Abstract
Sequencing-based, massively parallel genetic assays have revolutionized our ability to quantify the relationship between many genotypes and a phenotype of interest. Unfortunately, variant library expression platforms in mammalian cells are far from ideal, hindering the study of human gene variants in their physiologically relevant cellular contexts. Here, we describe a platform for phenotyping variant libraries in transfectable mammalian cell lines in two steps. First, a landing pad cell line with a genomically integrated, Tet-inducible cassette containing a Bxb1 recombination site is created. Second, a single variant from a library of transfected, promoter-less plasmids is recombined into the landing pad in each cell. Thus, every cell in the recombined pool expresses a single variant, allowing for parallel, sequencing-based assessment of variant effect. We describe a method for incorporating a single landing pad into a defined site of a cell line of interest, and show that our approach can be used generate more than 20 000 recombinant cells in a single experiment. Finally, we use our platform in combination with a sequencing-based assay to explore the N-end rule by simultaneously measuring the effects of all possible N-terminal amino acids on protein expression.
INTRODUCTION
Massively parallel genetic assays have revolutionized our ability to quantify the relationship between genotype and phenotype (1). In a massively parallel genetic assay, tens or hundreds of thousands of variants are introduced into a model system, a selection pressure is applied, and high-throughput sequencing is used to score each variant based on changes in frequency during selection. Using this approach, we can now measure the effect of all possible gene deletions in a genome (2,3) or all possible single mutants of a protein (4). Massively parallel genetic assays require that each cell or organism contain a defined genetic alteration, which must remain stable throughout the experiment. In some experimental systems, meeting these requirements is relatively simple. For example, bacteria and yeast can be transformed with a single plasmid per cell. However, these models are not ideal for one of the main applications of massively parallel genetic assays: understanding the effects of genetic variation on humans.
Cultured human cells are preferable, but no existing method of introducing variants yields a single, stable variant per cell at the required scale. The simplest option, plasmid transfection, results in the unstable introduction of hundreds or thousands of plasmids into each cell. Lentiviral transduction at low multiplicities of infection is a better choice, resulting in stable integration of a single transgene in some cells (5). However, the random nature of viral integration results in widely varying expression levels (6) that increase noise and confound comparisons. Furthermore, lentiviral vectors are pseudo-diploid, exhibiting significant recombination prior to integration (7). They are thus incompatible with strategies using short barcode identifiers to represent larger sequences of interest, and instead rely on sequencing the entire variable region that was introduced (8,9). CRISPR/Cas9 based approaches avoid these problems, but are limited by the efficiency and precision of the host DNA repair machinery, the inability to barcode variants or finely control expression, and reliance on existing haploid sequences within cells (10). Furthermore, neither lentiviral transduction nor CRISPR/Cas9 knock-in are suitable for the insertion of large transgenes: lentiviral vector transgenic payloads are limited to a few kilobases due to decreased titer stemming from viral packaging limits (11) while homology directed repair is inefficient for large inserts (12). Thus, a new experimental framework is needed to realize the potential of massively parallel genetic assays in human cells.
Site-specific recombinases provide an attractive means for expressing genomically integrated, single copies of transgenes in cultured human cells. Recombinase-based approaches are not limited by the size of the transgenic payload; in fact, a recent study demonstrated single-copy genomic insertion of a 27 kb synthetic gene circuit in HEK 293T cells (6). Commercially available Flp-In and Jump-In recombination platforms use the Flp and R4 recombinases, respectively, and have been used to genomically insert transgenes for over a decade (13). Unfortunately, these commercially available recombinase systems have low recombination rates (6), necessitating the use of antibiotic selections to recover the rare recombinant cells (Supplementary Table S1) (14,15). Furthermore, tyrosine recombinases like Flp are reversible, leading to repeated cycles of recombination and excision. However, serine recombinases are promising because only a single recombination event can occur for a given site, which cannot be reversed in the absence of exogenously provided directionality factors (16). Bxb1 is an ideal serine recombinase due to its high recombination rate and junction fidelity (14,15). Furthermore, the human genome reportedly lacks Bxb1 recombination sites (17), so human-derived cells can readily be engineered to contain a single Bxb1 site at a defined locus. In fact, Bxb1 has been used to recombine single transgenes or small libraries into cultured human-derived cells (6,18,19). Thus, a Bxb1 recombinase-based approach could be used to express large transgene libraries in human-derived cells, improving massively parallel genetic assays.
Here, we describe a Bxb1-based platform that can be used to express libraries of tens of thousands of transgenes in a transfectable cell line. Our platform is specifically tailored to the requirements of deep mutational scanning, a massively parallel genetic assay for protein variants (4). We designed a Bxb1-based ‘landing pad,’ which prevents expression of randomly integrated or incorrectly recombined transgenes. Furthermore, the landing pad enables facile recovery of cells harboring correctly recombined transgenes. We engineered cell lines harboring a single copy of the Bxb1 landing pad. We used a series of validation steps we developed to ensure these lines express only a single variant per cell, a feature required for massively parallel genetic assays. We demonstrate that Bxb1 landing pad cells can be used to generate large libraries harboring over twenty thousand unique transgenes, ∼1000-fold more than previous efforts. We also show that transgene frequency can be accurately quantified with high-throughput sequencing. Finally, we use our platform to study the N-end rule in HEK 293T cells, measuring the relative effects on expression of all possible codons at the N-terminus of GFP.
MATERIALS AND METHODS
Generation of genome engineering and landing pad expression plasmids
The dAAVS1–TetBxb1BFP plasmid was created by using the plasmid backbone and homology arms from AAV–CAGGS–EGFP (Addgene #22212), rtTA3G transactivator from pLenti–CMV–rtTA3-Blast (Addgene #26429), mTagBFP2 sequence from mTagBFP2–Actin-7 (Addgene #55273), and the Tet-responsive promoter from pLVX–TRE3G–mCherry (Clontech). Deletion of internal sequences from dAAVS1–TetBxb1BFP was used to create the dAAVS1–rtTA3G and dAAVS1-Dummy plasmids. The dAAVS1–TetBxb1BFP–BC plasmids were created by adding 15 degenerate nucleotides between the Tet-responsive promoter and the upstream BGH terminator. AAVS1 genomic disruption to generate HEK 293T candidate landing pad clones was performed either with TALENs (Addgene #59025 and #59026) or pSpCas9-2A-GFP (Addgene #48138) and AAVS1 gRNA T2 (Addgene #41818). The Bxb1 expression vector pCAG–NLS–HA–Bxb1 (Addgene #51271) was used to express Bxb1 and stimulate recombination. The attB-mCherry, attB-EGFP, attB-Ub3kGiM and attB-3kGiM recombination plasmids were created using coding sequences from p2attPC (Addgene #51547), pIRES2-DsRed-Express (Clontech), and AAV–CAGGS–EGFP. Other recombinant DNA elements were generated by oligonucleotide synthesis (Integrated DNA Technologies). All molecular cloning steps were performed with Gibson assembly (20). All primer and plasmid sequences can be found in Supplementary Table S2 and Supplemental Text 1.
Cell culture, plasmid transfection and landing pad clone generation
All cell culture reagents were purchased from ThermoFisher Scientific unless otherwise noted. HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Cells were passaged by detachment with Trypsin–EDTA 0.25%. To generate landing pad candidate clones 1 through 24, we transfected cells with equal amounts of AAVS1 TALEN expression plasmids, dAAVS1–TetBxb1BFP, dAAVS1–rtTA3G (unbarcoded) and dAAVS1-Dummy. To generate landing pad candidate clones 25 through 48 we transfected cells with equal amounts of pSpCas9–2A–GFP, AAVS1 gRNA T2, dAAVS1–TetBxb1BFP–BC (barcoded), and dAAVS1–Dummy. Clone 4 passed all validation tests aside from internal barcode sequencing, since it was created with unbarcoded landing pad repair template. We refer to clone 4, which was used for the bulk of experiments, as HEK 293T TetBxb1BFP.
Long-term passage of HEK 293T TetBxb1BFP cells was performed with media supplemented with 2 μg/ml doxycycline (Sigma-Aldrich). Cells were switched to doxycycline-free media at least one day before recombination with attB plasmid. For the recombination optimization experiments shown in Figure 3, 1.5 × 105 cells were transfected with 1 μg plasmid DNA using 2 μl Fugene6 (VWR) in a 12-well plate. In all other cases, recombination reactions were performed by transfecting 5 × 105 cells with 3 μg plasmid DNA using 6 μl of Fugene6 in a six-well plate.
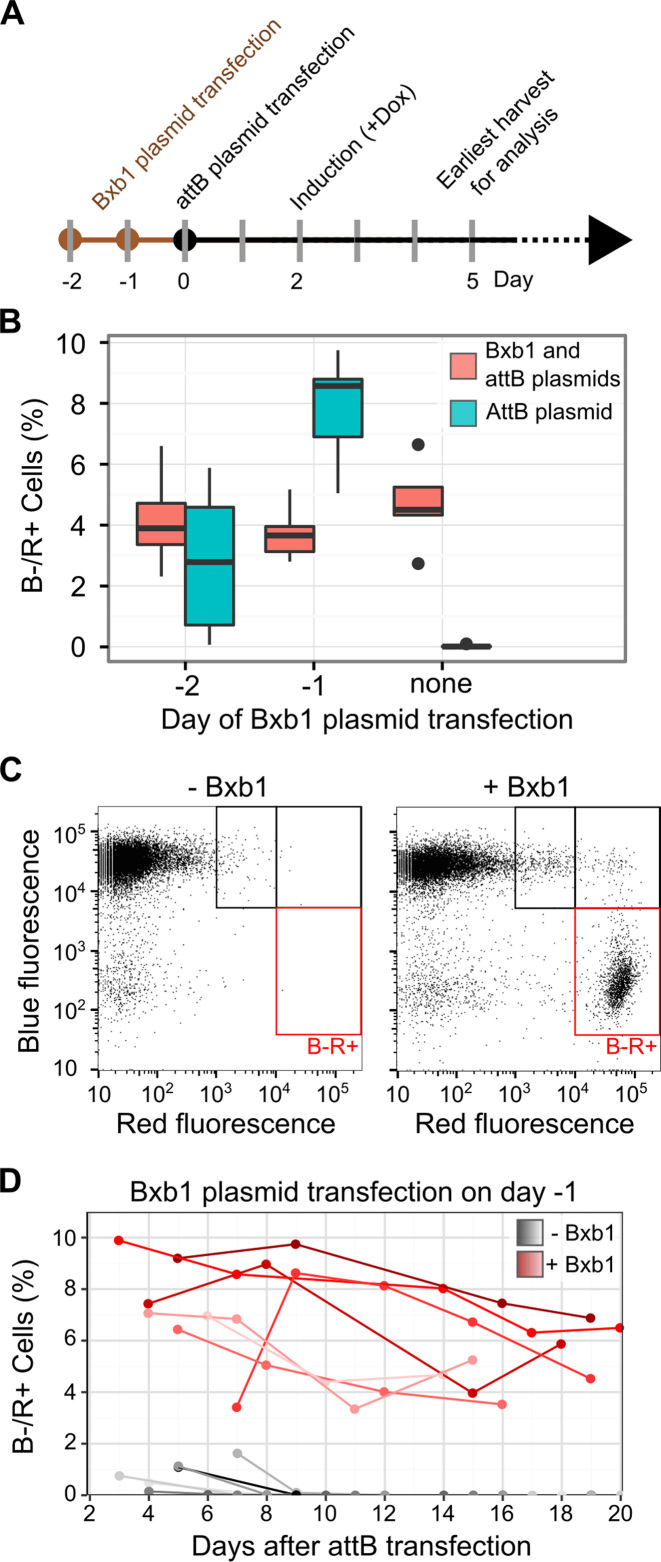
Figure 3.
Optimization of plasmid transfection and transgene expression. (A) A timeline of plasmid transfection, Tet promoter induction, and fluorescent reporter analysis by flow cytometry is shown. (B) HEK 293T TetBxb1BFP cells were transfected with Bxb1 expression plasmid one or two days before transfection with either attB-mCherry recombination plasmid alone (blue) or a 1:1 mixture of attB-mCherry and Bxb1 expression plasmids (red). Two days after attB-mCherry transfection, expression was induced with doxycycline. Three days later, flow cytometry was used to assess the fraction of BFP–/mCherry+ cells. Boxplots represent the results of seven replicates. (C) A representative scatterplot of red and blue fluorescence of HEK 293T TetBxb1BFP cells transfected with attB-mCherry, either with or without the Bxb1 expression plasmid, is shown. Black boxes denote cells likely exhibiting leaky plasmid expression or expression following random transgene integration behind a promoter. Red boxes denote BFP–/mCherry+ cells where the attB-mCherry plasmid has successfully recombined into the landing pad. (D) The percentage of BFP–/mCherry+ HEK 293T TetBxb1BFP cells after transfection with the attB-mCherry recombination plasmid as measured by flow cytometry is shown. Cells were transfected in the presence (red lines) or absence (gray lines) of previously transfected Bxb1 recombinase expression vector. Lines represent BFP–/mCherry+ percentages over time for seven replicates.
PCR validation of landing pad insertion
Genotyping PCR was performed in a 20 μl total reaction volume, with 10 μl Hifi Hotstart ReadyMix (Kapa Biosystems), 0.3 μM each primer, and 25–100 ng of genomic DNA. Reactions conditions: 95°C 3΄, 98°C 20”, 65°C 15”, 72°C 30” to 3’, repeat 34 times, 72°C 6’, 4°C hold. Reactions products were visualized using SYBR-Safe (ThermoFisher Scientific) on 1% TBE/agarose gels. Primers can be found in Supplementary Table S2.
Barcode library generation, two-step amplification, sequencing and data analysis
To generate the degenerate 18-nucleotide barcode library, a double stranded barcoding oligo was first created by filling in JJS_BC_F1.1 bound to JJS_BC_R primer using Klenow polymerase (New England Biolabs). This double-stranded DNA was ligated into the kanamycin-resistant plasmid attB_GPiM upstream of the IRES element using directional cloning with the EcoRI-HF and SacII restriction enzymes (New England Biolabs). The ligated product was cleaned with a Zymo clean and concentrator kit (Zymo Research), and electroporated into NEB 10-beta Electrocompetent Escherichia coli (New England Biolabs), yielding an estimated 2 million unique transformants as determined by dilution plating and colony count. The library was grown in LB supplemented with 50 ng/μl kanamycin, and plasmid DNA was extracted with a Qiagen Midiprep Kit. The resulting plasmid was labeled attB-GPiM-N18-2M.
HEK 293T TetBxb1BFP cells were transfected with pCAG-NLS-HA-Bxb1, followed the next day by attB-GPiM-N18-2M. Transfected cells were induced with 2 μg/ml doxycycline, and recombinant BFP–/mCherry+ cells were recovered by flow sorting 11 days later. Approximately 2.5% of the cells transfected with attB-GPiM-N18-2M were BFP-/mCherry+ after 11 days. 27 500 BFP–/mCherry positive cells were sorted, and allowed to expand for a week before harvesting. Genomic DNA was extracted from these cells using a DNeasy Kit (Qiagen). 50 μl first-round PCR reactions were each prepared with a final concentration of ∼50 ng/μl input genomic DNA, 1× Kapa Hifi ReadyMix, and 0.25 μM of the KAM499/JJS_501a primers. The reaction conditions were 95°C 5’, 98°C 20”, 65°C 15”, 72°C 90”, repeat seven times, 72°C 2’, 4°C hold. Two of these reactions were combined to form the 5 μg samples shown in Figure 4C, and eight reactions were combined to form the 20 μg samples. The resulting amplified DNA was bound to AMPure XP (Beckman Coulter), cleaned, and eluted with 35 μl or 140 μl water for the 5 and 20 μg reactions, respectively. Forty percent of the eluate was mixed with 2× Kapa Hifi ReadyMix and a final concentration of 125 nM JJS_seq_F and 125 nM of one of the indexed reverse primers, JJS_seq_R1a through JJS_seq_R12a. Reaction conditions for the second round PCR were 95°C 5’, 98°C 20”, 65°C 15”, 72°C 90”, repeat 14 times, 72°C 2’, 4°C hold. Reactions were cleaned with a Zymo Clean and Concentrator Kit (Zymo Research), and amplicons were extracted after separation on a 1% TBE/agarose gel using a Quantum Prep Freeze ‘N Squeeze DNA Gel Extraction Kit (Bio-Rad). Extracted amplicons were quantified using a Qubit dsDNA HS Assay Kit (Life Technologies) and sequenced on a NextSeq using a NextSeq 500/550 High Output v2 75 cycle kit (Illumina), using primers JJS_read_1, JJS_index_1, and JJS_read_2.
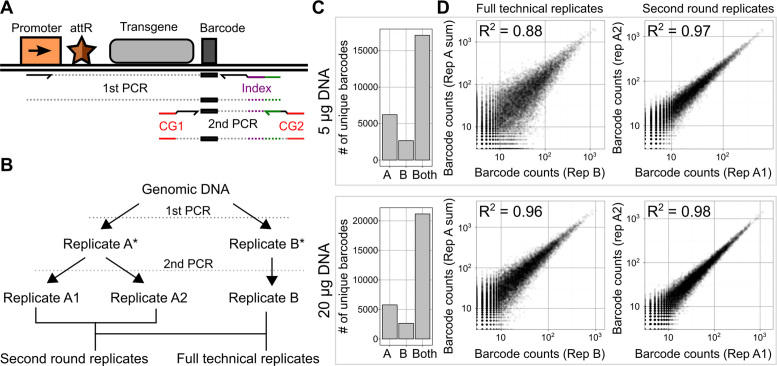
Figure 4.
Creating and quantifying a large library of recombinants using the landing pad. (A) A schematic of the sequential PCR strategy used to selectively amplify a recombined barcode and generate a sequencing-compatible amplicon is shown (CG: Illumina cluster generating sequences, attR: recombination junction). (B) A schematic of technical replicate amplifications performed to characterize precision in preparation and sequencing of recombined barcodes is shown. Sample intermediates are denoted by an asterisk. (C) High throughput sequencing of amplicons generated with either 5 or 20 μg of input DNA revealed the number of unique barcodes observed multiple times only in full technical replicate A, only in full technical replicate B, or in both replicates (left). (D) Scatterplots of the counts of the unique barcodes appearing in both full technical replicates (center) or both second round replicates (right) are shown. R2 values represent squared Pearson correlation coefficients.
Paired sequencing reads were de-multiplexed with bcl2fastq and joined using the fastq-join tool within the ea-utils package (http://expressionanalysis.github.io/ea-utils/). Enrich2 was used to count barcodes that contained no Ns and that had minimum Phred-scaled quality score of at least 20 at each base (http://github.com/FowlerLab/Enrich2). The Enrich2 configuration file is provided as Supplementary File 1. An R Markdown file containing subsequent analyses is provided as Supplementary File 2. We established a sample-specific cutoff for removing barcode sequences arising from sequencing error. Our cutoff was based on the read depth of the most frequently observed barcodes and an assumed error rate of ∼1%, which is a conservatively high estimation. For each sample, we determined the median read depth of the most frequently sequenced 5% of barcodes. For example, in replicate A1 of the 20 μg sample, the high-frequency barcodes had a median read depth of 143. At a 1% error rate, the median high-frequency, 18 nucleotide barcode in this sample is expected to produce ∼26 erroneous barcode sequences. Assuming that mutations are spread uniformly across the 54 possible single nucleotide mutations that could occur, each erroneous barcode sequence arising from a high-frequency barcode in this sample is expected to be present less than once. We required that barcodes must have been present two full counts above this sample-specific estimate to be included in our analysis. We repeated this procedure for all six samples and the minimum read cutoffs were two, three and three reads for the A1, A2 and B technical replicates for the 5 μg input, and two, three and two reads for the A1, A2 and B technical replicates for the 20 μg input. We note that this analysis is conservative because the average per-base quality score was much higher than 20, and thus likely that the error rate was <1% error rate used in our analysis. Raw barcode counts are reported in Supplementary Data 1.
Simultaneous measurement of all possible N-terminal codons on EGFP expression
The EGFP N-terminal degenerate codon library plasmid was created by amplifying attB-Ub3kGiM plasmid with primers KAM1077 and KAM1078, and ligating the overlapping terminal sequences with Gibson assembly. Colony counting following electroporation revealed an estimated ∼25 000 unique transformants. The resulting plasmid library was labeled attB-Ub3kGiM-N3. HEK 293T TetBxb1BFP cells were transfected with pCAG–NLS–HA–Bxb1, followed the next day by the attB–Ub3kGiM–N3 plasmid library. Seven days later, recombinant BFP–/mCherry+ cells from three independent transfections were sorted (N ≥ 5300 cells each) on an Aria III FACS machine. Seven days later, the recombinant cells were sorted into four equally populated bins based on EGFP/mCherry fluorescence ratio (see Supplementary Table S3). Cells in each bin were expanded separately for an additional seven days. Genomic DNA was extracted from the expanded cells in each bin using a DNeasy Kit (Qiagen). High throughput sequencing compatible amplicons were prepared by amplifying 4 μg of genomic DNA in a 50 μl reaction with 25 μl Kapa Hifi Hotstart ReadyMix, 1 μM KAM1016 forward primer and 1 μM reverse indexing primer (KAM1017 through KAM1024). Reactions conditions: 95°C 3’, 98°C 20”, 65°C 15”, 72°C 15”, repeat 24 times, 72°C 6’, 4°C hold. Amplicons were extracted after separation on an 1% TBE/agarose gel using a Quantum Prep Freeze ‘N Squeeze DNA Gel Extraction Kit (Bio-Rad). Extracted amplicons were quantified using a Qubit dsDNA HS Assay Kit (Life Technologies) and sequenced on a NextSeq using a NextSeq 500/550 High Output v2 75 Cycle Kit (Illumina) with the KAM1114 and KAM1026 primers.
Sequencing reads were de-multiplexed with bcl2fastq. Enrich2 was used to count variant codons that contained no Ns and that had minimum Phred-scaled quality score of at least 20 at each base (http://github.com/FowlerLab/Enrich2). The Enrich2 configuration file is provided as Supplementary File 1. An R Markdown file containing subsequent analyses is provided as Supplementary File 2. To calculate steady state expression scores, read counts from technical replicates within each experiment were added together. Then, individual variant counts within a bin were divided by the total number of counts in the bin to produce a fractional count. Steady state expression scores for each variant were calculated by multiplying the fractional counts in each bin with integer bin weights ranging from one for the lowest EGFP/mCherry fluorescence ratio to four for the highest ratio. Sequencing counts are reported in Supplementary Data 2. Mean expression scores and 95% confidence intervals derived from the four replicates are reported in Supplementary Data 3.
Flow cytometry and fluorescence activated cell sorting (FACS)
Cells were detached with trypsin, and resuspended in PBS containing 5% serum. Cells were fixed by pelleting at 300 × g, followed by resuspension in PBS containing 4% formaldehyde at 4°C for 10 min. Analytical flow cytometry was performed with a BD LSRII flow cytometer. BFP was excited with a 405 nm laser, and emitted light was collected after passing through a 450/50 nm band pass filter. EGFP was excited with a 488 nm laser, and emitted light was collected after passing through 505 nm long pass and 530/30 nm band pass filters. mCherry was excited with a 561 nm laser, and emitted light was collected after passing through 595 nm long pass and 610/20 nm band pass filters. Cells were sorted using a BD Aria III FACS machine equipped with an 85 μm nozzle. Filter sets were identical to those described for the LRSII, with the exception of mCherry emission which was detected using 600 nm long pass and 610/20 band pass filters. Before analysis of fluorescence, live, single cells were gated using FSC-A and SSC-A (for live cells) and FSC-A and FSC-H (for single cells). We gated for fluorescing cells such that they had fluorescence values at least 10–100 times higher than the median fluorescence value of the negative or control population, depending on the cutoff stringency desired in the experiment. For landing pad clone isolation, individual BFP+ cells were directly sorted into wells of 96-well plates. Four-way library sorting was performed by creating a FITC/PE-Texas Red ratiometric parameter in the BD FACSDIVA software, creating a histogram based on this ratio, and creating four immediately adjacent gates dividing the cells in the library into quartiles.
Analysis of ubiquitin cleavage by western blotting
HEK 293T TetBxb1BFP cells were transfected with the pCAG–NLS–HA–Bxb1 expression vector and attB–Ub3kGiM plasmids encoding the indicated ubiquitin-fused EGFP N-terminal variants. Two days after transfection, the cells were switched to Dox-containing media. BFP–/mCherry+ recombinant cells were obtained by flow sorting ten days after transfection, and allowed to expand for an additional seven days. These recombinant cells were then incubated with lysis buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100, and Protease Inhibitor Cocktail (Sigma-Aldrich)) for 10 min at 4°C, and pelleted at 21 000 × g for 5 min. The supernatant was collected, and protein concentration was determined by DC Protein assay (Bio-Rad) against a standard curve of bovine serum albumin. 18 μg of protein was loaded per well of a NuPAGE 4–12% Bis–Tris gel (Invitrogen) in MOPS buffer, using Spectra Multicolor Broad Range Protein Ladder (ThermoFisher Scientific) for size comparison, and transferred to a PVDF membrane. Western blotting was performed using a 1:4000 dilution of anti-GFP antibody (11814460001; Roche), followed by detection with a 1:10 000 dilution of anti-mouse-HRP (NA931V; GE Healthcare), or a 1:5000 dilution of anti-beta-actin–HRP (ab8224; Abcam).
RESULTS
Generation of Cells with Single Landing Pads
We developed an efficient workflow for generating and validating Bxb1 landing pad cell lines. First, the landing pad is inserted at the desired locus using Cas9- or TALEN-mediated homology directed repair (Figure 1A). The landing pad encodes a self-contained Tet-inducible system, with a 3’ CMV promoter driving transcription of a rtTA3G Tet-activator coding element (Figure 1B). Upstream, a Tet-inducible promoter drives expression of an mTagBFP2 blue fluorescent protein (BFP) reporter. A Bxb1 attP recombination site is located between the Tet inducible promoter and the BFP reporter, allowing for subsequent site-specific recombination of a transgenic cassette whose expression is driven by the Tet-inducible promoter after recombination. A key advantage of this arrangement is that the transgene is only expressed when correctly recombined into the landing pad. Additionally, recombinant cells are marked by loss of BFP expression.
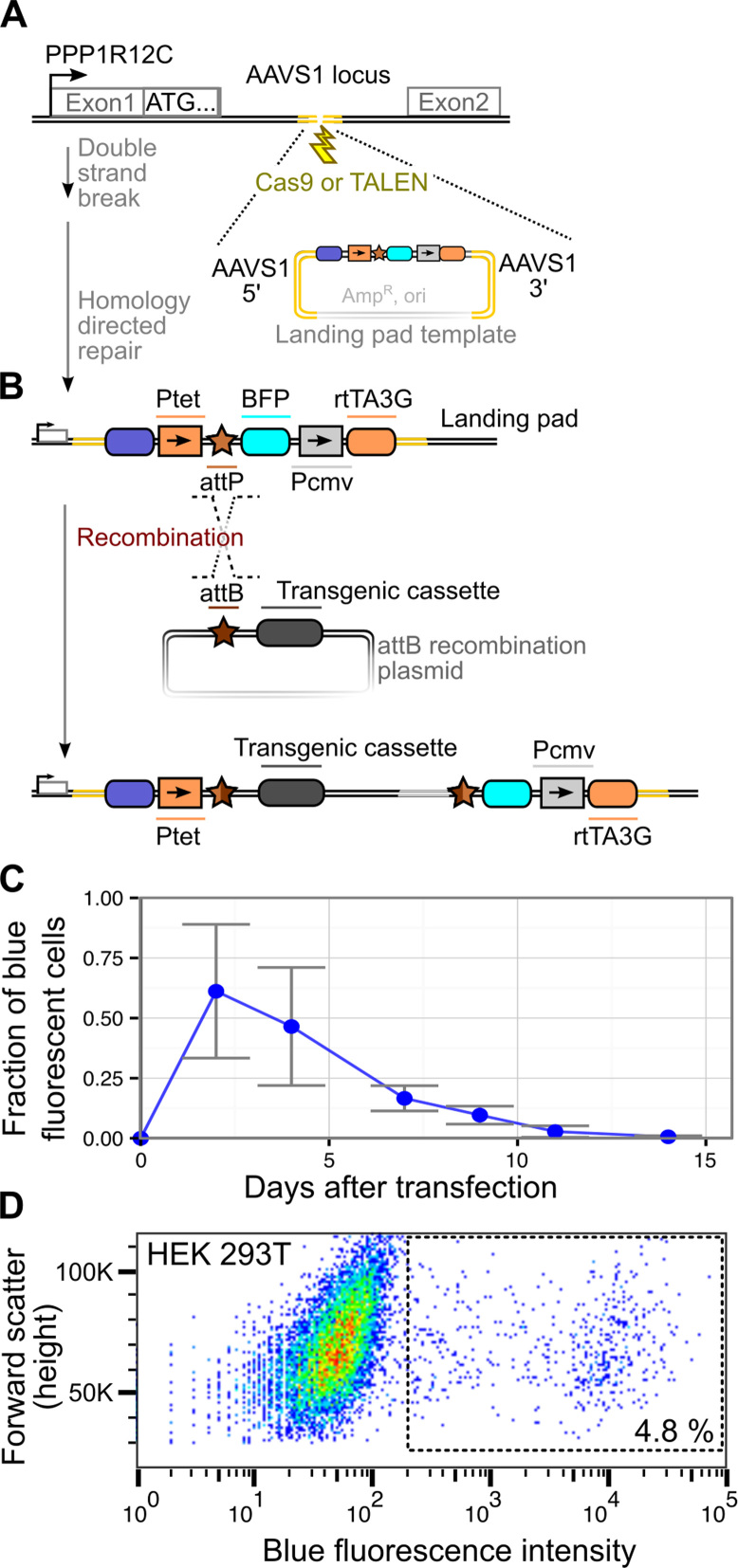
Figure 1.
A Bxb1 recombinase-based platform for transgene library expression. (A) Disruption of the AAVS1 locus on chromosome 19 stimulates genomic landing pad insertion through homology directed repair. (B) The genomically integrated landing pad is subsequently used to efficiently integrate a transgenic cassette using the site-specific Bxb1 recombinase. (C) Depletion of BFP+ cells over time after transfection of the landing pad plasmid shown in panel B, as determined by flow cytometry. Cells were considered BFP+ if their BFP fluorescence was greater than that of untransfected cells. Loss of BFP signal over time is largely reflective of dilution or degradation of transfected plasmid. Error bars reflect standard error (N = 3). (D) After three weeks of post-transfection growth with additional sorts for BFP+ cells on days 5 and 13, BFP fluorescence was measured by flow cytometry. Cells within the box are likely to contain genomic landing pad insertions. Color indicates point density from low (blue) to high (red).
To create landing pad cell lines, we transfected HEK 293T cells with AAVS1-disrupting and landing pad repair template plasmids (see Materials and Methods). Transfected cells were passaged in the presence of doxycycline to induce BFP reporter expression and monitor decay of the repair template plasmid over time. After ∼2 weeks, the fraction of BFP positive cells was <1%, suggesting that most transfected plasmid had been diluted or degraded (Figure 1C). Thus, we surmised that that the remaining BFP positive cells likely harbored stably integrated landing pads. We waited an additional week, and sorted single BFP positive cells into individual wells of a 96-well plate to isolate clonal integrants. In a separate experiment, we found that a series of sorts enriching for BFP positive cells during plasmid dilution (∼1 and 2 weeks after transfection) yielded a larger proportion of BFP positive cells (∼5%) at three weeks (Figure 1D).
Massively parallel genetic assays require a strict linkage between genotype and phenotype. Thus, identifying a clonal cell line harboring only a single copy of the landing pad at the intended locus is critical. Clones with multiple landing pads can arise owing to insertion events at multiple copies of the desired locus or random genomic integration of the landing pad plasmid. Screening for multiple insertions is complicated by the fact that many commonly used cell lines are aneuploid. For example, the HEK 293T cells used here are hypotriploid (21). Therefore, we developed a toolkit of simple assays that can be used to identify candidate clones that have a single landing pad located at the desired locus.
First, we made a qualitative visual assessment of BFP positivity for each candidate HEK 293T clone using fluorescence microscopy. Clones with apparently homogenous BFP fluorescence levels were retained, whereas clones that exhibited variable levels of BFP fluorescence were discarded. Next, PCR-based genotyping was used to analyze each candidate clone. To aid interpretation of the genotyping test results, we used a mixture of homology repair templates in our initial transfection. This mixture included the landing pad template, an rtTA3G template and a small dummy template (Figure 2A). Amplification with a genome-specific primer and a landing pad-specific primer revealed clones with a landing pad insertion at the AAVS1 locus. Amplification with genome-specific primers flanking the desired insertion site confirmed that the other copies of the AAVS1 locus contained either no insert, the rtTA3G insert or the dummy insert. The genotyping test enables unambiguous identification of clones with single AAVS1 landing pad insertions in cell lines with two or three copies of the target locus. For example, HEK 293T Clone 4 exhibited a clearly interpretable PCR amplicon pattern, with an unmodified copy of the AAVS1 locus, a copy modified with a dummy repair template, and a single landing pad insertion (Figure 2B and Supplementary Figure S1). Candidate clones that passed the genotyping test, including HEK 293T Clone 4, were Sanger sequenced to confirm that the entire landing pad sequence and insertion junctions were correct.
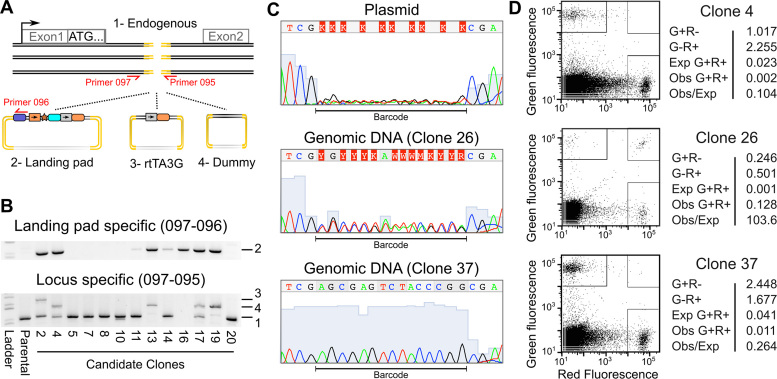
Figure 2.
Validation of single landing pad genomic insertion at the desired locus. (A) A schematic illustrating the three homology repair templates and the primer pairs used to identify their insertion is shown. (B) Agarose gels of the products of the landing pad specific (top panel) or AAVS1 locus specific PCR amplification (bottom panel) are shown. Numbers indicate expected band sizes for the unmodified locus (1), landing pad insertion (2), rtTA3G insertion (3) or dummy insertion (4). (C) Sanger sequencing traces are shown for the degenerate barcode sequence in the landing pad homology repair template plasmid preparation and for two candidate landing pad clones. (D) Candidate landing pad clones were transfected with a mixture of attB-EGFP and attB-mCherry recombination plasmids and the Bxb1 expression plasmid. The green and red fluorescence levels for three transfected candidate clones are shown (left panels). Boxes indicate cells considered EGFP+, mCherry+ or EGFP+/mCherry+. The percentages of live cells in each category along with the expected number of EGFP+/mCherry+ cells based on the number of EGFP+ and mCherry+ cells are shown (right panels).
PCR-based genotyping can identify clones with a single landing pad insertion at the desired locus, but cannot determine if the landing pad has been incorporated elsewhere in the genome. Thus, we developed two orthogonal approaches to ensure that candidate clones contained only a single landing pad insertion. First, we used a degenerate barcode included in the landing pad to identify candidate clones with a single insertion. We sequenced this barcode in candidates that passed the BFP expression and genotyping tests. Candidates with multiple landing pad insertions, such as Clone 26, can be readily identified because they harbor multiple barcodes (Figure 2C). Candidates with single landing pad insertions, such as Clone 37, have a single barcode (Figure 2C). Second, we used a fluorescence-based functional assay to determine if candidate clones could integrate more than one attB recombination plasmid per cell. Each candidate was transfected with a 1:1 mixture of attB-EGFP and attB-mCherry plasmids, along with a plasmid encoding Bxb1 recombinase. Then, EGFP and mCherry fluorescence levels were determined by flow cytometry (Figure 2D). In candidates with multiple landing pads, the percentage of EGFP+/mCherry+ cells greatly exceeded the expected value, which is the product of the percentage of each population of single positive cells (Figure 2D, middle). Candidates with a single landing pad had far fewer EGFP+/mCherry+ cells than expected (Figure 2D; top, bottom). Thus, our landing pad insertion workflow can be used to generate and validate clonal cell lines containing a single landing pad at the desired locus. We used Clone 4, referred to as HEK 293T TetBxb1BFP, hereafter.
Optimization of recombination efficiency and expression at the landing pad
Massively parallel genetic assays require generating tens of thousands of genetically altered cells and can demand high expression to maximize signal. Thus, we determined the optimal conditions for recombination of plasmids bearing a Bxb1 attB site at the landing pad, and for induction of integrated transgene expression. Recombination of a plasmid bearing an attB sequence requires the catalytic activity of the Bxb1 recombinase. Previous Bxb1 recombination protocols call for simultaneous transfection of Bxb1 expression and attB recombination plasmids (6,18,19). We suspected that expression of Bxb1 recombinase prior to recombination plasmid transfection could enhance recombination efficiency, facilitating increased library size.
To test this hypothesis, we transfected HEK 293T TetBxb1BFP cells with Bxb1 expression plasmid prior to transfection with either the attB-mCherry recombination plasmid alone or a mixture of the attB-mCherry recombination and Bxb1 expression plasmids (Figure 3A). We found that cells concomitantly transfected with a mixture of Bxb1 expression and attB-mCherry recombination plasmids yielded ∼4% recombined, BFP–/mCherry+ cells regardless of whether Bxb1 expression plasmid had been transfected beforehand (Figure 3B). However, when the Bxb1 expression plasmid was transfected a day before, the attB-mCherry recombination plasmid efficiency doubled to ∼8%.
Having optimized recombination efficiency, we next assessed the expression kinetics of the Tet-inducible promoter in HEK 293T TetBxb1BFP cells. We found that it took two days after induction of expression with doxycycline for most cells to become BFP+ (Supplementary Figure S2A). Upon removing doxycycline, we found that it took approximately five days for the existing BFP transcript and protein to decay, rendering most cells BFP– (Supplementary Figure S2B). Thus, cells should be analyzed for reporter fluorescence roughly two days after induction or five days after shutoff.
A key feature of our platform is the absence of a promoter on the transgene recombination plasmid. This arrangement should prevent transgene expression in the absence of correct recombination at the landing pad. To test this assumption, we transfected the attB-mCherry recombination plasmid in the absence of the Bxb1 expression plasmid. Although the attB-mCherry plasmid lacks a promoter, we still observed low-level mCherry expression (Figure 3C, left). However, none of these modestly mCherry+ cells were BFP- because they retained an intact BFP reporter at the landing pad. On the other hand, recombinant cells generated by transfecting Bxb1 expression and attB-mCherry plasmids had high levels of mCherry expression and also lost BFP expression (Figure 3C, right). Furthermore, the low level of plasmid-derived mCherry fluorescence observed in the absence of Bxb1 decays approximately five days after transfection whereas the high level of mCherry fluorescence observed after recombination is stable for at least twenty days (Figure 3D). Thus, our landing pad platform is capable of producing recombinant cells with high levels of transgene expression that are stable over several weeks. An important caveat is that expression of transgenes with significant fitness costs would likely not be stable over long periods of time.
Generating and quantifying a large library of recombinant cells
To determine whether we could create a large library of recombinant cells and measure the frequency of each library member accurately, we created an attB recombination plasmid harboring a short, degenerate barcode. We recombined this plasmid into HEK 293T TetBxb1BFP cells and collected ∼25 000 BFP–/mCherry+ recombinant cells.
Accurately determining the frequency of each member of a large library using deep sequencing can be technically challenging. The transfection procedure we used to generate the recombinant cells introduces thousands of plasmids into each transfected cell (22), which decay over a period of weeks (Figure 1C). Accordingly, amplification using primers specific to the transfected recombination plasmid, even weeks after transfection, would likely result in significant background arising from residual transfected plasmid. Thus, we employed a nested two-step amplification strategy where selective amplification for genomically integrated plasmids using a primer pair spanning the recombination junction was followed by amplification of a smaller Illumina-compatible amplicon (Figure 4A). Importantly, the second amplification is selective for the first amplification product because it relies on primer hybridization to a novel sequence introduced during the first amplification.
High throughput sequencing of amplicons from our recombined library produced using this procedure revealed nearly 21 000 unique barcodes (Figure 4B and C). We required that each barcode be observed multiple times in both replicates, placing a conservative lower bound on the number of unique barcodes in the library. PCR amplification can introduce significant bias, particularly when template concentrations are low (23), so we compared barcode counts between replicate amplifications. As expected, increasing input template DNA yielded an increase in the number of barcodes that were observed in both technical replicates (Figure 4C). Increasing input template also resulted in greater correlation of barcode counts between replicates. We conclude that this increase in reproducibility occurred during the first amplification, because barcode frequencies in replicates of the second amplification were highly correlated at both template concentrations (Figure 4D). Thus, our landing pad can be used to rapidly and efficiently create large, stable libraries of recombinant cells, which can be accurately quantified using high throughput sequencing.
Simultaneous measurement of all possible N-terminal codons on protein stability
To demonstrate the strong, single-cell genotype–phenotype link forged by our platform, we characterized the effects of all possible N-terminal codons on steady state protein expression in mammalian cells. The N-end rule is an evolutionarily conserved pattern of intracellular protein degradation dictated by the identity of the N-terminal amino acid (24). While most proteins retain their N-terminal methionine, a subset of proteins are cleaved or processed to reveal new N-terminal amino acids. Destabilizing N-terminal amino acids are recognized by N-recognin proteins, which target the protein for degradation. In eukaryotes, the N-end rule has two branches. In the classic Arg/N-end rule, N-terminally arginylated amino acids, as well as a subset of unmodified amino acids, are targeted. In the more recently discovered Ac/N-end rule, small uncharged amino acids that have been co-translationally acetylated are targeted (25).
The effects of different N-terminal amino acids have been measured using pulse-chase experiments where the rate of disappearance of a query protein is quantified by radiolabeling or western blot (26). However, these commonly employed methods cannot be readily parallelized within intact cells. We developed a parallizable N-end rule assay based on an N-terminal ubiquitin fusion to a fluorescent protein. Here, with the exception of proline, co-translational removal of ubiquitin by deubiquitinating enzymes efficiently exposes a new N-terminal amino acid (27) that dictates the steady state expression level of the fluorescent protein. We designed an attB recombination plasmid carrying this ubiquitin fusion assay cassette. Here, ubiquitin was N-terminally fused to EGFP upstream of an IRES–mCherry sequence. mCherry serves as a marker of recombination and a reference reporter to control for cell-to-cell variation in expression (Figure 5A). A library of N-terminal EGFP variants could be flow sorted based on their EGFP/mCherry fluorescence ratio and then sequenced to reveal the steady state expression of each variant.
Figure 5.
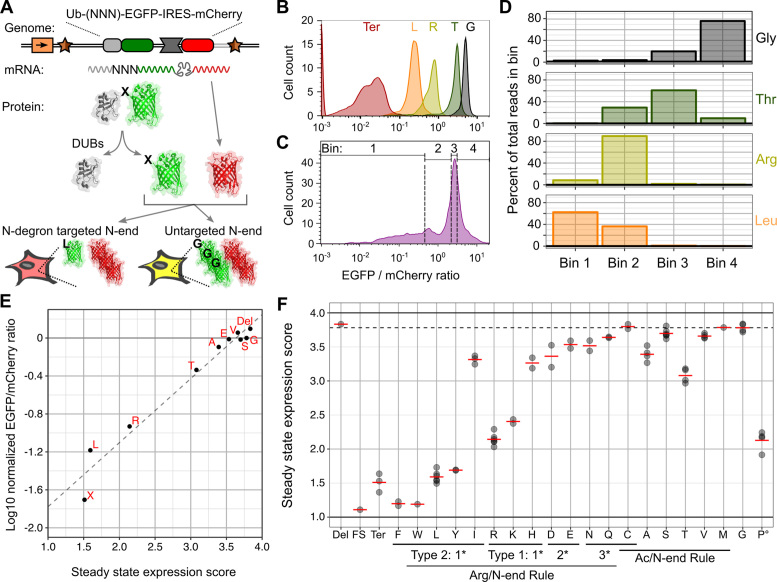
Parallel assessment of the effects of all possible N-terminal codons on protein expression. (A) A schematic of the N-terminal ubiquitin fusion assay is shown. Ubiquitin was fused to the N-terminus of EGFP and the first codon of EGFP was mutated. To control for cell-to-cell variability, the fusion protein was expressed from a transcript that also expressed mCherry. Removal of ubiquitin by cellular deubiquitinases exposed the N-terminus of EGFP, and the EGFP/mCherry ratio reveals EGFP N-terminal variant expression level. DUBs: deubiquitinating enzymes. (B) Flow cytometry was used to determine the EGFP/mCherry fluorescence ratio for clonal cells expressing ubiquitin fusion assay constructs encoding EGFP with an N-terminal leucine, arginine, threonine, glycine or stop. Smoothed histograms of the EGFP/mCherry fluorescence ratio distribution are shown for each variant (N ≥ 250 BFP–/mCherry+ cells). (C) The N-terminal codon of EGFP was fully randomized, and this library was flow sorted into four bins based on EGFP/mCherry fluorescence ratio. Each bin contained an equal number of cells. A smoothed histogram of the library sort is shown, with bars indicating bin boundaries (N = 7017 BFP–/mCherry+ cells). (D) Each bin was deeply sequenced and the number of reads per amino acid in each bin was determined. The distributions of read counts among the four bins for the four EGFP N-terminal variants in panel B are shown. (E) Using the distribution of read counts across bins, steady state expression scores were calculated for every EGFP N-terminal codon, including stops (X), a deletion of the first codon (Del) and variants with a single nucleotide insertion within the mutagenized codon (FS, frameshift). Ten variants’ steady state expression scores are plotted on the x-axis. These variants’ individually measured log-transformed EGFP/mCherry fluorescence ratios, normalized to glycine, are plotted on the y-axis (mean of two or more experiments). The dashed line shows the results of linear regression (Pearson R2 = 0.96). (F) Steady state expression scores for all variants are shown. Points indicate each codon's score, calculated from four experiments, and red lines indicate the mean scores of all codons for each amino acid. The dashed line indicates glycine's score. The ° symbol indicates that ubiquitin-proline fusions are incompletely cleaved, so assay results are not reflective of the effect of N-terminal proline on stability.
We confirmed that ubiquitin was removed from EGFP by western blotting, which revealed a single band at the expected size (Supplementary Figure S3A). Next, we determined whether the ubiquitin fusion assay could recapitulate known N-terminal amino acid effects: leucine and arginine are profoundly destabilizing, threonine is moderately destabilizing and glycine has no effect (24,26). We created these N-terminal EGFP variants in our ubiquitin fusion assay cassette and recombined them into HEK 293T TetBxb1BFP cells. Flow cytometry confirmed that the presence of either leucine or arginine at the N-terminal position reduced steady state EGFP expression to a level approaching that of a premature stop codon (Figure 5B). Insertion of threonine only slightly diminished steady state EGFP levels compared to glycine. The loss of steady state expression we observed was not due to disruption of EGFP folding or function, because these amino acid insertions had minimal effects when ubiquitin was replaced by a conventional initiating methionine (Supplementary Figure S3B).
Next, we created a library of plasmids containing a degenerate NNN codon between the ubiquitin and EGFP coding sequences, thus producing all possible N-terminal amino acids upon ubiquitin removal. Recombinant HEK 293T TetBxb1BFP cells containing this library were sorted into four equally populated bins spanning the spectrum of observed EGFP/mCherry fluorescence ratios (Figure 5C). Genomic DNA was harvested, amplified as described above and subjected to high throughput sequencing (Supplementary Table S3). The number of reads for each codon within a bin was highly reproducible between technical replicates (Supplementary Figure S3C). For leucine, arginine, threonine and glycine, the distribution of read counts across bins reflected the individually determined EGFP/mCherry intensity distributions (Figure 5D; compare to B, C). From the read count data, we calculated a steady state expression score for each codon and amino acid (see Methods; Supplementary Figure S3D). These parallel expression scores are highly correlated with individual expression measurements (Pearson's R2 = 0.96, Figure 5E).
Our results are largely consistent with previous studies of the effect of N-terminal amino acids in mammalian cells. As expected, library members encoding stop codons or frameshift mutations had low expression scores, and degenerate codons were highly correlated (Figure 5F). All large, hydrophobic type 2 Arg/N-end rule substrates substantially decreased expression, except isoleucine, which had a modest effect. Positively charged primary type 1 Arg/N-end rule substrates also decreased expression, although the effect of histidine was modest. The secondary and tertiary type 1 Arg/N-end rule substrates caused small decreases in expression. Cysteine, which requires oxidation to be rendered destabilizing, did not affect expression. Potential Ac/N-end rule substrates generally had little effect on expression, with the exception of alanine and threonine. In fact, threonine resulted in a 2-fold decrease in expression (Figure 5E). These results demonstrate that the landing pad platform can be used to rapidly and accurately measure single-cell phenotypes that arise from expression of a library of nucleic acid variants present in a mixed population of cells.
DISCUSSION
To facilitate massively parallel genetic assays in cultured human cells, we improved upon existing site-specific recombinase technologies to create a variant expression platform that can be used to study a variant library of any transgenic sequence of interest in mammalian cells. In principle, our landing pad platform could be used in any transfectable cell type, which is important because many phenotypes of interest are likely to depend on cell context. We demonstrated that the landing pad platform can be used to generate tens of thousands of recombinant cells, and we used the platform to measure the effects of all possible N-terminal amino acid substitutions on steady state expression.
Generating a clonal line that contains a single landing pad insertion at the desired locus is a critical step. Many previous implementations of recombinase-based approaches employed Southern blotting to validate single landing pad insertion at the desired locus (6). Southern blotting is technically challenging, and traditionally requires working with radioactivity. Therefore, we developed a battery of validation assays that use commonly available, easily interpretable techniques: PCR, agarose gel electrophoresis, Sanger sequencing and flow cytometry.
Insertion of the landing pad into cell lines that are difficult to transfect or grow may be facilitated using a splice acceptor-2A-puromycin resistance cassette that we inserted at the 5’ end of the landing pad. Expression of this resistance cassette from the upstream, ubiquitiously expressed PPP1R12C promoter confers antibiotic resistance on cells with a correct landing pad insertion (28). This cassette was not needed to generate HEK 293T TetBxb1BFP cells, but may be helpful for cell lines that are more difficult to engineer. Conveniently, in cases where cell type is not critical, the HEK 293T TetBxb1BFP cells we developed and validated are ready for use.
In this work, we engineered the landing pad into the AAVS1 locus. We chose this site because it is present in most human-derived cells, generally transcriptionally active, and easy to amplify and sequence. Thus, it is a convenient and easy-to-work with site for manipulation and transgene expression. However, the landing pad could be integrated at other defined sites using the approach we outline. Alternatively, the landing pad could be introduced using a lentiviral vector, which would result in random incorporation in the genome.
Our platform employs Bxb1 recombinase to insert a transgenic plasmid library into the engineered landing pad. Of the known site-specific DNA recombinases, Bxb1 has the highest efficiency and fidelity in mammalian cells (6,14). Bxb1 has been described as having no observed pseudo-site insertion in HEK 293T cells (17), although only low-resolution methods like karyotyping have been used to validate this assertion. Nevertheless, our experimental workflow is robust to off-target integration because transgene expression is dependent on correct recombination at the landing pad. Additionally, we employ a dual fluorescent reporter system that facilitates removal of cells expressing the transgene in the absence of correct recombination. Indeed, our N-end rule experiment demonstrates that our platform yields highly reproducible results, proving that it can be used to express a single transgene variant per cell.
Bxb1 recombination inserts the entire attB-containing plasmid into the landing pad, thus retaining transgene-adjacent sequences including bacterial sequences necessary for plasmid propagation. These sequences might impact transgene expression. Dual integrase cassette exchange, in which two serine integrases with different specificities precisely define the inserted sequence, might overcome this problem (18). We attempted to employ this strategy, but it was too inefficient to produce large libraries of transgenic cells. Regardless, the presence of plasmid DNA does not appear to affect transgene expression, which was largely stable in our HEK 293T TetBxb1BFP cells. However, we did observe a small decrease in the fraction of cells expressing an mCherry transgene from the landing pad over a period of three weeks. This decrease could be due to silencing arising from plasmid sequence or be due to a slight fitness cost associated with transgene expression. Future versions of the landing pad could incorporate insulators (29) or use minicircle recombination templates (30) in place of a large insertion plasmid to further stabilize expression.
A challenging aspect of massively parallel genetic assays in mammalian cells is quantitative amplification and sequencing of single copy insertions of transgenic sequences in a sea of genomic and transfected plasmid DNA. Using the selective amplification strategy we developed, quantification of moderately-sized libraries like our N-end rule library is exceptionally reproducible. Importantly, we also achieved high reproducibility for a library of over 20 000 unique variants. Here, using large amounts of input template DNA circumvented stochastic effects stemming from inefficient amplification. Alternative amplification protocols, such as the use of unique molecular identifiers (31), will also likely increase quantification accuracy.
Our N-end rule experiment demonstrates some of the advantages of using our landing pad platform for making parallel measurements. Individual assessment of the 64 possible N-terminal codons would be unwieldy. Accordingly, this is the first time that all 64 codons representing the 20 possible amino acids have been simultaneously tested for their contributions to the N-end rule in human-derived cells. Parallel assessment is preferable because it facilitates accurate comparisons between all samples. Our results are mostly consistent with previous measurements of the effect of N-terminal amino acids. However, we find that alanine and threonine, which are Ac/N-end rule substrates thought to be inert, are moderately destabilizing. The details of Ac/N-end rule in the mammalian cell cytosol are not well understood (32), and it remains to be seen why these two amino acids are destabilizing.
We also find that histidine and isoleucine, which are Arg/N-end rule substrates thought to be highly destabilizing, have only a moderate effect. The moderate effect of histidine relative to arginine and lysine might be explained by the lower affinity of the UBR recognition domain of the N-recognins UBR1 and UBR2 for histidine (33). The moderate effect of isoleucine might also be related to N-recognin substrate recognition. The N-domains of eukaryotic UBR1 and UBR2 are responsible for recognizing type 2 N-degrons including isoleucine (34). These domains are structurally related to the bacterial ClpS protein, which only recognizes N-terminal tyrosine, tryptophan, phenylalanine and leucine. Interestingly, a point mutation in the binding pocket of ClpS extends recognition to terminal isoleucine and valine residues (35). Thus, the ancestral binding pocket architecture is less accommodating of isoleucine, but can evolve to recognize it. The lack of isoleucine recognition for ClpS might be related to the modest effect of isoleucine in our assay. In conclusion, the differences in destabilization we observe reflect the nuances of substrate recognition in the different N-end rule pathways, and are highlighted by our parallel measurements.
Compared to other recombinase-based systems, our platform is more efficient, more portable, inducible, and allows single-variant expression through irreversible and directional transgene integration while preventing expression of unintegrated transgenes (Supplementary Table S1). Commercially available Flp-in and Jump-in cell lines suffer from low recombination rates, and require recombination plasmids to have their own inducible promoters, necessitating prolonged cell passages to allow transfected plasmids to disappear. Indeed, a recent study using Flp-In T-REx cells to study a library of several thousand variants required roughly 200 million cells on 45 large plates and multiple weeks of passaging (36). Our platform could produce the same library using a few hundred thousand cells on one small plate in less than a week. PhiC31-based platforms solve some of these problems, but have much lower recombination rates than Bxb1. The use of PhiC31-based platforms in massively parallel genetic assays is also complicated by the existence of endogenous PhiC31 pseudo-attP sites in the human genome that can result in the integration of multiple transgenes per cell. Existing Bxb1-based systems lack the inducible expression and cytometry-optimized features necessary for many massively parallel genetic assays like deep mutational scanning. Lastly, the additional tools we provide make our platform portable to other cell types and ready-to-use for high-throughput, sequencing-based phenotypic interrogation of variant libraries.
In summary, we improve massively parallel genetic assays in mammalian cells by offering an alternative to lentiviral or homology-directed repair-mediated library generation. Specifically, our platform enables facile generation of large libraries of transgenic cells with controllable, homogenous transgene expression from a user-defined locus. We provide a series of assays that can be used to vet candidate landing pad cell lines; in theory, our platform could be used to study any transgenic sequence of interest in any transfectable cell line. Once a cell line has been created and vetted using the tools we describe, it is compatible with almost any fluorescence- or growth-based phenotypic assay where each variant can be scored simultaneously with high throughput sequencing. Each phenotypic assay requires appropriate controls to demonstrate, as we did in for our N-end rule assay, that variants of known effect produce the expected phenotype (for discussion of massively parallel genetic assay design see (37–39)). Our platform represents an advance for massively parallel genetic assays ranging from the comprehensive study of variation of a nucleotide or protein sequence to the simultaneous investigation of diverse elements such as cDNA expression screening. Our platform could eventually enable many cellular phenotypes to be quantified for each variant in a large library, thus facilitating a deeper understanding of the relationship between genotype and phenotype.
ACCESSION NUMBERS
Illumina sequencing data has been submitted to the NCBI Sequence Read Archive (SRA) under SRA Study accession SRP095490.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lea Starita for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [1R01GM109110 and 5R24GM115277 to D.M.F.]; American Cancer Society [PF-15-221-01 to K.A.M.]; National Institutes of Health Interdisciplinary Training Grant in Cancer [2T32CA080416]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gasperini M., Starita L., Shendure J.. The power of multiplexed functional analysis of genetic variants. Nat. Protoc. 2016; 11:1782–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shalem O., Sanjana E.N., Hartenian E., Zhang F.. Genome-Scale CRISPR-Cas9 Knockout. Science. 2014; 343:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang T., Wei J.J., Sabatini D.M., Lander E.S.. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014; 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fowler D.M., Fields S.. Deep mutational scanning: a new style of protein science. Nat. Methods. 2014; 11:801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noderer W.L., Flockhart R.J.. Quantitative analysis of mammalian translation initiation sites by FACS‐seq. Mol. Syst. Biol. 2014; 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duportet X., Wroblewska L., Guye P., Li Y., Eyquem J., Rieders J., Rimchala T., Batt G., Weiss R.. A platform for rapid prototyping of synthetic gene networks in mammalian cells. Nucleic Acids Res. 2014; 42:13440–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schlub T.E., Smyth R.P., Grimm A.J., Mak J., Davenport M.P.. Accurately measuring recombination between closely related HIV-1 genomes. PLoS Comput. Biol. 2010; 6:e1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brenan L., Andreev A., Cohen O., Pantel S., Kamburov A., Cacchiarelli D., Persky N.S., Zhu C., Bagul M., Goetz E.M. et al. . Phenotypic characterization of a comprehensive set of MAPK1/ERK2 missense mutants. Cell Rep. 2016; 17:1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Majithia A.R., Tsuda B., Agostini M., Gnanapradeepan K., Rice R., Peloso G., Patel K.A., Zhang X., Broekema M.F., Patterson N. et al. . Prospective functional classification of all possible missense variants in PPARG. Nat. Genet. 2016; 48:1570–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Findlay G.M., Boyle E.A., Hause R.J., Klein J.C., Shendure J.. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014; 513:120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar M., Keller B., Makalou N., Sutton R.E.. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001; 12:1893–905. [DOI] [PubMed] [Google Scholar]
- 12. He X., Tan C., Wang F., Wang Y., Zhou R., Cui D., You W., Zhao H., Ren J., Feng B. et al. . Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016; 44:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee D., Park J.W., Kim Y., Kim J., Lee Y., Kim J., Kim J.. Toward a functional annotation of the human genome using artificial transcription factors. Genome Res. 2003; 13:2708–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu Z., Thomas L., Davies B., Chalmers R., Smith M., Brown W.. Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol. 2013; 13:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamaguchi S., Kazuki Y., Nakayama Y., Nanba E., Oshimura M., Ohbayashi T.. A method for producing transgenic cells using a multi-integrase system on a human artificial chromosome vector. PLoS One. 2011; 6:e17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rutherford K., Van Duyne G.D.. The ins and outs of serine integrase site-specific recombination. Curr. Opin. Struct. Biol. 2014; 24:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Russell J.P., Chang D.W., Tretiakova A., Padidam M.. Phage Bxb1 integrase mediates highly efficient site-specific recombination in mammalian cells. Biotechniques. 2006; 40:460–464. [DOI] [PubMed] [Google Scholar]
- 18. Zhu F., Gamboa M., Farruggio A.P., Hippenmeyer S., Tasic B., Schüle B., Chen-Tsai Y., Calos M.P.. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 2014; 42:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mulholland C.B., Smets M., Schmidtmann E., Leidescher S., Markaki Y., Hofweber M., Qin W., Manzo M., Kremmer E., Thanisch K. et al. . A modular open platform for systematic functional studies under physiological conditions. Nucleic Acids Res. 2015; 43:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O., Iii C.A.H., America N.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 21. Lin Y.-C., Boone M., Meuris L., Lemmens I., Van Roy N., Soete A., Reumers J., Moisse M., Plaisance S., Drmanac R. et al. . Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 2014; 5:4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cohen R.N., van der Aa M.A., Macaraeg N., Lee A.P., Szoka F.C.. Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J. Control. Release. 2009; 135:166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kebschull J.M., Zador A.M.. Sources of PCR-induced distortions in high-throughput sequencing data sets. Nucleic Acids Res. 2015; 43:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011; 20:1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gibbs D.J., Bacardit J., Bachmair A., Holdsworth M.J.. The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 2014; 24:603–611. [DOI] [PubMed] [Google Scholar]
- 26. Gonda D.K., Bachmair a, Wünning I., Tobias J.W., Lane W.S., Varshavsky A.. Universality and structure of the N-end rule. J. Biol. Chem. 1989; 264:16700–16712. [PubMed] [Google Scholar]
- 27. Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005; 399:777–799. [DOI] [PubMed] [Google Scholar]
- 28. Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B. et al. . Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009; 27:851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Phillips-Cremins J., Corces V.. Chromatin insulators: linking genome organization to cellular function. Mol. Cell. 2013; 50:461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen Z.-Y., He C.-Y., Kay M.A. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum. Gene Ther. 2005; 16:126–131. [DOI] [PubMed] [Google Scholar]
- 31. Kivioja T., Vähärautio A., Karlsson K., Bonke M., Enge M., Linnarsson S., Taipale J.. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods. 2011; 9:72–74. [DOI] [PubMed] [Google Scholar]
- 32. Park S.-E., Kim J.-M., Seok O.-H., Cho H., Wadas B., Kim S.-Y., Varshavsky A., Hwang C.-S.. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science. 2015; 347:1249–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matta-Camacho E., Kozlov G., Li F.F., Gehring K.. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat. Struct. Mol. Biol. 2010; 17:1182–1187. [DOI] [PubMed] [Google Scholar]
- 34. Tasaki T., Zakrzewska A., Dudgeon D.D., Jiang Y., Lazo J.S., Kwon Y.T.. The substrate recognition domains of the N-end rule pathway. J. Biol. Chem. 2009; 284:1884–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang K.H., Roman-Hernandez G., Grant R.A., Sauer R.T., Baker T.A.. The molecular basis of N-end rule recognition. Mol. Cell. 2008; 32:406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wissink E.M., Fogarty E.A., Grimson A.. High-throughput discovery of post-transcriptional cis-regulatory elements. BMC Genomics. 2016; 17:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fowler D.M., Stephany J.J., Fields S.. Measuring the activity of protein variants on a large scale using deep mutational scanning. Nat. Protoc. 2014; 9:2267–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peterman N., Levine E.. Sort-seq under the hood: implications of design choices on large-scale characterization of sequence-function relations. BMC Genomics. 2016; 17:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kowalsky C.A., Klesmith J.R., Stapleton J.A., Kelly V., Reichkitzer N., Whitehead T.A.. High-resolution sequence-function mapping of full-length proteins. PLoS One. 2015; 10:e0118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.