Abstract
Accurate chromosome segregation is essential for every living cell as unequal distribution of chromosomes during cell division may result in genome instability that manifests in carcinogenesis and developmental disorders. Irc5 from Saccharomyces cerevisiae is a member of the conserved Snf2 family of ATP-dependent DNA translocases and its function is poorly understood. Here, we identify Irc5 as a novel interactor of the cohesin complex. Irc5 associates with Scc1 cohesin subunit and contributes to cohesin binding to chromatin. Disruption of IRC5 decreases cohesin levels at centromeres and chromosome arms, causing premature sister chromatid separation. Moreover, reduced cohesin occupancy at the rDNA region in cells lacking IRC5 leads to the loss of rDNA repeats. We also show that the translocase activity of Irc5 is required for its function in cohesion pathway. Finally, we demonstrate that in the absence of Irc5 both the level of chromatin-bound Scc2, a member of cohesin loading complex, and physical interaction between Scc1 and Scc2 are reduced. Our results suggest that Irc5 is an auxiliary factor that is involved in cohesin association with chromatin.
INTRODUCTION
Genomic instability is a common feature of many cancers and developmental disorders. Faithful transmission of genetic information is thus of great importance as disturbances in DNA replication and chromosome segregation are the main factors that drive genomic instability. To ensure chromosome bi-orientation and equal division newly replicated sister chromatids are tethered together by a multi-protein complex called cohesin (1,2). The cohesin complex consists of three essential core components: two structural maintenance of chromosome proteins (SMC), Smc1 and Smc3, and non-SMC protein Scc1 (Rad21 in humans). Each SMC subunit is formed of N-terminal region containing Walker A motif, two long coiled-coil regions that are separated by hinge domain and C-terminal region containing Walker B motif. Coiled-coil self-folding brings N- and C-terminus into close proximity creating functional ATPase named the head domain (3). Smc1 and Smc3 form a heterodimer through an interaction between hinge and head domains (4). Scc1 creates a molecular bridge between Smc1 and Smc3 by interacting through its N-terminal part with a coiled-coil domain of Smc3 just above the Smc3 ATPase domain and by binding the Smc1 ATPase domain through its C-terminal part (5). These three proteins create a structure that entraps sister chromatids and holds them together (6,7). The cohesin complex has also other essential, stably associated regulatory subunits, Scc3 (SA1 and SA2 in humans) and Pds5 that interact with C-terminal and N-terminal part of Scc1, respectively (8). Another subunit called Wpl1 (WapI in Schizosaccharomyces pombe and humans) was shown to be unstably bound to cohesin through a complex interaction with Pds5, Scc3 and Smc3 ATPase domain (8,9).
Cohesins are loaded onto chromatin in late G1 by the Scc2/Scc4 loading complex (10). It has been proposed that Scc2/Scc4 induces cohesin ring folding that causes ATP hydrolysis. This weakens interaction between Smc1 and Smc3 head domains and triggers WapI-dependent dissociation of Scc1 from Smc3 allowing DNA entrapment (4). Cohesins become stably associated with chromatin in S phase through cohesion establishment factor Eco1 that acetylates two lysine residues, namely K112 and K113, on the Smc3 ATPase. This preserves the interactions between Smc1 and Smc3 heads and hinders WapI-dependent disruption of Scc1-Smc3 interface preventing cohesin removal from chromosomes (4,11,12). Smc3 acetylation together with Scc3 and Pds5 activities ensure stable cohesion between sister chromatids in G2 (13,14) until the onset of anaphase. Thereupon, the separase Esp1 cleaves Scc1 triggering Smc3 deacetylation by Hos1 and cohesin release from DNA, allowing proper chromatid separation and segregation (15–17).
Snf2-like proteins are ATP-dependent DNA translocases that slide on double-stranded DNA generating superhelical torsion that enables chromatin remodeling and protein removal from DNA (18). It has been demonstrated that chromatin remodelers play an important role in cohesion. In human cells, SNF2h complex was proposed to mediate cohesin loading onto chromatin (19). The similar activity was proposed for the RSC complex in Saccharomyces cerevisiae (20). Recently, it has been shown that RSC recruits Scc2/Scc4 to chromatin and at the same time, together with the loading complex, maintains nucleosome-free regions at Scc2/Scc4 binding sites creating the proper chromatin environment for cohesin loading (21). Irc5 (Increased Recombination Centers 5) is a putative Snf2-like DNA translocase from S. cerevisiae of yet unknown function (18). Irc5 was identified in a genome-wide screen for proteins involved in DNA damage response. Deletion of the IRC5 gene causes increased levels of spontaneous Rad52 foci and interhomolog recombination rates (22). In Neurospora crassa, mutation of Irc5 homolog, Mus30, results in sensitivity to several DNA damaging agents (23). On the other hand, human homolog of Irc5, the LSH/HELLS protein, has been reported to play a significant role in regulation of DNA methylation and DNA double-strand break repair (24,25).
Here, we show that lack of Irc5 causes decreased cohesin accumulation on chromatin resulting in premature sister chromatid separation and instability of rDNA region. We demonstrate that Irc5 interacts with the Scc1 cohesin subunit and that translocase activity of Irc5 is crucial for efficient accumulation of cohesin on the chromatin. Importantly, mutation of IRC5 also results in decreased recruitment of Scc2 to chromatin and impaired interaction between Scc2 and Scc1. Our results suggest that the DNA translocase Irc5 is important for cohesin association with chromosomes in yeast.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains used in this study are isogenic derivatives of W303 and are listed in Supplementary Table S1. Yeast cells were cultured in standard media at 28°C unless indicated otherwise. Deletion mutants were constructed using either PCR-based replacement method (26) or by genetic crossing of relevant mutants followed by tetrad dissection. Plasmids used in this work are detailed in Supplementary Table S2. The icr5DAEA mutant allele was generated using Quick Change Site-Directed Mutagenesis Kit (Stratagene) and verified by DNA sequencing.
DNA damage sensitivity tests
Mid-log phase cultures of relevant strains were 10-fold serially diluted and spotted onto YPD plates containing various concentrations of genotoxic agents.
RNA extraction, reverse transcription and quantitative PCR (qPCR)
To measure RSC8 mRNA level, total mRNA was isolated using RNeasy Mini Kit (Qiagen). Subsequently, 3 μg of RNA were treated with DNaseI (RNase-Free, Fermentas) following the manufacturer's instruction. Reverse transcription was performed with 1.5 μg of purified RNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instruction. qPCR reactions were done using LightCycler 480 System (Roche Applied Science) with 2xPCRMaster Mix SYBR Kit (A&A Biotechnology) using 1 μg of cDNA and the RSC8 primer set (Supplementary Table S3) in a total volume of 20 μl. All results were standardized using the reference gene IPP1, with the IPP1 primer set (Supplementary Table S3). The following conditions of amplifications were applied: 1 min at 95°C; 45 cycles of 10 s at 95°C, 10 s at 55°C and 20 s at 72°C. Measurements of RSC8 mRNA were repeated three times and every sample was used for qPCR twice.
Cohesion assay
To visualize premature sister chromatid cohesion with the TetO/TetR-GFP system, mid-log cells were arrested either in G1 with 5 μM α-factor or in G2/M with 15 μg/ml of nocodazole. The G1 and G2/M block was monitored microscopically by the formation of shmoos and large-budded cells, respectively. Next, cells were fixed with 4% paraformaldehyde, resuspended in SK buffer (1 M sorbitol, 0.05 M K2PO4) and stained with DAPI to visualize the nuclear DNA and to confirm preanaphase arrest. Cells were observed with the Axio Imager M1 epifluorescence microscope (Carl Zeiss, Germany) equipped with a 100x immersion oil objective (Plan-Neofluar 1006/1.30), the GFP and DAPI filter set and differential interference contrast (DIC). Z-stacked images were collected using AxioCam MRc digital color camera and processed with AxioVision 4.5 software. The number of GFP foci in mononuclear cells was scored for 300 cells in at least three independent experiments.
Analysis of rDNA stability
Genomic DNA was isolated using CHEF Genomic DNA Plug Kit (BioRad). Next, agarose-embedded DNA was digested with BamHI to excise rDNA region from chromosome XII and subjected to pulsed-field gel electrophoresis (PFGE) performed with CHEF-DR®III Pulsed Field Electrophoresis System (BioRad) at 6 V/cm, 14°C, 96° switch angle, for 24 h at a linear pulse of 1–2 min followed by Southern blot analysis using a probe against 35S rDNA.
Recombination assay
The rate of rDNA recombination was measured by scoring CanR Ade− colonies arising as a result of loss of CAN1-ADE1 marker genes inserted between rDNA repeats according to Burgess et al. (27).
Yeast two-hybrid assay (Y2H)
Matchmaker Two-Hybrid System 3 (Clontech) was used to perform yeast two-hybrid assay according to manufacturer's conditions. Indicated proteins were expressed in AH109 strain. Yeast were grown in the presence of 2 mM 3-amino-1,2,3-triazole. Plasmids used for Y2H are listed in Supplementary Table S2.
Co-immunoprecipitation (CoIP)
Native extracts for immunoprecipitation were prepared from 4 × 108 cells. Cell pellets were resuspended in IP buffer (20 mM Tris pH 8, 140 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA) containing 1 mM PMSF and protease inhibitors (Sigma-Aldrich, P8215). Next, cells were lysed with glass beads in a bead beater at 4°C. To exclude the possibility that interactions between proteins are mediated by DNA, cleared whole cell extracts (WCE) were first incubated with 300 μg/ml ethidium bromide for 3 h and then with anti-HA (Roche, 12CA5) or anti-Pk (Serotec, SV5-Pk1) antibody at 4°C overnight. Antibodies were captured with Protein G Dyneabeads (Invitrogen). After four washes with IP buffer proteins were eluted with Laemmli buffer (2% SDS, 20% glycerol, 120 mM Tris pH 6.8, 4% 2-mercaptoethanol, 0.01% bromophenol blue). 5% of WCE volume used for immunoprecipitation (input) together with CoIP eluates were resolved on Mini Protean TGX gels (BioRad) and transferred onto nitrocellulose membranes. Ponceau S stained membrane was cut and each fragment was incubated with appropriate antibody. FLAG-tagged proteins were detected with anti-FLAG antibodies (Sigma-Aldrich, F3165).
Chromatin-binding assays
To estimate the level of chromatin-bound Scc2-9Pk the chromatin fraction was extracted as previously described in details (28). To validate the fractionation every sample was analyzed by Western blot to detect cytoplasmatic (glucose-6-phosphate dehydrogenase antibody, anti-G6PDH, Sigma-Aldrich, A9521-1VL) and chromatin-bound (histone H2A, anti-H2A, 07-146 Millipore) proteins as markers. For quantification, the level of Scc2-9Pk bound to chromatin was normalized to histone H2A used as an internal loading control. Band quantification was performed using Image Lab software (Bio-Rad).
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed as described in Bennett et al. (29) with minor modifications. 4 × 108 cells were cross-linked with 1% formaldehyde for 20 min followed by 5 min incubation with 150 mM glycine. Next, cells were washed with ice-cold TBS and resuspended in 600 μl FA-lysis buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA) with addition of 1 mM PMSF and protease inhibitors (Sigma-Aldrich, P8215). Cells were lysed with glass beads in a bead beater at 4°C and then cell lysates were transferred to new tubes and sonicated to yield an average DNA size of 500 bp. Next, lysates were clarified by centrifugation, transferred to new tubes and additional 600 μl of FA-Lysis buffer was added. For IP reactions 4 μg of appropriate antibody was added to 500 μl of chromatin lysates and incubated at 4°C overnight. DNA-protein complexes were captured with Protein G Dyneabeads (Invitrogen) and sequentially washed with FA-lysis buffer, FA-500 buffer (50 mM HEPES–KOH pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA), LiCl wash buffer (10 mM Tris–HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA) and TE (10 mM Tris–HCl pH 7.5, 1 mM EDTA) on a rotator for 5 min each time. Complexes were then eluted with Elution Buffer (50 mM Tris–HCl pH 7.5, 1% sodium dodecylsulfate, 10 mM EDTA) by incubation at 65°C for 10 min. For input samples, 10 μl of cleared chromatin lysate were diluted in 490 μl of TE buffer and together with IP samples treated with 0.2 mg/ml Proteinase K for 5 h at 42°C followed by overnight incubation at 65°C. All samples were phenol–chloroform purified and ethanol precipitated in the presence of 20 μg/ml of glycogen. qPCR reactions were performed using both IP and input samples as templates, 2xPCRMaster Mix SYBR kit (A&A Biotechnology) and LightCycler 480 System (Roche Applied Science) in a total volume of 15 μl. Primers used for qPCR are listed in Supplementary Table S3. The following conditions of amplifications were applied: 1 min at 95°C; 45 cycles of 10 s at 95°C, 10 s at 56°C and 22 s at 72°C. The percentage (% input) value for each sample was calculated as follows: ΔCT [normalized ChIP] = CT [ChIP] − {CT [Input] − log2 (dilution factor)} and Input % = 100/2ΔCT [normalized ChIP]. The % input value represents the enrichment of protein at the specific locus and is normalized to ACT1 reference gene. All ChIP experiments were performed at least three times. qPCR reactions were performed three times for each sample.
RESULTS
Disruption of IRC5 causes increased sensitivity to MMS
To investigate the potential role of Irc5 in preservation of genomic integrity we first deleted the whole IRC5 open reading frame (ORF) in wild type diploid strain followed by tetrad dissection. We found that spores lacking IRC5 show slow growth phenotype (Supplementary Figure S1A). Moreover, the irc5Δ mutant exhibited increased sensitivity to genotoxins like camptothecin (CPT), phleomycin (PM), hydroxyurea (HU) and methyl methanesulfonate (MMS) (Supplementary Figure S1B). Next, we cloned the IRC5 gene on a centromeric plasmid and performed complementation test. It turned out that IRC5 on a plasmid failed to complement growth defect of irc5Δ mutant as well as increased sensitivity to CPT, PM, HU and MMS (Supplementary Figure S1B). The IRC5 gene is located on chromosome VI 194 bp upstream from the RSC8 gene which is positioned on the Crick strand and expressed in the opposite direction (Supplementary Figure S1C). Because deletion of RSC8 encoding a subunit of RSC chromatin remodeling complex is lethal (30), we hypothesized that the observed fitness defect in the irc5Δ mutant may result from downregulation of RSC8 transcription. To test this, we measured RSC8 mRNA levels in wild type and irc5Δ cells. We found that irc5Δ had ∼70% less of RSC8 mRNA than wild type strain confirming that IRC5 deletion interferes with RSC8 transcription (Supplementary Figure S1D). Next, we transformed the irc5Δ mutant with plasmids carrying DNA fragments of chromosome VI containing both IRC5 and RSC8 genes or the RSC8 gene alone (Supplementary Figure S1B,C). The RSC8 gene expressed from the plasmid fully complemented slow growth and increased sensitivity to HU, CPT and PM in the irc5Δ mutant. Importantly, only the plasmid bearing both IRC5 and RSC8 reversed sensitivity to MMS to wild type level (Supplementary Figure S1B). This suggests that not only Rsc8 but also Irc5 is needed for coping with MMS-induced DNA damage.
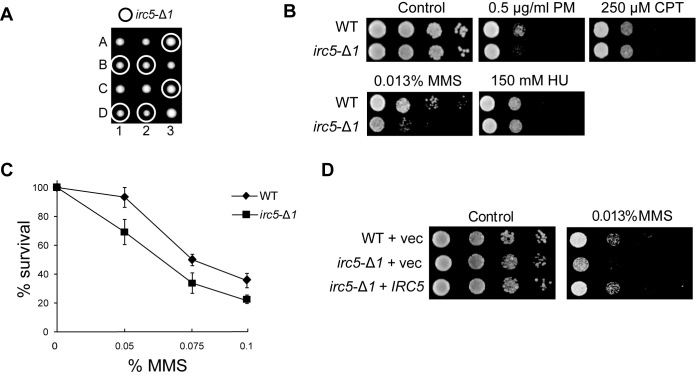
To obtain a mutant strain in which the IRC5 gene is disrupted without affecting RSC8 transcription, we deleted a 3΄ end of the IRC5 ORF from 901 to 2562 bp (relative to start codon). This fragment corresponds to amino acid residues from 235 to 853, which include the sequence coding SNF2_N and Helic_C domains that are crucial for DNA translocase activity (18). The resulting irc5-Δ1 mutant grew normally (Figure 1A) and had wild type level of RSC8 mRNA (Supplementary Figure S1D). Next, we analyzed irc5-Δ1 sensitivity to DNA damaging agents. The irc5-Δ1 mutant was not sensitive to CPT, PM and HU (Figure 1B). However, it was mildly sensitive to MMS when chronically treated on the plates compared to known MMS sensitive mutants like rad9Δ and srs2Δ (Figure 1B and Supplementary Figure S2). The irc5-Δ1 mutant also showed decreased survival after short-term acute exposure to MMS in liquid media (Figure 1C). To confirm that the increased sensitivity of irc5-Δ1 mutant to MMS was indeed the result of IRC5 disruption we performed a complementation test. We found that IRC5 on a plasmid restored resistance to MMS to wild type level (Figure 1D). Thus, in the subsequent set of experiments we used the irc5-Δ1 mutant instead of irc5Δ.
Figure 1.
Lack of Irc5 results in mild sensitivity to MMS. (A) The irc5-Δ1 mutant does not exhibit growth defect. Disruption of 3΄ end of IRC5 ORF was introduced into diploid wild type strain followed by sporulation and tetrad dissection. (B–D) The irc5-Δ1 mutation causes increased sensitivity to MMS. Wild type and the irc5-Δ1 mutant were analyzed by spot assay (B, D) and exposed to high concentrations of MMS for 1 h to determine survival rate (C). The bar represents the mean value ± standard error of mean.
Irc5 is important for sister chromatid cohesion
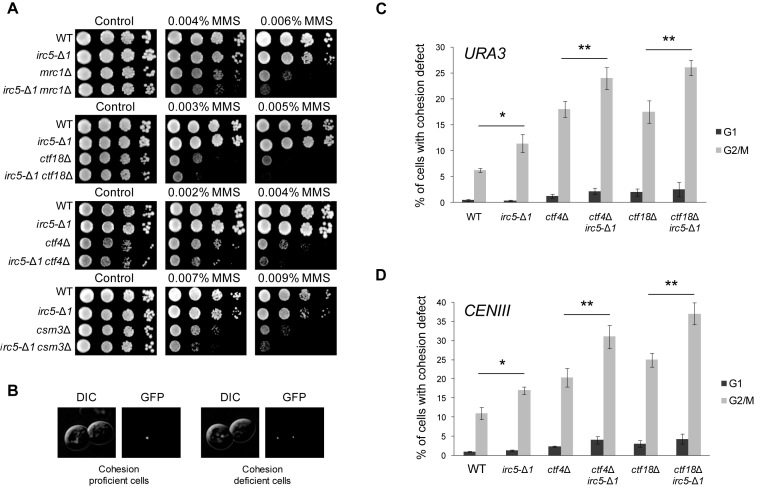
It has been demonstrated that chromatin remodelers of the Snf2 family play crucial roles in many processes that help to preserve genome stability (31–35). To determine in which pathway Irc5 works, we performed epistasis analysis between IRC5 and several genes involved in various aspects of DNA metabolism. irc5-Δ1 showed no genetic interactions with genes encoding DNA damage checkpoint activators (MEC1, RAD9, RAD24), genes involved in DNA damage tolerance (RAD18, RAD5, SGS1) or RAD27 nuclease. On the other hand, simultaneous disruption of IRC5 together with genes involved in homologous recombination (RAD52, RAD59, RAD51, RAD57) and encoding DNA helicase SRS2 and DNA nuclease MUS81 caused increased sensitivity to MMS compared to single mutants (data not shown). Interestingly, irc5-Δ1 showed negative genetic interactions also with MRC1, CTF18, CTF4 and CSM3 that encode proteins mediating replication fork stability and progression (Figure 2A). Interestingly, all of these genes were identified as accessory factors required for sister chromatid cohesion (SCC) establishment and divided into two groups based on genetic interactions between them. Genes without additive cohesion defect were classified into the same pathway, while genes that once combined show higher levels of premature chromatid separation were assigned to different cohesion pathways. First group contains Csm3, Tof1, Ctf4 and Chl1 whereas the second one includes Ctf8, Ctf18, Ddc1 and Mrc1 (36). Moreover, it has been previously reported that survival of cohesin mutants strongly depends on replication fork mediators (37). Taking our data into account, we decided to investigate the potential role of Irc5 as a cohesion factor.
Figure 2.
Lack of Irc5 results in premature sister chromatid separation. (A) Negative genetic interactions between irc5-Δ1 and deletion mutants lacking non-essential cohesion factors (mrc1Δ, ctf18Δ, ctf4Δ, csm3Δ). Indicated single and double mutants were analyzed by spot assay in the presence of MMS. (B) Representative pictures of sister chromatid proficient and deficient cells. (C, D) Irc5 plays a role in sister chromatid cohesion. Wild type and indicated mutant strains were arrested in G1 with α-factor or in G2/M with nocodazole. The number of GFP spots corresponding to URA3 (C) and CENIII (D) loci was scored for at least 300 cells in three independent experiments. Asterisks indicate statistically significant differences (*P < 0.05,**P < 0.01; two-tailed Student's t test). The bar represents the mean value ± standard error of mean.
Strains lacking genes involved in cohesion are characterized by premature sister chromatid separation (36,38) so we seek to determine whether the irc5-Δ1 mutant displays such phenotype. To do this, we performed a cohesion assay employing strains in which Tet operator (TetO) repeats are integrated at the URA3 locus or near the centromere of chromosome III and Tet repressor is tagged with GFP (TetR-GFP). When sister chromatids are tightly cohered, Tet-O arrays coated by TetR-GFP appear as a single fluorescent dot (Figure 2B). Precocious sister chromatids separation is manifested by the appearance of two fluorescent dots (Figure 2B). Tested yeast strains were arrested at G2/M with nocodazole or in G1 with α-factor to exclude the possibility that differences between strains are caused by preexisting aneuploidy. Next, the number of GFP foci per cell was determined. When we examined sister chromatid cohesion at URA3 locus we found that in wild type, two GFP foci were evident in 6% of cells while in the irc5-Δ1 strain 12% of cells displayed premature sister chromatid separation suggesting that Irc5 is important for SCC along chromosome arms (Figure 2C). Consistent with the results of cohesion assay at the URA3 locus, disruption of IRC5 also led to significant premature loss of cohesion at the centromere of chromosome III (Figure 2D). Importantly, these results were also confirmed using a strain in which metaphase arrest was induced by depletion of CDC20 and cohesion was examined at the centromere of chromosome IV in the presence of intact mitotic spindle (Supplementary Figure S3). To determine in which SCC pathway Irc5 works, we constructed double mutants devoid of IRC5 and CTF4 or CTF18 genes that represent parallel pathways for sister chromatid establishment and performed sister chromatid assay at both URA3 and CEN3 loci. In agreement with MMS sensitivity results (Figure 2A), loss of Irc5 exacerbated cohesion defect of both ctf4Δ as well as ctf18Δ cells at centromere and chromosome arm locations suggesting that Irc5 plays a role in both SCC pathways (Figure 2C and D).
Irc5 interacts with Scc1 cohesin subunit and promotes efficient cohesin binding to chromatin
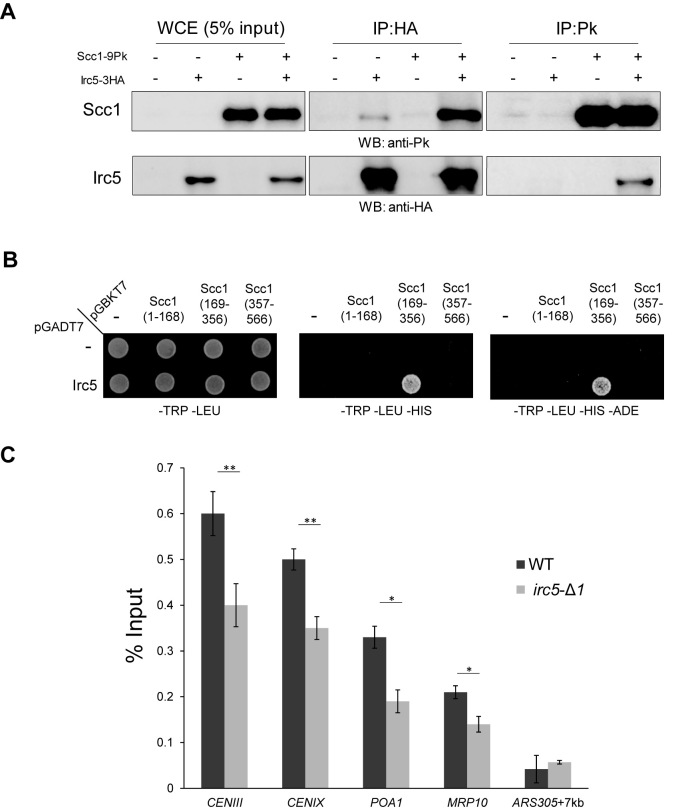
As our data show that Irc5 is needed for efficient sister chromatid cohesion we asked whether it may directly interact with components of cohesin complex. First, we fused 3HA tags to the C-terminus of Irc5. Irc5-3HA cells were grown in YPD medium until they reached mid-log phase and then they were collected for protein preparation and Western blot analysis with anti-HA antibodies. Surprisingly, we detected a band with a molecular weight (MW) of about 120 kDa instead of predicted 100 kDa for Irc5-3HA (Supplementary Figure S4A). Interestingly, after immunoprecipitation of Irc5-3HA, two bands could be detected – one migrating with a molecular weight of about 120 kDa as above and the much weaker second band that was detected at ∼90 kDa (Supplementary Figure S4B). Importantly, using mass spectrometry analysis we confirmed the presence of Irc5 protein in both bands (data not shown). These results suggest that Irc5 occurs in two forms and that the heavier is the predominant one. We also tagged the N-terminus of Irc5 with 7HA and expressed it under the control of GAL1 promoter. Under inducible conditions we were able to immunoprecipitate only one form of Irc5 migrating approximately at 128 kDa (Supplementary Figure S4C). This result suggests that the ∼90 kDa form of Irc5 might be a product of proteolytic cleavage of the N-terminal part of Irc5. Nevertheless, the significance of this modification remains to be established.
To investigate whether Irc5 interacts with cohesin subunits we performed co-immunoprecipitation (CoIP) assay. We found that Irc5 co-purifies with Scc1. Scc1 also associated with the immunoprecipitated Irc5 (Figure 3A). Importantly, it seems unlikely that the interaction between Irc5 and Scc1 was mediated by DNA since addition of ethidium bromide does not interfere with observed binding. We noticed that while Scc1 co-purified efficiently with Irc5, relatively small amount of Irc5 precipitated with Scc1 (Figure 3A). To clarify this, we compared the efficiency of Irc5-3HA and Scc1-9Pk immunoprecipitation. After IP reaction (Post-IP) only minor levels of Irc5-3HA remained in the whole cell extracts (WCE) showing that most of Irc5 was pulled down. On the other hand, majority of Scc1 was still present in WCE suggesting that the minor fraction of the total Scc1 was precipitated (Supplementary Figure S5). We conclude that the low levels of Irc5 that co-purifies with Scc1 may be at least partially caused by incomplete immunoprecipitation of the latter. Moreover, to examine the specificity of the interaction between Irc5 and Scc1 we performed yeast two-hybrid assay. We confirmed that Irc5 associates with Scc1. Additionally, we mapped Irc5-Scc1 interaction site to the central part of Scc1 (Figure 3B). Next, we examined whether cohesin binding to chromosomes is affected when Irc5 is absent. For this purpose we used chromatin immunoprecipitation (ChIP) to evaluate the cohesin subunit Scc1 binding to the centromeres and known cohesin binding sites. We found that the cohesin levels were reduced at the centromeres (CENIII and CENIX) as well as the chromosome arms (MRP10 and POA1) suggesting that the absence of Irc5 has a negative impact on cohesin binding to chromosomes (Figure 3C).
Figure 3.
Irc5 interacts with the Scc1 cohesin subunit. (A) Irc5-3HA and Scc1-9Pk were immunoprecipitated with anti-HA or anti-Pk antibody, respectively, and analyzed by Western blot. Protein levels in the whole cell extract (WCE) (5% of the input) are also shown. (B) Irc5 binds to the central part of Scc1 (aa 169–356) as shown by yeast two-hybrid assay. (C) Reduced levels of the Scc1 cohesin subunit at centromeres and chromosome arms in irc5-Δ1 cells. ChIP was performed in logarithmically growing wild type and irc5-Δ1 cells. Genomic regions corresponding to centromeres on chromosomes III and IX as well as cohesin binding sites on chromosome arms (MRP10, POA1) were amplified in qPCR. Asterisks indicate statistically significant differences (*P < 0.05,**P < 0.01; two-tailed Student's t test). The bar represents the mean value ± standard error of mean.
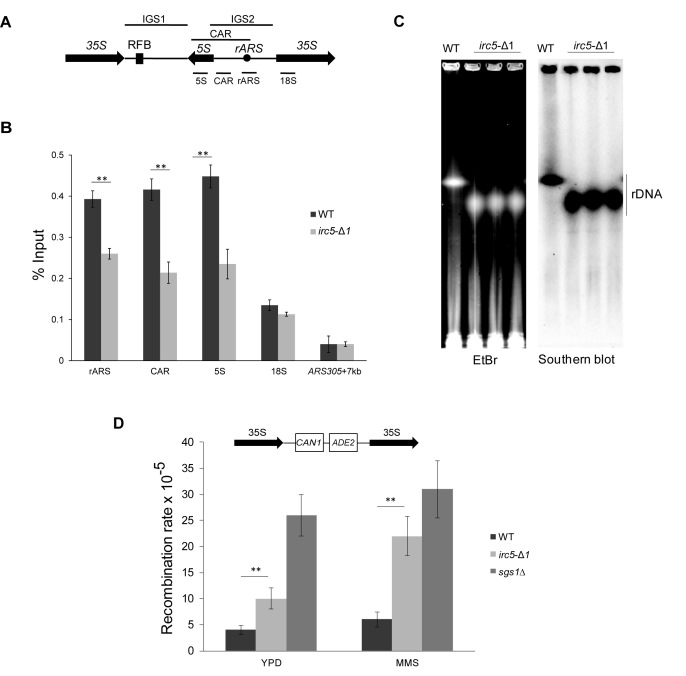
Irc5 contributes to rDNA stability
In S. cerevisiae rDNA is located on the chromosome XII where it forms a cluster of ∼150 copies of genes encoding ribosomal RNA. Every copy consists of two intergenic spacers (IGS1 and IGS2) and two transcription units containing 35S and 5S rDNA (Figure 4A). Because of the highly repetitive nature, rDNA is vulnerable to the rDNA gene copy loss through the recombination based mechanisms (39). To maintain the proper amount of rDNA gene copies cohesins bind to chromatin located in IGS2 (CAR) and impose equal sister-chromatid exchange (40). Depletion of cohesins in the rDNA region increases recombination rate leading to a rDNA gene copy loss (40,41). To determine the role of Irc5 in the rDNA maintenance, we examined the level of Scc1 occupancy in the rDNA cohesin binding region (rARS, CAR, 5S) and a control locus 18S in wild type and irc5-Δ1 cells. Cells lacking Irc5 had ∼40% less cohesins associated with rDNA compared to wild type (Figure 4B). This was accompanied with the loss of rDNA repeats (Figure 4C) and increased recombination in the rDNA array especially when the irc5-Δ1 mutant was treated with replication blocking agent MMS (Figure 4D). These results provide an additional piece of evidence suggesting Irc5 role in cohesin binding to chromatin.
Figure 4.
Cohesion defect in irc5-Δ1 cells causes rDNA instability. (A) Schematic representation of rDNA region in S. cerevisiae. Fragments amplified by ChIP primer sets (5S, CAR, rARS, 18S) are indicated as short bars below the scheme of rDNA region. IGS1, intergenic spacer 1, IGS2, intergenic spacer 2, RFB, replication fork barrier, CAR, cohesion-associated region. (B) Disruption of IRC5 reduces the Scc1 cohesin subunit levels at rDNA region. Mid-log phase wild type and irc5-Δ1 cells were used for ChIP. Primer sets (5S, CAR, rARS) covering rDNA cohesin association region (CAR) were used together with the 18S primer set covering more distal region. Asterisks indicate statistically significant differences (*P < 0.05,**P < 0.01; two-tailed Student's t test). The bar represents the mean value ± standard error of mean. (C) Lack of Irc5 causes loss of rDNA copies. Whole chromosomes were isolated from wild type and three independent irc5-Δ1 clones, digested with BamHI to excise rDNA region and subjected to PFGE followed by Southern blot analysis to detect rDNA region. (D) Elevated levels of recombination at rDNA region in irc5-Δ1 cells. Mid-log wild type and irc5-Δ1 cells were treated with 0.01% MMS for 1 h or left untreated. sgs1Δ was used as a positive control of mutant with increased levels of rDNA recombination. The bar represents the mean value ± standard error of mean.
Translocase activity of Irc5 is important for efficient cohesin accumulation on chromatin
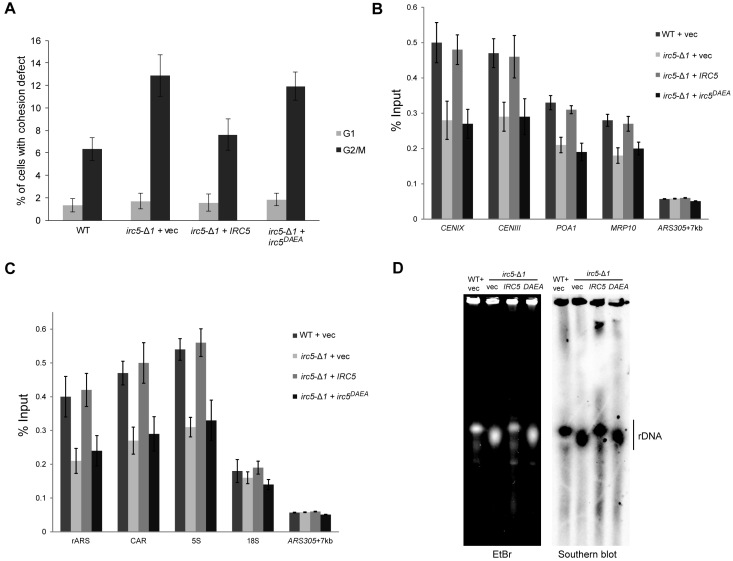
To determine the contribution of the ATP-driven translocase activity of Irc5 in sister chromatid cohesion we mutated two highly conservative amino acid residues (D352A, E353A) in the SNF2_N domain to obtain ATPase deficient variant (irc5DAEA) (42). Importantly, irc5DAEA was stably expressed at levels equal to wild type (Supplementary Figure S6). First, we analyzed irc5DAEA mutant sensitivity to MMS. Yeast cells expressing irc5DAEA behaved like the irc5-Δ1 mutant showing increased sensitivity to MMS (Supplementary Figure S7). This suggests that ATP-dependent translocase activity of Irc5 is required for MMS tolerance. To determine if translocase activity of Irc5 is important for proper sister chromatid cohesion, we performed cohesion assay. We found that irc5-Δ1 cells expressing irc5DAEAmutant had a similar cohesion defect as irc5-Δ1 with control vector (Figure 5A). Moreover, irc5DAEA mutant caused the same reduction of cohesin level at centromeres and chromosome arms as IRC5 disruption (Figure 5B). Finally, we showed that lack of translocase activity also reduces the levels of cohesin at rDNA (Figure 5C) leading to rDNA copy loss (Figure 5D). Taken together, these results indicate that translocase activity of Irc5 is required for cohesin accumulation on chromatin.
Figure 5.
The role of Irc5 translocase activity in cohesion. (A) irc5DAEA shows cohesion defect. Wild type and indicated mutant strains were arrested in G1 with α-factor or in G2/M with nocodazole. The number of GFP spots corresponding to URA3 loci was scored for at least 300 cells in three independent experiments. Differences between WT + vec and irc5-Δ1 + vec, WT+vec and irc5DAEA as well as irc5-Δ1 + IRC5 and irc5-Δ1 + vec, irc5-Δ1 + IRC5 and irc5DAEA were statistically significant (P < 0.05; two-tailed Student's t test). (B) Reduced levels of the Scc1 cohesin subunit at centromeres and chromosome arms in irc5DAEA cells. ChIP was performed in logarithmically growing wild type and irc5-Δ1 cells carrying indicated plasmids. Genomic regions corresponding to centromeres on chromosomes III and IX as well as cohesin binding sites on chromosome arms (MRP10, POA1) were amplified in qPCR. Differences between WT + vec and irc5-Δ1 + vec, WT + vec and irc5DAEA as well as irc5-Δ1 + IRC5 and irc5-Δ1 + vec, irc5-Δ1 + IRC5 and irc5DAEA were statistically significant (P < 0.05 for POA1 and MRP10 ChIP, P < 0.01 CENIX and CENIII ChIP; two-tailed Student's t test). The bar represents the mean value ± standard error of mean. (C) Lack of Irc5 translocase activity causes reduction in the Scc1 cohesin subunit levels at rDNA region. Mid-log phase wild type and irc5-Δ1 cells bearing indicated plasmids were used for ChIP. Primer sets (5S, CAR, rARS) covering rDNA cohesin association region (CAR) were used together with the 18S primer set covering more distal region. Differences between WT + vec and irc5-Δ1 + vec, WT + vec and irc5DAEA as well as irc5-Δ1 + IRC5 and irc5-Δ1 + vec, irc5-Δ1 + IRC5 and irc5DAEA were statistically significant (P < 0.01; two-tailed Student's t test). The bar represents the mean value ± standard error of mean. (D) The D352A, E353A mutation causes loss of rDNA copies. Whole chromosomes were isolated from indicated strains, digested with BamHI and subjected to PFGE followed by Southern blot analysis to detect rDNA region.
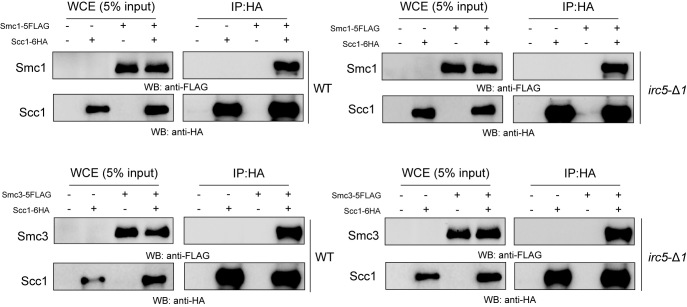
Stable cohesin ring formation in cells lacking Irc5
Cohesin ring consists of three essential core subunits: Smc1, Smc3 and Scc1. These proteins form a structure that entraps sister chromatids and holds them together (6,7). We hypothesized that decreased level of cohesin bound to chromatin in cells lacking Irc5 may be caused by disrupted interaction between Scc1, Smc1 and Smc3. To test this, we decided to perform CoIP assay between Scc1 and Smc1 as well as Scc1 and Smc3 (Figure 6). We found that all interactions studied were unaffected by IRC5 disruption. We infer from this result that Irc5 does not participate in cohesin ring formation or stability (Figure 6).
Figure 6.
The interaction between core cohesin subunits is not altered in cells lacking IRC5. Scc1-6HA was immunoprecipitated with anti-HA antibody and analyzed by western blot using anti-FLAG or anti-HA antibodies. Protein levels in the whole cell extract (WCE) (5% of the input) are also shown.
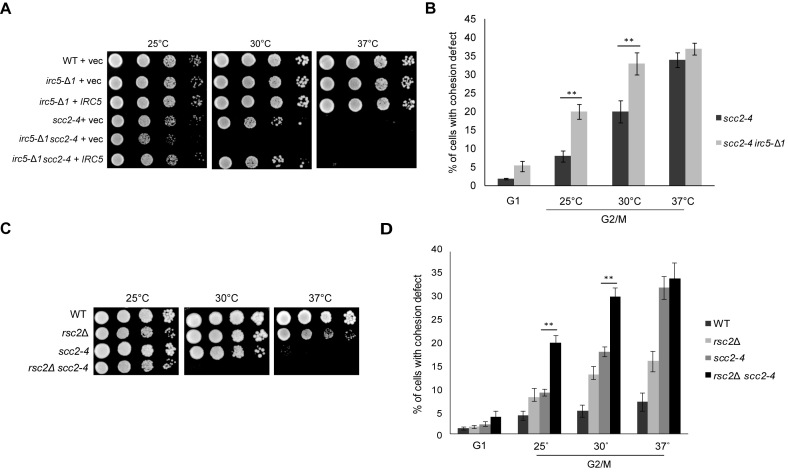
IRC5 shows genetic interaction with SCC2 cohesin loading complex subunit.
Cohesins are deposited onto chromatin through a complex interaction between the cohesin subunits, the Scc2/Scc4 cohesin loader complex and the RSC chromatin remodeler (7,20,21). Thus, we sought to determine the potential interaction between Irc5 and Scc2/Scc4. First, we performed epistasis analysis between SCC2 and IRC5. Because deletion of SCC2 is lethal we used a temperature-sensitive allele scc2-4 (43). The scc2-4 irc5-Δ1 double mutant grew slower compared to respective single mutants at permissive temperature (25°C) and failed to grow at semi-permissive temperature (30°C) (Figure 7A). Importantly, IRC5 on a plasmid restored the grow of scc2-4 irc5-Δ1 mutant at 30°C to the scc2-4 level, indicating that observed lethality was indeed the result of IRC5 disruption (Figure 7A). Next, we performed cohesion assay and found that at 25°C about 20% of scc2-4 irc5-Δ1 cells displayed cohesion defect compared to 8% in scc2-4. At 30°C, ∼33% of scc2-4 irc5-Δ1 cells showed premature separation of chromatids while in the scc2-4 mutant it was 20% (Figure 7B). Interestingly, disruption of RSC chromatin remodeling complex also led to scc2-4 mutant lethality ((44) and Figure 7C) and increased cohesion defect (Figure 7D). To investigate whether Irc5 binds to cohesin loader subunits Scc2 and Scc4 we performed CoIP assay. We found no evidence for physical interaction between Irc5 and Scc2 or Scc4 (data not shown). Together, these results suggest that Irc5 may promote cohesion in parallel to Scc2/Scc4.
Figure 7.
Irc5 and RSC complex are important for scc2-4 survival (A) Irc5 is required for scc2-4 viability. 10-fold dilutions of strains transformed with indicated plasmids were spotted on plates and incubated at permissive (25°C), semi-permissive (30°C) or restrictive temperature (37°C). (B) The irc5-Δ1 mutation increases cohesion defect of scc2-4 cells. To perform cohesion assay at URA3 locus, mid-log scc2-4 and scc2-4 irc5-Δ1 cells pre-cultured at 25°C were arrested in G1 or G2/M at 25°C, 30°C or 37°C. The number of GFP spots was scored for at least 300 cells in three independent experiments. Asterisk indicate statistically significant differences (**P < 0.01; two-tailed Student's t test). The bar represents the mean value ± standard error of mean. (C) Impact of RSC2 deletion on scc2-4 survival. 10-fold dilutions of indicated strains were spotted on plates and incubated at permissive (25°C), semi-permissive (30°C) or restrictive temperature (37°C). (D) Deletion of RSC2 exacerbates cohesion defect of scc2-4 cells. To perform cohesion assay at URA3 locus, mid-log WT, rsc2Δ, scc2-4 and rsc2Δ scc2-4 cells pre-cultured at 25°C were arrested in G1 or G2/M at 25°C, 30°C or 37°C. The number of GFP spots was scored for at least 300 cells in three independent experiments. Asterisk indicate statistically significant differences (**P < 0.01; two-tailed Student's t test). The bar represents the mean value ± standard error of mean.
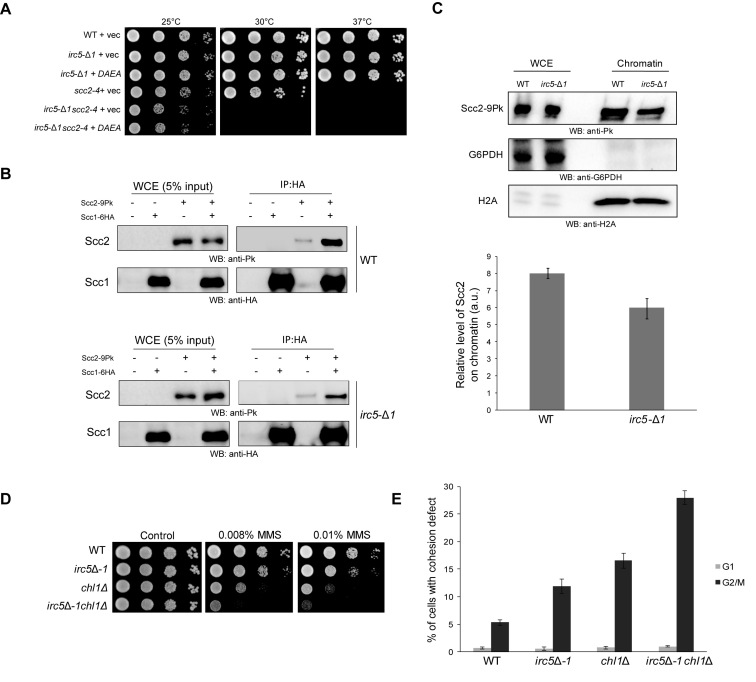
Irc5 promotes Scc2/Scc4 association with chromatin and interaction with Scc1
Cohesin loading is a complex process that requires optimal chromatin environment (7,20,21). As our results showed that chromatin remodeling activity of Irc5 is required for cohesin accumulation on chromatin (Figure 5B and C) we speculated that Irc5 may provide such conditions. First, we investigated if Irc5 translocase-deficient mutant supports scc2-4 irc5-Δ1 viability. The irc5DAEA mutant allele failed to complement scc2-4 irc5-Δ1 lethality at semi-permissive temperature suggesting that translocase activity of Irc5 is essential for scc2-4 viability at 30°C (Figure 8A). It was shown that cohesin loading is mediated by numerous contacts between cohesin subunits and cohesin loader complex (7) so we seek to determine if lack of Irc5 disrupts these interactions. Because we showed that Irc5 interacts with Scc1 (Figure 3A,B) we examined the association of Scc1 with Scc2. Lack of Irc5 reduced the interaction between Scc1 and Scc2 (Figure 8B). Next, we examined whether Irc5 is important for Scc2 binding to chromatin using chromatin-binding assay. We found that in irc5-Δ1 cells less Scc2 was bound to chromatin compared to wild type (Figure 8C). These results suggest that Irc5 may promote cohesion loading by facilitating association of the Scc2/Scc4 cohesin loader onto chromatin and supporting its interaction with cohesin.
Figure 8.
Irc5 is important for Scc2 interaction with chromatin. (A) Translocase activity of Irc5 is important for scc2-4 survival. Ten-fold dilutions of strains transformed with indicated plasmids were spotted on plates and incubated at permissive (25°C), semi-permissive (30°C) or restrictive temperature (37°C). (B) Lack of Irc5 disrupts interaction between Scc1 and Scc2. Scc1-6HA was immunoprecipitated with anti-HA and analyzed by Western blot using anti-HA and anti-Pk antibodies. Protein levels in the whole cell extract (WCE) (5% of the input) are also shown. (C) Irc5 mediates Scc2 binding to chromatin. Whole cell extracts and chromatin fractions were isolated from mid-log wild type and irc5-Δ1 cells and analyzed by western blot. For quantification, the level of Scc2 present in chromatin fraction was normalized to the level of histone H2A used as an internal loading control. The bar represents the mean value ± standard error of mean. (D) Negative genetic interactions between irc5-Δ1 and chl1Δ mutant. Indicated single and double mutants were analyzed by spot assay in the presence of MMS. (E) Disruption of IRC5 increases cohesion defect of chl1Δ mutant. Wild type and indicated mutant strains were arrested in G1 with α-factor or in G2/M with nocodazole. The number of GFP spots corresponding to URA3 loci was scored for at least 300 cells in three independent experiments. Differences between irc5-Δ1, chl1Δ and irc5-Δ1chl1Δ were statistically significant (P < 0.01; two-tailed Student's t test). The bar represents the mean value ± standard error of mean.
Finally, we analyzed the interactions between Irc5 and Chl1 DNA helicase that was proposed to be required for Scc2/Scc4 association with chromatin. Deletion of CHL1 results in decreased Scc2 levels on the chromatin, premature sister-chromatid cohesion and increased DNA damage sensitivity (45). We found that disruption of IRC5 resulted in increased sensitivity of chl1Δ mutant to MMS (Figure 8D). Moreover, loss of Irc5 exacerbated cohesion defect of chl1Δ cells (Figure 8E). This suggests that Chl1 and Irc5 act in separate cohesion pathways.
DISCUSSION
In this study, we identified a Snf2-related DNA translocase Irc5 as a novel interactor of cohesin. We show that disruption of IRC5 led to cohesin loss at centromeres as well as along chromosome arms (Figure 3C), a phenotype that is shared with mutants lacking genes essential for cohesin loading like Scc2, Scc3 or Chl1 (7,10,45,46) or chromatin remodelers (19–21). Consequently, disruption of IRC5 is accompanied by mild premature sister chromatid separation also at both locations (Figure 2C, D and Supplementary Figure S3). It is not clear why Irc5 loss results only in relatively weak cohesion defect. One possible explanation is that other chromatin remodelers can largely compensate for IRC5 deletion. On the other hand, it was shown that the reduction of intracellular cohesin concentrations to about 13% of wild type levels do not cause premature sister chromatid separation either at centromeres or chromosome arms. Interestingly, the same conditions strongly impact the rDNA stability and result in increased sensitivity to DNA damaging agents (41). These results indicate that some cohesin functions can be executed even at low levels of chromatin-bound cohesins while other require wild type cohesin levels on chromatin. Thus, Irc5 may be important for sister chromatid cohesion only in a subset of genomic locations or certain conditions. In agreement with these observations, we showed that Irc5 plays an important role in maintaining integrity of the rDNA region. IRC5 disruption caused significant reduction of cohesin bound to chromatin in the rDNA (Figure 4B) resulting in increased rates of unequal sister chromatid recombination that is favored in the absence of cohesin in the rDNA region (40,41) (Figure 4D). This led to rDNA repeats loss and destabilization of this region in irc5-Δ1 (Figure 4C). We also showed that Irc5 and its translocase activity are required for MMS tolerance and rDNA stability during replication stress (Figures 1B, C, 4D, Supplementary Figure S7). It was reported that cohesin accumulate at early origins upon MMS and HU treatment and cohesin-deficient cells exhibit strong defects in replication recovery after replication fork arrest resulting in increased sensitivity to these agents (47). It is possible that increased sensitivity of irc5-Δ1 cells to MMS is a result of reduced cohesin levels at stalled replication forks.
Cohesins are loaded onto chromatin by Scc2/Scc4 complex that interacts with cohesin ring at multiple sites within Smc1, Smc3, Scc1 and Scc3 cohesin subunits (7). It was also reported that RSC chromatin remodeling complex is essential for cohesin association with chromatin. RSC was shown to recruit cohesin loading complex to nucleosome-free regions that RSC itself helps to maintain (21). Interestingly, IRC5 shows strong genetic interaction with the Scc2/Scc4 cohesin loading complex. We demonstrate that survival of scc2-4 mutant at semi-permissive conditions depends on Irc5 presence (Figure 7A) and that disruption of IRC5 in scc2-4 background increases the level of premature sister chromatid separation (Figure 7B). Interestingly, RSC complex is also required for scc2-4 survival ((44) and Figure 7C). Moreover, lack of Irc5 led to decreased accumulation of Scc2 on chromatin (Figure 8C) and reduced interaction between Scc1 and Scc2 (Figure 8B). Importantly, we also show that translocase activity of Irc5 is important for proper cohesin accumulation on chromatin (Figure 5B, C) and scc2-4 survival (Figure 8A). These results indicate that, similarly to the RSC complex (21), Irc5 might change chromatin structure to create optimal environment for Scc2/Scc4-dependent cohesin loading. Overall, these observations underscore the role of chromatin remodeling DNA translocases as key factors that promote cohesion alongside with Scc2/Scc4 complex and Scc3. Our model predicts that lack of IRC5 results in decreased levels of cohesin loaded onto chromatin. Therefore, another factor that impairs cohesin loading should produce synergistic effects resulting in decreased cohesin occupancy on chromosomes and increased level of premature sister chromatid separation. In agreement with this assumption scc2-4 mutation or chl1Δ deletion together with IRC5 disruption exacerbates cohesion defects (Figures 7B and 8E). If the level of cohesin loaded on chromatin reaches breaking point, cells do not survive as seen for the scc2-4 irc5-Δ1 or scc2-4 rsc2Δ mutants (Figure 7A and C). Such interpretation would also explain genetic interactions between irc5-Δ1 allele and deletion mutants in both groups of non-essential cohesin establishment factors like ctf4Δ and ctf18Δ (Figure 2A, C, D). Simultaneous disruption of cohesion establishment and cohesin loading process would likely produce synergistic effects manifested by increased level of premature sister chromatid separation and increased sensitivity to DNA damage. Alternatively, but not exclusively, lack of cohesion-independent functions of ctf4Δ and ctf18Δ that are also required for MMS tolerance, may be partially responsible for increased sensitivity of double mutants.
Interestingly, we showed that, like other chromatin remodelers (19,20), Irc5 associates directly with Scc1 cohesin subunit (Figure 3A and B). It is not clear however, what is the exact role of this interaction. It seems that RSC is crucial for Scc2/Scc4 interaction with chromatin, placing its role in an upstream process of chromatin structure preparation for cohesin loading (21). At this stage interaction with cohesin may not be required. We propose that chromatin remodelers may play a dual role in sister chromatid cohesion: first, they can prepare chromatin for Scc2/Scc4-dependent cohesin loading, and second, they can be recruited to cohesin after its loading for example to help to pass different obstacles like nucleosomes or DNA-bound proteins and enable efficient cohesin translocation along chromosomes. In sum, our results suggest that Irc5 enables cohesin association with chromatin through its translocase activity. Lack of IRC5 causes suboptimal conditions for Scc2/Scc4 binding to chromatin and results in disrupted interaction of cohesin loading complex with cohesin. This leads to decreased cohesin levels at centromeres, chromosome arms and rDNA, leading to premature sister chromatid separation and rDNA instability.
Mutations in SNF2-N or Helic_C domains of human Irc5 homolog LSH/HELLS are found in acute myelogenous and lymphoblastic leukemia cells (48) as well as Immunodeficiency, Centromeric Instability and Facial Anomalies (ICF) syndrome patients (49). Interestingly, null mutation of LSH in mouse leads to either postnatal lethality or growth retardation and premature aging (50). On the other hand, mutations in cohesin or cohesin loading complex subunits result in Cornelia de Lange syndrome and Roberts syndrome manifested by craniofacial and limb malformations together with intellectual and cognitive retardation (51,52). Recently, mutations in cohesin subunits and cohesin regulatory proteins have been found in several cancers (2). Our finding that Irc5 is involved in cohesion raises a possibility that LSH may play a similar role in humans and that some diseases caused by LSH/HELLS mutations may result from cohesion defects.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jennifer Cobb, Adele Marston, Simonetta Piatti, Rodney Rothstein and Frank Uhlmann for providing strains.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre (Poland) [2013/11/D/NZ2/02696 to I.L.]. Funding for open access charge: National Science Centre (Poland).
Conflict of interest statement. None declared.
REFERENCES
- 1. Peters J.M., Tedeschi A., Schmitz J.. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008; 22:3089–3114. [DOI] [PubMed] [Google Scholar]
- 2. Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat. Rev. Cancer. 2014; 14:389–393. [DOI] [PubMed] [Google Scholar]
- 3. Marston A.L. Chromosome segregation in budding yeast: sister chromatid cohesion and related mechanisms. Genetics. 2014; 196:31–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murayama Y., Uhlmann F.. DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell. 2015; 163:1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gligoris T.G., Scheinost J.C., Bürmann F., Petela N., Chan K.L., Uluocak P., Beckouët F., Gruber S., Nasmyth K., Löwe J.. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science. 2014; 346:963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haering C.H., Farcas A.M., Arumugam P., Metson J., Nasmyth K.. The cohesin ring concatenates sister DNA molecules. Nature. 2008; 454:297–301. [DOI] [PubMed] [Google Scholar]
- 7. Murayama Y., Uhlmann F.. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014; 505:367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kulemzina I., Schumacher M.R., Verma V., Reiter J., Metzler J., Failla A.V., Lanz C., Sreedharan V.T., Rätsch G., Ivanov D.. Cohesin rings devoid of Scc3 and Pds5 maintain their stable association with the DNA. PLoS Genet. 2012; 8:e1002856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chatterjee A., Zakian S., Hu X.W., Singelton M.R.. Structural insights into the regulation of cohesion establishment by Wpl1. EMBO J. 2013; 32:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., Nasmyth K.. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000; 5:243–254. [DOI] [PubMed] [Google Scholar]
- 11. Unal E., Heidinger-Pauli J.M., Kim W., Guacci V., Onn I., Gygi S.P., Koshland D.E.. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008; 321:566–569. [DOI] [PubMed] [Google Scholar]
- 12. Zhang J., Shi X., Li Y., Kim B.J., Jia J., Huang Z., Yang T., Fu X., Jung S.Y., Wang Y. et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008; 31:143–151. [DOI] [PubMed] [Google Scholar]
- 13. Chan K.L., Gligoris T., Upcher W., Kato Y., Shirahige K., Nasmyth K., Beckouët F.. Pds5 promotes and protects cohesin acetylation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13020–13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roig M.B., Löwe J., Chan K.L., Beckouët F., Metson J., Nasmyth K.. Structure and function of cohesin's Scc3/SA regulatory subunit. FEBS Lett. 2014; 588:3692–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uhlmann F., Lottspeich F., Nasmyth K.. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999; 400:37–42. [DOI] [PubMed] [Google Scholar]
- 16. Beckouët F., Hu B., Roig M.B., Sutani T., Komata M., Uluocak P., Katis V.L., Shirahige K., Nasmyth K.. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol. Cell. 2010; 39:689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borges V., Lehane C., Lopez-Serra L., Flynn H., Skehel M., Ben-Shahar T.R., Uhlmann F.. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol. Cell. 2010; 39:677–668. [DOI] [PubMed] [Google Scholar]
- 18. Flaus A., Owen-Hughes T.. Mechanisms for ATP-dependent chromatin remodelling: the means to the end. FEBS J. 2010; 278:3579–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hakimi M.A., Bochar D.A., Schmiesing J.A., Dong Y., Barak O.G., Speicher D.W., Yokomori K., Shiekhattar R.. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002; 418:994–998. [DOI] [PubMed] [Google Scholar]
- 20. Huang J., Hsu J.M., Laurent B.C.. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell. 2004; 13:739–750. [DOI] [PubMed] [Google Scholar]
- 21. Lopez-Serra L., Kelly G., Patel H., Stewart A., Uhlmann F.. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat. Genet. 2014; 46:1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alvaro D., Lisby M., Rothstein R.. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007; 3:e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Basenko E.Y., Kamei M., Ji L., Schmitz R.J., Lewis Z.A.. The LSH/DDM1 Homolog MUS-30 is required for genome stability, but not for DNA methylation in Neurospora crassa. PLoS Genet. 2016; 12:e1005790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burrage J., Termanis A., Geissner A., Myant K., Gordon K., Stancheva I.. The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J. Cell Sci. 2012; 125:5524–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ren J., Briones V., Barbour S., Yu W., Han Y., Terashima M, Muegge K.. The ATP binding site of the chromatin remodeling homolog Lsh is required for nucleosome density and de novo DNA methylation at repeat sequences. Nucleic Acids Res. 2015; 43:1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wachn A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 27. Burgess R.C., Rahman S., Lisby M., Rothstein R., Zhao X.. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell Biol. 2007; 27:6153–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keogh M.C., Mennella T.A., Sawa C., Berthelet S., Krogan N.J., Wolek A., Podolny V., Carpenter L.R., Greenblatt F., Baetz K. et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006; 20:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bennett G., Papamichos-Chronakis M., Peterson C.L.. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat. Commun. 2013; 4:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Treich I., Carlson M.. Interaction of a Swi3 homolog with Sth1 provides evidence for a Swi/Snf-related complex with an essential function in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997; 17:1768–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vincent J.A., Kwong T.J., Tsukiyama T.. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 2008; 15:477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 78:273–304. [DOI] [PubMed] [Google Scholar]
- 33. Nimii A., Chambers A.L., Downs J.A., Lehmann A.R.. A role for chromatin remodellers in replication of damaged DNA. Nucleic Acids Res. 2012; 40:7393–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seeber A., Hauer M., Gasser S.M.. Nucleosome remodelers in double-strand break repair. Curr. Opin. Genet. Dev. 2013; 23:174–184. [DOI] [PubMed] [Google Scholar]
- 35. Lee L., Rodriguez J., Tsukiyama T.. Chromatin remodeling factors Isw2 and Ino80 regulate checkpoint activity and chromatin structure in S phase. Genetics. 2015; 199:1077–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu H., Boone C., Brown G.W.. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics. 2007; 176:1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McLellan J.L., O’Neil N.J., Barrett I., Ferree E., van Pel D.M., Ushey K., Sipahimalani P., Bryan J., Rose A.M., Hieter P.. Synthetic lethality of cohesins with PARPs and replication fork mediators. PLoS Genet. 2012; 8:e1002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mayer M.L., Pot I., Chang M., Xu H., Aneliunas V., Kwok T., Newitt R., Aebersold R., Boone C., Brown G.W. et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell. 2004; 15:1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kobayashi T. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell. Mol. Life. Sci. 2011; 68:1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., Nomura M.. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rDNA genes in yeast. Cell. 2004; 117:441–453. [DOI] [PubMed] [Google Scholar]
- 41. Heidinger-Pauli J.M., Mert O., Davenport C., Guacci V., Koshland D.. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr. Biol. 2010; 20:957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richmond E., Peterson C.L.. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996; 24:3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Michalis C., Ciosk R., Nasmyth K.. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997; 91:35–45. [DOI] [PubMed] [Google Scholar]
- 44. Baetz K., Krogan N., Emili A., Greenblatt J, Hieter P. The ctf13-30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 2004; 24:1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rudra S, Skibbens R.V.. Chl1 DNA helicase regulates Scc2 deposition specifically during DNA-replication in Saccharomyces cerevisiae. PLoS One. 2013; 8:e75435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orgil O., Matityahu A., Guacci V., Koshland D., Onn I.. A conserved domain in the Scc3 subunit of cohesin mediates the interaction with both Mcd1 and the cohesin loader complex. PLoS Genet. 2015; 11:e1005036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tittel-Elmer M., Lengronne A., Davidson M.B., Bacal J., François P., Hohl M., Petrini J.H., Pasero P., Cobb J.A.. Cohesin association to replication sites depends on Rad50 and promotes fork restart. Mol. Cell. 2012; 48:98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee D.W., Zhang K., Ning Z.Q., Raabe E.H., Tintner S., Wieland R., Wilkins B.J., Kim J.M., Blough R.I., Arceci R.J.. Proliferation-associated SNF2-like gene (PASG): a SNF2 family member altered in leukemia. Cancer Res. 2000; 60:3612–3622. [PubMed] [Google Scholar]
- 49. Thijssen P.E., Ito Y., Grillo G., Wang J., Velasco G., Nitta H., Unoki M., Yoshihara M., Suyama M., Sun Y. et al. Mutations in CDCA7 and HELLS cause immunodeficiency-centromeric instability-facial anomalies syndrome. Nat. Commun. 2015; 6:7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun L.Q., Lee D.W., Zhang Q., Xiao W., Raabe E.H., Meeker A., Miao D., Huso D.L., Arceci R.J.. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes. Dev. 2004; 18:1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vega H. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesin. Nat. Genet. 2005; 37:468–470. [DOI] [PubMed] [Google Scholar]
- 52. Liu J., Krantz I.D.. Cornelia de Lange syndrome, cohesin, and beyond. Clin. Genet. 2009; 76:303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.