Abstract
RNA-binding proteins (RBPs) are involved in mRNA splicing, maturation, transport, translation, storage and turnover. Here, we identified ACOT7 mRNA as a novel target of human WIG1. ACOT7 mRNA decay was triggered by the microRNA miR-9 in a WIG1-dependent manner via classic recruitment of Argonaute 2 (AGO2). Interestingly, AGO2 was also recruited to ACOT7 mRNA in a WIG1-dependent manner in the absence of miR-9, which indicates an alternative model whereby WIG1 controls AGO2-mediated gene silencing. The WIG1–AGO2 complex attenuated translation initiation via an interaction with translation initiation factor 5B (eIF5B). These results were confirmed using a WIG1 tethering system based on the MS2 bacteriophage coat protein and a reporter construct containing an MS2-binding site, and by immunoprecipitation of WIG1 and detection of WIG1-associated proteins using liquid chromatography-tandem mass spectrometry. We also identified WIG1-binding motifs using photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation analyses. Altogether, our data indicate that WIG1 governs the miRNA-dependent and the miRNA-independent recruitment of AGO2 to lower the stability of and suppress the translation of ACOT7 mRNA.
INTRODUCTION
MicroRNAs (miRNAs), ∼22-nt small non-coding RNAs, are found in a wide range of eukaryotes and play important roles in a variety of cellular processes (1–3). MiRNA associates with Argonaute (AGO) and other proteins to form the RNA-induced silencing complex (RISC), which functions in post-transcriptional gene silencing (2,4–6). The AGO protein family is a class of conserved essential molecules that are involved in diverse modes of small RNA-mediated gene silencing (2,7–10). Among the mammalian AGO protein family, only AGO2 has catalytic activity for slicing. Functions of the non-catalytic members AGO1, AGO3 and AGO4 appear to be at least partially redundant (11). As a consequence of AGO2 binding, the mRNA is sliced. On the other hand, for target mRNA repression, AGO proteins directly interact with a member of the GW protein family on the target mRNA. AGO-GW182 induces mRNA deadenylation by recruiting the deadenylase CCR4–NOT complex, subsequently triggering destabilization or translational repression of target mRNAs (5,11,12). MiRNAs in RISC generally bind to 3΄ untranslated regions (UTRs) of target mRNAs through base-pairing via a ∼7-nt complementary ‘seed’ region (5,13). However, it has been suggested that seed complementarity alone is not always sufficient to identify mRNA targets (14). Recent reports have revealed extensive AGO-bound mRNAs with no complementarity to miRNA seeds in AGO-crosslinking and immunoprecipitation (AGO-CLIP) experiments (15–18). Moreover, enrichment of a G-rich motif in sequences crosslinked with AGO2 was observed in the presence and absence of mature miRNAs (19). Thus, it remains unclear whether or not AGO proteins are RNA-binding proteins (RBPs) with their own specificities for mRNA sequence recognition.
RBPs critically contribute to the regulation of gene expression by affecting all aspects of RNA metabolism (20). In addition to mediating alternative splicing and polyadenylation of mRNAs, RBPs affect the stability and translation efficacy of target mRNAs, thereby controlling protein expression patterns in time and space (21,22). RBPs also facilitate mRNA quality control by recruiting specialized mRNA surveillance factors and are essential for proper microtubule-dependent localization of transcripts (23,24). Recent genetic, proteomic and animal model studies have suggested that RBPs may be involved in many human diseases, including cancers, neurological disorders and muscular atrophies (25–28). The functional diversity of RBPs reflects a corresponding diversity in the RNA-binding modules that are responsible for their recognition of target RNAs (26,29). Recently, several groups suggested that more specific miRNA–RBP interactions may be involved in the regulating the binding of miRNAs to target RNA motifs under various physiological conditions and in response to external stimuli (3,30–33). RBP binding induces local changes in the secondary structure of the mRNA 3΄UTR, creating or hiding sites for miRNA interaction and thereby modulating miRNA-mediated gene expression positively or negatively (30,34–36). RNA-binding activities of RBPs can be rapidly modulated by changes in RBP abundance, subcellular localization and post-translational modifications (3,37). However, we do not yet fully understand the mechanisms by which RBPs bind to target RNAs, or the consequences of such binding.
In this report, we identified ACOT7 mRNA as a novel WIG1 target. Interestingly, our findings reveal that the AGO2–GW182 complex exists on ACOT7 mRNA despite miRNA dysregulation due to WIG1 guidance. We also found that the translational status of ACOT7 mRNA is suppressed through the association of WIG1 with the general translation initiation factor 5B (eIF5B).
MATERIALS AND METHODS
Cell culture
MCF7 cells and HEK293T cells were grown in Dulbecco's modified Eagle's medium (WelGENE, Daegu, Korea). DICER knockout (−/−) HCT116 cell line, which was developed by Kim et al. (38), was purchased from the Korean Collection for Type Cultures (KCTC), Korea Research institute of Bioscience and Biotechnology (KRIBB), Korea. HCT116 parent cells and DICER knockout (−/−) HCT116 cell lines were grown in McCoy's 5A medium (WelGENE). Cells were cultured with 10% Fetal Bovine Serum (Lonza Group Ltd., Basel, Switzerland) and 1% penicillin and streptomycin solution (WelGENE) at 37°C in a 5% CO2 incubator.
Immunoprecipitation and biotin pulldown
Immunoprecipitation was performed as described previously (36). Briefly, Con Si- and WIG1 Si-transfected MCF7 cells were transfected with biotin-miR-9 or -miR-101 (Bioneer Inc., Daejeon, Korea) for streptavidin pulldown. After 2 days, harvested cells were rinsed and sonicated in NET-2 buffer. Yeast tRNA-coated streptavidin beads (Sigma-Aldrich, St Louis, MO, USA) were used to collect the biotinyl-miRNAs, and bead-bound RNAs were amplified using RT-sqPCR.
Biotinylated DNA probe-RNA hybridization and pulldown assay
Biotinylated DNA probe-RNA hybridization and biotinylation-streptavidin precipitation assay were performed with some modifications as previously described (39). Briefly, p3xFlag-WIG1 and pCK-Flag-AGO2-transfected MCF7 cells were transfected with pcFLuc or pcFLuc-ACOT7-3΄UTR-Wt or pcFLuc-ACOT7-3΄UTR-Mut. After 2 days, harvested cells were rinsed and sonicated in NET-2 buffer. Biotinyl-DNA probe and yeast tRNA-coated streptavidin beads (Sigma-Aldrich) were used to collect miRNAs, and collected miRNAs in precipitates were amplified using RT-sqPCR.
Plasmid construction
Construction details for p3xFlag-WIG1, pCK-Flag-AGO2 and pCMV-Myc-WIG1 were previously reported (36). pcFLuc-EV was generated with ligation of HindIII/XbaI-digested pGL2-basic (Promega Corp., Madison, WI, USA) fragment into pcDNA3 (Invitrogen Corp., Carlsbad, CA, USA). pcFLuc-ACOT7-3΄UTR-Wt or -Mut plasmids were constructed with ligation of XbaI-digested fragments containing ACOT7 3΄UTR (NM_007274) or mutated miR-9-binding sites (172C → A, 176G→ Δ) using gBlock (IDT Corp., Coralville, IA, USA) into pcFLuc plasmid. pcFLuc-MS2BS-ACOT7-3΄UTR-Wt or -Mut plasmids were constructed with ligation of BglII/XhoI-digested polymerase chain reaction (PCR) fragment amplified from plasmid pcFLuc-ACOT7-3΄UTR (Korea Human Gene Bank, Daejeon, Korea) using primers listed in Supplementary Table S1 into BglII/XhoI fragments of pcFLuc-ACOT7-3΄UTR-Wt or -Mut. To generate pMS2-HA plasmid, NheI/HindIII-digested pEGFP-C1 (Clontech Laboratories, Inc., Mountain view, CA, USA) was ligated with a NheI/HindIII fragment containing MS2-HA sequences using gBlock (IDT Corp.).
To construct pFL-ACOT7-3΄UTR-Wt or -Mut, pFL plasmid was inserted into an XbaI-digested pcFLuc-ACOT7-3΄UTR-Wt or -Mut fragment, respectively. To generate pCrPV-FL-ACOT7-3΄UTR-Wt or -Mut, HindIII fragment of pFL-ACOT7-3΄UTR-Wt or -Mut was ligated with a HindIII/EcoRI-digested PCR fragment amplified from php/CrPV/R-Gl plasmid and an EcoRI/HindIII-digested PCR fragment amplified from pFL-ACOT7-3΄UTR-Wt or -Mut plasmid using primers listed in Supplementary Table S1. To construct pEMCV-FL-ACOT7-3΄UTR-Wt or -Mut, pCrPV-FL-ACOT7-3΄UTR-Wt or -Mut was digested with BglII/EcoRI and ligated with a BglII/EcoRI-digested PCR fragment amplified from plasmid php/EMCV/R-Gl using primers listed in Supplementary Table S1.
To construct ACOT7-3΄UTR-BS Mut constructs, XbaI-digested fragments using gBlock (IDT Corp.) lacking putative WIG1-binding sites from the photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) analysis were inserted into pcFLuc or pFL plasmid. pFL-8BS construct containing 8-repeated MS2 protein-binding sites, php/EMCV/R-Gl and php/CrPV/R-Gl were kindly provided by Dr Lynne E. Maquat at University of Rochester, Rochester, NY, USA (40,41). pIRES-Flag/HA-GFP, -AGO2 and -AGO2 PAZ9 were kindly provided by Dr Sven Diederichs at the German Cancer Research Center (DKFZ) (42).
RNA interference and plasmid transfection
Cells were transiently transfected with the indicated plasmids, 100 nM in vitro-synthesized siRNAs (Bioneer Inc.) or miRNA mimetics (Bioneer Inc.), using either Lipofectamine 2000 (Invitrogen) or RNAi-MAX (Invitrogen) according to the manufacturer's instructions. The Con-, WIG1 (or WIG1 #1), WIG1 #2, WIG1 #3 and AGO2 (or AGO2-#1) specific siRNA (Si) sequences and anti-miR-106b sequences were used as described previously (36). AGO1, AGO2 #2, AGO2 #3, AGO3, AGO4, DICER (or DICER #1), DICER #2, DICER #3 and GW182 specific siRNA sequences used in this study were 5’-r(GAGAAGAGGUGCUCAAGAA)(dTdT)-3’, 5’-r(GCACGGAAGUCCAUCUGAA)(dTdT)-3’, 5’-r(UAAUACAUCUUUGUCCUGC)(dTdT)-3’, 5’-r(GAAAUUAGCAGAUUGGUAA)(dTdT)-3’, 5’-r(GGCCAGAACUAAUAGCAAU)(dTdT)-3’, 5’-r(UAAAGUAGCUGGAAUGAUG)(dTdT)-3’, 5’-r(UGCUUGAAGCAGCUCUGGA)(dTdT)-3’, 5’-r(GGAAGAGGCUGACUAUGAA)(dTdT)-3’ and 5’-r(UAGCGGACCAGACAUUUCU)(dTdT)-3’, respectively.
The miR-9-5p mimetic, miR-141-3p mimetic, miR-200a-3p mimetic, miR-101-3p mimetic, miR-142-3p mimetic, miR-Control, anti-miR-Control and anti-miR-9-5p sequences were 5’-r(UCUUUGGUUAUCUAGCUGUAUGA)-3’, 5’-r(UAACACUGUCUGGUAAAGAUGG)-3’, 5’-r(UAACACUGUCUGGUAACGAUGU)-3’, 5’-r(UACAGUACUGUGAUAACUGAA)-3’, 5’-r(UGUAGUGUUUCCUACUUUAUGGA)-3’, 5’-r(CCUACGCCACCAAUUUCGU)-3’, 2-methyl-O-labeled 5’-r(CCUACGCCACCAAUUUCGU)-3’ and 2-methyl-O-labeled 5’-r(UCAUACAGCUAGAUAACCAAAGA)-3’, respectively. After transfection, cells were collected and total protein and RNA were prepared for analysis.
Reverse transcription (RT) followed by semi-quantitative (sq)PCR or real-time, quantitative (q)PCR analysis
RT-sqPCR analysis was performed as described previously (36). Total RNA was isolated from cells using TRIzol reagent (Invitrogen); after reverse transcription (RT), PCR was performed using gene-specific primers (see below) and α-[32P]-dNTP (Perkin-Elmer NEN, Wellesley, MA, USA). Radioisotope-labeled PCR products were separated on 5% polyacrylamide gels and quantified using the Quantity One software package (Bio-Rad Laboratories, Hercules, CA, USA). A standard curve of intensity versus RNA quantity was generated by 2-fold serial dilutions of purified RNAs, and relative amounts of radioisotope-labeled PCR products were determined from the standard curve. RT-qPCR was performed using DNA Engine Thermal Cycler with a Chromo4 Real-time PCR Detector (Bio-Rad Laboratories) and the following mRNA-specific primers: for ACOT7, 5’-CTGCACCCTGCACGGCTTTG-3’ (sense) and 5’-CGGAAGCTGTGACGATGTTG-3’ (antisense) or 5’-TCTCCCATGTGCATCGGTG-3’ (sense) and 5’-TTTTCGGACATCACGTTGACC-3’ (antisense); for ANKRD11, 5’-CCAACAACGGGCACTACAAG-3’ (sense) and 5’-CGTCTTCCTCTTCTGAGCTC-3’ (antisense); and for actin, 5’-CAAGAGATGGCCACGGCT-3’ (sense) and 5’-TCCTTCTGCATCCTGTCGGC-3’ (antisense). Actin served as a negative or loading control. Primers for detection of FL were 5’-AATACGACTCACTATAGGCA-3’ (sense) and 5’-TCACTCTAGAGGATAGAATC-3’ (antisense); FL, 5’-CAACACCCCAACATCTTCG-3’ (sense) and 5’-CTTTCCGCCCTTCTTGGCC-3’ (antisense); and for RL, 5’-TGA TCCAGAACAAAGGAAAC-3’ (sense) and 5’-CTTATCTTGATGCTCATAGC-3’ (antisense). RL served as a negative or loading control.
Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) analysis
PAR-CLIP was performed with some modifications as previously described (43). Briefly, Flag-WIG1 was expressed in HEK 293T cells. Cells were incubated in medium supplemented with 100 mM 4SU for 16 h. Cells were crosslinked by ultraviolet light at 365 nm in a Spectrolinker XL-1500 UV crosslinker. After cells were sonicated and treated with 1 U/ml of RNase T1 (Fermentas, Burlington, Canada), Flag-WIG1 was immunoprecipitated with monoclonal anti-FLAG antibodies (M2, Sigma-Aldrich) bound to Protein G-conjugated Agarose. RNA residing in the immunoprecipitate was further trimmed with 100 U/ml of RNase T1. Beads were washed in lysis buffer and resuspended in one bead volume of dephosphorylation buffer. RNA was dephosphorylated and radioactively labeled with [γ-32P]-ATP using poly-nucleotide kinase. The protein–RNA complexes were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis, after which migrating radioactive bands were transferred to a NC membrane. Protein was subjected to western blotting or removed by digestion in proteinase K buffer in the presence of 1 mg/ml of proteinase K (Sigma-Aldrich). RNA was extracted by using Trizol (Invitrogen), converted into a cDNA library and sequenced using an Illumina platform. Adapters were removed, and the processed reads were aligned to the reference genome (GRCh37/hg19) by the Bowtie algorithm (1.1.2), allowing for two alignment errors (mutation, insertion or deletion). For analysis, PARalyzer settings were set to require a minimum of five sequence reads per group and no non-conversion mismatches per sequence read after masking for repeat elements using RepeatMasker. A PARalyzer-defined group was considered as a binding site only if it had a T-to-C mutation rate of 0.25, contained more than five sequence reads with T-to-C conversions and had two or more distinct crosslinking sites. The groups and clusters were annotated with hg19 Ensembl v72 database. To approximate binding intensity using crosslinked read frequencies, raw sequence reads (rather than non-redundant sequence reads) were counted. The problem of PCR amplification bias was avoided by limiting the number of PCR cycles used for cDNA amplification to stay within the exponential amplification phase of the PCR reaction.
RESULTS
WIG1 is critical to regulation of ACOT7 mRNA level via miR-9-mediated mRNA decay
In a previous work, we demonstrated that WIG1 binds to p21 (CDKN1A) mRNA and facilitates RISC recruitment to degrade p21 mRNA (36). In this work, we identified additional WIG1 target mRNAs by ribonucleoprotein immunoprecipitation (RIP) and microarray analysis (RIP-chip; Supplementary Table S2). Among the list of putative WIG1 target mRNAs, we confirmed physical interaction between WIG1 and ACOT7 mRNA using RIP and RT followed by semiquantitative polymerase chain reaction (sqPCR) (Figure 1A). Specificity of the RIP experiment was confirmed by negative and positive controls (Supplementary Figure S1). Depletion and overexpression of WIG1 increased and decreased ACOT7 mRNA and ACOT7 protein levels, respectively (Figure 1B).
Figure 1.
WIG1 regulates ACOT7 mRNA levels via miR-9-mediated mRNA decay. (A) Association of WIG1 with ACOT7 mRNA. MCF7 cells were harvested 2 days after transfection with p3xFlag-WIG1, and RIP analysis was performed using an anti-Flag M2 affinity gel. Immunoprecipitated proteins were subjected to immunoblotting using an anti-Flag antibody (top); RNAs were radiolabeled by RT-PCR using ACOT7-specific primers (bottom), and band intensities were quantified using Quantity One software. The four leftmost lanes represent 2-fold serial dilutions of RNA and confirm that RT-PCR was semi-quantitative. Data are expressed as the means ± SD from three independent experiments. (B) Effects of WIG1 depletion on ACOT7 mRNA and protein levels. MCF7 cells were transfected with WIG1 Si, and qRT-PCR and western blotting analyses were performed. Ectopic expression of WIG1 downregulates ACOT7 mRNA and protein levels. MCF7 cells transfected with p3xFlag-WIG1 construct were harvested and analyzed as described in the left panel. (C) Binding of AGO2 to ACOT7 mRNA in a WIG1-dependent manner. HEK 293T cells were harvested 2 days after transfection with either WIG1 or AGO2 Si, after which an RNP-IP assay was performed using anti-AGO2 or anti-WIG1 antibody, respectively. Immunoprecipitated proteins were subjected to immunoblotting using anti-AGO2 and anti-WIG1 antibody (left). Isolated RNAs were performed with qRT-PCR for quantification of ACOT7 mRNA levels (right). Actin mRNA and protein were used as a negative or normalized control. (D) Effects of miR-9 on ACOT7 mRNA and ACOT7 protein levels in WIG1-depleted MCF7 cells. Cell lysates were prepared 2 days after transfection of miR-9 into WIG1 Si-transfected cells and subjected to western blot analysis (top). RNAs were isolated, and RT-qPCR analysis was used to quantify ACOT7 mRNA levels (bottom). (E) Effects of miR-9 on ACOT7 mRNA and protein levels in WIG1-overexpressing MCF7 cells. p3xFlag-WIG1 plasmids were transfected into miR-9-transfected cells; 2 days later, protein levels were assessed by immunoblot analysis (top) and ACOT7 mRNA levels by RT-qPCR analysis (bottom). (F) WIG1-dependent binding of miR-9 to ACOT7 mRNA. Cell lysates were harvested and pulled-down with streptavidin beads 2 days after transfection with biotin-miR-9 in WIG1-depleted HEK 293T cells. Pulled-down RNA was amplified by RT-sqPCR using ACOT7-specific primers and quantified. Actin mRNA levels served as internal controls. The four leftmost lanes represent sequential 2-fold dilutions of RNA. Data are expressed as the means ± SD from three independent experiments. (G) Presence of miR-9 on target ACOT7 mRNA under WIG1-overexpressing conditions. Cell lysates were harvested and precipitated with streptavidin beads 2 days after transfection with biotin-miR-9 in WIG1-overexpressing HEK 293T cells. Precipitated RNAs were analyzed as described in panel F. *P < 0.05; **P < 0.01; #P > 0.05.
Next, we determined whether or not binding of AGO2 to ACOT7 mRNA is dependent on WIG1 and vice versa in WIG1- and AGO2-depleted cells. While binding of AGO2 to ACOT7 mRNA was found to be dependent on WIG1, binding of WIG1 to ACOT7 mRNA was not dependent on AGO2 (Figure 1C). We identified miR-9-5p, miR-141-3p and miR-200a-3p as microRNAs putatively interacting with ACOT7 3΄UTR (http://www.targetscan.org v7.0; Supplementary Figure S2A). Among them, miR-9-5p (hereafter miR-9) was highly expressed compared to miR-141-3p or miR-200a-3p and showed high affinity for ACOT7 mRNA, which required the presence of WIG1 (Supplementary Figure S2B). Transfection of the miR-9 mimic alone lowered ACOT7 mRNA and protein levels (Figure 1D). WIG1 depletion abrogated miR-9-induced reduction of ACOT7 mRNA and protein levels (Figure 1D). No off-target effects of three independent WIG1 Si at lower concentration were observed (Supplementary Figure S3). In WIG1-overexpressing cells, transfection of miR-9 further reduced ACOT7 mRNA and protein levels compared to miR-Control (Con)-transfected cells (Figure 1E). WIG1 overexpression promoted suppression of ACOT7 mRNA and protein levels by miR-9 (Figure 1E). In addition, we observed a synergistic effect of WIG1 and AGO2 co-overexpression on ACOT7 mRNA and protein levels (Supplementary Figure S4).
To further characterize this interaction, biotinylated miR-9 mimic and streptavidin-conjugated beads were used to pulldown miR-9-interacting mRNAs from WIG1-depleted and WIG1-overexpressing cells. As shown, ACOT7 mRNA was detected by RT-sqPCR analysis of the pulldown material (Figure 1F and G). Importantly, ACOT7 mRNA failed to precipitate with miR-9 in the absence of WIG1 (Figure 1F), whereas it was highly abundant in biotin-miR-9 pulldowns using lysates from WIG1-overexpressing cells (Figure 1G). ANKRD11 mRNA, an additional putative target of WIG1 (Supplementary Table S2), was also validated (Supplementary Figures S1 and 5). As shown, WIG1 contributed to regulation of ANKRD11 mRNA levels by mediating target-specific binding of miRNAs miR-142-3p or miR-101-3p, ultimately leading to miRNA-mediated decay of ANKRD11 mRNA (Supplementary Figure S5D–F). These results demonstrate that WIG1 is essential for regulation of the levels of target transcripts ACOT7 and ANKRD11 through miRNA-mediated mRNA decay.
WIG1 binds to ACOT7 mRNA 3΄UTR and recruits AGO2 under miRNA deficiency
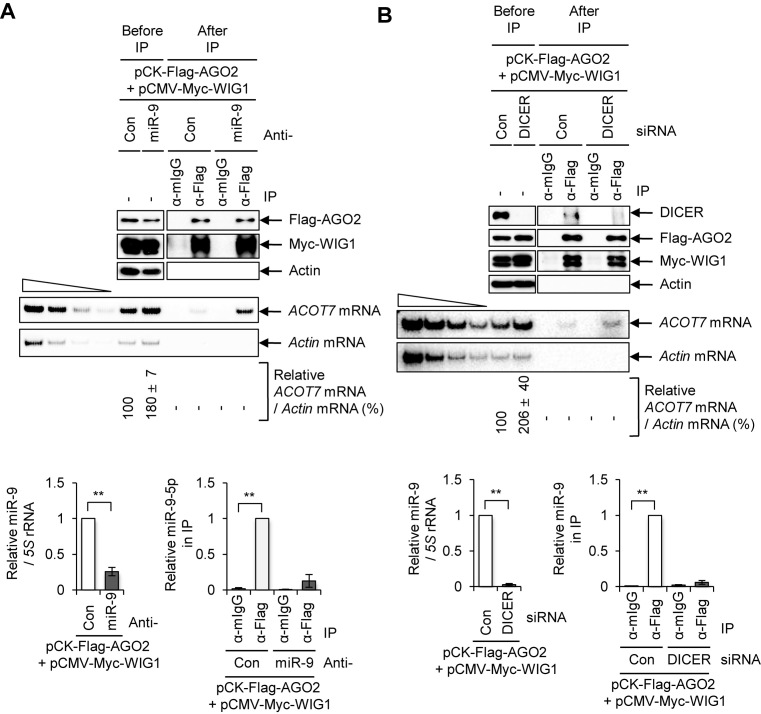
It is generally accepted that miRNAs mediate binding between AGO2 and complementary sequences in target mRNAs (2,5,8,9,11). Thus, we investigated whether or not the interaction of WIG1–AGO2 with ACOT7 mRNA depends on miRNA and target complementary sequences. First, we examined binding of the WIG1–AGO2 complex to ACOT7 mRNA after anti-miR-9 transfection (Figure 2A). We previously demonstrated that WIG1 directly associates with AGO2 (36) and the WIG1–AGO2 complex bound to ACOT7 mRNA target in the absence of miR-9 (Figure 2A). After DICER silencing, we confirmed disruption of the DICER-dependent miRNA maturation pathway using positive and negative control miRNAs (Supplementary Figure S6). We observed comparable interaction of WIG1–AGO2 with ACOT7 mRNA in both Con siRNA (Si)- and DICER Si-transfected cells (Figure 2B), suggesting that microRNAs were dispensable for WIG1 binding to ACOT7 mRNA.
Figure 2.
Inhibition of miRNA processing and disruption of ACOT7-3΄UTR miR-9-binding site are not sufficient to block AGO2-WIG1 binding to ACOT7 mRNA. (A) Effect of anti-miR-9-5p on AGO2-WIG1 binding to ACOT7 mRNA. HEK 293T cell lysates were harvested and precipitated with anti-Flag antibody 2 days after co-transfection of pCK-Flag-AGO2 and pCMV-Myc-WIG1 plasmids in combination with anti-miR-9. From RNP-IP, precipitated proteins were subjected to Western blotting (top). RNAs in precipitates were radiolabeled with RT-PCR using ACOT7-specific primers (bottom). Actin mRNA served as a negative control. Band intensity was quantified using Quantity One software. The four leftmost lanes represent 2-fold serial dilutions of RNA, and confirm RT-PCR was semi-quantitative. Relative level of miR-9 in anti-miR-9-treated cells was normalized to that of a 5S rRNA internal control, relative to anti-miR-Con. Fold enrichment of miR-9 in Flag-AGO2 immunoprecipitates was assessed after treatment with anti-miR-9, relative to anti-miR-Con. Data are presented as means ± SD from three independent experiments. (B) Binding of AGO2-WIG1 to ACOT7 mRNA in DICER-silenced cells. Immunoprecipitation was performed with anti-Flag antibody 2 days after co-transfection of pCK-Flag-AGO2 and pCMV-Myc-WIG1 in DICER Si-transfected HEK 293T cells. Precipitated proteins were subjected to western blotting (top). RNAs in precipitates were subjected to RT-PCR using ACOT7 mRNA-specific primers (bottom). Relative level of miR-9 in DICER-treated cells was normalized to that of a 5S rRNA internal control, relative to anti-miR-Con. Fold enrichment of miR-9 in Flag-AGO2 immunoprecipitates was assessed after treatment with DICER Si, relative to anti-miR-Con. Data are presented as means ± SD from three independent experiments. *P < 0.05; **P < 0.01; #P > 0.05.
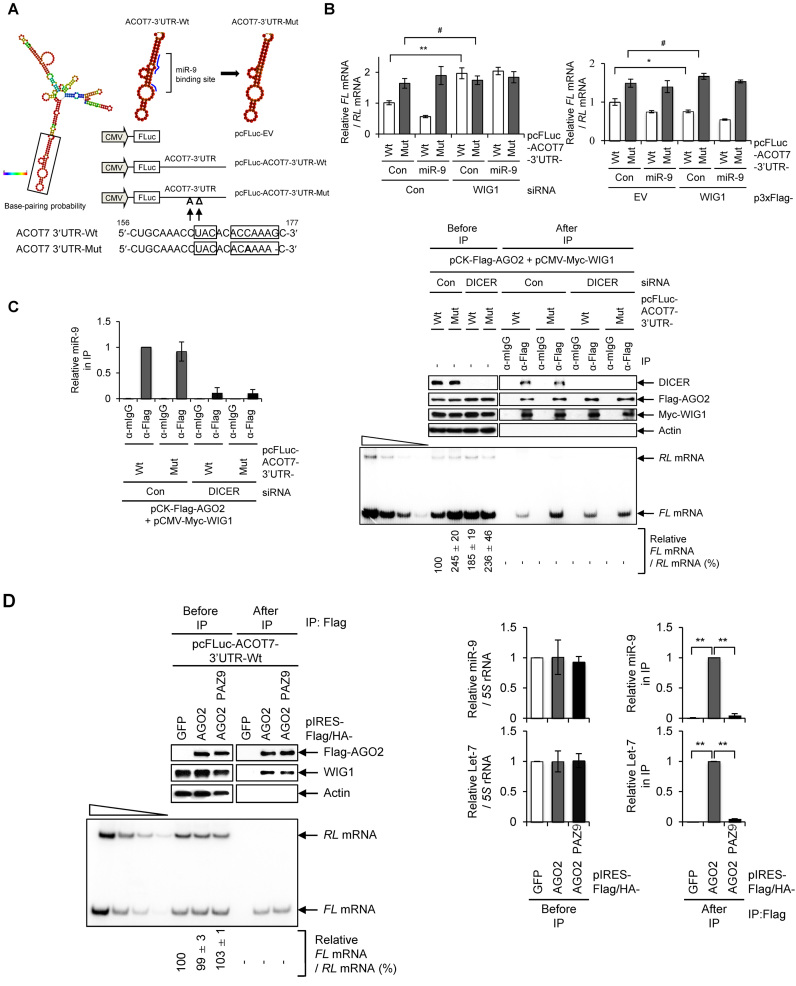
To further study this possibility, we tested a mutant (Mut) luciferase reporter construct bearing disruption of the miR-9 seed region within the full-length ACOT7 3΄UTR (pcFLuc-ACOT7-3΄UTR-Mut; Figure 3A). In contrast to changes in the level of wild-type (Wt) ACOT7 3΄UTR reporter mRNA after WIG1 depletion, ACOT7 3΄UTR Mut reporter mRNA was not responsive to alterations in the levels of WIG1 or miR-9 (Figure 3B, left). We observed similar changes in reporter mRNA levels in WIG1-overexpressing cells (Figure 3B, right). In addition, we observed that the WIG1–AGO2 complex bound to both ACOT7 3΄UTR Wt mRNA and ACOT7 3΄UTR Mut mRNA in both Con Si- and DICER Si-transfected cells (Figure 3C). These results further indicate that the WIG1–AGO2 complex was capable of binding to the ACOT7 3΄UTR in the absence of miR-9 or after disrupting the miR-9 complementary sequence. However, the possibility remains that other miRNAs could guide AGO2 through DICER-independent miRNA biogenesis (44,45). To test this possibility, we transfected AGO2 PAZ9 mutant, which contains nine point mutations within the AGO2 PAZ domain (42,46–48). Although AGO2 PAZ9 mutant showed reduced interactions with miRNAs (Figure 3D, right), the WIG1–AGO2 PAZ9 complex was capable of binding to the ACOT7 3΄UTR (Figure 3D, left) without changes in miR-9 levels (Figure 3D, middle). Altogether, these results indicate that WIG1 is indispensable for AGO2 binding to the ACOT7 3΄UTR, regardless of the miRNA loading ability of AGO2 or the presence of miRNA complementary sequence on the mRNA.
Figure 3.
WIG1 binds to ACOT7 mRNA 3΄UTR and recruits AGO2 despite disruption of miR-9 site, inhibition of miRNA synthesis or loss of miRNA. (A) Cloning strategy for disrupting miR-9 target sequence on ACOT7 3‘UTR with stem loops as predicted by RNAfold software. ACOT7 3΄UTR containing one substitution (172C → A) and one deletion (176G → Δ) in miR-9 target site was cloned into pcFLuc-empty vector (EV); Wt, wild-type; Mut, mutant. pcFLuc-EV was used as control. Base-pair probability is color-coded. (B) Effect of miR-9 on ACOT7-3΄UTR-Wt and -Mut reporters after modulation of WIG1 levels. Cells were transfected with either WIG1 Si or p3xFlag-WIG1 (Flag-Wig), followed by either pcFLuc-ACOT7-3΄UTR-Wt or -Mut in combination with miR-9. pRL-CMV was used as a reference plasmid. RNAs were isolated 2 days after transfection and measured by RT-qPCR analysis. FL mRNA levels were normalized to those expressed from pcFLuc-EV mRNA; RT-PCR intensities represent the means ± SD from three independent experiments. (C) Binding of AGO2 to ACOT7 3΄UTR containing mutations in miR-9 target sites. HEK 293T cells were transfected with DICER Si and then transfected with either pcFLuc-ACOT7-3΄UTR-Wt or -Mut reporter plasmids, pCK-Flag-AGO2 and pCMV-Myc-WIG1 in combination with pRL-CMV reference plasmid. Left, following RIP, precipitated proteins were assessed by western blot analysis to identify the indicated proteins (top), RNAs were radiolabeled using RT-PCR with FL and RL mRNA-specific primers and band intensities were quantified using Quantity One software (bottom). The four leftmost lanes represent sequential 2-fold dilutions of RNA. Left, miR-9 levels in Flag-AGO2 immunoprecipitates, as assessed by RT-qPCR analysis using pcFLuc-ACOT7-3΄UTR-Wt and -Mut reporters. Data represent the means ± SD from three independent experiments. (D) MicroRNA binding-defective AGO2 PAZ9 mutant binds to ACOT7 mRNA in the presence of WIG1. HEK 293T cells were transfected with pcFLuc-ACOT7-3΄UTR-Wt in combination with either pIRES-Flag/HA-AGO2 or -AGO2 PAZ9, after which RIP was performed using anti-Flag antibody. Precipitated proteins were assessed by western blot analysis (top), and precipitated RNAs were assessed by RT-qPCR analysis (bottom). pRL-CMV was used as a negative control for IP efficiency. Levels of miRNAs extracted from pre-immune lysates (Before IP) or immunoprecipitates (After IP) were assessed by RT-qPCR (right). Data represent the means ± SD from three independent experiments. *P < 0.05; **P < 0.01; #P > 0.05.
WIG1 suppresses translational initiation through the association of WIG1–AGO2–GW182 complex with eIF5B
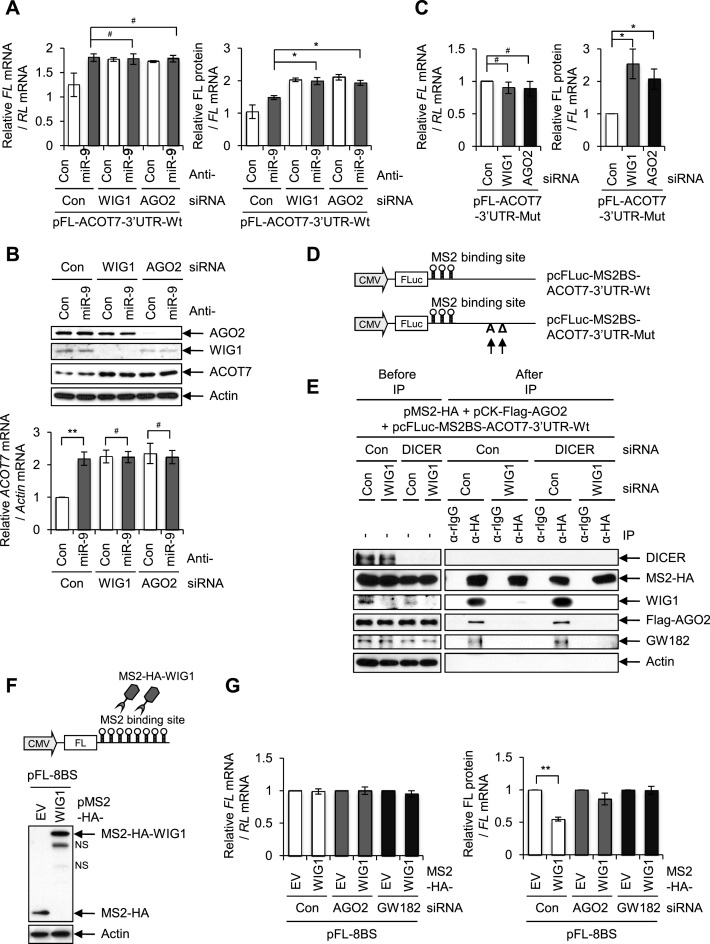
Since the WIG1–AGO2 complex was shown to be capable of binding to the ACOT7 3΄UTR independent of miRNA, we examined the role of the WIG1–AGO2 complex in gene silencing under such conditions. After transfection of either pFL-ACOT7-3΄UTR-Wt or pFL-ACOT7-3΄UTR-Mut, we calculated translational efficiency using dual luciferase assay and RT-qPCR detection of RL and FL mRNAs (Figure 4A). Translation was enhanced with increasing levels of FL-ACOT7-3΄UTR-Wt mRNA in the absence of miR-9 (Figure 4A, right). It was further enhanced when either WIG1 or AGO2 was additionally depleted in the absence of miR-9 (Figure 4A, right), although there was no change in FL-ACOT7-3΄UTR-Wt mRNA level (Figure 4A, left). Consistent results were observed when we examined endogenous ACOT7 mRNA and protein levels under the same conditions (Figure 4B). In pFL-ACOT7-3΄UTR-Mut-transfected cells, translational efficiencies were enhanced in the absence of either WIG1 or AGO2 (Figure 4C, right) with no change in reporter mRNA levels (Figure 4C, left).
Figure 4.
WIG1 tethering suppresses ACOT7 protein synthesis in AGO2-GW182 dependency. (A) Relative translational efficiency of ACOT7 3΄UTR reporters in the presence of anti-miR-9 in WIG1- or AGO2-depleted cells. MCF7 cells were transfected with either WIG1 Si or AGO2 Si as well as with pFL-ACOT7-3΄UTR-Wt and anti-miR-9 in combination with reference plasmid pRL-CMV. Cells were harvested 2 days after transfection, and FL and RL mRNAs were quantified using RT-qPCR. FL mRNA levels were normalized to RL mRNA levels (left). Translation rates were determined as the ratios of FL protein (luciferase activity, normalized to RL) to FL mRNA (normalized to RL mRNA). (B) Endogenous ACOT7 mRNA and protein levels in the presence of anti-miR-9 in WIG1- or AGO2-depleted cells. Lysates were prepared as explained in Figure 4A, followed by western blot analysis. Total RNA was subjected by qRT-PCR using ACOT7-specific primers. Actin was used as a loading control. (C) MCF7 cells were transfected with either WIG1 Si or AGO2 Si in combination with pFL-ACOT7-3΄UTR-Mut and pRL-CMV reference plasmid. FL mRNA levels and translation rates were determined as explained in panel A. (D) Diagrams of reporter vectors encoding ACOT7 mRNA 3΄UTR-Wt and -Mut with three copies of MS2-binding site (BS): pcFLuc-MS2BS-ACOT7-3΄UTR-Wt and -Mut. pcFLuc-EV was used as a reference control. (E) WIG1-dependent recruitment of AGO2-GW182 to 3΄UTR of ACOT7 mRNA in the absence of miRNA processing. HEK 293T cells were transfected with DICER Si and WIG1 Si and pcFLuc-MS2BS-ACOT7-3΄UTR-Wt reporter plasmid in combination with pCK-Flag-AGO2 and pMS2-HA. Cells were harvested 2 days after transfection and subjected to IP and western blot analysis to detect the indicated proteins. (F) Diagrams of MS2-HA-WIG1 reporter vector encoding FL coding region with eight copies of MS2-binding site (BS). (G) MCF7 cells were transfected either AGO2 Si or GW182 Si, followed by pFL-8BS, pMS2-HA and pMS2-HA-WIG1 in combination with pRL-CMV reference plasmid. Cells were harvested 2 days after transfection, followed by western blot analysis. Reporter RNA levels and translation rates were determined as explained in panel A. Data are presented as means ± SD from three independent experiments. *P < 0.05; **P < 0.01; #P > 0.05.
We next investigated whether or not WIG1 helps recruit another RISC component, GW182, to the target mRNA regardless of miRNA loading capability. We co-transfected a reporter plasmid and an effector plasmid. Reporter plasmid encodes either FLuc-MS2BS-ACOT7-3΄UTR-Wt or -Mut, which expresses three copies of the MS2 coat protein-binding site (MS2BS) downstream of the reporter STOP codon (Figure 4D). An effector plasmid produces MS2-HA, which expresses the bacteriophage MS2 coat protein tagged with the polypeptide HA (40). When we measured relative reporter mRNA levels with these constructs in the presence and absence of WIG1, reporter mRNAs were destabilized in WIG1- and miRNA-dependent manners (Supplementary Figure S7A). Endogenous GW182 associated with the WIG1–AGO2 complex on the ACOT7-3΄UTR when miRNA biogenesis was impaired (DICER Si) and in the absence of miRNA complementary sequence (FLuc-MS2BS-ACOT7-3΄UTR-Mut) (Supplementary Figure S7B).
It is generally believed that mammalian miRNAs guide the RISC to target mRNAs through the seed-pairing rule (18). However, recent reports have demonstrated that members of the AGO family of proteins directly associate with mRNAs lacking seed complementarity with miRNAs (18,49–51). We observed that AGO2 had a direct binding preference for the ACOT7 3΄UTR, which was dependent on the presence of WIG1 but not miRNAs (Figure 4E); GW182 was also detected in WIG1-dependent and miRNA-independent manners (Figure 4E). Whereas WIG1 still bound to ACOT7 3΄UTR in AGO2-depleted cells, GW182 was not present in the immunoprecipitates (Supplementary Figure S7C). Together, these data further support the view that recruitment of AGO2 to target ACOT7 mRNA is dependent on WIG1 but not miRNAs. Furthermore, GW182 associated with AGO2 on target ACOT7 mRNA in the presence and absence of miRNAs in a WIG1-dependent manner (Figure 4E). No off-target effect of DICER Si was observed, as shown in Supplementary Figure S8. In a previous report, we demonstrated that p21 mRNA is a novel target of the WIG1–AGO complex (36). Here, WIG1 was found to be involved in translational suppression of p21 mRNA independently of miR-106b (Supplementary Figure S9A).
To assess the combined effects of WIG1, AGO2 and GW182 on translational suppression, we generated a MS2-HA-WIG1 tethering construct on pFL-8BS reporter mRNA containing the full-length firefly luciferase (FL) coding region and MS2 protein-binding sites beyond the terminal codon (Figure 4F). Fusion protein levels in cells transfected with pMS2-HA-WIG1 were detected by Western blot analysis (Figure 4G). MS2-HA-WIG1 reduced the efficiency of translation from FL-8BS mRNA (Figure 4G, right, lane 1 versus 2) without altering mRNA levels (Figure 4G, left). Since GW182 interacts with AGO proteins via a specific AGO-interaction domain (6,52–55), we hypothesized that the tethered WIG1 might also require recruitment of GW182. Thus, we examined the extent of translational repression by tethered WIG1 upon depletion of either AGO2 or GW182 (Figure 4G and Supplementary Figure S9B); as shown, depletion of either AGO2 or GW182 rescued loss of translational efficiency mediated by tethered WIG1 (Figure 4G, right, lane 3 versus 4 and lane 5 versus 6). No off-target effect of AGO2 Si was observed, as shown in Supplementary Figure S10. In the absence of WIG1, AGO2–GW182 complex was not found in the ACOT7 3’UTR (Supplementary Figure S11A). However, AGO2–GW182 interaction was still observed in the absence of endogenous WIG1 (Supplementary Figure S11B). These data are consistent with a model whereby WIG1 recruits AGO2–GW182 to a target mRNA through direct interaction between WIG1 and AGO2 in an miRNA-independent manner, resulting in translational suppression.
We additionally examined whether or not WIG1 could interact with other human AGO proteins. AGO1 was detectable with GW182 in WIG1 immunoprecipitates (Supplementary Figure S12A). The AGO1–GW182 complex was not found on the ACOT7 3’UTR in the absence of WIG1 (Supplementary Figure S11A). However, AGO1-GW182 interaction per se was present in the absence of endogenous WIG1, as previously shown with AGO2 (Supplementary Figure S11B). In addition, GW182 was barely detectable on the ACOT7 3’UTR in either AGO1- or AGO2-depleted cells (Supplementary Figure S12B). In contrast to AGO2, AGO1 depletion affected the ACOT7 protein level but not mRNA level and deadenylation (Supplementary Figure S12C and D). From these results, we suggest that AGO1 and AGO2 might play different roles in the regulation of target gene expression with WIG1 dependency.
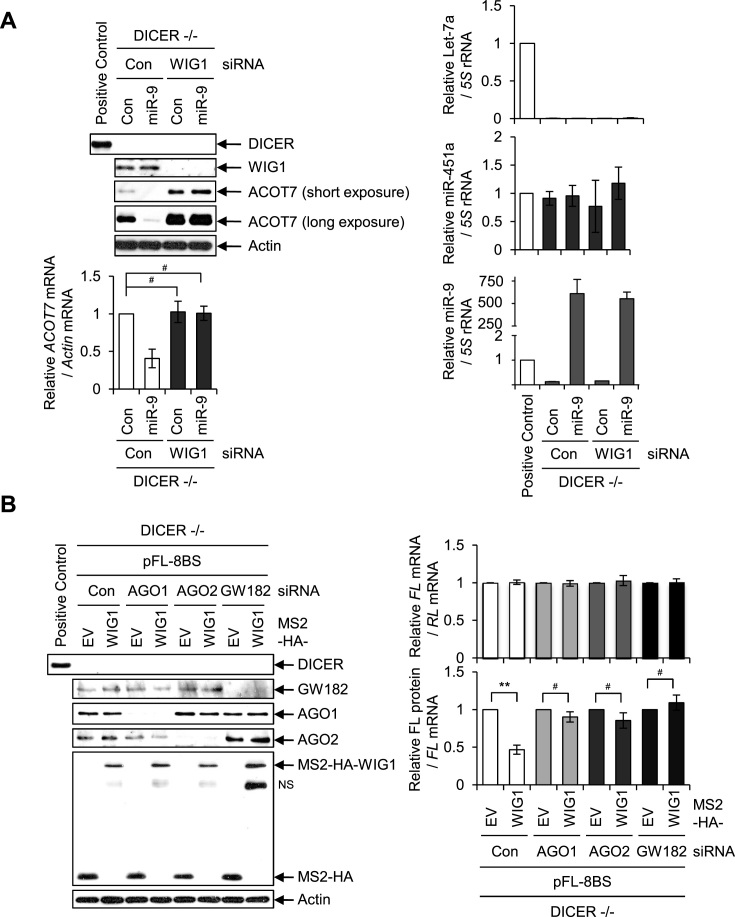
We further demonstrated the combined effect of WIG1, AGO1/2 and GW182 on translational suppression in human DICER −/− cells developed by Kim et al. (38). Transfection of miR-9 mimic alone lowered ACOT7 mRNA and protein levels in DICER −/− cells (Figure 5A, left, lane 1 versus 2). MiR-9 levels were observed with positive and negative controls in DICER −/− cells (Figure 5A, right). Upon WIG1 depletion, ACOT7 protein levels evidently increased in DICER −/− cells, regardless of miR-9 transfection (Figure 5A). However, WIG1 did not suppress protein expression after AGO1, AGO2 or GW182 depletion in DICER −/− cells (Figure 5B). Altogether, we observed that WIG1 is indispensable to regulation of ACOT7 translation in an miRNA-independent manner in DICER −/− cells.
Figure 5.
Effects of WIG1 on ACOT7 mRNA and protein levels in human HCT116 DICER knockout (−/−) cell line. (A) WIG depletion induces upregulation of ACOT7 protein levels without alteration of mRNA levels, regardless of miR-9, in DICER −/− cells. Cells were harvested at 2 days after miR-9 transfection and subjected to western blot analysis. RT-qPCR analysis was used to quantify specific RNA levels. Let-7a and miR-451a were used as positive and negative controls for DICER-dependent miRNA-maturation products, respectively. (B) WIG1 tethering in the absence of AGO1, AGO2 or GW182 induces translational suppression in DICER −/− cells. pFL-8BS reporters in combination with pMS2-HA plasmids were transfected in AGO1, AGO2 or GW182 Si-transfected DICER −/− cells. Two days after transfection, cell lysates analyzed with western blotting and RT-qPCR. Translation rate was measured as the ratio of FL protein to mRNA. HCT116 parental cells were used as a positive control regarding DICER −/− cells. Data are represented as the means ± SD from three independent experiments. **P < 0.01; #P > 0.05.
To identify which translation step is regulated by the WIG1–AGO2–GW182 complex, we generated several 5’UTR reporter constructs with different internal ribosome entry sites (IRESs) (Figure 6A). The cricket paralysis virus (CrPV) IRES requires no initiation factor, binds directly to ribosomal subunits and promotes elongation, whereas the encephalomyocarditis virus (EMCV) IRES requires all initiation factors apart from eIF4E (56). Furthermore, a hairpin structure upstream of each IRES blocks initiation of ribosomes at the cap to ensure translational initiation only at the IRES (57,58). MCF7 cells were transfected with each reporter construct containing WIG1 Si, after which mRNA decay and translational efficiency were evaluated. Stabilization of EMCV IRES- and CrPV IRES-ACOT7-3΄UTR-Wt reporter mRNAs was observed in WIG1-depleted cells (Figure 6B, top). However, in contrast to cap- and EMCV IRES-dependent translational efficiencies, CrPV IRES-dependent translational efficiency was not elevated in WIG1-depleted cells (Figure 6B, bottom). This indicates that WIG1-mediated translational repression was not dependent on elongation blockage, ribosome drop-off or polypeptide degradation.
Figure 6.
WIG1 prevents eIF5B assembly during translational initiation of 60S ribosomal subunit joining via AGO2-GW182 recruitment under miRNA deficiency. (A) Schematic representations of IRES constructs encoding FL with ACOT7 mRNA 3΄UTR. pFL-EV was used as a normalization control. (B) WIG1 suppresses translational initiation in the ACOT7-3΄UTR. MCF7 cells were transfected with WIG1 Si and pFL-ACOT7-3΄UTR-Wt, pEMCV-FL-ACOT7-3΄UTR-Wt or pCrPV-FL-ACOT7-3΄UTR-Wt in combination with pRL-CMV reference plasmid. Cells were harvested 24 h after transfection, and FL mRNA and (normalization control) RL mRNA were quantified by RT-qPCR analysis (top). Normalized level of each IRES reporter mRNA in the presence of each Con Si was set as 1. Translation rates were calculated as explained in Figure 4A. FL protein levels (FL activity) were normalized to RL levels (RL activity) (bottom); normalized levels of FL/RL activity in the presence of Con Si were set as 1. (C) MiR-9 is not required for WIG1-mediated translational initiation of ACOT7 mRNA. MCF7 cells were transfected with WIG1 Si and pFL-ACOT7-3΄UTR-Mut, pEMCV-FL-ACOT7-3΄UTR-Mut or pCrPV-FL-ACOT7-3΄UTR-Mut in combination with pRL-CMV reference plasmid. Cells were harvested 24 h after transfection and FL mRNA and RL mRNA (for normalization) were quantified RT-qPCR (top). Normalized levels of each IRES reporter mRNA in the presence of each Con Si was set as 1. Translation rate was determined as the ratio of normalized FL protein to FL mRNA (as defined in Figure 4A); normalized level of FL activity in the presence of Con Si was defined as 1. Data are represented as the means ± SD from three independent experiments. **P < 0.01. (D) RNA-independent interaction between WIG1 and eIF5B. HEK 293T cell lysates were obtained from WIG1-overexpressing HEK 293T cells. After immunoprecipitation using anti-Flag M2 affinity gel, proteins in the IP material were detected by western blot analysis (top). RT-qPCR detection of Actin mRNA was performed to assess completion of RNase A digestion (bottom). (E) EIF5B was preferentially recruited to WIG1-target mRNAs and specifically associates with AGO2-GW182. After transfection of HEK 293T cells with either AGO2 Si or GW182 Si and subsequent transfection with p3xFlag-WIG1, anti-Flag was used for immunoprecipitation, and the indicated proteins were detected by immunoblotting.
To test whether or not WIG1-mediated translational repression requires miRNA, we tested translational regulation of IRES-ACOT7-3΄UTR-Mut reporters under WIG1-depleted conditions (Figure 6C). In the absence of WIG1, we observed no difference between IRES-ACOT7-3΄UTR-Mut reporter mRNAs and cap-dependent ACOT7-Mut reporter mRNA (Figure 6C, top). Consistent with the above results (Figure 6B, bottom), translation of CrPV IRES reporter was unaffected by WIG1 depletion (Figure 6C, bottom). Next, we sought to identify which WIG1-interacting molecules might be involved in the translational initiation step. Using liquid chromatography-tandem mass spectrometry (LC-MS/MS), we detected eIF5B as a novel WIG1-interacting protein (Supplementary Figure S13A and B). We confirmed that WIG1 and eIF5B interacted directly and were not tethered by RNA, as their association was insensitive to RNase A digestion (Figure 6D).
To assess the specificity of the WIG1–eIF5B interaction, we examined interactions under either AGO2- or GW182-depleted conditions. The WIG1–eIF5B interaction was not detected in Flag-WIG1 immunoprecipitates upon depletion of either AGO2 or GW182 (Figure 6E), indicating that eIF5B associated with WIG1 through the AGO2–GW182 interaction. Collectively, these data provide evidence that WIG1-mediated translational suppression occurs at the translational initiation step through binding of the WIG1–AGO2–GW182 complex with eIF5B, regardless of miRNA-mediated translational control.
WIG1-binding motifs identified by PAR-CLIP analysis are critical for translational suppression
To identify binding motifs of WIG1 on the ACOT7-3΄UTR, we conducted PAR-CLIP analysis using anti-WIG1 antibody. After RNase T1 digestion and separation of crosslinked WIG1–RNA complexes, we performed RNA sequencing analysis of RNA bound to WIG1 (Figure 7A). A list of the cluster sequences and three highest-scoring motifs identified from the WIG1 PAR-CLIP analysis are shown (Supplementary Table S3 and Figure 7B).
Figure 7.
Identification of WIG1-binding sequence by PAR-CLIP analysis. (A) 5’-end 32P-labeled RNA-Flag-WIG1 immunoprecipitates were prepared from RNase T1-treated lysates in the presence or absence of 100 mM 4-thiouridine (4SU) photoactivatable nucleoside and crosslinked with UV 365 nm (right). Western blot analysis using anti-Flag antibody (left). (B) Representative RNA recognition elements (RREs) of the top three significantly enriched motifs derived from motif analysis of WIG1 PAR-CLIP. (C) Cloning strategy for disruption of putative WIG1 PAR-CLIP site on the 3΄UTR of ACOT7 mRNA with secondary structure predicted by RNAfold software. ACOT7-3΄UTR-BS Mut lacking two putative WIG1-binding sites (BS) from the PAR-CLIP analysis was cloned into pcFLuc vector. Base-pairing probabilities are color-coded. (D) Deletion of putative WIG1-binding sites disrupts WIG1 binding to the ACOT7 3΄UTR. HEK 293T cells were transfected with either pcFLuc-ACOT7-3΄UTR-Wt or -BS-Mut in combination with p3xFlag-WIG1 and pRL-CMV as a reference plasmid. After RIP using anti-Flag antibody, western blot analysis was performed using anti-Flag antibody (top) and RL and FL mRNAs RNAs were analyzed by quantification of radiolabeled RT-PCR products using Quantity One software (bottom). The four leftmost lanes represent sequential 2-fold dilutions of RNA. pRL-CMV were used as a negative control. The relative ratio of RT-sqPCR intensity represents the mean ± SD from three independent experiments. (E) Relative translational efficiency of ACOT7-3΄UTR reporters in WIG1-depleted cells. Diagrams of reporter vectors encoding ACOT7 mRNA 3΄UTR-BS-Mut with full-length FL coding region. pFL-EV or pFL-ACOT7-3΄UTR-Wt was used as normalization control or positive control, respectively. Two days after transfection of MCF7 cells with WIG1 Si and pFL-ACOT7-3΄UTR reporters (pFL-EV, -Wt, -BS Mut) and pRL-CMV reference plasmid, RNA was isolated and FL mRNA and RL mRNA (normalization control) quantified by RT-qPCR analysis (left). Protein levels were determined with luciferase activity assay (right), and translation rates were determined as explained in Figure 4A. **P < 0.01.
To investigate WIG1 binding to putative binding sequences from the PAR-CLIP analysis, we generated a reporter construct containing deletions of putative WIG1-binding sequences in the ACOT7 3΄UTR (pcFLuc-ACOT7-3΄UTR-BS Mut; Figure 7C). Physical interaction of WIG1 with ACOT7-3΄UTR-Wt reporter mRNA but not ACOT7-3΄UTR-BS Mut reporter mRNA was observed in WIG1 immunoprecipitates, (Figure 7D). In addition, reporter mRNA level and translational efficiency were enhanced in pFL-ACOT7-3΄UTR-BS Mut-transfected cells compared to ACOT7-3΄UTR-Wt reporter in Con Si-transfected cells (Figure 7E, lane 1 versus 2). In contrast, reporter mRNA level and translational efficiency were the same after WIG1 depletion in ACOT7-3΄UTR-Wt- and -BS Mut-transfected cells (Figure 7E, lane 3 versus 4), indicating that translation driven by ACOT7-3΄UTR- BS Mut was unaffected by WIG1 depression.
ACOT7 mRNA and protein levels are tuned in a WIG1-dependent manner following irradiation
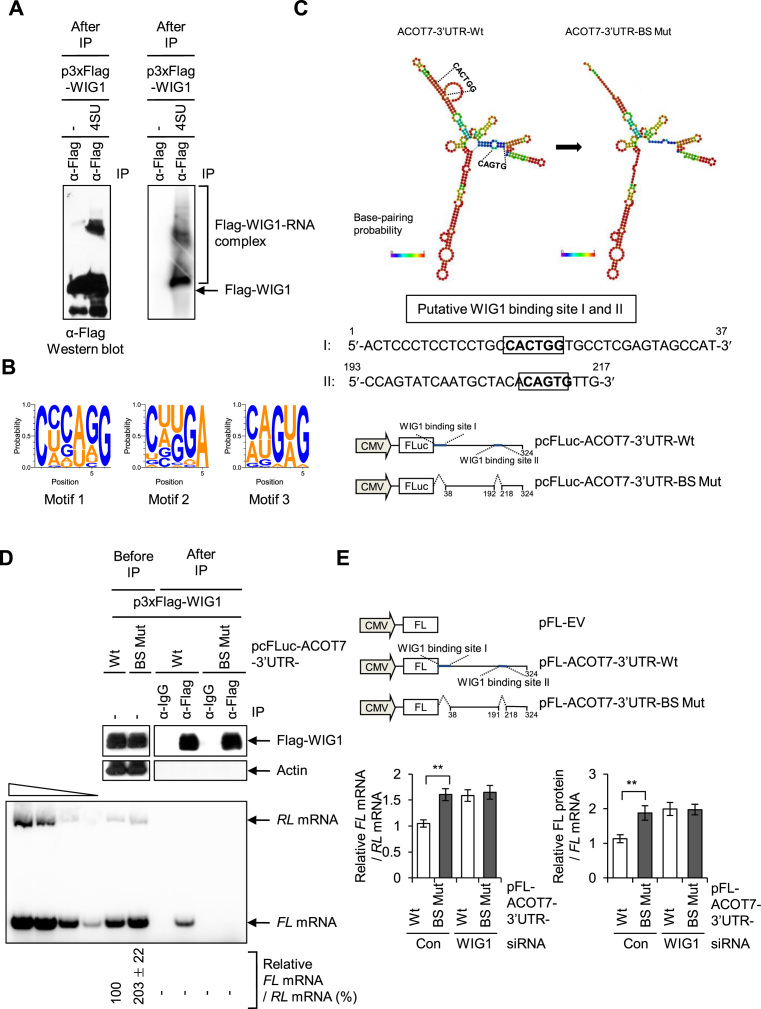
Since WIG1 expression is induced by the transcription factor p53 (59), we examined molecular statuses of p53, WIG1 and ACOT7 under p53-activating conditions. Ionizing radiation (IR) triggered p53 accumulation, WIG1 induction and ACOT7 downregulation in a dose-dependent manner (Figure 8A). Upon depletion of WIG1 in irradiated cells, we failed to observe IR-induced downregulation of ACOT7 mRNA and ACOT7 protein levels (Figure 8B, lane 2 versus 4). Furthermore, we observed that treatment with anti-miR-9 increased ACOT7 mRNA levels regardless of IR treatment (Figure 8C, bottom, lane 2 versus 4); however, under such conditions, IR exposure did not affect ACOT7 protein level, potentially indicating an effect of translational suppression due to p53-mediated WIG1 induction (Figure 8C, top, lane 2 versus 4). These results indicate that WIG1 fine-tuned ACOT7 gene expression via miRNA-dependent and -independent mechanisms under stress conditions. Altogether, our findings indicate that WIG1 plays a role in mRNA silencing, as seen for ACOT7 mRNA, via two alternative mechanisms depending on mRNA availability: (i) in the presence of miRNA, WIG1 mediates target ACOT7 mRNA decay and (ii) in the absence of miRNA, WIG1 is involved in ACOT7 mRNA translational suppression (Figure 8D).
Figure 8.
Regulation of ACOT7 mRNA and protein levels in a WIG1-dependent manner following irradiation. (A) MCF7 cells were harvested after exposure to 2, 4 or 6 Gy of ionizing radiation (IR) and subjected to immunoblotting for detection of p53, WIG1 and ACOT7. (B) Cells were transfected with Con Si or WIG1 Si, exposed to 6 Gy of IR, incubated for 24 h and then harvested for immunoblot and RT-PCR analyses. (C) After transfection with anti-miR-9, MCF7 cells were exposed to IR and then harvested for immunoblot and RT-PCR analyses. Actin was used as an internal control (A–C). Each bar in the graphs represents the mean ± SD compared to control cells from three independent experiments (B and C). *P < 0.05; **P < 0.01; #P > 0.05. (D) Proposed model for novel dual role of WIG1: miRNA-dependent mRNA decay and miRNA-independent translational suppression. RNA-binding protein WIG1 is a novel dual repressor of gene silencing. WIG1 regulates target mRNA stability in an miRNA-dependent manner and suppresses translational initiation in an miRNA-independent manner.
DISCUSSION
In the present study, we identified ACOT7 mRNA as a novel WIG1 target using different RNA–protein assays. ACOT7 mRNA expression was regulated by miR-9-mediated decay in a WIG1-dependent manner. Interestingly, ACOT7 expression was also regulated in an miRNA-independent manner via the WIG1–AGO2–GW182 complex, which suppressed ACOT7 translation. In this paradigm, WIG1 suppressed initiation of translation by associating with the general translation initiation factor eIF5B. Furthermore, we identified WIG1 putative binding RNA motifs using PAR-CLIP and confirmed the critical role of these motifs in the regulation of ACOT7 gene silencing by WIG1.
Importantly, miRNA-mediated gene regulation fundamentally affects various cell functions (9). Key molecular events such as translational inhibition, deadenylation and mRNA decay are primarily coupled with miRNA mediated-mRNA silencing (10). MiRNA-mediated mRNA repression is triggered through molecular interactions between miRNA and the target mRNA 3΄UTR. However, recent experiments revealed that extensive AGO-associated mRNAs lack seed complementarity with miRNAs (15,18,19), and AGO–mRNA interactions also occur in the absence of miRNAs (6). Thus, there is a need for a new model to explain AGO recruitment of mRNA targets without miRNA guidance (11). A previous report revealed the novel function of AGO proteins as RBPs with specific sequence preferences (18). The RNA motif for AGO recognition was identified in a structurally accessible region from PAR-CLIP data. The motif established a role for AGO in recognition of its own targets without miRNA guidance. Other reports suggested that AGO proteins recruit mRNA targets by guiding RBPs without the guidance of miRNAs (19,49,51).
Accordingly, RBPs might help to recruit AGO to target RNAs via protein–protein interactions, after which AGO protein may facilitate gene silencing via interactions with proteins in the GW family (46). Furthermore, mature miRNAs are not produced in DICER knock-out mouse embryonic stem cells (ESCs) or DICER-deficient mouse embryonic fibroblasts; in both of these cases, AGO proteins can still associate with mRNA targets (19). In addition, in Caenorhabditis elegans and human cells, the RBP Pumilio (PUF) binds to AGO proteins on target mRNAs and the PUF-AGO protein and eukaryotic elongation factor 1α (eEF1A) complex represses translation elongation in vitro (49). In Drosophila melanogaster, the RBP Smaug recruits AGO1 to mRNAs independently of miRNAs and causes the target mRNA nanos to silence gene expression (51). Despite the fact that AGO proteins are recruited by RBPs to mRNAs in an miRNA-independent manner, it would be difficult to demonstrate that recruitment of AGOs is directly involved in RBP-mediated mRNA translational inhibition. Whether or not miRNAs bind to AGOs also remains unclear in this model.
We observed that WIG1-dependent AGO2 targeting triggers miRNA-mediated mRNA decay. In addition, we observed that WIG1 facilitates AGO2 binding to target mRNA independent of the presence of miRNA, ultimately leading to translation repression via formation of the WIG1–AGO2–GW182–eIF5B complex. The essential translation factor eIF5B assists the 60S ribosomal subunit joining in translation initiation (60,61). We observed that the WIG1–AGO2–GW182–eIF5B complex was abrogated and that translational inhibition was rescued upon either AGO2 or GW182 depletion. These results support the notion that the WIG1–AGO2 interaction is critical for WIG1-mediated translational inhibition. Our results strongly support a model in which RBPs guide the recruitment of AGO to mRNA through protein–protein interactions independently of miRNA. The contribution of WIG1 to target mRNA silencing may, therefore, be distinct and more intractable than miRNA-mediated mRNA silencing; however, if no miRNAs are available or the miRNA processing pathway is defective, WIG1 may adopt an alternative pathway in order to fine-tune gene silencing. In fact, this dual property may be evolutionarily beneficial for regulation of gene expression to avoid detrimental cellular outcomes resulting from abnormal gene expression. Collectively, our findings suggest that WIG1 is a novel dual repressor of gene silencing that regulates target mRNA stability in an miRNA-dependent manner and suppresses translational initiation in an miRNA-independent manner.
WIG1 is a p53-inducible RNA-binding zinc-finger protein identified in almost all species and cell types (59,62,63). Several studies reported that WIG1 is involved in cell cycle arrest and cell death through stability regulation of mRNAs such as p53, N-myc, FAS and 14-3-3σ (64–66). Our previous work showed that WIG1 knockdown leads to cellular senescence by controlling p21 mRNA stability (36). We provided evidence that WIG1 plays a critical role in tumor progression in a murine xenograft model and human lung cancer tissues. Other study revealed the role of WIG1 in the maintenance of ESC through genome-wide RNAi screening (67). Another group recently showed that WIG1 is involved in the pathophysiology of Huntington's disease (68). WIG1 preferentially increases stability of mutant Huntingtin transcripts through higher affinity binding to mutant than normal counterparts. Therefore, these findings suggest that WIG1 could be a promising target molecule in relation to clinical applications.
Supplementary Material
ACKNOWLEDGEMENTS
The pFL-8BS, php/EMCV/R-Gl and php/CrPV/R-Gl constructs were kindly provided by Dr Lynne E. Maquat. pIRES-Flag/HA-GFP, -AGO2 and -AGO2 PAZ9 constructs were kindly provided by Dr Sven Diederichs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Basic Science Research Program [2014R1A2A1A11051988]; Nuclear Research and Development Program [2012-M2B2B1-2012055637]; Medical Research Center (MRC) through the National Research Foundation (NRF) funded by the Korean government (MSIP) [201409392]; NIA-IRP, NIH (to S.D., D.D., J.L.M., M.G.) [Z01-AG000511-19]. Funding for open access charge: Basic Science Research Program through the National Research Foundation (NRF) funded by the Korean government (MSIP) [2014R1A2A1A11051988].
Conflict of interest statement. None declared.
REFERENCES
- 1. Yi R., Qin Y., Macara I.G., Cullen B.R.. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003; 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carthew R.W., Sontheimer E.J.. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009; 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Kouwenhove M., Kedde M., Agami R.. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer. 2011; 11:644–656. [DOI] [PubMed] [Google Scholar]
- 4. Kim V.N., Han J., Siomi M.C.. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009; 10:126–139. [DOI] [PubMed] [Google Scholar]
- 5. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frohn A., Eberl H.C., Stohr J., Glasmacher E., Rudel S., Heissmeyer V., Mann M., Meister G.. Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol. Cell Proteomics. 2012; 11:1442–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004; 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 8. Meister G., Tuschl T.. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004; 431:343–349. [DOI] [PubMed] [Google Scholar]
- 9. Höck J., Meister G.. The Argonaute protein family. Genome Biol. 2008; 9:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Djuranovic S., Nahvi A., Green R.. A parsimonious model for gene regulation by miRNAs. Science. 2011; 331:550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 12. Huntzinger E., Izaurralde E.. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011; 12:99–110. [DOI] [PubMed] [Google Scholar]
- 13. Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P.. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007; 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lal A., Navarro F., Maher C.A., Maliszewski L.E., Yan N., O’Day E., Chowdhury D., Dykxhoorn D.M., Tsai P., Hofmann O. et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3΄UTR microRNA recognition elements. Mol. Cell. 2009; 35:610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chi S.W., Zang J.B., Mele A., Darnell R.B.. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009; 460:479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Helwak A., Kudla G., Dudnakova T., Tollervey D.. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013; 153:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., Kim T., Nutiu R., Ray D., Hughes T.R., Zhang Z.. Identifying mRNA sequence elements for target recognition by human Argonaute proteins. Genome Res. 2014; 24:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leung A.K., Young A.G., Bhutkar A., Zheng G.X., Bosson A.D., Nielsen C.B., Sharp P.A.. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 2011; 18:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hieronyms H., Silver P.A.. A system view of mRNP biology. Genes Dev. 2004; 18:2845–2860. [DOI] [PubMed] [Google Scholar]
- 21. Dai W., Zhang G., Makeyev E.V.. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 2012; 40:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huot M.E., Vogel G., Zabarauskas A., Ngo C.T., Coulombe-Huntington J., Majewski J., Richard S.. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol. Cell. 2012; 46:187–199. [DOI] [PubMed] [Google Scholar]
- 23. Lykke-Anderson J. mRNA quality control: Marking the message for life or death. Curr. Biol. 2001; 11:R88–R91. [DOI] [PubMed] [Google Scholar]
- 24. Hurst S., Talbot N.J., Stebbings H.. A staufen-like RNA-binding protein in translocation channels linking nurse cells to oocytes in Notonecta shows nucleotide-dependent attachment to microtubules. J. Cell Sci. 1999; 112:2947–2955. [DOI] [PubMed] [Google Scholar]
- 25. Wang G.S., Cooper T.A.. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007; 8:749–761. [DOI] [PubMed] [Google Scholar]
- 26. Lukong K.E., Chang K.W., Khandjian E.W., Richard S.. RNA-binding proteins in human genetic disease. Trends Genet. 2008; 24:416–425. [DOI] [PubMed] [Google Scholar]
- 27. Bagni C., Tassone F., Neri G., Hagerman R.. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J. Clin. Invest. 2012; 122:4314–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wurth L. Versatility of RNA-binding proteins in cancer. Comp. Funct. Genomics. 2012; 2012:178525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J.A., Elkon R., Agami R.. A pumilio-induced RNA structure switch in p27-3΄ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010; 12:1014–1020. [DOI] [PubMed] [Google Scholar]
- 31. Elcheva I., Goswami S., Noubissi F.K., Spiegelman V.S.. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol. Cell. 2009; 35:240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H.H., Kuwano Y., Srikantan S., Lee E.K., Martindale J.L., Gorospe M.. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009; 23:1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jafarifar F., Yao P., Eswarappa S.M., Fox P.L.. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011; 30:1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J.. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005; 120:623–634. [DOI] [PubMed] [Google Scholar]
- 35. Léveillé N., Elkon R., Davalos V., Manoharan V., Hollingworth D., Oude Vrielink J., le Sage C., Melo C.A., Horlings H.M., Wesseling J. et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat. Commun. 2011; 2:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim B.C., Lee H.C., Lee J.J., Choi C.M., Kim D.K., Lee J.C., Ko Y.G., Lee J.S.. Wig1 prevents cellular senescence by regulating p21 mRNA decay through control of RISC recruitment. EMBO J. 2012; 31:4289–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adeli K. Translational control mechanisms in metabolic regulation: critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. Am. J. Physiol. Endocrinol. Metab. 2011; 301:E1051–E1064. [DOI] [PubMed] [Google Scholar]
- 38. Kim Y.K., Kim B., Kim V.N.. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M.. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012; 47:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim Y.K., Furic L., Desgroseillers L., Maquat L.E.. Mammalian Staufen1 recruits Upf1 to specific mRNA 3΄UTRs so as to elicit mRNA decay. Cell. 2005; 120:195–208. [DOI] [PubMed] [Google Scholar]
- 41. Isken O., Kim Y.K., Hosoda N., Mayeur G.L., Hershey J.W., Maquat L.E.. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008; 133:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Winter J., Link S., Witzigmann D., Hildenbrand C., Previti C., Diederichs S.. Loop-miRs: active microRNAs generated from single-stranded loop regions. Nucleic Acids Res. 2013; 41:5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoon J.H., De S., Srikantan S., Abdelmohsen K., Grammatikakis I., Kim J., Kim K.M., Noh J.H., White E.J., Martindale J.L. et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 2014; 5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J.. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010; 465:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010; 328:1694–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu J, Rivas F.V., Wohlschlegel J., Yates J.R. 3rd, Parker R., Hannon G.J.. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005; 7:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Diederichs S., Haber D.A.. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007; 131:1097–1108. [DOI] [PubMed] [Google Scholar]
- 48. Gu S., Jin L., Huang Y., Zhang F., Kay M.A.. Slicing-independent RISC activation requires the argonaute PAZ domain. Curr. Biol. 2012; 22:1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Friend K., Campbell Z.T., Cooke A., Kroll-Conner P., Wickens M.P., Kimble J.. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 2012; 19:176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karginov F.V., Hannon G.J.. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013; 27:1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pinder B.D., Smibert C.A.. microRNA-independent recruitment of Argonaute 1 to nanos mRNA through the Smaug RNA-binding protein. EMBO Rep. 2013; 14:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E.. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006; 20:1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ding L., Han M.. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007; 17:411–416. [DOI] [PubMed] [Google Scholar]
- 54. Yao B., La L.B., Chen Y.C., Chang L.J., Chan E.K.. Defining a new role of GW182 in maintaining miRNA stability. EMBO Rep. 2012; 13:1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pfaff J., Meister G.. Argonaute and GW182 proteins: an effective alliance in gene silencing. Biochem. Soc. Trans. 2013; 41:855–860. [DOI] [PubMed] [Google Scholar]
- 56. Meijer H.A., Kong Y.W., Lu W.T., Wilczynska A., Spriggs R.V., Robinson S.W., Godfrey J.D., Willis A.E., Bushell M.. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013; 340:82–85. [DOI] [PubMed] [Google Scholar]
- 57. Gregorio E.D., Preiss T., Hentze M.W.. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999; 18:4865–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Holbrook J.A., Neu-Yilik G., Gehring N.H., Kulozik A.E., Hentze M.W.. Internal ribosome entry sequence-mediated translation initiation triggers nonsense-mediated decay. EMBO Rep. 2006; 7:722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Israeli D., Tessler E., Haupt Y., Elkeles A., Wilder S., Amson R., Telerman A., Oren M.. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J. 1997; 16:4384–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pestova T.V., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U.. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000; 403:332–335. [DOI] [PubMed] [Google Scholar]
- 61. Jackson R.J., Hellen C.U., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Varmeh-Ziaie S., Okan I., Wang Y., Magnusson K.P., Warthoe P., Strauss M., Wiman K.G.. Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene. 1997; 15:2699–2704. [DOI] [PubMed] [Google Scholar]
- 63. Vilborg A., Bersani C., Wilhelm M.T., Wiman K.G.. The p53 target Wig-1: a regulator of mRNA stability and stem cell fate?. Cell Death Differ. 2011; 18:1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vilborg A., Glahder J.A., Wilhelm M.T., Bersani C., Corcoran M., Mahmoudi S., Rosenstierne M., Grandér D., Farnebo M., Norrild B. et al. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:15756–15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vilborg A., Bersani C., Wickström M., Segerström L., Kogner P., Wiman K.G.. Wig-1, a novel regulator of N-Myc mRNA and N-Myc-driven tumor growth. Cell Death Dis. 2012; 3:e298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bersani C., Xu L.D., Vilborg A., Lui W.O., Wiman K.G.. Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14-3-3σ. Oncogene. 2014; 33:4407–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chia N.Y., Chan Y.S., Feng B., Lu X., Orlov Y.L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.S. et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010; 468:316–320. [DOI] [PubMed] [Google Scholar]
- 68. Kim S.H., Shahani N., Bae B.I., Sbodio J.I., Chung Y., Nakaso K., Paul B.D., Sawa A.. Allele-specific regulation of mutant Huntingtin by Wig1, a downstream target of p53. Hum Mol Genet. 2016; 25:2514–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.