. Although already highly functional, there are several areas in which further enhancements are possible. One unexplored area is the TPA buffer composition. Salt concentration and chemical additives can have large effects on DNA annealing and hybridization, both of which are key mechanisms in TPA. We tested two common PCR additives (betaine and 1,2-propanediol), but neither led to improved TPA performance under the conditions tested (data not shown). Nonetheless, it seems unlikely that the ubiquitous CutSmart buffer, which we have used out of experimental expediency, is the optimal buffer and a systematic test of salt concentrations and additives will likely find a superior solution. As TPA makes use of E. coli to phosphorylate and seal the numerous nicks on the transformed plasmid, using different strains may also affect assembly efficiency and fidelity. This has not been thoroughly investigated in this study.
Due to the narrow optimal hybridization temperature window, it is important to choose a set of overlaps that have a narrow TM range (±1°C). However, the exact TM can be very flexible, ranging from 35 to 60°C, giving us many ways to minimize the TM range of the set. The constraint and flexibility created an optimization problem that a computational design tool can help to address. While we expect TPA to work for TM <35°C, we find it difficult to unify overlap TM when the overlap regions become too short. Furthermore, we suspect that the tolerance to DNA fragments with secondary structures (promoters and such) may come, in part, from the high hybridization temperature.
Although TPA can assemble larger plasmid (31 kb in a five-fragment assembly) than most in vitro DNA assembly methods, it suffers from the same drawbacks as others that have pushed against the size limit. For example, the assembly capability, as measured by efficiency and fidelity, drops sharply with increasing plasmid size (6,11). Moving from a five-fragment 7 kb assembly to a five-fragment 31 kb assembly, TPA has experienced a 1000-fold decrease in efficiency. This change can be partially attributed to the a decrease in transformation efficiency, which is about 4-folds when plasmid size increases from 7 to 31 kb (26), as well as a decrease in circularization rate, which decreases by about 20-folds (27). These two factors accounted for about an 80-folds decrease in efficiency, leaving about 12-folds unaccounted for. Other less quantifiable factors such as PCR quality and secondary structure may have played a role, but they have not been investigated in this study.
The interpretation of the fidelity trends is more nuanced. Fidelity as measured by zeaxanthin production has decreased only slightly with increasing fragment number (Figure 3B). Fidelity as measured by restriction digest has decreased significantly with increasing plasmid size (Table 3). Fidelity as measured by junction sequencing does not appear to vary significantly with increasing plasmid size, but the error rate is above the PCR error baseline (Table 3). These observations can be reconciled by a reaction model that has the following characteristics. Illegitimate circularization that causes gross rearrangement happens at a low but steady frequency. As such, when transformation efficiency is high, gross rearrangement is rarely detectable. Only when transformation efficiency is low, such as in our 21 and 31 kb assemblies, do they become significant. Hybridization junction errors that do not lead to gross rearrangement are more common, and it appears to occur randomly with around 6.8% probability per junction. Given the junction error rate, we should expect only 49% junction error-free plasmids in a 10-fragment assembly. However, since most of our junctions are in non-coding regions, they are much more tolerant to small (1–2 bp) substitutions and indels. Even when a mutation happens within the coding region, we estimate that there is only around 17% chance that the mutation inactivates the pathway (Supplementary Discussion). Nonetheless, based on these observations, we recommend placing junctions within non-coding regions if possible.
Although DNA polymerase's effect on sequence-level fidelity is significant, especially for large plasmids, it is a problem common to all assembly methods that use PCR-prepared fragments. Using high fidelity DNA polymerase and minimizing the number of PCR cycles can reduce PCR error, provided that the fragment can still be specifically amplified. Other than the need to make blunt-end PCR products, TPA does not impose any additional requirement on the DNA polymerase. Therefore, any proof-reading DNA polymerase should work with TPA.
Through our analysis of TPA and GA junctions, junction errors appear to be a prevalent and important but understudied aspect of assembly fidelity. While TPA's junction errors seem to be randomly distributed, GA's junction errors have showed context dependence. One particular GA junction has error in eight of eight sequenced clones, more than two standard deviations from the average rate of GA's junction errors. Due to the lack of data, we do not have any explanation as to why this particular junction is so challenging for GA. However, this observation suggests that, under certain sequence context, it may be difficult to find a junction-perfect GA clone. The average rate of TPA's junction errors is about 6.8% whereas the average rate of GA's junction errors is about 28%.
The TPA non-enzymatic DNA assembly method is scarless and generally sequence independent. We have demonstrated its capability by assembling a 7 kb plasmid from 10 fragments at close to 80% fidelity, and a 31 kb plasmid from five fragments at around 50% fidelity. Assembling more than 10 fragments is likely possible for constructing smaller plasmids, while assembling a plasmid larger than 31 kb is likely possible using fewer fragments. The 31 kb plasmid has four Saccharomyces cerevisiae TEF1 promoters and two S. cerevisiae ADH1 terminators (Supplementary Figure S3), which suggests that TPA can tolerate repetitive sequences. TPA compares favorably to GA—being capable of assembling more fragments, larger plasmids, high GC fragments and DNA fragments with secondary structures such as promoters. It does call for a more complicated experimental procedure, requiring twice the number of primers and PCR reactions. At the bench top scale, TPA makes a good alternative to GA when the construction becomes too challenging for the latter. In fact, TPA's long primers are GA compatible, so one can always try GA before ordering the second set of primers to attempt TPA. However, the GA primers should be designed using the TPA guideline since TPA is much more sensitive to overlap TM than GA is to overlap length. At the robotic biological foundry scale, TPA makes an attractive alternative option as a non-enzymatic assembly can potentially simplify workflow, minimize human intervention and increase throughput. All in all, TPA will be an invaluable addition to a synthetic biologist's toolbox.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Agency for Science, Technology and Research (A*STAR), Singapore for supporting the project in the Metabolic Engineering Research Laboratory (MERL) through the Visiting Investigator Programme to H.Z.
Authors contribution: J.L. and Z.L. conceived and designed the experiments. J.L., Z.L. and X.Z.L. performed the experiments. J.L., Z.L., X.Z.L., E.L.A. and H.Z. analyzed the data, wrote and revised the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agency for Science, Technology and Research (A*STAR), Singapore. Funding for open access charge: A*STAR Visiting Investigator Programme (H.Z.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Ellis T., Adie T., Baldwin G.S.. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr. Biol. 2011; 3:109–118. [DOI] [PubMed] [Google Scholar]
- 2. Cobb R.E., Ning J.C., Zhao H.. DNA assembly techniques for next-generation combinatorial biosynthesis of natural products. J. Ind. Microbiol. Biot. 2014; 41:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chao R., Yuan Y., Zhao H.. Recent advances in DNA assembly technologies. FEMS Yeast Res. 2015; 15:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roth T.L., Milenkovic L., Scott M.P.. A rapid and simple method for DNA engineering using cycled ligation assembly. PLoS One. 2014; 9:e107329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nielsen M.T., Madsen K.M., Seppala S., Christensen U., Riisberg L., Harrison S.J., Møller B.L., Nørholm M.H.. Assembly of highly standardized gene fragments for high-level production of porphyrins in E. coli. ACS Synth. Biol. 2014; 4:274–282. [DOI] [PubMed] [Google Scholar]
- 6. Yuan Y., Andersen E., Zhao H.. A flexible and versatile strategy for construction of large biochemical pathways. ACS Synth. Biol. 2015; 5:46–52. [DOI] [PubMed] [Google Scholar]
- 7. Briggs A.W., Rios X., Chari R., Yang L., Zhang F., Mali P., Church G.M.. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012; 40:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 9. Engler C., Gruetzner R., Kandzia R., Marillonnet S.. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009; 4:e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bitinaite J., Rubino M., Varma K.H., Schildkraut I., Vaisvila R., Vaiskunaite R.. USER™ friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 2007; 35:1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kok S.D., Stanton L.H., Slaby T., Durot M., Holmes V.F., Patel K.G., Platt D., Shapland E.B., Serber Z., Dean J.. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth. Biol. 2014; 3:97–106. [DOI] [PubMed] [Google Scholar]
- 12. Shao Z., Zhao H.. DNA assembler: a synthetic biology tool for characterizing and engineering natural product gene clusters. Methods Enzymol. 2011; 517:203–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stevenson J., Krycer J.R., Phan L., Brown A.J.. A practical comparison of ligation-independent cloning techniques. PLoS One. 2013; 8:e83888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Storch M., Casini A., Mackrow B., Fleming T., Trewhitt H., Ellis T., Baldwin G.S.. BASIC: a new biopart assembly standard for idempotent cloning provides accurate, single-tier DNA assembly for synthetic biology. ACS Synth. Biol. 2015; 4:781–787. [DOI] [PubMed] [Google Scholar]
- 15. Hillson N.J. Koeppl H. DNA assembly method standardization for synthetic biomolecular circuits and systems. Design and Analysis of Biomolecular Circuits. 2011; Verlag, Berlin: Springer; 295–314. [Google Scholar]
- 16. Liang J., Chao R., Abil Z., Bao Z., Zhao H.. FairyTALE: a high-throughput TAL effector synthesis platform. ACS Synth. Biol. 2013; 3:67–73. [DOI] [PubMed] [Google Scholar]
- 17. Quan J., Tian J.. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009; 4:e6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oh S., Jokhadze G., Duong T., Bostick M., Farmer A.A.. Optimized In-Fusion cloningsystem demonstrates high efficiency and accuracy of multi-fragment cloning. Synthetic Biology: Engineering, Evolution & Design. 2014; Conference poster. [Google Scholar]
- 19. Fu C., Donovan W.P., Shikapwashya-Hasser O., Ye X., Cole R.H.. Hot fusion: an efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PloS One. 2014; 9:e115318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M.Z., Elledge S.J.. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007; 4:251–256. [DOI] [PubMed] [Google Scholar]
- 21. Zhang Y., Werling U., Edelmann W.. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012; 40:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kadkhodaei S., Memari H.R., Abbasiliasi S., Rezaei M.A., Movahedi A., Shun T.J., Ariff A.B.. Multiple overlap extension PCR (MOE-PCR): an effective technical shortcut to high throughput synthetic biology. RSC Adv. 2016; 6:66682–66694. [Google Scholar]
- 23. Tillett D., Neilan B.A.. Enzyme-free cloning: a rapid method to clone PCR products independent of vector restriction enzyme sites. Nucleic Acids Res. 1999; 27:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi S., Liang Y., Zhang M.M., Ang E.L., Zhao H.. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab. Eng. 2016; 33:19–27. [DOI] [PubMed] [Google Scholar]
- 25. Shi S., Si T., Liu Z., Zhang H., Ang E.L., Zhao H.. Metabolic engineering of a synergistic pathway for n-butanol production in Saccharomyces cerevisiae. Sci. Rep. 2016; 6:25675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsumura A.B.A.I. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983; 166:557–580. [DOI] [PubMed] [Google Scholar]
- 27. Vologodskii A. Equilibrium large-scale conformational properties of DNA. Biophysics of DNA. 2015; Cambridge: Cambridge University Press; 108–110. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.