Abstract
The poly-A specific ribonuclease (PARN), initially characterized for its role in mRNA catabolism, supports the processing of different types of non-coding RNAs including telomerase RNA. Mutations in PARN are linked to dyskeratosis congenita and pulmonary fibrosis. Here, we show that PARN is part of the enzymatic machinery that matures the human 18S ribosomal RNA (rRNA). Consistent with its nucleolar steady-state localization, PARN is required for 40S ribosomal subunit production and co-purifies with 40S subunit precursors. Depletion of PARN or expression of a catalytically-compromised PARN mutant results in accumulation of 3΄ extended 18S rRNA precursors. Analysis of these processing intermediates reveals a defect in 3΄ to 5΄ trimming of the internal transcribed spacer 1 (ITS1) region, subsequent to endonucleolytic cleavage at site E. Consistent with a function of PARN in exonucleolytic trimming of 18S-E pre-rRNA, recombinant PARN can process the corresponding ITS1 RNA fragment in vitro. Trimming of 18S-E pre-rRNA by PARN occurs in the nucleus, upstream of the final endonucleolytic cleavage by the endonuclease NOB1 in the cytoplasm. These results identify PARN as a new component of the ribosome biogenesis machinery in human cells. Defects in ribosome biogenesis could therefore underlie the pathologies linked to mutations in PARN.
INTRODUCTION
The detailed study of the different pathways leading to the production of the two human ribosomal subunits has attracted increasing interest over the past decade, in large part due to the discovery of a growing number of human disorders in which ribosome production is altered. These so-called ribosomopathies can be caused by haploinsufficiency of genes encoding ribosomal proteins (RPs). Hence, mutations in several RP genes have been identified so far in Diamond Blackfan anemia, while 5q− syndrome and isolated congenital asplenia are due to mutations in RPS14 and RPSA, respectively (1,2). Furthermore, mutations in several genes encoding subunits of RNA polymerase I and III alter rDNA transcription and cause Treacher-Collins syndrome (3). Likewise, mutation of ribosome biogenesis factors (RBFs) involved in various steps of this process can lead to disease by impairing ribosome production, as exemplified by the mutations in SBDS causing Shwachman-Diamond syndrome (4).
Despite considerable progress, a molecular mechanistic understanding of the ribosomal RNA maturation pathways in human cells is still missing, including the identification of all the ribonucleases involved in production of the different RNA processing intermediates. Three of the four ribosomal RNAs (rRNAs) derive from a common precursor called 47S pre-rRNA. It contains the 18S, 5.8S and 28S rRNA sequences, which are flanked by RNA segments that are gradually removed by the sequential action of several endo- and exonucleases (Supplementary Figure S1) (5,6). Interestingly, almost all endonucleolytic cleavages in mammalian pre-rRNAs are followed by subsequent exonucleolytic processing. XRN2 is the only 5΄-3΄ exonuclease implicated in the human rRNA processing pathway to date (7–9). In contrast, 3΄-5΄ exonucleases appear more diverse: in addition to the two catalytic subunits of the nuclear exosome, DIS3 and RRP6 (8,10,11), the proteins ERI1/3’hExo (12) and ISG20L2 (13) have been implicated in formation of the 3΄ end of mammalian 5.8S rRNA. Noticeably, the action of several 3΄-5΄ exoRNases in the 18S rRNA processing pathway of human cells adds to a growing list of evolutionary divergences of ribosome biogenesis between mammals and fungi (6,11).
Production of the two ribosomal subunits in human follows separate pathways after cleavage of the pre-rRNA within the internal transcribed spacer 1 (ITS1) at site 2 (Supplementary Figure S1). The resulting precursors to the 40S ribosomal subunit bear an ITS1 extension of around 950 nt following the 3΄ end of the 18S rRNA that is gradually removed in the course of 40S subunit maturation (14,15). This ITS1 extension is first processed in the nucleolus by exonucleolytic digestion up to the boundary of a highly conserved domain within ITS1 (8,9). It then undergoes an endonucleolytic cleavage at site E, yielding the 18S-E pre-rRNA, which bears a 78 or 81 nucleotide extension at the 3΄ end of 18S rRNA (8,16). Concomitant with nucleolar release, nucleoplasmic maturation and nuclear export of pre-40S particles, exonucleolytic trimming of this ITS1 trailer starts in the nucleus and continues in the cytoplasm (8). Finally, NOB1, the homolog of the yeast endonuclease Nob1p (17,18), generates the mature 3΄ end of the 18S rRNA in the cytoplasm (8,9). This combined action of endo- and exoRNases differs compared to 18S rRNA synthesis in baker's yeast, in which ITS1 is cleaved by two successive endonucleolytic cleavages at site A2 and D (6).
Here, we identify the poly(A) ribonuclease (PARN) as the enzyme ensuring 3΄-5΄ exonucleolytic processing of the 18S-E pre-rRNA. PARN owes its name to its deadenylase activity and has been extensively studied for its role in poly(A) tail shortening in the context of mRNA turnover (19,20). However, the functional repertoire of PARN has meanwhile been extended to the degradation and maturation of different types on non-coding RNAs, including scaRNAs and box H/ACA snoRNAs (21), the human telomerase RNA component (22–24), miRNAs (25,26) and piRNAs (27,28). Since we had previously identified PARN as a component of human pre-40S particles (29), we set out to test whether PARN assists 40S ribosomal subunit biogenesis. Now, we demonstrate that PARN functions in the nuclear 3΄-5΄ trimming of 18S-E pre-rRNA after cleavage at site E, revealing an unexpected role for PARN in processing a highly GC-rich RNA substrate. PARN-mediated exonucleolytic shortening of 18S-E pre-rRNA occurs in the nucleus and promotes the subsequent final 3΄ end maturation of 18S rRNA by the endonuclease NOB1 in the cytosol. Together with the nucleolar localization of PARN, our data suggest that nuclear pre-rRNA processing is one of the major functions of this 3΄-5΄ exonuclease. Since mutations in PARN were recently associated to impaired telomere maintenance in patients suffering from dyskeratosis congenita (30–32) and familial pulmonary fibrosis (33), PARN emerges as a new protein potentially bridging telomere maintenance and ribosome biogenesis in congenital diseases.
MATERIALS AND METHODS
Expression plasmids and purification of recombinant enzymes
Human PARN was cloned in the eukaryotic expression vector pcDNA5/FRT/TO-HASt. Four adjacent amino acids in the PARN sequence (I113, D114, F115, L116) were silently mutated with the Q5 site-directed mutagenesis kit (New England Biolabs) in order to abolish siRNA PARN-1 activity (PARN-1_siMut primer pair; Supplementary Table S1). This sequence was further mutated to generate a catalytically-dead form of PARN (H337A) (PARN-H377A primer pair). Plasmids were purified with endotoxin-free kits (Qiagen).
His-tagged versions of wild-type PARN and its catalytic mutant (H377A) were cloned in the prokaryotic expression vector pET28b(+) and expressed at 25°C in Escherichia coli Rosetta cells (Novagen) induced with 1 mM IPTG. Cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 3 mM MgCl2, 5% (v/v) glycerol; 35 ml/1.5 l culture) and lysed by sonication. The lysate, ultra-centrifuged at 55 000 rpm for 1 h with a Ti70 rotor, was then supplemented with 20 mM imidazole and 2 mM β-mercaptoethanol and incubated at 4°C for 1 h with Ni-NTA agarose beads (Qiagen). After washing the beads with lysis buffer containing imidazole and β-mercaptoethanol, the bound proteins were eluted with 400 mM imidazole in lysis buffer (0.5 ml fractions). The purified proteins were rebuffered to 20 mM Tris-HCl, pH 7.8, 25 mM KCl, 5 mM MgCl2, 5% glycerol and loaded onto a HiTrap Q HP ion exchange column (GE Healthcare), equilibrated with ion exchange buffer (IEB; 20 mM Tris-HCl, pH 7.8, 25 mM KCl, 5 mM MgCl2, 5% (v/v) glycerol). The column was washed with IEB containing 50 mM KCl, prior to protein elution with IEB containing 300 mM KCl.
In vitro processing assays
The DNA template for in vitro transcription was generated by polymerase chain reaction (PCR) using the ‘MHR 11.9’ cosmid containing the human rDNA unit as a template, with the forward primer OHA416 carrying the T7 promoter followed by a region homologous to helix 45 of the 18S rRNA and the reverse primer OHA418 bearing a region of complementary to the ITS1 sequence up to position +78 immediately followed by a BtsI restriction site. PCR reactions were carried out using Phusion HF DNA polymerase (Thermo Scientific) following the recommended guidelines for amplification of GC-rich sequences. PCR fragments were purified from crude PCR reactions by phenol/chloroform extraction and ethanol precipitation, digested by BtsI to cleave the transcription template right after the position corresponding to ITS1+78 and gel purified. In vitro transcription was carried out using T7 RNA polymerase (Promega) in the presence of α32P-rCTP (800 Ci/mmol, 10 mCi/ml; PerkinElmer) following the guidelines for the synthesis of high specific activity RNA probes. DNA templates were then eliminated using RQ1 RNase-free DNase (Promega) and the resulting transcripts were purified by phenol/chloroform extractions and ethanol precipitation in the presence of 0.3 M ammonium acetate, pH 5.2. Radiolabeled RNA substrates were diluted at 5000 cpm/μl in a buffer containing 50 mM HEPES-KOH, pH 7.5, and 5 mM MgCl2, denatured for 3 min at 95°C in a PCR machine and slowly renatured by ramping down temperature to 30°C at a rate of 0.1°C/s. RNAs were then diluted 2-fold with 30°C-prewarmed 2 × Processing Buffer containing 200 mM NaCl and 0.2 μg/μl BSA (New England Biolabs). Wild-type PARN or the H377A catalytic mutant were added to a final protein concentration of 10 mM. Reaction samples corresponding to ∼50 000 cpm were taken at increasing incubation times, and extracted with a phenol/chloroform mixture to stop the reaction. The RNAs were precipitated with ethanol and separated on a 6% denaturing gel (acrylamide:bisacrylamide (19:1); 8M urea, 1 × TBE (Tris 89 mM, boric acid 89 mM, EDTA 2 mM) buffer). After migration, the gel was dried, exposed to a PhosphorImager screen and radioactive signals were revealed using a Typhoon Trio PhosphorImager (GE Healthcare).
Cell culture and treatments with inhibitors or siRNA duplexes
HeLa and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1 mM sodium pyruvate. HeLa K cells inducibly expressing RPS2-YFP have been described previously (34). HEK293 FlpIn TRex cell lines expressing HASt-GFP, ENP1-StHA, HASt-DIM2, HASt-LTV1 and RIO1 (D324A)-StHA have been described previously (29). Polyclonal HEK293 FlpIn TRex cell lines expressing HASt-PARN and HASt-PARN-H377A were generated as described previously (29).
Inhibition of nucleo-cytoplasmic transport was performed for 2 h with 20 nM leptomycin B (LC laboratories), and RNA polymerase I abortive transcripts were generated in the presence of 50 ng/ml actinomycin D (Sigma). To knock down expression of the corresponding human protein genes, each siRNA duplex (Eurogentec) (listed in Supplementary Table S2) was added at a final concentration of 500 nM to 200 μl of cell suspension (50 × 106 cells/ml diluted in Na phosphate buffer, pH 7.25, containing 250 mM sucrose and 1 mM MgCl2). After electro-transformation at 240 V (35), cells were plated and collected 48 h later. Alternatively, cells were electro-transformed a second time after 48 h and collected 2 days later for the following genes: PARN (siRNA PARN-1 was used unless specified; siRNA PARN-6); NOB1 (siRNAs NOB1-1Q and NOB1-2Q); PAPD5 (siRNAs PAPD5-5 and PAPD5-6); PAPD7 (siRNAs PAPD7-3 and PAPD7-5). Control cells were electro-transformed with a scramble siRNA. Knockdown efficiency of siRNAs was assessed by quantitative PCR, using GAPDH as an internal control (36). Rescue experiments were performed at day 4 with Jetprime transfection reagent (Polyplus) at a 3:1 ratio, using the empty vector (pcDNA5-HASt) as a negative transfection control. Cells were harvested 30 h post-transfection.
Pulse chase analysis
HeLa cells were electro-transformed twice with siRNAs as described above and grown in 12-well plates. Cells were incubated in methionine-free DMEM (Invitrogen) for 30 min at 37°C, labeled for 15 min with L-methyl 3H methionine (50 mCi/ml) and rinsed twice with DMEM containing unlabeled methionine. They were then incubated for 0 min (i.e. immediately rinsed with ice-cold phosphate buffered saline (PBS)), 30 min, 1 h, 1 h 30 min, 2 h or 2 h 30 min in DMEM containing methionine. After the corresponding chase time, cells were rinsed twice with ice-cold PBS and immediately lysed with 1 ml Trizol reagent (Invitrogen). Total RNAs were extracted with Tri reagent as described below and their concentration was determined with a Qubit fluorometer (Thermo Fisher Scientific). RNA samples were separated on a 1.2% agarose gel and passively transferred to a nylon membrane as described below. The membrane was exposed for 4–6 days to Biomax KODAK TM MS films through a KODAK TM BioMax TranScreen LE.
Cell fractionation and analysis of ribosomes by sucrose density gradient centrifugation
Cells were detached by trypsin treatment, washed and mechanically disrupted with a Dounce homogenizer as described (36). After centrifugation at 1000 g for 10 min at 4°C, the supernatant (cytoplasmic fraction) was recovered and the nuclei-containing pellet was washed and purified by centrifugation on a sucrose cushion. For nuclear RNA analysis, the pellet was resuspended in Tri reagent and RNAs were extracted. Alternatively, the pellet of nuclei was diluted in buffer B (25 mM Tris-HCl, pH 7.5, 100 mM KCl, 1 mM NaF, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.05% NP-40, 1 mM dithiothreitol (DTT), 40 μg/ml phenylmethylsulfonyl fluoride (PMSF)) and sonicated for 2 min with a Bioruptor (Diagenode) (37). The lysate was centrifuged and the protein content of the supernatant was estimated by Bradford assay. The nuclear fractions thus obtained were separated by centrifugation with an SW41 rotor (Optima L100XP ultracentrifuge; Beckman Coulter) at 38 000 rpm and 4°C during 200 min on 10–30% sucrose gradients containing 10 mM Tris-HCl, pH 7.5, 100 mM KCl, 2 mM EDTA, 1 mM DTT. For polysome analyses, HeLa cells were treated with 100 μg/ml cycloheximide (Sigma-Aldrich) for 10 min. The cytoplasmic fractions were prepared on ice as described above, except that cycloheximide was added to all buffers (36). A sample volume containing 1 mg of total proteins was loaded on a 10–50% (wt/wt) sucrose gradient and the tubes were centrifuged at 36 000 rpm for 105 min. The gradient fractions were collected at OD254 nm with a Foxy Jr. gradient collector (Teledyne Isco). For RNA isolation from sucrose fractions, the samples were first centrifuged on Vivaspin 500 concentrators (Sartotrius Stedim Biotech) before RNA extraction with Tri reagent.
Isolation of total RNAs and characterization of the 3΄ extremity of 18S-E pre-rRNAs
RNAs were extracted from cells with Tri reagent, followed by phenol/chloroform extraction of the aqueous phase and isopropanol precipitation. Quantifications were performed at 260 nm with a Nanodrop spectrophotometer. The 3΄ extremities of 18S-E pre-rRNAs were analyzed by rapid amplification of cDNA 3΄ ends (3΄RACE) (8,38). Primer ITS1-Hs-RACE, which spans the 18S-ITS1 junction up to nt 13 in the ITS1, was used for PCR amplification. The amplified fragments were sub-cloned, automatically sequenced and aligned with Jalview software (8). For RNase H cleavage assays, 4 μg total RNAs were denatured at 95°C for 3 min with a reverse probe hybridizing in the 3΄ end of 18S rRNA (primers RNaseH_1 or RNaseH_2; 1 μl at 100 μM) (39). After annealing by cooling down to room temperature for 10 min, the reaction mixture was diluted to 30 μl with a reaction mix containing 1 × RNase H reaction buffer, 65 μM DTT, 0.5 U/μl RNasin (Promega) and 50 U RNase H (New England Biolabs), and incubated at 37°C for 30 min. The reaction was then blocked by addition of 0.3 M sodium acetate, pH 5.2 and 0.2 mM EDTA, and the RNAs were recovered by ethanol precipitation after phenol extraction.
RNA analysis by northern blot
In order to analyze the precursors to the 28S and 18S rRNAs, total RNA samples (3 μg/lane) were dissolved in formamide and separated on a 1.2% agarose gel containing 1.2% formaldehyde and 1 × Tri/Tri buffer (30 mM triethanolamine, 30 mM tricine, pH 7.9). Smaller RNAs were either separated on a 6% (snoRNAs) or 12% (RNase H assays) polyacrylamide gel (19:1) in 1 × TBE buffer containing 7 M urea. RNAs were transferred to Hybond N+ nylon membrane (GE Healthcare, Orsay, France) and cross-linked under UV light. Membrane hybridization with radiolabeled oligonucleotide probes was performed as described (8). Signals were acquired with a Typhoon Trio PhosphorImager and quantified using the MultiGauge software. The sequences of all the probes used in the present study are listed in Supplementary Table S3.
Western blots
Whole-cell protein extracts were prepared with ice-cold cell lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1% Triton X-100, 200 mM NaCl, 1 × Complete EDTA-free protease inhibitor cocktail (Roche Diagnostics). For protein extraction from sucrose gradients, the sucrose was removed from the fractions with Vivaspin 500 concentrators. The protein concentration was determined using a Bradford assay kit (BioRad). Protein extracts (17 μg) were separated using a 4–12% gradient Bis-Tris gel (Life Technologies), transferred onto a nitrocellulose membrane (GE Healthcare) and incubated with primary antibodies (see below), before incubation with the appropriate secondary antibodies. Immuno-detection was performed using a home-made Luminol solution.
Antibodies
The anti-PARN antibody for western blot analysis was raised against purified, recombinant His-PARN and affinity purified with the antigen coupled to SulfoLink beads (Thermo Fisher Scientific). The anti-C21orf70 antibody was raised against a C-terminal peptide (C-GQTLARQMQLEDGGQL) and purified as described above. The antibody against RPL23a has been raised as previously described (40). The antibodies against DIM2, ENP1, NOB1, RPS3 (34), NOC4L, RRP12, RPS19 (29) and TSR1 (41) have been described previously. Anti-PARN for immunofluorescence was purchased from Abcam (ab89831), anti-β-actin and anti-α-tubulin from Sigma Aldrich (A1987 and T5168), anti-CK1δ and anti-HNRNPC from Abcam (AF12G4 and ab10294), anti-CK1ε (610445), anti-HA from Covance (MMS-101P) and anti-RPL10 from Santa Cruz Biotechnologies, Inc. (sc-798). Secondary antibodies for immunofluorescence were purchased from Invitrogen.
Tandem affinity purification
Preparation of cell extracts and tandem affinity purification was performed as described previously (29). In short, expression of the bait protein in stable HEK293 cells was induced for 24 h with tetracycline before cells were detached with PBS containing 0.5 mM EDTA and harvested by centrifugation. Cells were lysed using a Dounce homogenizer in 10 mM Tris-HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.5% NP-40 (Fluka) containing protease and phosphatase inhibitors. Lysates were cleared by centrifugation (4500 g, 12 min, 4°C) and incubated with StrepTactin beads (IBA) for 30 min at 4°C while rotating. Beads were washed three times with tandem affinity purification (TAP) buffer (10 mM Tris-HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2, protease and phosphatase inhibitors) and eluted three times with 300 μl TAP buffer containing 2.5 mM d-desthiobiotin (Sigma). Eluates were then incubated with HA-agarose (Sigma) for 1 h at 4°C while rotating. After washing twice with TAP buffer and once with 50 mM Tris pH 7.5, 2 mM MgCl2, bound proteins were eluted with 50 μl 2× sodium dodecyl sulphate (SDS) sample buffer without DTT in Mobicol spin columns (MoBiTec). Before further analysis, samples were supplemented with 50 mM DTT.
Immunofluorescence
For immunofluorescence analysis, cells were fixed in PBS containing 4% paraformaldehyde and permeabilized in 0.1% Triton X-100, 0.02% SDS in PBS for 5 min. All subsequent steps were performed as described previously (34). Observations were made with an inverted microscope (IX-81; Olympus) equipped with an ORCA Flash 4.0 camera (Hamamatsu) or with a Leica SP2 AOBS confocal scanning microscope.
RESULTS
PARN is associated with early pre-40S particles
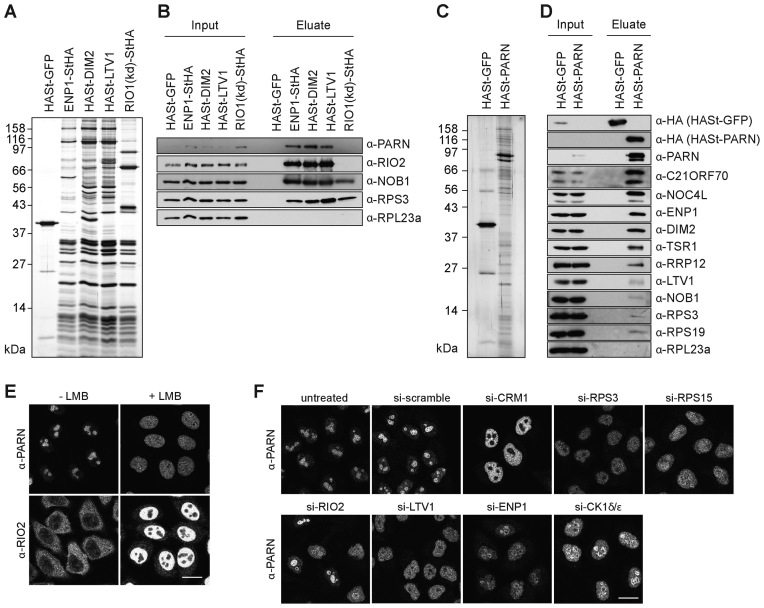
PARN has previously been co-purified with pre-40S particles isolated by TAP of the nuclear 40S ribosomal subunit biogenesis factor NOC4 (29). In order to better define which pre-ribosomal particles contain PARN, we used several RBFs as baits to purify 40S subunit precursors at various maturation stages (Figure 1A). In agreement with our previous results, PARN was co-isolated with tagged ENP1, DIM2 or LTV1, which associate with both nuclear and cytoplasmic pre-40S particles, but not with a tagged, kinase-dead derivative of RIO1 (RIO1(kd)), which is a cytoplasmic RBF (Figure 1B) (41,42). Conversely, TAP of tagged PARN enriched a series of predominantly nuclear RBFs including NOC4, RRP12, ENP1, DIM2 and TSR1 (Figure 1C and D). Notably, LTV1 and especially NOB1, which only join the nuclear pre-40S particles at a late maturation step, were only poorly enriched in these pull-downs. These data support the association of PARN with nuclear precursors to the small ribosomal subunit, consistent with its prominent nucleolar localization (Figure 1E and Supplementary Figure S2A), and raised the possibility that PARN could function as a nuclear, 40S-specific RBF.
Figure 1.
Characterization of PARN association with pre-40S particles and PARN localization. (A) TAPs from HEK293 cells expressing HASt-GFP (negative control) or the tagged 40S-specific RBFs ENP1-StHA, HASt-DIM2, HASt-LTV1 and RIO1(kd)-StHA. Eluates were analyzed by SDS-PAGE and silver staining. (B) Western blot analysis of the TAP experiment in (A) using the indicated antibodies against PARN, RPs and the 40S RBFs RIO2 and NOB1. (C) TAP from HEK293 cells expressing HASt-GFP (negative control) or HASt-PARN. (D) Western blot analysis of the TAP experiment in (C) using the indicated antibodies against the bait (α-HA), PARN, RPs and various 40S RBFs. (E) IF analysis of PARN and RIO2 upon inhibition of CRM1-dependent nucleocytoplasmic transport by leptomycin B (20 nM; 2 h). Relocalization of the shuttling RBF RIO2 from the cytoplasm to the nucleus served as positive control. Scale bar, 20 μm. (F) IF analysis of PARN after depletion of RPSs, the export factor CRM1 and 40S RBFs (RIO2, LTV1, ENP1, CK1δ/ε). Scale bar: 20 μm.
A number of nucleolar 40S-specific RBFs, including ENP1, DIM2, RRP12, LTV1 and TSR1, accompany pre-40S particles from the nucleolus via the nucleoplasm to the cytosol (34,41,43). Like shuttling nucleolar RBFs, PARN redistributed from the nucleolus to the nucleoplasm upon inhibition of pre-40S particle export by leptomycin B (Figure 1E). A similar phenotype was observed upon knockdown of RPS15 or CRM1 with siRNAs (Figure 1F), which is known to restrain pre-40S particles in the nucleus (43,44). To address whether PARN would accompany pre-40S particles into the cytoplasm, we depleted additional RPs and several RBFs. Knocking down RBFs involved in the large subunit pathway (LSG1 and eIF6) had no effect on PARN localization (Supplementary Figure S2B). In contrast, depletion of RPS3 or of several RBFs (ENP1, CK1δ/ε, RIO2 or LTV1) implicated in the small subunit pathway (34,36,41,45) led to the relocation of PARN from the nucleoli to the nucleoplasm (Figure 1F and Supplementary Figure S2C), although knockdown of CK1δ/ε, RIO2, LTV1 or RPS3 is known to trigger cytoplasmic 40S subunit maturation defects (34,41,45). Thus, independent of the nature of the pre-40S particle assembly defect, PARN never accumulated in the cytoplasm. These data suggest that PARN is released from pre-40S particles before their nuclear export, although we cannot exclude that PARN is rapidly recycled after pre-40S particle export by an as yet unknown pathway.
PARN is required for exonucleolytic processing of 18S-E pre-rRNA
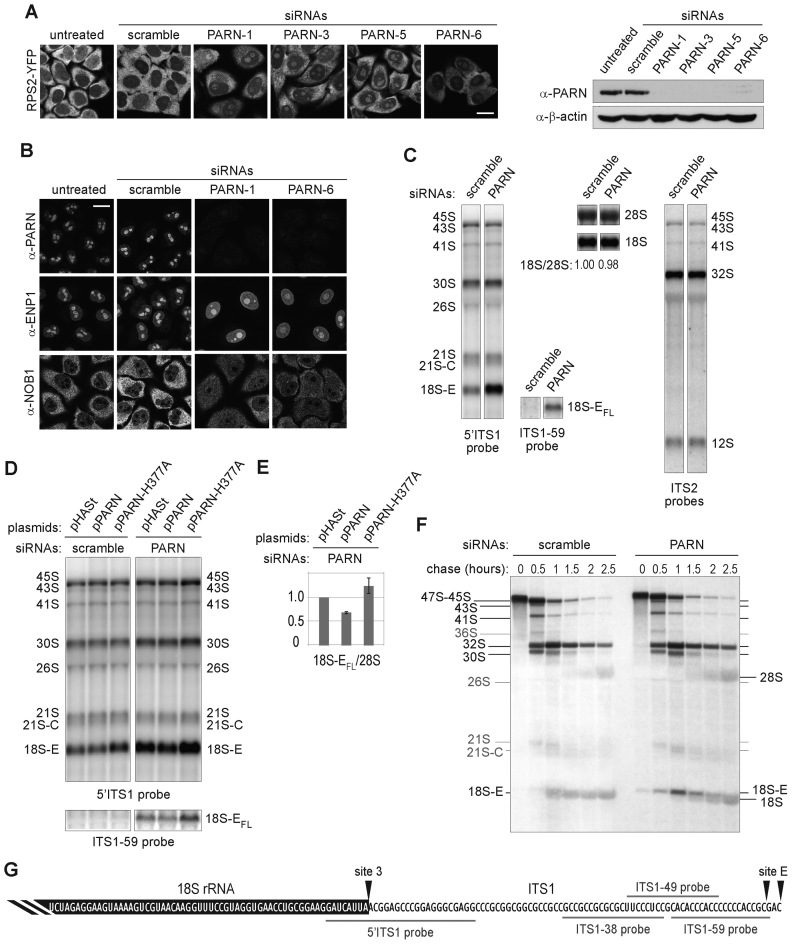
To investigate the potential role of PARN in 40S subunit biogenesis, we analyzed how its depletion affected the tetracycline-inducible 40S subunit biogenesis reporter RPS2-YFP (34). Knockdown of PARN with four different siRNAs induced nucle(ol)ar accumulation of the reporter, indicative of 40S subunit biogenesis defects (Figure 2A). Further, PARN depletion led to changes in the steady-state localization of the pre-40S particle RBFs ENP1 and NOB1, inducing nucleoplasmic accumulation of ENP1 and also an increase in the nucleoplasmic levels of NOB1, associated with a striking decrease in its cytoplasmic levels (Figure 2B). These results suggest that PARN may promote nuclear steps of pre-40S particle biogenesis.
Figure 2.
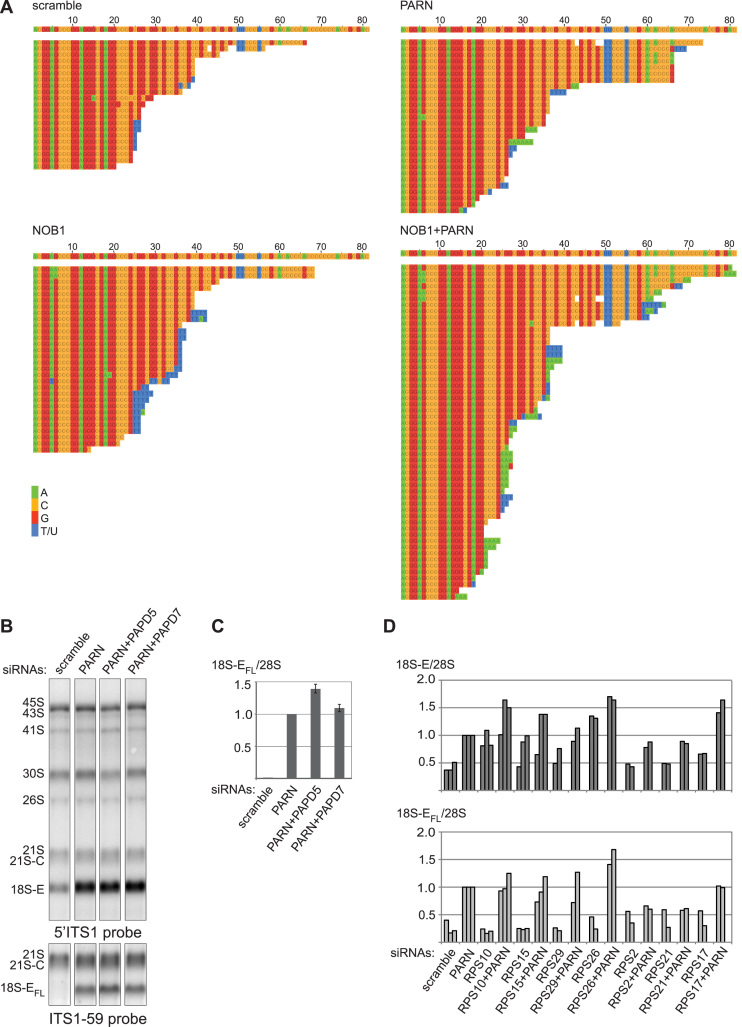
Impact of PARN depletion on the processing of pre-rRNAs. (A) Subcellular localization of the 40S reporter RPS2-YFP after depletion of PARN with four different siRNAs and western blot of the corresponding samples using the indicated antibodies. (B) IF analysis of PARN and the 40S RBFs ENP1 and NOB1 after depletion of PARN with two siRNAs. Scale bar, 20 μm. (C) Total RNAs (3 μg/lane) extracted from HeLa cells treated either with an siRNA without target (scramble), or with siRNA PARN-1 were analyzed by northern blot. The nylon membrane was hybridized with the 5΄ITS1 probe revealing all the precursors to the 18S rRNA, the ITS1-59 probe recognizing the full-length forms of 18S-E pre-rRNA generated after cleavage at site E (18S-EFL pre-rRNAs), a mixture of ITS2 probes revealing the precursors to the 5.8S and 28S rRNAs or 18S and 28S probes. (D) HeLa cells treated with a scramble siRNA or with the siRNA PARN-1 were rescued with plasmids bearing either a HASt-tagged version of PARN silently mutated to render it insensitive to siRNA PARN-1 (pPARN), or a catalytically inactive PARN mutant (pPARN-H377A). The empty plasmid (pHASt) was used as a control. Total RNA extracts were analyzed by northern blot and revealed with the 5΄ITS1 probe. Western blot analysis of PARN expression is displayed in Supplementary Figure S3. (E) 18S-EFL precursors revealed with the ITS1-59 probe were quantified relative to 28S rRNA (average +/− standard deviation for three independent rescue experiments). (F) Neo-synthesized RNAs from HeLa cells transfected for 96 h with the corresponding siRNAs were pulse-labeled with L-[methyl-3H]-methionine. Cells were harvested after the indicated chase times. Total RNAs extracted from these cells were separated by electrophoresis, transferred to a nylon membrane and labeled RNAs were detected by autoradiography. (G) Schematic representation of the 3΄ end of 18S-EFL pre-rRNA. The 3΄ terminus of the 18S rRNA is highlighted in black. It is followed by 81 nt of the ITS1, representing the 3΄ end of the longest form of 18S-E pre-rRNA released after cleavage at site E (18S-EFL pre-rRNAs), while the alternative cleavage site releases an 18S-E pre-rRNA bearing a 78 nt extension in 3΄. The positions of the ITS1 probes used for northern blot experiments are shown in gray.
Since PARN is an exonuclease, we next assessed its potential role in pre-rRNA processing by loss-of-function experiments (Figure 2C). Knockdown of PARN provoked a marked accumulation of 18S-E pre-RNAs, as analyzed by northern blotting, while earlier precursors seemed unaffected (Figure 2C). 18S-E pre-RNAs are produced by cleavage at site E and initially bear a 3΄ ITS1 segment of 78 or 81 nt in length, which is then subjected to exonucleolytic trimming in the nucleus and cytoplasm (Supplementary Figure S1) (8). A probe hybridizing to nt 59–78 of ITS1 (probe ITS1-59, schematized on Figure 2G) revealed a substantial accumulation of full length 18S-E (18S-EFL) pre-rRNAs in PARN-depleted cells (Figure 2 C and D). In contrast, these species were very sparse in cells treated with a scramble siRNA, reflecting their rapid processing into shorter 18S-E precursors. The 18S-E pre-rRNA processing defect observed in PARN-depleted cells was partially rescued by exogenous expression of PARN, which decreased the level of 18S-EFL pre-rRNAs by ∼30% when compared to cells transfected with an empty vector (Figure 2D, right panels and Figure 2E). On the contrary, expression of a catalytically inactive PARN-H377A mutant aggravated the accumulation of 18S-EFL pre-rRNAs relative to control cells. Histidine 377 is required in the active site for orientation and activation of the nucleophilic water (46). The sole over-expression of PARN-H377A also induced accumulation of 18S-E pre-rRNAs (Figure 2D, left panel), albeit to a lesser extent than PARN depletion. We did not detect increased levels of 18S-EFL pre-rRNA in this case. We hypothesize that the mutant does not fully compete with endogenous PARN, which still allows partial digestion of the 3΄ end of 18S-EFL pre-rRNA. Consistent with a dominant-negative effect of the PARN mutant, TAP experiments performed with the tagged PARN-H377A mutant yielded similar protein profiles as wild-type PARN, indicating that PARN associates with pre-40S particles independently of its catalytic activity (Supplementary Figure S2D and E). Altogether, these results demonstrate that the exonucleolytic activity of PARN is required for efficient processing of 18S-E pre-rRNAs into 18S rRNA.
To further substantiate a role of PARN in 18S rRNA production, we performed a pulse-chase analysis of rRNA synthesis with L-[methyl-3H]-methionine (Figure 2F). Compared to control cells, PARN depletion induced a marked delay in 18S rRNA formation, while 28S rRNA accumulation appeared unaffected. After 1 h of chase in control cells, the 18S and 18S-E species appeared as one thick band. This pattern was only seen after 2 h of chase in PARN-depleted cells. In addition, PARN-depleted cells displayed the marked and transient accumulation of a precursor migrating above the 18S rRNA, which was barely observed in control cells. This band was clearly different from the 18S-E pre-rRNA accumulating in NOB1-depleted cells (Supplementary Figure S4). In that case, the 18S-E pre-rRNA was difficult to distinguish from the 18S rRNA under similar electrophoresis conditions, as expected from the accumulation of short forms of 18S-E pre-rRNA (see Figure 5C). These data show that PARN depletion strongly delays 18S rRNA production and enforce the conclusion that PARN is required for efficient processing of the long forms of 18S-E pre-rRNA.
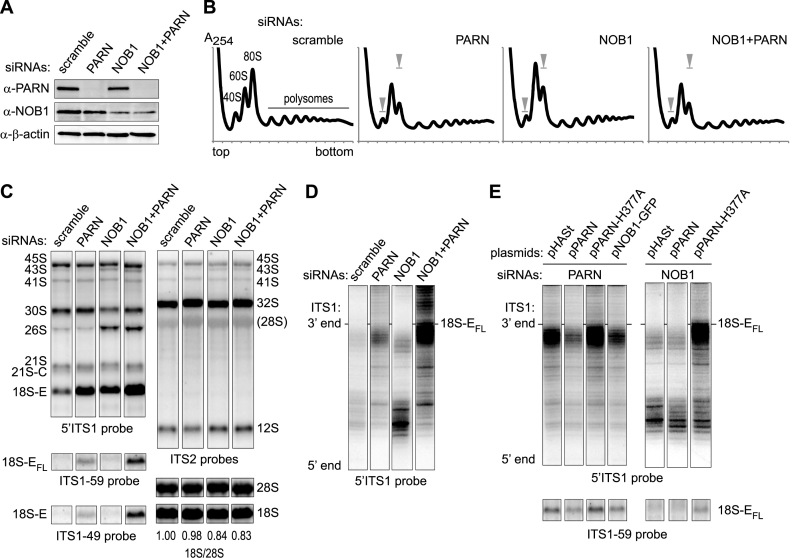
Figure 5.
Effects of the co-depletion of NOB1 and PARN on ITS1 trimming. (A) Western blot analysis of proteins from HeLa cells treated for 96 h with the indicated siRNAs: scramble, PARN-1 or/and a mixture of NOB1-3, NOB-1Q and NOB-2Q. (B) Cytoplasmic fractions from the corresponding samples were analyzed on 10–50% sucrose gradients. The summits of the 40S and 80S peaks in the control are symbolized by gray arrow heads in the other samples. (C) Total RNAs (3 μg/lane) from HeLa cells treated with the indicated siRNAs were analyzed by northern blot and hybridized with the indicated probes. (D and E) RNase H assays were performed on 4 μg total RNAs. After separation on a 12% polyacrylamide gel containing urea, 18S-E precursors were evidenced with a 5΄ITS1 probe. (D) Analysis of 18S-E pre-rRNA 3΄ ends in RNA samples from cells treated with the same siRNAs as in (B) and (C). (E) RNase H assays were conducted on RNAs extracted from PARN-depleted cells after over-expression of exogenous PARN constructs as described in Figure 2D, or of a GFP-tagged version of NOB1 (pNob1-GFP). Similar assays were conducted on NOB1-depleted cells expressing exogenous PARN. The fragment distributions were compared to the global levels of 18S-EFL revealed by the ITS1-59 probe in the corresponding samples.
PARN can process the ITS1 in vitro
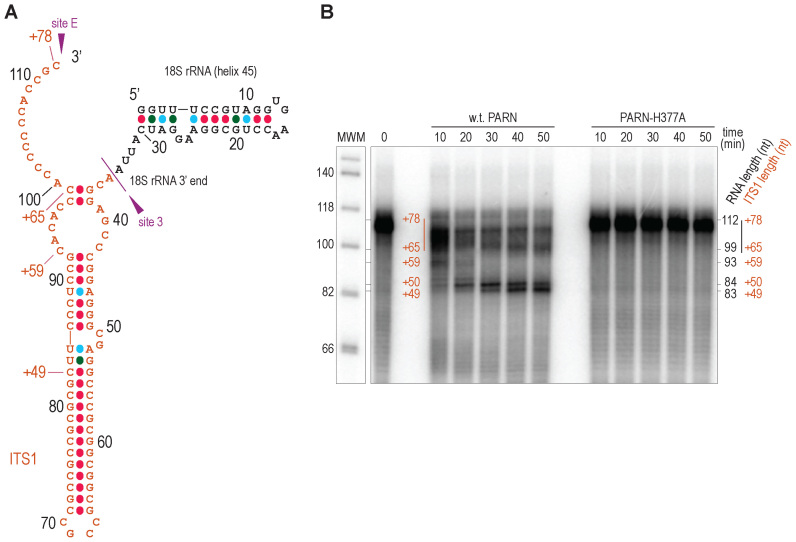
The data presented above prompted us to investigate whether PARN is able to process the highly GC-rich ITS1 segment of 18S-E precursors. PARN has previously been shown to efficiently digest adenosines, but to be less processive on uridines and cytidines, and to display little reactivity towards guanosines (47). To directly monitor PARN's exonuclease activity, we analyzed the in vitro degradation of a synthetic RNA transcript comprising the last helix of human 18S rRNA (H45), followed by the ITS1 segment contained in 18S-EFL (Figure 3A). This substrate was indeed partially trimmed by recombinant human PARN. RNA digestion was not due to contaminating E. coli RNases since it was not observed with the catalytic mutant PARN-H377A purified under the same conditions (Figure 3B). A time-course experiment revealed the gradual processing of the substrate by wild-type PARN, yielding a prominent product of around 83 nt in length, corresponding to a 49-nt ITS1 extension. This coincides with a highly GC-rich RNA segment located in a predicted stem loop (Figure 3A) (48). These data show that PARN has the ability to progress through the ITS1, although its progression is impaired by long GC stretches. Importantly, the lengths of these in vitro digestion intermediates were in good agreement with those observed in 3΄RACE experiments in control cells (8), suggesting a common digestion pattern in vitro and in vivo.
Figure 3.
In vitro processing of ITS1 by recombinant PARN. (A) Predicted secondary structure of the 112-nt long, in vitro-transcribed RNA substrate used in in vitro assays. It comprises helix 45 of 18S rRNA, followed by the ITS1 sequence up to the distal position of site E (nt +78 with respect to the 3΄ end of mature 18S rRNA). The total nucleotide length is indicated with black digits, while the length of the ITS1 sequence is annotated with a "+" sign. (B) The radiolabeled transcript described in (A) was denatured at 95°C, renatured by slowly ramping down temperature to 30°C and then incubated with wild-type PARN or the catalytic H377A mutant. After the indicated incubation times, the reactions were stopped by phenol/chloroform extraction. The RNAs were purified and analyzed on a 12% polyacrylamide gel in denaturing conditions. As an unprocessed control, the in vitro transcript was incubated for 40 min in the same conditions except that PARN was omitted (0). MWM: DNA molecular weight marker. The ITS1 lengths of the different processing intermediates (annotated with a "+" sign) were deduced from the total length of the corresponding RNA molecules inferred from comparison with the molecular weight marker.
PARN is coupled to a poly(A) polymerase for processing of the 18S-E pre-rRNA
Processing of snoRNAs by PARN is primed by the activity of a poly-(A) polymerase, PAPD5, which oligo-adenylates the 3΄ end of the RNA substrate (21). In order to assess whether 18S-E pre-rRNA processing by PARN involves addition of untemplated nucleotides in vivo, the 3΄ end of the 18S-E species was analyzed by 3΄RACE after loss of PARN and/or NOB1, the endonuclease responsible for the ultimate cleavage of 18S-E precursors at site 3 (8,9,42). Knockdown of NOB1 produced short forms of 18S-E pre-rRNAs bearing oligo(U) extensions (Figure 4A), which were shown to be added in the cytoplasm (8). PARN depletion resulted in longer 3΄ ends, some of which displayed short oligo(A) or oligo(U) tails. When PARN and NOB1 were co-depleted, this phenotype was amplified, with a higher abundance of oligo(A) tails. These additional nucleotides were already present immediately after cleavage at site E, as one of the longest fragments ending at nt +78 bore 3 untemplated adenosines at its 3΄ end. These data suggest that PARN activity could be coupled with that of a nuclear poly(A) polymerase, as previously observed in other maturation pathways involving this enzyme (21–23). In support of this hypothesis, co-depletion of PAPD5 with PARN slightly increased 18S-EFL accumulation (Figure 4B and C), although PAPD5 knockdown was only partial (data not shown). In contrast, no effect was seen upon co-depletion of PAPD7 (Figure 4B and C). PAPD5 may thus be a good candidate to stimulate 18S-E pre-rRNA processing by PARN.
Figure 4.
Modification of the 3΄ ends of 18S-E precursors. (A) Alignment of the sequences obtained by 3΄ RACE for 18S-E pre-rRNAs from cells treated with the indicated siRNAs. The first sequence corresponds to that of the ITS1 (Genbank accession number: X17624.1) up to position +81, corresponding to the distal position of site E. Some sequences display short stretches of untemplated nucleotides (T/U: blue; A: green). (B and C) In order to assess the potential role of two nuclear poly(A) polymerases (PAPD5 and PAPD7) for PARN function, total RNAs from HeLa cells treated with the corresponding siRNAs were analyzed by northern blot. (C) Level of 18S-EFL pre-rRNAs relative to 28S rRNA in each sample (average +/− standard deviation from three (PARN, PAPD5) or two (PAPD7) independent experiments). (D) Effects of PARN co-depletion with RPSs essential for 18S-E pre-rRNA maturation were assessed by northern blot and revealed with ITS1 probes (see Supplementary Figure S3 for corresponding northern blots). The levels of 18S-E (5΄ITS1 probe; black) or 18S-EFL pre-rRNAs (ITS1-59 probe; gray) relative to 28S rRNA from two or three independent experiments were represented as individual bars for each siRNA treatment.
PARN is primarily involved in pre-rRNA processing, not in quality control
The data above strongly argue for a role of PARN in 18S-E pre-rRNA processing. However, poly(A) addition was also shown to label misprocessed pre-rRNAs for degradation by the exosome through a surveillance mechanism (49–52). We hypothesized that if PARN were involved in a nuclear quality control mechanism, co-depletion of PARN and RPSs essential for 18S-E rRNA maturation should strongly aggravate the accumulation of 18S-E pre-rRNAs observed upon depletion of each protein alone. Hence, we individually knocked down RPSs required for nuclear export and final processing of the 18S-E pre-rRNA (36,53). Depletion of these RPSs induced a moderate accumulation of 18S-E pre-rRNA, which was lower than in PARN-depleted cells (Figure 4D and Supplementary Figure S5A; 5΄ITS1 probe). Importantly, in most instances, co-depletion of PARN with the RPSs only led to a mild accumulation of 18S-E pre-rRNA, which remained lower than upon knockdown of PARN alone. In addition, knockdown of these RPSs did not promote accumulation of the 18S-EFL species relative to the control (Figure 4D) and co-depleting PARN never resulted in a significant increase of 18S-EFL pre-rRNAs compared to the sole depletion of PARN. The only exception was RPS26, whose co-depletion with PARN accumulated 18S-EFL pre-rRNAs to a level comparable to the cumulated amounts of individual depletions (Figure 4D). Interestingly, the 18S rRNA 3΄-end nests onto RPS26 in the human 40S ribosomal particle (54), which might explain why knockdown of RPS26 combined with a delayed ITS1 trimming induced by PARN depletion is more deleterious for the 3΄ end of 18S rRNA. Although the above results did not plead for PARN involvement in a quality control mechanism, we further investigated this point by inducing earlier processing defects accumulating 30S pre-rRNAs. Compared to RPS11, RPS23 or RPS24 depletion, accumulation of this precursor was not further increased by PARN knockdown (Supplementary Figure S5B). Finally, actinomycin D treatment was used to release abortive RNA polymerase I transcripts, which were shown to be poly-adenylated in mouse cells (51). For this, control cells were compared to cells depleted of PARN or RPS26, or co-depleted of these two proteins (Supplementary Figure S5C). PARN depletion never led to an increased accumulation of abortive transcripts compared to the corresponding controls. Taken together, these data show no evidence that PARN participates in the degradation of 18S rRNA precursors at early, intermediate or late steps of 40S ribosomal subunit production. Therefore, we conclude that the primary function of PARN in ribosome biogenesis is its involvement in pre-rRNA processing.
PARN acts in the nucleus, upstream of NOB1
We then examined the functional relationship between PARN and NOB1. Sucrose gradient analysis showed a marked decrease of free 40S subunits and a concomitant increase of 60S subunits upon PARN depletion, while 80S monosomes and polysomes were less abundant compared to the control (Figure 5A and B). The polysome profiles obtained with cells depleted of PARN were similar to those obtained upon knockdown of NOB1 or knockdown of both NOB1 and PARN, consistent with the two proteins acting sequentially in the pre-40S maturation pathway.
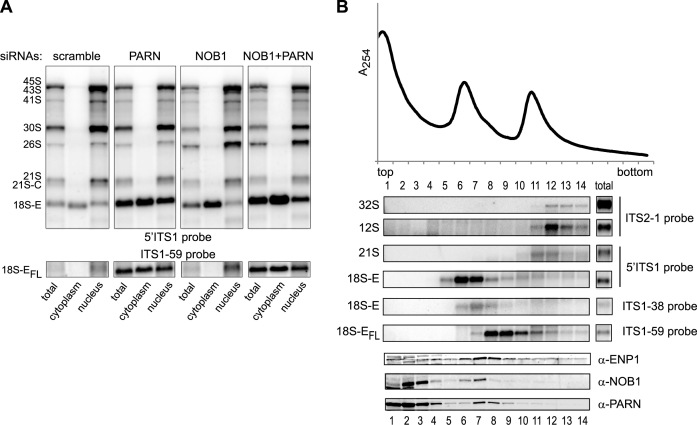
In northern blot analyses, NOB1 knockdown resulted in the accumulation of short forms of 18S-E pre-rRNAs, but not of 18S-EFL (Figure 5C). This was confirmed by pulse-chase experiments performed in NOB1-depleted cells (Supplementary Figure S4; to be compared to scramble and PARN siRNAs). Nevertheless, NOB1 depletion strongly enhanced the accumulation of long forms of 18S-E pre-rRNA in the absence of PARN (Figure 5C). This was further characterized by assessing 18S-E processing intermediates by RNase H assays with a probe hybridizing to the 3΄ domain of the 18S rRNA. In control cells, we observed bands migrating slightly below the 18S-EFL precursors, together with a ladder of much shorter forms (Figure 5D). NOB1 depletion resulted in a strong increase of short 18S-E precursors, consistent with 3΄ RACE analyses (Figure 4A) (8). In contrast, PARN depletion led to a marked accumulation of long precursors migrating just below the 18S-EFL pre-rRNAs. Re-expression of PARN rescued this phenotype to a large extent, while the PARN-H377A mutant aggravated the build-up of long forms of 18S-E (Figure 5E). Importantly, the combined knockdown of PARN and NOB1 resulted in the accumulation of long forms of 18S-E pre-rRNAs but not of short ones (Figure 5D and Supplementary Figure S4). This phenotype was also observed upon over-expression of catalytically-dead PARN-H377A in NOB1-depleted cells (Figure 5E). This epistatic relationship of PARN to NOB1 indicates that PARN acts upstream of NOB1. Moreover, co-depletion of PARN and NOB1 yielded a much stronger accumulation of 18S-EFL species than the sole knockdown of PARN. This suggests that NOB1 can also cleave untrimmed 18S-E pre-rRNAs, albeit less efficiently. In accordance with this assumption, over-expression of NOB1 in PARN-depleted cells led to a decreased amount of 18S-EFL pre-rRNA (Figure 5E).
To further establish where these processing steps take place in the cell, we examined the intracellular localization of these 18S rRNA precursors by cell fractionation followed by northern blot analysis (Figure 6A). As expected from previous results, loss of NOB1 resulted in a marked accumulation of 18S-E pre-rRNA in the cytoplasm, but not in the nucleus, compared to control cells. The cytoplasmic fraction was devoid of long 18S-E pre-rRNAs, which supports the notion that the 3΄ extremity of 18S-EFL precursors is trimmed by PARN in the nucleus following cleavage at site E. Consistent with this idea, the 18S-EFL pre-rRNA accumulated in the nuclear fraction of PARN-depleted cells. However, this species was also strongly detected in the cytoplasm, indicating that untrimmed 18S-E pre-rRNA can be exported from the nucleus. Upon co-depletion of PARN and NOB1, the level of 18S-EFL species further increased in the cytoplasm compared to PARN-depleted cells, but not in the nucleus. Taken together, these data indicate that PARN processes the 18S-E pre-rRNA in the nucleus upstream of cleavage by NOB1 in the cytoplasm. In addition, they show that trimming by PARN is necessary to ensure the rapid flow of 18S-E pre-rRNA out of the nucleus and efficient 18S rRNA production by NOB1 in the cytoplasm.
Figure 6.
Intracellular localization of 18S rRNA precursors. (A) Cell fractionation was performed on HeLa cells treated with the indicated siRNAs. Northern blot analysis of total RNAs (3 μg/lane) were compared to the contents of the cytoplasmic (4.5 μg/lane) and nuclear fractions (1.5 μg/lane). (B) A nuclear extract prepared from HeLa cells was analyzed on a 10–30% sucrose gradient. The RNA content of each fraction was analyzed by northern blot to identify 28S and 5.8S precursors (ITS2-1 probe) and 18S precursors (5΄ITS1, ITS1-38 and ITS1-59 probes). Western blot analyses were performed with antibodies directed against ENP1/Bystin, NOB1 and PARN.
We next examined the relative timing of PARN and NOB1 association with the pre-40S particles. To do so, we separated pre-ribosomes contained in nuclear extracts on sucrose gradient (Figure 6B). Two peaks were detected, corresponding to late pre-40S particles containing 18S-E pre-rRNAs (fractions 6–9) and to a mixture of pre-40S- and pre-60S precursors (fractions 10–12). The 18S-E species spread over fractions 5–10, following a ‘processing gradient’, with the 18S-EFL pre-rRNAs being detected in the denser fractions 8–10, the intermediate 18S-E species mostly in fraction 7 and the bulk of 18S-E pre-rRNAs in lighter fractions 5–7. NOB1 peaked in fractions 6–7, which contained the trimmed 18-E species and only a small fraction of it overlapped with 18S-EFL in denser fractions. ENP1/Bystin, an RBF found in cytoplasmic and nucle(ol)ar pre-40S particles was present in the same fractions as NOB1, but was also clearly detected in denser fractions containing 18S-EFL or earlier precursors. PARN in turn was most abundant in fractions 7 and 8, with only a small amount present in fraction 6. It co-migrated with full-length or partially trimmed 18S-E RNAs in fractions 7–9, as expected for a role after cleavage at site E. These data suggest that PARN is recruited to nuclear pre-40S particles before NOB1 and dissociates from the particles prior to nuclear export, which corroborates the low level of NOB1 co-purification with TAP-tagged PARN reported above (Figure 1B).
Taken together, these results establish that PARN and NOB1 are sequentially involved in the processing of the 18S rRNA 3΄ end, with PARN acting in the nucleus, upstream of NOB1 and facilitating nuclear export of the pre-40S particles.
DISCUSSION
In recent years, a growing number of evidence has revealed the function of PARN in the production and the stability of several noncoding RNAs. Our data further broaden the involvement of PARN in ncRNA maturation to 18S rRNA biogenesis. Given the high rate of rRNA maturation, it is likely that the function of PARN in ribosome biogenesis represents its major activity. So far, RRP6 has been the only 3΄-5΄ exoRNase identified in the maturation of the 18S rRNA 3΄ end. It was proposed to act after cleavage at site 2 to shorten the 21S species into 21S-C pre-rRNAs (8,9). This study now shows that PARN is the nuclear enzyme that trims the full-length 18S-E pre-rRNA into shorter precursors after cleavage at site E (Figure 7). Several lines of evidence indicate that the activity of PARN in ribosome biogenesis is restricted to the nucleus. The preferential interaction of PARN with factors that are incorporated into nucleolar pre-ribosomes (NOC4, Bystin/ENP1, DIM2, TSR1) indicates that it is recruited at a nucleolar step. This hypothesis is supported by the abundance of PARN in nucleoli. In contrast, PARN poorly co-purifies with NOB1, which is mainly present in late nucleoplasmic and cytoplasmic pre-40S particles. Along this line, we found no condition under which PARN accumulated in the cytoplasm upon impairment of late 40S subunit maturation steps. These data suggest that PARN is released from pre-40S particles before nuclear export. This conclusion is supported by the predominant association of PARN with pre-40S particles containing long ITS1 tails, as they occur in the nucleus, and its low abundance in sucrose gradient fractions containing further matured pre-18SE rRNA species along with NOB1 and Bystin/ENP1. We have previously demonstrated that 3΄ trimming of the 18S-E pre-rRNAs starts in the nucleus, but continues in the cytoplasm (8). We therefore postulate that another cytoplasmic exonuclease remains to be identified, which might involve the oligo-uridylation of the 18S-E pre-rRNA 3΄ end (8). During the final writing of this manuscript, Ishikawa et al. published results demonstrating a similar activity of PARN in 18S-E pre-rRNA processing, but proposed a delayed timing of PARN action during the last steps of the 18S-E maturation in the nucleus and the cytoplasm (55). In contrast, our data lead us to conclude that PARN acts in the nucleus before nuclear export of the pre-40S particles and processes the 3΄ end of the 18S-E pre-rRNA immediately after cleavage at site E (Figure 7).
Figure 7.
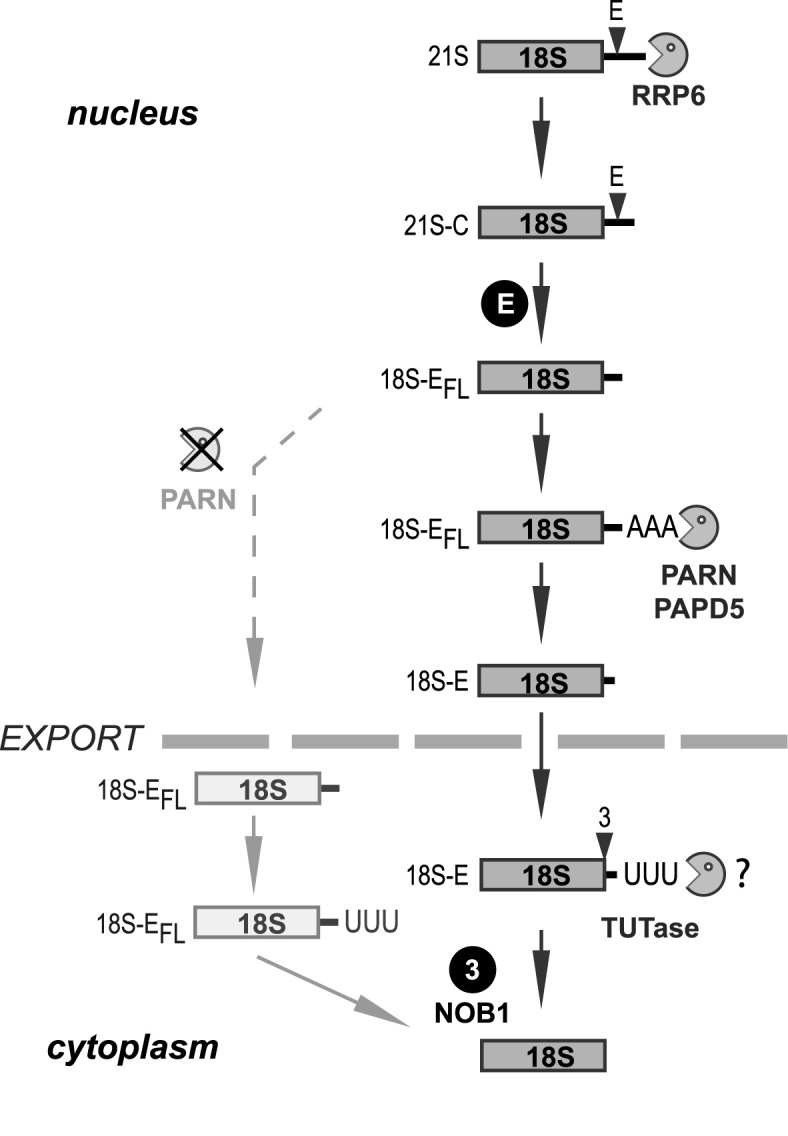
Model of the involvement of PARN in human 18S-E pre-rRNA maturation. The data presented in this manuscript lead us to propose that PARN ensures the trimming of the 18S-E pre-rRNA 3΄ end upstream of nuclear export. Its action is stimulated by oligo-adenylation of the 18S-E pre-rRNA, which could be catalyzed by PAPD5. Since the 18S-E pre-rRNA is further trimmed in the cytoplasm (8), we postulate the existence of another 3΄-5΄ exonuclease playing this role. Addition of untemplated uridines at the 3΄ end might also assist this process. Trimming by PARN is not strictly required for nuclear export of pre-40S particles, but cleavage by NOB1 in the cytoplasm appears to be strongly delayed when PARN is absent.
Because of the high GC content of the ITS1 and the putative presence of a strong secondary structure (see Figure 3A), the 3΄ end of the 18S-E pre-rRNA was an unexpected substrate for PARN. PARN was previously characterized as a highly processive RNase with a marked preference for A-rich unstructured substrates (20). Nevertheless, we show here that PARN can process to some extent the structured and GC-rich 3΄-end of the 18S-E pre-rRNA in vitro. The oligo-adenylation of the 18S-E pre-rRNA observed upon PARN depletion strongly suggests that the action of a poly(A) polymerase might be associated to PARN function, likely to prime its exonuclease activity. Our data show that addition of untemplated adenosines could be catalyzed by the poly(A) polymerase PAPD5/TRF4-2, which has already been associated with PARN action (21–23). It is located in the nucleus of human cells (56), where it adenylates aberrant rRNA transcripts targeted for degradation by the TRAMP complex (51,52). We mostly found untemplated adenosines after G or C residues, which are less efficiently hydrolyzed by PARN (19,47,57). This suggests that oligo-adenylation is required to stimulate PARN activity on these unfavorable substrates. One might speculate that the slowing-down of PARN exonucleolytic action by GC-rich segments could allow sequential conformational changes to take place during these processing steps and thus contribute to the coordinated release and recruitment of RBFs in nuclear pre-40S particles prior to their export.
PARN and NOB1 are both involved in the final processing of the 3΄ end of the 18S pre-rRNA after cleavage at site E, which raises the question of their functional coordination and physical interactions. The preferential association of PARN with early pre-40S RBFs (Figure 1B) together with the absence of NOB1 in the nucleolus indicates that PARN recruitment into pre-ribosomes precedes that of NOB1, a conclusion also supported by our analysis of nuclear pre-40S particles on sucrose gradient (Figure 6B). One attractive hypothesis is that trimming of the ITS1 by PARN would clear NOB1's binding site in the vicinity of the 18S rRNA 3΄ end, thereby promoting NOB1 association to pre-40S particles. Further studies, including structural analyses of isolated particles, will be needed to answer this question. In baker's yeast, CRAC data have revealed that Nob1p occupies different binding sites during the 18S rRNA maturation process (58,59). It has been postulated that Nob1p first binds to helix 40 in the head domain of the yeast pre-40S particle, and then relocates between the back of the head and the platform to a region overlapping the 3΄ end of the 18S rRNA and the ITS1 (60). This repositioning would allow Nob1p to access site D and perform the final endonucleolytic cleavage on the 20S pre-rRNA of yeast cells (59,61,62). Similarly, we have proposed that NOB1 needs to be repositioned relative to the 18S rRNA 3΄ terminus in human late pre-40S particles to allow endonucleolytic cleavage (53). After cleavage at site E, trimming of the 18S-EFL by PARN could be the first act of this highly regulated multistep remodeling of the pre-40S particle that gradually prepares final cleavage of the 3΄ end of the 18S rRNA by NOB1.
PARN mutations have been related to rare syndromes displaying bone marrow failure, neurological defects and pulmonary fibrosis (30,31,33), among which the Hoyeraal-Hreidarsson syndrome (HHS) (31,32), a severe form of dyskeratosis congenita (63). These diseases were primarily attributed to telomere shortening. The present demonstration of PARN involvement in 18S rRNA maturation makes it the third protein involved in dyskeratosis congenita to associate functions in ribosome biogenesis and telomere elongation, together with dyskerin and NHP2. We therefore predict that patients affected by mutations in PARN will display impairment in ribosome formation. Consistent with this assumption, sucrose gradient analysis of polysomes in such a patient has revealed an impaired 40S production, which could result from a defect in 18S rRNA maturation (30). PARN emerges as a key protein at the cross-road of several cellular pathways related to genome maintenance and gene expression: ribosome biogenesis, telomere maintenance, mRNA stability, synthesis and catabolism of small non-coding RNAs, DNA damage response. Depending on the kind of mutation affecting PARN, the nature and the severity of the disease may be connected to the degree of dysfunction of these different processes.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Drs Aline Marnef and Tamas Kiss (LBME, Toulouse) for the gift of snoRNA and snRNA probes, to Dr Marie-Pierre Rols (IPBS, Toulouse) for providing her electro-transformation protocol, to Dr Franziska Wandrey (ETH, Zürich) for the Nob1-GFP construct and to Drs Katherine Sloan and Markus Bohnsack (Institute for Molecular Biology, Göttingen) for the RNase H digestion assay protocol.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; University of Toulouse-Paul Sabatier; Agence Nationale de la Recherche [RIBOCRASH 10-BLAN-1115.1 to P.E.G., RIBOMAN ANR15-CE12-0001-02 to P.E.G.]; Association pour la Recherche contre le Cancer [PJA20131200432 to M.F.O’D.]; Swiss National Science Foundation [31003A_166565, NCCR ‘RNA&Disease’ to U.K.]. Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ellis S.R., Gleizes P.E.. Diamond Blackfan anemia: ribosomal proteins going rogue. Semin. Hematol. 2011; 48:89–96. [DOI] [PubMed] [Google Scholar]
- 2. Danilova N., Gazda H.T.. Ribosomopathies: how a common root can cause a tree of pathologies. Dis. Model. Mech. 2015; 8:1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dauwerse J.G., Dixon J., Seland S., Ruivenkamp C.A., van Haeringen A., Hoefsloot L.H., Peters D.J., Boers A.C., Daumer-Haas C., Maiwald R. et al. . Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 2011; 43:20–22. [DOI] [PubMed] [Google Scholar]
- 4. Wong C.C., Traynor D., Basse N., Kay R.R., Warren A.J.. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011; 118:4305–4312. [DOI] [PubMed] [Google Scholar]
- 5. Mullineux S.T., Lafontaine D.L.. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand. Biochimie. 2012; 94:1521–1532. [DOI] [PubMed] [Google Scholar]
- 6. Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E.. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA. 2015; 6:225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang M., Pestov D.G.. 5΄-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res. 2011; 39:1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Preti M., O’Donohue M.F., Montel-Lehry N., Bortolin-Cavaille M.L., Choesmel V., Gleizes P.E.. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res. 2013; 41:4709–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sloan K.E., Mattijssen S., Lebaron S., Tollervey D., Pruijn G.J., Watkins N.J.. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 2013; 200:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schilders G., van Dijk E., Pruijn G.J.. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007; 35:2564–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L.. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013; 51:539–551. [DOI] [PubMed] [Google Scholar]
- 12. Ansel K.M., Pastor W.A., Rath N., Lapan A.D., Glasmacher E., Wolf C., Smith L.C., Papadopoulou N., Lamperti E.D., Tahiliani M. et al. . Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat. Struct. Mol. Biol. 2008; 15:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coute Y., Kindbeiter K., Belin S., Dieckmann R., Duret L., Bezin L., Sanchez J.C., Diaz J.J.. ISG20L2, a novel vertebrate nucleolar exoribonuclease involved in ribosome biogenesis. Mol. Cell. Proteomics. 2008; 7:546–559. [DOI] [PubMed] [Google Scholar]
- 14. Idol R.A., Robledo S., Du H.Y., Crimmins D.L., Wilson D.B., Ladenson J.H., Bessler M., Mason P.J.. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol. Dis. 2007; 39:35–43. [DOI] [PubMed] [Google Scholar]
- 15. Carron C., O’Donohue M.F., Choesmel V., Faubladier M., Gleizes P.E.. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Research. 2011; 39:280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wells G.R., Weichmann F., Colvin D., Sloan K.E., Kudla G., Tollervey D., Watkins N.J., Schneider C.. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res. 2016; 44:5399–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fatica A., Tollervey D., Dlakic M.. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA. 2004; 10:1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pertschy B., Schneider C., Gnadig M., Schafer T., Tollervey D., Hurt E.. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 2009; 284:35079–35091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinez J., Ren Y.G., Thuresson A.C., Hellman U., Astrom J., Virtanen A.. A 54-kDa fragment of the Poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting Poly(A)-specific 3΄ exonuclease. J. Biol. Chem. 2000; 275:24222–24230. [DOI] [PubMed] [Google Scholar]
- 20. Virtanen A., Henriksson N., Nilsson P., Nissbeck M.. Poly(A)-specific ribonuclease (PARN): an allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit. Rev. Biochem. Mol. Biol. 2013; 48:192–209. [DOI] [PubMed] [Google Scholar]
- 21. Berndt H., Harnisch C., Rammelt C., Stohr N., Zirkel A., Dohm J.C., Himmelbauer H., Tavanez J.P., Huttelmaier S., Wahle E.. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012; 18:958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moon D.H., Segal M., Boyraz B., Guinan E., Hofmann I., Cahan P., Tai A.K., Agarwal S.. Poly(A)-specific ribonuclease (PARN) mediates 3΄-end maturation of the telomerase RNA component. Nat. Genet. 2015; 47:1482–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen D., Grenier St-Sauveur V., Bergeron D., Dupuis-Sandoval F., Scott M.S., Bachand F.. A polyadenylation-dependent 3΄ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep. 2015; 13:2244–2257. [DOI] [PubMed] [Google Scholar]
- 24. Tseng C.K., Wang H.F., Burns A.M., Schroeder M.R., Gaspari M., Baumann P.. Human telomerase RNA processing and quality control. Cell Rep. 2015; 13:2232–2243. [DOI] [PubMed] [Google Scholar]
- 25. Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y.. Poly(A)-specific ribonuclease mediates 3΄-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013; 5:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Katoh T., Hojo H., Suzuki T.. Destabilization of microRNAs in human cells by 3΄ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 2015; 43:7521–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Izumi N., Shoji K., Sakaguchi Y., Honda S., Kirino Y., Suzuki T., Katsuma S., Tomari Y.. Identification and functional analysis of the pre-piRNA 3΄ trimmer in silkworms. Cell. 2016; 164:962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang W., Tu S., Lee H.C., Weng Z., Mello C.C.. The RNase PARN-1 trims piRNA 3΄ ends to promote transcriptome surveillance in C. elegans. Cell. 2016; 164:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wyler E., Zimmermann M., Widmann B., Gstaiger M., Pfannstiel J., Kutay U., Zemp I.. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA. 2011; 17:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dhanraj S., Gunja S.M., Deveau A.P., Nissbeck M., Boonyawat B., Coombs A.J., Renieri A., Mucciolo M., Marozza A., Buoni S. et al. . Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN). J. Med. Genet. 2015; 52:738–748. [DOI] [PubMed] [Google Scholar]
- 31. Tummala H., Walne A., Collopy L., Cardoso S., de la Fuente J., Lawson S., Powell J., Cooper N., Foster A., Mohammed S. et al. . Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest. 2015; 125:2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burris A.M., Ballew B.J., Kentosh J.B., Turner C.E., Norton S.A., Giri N., Alter B.P., Nellan A. NCI DCEG Cancer Genomics Research Laboratory, NCI DCEG Cancer Sequencing Working Group et al. . Hoyeraal-Hreidarsson Syndrome due to PARN Mutations: fourteen years of follow-up. Pediatr. Neurol. 2016; 56:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stuart B.D., Choi J., Zaidi S., Xing C., Holohan B., Chen R., Choi M., Dharwadkar P., Torres F., Girod C.E. et al. . Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat. Genet. 2015; 47:512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zemp I., Wild T., O’Donohue M.F., Wandrey F., Widmann B., Gleizes P.E., Kutay U.. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 2009; 185:1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paganin-Gioanni A., Bellard E., Escoffre J.M., Rols M.P., Teissie J., Golzio M.. Direct visualization at the single-cell level of siRNA electrotransfer into cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10443–10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O’Donohue M.F., Choesmel V., Faubladier M., Fichant G., Gleizes P.E.. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 2010; 190:853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strezoska Z., Pestov D.G., Lau L.F.. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 2000; 20:5516–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kiss T., Filipowicz W.. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993; 12:2913–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haag S., Kretschmer J., Bohnsack M.T.. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA. 2015; 21:180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pool M.R., Stumm J., Fulga T.A., Sinning I., Dobberstein B.. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002; 297:1345–1348. [DOI] [PubMed] [Google Scholar]
- 41. Zemp I., Wandrey F., Rao S., Ashiono C., Wyler E., Montellese C., Kutay U.. CK1delta and CK1epsilon are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J. Cell Sci. 2014; 127:1242–1253. [DOI] [PubMed] [Google Scholar]
- 42. Widmann B., Wandrey F., Badertscher L., Wyler E., Pfannstiel J., Zemp I., Kutay U.. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell. 2012; 23:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rouquette J., Choesmel V., Gleizes P.E.. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005; 24:2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomas F., Kutay U.. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003; 116:2409–2419. [DOI] [PubMed] [Google Scholar]
- 45. Wild T., Horvath P., Wyler E., Widmann B., Badertscher L., Zemp I., Kozak K., Csucs G., Lund E., Kutay U.. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010; 8:e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ren Y.G., Martinez J., Virtanen A.. Identification of the active site of poly(A)-specific ribonuclease by site-directed mutagenesis and Fe(2+)-mediated cleavage. J. Biol. Chem. 2002; 277:5982–5987. [DOI] [PubMed] [Google Scholar]
- 47. Henriksson N., Nilsson P., Wu M., Song H., Virtanen A.. Recognition of adenosine residues by the active site of poly(A)-specific ribonuclease. J. Biol. Chem. 2010; 285:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coleman A.W. Analysis of mammalian rDNA internal transcribed spacers. PLoS One. 2013; 8:e79122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D.. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005; 121:713–724. [DOI] [PubMed] [Google Scholar]
- 50. Dez C., Houseley J., Tollervey D.. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006; 25:1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shcherbik N., Wang M., Lapik Y.R., Srivastava L., Pestov D.G.. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010; 11:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sloan K.E., Bohnsack M.T., Schneider C., Watkins N.J.. The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA. 2014; 20:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Larburu N., Montellese C., O’Donohue M.F., Kutay U., Gleizes P.E., Plisson-Chastang C.. Structure of a human pre-40S particle points to a role for RACK1 in the final steps of 18S rRNA processing. Nucleic Acids Res. 2016; 44:8465–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R.. Structures of the human and Drosophila 80S ribosome. Nature. 2013; 497:80–85. [DOI] [PubMed] [Google Scholar]
- 55. Ishikawa H., Yoshikawa H., Izumikawa K., Miura Y., Taoka M., Nobe Y., Yamauchi Y., Nakayama H., Simpson R.J., Isobe T.. Poly(A)-specific ribonuclease regulates the processing of small-subunit rRNAs in human cells. Nucleic Acids Res. 2016; 45:3437–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., Jensen T.H.. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 2011; 43:624–637. [DOI] [PubMed] [Google Scholar]
- 57. Korner C.G., Wormington M., Muckenthaler M., Schneider S., Dehlin E., Wahle E.. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998; 17:5427–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Granneman S., Petfalski E., Swiatkowska A., Tollervey D.. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010; 29:2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Turowski T.W., Lebaron S., Zhang E., Peil L., Dudnakova T., Petfalski E., Granneman S., Rappsilber J., Tollervey D.. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 2014; 42:12189–12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lamanna A.C., Karbstein K.. Nob1 binds the single-stranded cleavage site D at the 3΄-end of 18S rRNA with its PIN domain. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:14259–14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lamanna A.C., Karbstein K.. An RNA conformational switch regulates pre-18S rRNA cleavage. J. Mol. Biol. 2011; 405:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lebaron S., Schneider C., van Nues R.W., Swiatkowska A., Walsh D., Bottcher B., Granneman S., Watkins N.J., Tollervey D.. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012; 19:744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Glousker G., Touzot F., Revy P., Tzfati Y., Savage S.A.. Unraveling the pathogenesis of Hoyeraal-Hreidarsson syndrome, a complex telomere biology disorder. Br. J. Haematol. 2015; 170:457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.