Abstract
Functionalized nanoparticles have seen valuable applications, particularly in the delivery of therapeutic and diagnostic agents in biological systems. However, the manufacturing of such nano-scale systems with the consistency required for biological application can be challenging, as variation in size and shape have large influences in nanoparticle behavior in vivo. We report on the development of a versatile nano-scaffold based on the modular functionalization of a DNA G-quadruplex. DNA sequences are functionalized in a modular fashion using well-established phosphoramidite chemical synthesis with nucleotides containing modification of the amino (N2) position of the guanine base. In physiological conditions, these sequences fold into well-defined G-quadruplex structures. The resulting DNA nano-scaffolds are thermally stable, consistent in size, and functionalized in a manner that allows for control over the density and relative orientation of functional chemistries on the nano-scaffold surface. Various chemistries including small modifications (N2-methyl-guanine), bulky aromatic modifications (N2-benzyl-guanine), and long chain-like modifications (N2-6-amino-hexyl-guanine) are tested and are found to be generally compatible with G-quadruplex formation. Furthermore, these modifications stabilize the G-quadruplex scaffold by 2.0–13.3 °C per modification in the melting temperature, with concurrent modifications producing extremely stable nano-scaffolds. We demonstrate the potential of this approach by functionalizing nano-scaffolds for use within the biotin–avidin conjugation approach.
INTRODUCTION
Engineered nanoparticles have found promising applications as therapeutic delivery vehicles (1–3) and biological imaging agents (4,5). Nanoparticle systems offer numerous therapeutic advantages, including targeted delivery approaches (4) and controlled-release of drug payloads (2). Despite these advantages, the routine use of nanoparticles in medicine is hampered by a number of outstanding challenges (6). Particularly, subtle changes to nanoparticle dimension/composition can heavily influence pharmacokinetic properties, and ultimately efficacy and toxicity (4,7).
An attractive approach to nanosystems engineering involves nucleic acid-based nanostructures and nanodevices. G-quadruplexes are four-stranded structures formed by the cyclical assembly of guanine bases in guanine-rich nucleic acid sequences (Figure 1) (8). These structures have garnered research attention owing largely to their various biological functions (9,10) and their promising roles as anti-cancer drug targets (11–14). From a pharmacological perspective, many biologically-active G-quadruplex-forming sequences have been identified, including sequences that display activity against cancer proliferation (15), HIV (16) and blood coagulation (17). These bead-like structures are useful molecular scaffolds, self-assembling and self-folding in an ion-dependent manner into thermally stable nano-scaffolds (18). A host of nanotechnology applications have also been demonstrated which employ the G-quadruplex architecture, including catalysts (19–21), sensing devices (22), imaging agents (21–23) and molecular electronics systems (24,25).
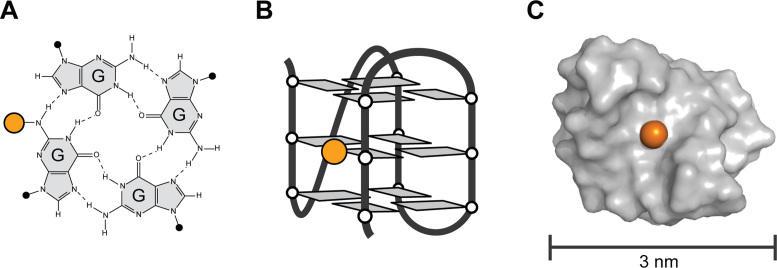
Figure 1.
N2-modification of G-quadruplex nano-scaffolds. (A) G-tetrad formed by the hydrogen bonding of four guanine bases. (B) Schematic folding of a single G-rich DNA strand into a G-quadruplex structure. (C) Surface representation of a G-quadruplex nano-scaffold. DNA is colored in gray. The location of a single N2-position is shown as an orange sphere.
Many of these applications are made possible by the incorporation of alternative, often functional, chemistries during DNA synthesis. One convenient point of modification is the 5΄- or 3΄-end of the DNA strand. For example, modification of strand termini can enhance the activity of G-quadruplex based aptamers (26) and enables the use of G-quadruplexes as FRET based sensors (22). However, such modifications are inherently restricted by the location and number of strand termini in the G-quadruplex molecule. Modifications can also be made to the loops of G-quadruplexes, which are segments of sequence not involved in the G-tetrad formation. However, the structure and stability of a G-quadruplex is highly sensitive to loop length and sequence composition (27–30). Even slight changes in loop base chemistry can alter the G-quadruplex conformations adopted (31). Another strategy for modification of G-quadruplex is to substitute chemically altered guanines bases into the G-tetrad core of the G-quadruplex (32). In principle, modification of the guanine base allows for functionalization at a desired location on the G-quadruplex nano-scaffold, and multiple modified guanines per tetrad layer can be accommodated. However, many types of guanine base analogs have been observed to destabilize and/or alter the fold of the G-quadruplex structure (33,34).
To preserve the G-quadruplex fold, effective modification of the guanine base requires the preservation of the cyclical hydrogen bonding of the guanine tetrad. In this respect, the C8 and N2 position of the guanine base are attractive locations as they do not disrupt G-tetrad hydrogen bonding and they allow for substituents to be projected into the groove of the G-quadruplex structure (Figure 1). Modification at the C8 position of the guanine base has been explored in depth (32), and its limitations are well understood. Notably, C8-modifications affect the stability of G-quadruplex formation in a manner dependent on the glycosidic conformation of the guanine base (35–37), This behavior limits the ability to functionalize at the C8-position, with only substitutions to guanines in a syn-conformation being effectively incorporated.
By comparison, there has been a notable lack research on modification at the N2-position (Figure 1A). In past decades, studies have reported that N2-modifications of therapeutic aptamers are capable of altering affinity toward their target protein (38,39) and that N2-modifications are capable of stabilizing a G-quadruplex-forming molecule with anti-HIV properties (39). More recently, N2-modified guanosines have been studied for their self-assembling properties (40–42). While these studies hint at the potential benefits of N2-guanine modification, a basic understanding of how N2-modifications affect G-quadruplex structure/stability, and what types of modifications are tolerated, is currently lacking.
In this work, we set out to characterize the effects of a variety of N2-modified guanines bases on the formation and stability of G-quadruplexes. Our results reveal that N2-modified guanines are versatile and potent stabilizers of the G-quadruplex architecture and can be substituted in a site-specific manner into a variety of locations on the G-quadruplex without disrupting its formation. Furthermore, multiple N2-modifications may be incorporated simultaneously leading to a highly stabilized G-quadruplex. We present the design of a ‘Ball-with-Hair’ construct containing multiple ‘chain-like’ N2-6-amino-hexyl-guanine chemistries serving as potential sites for nano-scaffold functionalization. We demonstrate the potential use of this tool by incorporating functional chain-like chemistries that bind protein through the biotin–avidin complex. The findings reported here illustrate a novel approach to generating highly stable and functional G-quadruplex nano-scaffolds through modular N2-guanine modification.
MATERIALS AND METHODS
Synthesis and sample preparation
DNA samples were synthesized on an Applied Biosystems 394 DNA Synthesizer (Foster City, CA, USA) and purified following Glen Research protocols. The modified phosphoramidite N2-6-amino-hexyl-guanine was purchased from Glen Research (Sterling, VA, USA). Modified phosphoramidites N2-methyl-guanine and N2-benzyl-guanine were purchased from Berry & Associates (Dexter, MI, USA). N2-dodecane amino-biotin guanosine phosphoramidite was purchased from ChemGenes (Wilmington, MA, USA). Purified DNA samples were dialyzed against H2O, ∼30 mM KCl and H2O before being dried and dissolved in solution containing KCl, potassium phosphate (KPi) buffer (pH 7), 20 μM 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) used as a chemical shift reference in NMR, and containing 10% D2O.
Ionic conditions
The effects of substitutions into the (3+1) Native G-quadruplex nano-scaffold were studied in a variety of salt concentrations including ‘low salt’ (1 mM KPi + 1 mM KCl), ‘moderate salt’ (10 mM KPi + 10 mM KCl) and ‘physiological salt’ (10 mM KPi + 130 mM KCl) environments. Moderate salt conditions were the default conditions used. Low salt conditions were used for technical reasons in order to aid in the comparison of thermal stabilities of constructs with multiple substitutions, as these constructs had high thermal stabilities. Physiological salt conditions were used to test the thermal stability of these constructs in high salt conditions more similar to those in the cellular environment.
NMR spectroscopy
NMR spectra were obtained on a 600 MHz Bruker spectrometer (Billerica, MA, USA) at a temperature of 25 °C using a jump-and-return type water suppression pulse sequence (43). Samples were annealed prior to recording of NMR spectra by heating to 100 °C for 2 min then cooling slowly at room temperature. Chemical shifts were internally referenced with 2,2-dimethyl-2-silapentane-5-sulfonate (D.S.S).
CD spectroscopy
CD spectra were recorded on a JASCO-815 spectropolarimeter (Tokyo, Japan). Samples contained a DNA strand concentration of ∼5 μM. Spectra were recorded at 20 °C in the range of 220–320 nm and averaged over 10 scans. Spectra were normalized through subtracting by the CD signal at 320 nm and dividing by the sample concentration determined by concurrent UV absorption measurements. Samples were annealed prior to recording of CD spectra by heating to 100 °C than cooling slowly at room temperature.
UV melting experiments
Thermal denaturing of constructs containing single N2-substitutions was studied in moderate salt conditions (10 mM KPi + 10 mM KCl) using UV spectroscopy. Melting experiments were performed on a JASCO V-650 UV–visible spectrophotometer (Tokyo, Japan). Samples containing ∼5 μM DNA strand concentration were first heated to 83 °C and cooled to 14 °C at a rate of 0.2 °C/min. The absorbance was sampled every 0.5 °C at 295 and 320 nm. At the end of the cooling cycle, samples were held for 10 min before being heated to 83 °C at the same rate. Melting curves were normalized first by subtraction at 320 nm and then further processed to create fraction folded curves (44). Melting temperatures (Tm) reported represent an average over heating and cooling cycles. Hysteresis of <2 °C (generally < 1.5 °C) was observed over all sequences.
CD melting experiments
CD melting experiments were performed for constructs containing multiple N2-modifications in low salt conditions (in 1 mM KCl and 1 mM KPi). CD melting experiments were also carried out for the N2-Ball-with-Hair construct in physiological salt conditions (10 mM KPi + 130 mM KCl). Spectra were recorded on a JASCO-815 spectropolarimeter (Tokyo, Japan). Samples contained a DNA strand concentration of ∼5 μM. Samples were heated to 95 °C (98 °C for N2-Ball-with-Hair sample) and cooled to 15 °C at a rate of 0.2 °C/min. Samples were then heated in a similar manner. Data was recorded every 0.5 °C at 290 nm and corrected by subtraction at 320 nm. Normalized melting curves are used to determine the melting temperature (Tm) of modified sequences (45), which is taken as an average of the heating and cooling curves. Hysteresis in a heating/cooling cycle was observed to be below 2 °C for all samples.
Visualization
Models of nano-scaffold structures were built based on a NMR model of the Native (3+1) G-quadruplex (PDB ID 2GKU) (46). For the N2-Ball-with-Hair sequence, guanine bases containing N2-6-amino-hexyl modifications in a straight orientation were fit to guanine bases within the NMR structure. The dihedral angles around the connection of the guanine base to the N2-6-amino-hexyl functional group (guanine(C2)–guanine(N2)–Hex(C1)–Hex(C2)) were then manually varied to project the N2-6-amino-hexyl into the groove. The illustrative models were visualized in Pymol.
Gel electrophoresis
Native PAGE was performed in a vertical polyacrylamide gel containing 20% polyacrylamide in the lower half and 10% polyacrylamide in the upper half. A running buffer containing 10 mM KCl and 10 mM KPi was used to promote G-quadruplex formation. Gels were visualized with Bio-Rad Fast Blast DNA stain. Samples contained 10 μM DNA/neutravidin with K+ concentrations of ∼20 mM.
RESULTS
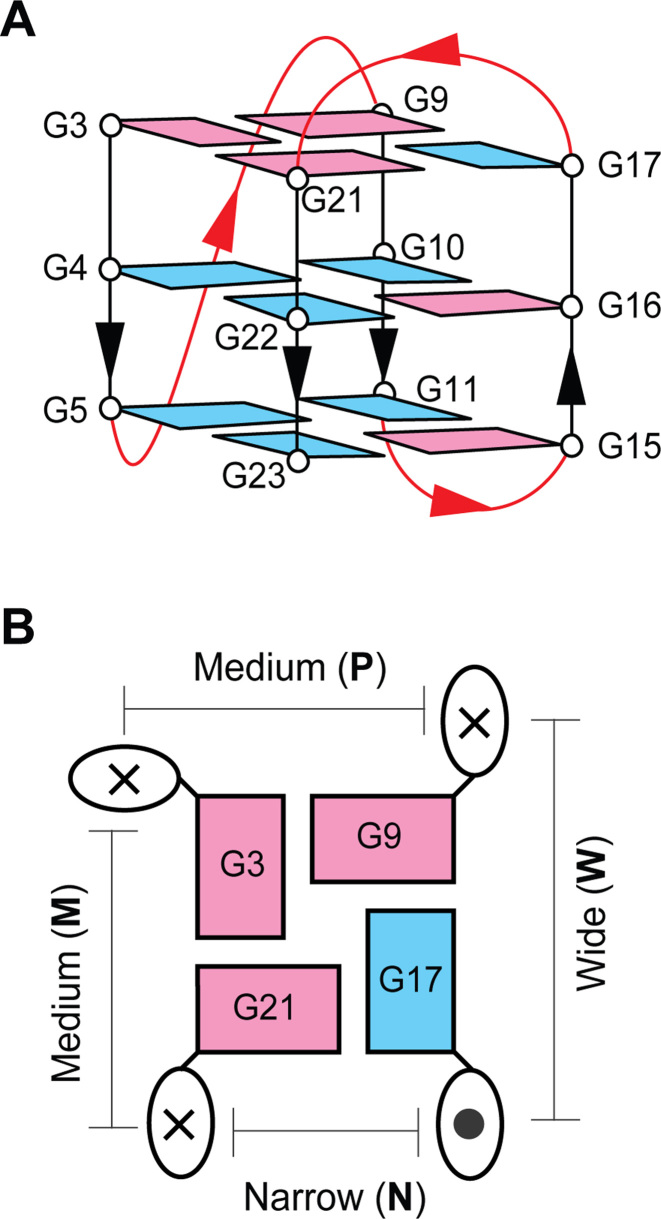
In this study, we investigate the effects of modifying two DNA sequences. Primarily, we investigate the sequence d[TT(GGGTTA)3GGGA], termed the ‘Native’ sequence, which is known to fold into a single well-defined G-quadruplex structure (Figure 2) (46). In this sequence, four segments of GGG assemble in three stacked G-tetrads which create the core of this G-quadruplex nano-scaffold (Figure 2A). Guanine bases within the core are characterized by their glycosidic orientation, with respect the sugar-backbone, as being in a syn or anti orientation. The strand directionality of the DNA sequence (5΄→3΄) reveals three strands pointing in the same direction and one strand oriented in the opposite direction, defining a (3+1) G-quadruplex topology. Connecting the corners of the G-tetrad core are segments of sequence that do not participate in G-tetrad formation termed ‘loops’. The Native G-quadruplex contains one propeller loop and two edge-wise loops. Grooves into which an N2-guanine modification would be projected (Figure 2B) are classified as wide (W), medium (M), or narrow (N) based upon their width or as a groove containing a propeller loop (P). The diverse structural features of this (3+1) G-quadruplex provide a useful scaffold to test the N2-functionalization of G-quadruplex nano-scaffolds.
Figure 2.
Native (3+1) G-quadruplex. (A) Folding schematic of the 24-nt Native sequence studied in this work. Guanine bases are colored based on their syn (magenta) or anti (cyan) glycosidic conformation. Non-guanine bases are omitted for clarity. (B) A schematic of the top G3·G21·G17·G9 tetrad. The 5΄→3΄ strand directionality is indicated as into (×) and out of (•) the page. Grooves are labeled based on their dimensions (W, N, M) or the presence of a propeller loop (P).
Secondarily, to generalize our understanding of the effects of N2 modification, we investigate a parallel-stranded G-quadruplex formed by the sequence d[TT(GGGT)4] (Supplementary Figure S7).
Formation of N2-modified G-quadruplex nano-scaffolds
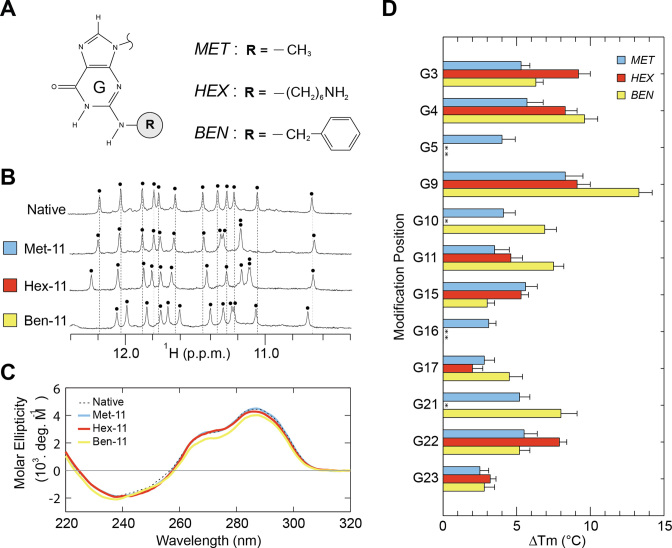
We systematically investigated the effects of N2-modifications on G-quadruplex structure and stability using multiple guanine analogs (Figure 3A). These modified bases included N2-methyl-guanine (MET) containing a small methyl group substitution, N2-benzyl-guanine (BEN) containing a bulky aromatic chemistry, and N2-6-amino-hexyl-guanine (HEX) containing a flexible ‘chain-like’ chemistry. These chemistries were chosen to explore a range of substituent sizes and flexibilities.
Figure 3.
G-quadruplex nano-scaffolds containing single N2-modifications. (A) N2-modified guanines explored in our systematic study. (B) Illustrative NMR and (C) CD spectra of G-quadruplexes containing a single N2-modification. Each black dot indicates an imino proton peak. (D) Change in the melting temperature (Tm) in moderate salt condition (10 mM KPi + 10 mM KCl) upon N2-modification. N2-modifed sequences which display multiple G-quadruplex conformations are indicated with asterisks (*).
The formation of G-quadruplex nano-scaffolds by N2-modified sequences was evaluated primarily by monitoring the imino proton region of 1H NMR spectra (Figure 3B; Supplementary Figures S1–S3), a region characteristic of G-quadruplex formation (47). Imino proton spectra may be used to evaluate the number of conformers in solution and similarity between spectra is a strong indicator that a similar structure has been adopted. Single-position N2-modifications of MET, BEN or HEX were made into each of the 12 guanines available for substitution in the native G-quadruplex (Table 1). G-quadruplex formation was also probed by CD spectroscopy (Figure 3C; Supplementary Figures S4–S6), which provides spectral patterns characteristic of specific G-quadruplex folding topologies.
Table 1. N2-modified DNA sequences.
| Name | Sequence (5΄- 3΄) | Number of modifications | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | MET | BEN | HEX | BIO | ||||||||||
| Native | TT | GGG | TTA | GGG | TTA | GGG | TTA | GGG | A | |||||
| Single-Position | ||||||||||||||
| Met-3 | TT | MGG | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Met-4 | TT | GMG | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Met-5 | TT | GGM | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Met-9 | TT | GGG | TTA | MGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Met-10 | TT | GGG | TTA | GMG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Met-11 | TT | GGG | TTA | GGM | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Met-15 | TT | GGG | TTA | GGG | TTA | MGG | TTA | GGG | A | 1 | 1 | |||
| Met-16 | TT | GGG | TTA | GGG | TTA | GMG | TTA | GGG | A | 1 | 1 | |||
| Met-17 | TT | GGG | TTA | GGG | TTA | GGM | TTA | GGG | A | 1 | 1 | |||
| Met-21 | TT | GGG | TTA | GGG | TTA | GGG | TTA | MGG | A | 1 | 1 | |||
| Met-22 | TT | GGG | TTA | GGG | TTA | GGG | TTA | GMG | A | 1 | 1 | |||
| Met-23 | TT | GGG | TTA | GGG | TTA | GGG | TTA | GGM | A | 1 | 1 | |||
| Ben-3 | TT | BGG | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Ben-4 | TT | GBG | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Ben-5 | TT | GGB | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Ben-9 | TT | GGG | TTA | BGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Ben-10 | TT | GGG | TTA | GBG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Ben-11 | TT | GGG | TTA | GGB | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Ben-15 | TT | GGG | TTA | GGG | TTA | BGG | TTA | GGG | A | 1 | 1 | |||
| Ben-16 | TT | GGG | TTA | GGG | TTA | GBG | TTA | GGG | A | 1 | 1 | |||
| Ben-17 | TT | GGG | TTA | GGG | TTA | GGB | TTA | GGG | A | 1 | 1 | |||
| Ben-21 | TT | GGG | TTA | GGG | TTA | GGG | TTA | BGG | A | 1 | 1 | |||
| Ben-22 | TT | GGG | TTA | GGG | TTA | GGG | TTA | GBG | A | 1 | 1 | |||
| Ben-23 | TT | GGG | TTA | GGG | TTA | GGG | TTA | GGB | A | 1 | 1 | |||
| Hex-3 | TT | HGG | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Hex-4 | TT | GHG | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Hex-5 | TT | GGH | TTA | GGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Hex-9 | TT | GGG | TTA | HGG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Hex-10 | TT | GGG | TTA | GHG | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Hex-11 | TT | GGG | TTA | GGH | TTA | GGG | TTA | GGG | A | 1 | 1 | |||
| Hex-15 | TT | GGG | TTA | GGG | TTA | HGG | TTA | GGG | A | 1 | 1 | |||
| Hex-16 | TT | GGG | TTA | GGG | TTA | GHG | TTA | GGG | A | 1 | 1 | |||
| Hex-17 | TT | GGG | TTA | GGG | TTA | GGH | TTA | GGG | A | 1 | 1 | |||
| Hex-21 | TT | GGG | TTA | GGG | TTA | GGG | TTA | HGG | A | 1 | 1 | |||
| Hex-22 | TT | GGG | TTA | GGG | TTA | GGG | TTA | GHG | A | 1 | 1 | |||
| Hex-23 | TT | GGG | TTA | GGG | TTA | GGG | TTA | GGH | A | 1 | 1 | |||
| Multiple | ||||||||||||||
| N2-One-Groove | TT | GGG | TTA | GMM | TTA | GGG | TTA | GGG | A | 2 | 2 | |||
| N2-Two-Groove | TT | HGG | TTA | HGG | TTA | GGG | TTA | GGG | A | 2 | 2 | |||
| N2-All-Groove | TT | HGG | TTA | HGM | TTA | MGG | TTA | GGG | A | 4 | 2 | 2 | ||
| N2-Ball-with-Hair | TT | HHG | TTA | HGH | TTA | HGH | TTA | GHH | A | 8 | 8 | |||
| Functional | ||||||||||||||
| Bio-22 | TT | GGG | TTA | GGG | TTA | GGG | TTA | GOG | A | 1 | 1 | |||
| Bio-11-22 | TT | GGG | TTA | GGO | TTA | GGG | TTA | GOG | A | 2 | 2 | |||
All 36 modified sequences demonstrated the G-quadruplex formation as determined by the presence of distinct peaks in the imino proton region 10–12 ppm of NMR spectra (Supplementary Figures S1–S3). A total of 30 sequences (83%) were observed to fold predominately into the same G-quadruplex conformation as the parent sequence (Figure 2) based on similarity of NMR and CD spectra (Figure 3B and C; Supplementary Table S2). This included 12 sequences containing a single-substitution of MET, 10 containing BEN, and 8 containing HEX. Classification by groove type revealed that sequences containing a substitution into the W, M, N and P grooves maintained the (3+1) G-quadruplex conformation in 8, 9, 6 and 7 sequences, respectively, out of 9 possible sequences.
To test the universality of this approach, we examined select N2-modifications into an additional nano-scaffold construct formed by the sequence d[T2(G3T)4] which folds into a structurally different parallel-type G-quadruplex (48) (Supplementary Table S3). G-quadruplex formation was again observed for all sequences via distinct peaks in imino proton NMR spectra (Supplementary Figure S7). Of the nine sequences tested, eight formed a single species with similar spectral characteristics to the parent d[T2(G3T)4] sequence. Taken collectively, these data suggest that N2-guanine modification is compatible with a variety of structural contexts in different G-quadruplex-forming sequences.
Enhanced thermal stability of N2-modified G-quadruplex nano-scaffolds
We proceeded to investigate how N2-guanine modifications affect the thermal stability of G-quadruplex nano-scaffolds. We focused our examination on the sequences that maintained the native (3+1) G-quadruplex structure (Supplementary Table S2). We use thermal denaturing (melting) experiments as a well-established approach to determining the relative thermal stability of different G-quadruplexes (44). Thermal stability is characterized by a melting temperature (Tm) which was determined for the Native and N2-modified constructs by monitoring UV absorption as a function of temperature.
All sequences that maintained the original (3+1) G-quadruplex conformation demonstrated enhanced thermal stability (Figure 3D, Supplementary Figures S8–S10), with a change in the melting temperature (ΔTm) observed in the range of +2.0 to +13.3 °C for each sequence containing a single N2-modification. Average ΔTm values of 4.6, 6.7 and 6.2 °C were observed upon modification with MET, BEN and HEX respectively. Further classification of ΔTm values by groove type revealed average ΔTm values of 4.5, 5.3, 5.0 and 8.4 °C for the W, M, N and P grooves respectively.
Design of highly stable nano-scaffolds
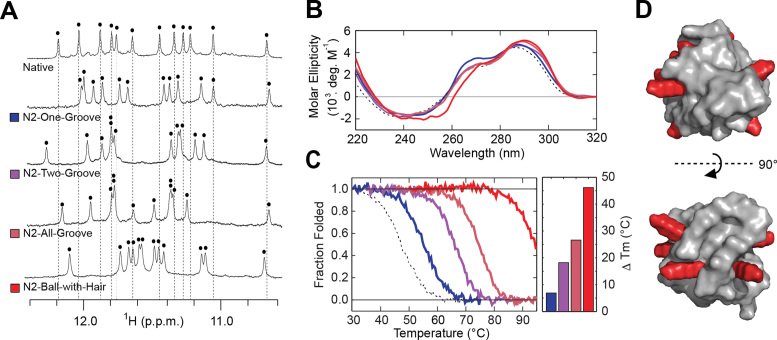
The stabilizing nature of single N2-modified guanine substitutions led us to design a number of sequences containing N2-modifications at multiple positions (Table 1). Modification of the same groove was examined in the N2-One-Groove construct containing two MET substitutions into the W groove at G10 and G11. Alternatively, the N2-Two-Groove construct tests the incorporation of two HEX into G3 of the W groove and G9 of the P groove. The modification of all four grooves by either HEX or MET is examined in the N2-All-Groove construct. The N2-Ball-with-Hair construct contains eight HEX modifications designed to provide flexible chemistries capable of extending outwards from the surface of the G-quadruplex nano-scaffold offering potential sites for functionalization (Figure 4D).
Figure 4.
G-quadruplex nano-scaffolds containing multiple N2-modifications. (A) NMR and (B) CD spectra of designed constructs containing multiple N2-modifications. Each black dot indicates an imino proton peak. (C) Thermal denaturing curves of designed sequences in low salt condition (1 mM KPi + 1 mM KCl) with quantification of change in the melting temperature (Tm). (D) Model of the N2-Ball-with-Hair construct, containing 8 HEX modifications. Flexible HEX modifications shown in elongated state.
The formation of G-quadruplex nano-scaffolds by the designed constructs was again examined by NMR and CD spectroscopy. All constructs were determined to form nano-scaffolds and to adopt the same (3+1) G-quadruplex conformation as the Native sequence based on similar 1H NMR and CD spectra (Figure 4A and B). The thermal stability was probed in the CD melting experiments. All of the G-quadruplex nano-scaffolds containing multiple N2-modifications displayed increased thermal stability (Figure 4C, Supplementary Table S4). The N2-One-Groove, N2-Two-Groove, and N2-All-Groove sequences displayed ΔTm values of +6.9, +18.2 and +26.6 °C respectively, roughly equivalent to the sum of ΔTm values gained from individual modifications observed in single N2-substitution studies. The N2-Ball-with-Hair construct demonstrated the highest thermal stability, with a ΔTm of +46.2 °C. The stability of theN2-Ball-with-Hair nano-scaffold was further tested in physiological salt condition (10 mM KPi + 130 mM KCl). Temperature-dependent CD spectra show >80% of the G-quadruplex signal intensity to remain at 93°C (Supplementary Figure S11), indicating a highly stable construct.
Functionalization of G-quadruplex nano-scaffolds
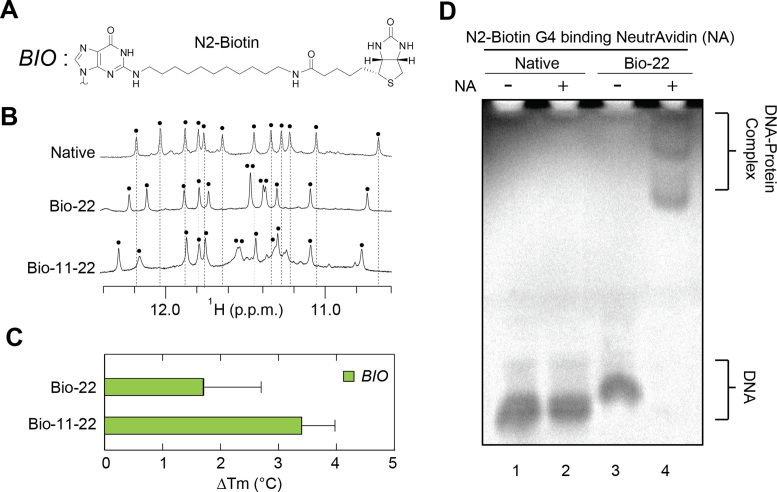
A major advantage of a G-quadruplex-based nano-scaffold approach is the ability to functionalize the system in a modular manner with geometric control over the number, position and orientation of functional chemistries. We set out to demonstrate the value of such an approach by incorporating a N2-Biotin (BIO) moiety (Figure 5A) into the Native sequence. Biotin is capable of binding the homotetramer NeutrAvidin (NA) protein (49), a derivative of the well-studied Avidin protein (50) employed in a variety of biotechnology applications (51).
Figure 5.
G-quadruplex nano-scaffolds containing biotin (BIO) N2-modifications. (A) Schematic of a BIO chemistry containing a functional biotin modification at the N2-position. (B) NMR spectra of BIO-modified G-quadruplex nano-scaffolds. Each black dot indicates an imino proton peak. (C) Change in the melting temperature (Tm) in moderate salt condition (10 mM KPi + 10 mM KCl) for BIO-modified sequences. (D) Non-denaturing PAGE of the Native and Bio-22 modified sequences in the presence and absence of the NeutrAvidin (NA) protein.
Sequences containing a single modification at G22 (Bio-22) and double modification at G11 and G22 (Bio-11-22) were synthesized (Table 1) and were observed to form G-quadruplexes in NMR spectra (Figure 5B). The similar pattern of spectra suggests the same (3+1) G-quadruplex conformation is adopted by functionalized sequences. Interestingly, the change in thermal stability for these functional BIO modifications is a modest +1.7 °C and +3.4 °C for a nano-scaffold containing one and two BIO modifications respectively (Figure 5C). Non-denaturing PAGE reveals that BIO-modified nano-scaffolds readily bind their NeutrAvidin (NA) protein target, indicated by the retarded migration of BIO-containing DNA in the presence of NA protein (Figure 5D). The presence of multiple bands can be attributed to the propensity of NA to assemble into tetramers (49).
DISCUSSION
The current work investigates the effects of N2-guanine modifications on the formation and stability of G-quadruplex nano-scaffolds. A systematic incorporation of MET, BEN and HEX chemistries into various positions of two different G-quadruplex constructs reveals that N2-modifications are generally compatible with G-quadruplex formation. When modifying the Native construct, 30 of 36 sequences adopted the same (3+1) G-quadruplex conformation as the parent sequence. This is determined by the observation of 12 dominant imino proton peaks indicating a single conformation has been adopted and the similarity of CD spectra. Although similar NMR spectral pattern was observed, the shifting of individual peaks upon modification is a common phenomenon that depends on the position and nature of the modified guanine base into a G-quadruplex scaffold (37,52). In the remaining six sequences, multiple species were observed. It is important to note that multiple species can arise from the destabilization of the original construct and/or from the stabilization of an alternative G-quadruplex structure. The smaller MET substitutions are found to be less disruptive than BEN or HEX substitutions. Modifications into the N or P grooves were more disruptive than those into M and W grooves. It is possible that the W and M grooves are more open and favorable environment for the introduction of N2-chemistries, compared to more constricted N grooves or P grooves where clashes with the neighboring backbone or loop may arise.
The description of G-quadruplex DNA acting as a nano-scaffold is fitting considering the properties of these structures. G-quadruplex forming sequences are often polymorphic, capable of adopting multiple structural conformations of various stoichiometry. Nonetheless, numerous G-quadruplex-forming sequences have been identified in which a single guanine-rich sequence self-folds in a predictable and well-behaved manner. The Native sequence explored in this work is one such sequence, in which a 24-nucleotide strand of DNA readily folds into a single G-quadruplex structure of known shape and size (46). The (3+1) G-quadruplex structure of the Native sequence (Figure 2) has a unique NMR chemical shift and CD spectral patterns which are maintained upon N2-modification (Figure 3B and C), indicating the same single structure is adopted by N2-modified sequences. Moreover, the Native and select modified sequences are found to exists mainly as a single monomeric species in non-denaturing gel electrophoresis experiments (Supplementary Figure S12). Collectively, these data imply that the N2-modified G-quadruplex nano-scaffolds studied in this work are monodisperse in character.
The size of G-quadruplex nano-scaffolds are dependent upon the sequence and the modifications employed. The (3+1) Native sequence folds into a G-quadruplex which is 3–4 nm in size depending on the diameter measured. Bulky BEN and chain-like HEX modifications protrude roughly 1 nm past the surface of the molecule. However, long modifications such as BIO are expected to be flexible and protrude up to 3 nm above the molecular surface.
N2-modifications to G-quadruplex are found to be highly stabilizing. The extent of stabilization is chemistry-dependent, with BEN > HEX > MET in terms of average stability enhancement. Stabilizing effects are also position-dependent. For example, modification to positions G4 and G9 are highly stabilizing while positions G17 and G23 show moderate Tm enhancement. The additive nature of N2-modifications leads to nano-scaffolds with high thermal stabilities when multiple modifications are incorporated. The stabilizing effects of N2-modifications observed here are generally larger than previously reported C8-modifications made to the same Native construct in similar conditions (37).
The generally stabilizing nature of N2-guanine modifications to G-quadruplex nano-scaffolds is an important conclusion of this work. We hypothesize that stabilizing effects may arise from a variety of factors: (i) Multiple G-quadruplex structures have been previously observed in which nucleobases are distinctly folded into the hydrophobic groove (53–55). It is likely that the projection of hydrophobic N2-modifications into the G-quadruplex benefits from a combination of favorable Van der Waals interactions with neighboring sugar-phosphate backbones and the exclusion of solvent from the hydrophobic groove. Non-polar solvent has been shown to be a driver of G-quadruplex structure compared to aqueous environments (56), suggesting that water depletion in the G-quadruplex groove may be stabilizing. (ii) More specifically, the N2-6-amino-hexyl (HEX) modification is likely to be positively charged at pH 7, comparable to the behavior of lysine (57). HEX modifications are expected to be flexible, and could fold back such that the charged NH3+ termini interact with the negatively charged phosphate backbone. The modest stabilization observed for BIO modifications compared to HEX modifications is likely due to the neutrality of the BIO substituent, and to an increase in entropy associated with chemical linker that is roughly three times larger than that of HEX. 3) Additionally, the substitution of a single amino proton with a bulkier chemistry (N2-modification) freezes the rotation about the N2-position. The Hoogsteen hydrogen bonding of the remaining amino proton can be visualized by the emergence of sharp peaks in the region of 9.0–10.0 ppm in NMR spectra of most modified sequences (Supplementary Figures S1–S3). It is unclear if the freezing of the amino bond plays a role in the observed stability enhancement.
The G-quadruplex nano-scaffolds studied in this work represent a novel approach to generating stable nano-scale particles. The functional BIO modifications explored here illustrate how these scaffolds may be functionalized and utilized in well-established conjugation approaches. In the N2-Ball-with-Hair construct, 8 chain-like HEX chemistries are successfully attached to the G-quadruplex nano-scaffold, demonstrating how multiple functional chemistries can be incorporated simultaneously. Our described approach to generating DNA nano-scaffolds from G-quadruplex structures produces DNA particles of consistent dimension and shape while controlling the position and density of chemistries on the nano-scaffold surface. Moreover, incorporating knowledge of G-quadruplex structure into nano-scaffold design allows one to control the relative orientation of modifications on the particle's surface. Importantly, the G-quadruplex nano-scaffolds described here are generated by the modular incorporation of modified phosphoramidites during DNA synthesis using standard solid phase synthesis approaches, allowing for nano-scaffold production that is simple and scalable.
CONCLUSION
We demonstrate that a variety of N2-guanine modifications are compatible with G-quadruplex formation. These modifications are incorporated during solid phase synthesis to yield nano-scaffolds that are consistent in size and shape. Modifications, in which the original parent structure is maintained, stabilize the G-quadruplex nano-scaffold by 2.0–13.3 °C per modification in the melting temperature. This effect is additive, with multi-modified nano-scaffolds demonstrating strongly enhanced thermal stability. The functional modification of nano-scaffolds with a N2-biotin chemistry allows the binding of avidin protein. G-quadruplex forming nucleic acids containing N2-modifications of the guanine base represent an easy and novel approach to generating functional and highly stable nano-scaffolds.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dang Khoa Pham and Man Ting Low for their participation in the early stages of this project, and Brahim Heddi and Vee Vee Cheong for their helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore National Research Foundation Investigatorship [NRF-NRFI2017-09]; Singapore Ministry of Education and Nanyang Technological University (to A.T.P.). Funding for open access charge: Nanyang Technological University .
Conflict of interest statement. A patent on the N2-modification of G-quadruplexes has been filed by Nanyang Technological University with both authors as inventors.
REFERENCES
- 1. Petros R.A., DeSimone J.M.. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug. Discov. 2010; 9:615–627. [DOI] [PubMed] [Google Scholar]
- 2. Giljohann D.A., Seferos D.S., Daniel W.L., Massich M.D., Patel P.C., Mirkin C.A.. Gold nanoparticles for biology and medicine. Angew. Chem. Int. Edit. 2010; 49:3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noorlander C.W., Kooi M.W., Oomen A.G., Park M.V., Vandebriel R.J., Geertsma R.E.. Horizon scan of nanomedicinal products. Nanomedicine. 2015; 10:1599–1608. [DOI] [PubMed] [Google Scholar]
- 4. Veiseh O., Gunn J.W., Zhang M.Q.. Design and fabrication of magnetic nanoparticles for targeted drug delivery and imaging. Adv. Drug. Deliver. Rev. 2010; 62:284–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boisselier E., Astruc D.. Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chem. Soc. Rev. 2009; 38:1759–1782. [DOI] [PubMed] [Google Scholar]
- 6. Desai N. Challenges in development of nanoparticle-based therapeutics. AAPS J. 2012; 14:282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Jong W.H., Borm P.J.A.. Drug delivery and nanoparticles: applications and hazards. Int. J. Nanomed. 2008; 3:133–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis J.T. G-quartets 40 years later: From 5΄-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004; 43:668–698. [DOI] [PubMed] [Google Scholar]
- 9. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel D.J., Phan A.T., Kuryavyi V.. Human telomere, oncogenic promoter and 5΄-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007; 35:7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Cian A., Lacroix L., Douarre C., Temime-Smaali N., Trentesaux C., Riou J.F., Mergny J.L.. Targeting telomeres and telomerase. Biochimie. 2008; 90:131–155. [DOI] [PubMed] [Google Scholar]
- 13. Monchaud D., Teulade-Fichou M.P.. A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008; 6:627–636. [DOI] [PubMed] [Google Scholar]
- 14. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jing N.J., Li Y.D., Xiong W.J., Sha W., Jing L., Tweardy D.J.. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004; 64:6603–6609. [DOI] [PubMed] [Google Scholar]
- 16. de Soultrait V.R., Lozach P.Y., Altmeyer R., Tarrago-Litvak L., Litvak S., Andreola M.L.. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002; 324:195–203. [DOI] [PubMed] [Google Scholar]
- 17. Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J.. Selection of single-stranded-DNA molecules that bind and inhibit human thrombin. Nature. 1992; 355:564–566. [DOI] [PubMed] [Google Scholar]
- 18. Gonzalez-Rodriguez D., van Dongen J.L.J., Lutz M., Spek A.L., Schenning A.P.H.J., Meijer E.W.. G-quadruplex self-assembly regulated by Coulombic interactions. Nat. Chem. 2009; 1:151–155. [DOI] [PubMed] [Google Scholar]
- 19. Chinnapen D.J.F., Sen D.. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wieland M., Hartig J.S.. Turning inhibitors into activators: a hammerhead ribozyme controlled by a guanine quadruplex. Angew. Chem. Int. Ed. 2006; 45:5875–5878. [DOI] [PubMed] [Google Scholar]
- 21. Tang Z., Goncalves D.P., Wieland M., Marx A., Hartig J.S.. Novel DNA catalysts based on G-quadruplex recognition. Chembiochem. 2008; 9:1061–1064. [DOI] [PubMed] [Google Scholar]
- 22. Juskowiak B. Analytical potential of the quadruplex DNA-based FRET probes. Anal. Chim. Acta. 2006; 568:171–180. [DOI] [PubMed] [Google Scholar]
- 23. Huang H., Suslov N.B., Li N.S., Shelke S.A., Evans M.E., Koldobskaya Y., Rice P.A., Piccirilli J.A.. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014; 10:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang Y.C., Sen D.. A contractile electronic switch made of DNA. J. Am. Chem. Soc. 2010; 132:2663–2671. [DOI] [PubMed] [Google Scholar]
- 25. Livshits G.I., Stern A., Rotem D., Borovok N., Eidelshtein G., Migliore A., Penzo E., Wind S.J., Di Felice R., Skourtis S.S. et al. . Long-range charge transport in single G-quadruplex DNA molecules. Nat. Nanotechnol. 2014; 9:1040–1046. [DOI] [PubMed] [Google Scholar]
- 26. Koizumi M., Koga R., Hotoda H., Momota K., Ohmine T., Furukawa H., Agatsuma T., Nishigaki T., Abe K., Kosaka T. et al. . Biologically active oligodeoxyribonucleotides - IX. Synthesis and anti-HIV-1 activity of hexadeoxyribonucleotides, TGGGAG, bearing 3΄- and 5΄-end-modification. Bioorg. Med. Chem. 1997; 5:2235–2243. [DOI] [PubMed] [Google Scholar]
- 27. Hazel P., Huppert J., Balasubramanian S., Neidle S.. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004; 126:16405–16415. [DOI] [PubMed] [Google Scholar]
- 28. Rachwal P.A., Brown T., Fox K.R.. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007; 581:1657–1660. [DOI] [PubMed] [Google Scholar]
- 29. Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010; 277:1107–1117. [DOI] [PubMed] [Google Scholar]
- 30. Avino A., Portella G., Ferreira R., Gargallo R., Mazzini S., Gabelica V., Orozco M., Eritja R.. Specific loop modifications of the thrombin-binding aptamer trigger the formation of parallel structures. FEBS J. 2014; 281:1085–1099. [DOI] [PubMed] [Google Scholar]
- 31. Phan A.T., Patel D.J.. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003; 125:15021–15027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sagi J. G-quadruplexes incorporating modified constituents: a review. J. Biomol. Struc. Dyn. 2013; 32:477–511. [DOI] [PubMed] [Google Scholar]
- 33. Gros J., Rosu F., Amrane S., De Cian A., Gabelica V., Lacroix L., Mergny J.L.. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007; 35:3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mekmaysy C.S., Petraccone L., Garbett N.C., Ragazzon P.A., Gray R., Trent J.O., Chaires J.B.. Effect of O6-methylguanine on the stability of G-quadruplex DNA. J. Am. Chem. Soc. 2008; 130:6710–6711. [DOI] [PubMed] [Google Scholar]
- 35. Dias E., Battiste J.L., Williamson J.R.. Chemical probe for glycosidic conformation in telomeric DNAs. J. Am. Chem. Soc. 1994; 116:4479–4480. [Google Scholar]
- 36. Xu Y., Noguchi Y., Sugiyama H.. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006; 14:5584–5591. [DOI] [PubMed] [Google Scholar]
- 37. Lech C.J., Cheow Lim J.K., Wen Lim J.M., Amrane S., Heddi B., Phan A.T.. Effects of site-specific guanine C8-modifications on an intramolecular DNA G-quadruplex. Biophys. J. 2011; 101:1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He G.X., Krawczyk S.H., Swaminathan S., Shea R.G., Dougherty J.P., Terhorst T., Law V.S., Griffin L.C., Coutre S., Bischofberger N.. N2- and C8-substituted oligodeoxynucleotides with enhanced thrombin inhibitory activity in vitro and in vivo. J. Med. Chem. 1998; 41:2234–2242. [DOI] [PubMed] [Google Scholar]
- 39. Koizumi M., Akahori K., Ohmine T., Tsutsumi S., Sone J., Kosaka T., Kaneko M., Kimura S., Shimada K.. Biologically active oligodeoxyribonucleotides. Part 12: N2-methylation of 2΄-deoxyguanosines enhances stability of parallel G-quadruplex and anti-HIV-1 activity. Bioorg. Med. Chem. Lett. 2000; 10:2213–2216. [DOI] [PubMed] [Google Scholar]
- 40. Liu X.Y., Kwan I.C.M., Wang S.N., Wu G.. G-quartet formation from an N-2-modified guanosine derivative. Org. Lett. 2006; 8:3685–3688. [DOI] [PubMed] [Google Scholar]
- 41. Kaucher M.S., Davis J.T.. N2, C8-disubstituted guanosine derivatives can form G-quartets. Tetrahedron Lett. 2006; 47:6381–6384. [Google Scholar]
- 42. Martic S., Liu X.Y., Wang S.N., Wu G.. Self-assembly of N-2-modified guanosine derivatives: Formation of discrete G-octamers. Chem.-Eur. J. 2008; 14:1196–1204. [DOI] [PubMed] [Google Scholar]
- 43. Plateau P., Gueron M.. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982; 104:7310–7311. [Google Scholar]
- 44. Mergny J.L., Phan A.T., Lacroix L.. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998; 435:74–78. [DOI] [PubMed] [Google Scholar]
- 45. Rachwal P.A., Fox K.R.. Quadruplex melting. Methods. 2007; 43:291–301. [DOI] [PubMed] [Google Scholar]
- 46. Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J.. Structure of the human telomere in K+ solution: An intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006; 128:9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adrian M., Heddi B., Phan A.T.. NMR spectroscopy of G-quadruplexes. Methods. 2012; 57:11–24. [DOI] [PubMed] [Google Scholar]
- 48. Do N.Q., Phan A.T.. Monomer-dimer equilibrium for the 5΄-5΄ stacking of propeller-type parallel-stranded G-quadruplexes: NMR structural study. Chem.-Eur. J. 2012; 18:14752–14759. [DOI] [PubMed] [Google Scholar]
- 49. Marttila A.T., Laitinen O.H., Airenne K.J., Kulik T., Bayer E.A., Wilchek M., Kulomaa M.S.. Recombinant NeutraLite Avidin: a non-glycosylated, acidic mutant of chicken avidin that exhibits high affinity for biotin and low non-specific binding properties. FEBS Lett. 2000; 467:31–36. [DOI] [PubMed] [Google Scholar]
- 50. Green N.M. Avidin and streptavidin. Methods Enzymol. 1990; 184:51–67. [DOI] [PubMed] [Google Scholar]
- 51. Laitinen O.H., Nordlund H.R., Hytonen V.P., Kulomaa M.S.. Brave new (strept)avidins in biotechnology. Trends Biotechnol. 2007; 25:269–277. [DOI] [PubMed] [Google Scholar]
- 52. Cheong V.V., Heddi B., Lech C.J., Phan A.T.. Xanthine and 8-oxoguanine in G-quadruplexes: formation of a G.G.X.O tetrad. Nucleic Acids Res. 2015; 43:10506–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lim K.W., Amrane S., Bouaziz S., Xu W.X., Mu Y.G., Patel D.J., Luu K.N., Phan A.T.. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009; 131:4301–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Y., Patel D.J.. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-Tetraplex. Structure. 1993; 1:263–282. [DOI] [PubMed] [Google Scholar]
- 56. Miyoshi D., Sugimoto N.. Molecular crowding effects on structure and stability of DNA. Biochimie. 2008; 90:1040–1051. [DOI] [PubMed] [Google Scholar]
- 57. Urry D.W., Peng S.Q., Gowda D.C., Parker T.M., Harris R.D.. Comparison of electrostatic-induced and hydrophobic-induced pKa shifts in polypentapeptides - the lysine residue. Chem. Phys. Lett. 1994; 225:97–103. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.