Abstract
Members of the DEAD-box family are often multifunctional proteins involved in several RNA transactions. Among them, yeast Saccharomyces cerevisiae Mss116 participates in mitochondrial intron splicing and, under cold stress, also in mitochondrial transcription elongation. Here, we show that Mss116 interacts with the mitoribosome assembly factor Mrh4, is required for efficient mitoribosome biogenesis, and consequently, maintenance of the overall mitochondrial protein synthesis rate. Additionally, Mss116 is required for efficient COX1 mRNA translation initiation and elongation. Mss116 interacts with a COX1 mRNA-specific translational activator, the pentatricopeptide repeat protein Pet309. In the absence of Mss116, Pet309 is virtually absent, and although mitoribosome loading onto COX1 mRNA can occur, activation of COX1 mRNA translation is impaired. Mutations abolishing the helicase activity of Mss116 do not prevent the interaction of Mss116 with Pet309 but also do not allow COX1 mRNA translation. We propose that Pet309 acts as an adaptor protein for Mss116 action on the COX1 mRNA 5΄-UTR to promote efficient Cox1 synthesis. Overall, we conclude that the different functions of Mss116 in the biogenesis and functioning of the mitochondrial translation machinery depend on Mss116 interplay with its protein cofactors.
INTRODUCTION
Mitochondria are eukaryotic organelles that house pathways essential for aerobic energy production by the oxidative phosphorylation (OXPHOS) system. As a remnant of their bacterial ancestry, mitochondria contain their own genome (mtDNA) and gene expression system. The mtDNA encodes only a few essential subunits of the OXPHOS system (8 in Saccharomyces cerevisiae, 13 in human), one mitoribosomal protein in yeast, and universally, two ribosomal RNAs (rRNAs) and a variable set of tRNAs. The remaining subunits of the OXPHOS system, the mitoribosome proteins and the accessory factors that modify RNA and proteins and help organize these complexes are all encoded by the nuclear genome. Therefore, efficient protein synthesis and complex assembly within this organelle require a careful coordination between the mitochondrial and nuclear genomes.
Translation of mitochondrial mRNAs occurs in mitoribosomes (1). High-resolution cryo-EM structures of the yeast, porcine and human mitoribosomal subunits and of the entire yeast and human mitoribosomes became recently available. They have uncovered new information regarding mitoribosome composition and function to illustrate their evolutionary divergence from their bacterial ancestors and their adaptation to synthesis of highly hydrophobic membrane proteins (2–5). The mitoribosome assembly process is poorly understood although recent work has contributed to identifying some novel factors involved, such as the DEAD-box helicases Mrh4 in yeast (6) and DDX28 and DHX30 in human cells (7,8). Similarly, the actual mitochondrial translation process remains to be fully deciphered, particularly translation initiation. Since the mammalian mitochondrial mRNAs entirely lack a 5΄-UTR (untranslated region), a Shine/Dalgarno interaction between the mRNA and the 12S rRNA in the small ribosomal subunit does not occur during mitochondrial translation. Instead, an accurate translation initiation at the correct AUG is promoted preferably, owing to the lack of secondary structure at or around the start codon (9). Yeast mitochondrial mRNAs also lack a typical Shine/Dalgarno element. However, yeast mitochondrial mRNAs have long 5΄-UTRs and mRNA-specific translational activators bind to this region, possibly owing to localization of the small ribosomal subunit near the translational start codon (10), and/or assisting in resolving mRNA secondary structures around this area. Moreover, the processes of mitochondrial gene expression are coupled. Abundant evidence supports the concepts of co-transcriptional mRNA splicing (in yeast), processing and translation or co-translational membrane insertion of mitochondrial polypeptides (11–13). In yeast, the mitochondrial ribosome forms distinct clusters called MIOREX (mitochondrial organization of gene expression (14)) a subset of which is engaged in a large complex with the nucleoid that couples transcription, mRNA maturation, translation and RNA decay. Similar submitochondrial compartments are also seen in human mitochondria and are termed RNA granules or mitochondrioli (7,8,15–17). However, the factors involved in channeling the mRNAs through the different steps of gene expression within these large mitoribosome neighborhoods and their mechanisms of action remain largely uncharacterized.
NTP-dependent DEAD-box helicases play fundamental roles in mitochondrial gene expression processes such as pre-mRNA splicing (yeast Mss116) and ribosome assembly (Mrh4 in yeast; DDX28 and DHX30 in human) (reviewed in (18,19)). Among them, the S. cerevisiae Mss116 is the best characterized functionally, structurally and mechanistically (20–23). Mss116 was first identified in a genetic screen for nucleus-encoded factors involved in splicing of the intron-containing transcripts COB and COX1 of the yeast genome (24). Subsequently, it has been extensively reported that Mss116 is required for efficient splicing of all mitochondrial group I and group II introns present in three mitochondrial genes, COX1, COB and 21S rRNA (25), and promotes mitochondrial pre-mRNA processing in an adenosine triphosphate (ATP)-dependent manner (22,25,26). Mss116 acts as an RNA chaperone (27–29) and, similar to other DEAD-box proteins, promotes duplex unwinding based on local destabilization of RNA helical regions (30). Recent in vitro experiments have also shown that Mss116 modulates the activity of mitochondrial RNA-polymerase during transcription elongation in an ATP-independent fashion (31). However, transcription-related functions of Mss116 in vivo are only supported by genetic data. They show that the effects of a null mss116 mutation on colony survival under cold stress can be suppressed by overexpression of each of the two factors involved in mitochondrial transcription in S. cerevisiae, either the single-subunit catalytic core of mtRNA-polymerase (RPO41) or the transcription factor 1 (MTF1, (31). Additional observations have also suggested a role for Mss116 in mitochondrial translation, as mss116-deleted strains carrying intronless mtDNA remain respiratory-deficient and synthesis of cytochrome c oxidase (COX) subunits may be attenuated (24,25). However, the mechanism/s by which Mss116 could influence or regulate mitochondrial translation remain unexplored.
During studies aiming to identify mitochondrial ribosome biogenetic factors, we found Mss116 as part of the mitoribosomal proteome. In the present study, we have used strains carrying intronless mtDNA to analyze the possible role/s of Mss116 in mitoribosome biogenesis and mitochondrial translation. We have demonstrated that Mss116 is required for efficient mitoribosome assembly and that it associates with the 54S large mitoribosome subunit (mtLSU) and fully assembled ribosomes. Importantly, we show that Mss116 is specifically required for COX1 mRNA translation initiation and elongation, at least in part by interacting with the COX1 mRNA-specific translational activator Pet309, modulating its stability and therefore its productive action on the 5΄-UTR of COX1 mRNA. Our results support the concept of coupling among mRNA transactions, from transcription to RNA splicing and translation.
MATERIALS AND METHODS
Yeast strains and media
All S. cerevisiae strains used are listed in Supplementary Table S1. The construction of strains expressing mutant alleles of mss116 and the composition of the standard culture medium used is defined in the Supplementary Material.
Peptide antibody against Mss116
We have used the services of Open Biosystems/Thermo Scientific (Huntsville, AL, USA) to generate an affinity-purified rabbit polyclonal peptide antibody against Mss116. The peptide, SRPRTRSREDDDEV, comprises amino acids 67–80 on Mss116. A list of all antibodies used in this study is presented in Supplementary Table S2.
Characterization of the mitochondrial respiratory chain and oxidative phosphorylation system
Endogenous cell respiration was assayed in whole cells in the presence of galactose using a Clark-type polarographic oxygen electrode from Hansatech Instruments (Norfolk, UK) at 24°C as described (32).
Mitochondria were prepared from the different strains as described (6) and used for spectrophotometric assays performed at 24°C to measure KCN-sensitive COX activity, antimycin A-sensitive NADH cytochrome c reductase, and succinate cytochrome c reductase activities and oligomycin-sensitive ATP synthase activity, as described (32). Total mitochondrial cytochrome spectra were obtained as reported (32).
The abundance of OXPHOS complexes was analyzed by Blue Native polyacrylamide gel electrophoresis (BN-PAGE) using a linear 3–12% gradient gel (33).
In vivo mitochondrial protein synthesis
Mitochondrial gene products were labeled with [35S]-methionine (7 mCi/mmol, Perkin Elmer) in whole cells at 30°C in the presence of 0.2 mg/ml cycloheximide to inhibit cytoplasmic protein synthesis (32). Equivalent amounts of total cellular proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane and exposed to Kodak X-OMAT X-ray film.
Sucrose gradients
The sedimentation properties in sucrose gradients of Mss116, Mrh4 and mitoribosomal proteins from total mitochondrial extracts or purified ribosomes were analyzed essentially as described (34).
Analysis of mitochondrial polysomes
Yeast cells were grown in medium supplemented with 4mg/ml chloramphenicol (CAP) for 3 h and in fresh medium for 1 h before proceeding with mitochondrial isolation as described (34–36). Mitochondria were further purified using a step sucrose gradient as described (37). Mitochondrial extracts were prepared in the presence of 0.8% sodium deoxycholate (NaDOC), 10 mM Tris–HCl pH. 7.5, 100 mM NH4Cl, 20 mM MgCl2 and 200 U of RNaseOUT (Thermo Fisher) and loaded onto 10 ml 10–30% sucrose gradients. The gradients were centrifuged in a SW41 Ti Rotor (Beckman) at 40,000 rpm during 1.5 h at 4°C. Subsequently, the gradients were fractionated using a BR-188 Density Gradient Fractionation System (Brandel). Aliquots of each of 28 fractions were used for immunoblot analysis of mitoribosomal markers. The rest was used for RNA extraction, and the presence of the COX1 andCOB mRNAs was analyzed by cDNA synthesis followed by real-time polymerase chain reaction (RT-PCR) amplification.
RNA analysis
Methods for RNA isolation, northern blot analyses and RT-PCR are described in the Supplementary Experimental Procedures.
Statistical analysis
All of the experiments were done at least in triplicate. The data are presented as the means ± S.D. of absolute values or percentages of control. The values obtained for wild-type (WT) and Δmss116 mutant strains for the different parameters studied were compared by Student's t-test. P < 0.05 was considered significant.
RESULTS
Mss116 co-purifies with Mrh4 and mitoribosomal proteins
Studies in yeast and human cells have demonstrated extensive interactions of the mitochondrial translation machinery with factors involved in various post-transcriptional steps of gene expression, including mitoribosome assembly, tRNA modification and mRNA processing and turnover (7,8,14–17). In yeast, all these proteins interact with the mitoribosome to form distinct mitochondrial organization of gene expression (MIOREX) complexes (14). Previous studies in our laboratory identified a MIOREX component, the DEAD-box protein Mrh4, as a mitoribosome assembly factor acting at late stages of the mitoribosome large subunit (mtLSU) assembly (1,6).
To further characterize the function of Mrh4, we aimed to identify the proteins that directly or indirectly interact with a functional 6xHis-tagged version of Mrh4. For this purpose, we followed two strategies, described in the Supplementary Material, to use high salt conditions to strip Mrh4 and associated proteins from the mitoribosome or to analyze Mrh4-interacting proteins in a strain devoid of mtDNA (ρo strain), and therefore of rRNAs and mitoribosomes (Supplementary Figure S1). In the two experiments, the Mrh4-interacting proteins were analyzed by mass spectrometry. Strains expressing non-tagged Mrh4 were used as negative controls. The Mrh4-containing complex that was stripped from the mitoribosome was enriched with the DEAD-box protein Mss116, the degradosome components (Suv3 and Dss1), the 21S rRNA methyltransferase Mrm1, the GTPase Mtg1 and a variable set of mtLSU and mtSSU riboproteins (Supplementary Table S3). The Mrh4-containing complex purified from ρ0 mitochondria was also enriched in Mss116, Suv3 and Dss1, with a different set of riboproteins (Supplementary Table S3). These results are consistent with the proteins that were identified in the MIOREX complex (14) and further reinforce the interactions of mitoribosome assembly factors, RNA processing and turnover enzymes with the mitoribosome.
In this report, we have focused on characterizing the role of Mss116 in mitochondrial translation and ribosome assembly in a yeast strain where mRNA splicing-related functions of Mss116 are dispensable. We do not expect that this role/s will overlap with the roles of Mrh4 because multiple copies of MRH4 do not suppress the respiratory defect in a Δmss116 mutant strain, and vice versa (Supplementary Figure S2).
Mss116 is required for mitochondrial OXPHOS biogenesis in a strain carrying stable intronless mtDNA
To study the possible role/s of Mss116 in mitoribosome biogenesis and mitochondrial translation, we engineered a Δmss116 strain carrying intronless mtDNA. Moreover, because mitochondrial ribosome or translation mutants have a tendency to lose their mtDNA, the Δmss116 strain was constructed in a background containing a previously reported episomal construct (SUP) overexpressing a mtDNA stabilizing factor (6). However, plasmid segregation experiments followed by mtDNA content determination showed that even in the absence of the mtDNA stabilizing factor, the percentage of mtDNA-containing cells (ρ+) after 20 generations at 30°C was consistently above 98%. Additionally, the mtDNA-encoded mitoribosomal small subunit (SSU) protein Var1 will not be synthesized if mitoribosome assembly or mitochondrial translation is impaired, potentially affecting the phenotype. To prevent the potential confounding effects of an Mss116 deletion with the effects of no Var1 translation, the Δmss116/SUP intronless strain was transformed with a construct that successfully relocates a recoded version of the VAR1 gene to the nucleus as described previously (38). Therefore, we performed all experiments in Δmss116/SUP mtDNA intronless strains expressing ectopic VAR1 (hereinafter Δmss116). Furthermore, to eliminate the occurrence of mitochondrial transcription elongation defects previously observed at low temperature (16°C) in Δmss116 intronless strains (31), all these experiments were conducted at 30°C.
The Δmss116 strain was found to grow extremely poorly in respiratory YPEG media (Figure 1A). Furthermore, it had normal levels of mitochondrial cytochromes c and b but markedly lowered cytochrome a, a3, the prosthetic groups of complex IV (CIV) or cytochrome c oxidase (COX) (Figure 1B). Consistently, endogenous cell respiration and mitochondrial NADH or ascorbate-TMPD oxidation were extremely lowered (Figure 1C). OXPHOS complex III (bc1 complex) activity was not affected, but oligomycin-sensitive ATPase activity decreased by ∼30% compared to the WT. Instead, COX activity was barely detectable in Δmss116 mitochondria (Figure 1C), in agreement with its markedly low levels of Cox1 and assembled CIV (Figure 1D and E).
Figure 1.
Mss116 is essential for OXPHOS system assembly and function. See also Supplementary Figure S1 and Table S1. (A) Growth test using serial dilutions of the indicated strains in complete fermentable (YPD) and respiratory (YEPG) media. Pictures were taken after 2 and 4 days of growth at 30°C. (B) Total mitochondrial cytochrome spectra from the WT and Δmss116 strains. (C) Assessment of the respiration and OXPHOS enzyme functions. Polarographic determination of endogenous cell respiration, and oxidation of NADH or Ascorbate-TMPD in isolated mitochondria. Spectrophotometric measurement of NCCR (NADH cytochrome c reductase), QCCR (Ubiquinol cytochrome c reductase, cytochrome c oxidase (COX) and oligomycin-sensitive ATPase. The bars indicate the means ± SD from at least three independent sets of measurements. (D) Steady-state levels of the indicated proteins estimated by immunoblot analyses of mitochondrial proteins. The Immunoblot for Cox1 is presented in duplicate, representing two exposure times. (E) Immunoblot analysis of OXPHOS complexes extracted from mitochondria using native conditions and separated by blue native (BN)-polyacrylamide gel electrophoresis.
Mss116 interacts with the 54S mtLSU in dissociated and assembled mitoribosomes
To proceed with biochemical experiments, we generated an antibody against an Mss116 peptide, which allowed localizing the protein in mitochondrial extracts (Figure 1D).
To probe for a possible interaction of Mss116 with the mitoribosome, proteins were extracted from isolated WT mitochondria using 0.8% Triton X-100 and 25 mM KCl and analyzed by sucrose gradient sedimentation. Mss116 co-sedimented with fully assembled ribosomes in extracts prepared in the presence of 0.5 mM MgCl2 and with dissociated mtLSU in extracts prepared in the presence of 5 mM ethylenediaminetetraacetic acid (EDTA) (Figure 2A and B), thus suggesting an interaction of Mss116 with the 54S mitoribosomal subunit. The interaction was disrupted in the presence of high salt concentrations, which facilitated the accumulation of Mss116 in a smaller complex, co-sedimenting with Mrh4 (Supplementary Figure S1). The Mss116-ribosome interaction was also lost when the extracts were incubated with high concentrations of RNase. The RNase treatment disrupted ribosomal integrity, which resulted in the accumulation of Mss116 as a smaller complex (not shown).
Figure 2.
Mss116 interacts with the mitoribosomal LSU and its absence affects mitoribosome biogenesis. (A and B) Sucrose gradient sedimentation analyses of mitoribosomal proteins and assembly factors on mitochondrial extracts prepared from the WT and Δmss116 strains in the presence of 0.8% Triton X-100 and the conditions stated including either Mg2+ (A) or ethylenediaminetetraacetic acid (EDTA) (B). (C) Steady-state levels of mitoribosomal proteins estimated by immunoblot analyses. Porin was used as a loading control. (D) Sucrose gradient sedimentation analyses of SSU and LSU in mitochondrial extracts from the indicated strains prepared in the presence of 5 mM EDTA. The fractions were used to measure total RNA concentration (bottom) and to analyze the distribution of Mss116 and the ribosomal proteins by immunoblotting (top).
Analysis of the steady-state levels of mitoribosome proteins indicated that the concentrations of three mtSSU and most mtLSU proteins tested were reduced in Δmss116 cells compared to the WT control. Regarding the mtLSU, late-assembly riboproteins such as bL32, uL16 and bL33 were most prominently reduced compared to the WT strain (Figure 2C). We noticed that the levels of Mrh4 were slightly increased, suggesting some compensatory mechanism.
The steady state levels of the mitoriboproteins suggest lowered mitoribosome abundance in Δmss116 mitochondria. However, the sedimentation pattern of the mitoribosome markers in sucrose gradients did not change between the Δmss116 and WT strains under either EDTA or Mg2+ extraction conditions (Figure 2A and B). Together, these data indicate that, although its role is not essential, Mss116 might participate in the biogenesis of the mtLSU, probably at an early step, which prevents the accumulation of large assembly intermediates. A potential early assembly intermediate could be degraded during sample processing. To minimize RNA degradation during sample manipulation, we deleted the NUC1 gene encoding a mitochondrial nuclease that is usually activated after solubilization, a previously reported strategy that does not produce any respiratory phenotype (14,39). Deletion of nuc1, however, did not help to stabilize any assembly intermediate in mitochondria purified from this strain (Figure 2D) thus suggesting that mitoribosome assembly intermediates may not accumulate in mitochondria in the absence of Mss116.
Mss116 is required for efficient overall mitochondrial protein synthesis and is nearly essential for COX1 mRNA translation
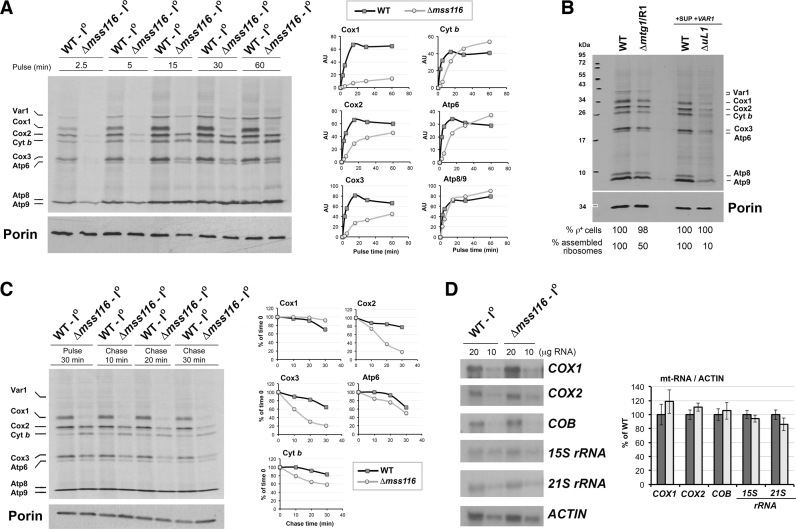
To test whether the OXPHOS biogenesis defect in the Δmss116 strain stems from a defect in mitochondrial gene expression, we performed in vivo mitochondrial translation experiments. Our results showed that Δmss116 cells can synthesize mitochondrial proteins albeit not efficiently (Figure 3A). As previously reported by other groups in different yeast backgrounds (24,25), the Δmss116 mutations most specifically affected synthesis of Cox1, Cox2 and Cox3 (Figure 3A). To assess better the effect of the Δmss116 mutation on the amount of Cox1/2/3 available for CIV assembly, the rate of incorporation of [35S]-methionine into the different translation products was measured at increasing pulse times. We found that the incorporation of [35S]-methionine into Cox1 in the mutant is <10% of WT, even at the earliest time points (Figure 3A). Although there was also less labeling of Cox2 and Cox3 in the mutant, the effect was not nearly as dramatic as with Cox1, particularly in the longer pulses. For cytochrome b and Atp6, although the rate of synthesis was lower than in the WT, in the longer pulses both proteins were labeled to a greater extent in the Δmss116 mutant (Figure 3A). The possibility exists that a decrease in mitoribosome amount might not affect all mRNAs to the same extent, depending on the timing of translation for the different mRNAs. In this case, the defect in COX1 mRNA translation in the absence of Mss116 could be due to decreased ribosome levels. However, in yeast mitoribosome assembly mutants, the synthesis rates of Cox1 and all other polypeptides are evenly decreased, as opposed to the Δmss116 mutant strain. This is illustrated in Figure 3B, where we have analyzed protein synthesis in two mitoribosome-deficient strains available in the laboratory. These data argue that the defect in COX1 mRNA translation in the absence of Mss116 is not a consequence of the decreased ribosome levels present in the Δmss116 mutant strain.
Figure 3.
Mitochondrial protein synthesis is altered in the absence of Mss116. (A) In vivo mitochondrial protein synthesis in the indicated strains following incorporation of [35S]-methionine into newly synthesized polypeptides as a function of time. Proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane and exposed to X-ray film. The graphs show the densitometry values obtained by using the histogram function of the Adobe Photoshop program on digitalized images. The values were normalized by the immunoblot signal of Porin and expressed relative to the control. Two independent repetitions did not differ by more than 5%. (B) In vivo mitochondrial protein synthesis as in panel (A), during 15 min, using the indicated strains. Immunoblotting for Porin was used as a loading control. Two mitoribosomal mutants were used. The strain Δmtg1/R1 carries a null allele of the mtLSU assembly factor MTG1 and a partial suppressor mutation in the 21S rRNA. The strain ΔuL1 carries a null allele of the mtLSU subunit uL1. For each mutant, the corresponding wild-type (WT) control strain was used. In all cases, the percentage of cells carrying mitochondrial DNA (ρ+) was near 100%. (C) Pulse-chase mitochondrial translation. Following a 30-min pulse, cold methionine and puromycin were added and the stability of newly-synthesized mitochondrial proteins was followed as a function of time at 30°C. Signals were quantified as in (A) and expressed as percentage of chase time zero. (D) Northern blot analyses of total RNA probed for COX1, COX2, COB and 15S and 21S rRNA. After processing, the membranes were exposed to X-ray film, and quantification of the signals was carried out as in panel (A). The values were normalized by the signal of ACT1 mRNA as the loading control and expressed relative to the control. Error bars represent the mean ± SD of three independent repetitions. *P < 0.05.
The poor labeling of Cox1 in the Δmss116 mutant strain could be due to an effect of the mss116 mutation on the rate of synthesis and/or turnover of the protein. However, the fact that incorporation of [35S]-methionine into Cox1 is undetectable after short pulses (2.5–5 min) argues against fast turnover. To further explore this point, the stability of newly translated mitochondrial products was examined by pulse–chase experiments. Cells were pulsed with [35S]-methionine at 30°C for 30 min in the presence of cycloheximide and chased at the same temperature for different time periods after adding puromycin and excess cold methionine. In the Δmss116 strain, both Cox2 and Cox3 degraded at a much faster rate, likely because they cannot be assembled into COX in the absence of normal amounts of Cox1, while the degradation rate of Cyt b and Atp6 was almost the same as in the WT strain (Figure 3C). Even though there was some degradation of Cox1 in the WT, the residual ∼10% of Cox1 in the mutant did not decrease much further during the chase (Figure 3C), likely because this entire portion is readily used to assemble the residual amount of COX.
Using these strains, we also demonstrated that the protein synthesis defect in Δmss16 cells is not due to a discrepancy in RNA levels, as both WT and Δmss116 mitochondria had similar amounts of mRNAs, including COX1 mRNA, and of 15S and 21S ribosomal RNAs (Figure 3D).
Mss116 is required for translation initiation of COX1 mRNA
The general decrease in mitochondrial protein synthesis rate in the absence of Mss116 can be explained by the lowered concentration of mitochondrial ribosomes. Therefore, despite the sequence and structural similarity of Mss116 with eIF4A (eukaryotic initiation factor-4A), a well characterized DEAD-box protein, Mss116 does not seem to affect general mitochondrial translation initiation, because only a subset of translation products, essentially Cox1 and at a lower extent Cox3, are deeply affected. Nevertheless, Mss116 could be targeted specifically to these mRNAs for their efficient translation initiation or to disrupt potential stable secondary structures to allow access of translation factors or ribosomes, as proposed previously (25).
To test the direct involvement of Mss116 in COX1 mRNA translation, we used strains carrying a mitochondrion-encoded reporter gene (40) in either WT or Δmss116. The strains are deleted for the nuclear ARG8 gene, and the reporter is a recoded version of ARG8, termed ARG8m, that encodes a matrix-localized biosynthetic enzyme, which allows us to look directly at mitochondrial translation by scoring the growth of yeast in the absence of arginine and accumulation of the reporter protein. In the strain XPM10b, the COX1 coding sequence was completely replaced by ARG8m (Δcox1::ARG8m) (Figure 4A) and in the strain XPM78a, ARG8m was introduced at the end of the 512-nt long intronless COX1 coding sequence [COX1 (1–512)::ARG8m] (Figure 4A). In these strains, ARG8 mRNA translation is dependent on the COX1 5΄- and 3΄-untranslated regions (UTRs). When carrying a WT allele of MSS116, Arg8m is synthesized in both strains (Figure 4B) and supports arginine-independent growth (Figure 4C). However, the Δmss116 mutation markedly attenuated Arg8m synthesis, particularly in the XPM78a strain where Cox1-Arg8 is virtually absent (Figure 4B), and therefore the strains were unable to grow in the absence of exogenous arginine (Figure 4C). These results indicated that Mss116 is directly required for efficient translation of COX1 mRNA.
Figure 4.
Mss116 is required for COX1 mRNA translation initiation and elongation. See also Supplementary Figures S2 and 3. (A) Scheme depicting the mitochondrial genotype of strains carrying mitochondrial ARG8 as a reporter of mRNA translation. (B) In vivo mitochondrial protein synthesis in the presence of cycloheximide using the strains presented in panel (A). Porin was used as a loading control. (C) Growth test using serial dilutions of the indicated strains in minimum media supplemented or not with arginine and in complete respiratory medium (YEPG). Pictures were taken after 2 days of growth at 30°C.
To further test whether Mss116 is required exclusively for COX1 mRNA translation initiation or plays additional roles in translation elongation, we introduced the mss116 deletion into the XPM171a strain (40). This strain is respiratory-competent and arginine-independent because it contains the ARG8m gene under the control of the COX1 promoter, the COX1 gene under the control of the COX2 promoter plus the COX2 3΄UTR and a WT COX2 gene (Figure 4A). The introduction of the Δmss116 mutation resulted in an Arg− phenotype (Figure 4C), confirming the translation initiation defect from the COX1 promoter. However, it also produced a respiratory-deficient phenotype (Figure 4C) owing to the lack of COX1 synthesis from the COX2 promoter (Figure 4B). These results indicate that Mss116 targets the COX1 mRNA coding sequence. To further assess the interactions of Mss116 with the COX1 mRNA 5΄- and 3΄-UTR, we used two strains carrying an ARG8m reporter construct flanked by either the 5΄-UTR of COX1 and the 3΄-UTR of COX2 (strain XPM271a) or vice versa (strain XPM304b). A Δmss116 mutation in these strains resulted in defective Arg8 synthesis (Figure 4B) and an Arg- growth phenotype (Figure 4C). These results suggest that Mss116 may have targets all along the COX1 mRNA sequence, including the 5΄- and 3΄-UTRs, thus making this protein important during COX1 mRNA translation initiation and elongation.
The COX1 mRNA translation defect in Δmss116 cells is not bypassed by excess of Mss51 or Pet309, the two known COX1 mRNA translational activators
In S. cerevisiae, translation of each mitochondrion-coded COX subunit mRNA is specifically activated by distinct nucleus-encoded translational activators (41). Cox1 synthesis is controlled by Mss51 and the PPR (pentatricopeptide repeat) protein Pet309, which act on the COX1 mRNA 5΄-UTR. Mss51 additionally interacts with newly synthesized Cox1 and plays an important role in COX1 mRNA translation regulation through a negative feedback loop that couples Cox1 synthesis to its assembly into COX (34,40,42–44). We and others have suggested that binding of Mss51 to the 5΄-UTR of COX1 mRNA could be necessary for optimal initiation of translation by Pet309, whereas the interaction of Mss51 with newly synthesized Cox1 may regulate elongation of the nascent polypeptide (40,45).
The role of Mss116 in COX1 mRNA translation is not essential, as traces of Cox1 are synthesized in the Δmss116 mutant strain. If Mss116 acts to enhance the efficiency of the process, one could expect that excess of Pet309 or Mss51 (WT or the constitutively active variant Mss51-F199I (34,42,46)) could bypass the Mss116 requirement. However, this was not the case regarding respiratory growth or Cox1 synthesis (Supplementary Figures S2 and 3). In a WT strain, Mss51 accumulates in a prominent 450 kDa Cox1 stabilization complex and a less abundant 120 kDa complex that serves as a reservoir of Mss51 competent for translation initiation (42,43,47). In COX mutants in which the 450 kDa complex is disrupted (e.g. cox1 mutants), Mss51 accumulates in the 120 kDa complex (43). In the Δmss116 mutant strain, Mss51 accumulates only in the 120 kDa complex (Supplementary Figure S3B), conceivably because of the virtual absence of newly synthesized Cox1, which could be in turn due to the inaccessibility of Mss51 to its place of action on the COX1 mRNA during translation initiation.
Together, these data agree with the ARG8-reporter assay results presented in the previous section and indicate that deficient Cox1 synthesis in the absence of Mss116 is not due to COX1 mRNA negative translational regulation but to an impairment in the actual process of translation.
Mss116 physically interacts with Pet309 and promotes its stability
Next, we investigated whether Mss116 genetically and/or physically interacts with COX1 mRNA translational activators, one of which could act as a co-factor to target Mss116 to its place of action of the COX1 mRNA 5΄UTR.
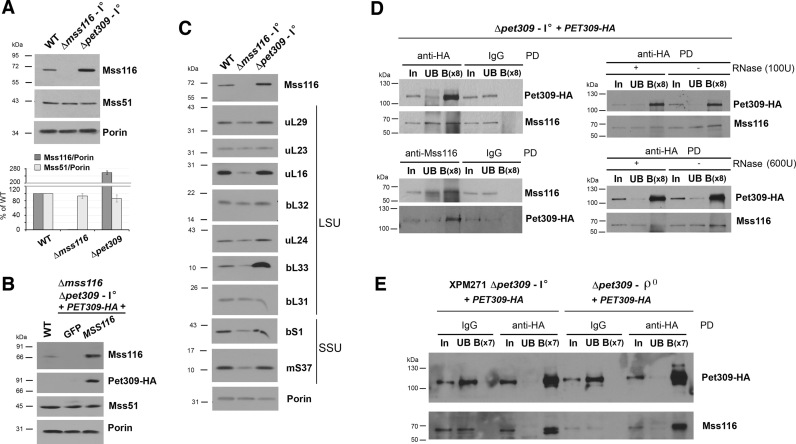
We started by estimating Mss116 levels in the presence or absence of Pet309 or Mss51. While Mss51 does not affect Mss116 levels and vice versa (Figure 5A and B), the absence of Pet309 results in 2.5-fold increased levels of Mss116 (Figure 5A), perhaps as a compensatory mechanism. Because an Ab against Pet309 is not available, we used a Δmss116Δpet309 strain expressing a functional hemagglutinin (HA)-tagged version of the protein from an integrative plasmid to show that Pet309 was barely detected in Δmss116 mitochondria (Figure 5B). This suggests that Pet309 and Mss116 might form a stable complex that stabilizes Pet309.
Figure 5.
Mss116 physically interacts with Pet309 and support its stability. See also Supplementary Figure S4. (A–C) Steady-state levels of (A) Mss116 and Mss51, (B) Mss116, Mss51 and HA-tagged Pet309, and (C) mitoribosomal proteins. In (B), a plasmid expressing GFP was used as a control. Mitochondrial proteins were separated by SDS-PAGE and analyzed by immunoblotting. Antibody–protein complexes were visualized by chemiluminescence and exposition to an X-ray film. The graph in panel (A) shows the densitometry values obtained by using the histogram function of the Adobe Photoshop program on digitalized images. The values were normalized by the signal of Porin as the loading control and are expressed relative to the control. Bars represent the average of three independent experiments ± SD. (D and E) Immunoprecipitation of Mss116 or Pet309-HA from mitochondrial extracts prepared from the indicated strains, using HA-conjugated magnetic beads. In (D), the extracts where either left untreated (left panel) or incubated with the indicated concentrations of RNase A for 30 min at 4°C (right panel). In, input; UB, unbound; B, bound. The concentration of B is 8-fold higher (x8) or 7-fold higher (x7) than that of UB.
Importantly, only the absence of Mss116 but not the absence of Pet309 limits the accumulation of mitoribosomes. This was concluded by estimating the steady-state levels of LSU mitoribosome proteins, which are similar in Δpet309 and in WT mitochondria (Figure 5C), with the exception of the late LSU assembly subunit bL33 (6), whose levels are several-fold enhanced. As reported (45), Δpet309 cells fail to synthesize Cox1 (Supplementary Figure S4) but synthesize normally all other mitochondrial polypeptides, which is expected given the normal accumulation of mitoribosomes in the absence of Pet309.
Mss116 does not physically interact with Mss51 in a stable manner because Mss51-GST pulldown experiments failed to detect Mss116 in the eluate (Supplementary Figure S5). On the contrary, both Mss116 and Pet309 were previously found to co-immunoprecipitate with the mitochondrial RNA polymerase (48), which could anticipate a physical interaction between the two proteins. To investigate whether Mss116 and Pet309 interact physically, we created a strain expressing Pet309-HA in the context of the nuc1 deletion. Mitochondrial extracts from this strain were prepared in the presence of 1% digitonin and used to confirm efficient and specific Pet309-HA immunoprecipitation with anti-HA-conjugated magnetic beads (Figure 5D, left panel). A fraction of Mss116 reproducibly co-immunoprecipitated with Pet309-HA, indicating a physical interaction between the two proteins. The interaction was confirmed by the reciprocal co-immunoprecipitation experiment (Figure 5D, left panel). Furthermore, the interaction was not disrupted by treatment with 100 U/μl RNase A and only partially by incubation in the presence of a large excess of 600 U/μl RNase A (Figure 5D, right panel). These results suggest that Mss116 and Pet309 physically interact, probably near the COX1 mRNA. They also suggest that Pet309 could act as an Mss116 co-factor to target the helicase to the COX1 mRNA. To further explore the COX1 mRNA determinants of the Mss116–Pet309 interaction, we performed HA-pulldown analyses using two strains modified to express Pet309-HA: a rho zero (ρ0) strain, devoid of mtDNA (and thus of COX1 mRNA), and the XPM271a strain where only the COX1 mRNA 5΄UTR is present but neither the coding sequence nor the 3΄UTR. In both cases, Mss116 efficiently co-immunoprecipitated with Pet309-HA (Figure 5E), thus indicating that the interaction between these two proteins can occur in an mRNA-independent manner.
Mitoribosome loading onto COX1 mRNA can occur in the absence of Mss116
A failure of translational activators to bind their specific RNA is expected to result in failure to load the mRNA correctly onto the mitoribosome (14). To assess whether the absence of Mss116 and Pet309 limits the mitoribosome-COX1 mRNA association, we used highly purified mitochondria from nuc1-deleted WT, Δpet309 and Δmss116 strains carrying intronless mtDNA. Yeast cells were grown in CAP for 3 h and in fresh medium for 1 h prior to mitochondrial isolation. The CAP treatment allows accumulation of cytoplasmically synthesized proteins required for mitoribosome assembly and mitochondrial gene expression, which results in a robust increase in the proportion of mito-polysomes and of mitochondrial protein synthesis (34,35). Mitochondrial extracts were prepared in the presence of 0.8% NaDOC, 10 mM Tris pH 7.5, 100 mM NH4Cl, 20 mM MgCl2 and 200 U of RNaseOUT, and fractionated in sucrose gradients. Aliquots of each fraction were used for immunoblot analysis of mitoribosomal markers. The rest was used for RNA extraction and the presence of the COX1 and COB mRNAs analyzed by cDNA synthesis followed by RT-PCR amplification (Figure 6A). Polysomes were detected in the three strains analyzed (Figure 6B–D). In WT mitochondria, similar amounts of COX1 and COB mRNAs were detected in the SSU, monosome and polysome fractions (Figure 6E). In Δpet309 mitochondria, intronless COX1 mRNA is stable but not translated (45,49) inasmuch as translation initiation requires the binding of Pet309 to the 5΄ UTR of COX1 mRNA. Our data show that Pet309 is not essential for mitoribosome loading onto COX1 mRNA and support its requirement for COX1 mRNA translation given that this mRNA is barely detected in the polysome fractions (Figure 6C and E). In the absence of Mss116, a larger proportion of the residual ∼50% of mitoribosome subunits present is detected in monosomes and polysomes (Figure 6D). This is probably because as seen for Δpet309, COX1 mRNA loading was hardly detected in the Δmss116 mitochondrial polysome fractions (Figure 6D and E).
Figure 6.
Mitoribosome loading onto COX1 mRNA can occur in the absence of Mss116, but the COX1 mRNA is not detected in polysomes. (A) Scheme depicting the experimental protocol for mito-polysome profiling. (B–D) Sucrose gradient sedimentation analyses of mitoribosomal subunits, monosomes and polysomes in extracts obtained from mitochondria isolated from the indicated strains, all (including the WT strain) carrying the mtDNA SUP- and ectopic VAR1 expressing plasmids. In each case, the upper panel is an immunoblot analysis of mitoribosome subunit markers. The Immunoblot for fractions 16–28 is presented in duplicate, with short (as for the blot with fractions 1–15) or longer exposure time. The graphs in the lower panels represent the relative amount of the indicated rRNAs and mRNAs in the gradient fraction, estimated by qPCR. (E) Relative mRNA loading into the mt-SSU, mitoribosomes and polysomes estimated using samples from sucrose gradient fractions in panels (B–D). For each strain, the relative mRNA percentage in each structure was calculated. The bars represent the average of three independent experiments ± SD.
The catalytic activity of Ms116 is required for its role in COX1 mRNA translation
We also asked whether the ATP-dependent helicase activity of Mss116 is necessary for its roles in COX1 mRNA translation. To investigate this question, we created Δmss116 strains that express WT Mss116, or variants carrying mutations in the ATP-binding and DEAD box conserved regions in Mss116 from centromeric plasmids (Figure 7A). Mutation of lysine K158 to alanine in the ATPase-A motif and a change of aspartate D267 or glutamate E268 in the DEAD box motif to alanine did not alter the stability of each Mss116 variant and, consequently, Pet309 also remained stable (Figure 7B). The mutations did not affect the ability of Mss116 to complex with Pet309 (Figure 7C) but abolished Mss116 function on COX1 mRNA translation (Figure 7D). These results demonstrate that the ATP-dependent helicase activity of Mss116 is necessary for its function in mitochondrial translation.
Figure 7.
The catalytic activity of Ms116 is required for its role in COX1 mRNA translation. (A) Diagram of the Mss116 protein depicting the conserved core with nine motifs characteristic of the DEAD-box subfamily of DExH/D-box proteins (black bars). Asterisks indicate the locations of Mss116 mutations analyzed in this work. (B) Steady-state levels of Mss116 and HA-tagged Pet309 in WT and Δmss116 and Δmss116Δpet309+PET309-HA strains expressing GFP as a negative control, WT MSS116 or the indicated mss116 variants. Porin was used as the loading control. (C) Immunoprecipitation of Pet309-HA from mitochondrial extracts obtained from the indicated strains, using HA-conjugated magnetic beads. Mss116 was also detected in the different fractions. In, input; UB, unbound; B, bound. (D) In vivo mitochondrial protein synthesis in the WT and Δmss116 (Δ) strains expressing GFP as a negative control, WT MSS116 or the indicated mss116 variants, following the incorporation of 35S-methionine into newly synthesized proteins (30-min pulse) in the presence of cycloheximide to inhibit cytoplasmic protein synthesis. Immunoblot for Porin was used as a loading control.
Prediction of Pet309 binding sites in the COX1 mRNA 5΄UTR by in silico analysis
To predict the potential binding site for Pet309 on the COX1 5΄-UTR, we performed an in silico analysis based on the work by Yagi et al. on the elucidation of the PPR code (50). Pet309 has 22 predicted PPR domains (51), expected to bind to 22 nt (52). Multiple studies have indicated that amino acids in the 2nd position and particularly those in the 5th and 35th positions of a PPR domain are the main contributors to nucleotide binding specificity (53–55). Yagi et al. analyzed 464 PPR motifs present in 33 proteins from Arabidopsis thaliana and Physcomitrella patens and, because these proteins have a known binding sequence on their cognate RNAs, the authors could assign a specific nucleotide to each PPR motif. Using these data, they assessed the frequency in which PPRs with a specific combination of amino acids in the 2nd, 5th and 35th positions, the 5th and 35th positions, or only the most specificity-relevant 35th position, bind each of the 4 nt and calculated the corresponding binding probabilities.
From the 22 predicted PPR motifs in Pet309, 13 have a combination of residues that fit the pattern reported by Yagi et al (Supplementary Table S4). For the 9 PPR motifs lacking the predicted amino acid combinations, we assigned equal binding probability for the 4 nt to obtain a degenerate sequence that was searched on the COX1 5΄UTR by using the software ‘Degenerate motif finder’ by Insilicase (http://www.insilicase.com/Web/DegenerateSites.aspx). We identified two sequences that fitted the degenerate pattern: AUUUAAUUAAUUUUUUUUUUUA, which spans from −76 to −55 of the COX1 5΄UTR (taking the adenine from the initiation codon as +1) and AUUUAAAUGAUUUUAAUAUAUA, which spans from −356 to −335 of the COX1 5΄UTR (Figure 8A).
Figure 8.
Roles of Mss116 in mitoribogenesis and COX1 mRNA translation. (A) Primary nucleotide sequence of the COX1 5΄-UTR with the two in silico-predicted Pet309-binding sites shaded in cyan, and the AUG translational start codon and downstream sequence in bold. (B) Model of the secondary structure of COX1 mRNA 5΄UTR. The mfold-quikfold program for RNA folding prediction based on thermodynamic parameters (56) was used to analyze the folding with minimum free energy (ΔG) of the COX1 mRNA 5΄UTR. The potential Pet309 binding sites from (A) in the COX1 mRNA 5΄UTR are shaded in blue. The broad Mss51 interaction site, previously determined by Y3H experiments (45) is indicated. (C) Schematics portraying the distinct roles of Mss116 in mitoribosome assembly and COX1 mRNA translation. To model the interaction between Mss116 and Pet309 we performed a docking analysis using the server GrammX (69). We used the crystallographic model of Mss116 co-purified with ssRNA (PDB: 4TZ6) (70), and a model of Pet309 obtained from the ITASSER server (71).
A model of the COX1 mRNA 5΄UTR structure, obtained with the mfold server (56), portrays the potential structural complexity of the molecule without considering RNA–protein interactions (Figure 8B) and highlights the requirement of RNA binding proteins, such as Mss116 and Pet309, to facilitate COX1 mRNA translational activation.
DISCUSSION
Mss116 is an S. cerevisiae DEAD-box helicase that plays a major role in splicing of group I and II introns (24,25,27,28,57). Furthermore, Mss116 has been shown to perform additional functions in transcription elongation (31) and mitochondrial translation in an intronless background (24,25). The observation of Mss116 interacting with the mitoribosome and the lack of information regarding the mechanism by which Mss116 activates mitochondrial translation led us to focus on this question further. Multitasking is not a unique property of Mss116 but is common among many DEAD/DXAH proteins (58). For example, several cytoplasmic mRNA-specific helicases that participate in translation initiation such as Ded1/DDX3 and RHA/DHX9, and maybe also Vasa/DDX4, all appear to play additional roles in transcription, pre-mRNA splicing, export from the nucleus and transport (58). This feature makes it difficult to separate a direct effect from the indirect effects of the proteins on translation initiation. Therefore, to study the role of Mss116 exclusively on mitochondrial translation we have bypassed its requirement for splicing by using strains devoid of introns and by performing our experiments at 30°C to avoid transcription elongation defects in the absence of Mss116.
Our studies demonstrate that Mss116 plays roles in the assembly of the mitochondrial translational machinery as well as in the translation process itself. The total amount of mitochondrial ribosomes is lowered in a Δmss116 strain. WT Mss116 interacts with the mtLSU and the monosome, suggesting roles in ribosome assembly and beyond. Ribosome assembly intermediates were not detected in the Δmss116 strain, which points toward a role of Mss116 at early stages of the mtLSU assembly process. The reduced mitoribosome content resulted in a decline in overall mitochondrial protein synthesis in the Δmss116 strain, with the synthesis of the three COX genes being mostly affected. However, the accumulation of newly synthesized Cox2 and Cox3 proteins is nearly equivalent to the WT levels over long pulses. On the contrary, synthesis of Cox1 is specifically and drastically affected.
Further, the use of a mitochondrial reporter gene helped us in disclosing multiple targets of Mss116 on the COX1 mRNA. One is in the 5΄UTR, which involves Mss116 in stimulation of COX1 mRNA translation initiation. The Mss116 function is likely related to the COX1 mRNA translational activator Pet309, with which it interacts and provides stability. Furthermore, Mss116 accumulates in the absence of Pet309, suggesting a compensatory effect and both proteins are necessary to engage mitoribosomes in the translation of the COX1 mRNA. In vivo, Pet309 binds to the COX1 mRNA, and this interaction is severely attenuated in the absence of Mss51 (39). Models of the secondary structure of the COX1 mRNA 5΄UTR region generated with the mfold program predicted the complexity of this region (Figure 8). Moreover, the in silico analysis presented here has disclosed potential binding sites for Pet309 in the COX1 mRNA 5΄UTR. These regions span from 55 to 76 and from 335 to 356 nt upstream of the AUG start codon. According to the mfold COX1 mRNA model, both regions are part of potential stem-loop secondary RNA structures (Figure 8). Thus, the presence of a helicase such as Mss116 seems indicated to disrupt stable secondary structures to allow access of Pet309 to the COX1 5΄UTR. Indeed, we have demonstrated that the helicase activity of Mss116 is essential for COX1 mRNA efficient translation. This is reminiscent of the function of the DEAD-box protein eIF4A, a component of the eIF4F complex, in eukaryotic translation. Owing to its localization on the cap, eIF4A may promote preinitiation complex loading by disrupting RNA secondary structures in the vicinity of the 5΄ terminus (59).
We could propose a model in which Mss116 helicase changes the COX1 mRNA conformation to allow Pet309 binding to its cognate sequence. However, inasmuch as Pet309, as do most PPR proteins, has mRNA recognition capacity but lacks obvious domains that could carry a catalytic activity, we favor a model in which Pet309 binds to the COX1 mRNA 5΄UTR and then acts as an adaptor for directing the role of other factors, such as Mss116 (Figure 8). Among the two potential Pet309 targets on the COX1 mRNA, the sequence from nucleotide −55 to −76 seems more plausible for Pet309 interaction than the sequence from nucleotide −335 to −356, because it is closer to the AUG start codon. In addition, this sequence is part of a possible binding region of Mss51. No in vivo interaction of Mss51 with the COX1 mRNA has been found (37), but Y3H experiments showed the interaction of the hydrophilic N-terminal portion of Mss51 with a COX1 mRNA 5΄UTR fragment of 250 nt upstream of the start codon (45). Mss116–Pet309 binding might induce a further conformational change in the RNA structure to facilitate translation initiation. Experiments in vitro with the chloroplast PPR10 protein demonstrated that it binds to single-stranded RNA to induce remodeling of the adjacent sequences to facilitate ribosome recruitment (52). Our experiments argue against a Mss116–Pet309 role in ribosome recruitment, because mitoribosome loading proceeds in the absence of any of these proteins, yet translation is impaired. Alternatively, binding of Mss116–Pet309 to the COX1 mRNA might allow access of Mss51 to the COX1 translation machinery, and/or assist the mitoribosome to correctly interact with the AUG start codon in the COX1 mRNA.
Mss116 has additional targets in the COX1 mRNA coding sequence and 3΄UTR, which suggests a role in COX1 mRNA translation elongation or termination. The nature of this interaction—whether Mss116 binds directly to the mRNA via a sequence or structural motif or interacts indirectly through elongation or release factors as done by S. cerevisiae cytoplasmic Dbp5 (60)—is yet to be disclosed and will be a matter of future investigations. However, it is tempting to speculate that the interaction of Mss116 with the mitoribosome places this helicase in an ideal location to scan and resolve secondary structures in the COX1 mRNA coding sequence to facilitate its translational elongation.
A standing question relates to how the roles of Mss116 in COX1 mRNA splicing, transcription and translation are coordinated. A key element could be Pet309 because, beyond its role as a translational activator, it ensures either the fidelity of splicing of COX1 mRNA introns or the stability of splicing intermediates (49). Therefore, the physical interaction between the two proteins and the stabilization of Pet309 in this complex could be essential to coordinate their multiple roles in COX1 gene expression. Pet309 was found to form a complex with other mRNA-specific translational activators, the transcription factor Nam1/Mtf2 and several message-specific RNA-binding proteins, including the RNA polymerase core Rpo41 (61). Furthermore, Mss116 modulates the activity of mitochondrial RNA-polymerase during transcription elongation in certain conditions (31). These functional interactions support the concept of mitochondrial transcriptional and translational coupling.
The role of Pet309 as an mRNA stabilizer and as an adaptor for other proteins is evolutionary conserved. The Pet309 human homolog is LRPPRC, a protein mutated in the French–Canadian variant of Leigh's syndrome that is characterized by cytochrome c oxidase deficiency (62). LRPPRC physically interacts with SLIRP forming a complex that co-stabilizes both proteins and mediates mt-mRNA stability (63,64). However, studies in vivo in mouse tissues have shown that LRPPRC and SLIRP have distinct roles within the mt-mRNA-stabilizing complex they form. LRPPRC is required for maintenance of polyadenylation of mtDNA-encoded mRNAs (65,66). Instead, SLIRP is required for proper association of mRNAs to the mitochondrial ribosome and efficient translation (67).
The multiple roles of Mss116 resemble other nuclear gene products involved in mitochondrial RNA processing that play a second role in mitochondrial biogenesis or function. Among them, Rpm2, the protein subunit of mitochondrial RNase P in S. cerevisiae, is particularly relevant. Rpm2 also has a yet undefined role in the translation of COX1 and COX2 mRNAs, which has been linked to ∼100 aa C-terminal portion of Rpm2 that shares 49% similarity with Pet309 (68).
In conclusion, our data illuminate new roles for Mss116 in mitochondrial gene expression: Mss116 is required for mitoribosome biogenesis and also acts as a COX1 mRNA-specific translation factor in yeast mitochondria. In the wider context, our results suggest the existence of additional mitoribosome-associated proteins involved in mRNA transactions that act in concert with translational factors to determine mitochondrial mRNA-specific translation. Furthermore, our results support the conclusion that the availability of cofactors allows RNA helicases to perform multiple functions, as shown here for Mss116 in the biogenesis and function of the mitochondrial translation machinery.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Brant Watson for critical reading of the manuscript. We thank Dr T. Fox (Cornell University NY, USA), Dr D. Markov (Rowan University, NJ, USA), Dr M. Ott (Stockholm University, Stockholm, Sweden) and Dr R. Stuart (Marquette University, WI, USA) for providing reagents.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH-RO1 Grants [GM105781, GM112179 to A.B.]; NIH-R35 Grant [GM118141 to A.B.]; MDA Grant [MDA-381828 to A.B.]; AHA Development Grant [14SDG20040003 to F.F.]; PAPIIT-UNAM Grant [IN204414 to X.P.-M.]; CONACyT Posdoctoral Fellowship (to A.Z.-O.); AHA Predoctoral Fellowships (to D.D., R.Z.). Funding for open access charge: NIH-R35 Grant [GM118141 to A.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1. De Silva D., Tu Y.T., Amunts A., Fontanesi F., Barrientos A.. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 2015; 14:2226–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amunts A., Brown A., Bai X., Llácer J.L., Hussain T., Emsley P., Long F., Murshudov G., Scheres S.H.W., Ramakrishnan V.. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014; 343:1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amunts A., Brown A., Toots J., Scheres S.H.W., Ramakrishnan V.. The structure of the human mitochondrial ribosome. Science. 2015; 348:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greber B.J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., Ban N.. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015; 348:303–308. [DOI] [PubMed] [Google Scholar]
- 5. Desai N., Brown A., Amunts A., Ramakrishnan V.. The structure of the yeast mitochondrial ribosome. Science. 2017; 355:528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Silva D., Fontanesi F., Barrientos A.. The DEAD-Box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metab. 2013; 18:712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tu Y.T., Barrientos A.. The human mitochondrial DEAD-box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep. 2015; 10:854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Antonicka H., Shoubridge E.A.. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015; 10:920–932. [DOI] [PubMed] [Google Scholar]
- 9. Scharff L.B., Childs L., Walther D., Bock R.. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 2011; 7:e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fox T.D. Hershey JWB, MB M, Sonenberg N. Translational Control. 1996; NY: Cold Spring Harbor Press: Cold Spring Harbor; 733–758. [Google Scholar]
- 11. Surovtseva Y.V., Shadel G.S.. Transcription-independent role for human mitochondrial RNA polymerase in mitochondrial ribosome biogenesis. Nucleic Acids Res. 2013; 41:2479–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keil M., Bareth B., Woellhaf M.W., Peleh V., Prestele M., Rehling P., Herrmann J.M.. Oxa1-ribosome complexes coordinate the assembly of cytochrome c oxidase in mitochondria. J. Biol. Chem. 2012; 287:34484–34493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hell K., Neupert W., Stuart R.A.. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001; 20:1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kehrein K., Schilling R., Moller-Hergt B.V., Wurm C.A., Jakobs S., Lamkemeyer T., Langer T., Ott M.. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 2015; 12:843–853. [DOI] [PubMed] [Google Scholar]
- 15. Barrientos A. Mitochondriolus: assembling mitoribosomes. Oncotarget. 2015; 6:16800–16801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Antonicka H., Sasarman F., Nishimura T., Paupe V., Shoubridge E.A.. The mitochondrial RNA-binding protein GRSF1 Localizes to RNA granules and Is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013; 17:386–398. [DOI] [PubMed] [Google Scholar]
- 17. Jourdain A.A., Koppen M., Wydro M., Rodley C.D., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., Martinou J.C.. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013; 17:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szczesny R.J., Wojcik M.A., Borowski L.S., Szewczyk M.J., Skrok M.M., Golik P., Stepien P.P.. Yeast and human mitochondrial helicases. Biochim. Biophys. Acta. 2013; 1829:842–853. [DOI] [PubMed] [Google Scholar]
- 19. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cao W., Coman M.M., Ding S., Henn A., Middleton E.R., Bradley M.J., Rhoades E., Hackney D.D., Pyle A.M., De La Cruz E.M.. Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins. J. Mol. Biol. 2011; 409:399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Del Campo M., Lambowitz A.M.. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol. Cell. 2009; 35:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mallam A.L., Del Campo M., Gilman B., Sidote D.J., Lambowitz A.M.. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012; 490:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mohr G., Del Campo M., Mohr S., Yang Q., Jia H., Jankowsky E., Lambowitz A.M.. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J. Mol. Biol. 2008; 375:1344–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seraphin B., Simon M., Boulet A., Faye G.. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989; 337:84–87. [DOI] [PubMed] [Google Scholar]
- 25. Huang H.R., Rowe C.E., Mohr S., Jiang Y., Lambowitz A.M., Perlman P.S.. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Niemer I., Schmelzer C., Borner G.V.. Overexpression of DEAD box protein pMSS116 promotes ATP-dependent splicing of a yeast group II intron in vitro. Nucleic Acids Res. 1995; 23:2966–2972. [PMC free article] [PubMed] [Google Scholar]
- 27. Del Campo M., Tijerina P., Bhaskaran H., Mohr S., Yang Q., Jankowsky E., Russell R., Lambowitz A.M.. Do DEAD-box proteins promote group II intron splicing without unwinding RNA. Mol. Cell. 2007; 28:159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mohr S., Matsuura M., Perlman P.S., Lambowitz A.M.. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:3569–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mallam A.L., Jarmoskaite I., Tijerina P., Del Campo M., Seifert S., Guo L., Russell R., Lambowitz A.M.. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12254–12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Q., Del Campo M., Lambowitz A.M., Jankowsky E.. DEAD-box proteins unwind duplexes by local strand separation. Mol. Cell. 2007; 28:253–263. [DOI] [PubMed] [Google Scholar]
- 31. Markov D.A., Wojtas I.D., Tessitore K., Henderson S., McAllister W.T.. Yeast DEAD box protein Mss116p is a transcription elongation factor that modulates the activity of mitochondrial RNA polymerase. Mol. Cell. Biol. 2014; 34:2360–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barrientos A., Korr D., Tzagoloff A.. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2002; 21:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diaz F., Barrientos A., Fontanesi F.. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using blue native gel electrophoresis. Curr. Protoc. Hum. Genet. 2009; doi:10.1002/0471142905.hg1904s63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barrientos A., Zambrano A., Tzagoloff A.. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004; 23:3472–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ibrahim N.G., Beattie D.S.. Regulation of mitochondrial protein synthesis at the polyribosomal level. J. Biol. Chem. 1976; 251:108–115. [PubMed] [Google Scholar]
- 36. Herrmann J.M., Stuart R.A., Craig E.A., Neupert W.. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J. Cell Biol. 1994; 127:893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meisinger C., Pfanner N., Truscott K.N.. Isolation of yeast mitochondria. Methods Mol. Biol. 2006; 313:33–39. [DOI] [PubMed] [Google Scholar]
- 38. Sanchirico M., Tzellas A., Fox T.D., Conrad-Webb H., Periman P.S., Mason T.L.. Relocation of the unusual VAR1 gene from the mitochondrion to the nucleus. Biochem. Cell Biol. 1995; 73:987–995. [DOI] [PubMed] [Google Scholar]
- 39. Zamudio-Ochoa A., Camacho-Villasana Y., Garcia-Guerrero A.E., Perez-Martinez X.. The Pet309 pentatricopeptide repeat motifs mediate efficient binding to the mitochondrial COX1 transcript in yeast. RNA Biol. 2014; 11:953–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perez-Martinez X., Broadley S.A., Fox T.D.. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003; 22:5951–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fontanesi F. Mechanisms of mitochondrial translational regulation. IUBMB Life. 2013; 65:397–408. [DOI] [PubMed] [Google Scholar]
- 42. Fontanesi F., Clemente P., Barrientos A.. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2011; 286:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fontanesi F., Soto I.C., Horn D., Barrientos A.. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell. Biol. 2010; 30:245–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perez-Martinez X., Butler C.A., Shingu-Vazquez M., Fox T.D.. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol. Biol. Cell. 2009; 20:4371–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zambrano A., Fontanesi F., Solans A., de Oliveira R.L., Fox T.D., Tzagoloff A., Barrientos A.. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2007; 18:523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soto I.C., Fontanesi F., Myers R.S., Hamel P., Barrientos A.. A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab. 2012; 16:801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mick D.U., Vukotic M., Piechura H., Meyer H.E., Warscheid B., Deckers M., Rehling P.. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 2010; 191:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Markov D.A., Savkina M., Anikin M., Del Campo M., Ecker K., Lambowitz A.M., De Gnore J.P., McAllister W.T.. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast. 2009; 26:423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Manthey G.M., McEwen J.E.. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995; 14:4031–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T.. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013; 8:e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lipinski K.A., Puchta O., Surendranath V., Kudla M., Golik P.. Revisiting the yeast PPR proteins–application of an Iterative Hidden Markov Model algorithm reveals new members of the rapidly evolving family. Mol. Biol. Evol. 2011; 28:2935–2948. [DOI] [PubMed] [Google Scholar]
- 52. Prikryl J., Rojas M., Schuster G., Barkan A.. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sc.i U.S.A. 2011; 108:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I.. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012; 8:e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H.W. et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013; 504:168–171. [DOI] [PubMed] [Google Scholar]
- 55. Shen C., Zhang D., Guan Z., Liu Y., Yang Z., Yang Y., Wang X., Wang Q., Zhang Q., Fan S. et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016; 7:11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Halls C., Mohr S., Del Campo M., Yang Q., Jankowsky E., Lambowitz A.M.. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 2007; 365:835–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Marintchev A. Roles of helicases in translation initiation: a mechanistic view. Biochim. Biophys. Acta. 2013; 1829:799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jarmoskaite I., Russell R.. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip. Rev. RNA. 2011; 2:135–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J.J., Seedorf M., Cole C.N., Krebber H.. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007; 315:646–649. [DOI] [PubMed] [Google Scholar]
- 61. Krause K., Lopes de Souza R., Roberts D.G., Dieckmann C.L.. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell. 2004; 15:2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F. et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Spahr H., Rozanska A., Li X., Atanassov I., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., Rackham O., Larsson N.G.. SLIRP stabilizes LRPPRC via an RRM-PPR protein interface. Nucleic Acids Res. 2016; 44:6868–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sasarman F., Brunel-Guitton C., Antonicka H., Wai T., Shoubridge E.A.. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010; 21:1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chujo T., Ohira T., Sakaguchi Y., Goshima N., Nomura N., Nagao A., Suzuki T.. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012; 40:8033–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Camara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K. et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012; 31:443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lagouge M., Mourier A., Lee H.J., Spahr H., Wai T., Kukat C., Silva Ramos E., Motori E., Busch J.D., Siira S. et al. SLIRP regulates the rate of mitochondrial protein synthesis and protects LRPPRC from degradation. PLoS Genet. 2015; 11:e1005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stribinskis V., Gao G.J., Ellis S.R., Martin N.C.. Rpm2, the protein subunit of mitochondrial RNase P in Saccharomyces cerevisiae, also has a role in the translation of mitochondrially encoded subunits of cytochrome c oxidase. Genetics. 2001; 158:573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tovchigrechko A., Vakser I.A.. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006; 34:W310–W314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mallam A.L., Sidote D.J., Lambowitz A.M.. Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. Elife. 2014; 3:e04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008; 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.