Abstract
Cells regulate biological responses in part through changes in transcription start sites (TSS) or cleavage and polyadenylation sites (PAS). To fully understand gene regulatory networks, it is therefore critical to accurately annotate cell type-specific TSS and PAS. Here we present a simple and straightforward approach for genome-wide annotation of 5΄- and 3΄-RNA ends. Our approach reliably discerns bona fide PAS from false PAS that arise due to internal poly(A) tracts, a common problem with current PAS annotation methods. We applied our methodology to study the impact of temperature on the Drosophila melanogaster head transcriptome. We found hundreds of previously unidentified TSS and PAS which revealed two interesting phenomena: first, genes with multiple PASs tend to harbor a motif near the most proximal PAS, which likely represents a new cleavage and polyadenylation signal. Second, motif analysis of promoters of genes affected by temperature suggested that boundary element association factor of 32 kDa (BEAF-32) and DREF mediates a transcriptional program at warm temperatures, a result we validated in a fly line where beaf-32 is downregulated. These results demonstrate the utility of a high-throughput platform for complete experimental and computational analysis of mRNA-ends to improve gene annotation.
INTRODUCTION
Tight control of gene expression is essential for proper cell homeostasis (1). Indeed the stability, localization and expression levels of a given mRNA are the result of an exquisite process involving both transcriptional and post-transcriptional regulatory mechanisms. Promoter sequences are the main regulators of transcription, while untranslated regions (UTRs) contribute greatly to post-transcriptional regulation. Precise mapping of transcription start sites (TSS) and 3΄ UTRs is therefore critical to understand gene regulation and transcript diversity, as the use of alternative promoters and cleavage and polyadenylation sites (PAS) can have profound impact on the molecular and cellular physiology of cells (2–6). To date, several strategies have been specifically designed for accurately mapping and quantifying 5΄- or 3΄-ends of transcripts (7–22). Although these methods have been used to accurately map TSS and 3΄ UTRs in cell lines, their applicability to in vivo or primary cell cultures has lagged because of the challenges of current methods. Current 5΄ library protocols are very labor intensive and require large amounts of starting material (7,8,10,12,17,20). For example, the most widely used method for TSS annotation is cap analysis of gene expression (CAGE) (23,24). CAGE requires 5 μg of RNA and takes 4 days of intensive work (25).
As opposed to 5΄ libraries, current 3΄-end library construction methods do not require large input amounts. On the contrary, some 3΄ library construction methods are the bases for building low input RNA-seq quantification libraries and even for single cell RNA sequencing (26,27). However, these methods are not optimal for annotation purposes, mainly because of their reliance on oligo(dT) primed cDNA (9,11,13–15,18,19,22). Oligo(dT) tends to also prime internal A-rich sequences that need to be computationally identified a posteriori (13,14,28–30). Although there are computational methods available for the detection of internal versus polyadenylated sites they inevitably miss some internal priming sites while excluding some bona fide 3΄-ends, as some cleavage sites are flanked with A-rich sequences (31). Alternative experimental approaches like PAS-seq or TAIL-seq rely on sequencing the actual poly(A) tail to avoid internal priming, but require both long reads and high sequencing depth and hence have high cost per sample (13,32). Finally, a recent approach (18) while greatly reducing the abundance of internal poly(A) tracts, requires long (>50 bp) poly(A) tails and hence precludes the annotation of mRNA isoforms with short poly(A) tails, which are very common in highly regulated mRNAs in many organisms, including mammals (33).
Here, we present a comprehensive approach for simple and accurate 5΄- and 3΄-end purification, which can be highly multiplexable. We applied our technology to explore the effect of temperature on TSS and 3΄-UTR usage in Drosophila melanogaster. This system showcases the strengths of our system: starting input material is lower than other available technologies, the genome is both compact and complex, with many genes overlapping their 5΄- and 3΄-ends and the experimental setting involves many samples and hence benefits from a multiplexed approach.
We applied our 5΄- and 3΄-end methods, which we named Exo-seq and RNaseH-seq, respectively, to D. melanogaster cultured at three different temperatures (18, 25 and 29°C). Our analysis revealed hundreds of novel TSS and PAS, and provided adjustments for many more. Surprisingly, we found that proximal PASs of genes expressing multiple PASs rely on a different motif from the canonical polyadenylation signal, suggesting a novel cis-regulatory element that control alternative polyadenylation choice. As previously reported, TSS mapping revealed a dichotomy in TSS types, with some transcripts having very precise transcription start sites, while others having a less defined start site that spans a 50-base window. To classify TSS types we employed the Gini coefficient, a widely used measurement by economists to measure the tendency of a distribution to concentrate in very few points (34). We use the Gini index to quantify the concentration of 5΄ reads on a single base rather than a region of the promoter. The distribution of the Gini coefficient is clearly bi-modal and we show that it leads to a straightforward classification of the promoters into the two types. Last but not least, we used the quantitative nature of our data to explore the impact of temperature on gene expression. Promoters of genes upregulated at warm temperatures (29°C) were enriched in DNA replication-related element factor (DREF)/boundary element association factor of 32 kDa (BEAF-32) DNA binding motifs. Knock-down of beaf-32 confirmed its role in the transcriptional response to high temperature.
These results demonstrate the utility of a straightforward, inexpensive and simple platform that offers a complete experimental and computational solution to mRNA-end annotation and uncover new 5΄ and 3΄ signals important for regulating gene expression.
MATERIALS AND METHODS
5΄-end RNA-Seq (Exo-seq)
Full protocol can be found under ‘supplementary protocols’, along with notes for further optimization and a scheme summarizing the various sequences added to the RNA fragment. In addition, the detailed protocol and any future improvements that will be made can be found at: http://garberwiki.umassmed.edu/Exoseq. In short, 100 ng − 1 μg of total RNA was poly(A) selected using oligo(dT) beads (ThermoFisher Scientific) following the manufacturer protocol. For the samples presented below 1 μg of total RNA was used, however we were able to successfully start with lower amounts of input (data not shown). Poly(A)+ RNA was then fragmented using Ambion Fragmentation buffer. Fragmented RNA was then cleaned-up using 2.5× volume on Solid Phase Reversible Immobilization (SPRI) beads (Agencourt), Polynucleotide Kinase (PNK) treated (T4 PNK, NEB), cleaned and incubated with Terminator Exonuclease (Epicenter). Reaction mixture was dephosphorylated with FastAP (ThermoFisher Scientific), cleaned (2.5× SPRI) and then ligated to linker 1 (5Phos/AXXXXXXXXAGATCGGAAGAGCGTCGTGTAG/3ddC/ using T4 RNA ligase I (NEB). XXXXXXXX is an internal barcode specific for each sample). Ligated RNA was cleaned-up by Silane beads (Dynabeads MyOne, ThermoFisher Sientific). Reverse Transcription (RT) was then performed, with a specific primer (5΄-CCTACACGACGCTCTTCC-3΄). Then, RNA–DNA hybrids were degraded by incubating the RT mixture with 10% 1M NaOH at 70°C for 12 min. pH was then normalized by addition of corresponding amount of 0.5M Sodium Acetate. The reaction mixture was cleaned up using Silane beads and we performed a second ligation, in which the 3΄-end of cDNA was ligated to linker 2 (5Phos/AGATCGGAAGAGCACACGTCTG/3ddC/) using T4 RNA ligase I. The sequences of linker 1 and linker 2 are partially complementary to the standard Illumina read 1 and read 2/barcode adapters, respectively. Reaction Mixture was cleaned up and polymerase chain reaction (PCR) enrichment was set up using enrichment primers 1 and 2 (5΄-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3΄, 5΄-CAAGCAGAAGACGGCATACGAGATXXXXXXXXGTGACTGGAGTTCAG ACGTGTGCTCTTCCGATCT-3΄, where XXXXXXX is a barcode sequence). A total of 10–14 cycles of enrichment were performed depending on the initial input amount of RNA. After cleanup with 0.75× volume of SPRI beads library was ready for characterization by Bioanalyzer.
3΄-end RNA-seq (RNaseH-seq)
Full protocol can be found under ‘supplementary protocols’, along with notes for further optimization and a scheme summarizing the various sequences added to the RNA fragment. In addition, the detailed protocol and any future improvements that will be made can be found at: http://garberwiki.umassmed.edu/Exoseq. In short, RNA was fragmented (Mg RNA fragmentation module, NEB), cleaned and poly(A) selected using oligo(dT) beads (ThermoFisher Scientific). Poly(A) tails were removed by the addition of oligo(dT) primer to the polyA+ fragments and RNAse H treatment (NEB). After RNA cleanup (Silane beads, ThermoFisher Scientific) RNA was dephosphorylated with FastAP (ThermoFisher Scientific), cleaned (2.5× SPRI) and then ligated to linker 1. Library preparation was continued as described above (for Exo-seq).
RNAseH- sequencing
Full protocol can be found under ‘supplementary protocols’. In short, RNA was fragmented (Mg RNA fragmentation module, NEB), cleaned, dephosphorylated with FastAP (ThermoFisher Scientific) and ligated to linker 1. RNA samples were then cleaned and pooled into a single tube. 3΄-end fragments were positively selected (using Poly(A)+ selection with oligo(dT) beads, ThermoFisher Scientific). Library preparation was continued as described above for Exo-seq, starting from the first strand (RT) step.
Full length RNA-sequencing
Full-length RNA libraries were done similarly to the described RNAseH- samples, with a difference in the order of the different steps; First, RNA was PolyA+ selected, then fragmented (using Mg-based fragmentation), cleaned (2.5× SPRI cleanup), FastAP treated, cleaned (Silane cleanup) and continued in library preparation (starting at first ligation of 3΄ adapter).
Annotating 5΄- and 3΄-ends at a single-base resolution
As with every mRNA-end sequencing technique, not all reads sequenced originate from the TSS (for Exo-seq) or the 3΄-end (for RNaseH-seq) of the gene. The transcript identification problem seeks to identify regions whose coverage is higher than expected by chance using a suitable null model. The null model we use assumes a uniform coverage of reads across the gene, and we seek to identify regions whose coverage is higher than expected by chance. This coverage is dependent on the expression level of the gene and hence we require a gene annotation set to compute this local null model. In this work we used the UCSC RefSeq gene set as the input. For each transcript, we calculate the number (pile-up) of read starts at each base position within the exons of the gene. We further extended both the 5΄ and 3΄ exons by 2 Kb and into the first intron to be able to detect start or ends beyond the annotation limits. For each (extended) transcript we first compute its null model by averaging all pileups within the transcript by the (extended) transcript length. To determine whether a pile-up is significantly higher than expected, we calculate the Z-score for each base using this null distribution model. Manual inspection of Z-scores suggested that six standard deviations were stringent enough to detect transcript ends. Hence, all pileups that were more than six standard deviations from the mean were tagged as significant and merged consecutive significant bases into peaks. In addition, to avoid false positives in lowly expressed genes, if the most covered peak in a gene had <10 reads, a higher threshold of Z-score was chosen. We implemented transcript end identification as a module in ESAT (“End Sequencing Analysis Toolkit”) which is open source and freely available to the public (https://github.com/garber-lab/ESAT (35)).
Detection and removal of internal poly(A)
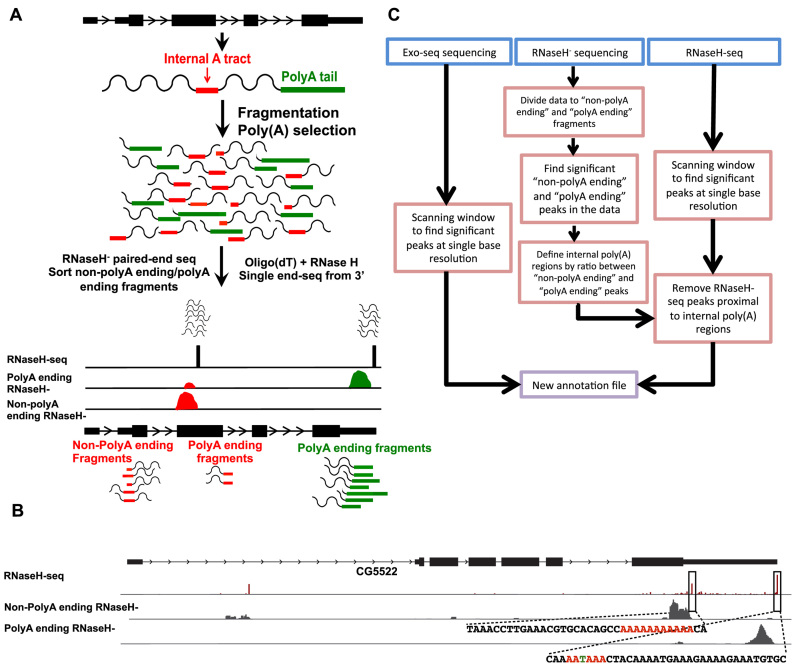
Because internal poly(A) tracts may occur anywhere within the sequenced fragment, and not necessarily at the 3΄-end, reads from RNaseH− libraries were sorted into two groups of ‘non-polyA ending’ and ‘polyA ending’ reads. The sorting was done with the last three bases of the 3΄ reads (which are the last bases of the poly(A)+ fragment). If those bases were only adenosines, the 5΄ read was regarded as a ‘polyA ending’ read, else it was regarded as a ‘non-polyA ending’ read. Then, each group was aligned to the genome. After alignment, we use ESAT to quantify significant peaks in the ‘polyA ending’ and ‘non-polyA ending’ aligned files (see below). A ‘non-polyA ending’ peak would be considered significant if the ratio of the number of reads in this peak to the number of reads in the ‘polyA ending’ peak that it overlaps is higher than a certain threshold (for this work we used the threshold of 0.9). All RNaseH-seq peaks that are <300 bp downstream from a ‘non-polyA ending’ peak were classified as false PAS and disqualified from further analysis, since 300 bp is the size of the sequenced fragments.
Quantification and analysis of all 5΄- and 3΄-end methods
For quantification purposes, we used the normalized counts obtained using the ESAT program (https://github.com/garber-lab/ESAT (35)). For the RNAseH− libraries we used ESAT to compute gene expression values by scanning the gene UTR and up to 2 Kb downstream of the annotated end for a maximally transcribed 500 bp window. The size of the window was selected based on the library insert size distribution. Once a maximally enriched window was identified, a gene was assigned all reads that fell within the (extended) loci, up to the maximally enriched window. For RNaseH-seq and Exo-seq, as the data is more concentrated and an accurate annotation was already defined, we assigned all the reads that fell within 50 bp upstream (for RNaseH-seq) or downstream (for Exo-seq) of the newly annotated PAS or TSS.
Fly samples
Drosophila Canton S (CS) wild-type flies were reared at 25°C on a standard diet (yeast: 38 g/l, yellow corn mill: 91 g/l, agar: 10 g/l, molasses: 8.7% v/v, propanoic acid (BioLab): 0.9% v/v, Tegasept solution (Sigma-Aldrich; 300 g/l in EtOH (BioLab)): 0.8% v/v)). Flies were kept in 12:12 light:dark cycles at 25°C. For RNA extraction, newborn flies were entrained for 3 days in 12:12 light:dark conditions at different temperatures (18, 25 or 29°C). Exo-seq samples were collected at Zeitgeber time (ZT) 3, ZT9, ZT15, ZT21 and RNaseH-seq samples were collected at ZT3 and ZT15. RNaseH− samples were collected at ZT3, ZT7, ZT11, ZT15, ZT19 and ZT23. RNA was extracted using Trizol (Invitrogen). ZT indicates the time in hours after the lights are on in a 12:12 light:dark cycle (e.g. ZT3, 3 h after lights on and ZT15, 3 h after lights are off). Full-length RNA-seq samples were collected at ZT3, ZT15 and ZT23 for flies entrained in 18 and 29°C and collected at ZT3 and ZT15 for flies entrained in 25°C.
RNA-sequencing
RNA was sequenced as paired-end samples, where 11 bases were read from the 3΄-end of the fragment (read 1; the first 8 bases are our internal barcode) and 40 bases were read from the 5΄-end of the fragment (read 2) for Exo-seq sequencing and RNaseH− sequencing. Then, a customized python script was used to sort the reads based on the barcode found on read 1, allowing for a maximum of one mismatch in the barcode. Read 1 was then discarded and read 2 was aligned as a single-end read. Reads mapping to ribosomal RNA were discarded by aligning all the reads to known Drosophila rRNA sequences with bowtie2 (36). The remaining reads were mapped to the Drosophila genome (dm3) with tophat2 (37) and set of known transcripts included in UCSC RefSeq known genes, allowing a maximum of two mismatches per read, discarding reads that were not uniquely mapped. For RNaseH-seq the analysis was similar, except we read only 50 bases (single read) from the 3΄-end, where the first 8 bases are our internal barcode. Also, since a few bases of the poly(A) can still remain on the read, if the read began with a sequence of adenines they were trimmed prior to alignment.
ChIP-seq
Chromatin immunoprecipitation (ChIP) was performed using the protocol described in (38), except that the antibody used was anti-H3K4met3 (abcam8580). DNA library preparation was performed as described (39) and libraries were sequenced using the Illumina sequencing platform. Significant peak regions were found using MACS peak calling software (40). Generation of the heatmap and average plot was done using ngs.plot (41), with the default parameters except –L 1000, -FL 300.
Comparison of methodologies to distinguish real PAS from internal poly(A) tracts
Each of the 8357 discovered PAS was classified as a true or false (internal poly(A) tract) PAS by two different methods: (i) Using RNAseH-seq followed by RNAseH− sequencing, and (ii) by the standard in silico method where a PAS is classified as internal if it has six consecutive A's directly downstream of it or if 7 out of the 10 downstream bases are A's (13). To validate and compare the obtained results we generated a second set of RNAseH− libraries, which we sequenced using 100 bases long pair-end reads.
These paired-end reads were mapped to the drosophila genome (dm3) using the STAR aligner (42), as it allows partial mapping of reads that end in a poly(A) tail, having those bases soft-clipped. STAR was run with default parameters with the addition of ‘—clip5pNbases 8,0’ to remove the internal barcode used for multiplexing the samples.
In order to use the long reads to classify each PAS as true or false, we counted the number of reads that span the PAS position but not necessarily end in the PAS position, and the number of reads that exactly end in the PAS position. In addition, out of the total number of reads that end in the PAS, we counted how many align to the genome without soft-clipping, and how many only align after soft-clipping at least six bases from the end of the read. We reasoned that for a true PAS, reads that span the PAS position will end in non-genomic adenosines, thus will have to be soft-clipped in order to align, and the genomic end position of the read after soft-clipping will exactly be the PAS position. For this analysis, we counted only read 1 (as it is the read spanning the PAS) with the correct strand annotation (map to the strand opposite of the transcript). However, when the PAS is in an A-rich genomic region, the STAR aligner can map bases of a possibly true poly(A) tail to the genome, even allowing a few mismatches. Thus, a read was counted as ending in the PAS position if the end position of the read was up to 10 bp upstream of the PAS, or if its end position was in the consecutive downstream adenosine homodimer. We defined the consecutive downstream adenosine homodimer as the region starting with the PAS and up to a maximum of 20 bp downstream that includes only adenosines, allowing for up to four non-adenosine bases. The choice of four non-adenosines was made after manually observing the number of mismatches that would still get a sequence of an adenosine homopolymer mapped to the downstream genomic region of the PAS.
Determination of genes regulated by Beaf-32
To down regulate beaf-32, we generated flies expressing a UAS–RNAi transgene (BDSC number 35642) under the control of the actin5C-gal4 promoter (Stock #25374, Bloomington Stock Center, Indiana, USA). We utilized actin5C-gal4 flies as control. Flies were reared at 25°C on a standard diet (yeast: 38 g/l, yellow corn mill: 91 g/l, agar: 10 g/l, molasses: 8.7% v/v, propanoic acid (BioLab): 0.9% v/v, Tegasept solution (Sigma-Aldrich; 300 g/l in EtOH (BioLab)): 0.8% v/v)). After eclosion flies were kept in 12:12 light:dark cycles at 25°C. Newborn flies were entrained for 3 days in 12:12 light:dark conditions at different temperatures (18, 29°C). RNA was extracted from their heads using TRI Reagent, Sigma Aldrich. (Dnase treated) RNA was fragmented in the presence of 10× FastAP buffer (ThermoFisher Scientific) at 94°C for 3 min and placed on ice. RNA was then dephosphorylated with FastAP, cleaned (2× SPRI) and ligated to linker 1. Library preparation was continued as described above, starting from linker 1 3΄ ligation (for Exo-seq).
Assessing reproducibility among biological replicates and various methods
To assess the reproducibility and compare various methods, we computed the pairwise correlation among samples. Number of reads that map to each transcript in each sample was computed using featureCounts (43). Pairwise correlation was computed on the log2 of the counts. To reduce noise, transcripts with <10 mapped reads in both samples were removed.
Differential expression analysis
Detection of differentially expressed genes between 18 and 29°C was done using the R package ‘DESeq’ (44), where all the samples from each temperature were used as replicates. An isoform was considered differentially expressed if it had an adjustedP-value of <0.01. Finding statistically significant enriched GO categories was done using the R package ‘GOstats’ (45).
The heatmap in Figure 4E was created using the R heatmap.2 function clustering on both columns and rows (Colv = T, Rowv = T) and scaling rows (scale = ‘row’). Only genes differentially expressed at an adjustedP-value < 0.01 were used. The heatmap in Figure 5A was created using GENE-E (https://software.broadinstitute.org/GENE-E/), using only genes differentially expressed at an adjusted P-value < 0.1 and absolute log2 fold change >0.75 in at least one of the pairwise comparisons (wild-type 18°C versus wild-type 29°C, beaf32−/− 18°C versus beaf32−/− 29°C, wild-type 18°C versus beaf32−/− 18°C, wild-type 29°C versus beaf32−/− 29°C).
Figure 4.
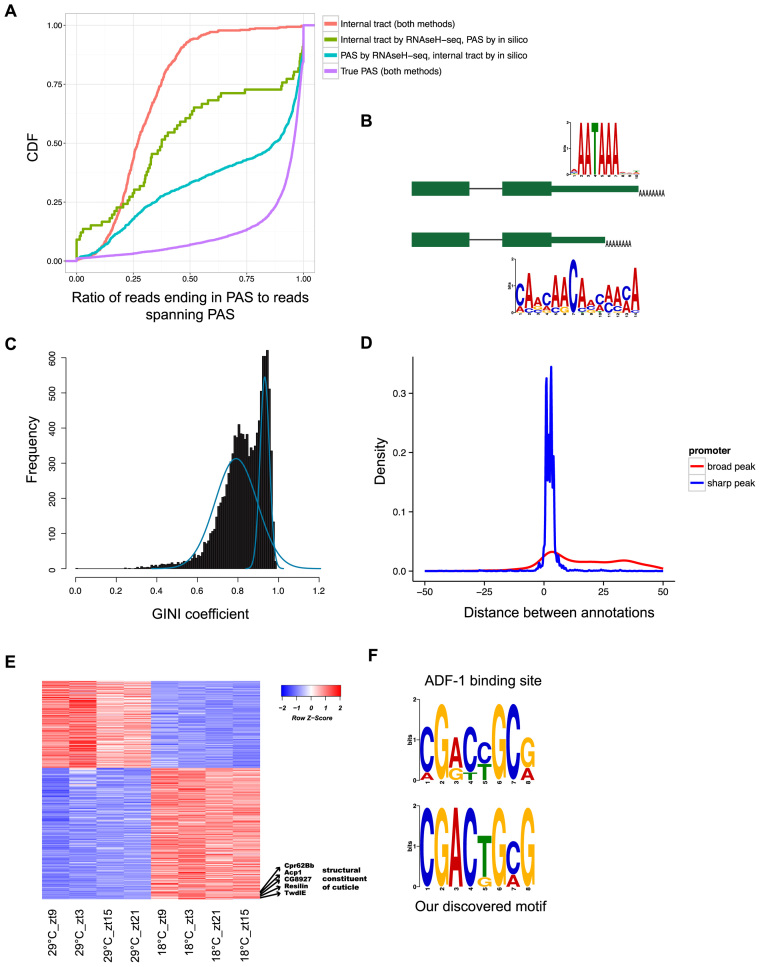
RNaseH-seq and Exo-seq uncover alternative end-RNA processing signals and enriched motifs. (A) Cumulative density function of the ratio of reads that end in a PAS to reads spanning the PAS in each one of the following groups: peaks classified as internal tracts both by RNaseH-seq and the common in silico method (red), peaks classified as true PAS by both methods (purple), peaks classified as true PAS by RNaseH-seq but as internal tracts in silico (light blue) and peaks classified as internal tracts by RNaseH-seq but as true PAS in silico (green). (B) Genes with alternative 3΄-ends in the last exon are enriched for different RNA motifs. The longer transcripts are enriched for the canonical cleavage and PAS while the shorter transcripts are enriched for a newly identified motif near the 3΄-end. (C) Histogram of the Gini coefficients in 50 base windows around predicted TSS, the curves (in blue) are the bi-modal fit of two Gaussian distributions. (D) Distribution of the distance to modEncode annotated TSS of Exo-seq-predicted TSSs having low Gini coefficient (<0.78, ‘broad’ promoter, red) and high Gini coefficient (>0.9, ‘sharp’ promoters, blue). (E) Heatmap of the Exo-seq expression values for 540 differentially expressed genes between 18 and 29°C. Several of the genes involved in cuticle formation are highlighted. (F) The known ADF-1 binding motif (top) compared to the motif found in the core promoter of genes upregulated in 18°C (bottom).
Figure 5.
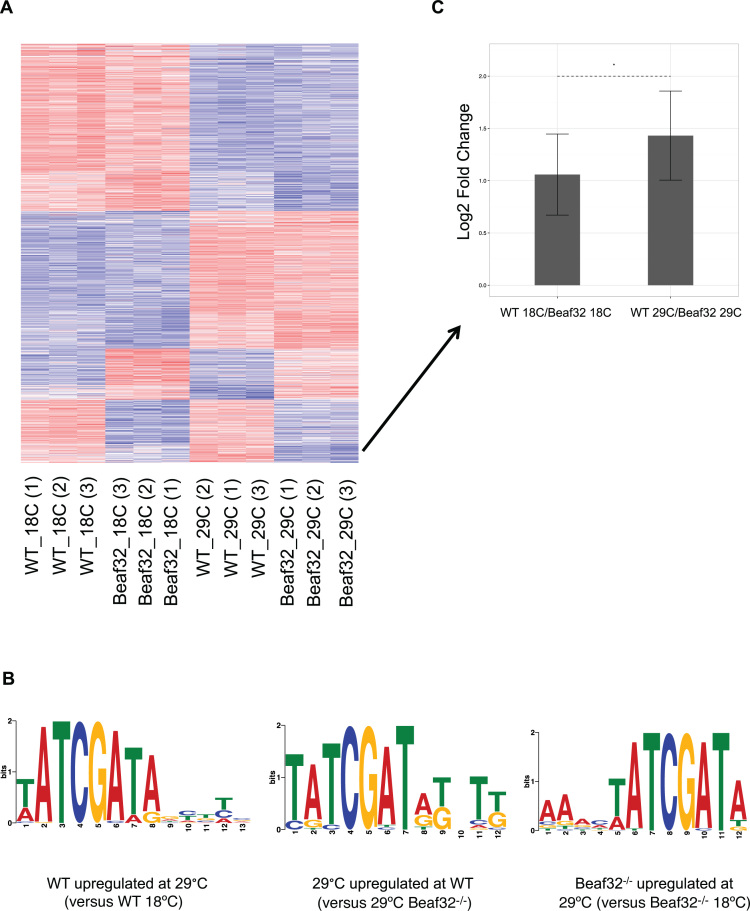
BEAF-32 is active at the transcriptional response at 29°C. (A) Heatmap of differentially expressed genes between 18 and 29°C at wild-type or beaf32−/− samples, or between wild-type and beaf32−/− samples at 18 or 29°C. A gene was included in the heatmap if it had an adjusted P-value < 0.1 and log2 fold change >0.75 in at least one of the pairwise comparison. (B) The motif enriched in genes upregulated at 29°C in wild-type flies versus 18°C wild-type (left), compared with the enriched motif in genes upregulated at 29°C wild-type versus 29°C beaf32−/− flies (middle) and the enriched motif in genes upregulated at 29°C beaf32−/− flies versus beaf32−/− at 18°C (right). (C) Log2 Fold Change at 18 and 29°C between the wild-type and beaf32−/− samples of the genes upregulated at 29°C wild-type versus 29°C beaf32−/− and harbor the BEAF-32 binding motif at their promoter. Arrow represents the location of most of those genes on the heatmap. Asterisk represents P-value < 0.05 between the distributions of the fold change using a t-test. Error bar represents the standard deviation.
Motif analysis
All motif analysis in this work was performed using MEME (46). The Search of enriched motifs in isoforms with two PAS in the last exon was performed on the last 50 bp upstream of the 3΄-end. In searching for motifs in the proximal UTRs the distal UTRs were used as a negative set and vice versa. For finding enriched motifs in the core promoters of genes upregulated in 18°C, we searched 100 bp upstream of the TSS, while the sequences 100 bp upstream of the TSS of genes upregulated in 29°C was used as a negative set and vice versa for genes upregulated in 29°C. Finding all instances of the motifs in our set of sequences was done with FIMO (47) and comparison to known motifs was performed with TOMTOM (48).
Promoter Gini coefficient
We measured the inequality or non-uniformity of the distribution of read start positions in the promoter region of each gene [−50, +50] using the Gini coefficient, which is the most widely used measure for inequality. The Gini coefficient range from zero to one, zero means that all the values are the same, while one means maximal inequality. In our case, a smaller Gini coefficient indicates that the peak is broad (read coverage is more uniform), while larger Gini coefficient indicates that the peak is sharp (read coverage is centered around one or few bases). Computation of the Gini coefficient was done in a custom python script that is available on demand. When plotted across all identified TSSs, the Gini coefficient distributes clearly as a bi-modal distribution. To estimate the two component distributions, we used an Expectation-Maximization (EM) algorithm to estimate parameters of two Gaussian mixture models over the distribution of Gini coefficients. Based on the mean of these distributions, every promoter with a Gini coefficient <0.78 was classified as ‘broad’ and every promoter with a Gini coefficient over 0.9 was classified as ‘sharp’.
Comparing Gini coefficient with SI index
In order to test the correlation between the Gini coefficient and the SI index suggested by (49), we calculated the Spearman correlation between the Gini coefficient and the ‘total_SI’ measurement of all promoters that overlap between the two studies, a total of 8140 promoters.
RESULTS
5΄ and 3΄ RNA-sequencing with Exo-seq and RNaseH-seq
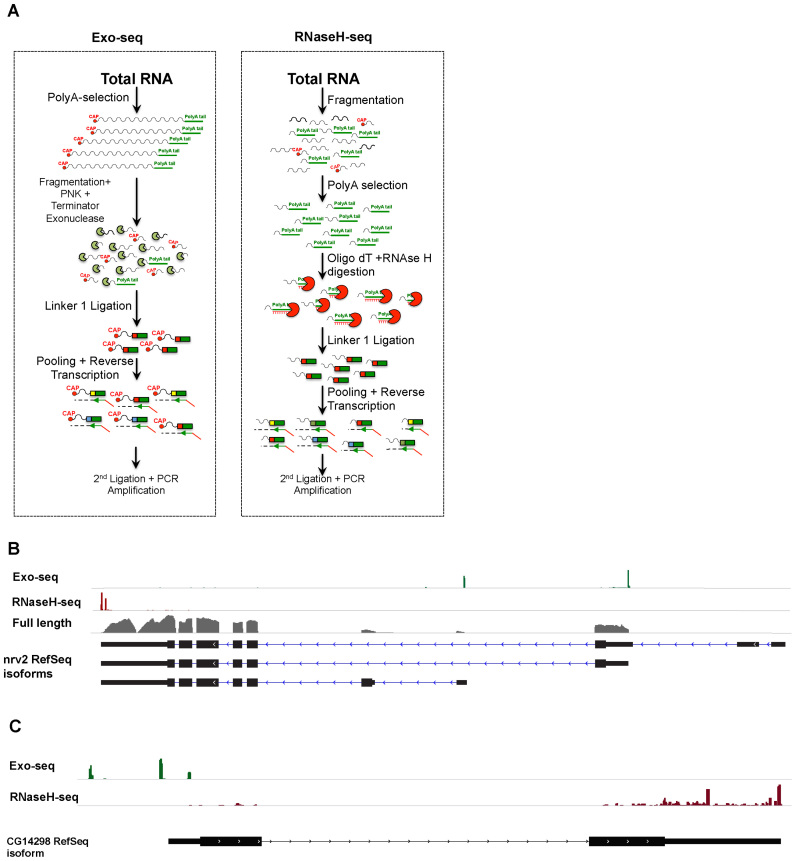
All RNAs transcribed by RNA Polymerase II (PolII) possess 5΄ CAP, which consists of an inverted Guanosine at their 5΄-end (50,51). The CAP is resistant to 5΄ exonucleases (52) and hence can be used to preferentially enrich for 5΄ RNA ends. To map TSSs of PolII-transcribed genes, we first digest poly(A)+ RNA into shorter fragments using metal-based fragmentation (e.g. Zn++) and treat the digested RNA with PNK. These two consecutive reactions assure the presence of a 5΄ phosphate in most RNA fragments except those protected by the CAP structure (Figure 1A, left panel). Non-CAP fragments are then digested using a 5΄ RNA exonuclease (Terminator Exonuclease), which degrades RNA from 5΄→ 3΄ provided that the RNA begins with a 5΄ phosphate (Figure 1A, left panel). This procedure allows us to enrich specifically for RNA that contains 5΄-CAPs. It should be noted that while this method exhibit a few similarities to a previously described method (21) our approach is cleaner and more accurate, as we require less PCR cycles and the fragment size is more homogenous. In addition, our stating material is significantly lower.
Figure 1.
Exo-seq and RNaseH-seq enrich 5΄ and 3΄ mRNA ends respectively. (A) Schematic representation of our method for isolating 5΄ (Exo-seq) and 3΄ (RNaseH-seq) transcript-ends. (B) An Integrative Genome Viewer plot showing data coverage of the nirvana-2 (nrv2) gene with Exo-seq and RNaseH-seq on RNA extracted from Drosophila heads cultured at three different temperatures (18, 25 and 29°C), together with full-length RNA-seq at 25°C. (C) An Integrative Genome Viewer plot of Exo-seq and RNaseH-seq of the CG14298 gene showing multiple (and previously unknown) TSS (upper track) and PAS (lower track).
To map the poly(A) 3΄-ends, we fragment total RNA as above and then select poly(A)-containing fragments using oligo(dT) beads. To sequence the exact location of the poly(A) site, we digest the selected RNA using RNase H, an enzyme that cleaves RNA in a RNA–DNA hybrid (53). This step both eliminates the poly(A) RNA and allows us to ligate an adaptor to the exact junction point just downstream of the cleavage and PAS (Figure 1A, right panel).
After isolating 5΄- or 3΄-ends, we generate RNA-seq libraries using a slight modification of a previously described multiplexed library construction method (see ‘Materials and Methods’ section, (54)). Briefly, we ligate a barcoded adapter at the 3΄-end of the fragments at the beginning of the library construction procedure. Once barcoded, all samples can be pooled so that all subsequent reactions can be performed on a single pool. Multiplexing from the first step diminishes the cost per library and decreases library-to-library variation as amplification (both during reverse transcription and during PCR) is carried out in the pooled sample. Our library generation methods result in an accurate transcript-end annotation, which is particularly important for genes with multiple TSSs and PASs (Figure 1B and C).
Because transcripts differ up to 1000-fold in their transcription levels, determining the exact RNA-ends using a single base ‘pile-up’ approach is a significant challenge. The challenge arises from the fact that differences in coverage between genes significantly affect the signal to noise ratio. Lowly expressed genes display low read coverage, making it difficult to distinguish a true pile-up from random noise. On the other hand, genes with high expression have easily distinguishable pile-ups, but also generate a larger number of reads in internal regions of the transcript. In this sense the pile-up calling needs a local, gene specific model that takes into account the gene expression level. Therefore, to accurately annotate transcript starts and ends we implemented an annotation algorithm that uses a local transcript specific coverage model to estimate the significance of any given read pile-up (see ‘Materials and Methods’ section).
Identification and elimination of internal poly(A) tracts in 3΄-end libraries
As with most oligo(dT) based polyadenylation enrichment methods, our methodology successfully identifies 3΄-ends, but also enriches for internal adenosine homopolymer tracts. To overcome this limitation, we devised an experimental—rather than a computational—procedure that allows us to determine whether an identified 3΄-end is derived from an internal poly(A) tract or a real 3΄-end. To do so, we generate a second library set from the same samples using a protocol for 3΄-end sequencing without RNase H digestion (RNaseH− sequencing, see ‘Materials and Methods’ section). Without the RNase H digestion, poly(A) tail or internal poly(A) tract are left intact. In these libraries, reads originating from bona fide poly(A) tails must all end in adenosines homopolymers (‘polyA ending’ reads). On the other hand, since fragmentation is random, many fragments originating from an internal poly(A) tract contain nucleotides other than adenosines at their 3΄-ends (‘non-polyA ending’ reads, Figure 2A).
Figure 2.
Experimental procedure for identifying internal poly(A) tracts. (A) Schematic representation of our strategy. RNaseH− reads are sorted to ‘non-polyA ending’ or ‘polyA ending’ reads based on the last few bases of the 3΄ fragments. Then, each group of reads is aligned to the genome separately. A genomic site is classified as a peak of ‘non-polyA ending’ reads based on the ratio of ‘non-polyA ending’ to ‘polyA ending’ aligned reads. (B) An Integrative Genome Viewer plot of the CG5522 gene. Upper track—RNaseH-seq aligned reads. Middle track—reads from RNaseH− libraries not ending with a poly(A) sequence (‘non-polyA ending’ reads). Lower track—reads from RNaseH− libraries ending with a poly(A) sequence (‘polyA ending’ reads). Sequences for the regions surrounding a real gene-end and an internal poly(A) are shown. Colored bases indicate key features: the internal poly(A) tract and the canonical cleavage and polyadenylation site (PAS). (C) Summary of the computational pipeline utilized for generating the new annotation incorporating the newly characterized 5΄ and 3΄ mRNA ends.
Indeed, RNaseH− reads originating from internal poly(A) sites have a high ratio of ‘non-polyA ending’ reads compared to ‘polyA ending’ reads. (Figure 2A and B). As a result, estimating this ratio using RNaseH− in combination with the single base resolution of RNaseH-seq libraries we can accurately annotate transcript ends (Figure 2C, see ‘Materials and Methods’ section).
Validation of the Exo-seq and RNaseH-seq methodologies
It has been observed that insects display specific strong behavioral responses to temperature changes (55). Interestingly, adaptation of activity patterns to temperature changes have been shown to be mediated by both transcriptional and post-transcriptional changes, such as alternative splicing of an exon in the 3΄ UTR of the gene period (55,56). However, it is unknown whether temperature produces global changes in the TSS or PAS of genes involved in the physiological response to temperature. In order to systematically explore the impact of temperature in isoform choice we applied Exo-seq and RNaseH-seq to RNA extracted from Drosophila heads entrained at three different temperatures (18, 25 and 29°C, see ‘Materials and Methods’ section).
We generated 12 Exo-seq samples (four for each temperature) resulting in a total of 96 million mapped reads (8 million reads per sample) and six RNaseH-seq samples (two for each temperature) totaling 23 million mapped reads (3.8 million reads per sample). Biological replicates showed very good reproducibility (R = 0.968–0.985 for Exo-seq and R = 0.892–0.947 for RNaseH-seq), consistent with full length RNA-seq collected under similar experimental conditions (R = 0.938–0.989) (‘Materials and Methods’ section and Supplementary Figure S1A). Importantly, our reproducibility is comparable to that of two biological replicates for CAGE experiment done on the same tissue by the modEncode project (57) (R = 0.92).
In addition, Exo-seq and RNAseH-seq exhibit a good correlation with the full-length RNA-seq data under the same biological conditions (R = 0.917–0.938 for Exo-seq and R = 0.853–0.898 for RNAseH-seq, Supplementary Figure S1B), demonstrating its ability to be used for quantification of transcript expression levels.
To annotate TSSs and PASs we pooled all the samples together to increase coverage depth. In all, application of RNaseH-seq to fly heads yielded a total of 7830 high confidence peaks mapping to 6980 genes, while Exo-seq identified 11293 high confidence transcription start sites in fly heads, mapping to 8753 genes.
As expected, we observe a strong 5΄ and 3΄ bias in the mRNA-end libraries, showing greater than 40-fold enrichment of the 5΄- and 3΄-ends in comparison to gene body as expected from libraries targeting transcript ends (see Figure 3A for comparison using our new annotation and Supplementary Figure S2 for comparison with the original (RefSeq) annotation).
Figure 3.
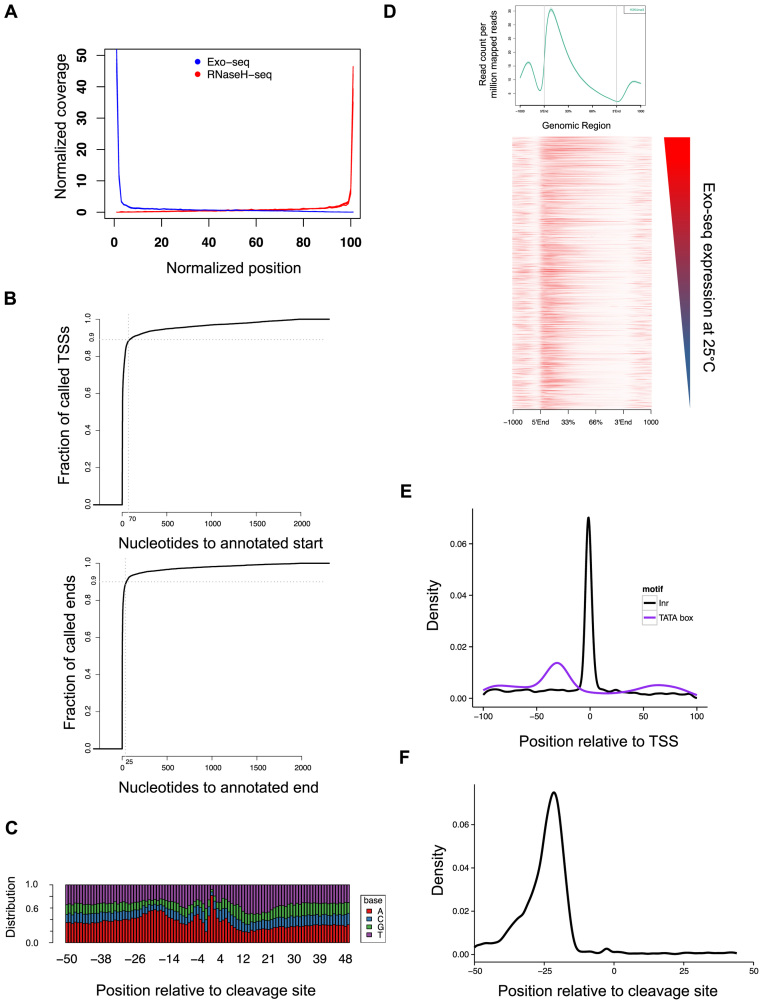
Validation of the Exo-seq and RNaseH-seq methods. (A) Exo-seq and RNaseH-seq show enrichment for annotated transcript ends. The graph represents the normalized coverage across all genes. The plot includes data from 12 Exo-seq libraries and 6 RNaseH-seq libraries. (B) Distance of the closest annotation determined by Exo-seq (upper panel) or RNaseH-seq (lower panel) to the published modEncdoe annotation. (C) Base distribution of 3΄-end regions of RNaseH-seq annotated genes that differ from the modEncode annotation in 1–5 bases. (D) Average profile (top) and heatmap of the distribution of H3K4me3 ChIP-seq reads (bottom). Average profile is based on read distribution across all gene annotations, while the heatmap presents the top 5000 highly expressed genes at 25°C, sorted by expression. (E and F) Distribution of the position of known motifs with respect to discovered transcript ends: promoter motifs TATAWA (TATA box) and TCAKT [initiation motif (Inr); E] and the canonical cleavage site AAUAAA (F).
Newly annotated mRNA-ends are consistent with previous annotations and H3K4me3 ChIP-seq data
To validate our calls, we compared our newly generated annotation with the published analysis from the modEncode project, which used different methods for 5΄- and 3΄-end identification (57). Indeed, our data is in strong agreement with these annotations (Figure 3B). Specifically, 49% of 5΄-ends and 56% of 3΄-ends generated by our data fall within 5 bp of the modEncode annotation. Moreover, 90% of the identified 5΄- and 3΄-ends are within 70 and 25 bases of the annotation, respectively. A close examination of the annotation where the 3΄-ends have a disagreement of just a few bases (1–5) revealed that over 81% of those annotated mRNA have adenines exactly at the end of the annotated mRNA (Figure 3C), which we cannot distinguish from adenines originating from the poly(A) tail. These results demonstrate the validity of our methodology to accurately detect known mRNA ends.
To further validate our results, we tested whether the annotated 5΄-ends are enriched for TSS-specific histone marks. To do so, we carried out two ChIP and DNA sequencing (ChIP-seq) experiments against tri-methylation at H3K4 at 25°C (see ‘Materials and Methods’ section). H3K4 tri-methylation is associated with TSS of actively transcribed genes (58). We find that 92.5% of the H3K4 marks are found within 50 bp of our newly defined TSS (Figure 3D), covering 69% of the annotated 5΄-ends.
We observed an enrichment of the transcriptional initiation (Inr) and TATA-box motifs at the identified TSS, as 9% of the annotations contain a TATA-box 27–34 bases upstream of the TSS and 13% have the initiation motif up to two bases of the annotated TSS (Figure 3E). This is consistent with previously reported data (57) in which 4.3% of transcripts have a TATA motif 24–32 bp upstream of the TSS and 12% of the transcripts have an Inr motif up to 1 bp of the TSS. Also, 45% of the 3΄-UTRs showed the canonical cleavage and polyadenylation motif 15–30 bases upstream of the cleavage site (Figure 3F).
RNaseH-seq outperforms current in silico approach for distinguishing internal poly(A) tracts from real PAS
Using RNaseH-seq we detected 8357 putative transcript ends of which we determined that 527 (6.3%) originated from internal poly(A) tracts using RNaseH− libraries sequenced at a depth similar to RNaseH-seq (average of 3 million reads per sample). This resulted in a total of 7830 high confidence 3΄-end sites (Supplementary Table S1), mapping to 6980 genes. Of these 7830 candidate PASs, 793 (10%) represents previously unannotated 3΄-isoforms (rather than an adjustment for a known isoform), as we find more than one significant peak for the same isoform. Our approach classified 152 previously annotated mRNA 3΄-ends as internal poly(A) tracts (Supplementary Table S2), suggesting that those 3΄-ends were miss-annotated.
We then compared our determination of internal poly(A) tracts to the commonly used computational method to detect false positives (13). The latter consists on classifying any peak followed by a few consecutive adenosines as an internal poly(A) tract. Out of the 527 PAS classified as internal poly(A) tracts by RNaseH-seq, 87% (458) are also classified as internal in silico. Surprisingly, 1075 of peaks classified as true PAS by RNaseH-seq are classified as internal tracts by the computational method.
In order to further distinguish between internal poly(A) tracks and PAS, we sequenced two new RNaseH− libraries, but this time using 100 bp paired-end reads. As before, reads originating from true PAS will end in a sequence of adenosines and reads originating from an internal poly(A) tract will most likely contain nucleotides other than adenosine at their 3΄-ends. Since we are reading 100 bases of the 3΄ end of the transcript (instead of just a few bases), we can use those reads to more accurately distinguish true PAS from internal adenosine tracts (see ‘Materials and Methods’ section). Reads originating from true PAS will map to the genome only after removing the sequence of adenosines from the end of the read and their genomic ending position will exactly be the PAS. On the other hand, reads originating from internal poly(A) tracts will not end in adenosines and their full sequence will be mapped to the genome and span, but not end, in the suggested PAS.
The longer pair-end reads provided direct evidence that the 458 peaks classified as internal tracts by our method and in silico are internal tracts and not true PAS. Indeed 93.6% (429) of peaks in this group have <50% of spanning reads also ending in the position of the peak (Figure 4A, red line). In addition, for 80% (366) of these peaks, more than 75% of reads ending in the PAS are mapped in full (i.e. without the need to trim bases from their end), thus they do not contain a non-genomic poly(A) tail (Supplementary Figure S3A, red line). On the other hand, the peaks classified as true PAS by both methods are enriched for sites where most spanning reads also end in the PAS (87% of peaks have at least 75% of mapped reads ending in the PAS position; Figure 4A, purple line), and most reads than end in the PAS had to be trimmed prior to alignment due to their poly(A) tail (for 89% of PAS in this group at least 75% of reads were trimmed prior to alignment; Supplementary Figure S3A, purple line).
Interestingly, in the group of 1075 PAS classified as true PAS by our method but as internal poly(A) tracts by the in silico approach, we see enrichment for peaks with properties similar to true PAS: most spanning reads also end in the PAS and most reads ending in the PAS had to be trimmed prior to alignment (Figure 4A, blue line and Supplementary Figure S3A, blue line). This shows that our methodology has increased sensitivity for the discovery of true PAS. It should be noted that in the same group of peaks, the proportion of PAS with a low ratio of reads ending in the PAS to reads spanning the PAS is higher compared to the group of PAS that both methods classify as a true peak. However, we believe that since those regions are rich in adenosines, it is not possible to discern a true poly(A) tail from genomic adenosines and require a different validation strategy (Supplementary Figure S3B). Finally, the set of 69 peaks classified as internal tracts by our method but not in silico are mostly comprised of peaks with a low ratio of ending reads to spanning reads and most PAS-ending reads map without trimming, indicating that those are indeed internal tracts. In sum, these results demonstrate that RNAseH-seq can efficiently distinguish between internal poly(A) tracts and real PAS.
RNaseH-seq discovers a novel motif associated with a subset of cleavage and polyadenylation sites
Among the 3΄-ends identified by RNaseH-seq, 419 genes had two alternative 3΄-ends within their 3΄-UTR (Supplementary Table S3). Surprisingly, motif enrichment analysis on both the proximal and distal 3΄-UTR isoforms revealed that while 80% of the distal 3΄-UTRs harbor a canonical cleavage and polyadenylation motif (AAUAAA), the same is not the case for proximal UTRs which are instead enriched for a CA-rich motif (CAVCAACAVMMAMA; Figure 4B) with 64 (15%) of the proximal UTRs harboring this new motif within 100 bases of the 3΄-end, 53 of which have not been previously annotated. This motif does not resemble any known landing site either for a miRNA or RNA binding protein. It should be noted that while the canonical polyadenylation motif is commonly found approximately 22 bp upstream of the cleavage site, the new motif does not display a preferred position upstream the mRNA end (Supplementary Figure S4A). Overall we see a strong correlation (0.88) between the expression levels of the short and long isoforms, and therefore cannot infer that there is a motif-dependent modulation of PAS usage in fly heads (Supplementary Figure S4B). GO enrichment analysis reveals significant enrichment for genes that are active in oocyte development (Supplementary Table S4). Interestingly, another CA-rich motif have been found enriched in testis (15), suggesting a tissue specific function of this motif.
TSS annotation accuracy depends on the promoter class
Of the 11293 high confidence transcription start sites (Supplementary Table S5) we identified 9% (1018) as novel. Notably, 19% (1636) of annotated genes show evidence of being transcribed from at least two different promoters, confirming the extensive use of alternative promoters in Drosophila (17,59).
While analyzing the Exo-seq data, we found that sequence coverage of 5΄-ends is variable, with some genes displaying a sharp peak at the TSS and others having a broader region (Supplementary Figure S5). These two types of TSSs have been previously described with other sequencing strategies and technologies (3,49). We quantified the broadness of Exo-seq peaks using the Gini coefficient, a statistic that measures the degree to which a region is uniformly covered by sequence reads. The Gini coefficient has been routinely used in economics to measure wealth inequality distribution, and more recently in biology (60,61) and hence offers an ideal measurement of the ‘broadness’ or ‘sharpness’ of TSS. We calculated the Gini coefficient for each TSS, taking 50 bp upstream and downstream of our annotated TSS (see ‘Materials and Methods’ section). Our measurement of the Gini coefficient is correlated with the SI index suggested by (49) (Spearman correlation 0.58, see ‘Materials and Methods’ section). Importantly, the Gini coefficients are consistent across experimental methods (Spearman correlation 0.635–0.648 between the Gini coefficients of Exo-seq and CAGE, comparable to Spearman correlation of 0.783 between the two CAGE biological replicates) and the Gini coefficients are not correlated with TSS expression (Pearson correlation −0.07 between the Gini coefficients and the log2 read count of each TSS). By calculating the Gini coefficient, the distribution of promoter spread is clearly bimodal (Figure 4C), allowing us to easily classify promoters as ‘sharp’ or ‘broad’.
We hypothesized that some of the inconsistencies between previous annotations and ours could be due to the subset of TSS that displays broader TSSs. To test this hypothesis we inspected how the sharpness of the peak correlates with the distance between Exo-seq and the modEncode data. Indeed, we find that for TSSs in ‘sharp’ promoters the identified TSS is similar at the single base resolution to the existing annotation. However, in ‘broad’ promoters the TSS identified by Exo-seq tend to disagree with existing annotations (Figure 4D). More specifically, while 75% of ‘sharp’ promoters are annotated within five bases of the existing annotation (median value of three bases), only 17% of the ‘broad’ promoters are annotated at this distance of the previous annotation (median value of 35 bases). Given that Exo-seq is intrinsically a different methodology than EST and mRNA sequencing used by previous annotation methods, we conclude that accurate annotations of TSS depend on the biological properties of the promoter and are not a results of technical biases.
Exo-seq reveals extensive transcriptional changes in response to temperature
As Exo-seq allows accurate quantification (Supplementary Figure S1B), we next used it to characterize gene expression changes in response to temperature adaptation. We performed differential gene expression analysis on Exo-seq expression (Supplementary Table S6) and identified a total of 540 differentially expressed TSS between flies entrained at 18 and 29°C, corresponding to 515 unique genes (Figure 4E; Supplementary Figure S6A and B; Supplementary Table S7). The differentially expressed genes are enriched for many diverse basic biological functions such as translation, transport and various metabolic processes (Supplementary Table S8). Interestingly, we find very high enrichment of genes involved in cuticle formation (including resilin) among genes upregulated at 18°C, which is consistent with previous reports showing that cuticle development and deposition is temperature dependent (55,62). We also find 123 genes with alternative TSSs exhibiting significant changes in only one isoform in response to temperature. Hence, in those cases, the transcriptional change relies only on one of the alternative promoters to achieve desired expression levels. Moreover, only one gene (emp) exhibited differential TSS expression in both temperatures (i.e. one TSS is significantly higher in 29°C while another TSS is significantly higher in 18°C, Supplementary Figure S6C), showing that a complete promoter switching is not a widespread phenomena in adjustment to different temperatures.
We next used motif discovery to investigate whether certain DNA cis-regulatory sequences may be specific to genes with temperature sensitive expression. We searched for enriched cis-regulatory motifs in the core promoter regions of the differentially expressed genes, up to 100 bp upstream of TSS. We find that genes with increased expression at 18°C are enriched (P < 10−2) in a motif with significant similarity to the binding site of ADF-1 (Adh transcription factor 1, P < 10−4, Figure 4F). This suggests that this transcription factor might play a key role in recovering homeostasis after changes in temperature. ADF-1 is a general transcription factor that have been shown to regulate many different functions such as dendrite growth (63) and olfactory memory (64) and indeed among upregulated genes at 18°C is Dopa decarboxylase(ddc), a well-characterized target of ADF-1 and a key regulator of Dopamine and Serotonin metabolism (65,66). Out of the 37 genes harboring an ADF-1 binding site in their promoter, three genes (Peritrophin-A, Cht2 and CG8927) are involved in cuticle formation, suggesting the involvement of Adf-1 as a regulator of cuticle deposition.
In addition, genes with increased expression at 29°C are enriched (P < 10−9) for the abundant (67) DNA-replication-related element (DRE; Supplementary Figure S7). DRE sequences are bound both by DREF and BEAF-32 (68). DREF is a key transcription factor in regulation of cell proliferation (69), while BEAF-32 binds a boundary sequence present in DRE (70), preventing the binding of DREF. Interestingly, the expression mRNA levels of those factors do not change significantly between 18 and 29°C, suggesting that their activity is regulated at the post-translational level (e.g. by protein phosphorylation). Another possibility is that changes in expression of co-factors activate the binding of those transcription factors. For example, the protein SRY-DELTA was shown to interact with BEAF-32 (71). Sry-delta is significantly upregulated at 29°C, thus changes in BEAF-32 binding could be due to changes in the expression of its co-factor.
In order to further investigate the role of BEAF-32 in the transcriptional response to temperature changes in Drosophila, we compared the head transcriptome of wild-type flies and flies in which we downregulated beaf-32 by RNAi (see ‘Materials and Methods’ section). We raised the flies at 25°C and transferred them to 18 and 29°C for three days, after which we collected them, isolated RNA from their heads and performed 3΄ RNA-seq (see ‘Materials and Methods’ section). We confirmed that beaf-32 mRNA was strongly downregulated by the knock-down by RT-PCR and in the sequencing data (Supplementary Figure S8, Table S9 and data not shown). Differential expression analysis of the new wild-type samples recapitulated the enrichment of the BEAF-32 motif in promoters of genes upregulated at 29°C (Figure 5A, B (left), Supplementary Table S10). Similarly, we observe enrichment of this motif at promoters of genes upregulated at 29°C in the wild-type samples compared to the beaf32−/− samples (Figure 5B (middle)) but not at genes upregulated at wild-type 18°C compared to beaf32−/− samples at 18°C. More specifically, we observe 39 genes significantly upregulated (adjustedP-value < 0.1 and log2 fold change >0.75) in the wild-type samples which harbor the BEAF-32 binding motifs at their promoter, while only 18 of them are also significantly upregulated at 18°C. In addition, the fold change difference of all 39 genes is significantly higher at 29°C compared to 18°C (Figure 5C). This validates the specificity of BEAF-32 binding at genes activated at 29°C. Interestingly, we also observe a similar yet not identical motif at the promoter of genes upregulated at 29°C compared to 18°C at the beaf32−/− samples (Figure 5B (right)). This motif is similar to a motif previously reported as enriched in DREF and BEAF-32 binding sites, while the motif found in promoters of the upregulated genes in wild-type samples is similar to a motif previously reported as enriched in BEAF-32 only binding sites (72). As the DRE motif is competitively bound both by BEAF-32 and DREF, we speculate that DREF activates those genes in the absence of BEAF-32. In addition, GO enrichment analysis shows that genes upregulated at 29°C at the beaf32−/− samples but not in the wild-type samples are enriched for metabolic processes, which is a hallmark of DREF regulated genes (72) (Supplementary Table S11). Thus, we demonstrate the involvement of BEAF-32 at the transcriptional response to temperature at 29°C but not at 18°C.
DISCUSSION
Here we described 5΄ and 3΄-RNA-sequencing protocols, together with a computational pipeline allowing for accurate annotation of transcript boundaries. The straightforwardness and simplicity of our methods make them an ideal complement of full RNA-seq libraries when a complete annotation of transcripts is necessary. An important example that requires accurate mapping of transcript boundary is functional analysis of non-coding RNAs where disruption or hindrance of the promoter is the only way to disrupt these transcripts without resorting to large deletions, which can confound interpretations (73).
Although our method can't simultaneously annotate both ends of the same transcript as previously reported (20), its lower input requirement and its easy multiplexed nature allow construction of both 5΄ and 3΄ libraries from the same sample and across multiple samples. By pooling and sequencing samples together our method can reduce both biological and technical noise. Another great advantage of the simplicity and modularity of the protocols is that they can be easily modified and optimized. For example, the protocol could be modified for using ligation primers containing unique identifiers to reduce PCR bias.
Accurate annotation of the 3΄-ends of mRNA with RNAseH-seq enabled us to discover a putative new CA-rich cleavage and polyadenylation motif that is enriched in genes relevant to oocyte development. While this motif have not been previously characterized, similar (yet not identical) CA-rich motifs have been reported upstream of proximal 3-UTRs in the Central Nervous System (CNS) and Testis (15). This motif can also be an extended form of the previously described CAAC motif, which was found in the 3΄-UTR of genes implicated in Notch signaling (74). Moreover, our experimental method for discerning true PAS from internal poly(A) tracts is significantly more sensitive than the common in silico method, suggesting that many true PAS are flanked with A-rich regions.
Transcriptional and post-transcriptional controls such as alternative splicing of 3΄-UTRs have been implicated in the response of Drosophila to changes in temperature (55,56,75). Our analysis revealed that genes that are upregulated in 18°C are enriched for the binding motif of ADF-1 in its core promoter, while genes that are upregulated in 29°C are enriched for the binding motif of DREF and BEAF-32. Indeed, a follow-up experiment validated the specificity of BEAF-32 activity in genes activated at 29°C but not at 18°C, demonstrating the role of BEAF-32 at the transcriptional response to warm (but not cold) temperatures. Interestingly, our data also suggests that in the absence of BEAF-32, the transcription factor DREF (which competitively binds to the BEAF-32 binding site) can activate genes upregulated at 29°C, compensating for the absence of BEAF-32. The fact that there is no significant change in the mRNA expression of those factors suggests that the regulatory mechanism is probably more complex and could also involve post-transcriptional mechanisms or co-factors. Also, a previous study have shown the involvement of Adf-1 in Polycomb-mediated chromatin repression (76), thus the regulation network could not only enhance transcription at 18°C, but could possibly repress transcription at 29°C.
The simplicity and straightforwardness of our approaches make them useful for going beyond traditional annotation methodologies. For example, we recently used our Exo-seq method to study parental imprinting in human cell lines (77). Parental imprinting involves a subset of genes that are expressed exclusively from one parental allele. Combining DNA methylation data with our Exo-seq method enabled the identification of tissue- and isoform-dependent imprinted genes and finding novel candidates for imprinted genes including novel promoters. This, along with the rest of our work presented here, demonstrate the great potential and variety of results we can achieve using Exo-seq and RNAseH-seq.
ACCESSION NUMBER
All sequencing data used in this work have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE60215.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Reut Ashwal-Fluss for help with some of the computational analysis and Patrick McDonel for help with protocol optimization.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
International Human Frontiers Science Program Organization [PG #31/2011 to S.K.]; European Research Council Consolidator Grant [ERC #647989 to S.K.]; Defense Advanced Research Project Agency [D13AP00074 to M.G., S.K.]; National Institute of Health [U01HG007910-01, UL1TR001453-01, 5U54HD082013-02 to M.G.). Funding for open access charge: European Research Council Consolidator Grant [ERC #647989 to S.K.].
Conflict of interest statement. Alexander A. Shishkin is an inventor on a massively multiplexed RNA-sequencing patent (publication number WO 2014152155 A1). The authors declare that they don't have any other conflict of interest.
REFERENCES
- 1. Yosef N., Regev A.. Impulse control: temporal dynamics in gene transcription. Cell. 2011; 144:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005; 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- 3. Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C.. The transcriptional landscape of the mammalian genome. Science. 2005; 309:1559–1563. [DOI] [PubMed] [Google Scholar]
- 4. Davuluri R. V, Suzuki Y., Sugano S., Plass C., Huang T.H.-M.. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008; 24:167–177. [DOI] [PubMed] [Google Scholar]
- 5. Elkon R., Ugalde A.P., Agami R.. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013; 14:496–506. [DOI] [PubMed] [Google Scholar]
- 6. de Klerk E., ’t Hoen P.A.C.. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015; 31:128–139. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Z., Dietrich F.S.. Mapping of transcription start sites in Saccharomyces cerevisiae using 5΄ SAGE. Nucleic Acids Res. 2005; 33:2838–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ni T., Corcoran D.L., Rach E.A., Song S., Spana E.P., Gao Y., Ohler U., Zhu J.. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods. 2010; 7:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ozsolak F., Kapranov P., Foissac S., Kim S.W., Fishilevich E., Monaghan A.P., John B., Milos P.M.. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010; 143:1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Plessy C., Bertin N., Takahashi H., Simone R., Salimullah M., Lassmann T., Vitezic M., Severin J., Olivarius S., Lazarevic D.. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat. Methods. 2010; 7:528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fu Y., Sun Y., Li Y., Li J., Rao X., Chen C., Xu A.. Differential genome-wide profiling of tandem 3΄ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011; 21:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanamori-Katayama M., Itoh M., Kawaji H., Lassmann T., Katayama S., Kojima M., Bertin N., Kaiho A., Ninomiya N., Daub C.O.. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res. 2011; 21:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shepard P.J., Choi E.A., Lu J., Flanagan L.A., Hertel K.J., Shi Y.. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011; 17:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haenni S., Ji Z., Hoque M., Rust N., Sharpe H., Eberhard R., Browne C., Hengartner M.O., Mellor J., Tian B.. Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 3΄-end-seq. Nucleic Acids Res. 2012; 40:6304–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smibert P., Miura P., Westholm J.O., Shenker S., May G., Duff M.O., Zhang D., Eads B.D., Carlson J., Brown J.B. et al. . Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012; 1:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun Y., Fu Y., Li Y., Xu A.. Genome-wide alternative polyadenylation in animals: insights from high-throughput technologies. J. Mol. Cell Biol. 2012; 4:352–361. [DOI] [PubMed] [Google Scholar]
- 17. Batut P., Dobin A., Plessy C., Carninci P., Gingeras T.R.. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013; 23:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., Park J.Y., Yehia G., Tian B.. Analysis of alternative cleavage and polyadenylation by 3΄ region extraction and deep sequencing. Nat. Methods. 2013; 10:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moqtaderi Z., Geisberg J. V, Jin Y., Fan X., Struhl K.. Species-specific factors mediate extensive heterogeneity of mRNA 3΄ ends in yeasts. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11073–11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pelechano V., Wei W., Jakob P., Steinmetz L.M.. Genome-wide identification of transcript start and end sites by transcript isoform sequencing. Nat. Protoc. 2014; 9:1740–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsumoto K., Suzuki A., Wakaguri H., Sugano S., Suzuki Y.. Construction of mate pair full-length cDNAs libraries and characterization of transcriptional start sites and termination sites. Nucleic Acids Res. 2014; 42:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai D.-P., Tan S., Kang Y.-N., Wu J., Ooi H.-S., Chen J., Shen T.-T., Qi Y., Zhang X., Guo Y. et al. . Genome-wide profiling of polyadenylation sites reveals a link between selective polyadenylation and cancer metastasis. Hum. Mol. Genet. 2015; 24:3410–3417. [DOI] [PubMed] [Google Scholar]
- 23. Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M.. CAGE: cap analysis of gene expression. Nat. Methods. 2006; 3:211–222. [DOI] [PubMed] [Google Scholar]
- 24. de Hoon M., Hayashizaki Y.. Deep cap analysis gene expression (CAGE): genome-wide identification of promoters, quantification of their expression, and network inference. Biotechniques. 2008; 44:627–632. [DOI] [PubMed] [Google Scholar]
- 25. Takahashi H., Lassmann T., Murata M., Carninci P.. 5[prime] end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012; 7:542–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hashimshony T., Wagner F., Sher N., Yanai I.. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012; 2:666–673. [DOI] [PubMed] [Google Scholar]
- 27. Jaitin D.A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A. et al. . Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014; 343:776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sheppard S., Lawson N.D., Zhu L.J.. Accurate identification of polyadenylation sites from 3΄ end deep sequencing using a naive Bayes classifier. Bioinformatics. 2013; 29:2564–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilkening S., Pelechano V., Järvelin A.I., Tekkedil M.M., Anders S., Benes V., Steinmetz L.M.. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013; 41:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ji G., Guan J., Zeng Y., Li Q.Q., Wu X.. Genome-wide identification and predictive modeling of polyadenylation sites in eukaryotes. Brief. Bioinform. 2015; 16:304–313. [DOI] [PubMed] [Google Scholar]
- 31. Nunes N.M., Li W., Tian B., Furger A.. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010; 29:1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang H., Lim J., Ha M., Kim V.N.. TAIL-seq: genome-wide determination of poly(A) tail length and 3΄ end modifications. Mol. Cell. 2014; 53:1044–1052. [DOI] [PubMed] [Google Scholar]
- 33. Meijer H.A., Bushell M., Hill K., Gant T.W., Willis A.E., Jones P., de Moor C.H.. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007; 35:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mikkelson G.M., Gonzalez A., Peterson G.D.. Economic inequality predicts biodiversity loss. PLoS One. 2007; 2:e444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Derr A., Yang C., Zilionis R., Sergushichev A., Blodgett D.M., Redick S., Bortell R., Luban J., Harlan D.M., Kadener S. et al. . End Sequence Analysis Toolkit (ESAT) expands the extractable information from single-cell RNA-seq data. Genome Res. 2016; 26:1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bartok O., Teesalu M., Ashwall-Fluss R., Pandey V., Hanan M., Rovenko B.M., Poukkula M., Havula E., Moussaieff A., Vodala S. et al. . The transcription factor Cabut coordinates energy metabolism and the circadian clock in response to sugar sensing. EMBO J. 2015; 34:1538–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I.. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 2013; 8:539–554. [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen L., Shao N., Liu X., Nestler E.. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 44. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Falcon S., Gentleman R.. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007; 23:257–258. [DOI] [PubMed] [Google Scholar]
- 46. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S.. Quantifying similarity between motifs. Genome Biol. 2007; 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoskins R.A., Landolin J.M., Brown J.B., Sandler J.E., Takahashi H., Lassmann T., Yu C., Booth B.W., Zhang D., Wan K.H. et al. . Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011; 21:182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shatkin A.J. Capping of eucaryotic mRNAs. Cell. 1976; 9:645–653. [DOI] [PubMed] [Google Scholar]
- 51. McCracken S., Fong N., Rosonina E., Yankulov K., Brothers G., Siderovski D., Hessel A., Foster S., Shuman S., Bentley D.L.. 5΄-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997; 11:3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Houseley J., Tollervey D.. The many pathways of RNA degradation. Cell. 2009; 136:763–776. [DOI] [PubMed] [Google Scholar]
- 53. Tadokoro T., Kanaya S.. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009; 276:1482–1493. [DOI] [PubMed] [Google Scholar]
- 54. Shishkin A.A., Giannoukos G., Kucukural A., Ciulla D., Busby M., Surka C., Chen J., Bhattacharyya R.P., Rudy R.F., Patel M.M. et al. . Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods. 2015; 12:323–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boothroyd C.E., Wijnen H., Naef F., Saez L., Young M.W.. Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 2007; 3:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Majercak J., Sidote D., Hardin P.E., Edery I.. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999; 24:219–230. [DOI] [PubMed] [Google Scholar]
- 57. Boley N., Stoiber M.H., Booth B.W., Wan K.H., Hoskins R.A., Bickel P.J., Celniker S.E., Brown J.B.. Genome-guided transcript assembly by integrative analysis of RNA sequence data. Nat. Biotechnol. 2014; 32:341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 59. Rach E.a, Yuan H.-Y., Majoros W.H., Tomancak P., Ohler U.. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 2009; 10:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wittebolle L., Marzorati M., Clement L., Balloi A., Daffonchio D., Heylen K., De Vos P., Verstraete W., Boon N.. Initial community evenness favours functionality under selective stress. Nature. 2009; 458:623–626. [DOI] [PubMed] [Google Scholar]
- 61. Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S.. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014; 505:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ito C., Goto S.G., Tomioka K., Numata H.. Temperature entrainment of the circadian cuticle deposition rhythm in Drosophila melanogaster. J. Biol. Rhythms. 2011; 26:14–23. [DOI] [PubMed] [Google Scholar]
- 63. Timmerman C., Suppiah S., Gurudatta B. V, Yang J., Banerjee C., Sandstrom D.J., Corces V.G., Sanyal S.. The Drosophila transcription factor Adf-1 (nalyot) regulates dendrite growth by controlling FasII and Staufen expression downstream of CaMKII and neural activity. J. Neurosci. 2013; 33:11916–11931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. DeZazzo J. nalyot, a mutation of the Drosophila myb-related adf1 transcription factor, disrupts synapse formation and olfactory memory. Neuron. 2000; 27:145–158. [DOI] [PubMed] [Google Scholar]
- 65. Livingstone M.S., Tempel B.L.. Genetic dissection of monoamine neurotransmitter synthesis in Drosophila. Nature. 303:67–70. [DOI] [PubMed] [Google Scholar]
- 66. England B.P., Heberlein U., Tjian R.. Purified Drosophila transcription factor, adh distal factor-1 (Adf-1), binds to sites in several Drosophila promoters and activates transcription. J. Biol. Chem. 1990; 265:5086–5094. [PubMed] [Google Scholar]
- 67. Ohler U., Liao G., Niemann H., Rubin G.M.. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002; 3, RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hart C.M., Cuvier O., Laemmli U.K.. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma. 1999; 108:375–383. [DOI] [PubMed] [Google Scholar]
- 69. Matsukage A., Hirose F., Yoo M.-A., Yamaguchi M.. The DRE/DREF transcriptional regulatory system: a master key for cell proliferation. Biochim. Biophys. Acta. 2008; 1779:81–89. [DOI] [PubMed] [Google Scholar]
- 70. Zhao K., Hart C.M., Laemmli U.K.. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995; 81:879–889. [DOI] [PubMed] [Google Scholar]
- 71. Rhee D.Y., Cho D.-Y., Zhai B., Slattery M., Ma L., Mintseris J., Wong C.Y., White K.P., Celniker S.E., Przytycka T.M. et al. . Transcription factor networks in Drosophila melanogaster. Cell Rep. 2014; 8:2031–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gurudatta B.V., Yang J., Van Bortle K., Donlin-Asp P.G., Corces V.G.. Dynamic changes in the genomic localization of DNA replication-related element binding factor during the cell cycle. Cell Cycle. 2013; 12:1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M. et al. . Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013; 2:e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lai E.C., Burks C., Posakony J.W.. The K box, a conserved 3΄ UTR sequence motif, negatively regulates accumulation of enhancer of split complex transcripts. Development. 1998; 125:4077–4088. [DOI] [PubMed] [Google Scholar]
- 75. Bartok O., Kyriacou C.P., Levine J., Sehgal A., Kadener S.. Adaptation of molecular circadian clockwork to environmental changes: a role for alternative splicing and miRNAs. Proc. Biol. Sci. 2013; 280:20130011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Orsi G.a, Kasinathan S., Hughes K.T., Saminadin-Peter S., Henikoff S., Ahmad K.. High-resolution mapping defines the cooperative architecture of Polycomb response elements. Genome Res. 2014; 24:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stelzer Y., Bar S., Bartok O., Afik S., Ronen D., Kadener S., Benvenisty N.. Differentiation of human parthenogenetic pluripotent stem cells reveals multiple tissue- and isoform-specific imprinted transcripts. Cell Rep. 2015; 11:308–320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.