ABSTRACT
Chromatin condensation during mitosis produces detangled and discrete DNA entities required for high fidelity sister chromatid segregation during mitosis and positions DNA away from the cleavage furrow during cytokinesis. Regional condensation during G1 also establishes a nuclear architecture through which gene transcription is regulated but remains plastic so that cells can respond to changes in nutrient levels, temperature and signaling molecules. To date, however, the potential impact of this plasticity on mitotic chromosome condensation remains unknown. Here, we report results obtained from a new condensation assay that wildtype budding yeast cells exhibit dramatic changes in rDNA conformation in response to temperature. rDNA hypercondenses in wildtype cells maintained at 37°C, compared with cells maintained at 23°C. This hypercondensation machinery can be activated during preanaphase but readily inactivated upon exposure to lower temperatures. Extended mitotic arrest at 23°C does not result in hypercondensation, negating a kinetic-based argument in which condensation that typically proceeds slowly is accelerated when cells are placed at 37°C. Neither elevated recombination nor reduced transcription appear to promote this hypercondensation. This heretofore undetected temperature-dependent hypercondensation pathway impacts current views of chromatin structure based on conditional mutant gene analyses and significantly extends our understanding of physiologic changes in chromatin architecture in response to hypothermia.
KEYWORDS: cohesin, condensin, DNA hypercondensation/compaction, heterochromatin, hyperthermia, rDNA condensation, transcription
Introduction
Chromatin condensation may have been the first dramatic cell cycle change observable to early microscopists and remains a topic both of fascination and clinical relevance. Chromatin condensation and the arrangement of chromosomes within the nuclear volume are collectively mediated by cohesin- and condensin-dependent DNA segment tetherings that occur in either cis (within a single DNA molecule) or trans (between separate DNA molecules) conformations.1,2 Recent evidence is consistent with the notion that cis DNA tetherings established during interphase are critical for the proper deployment of developmental transcription programs.3,4 Intriguingly, proper development likely also requires G1 trans tethers (here, between non-homologous chromosomes) that link together domains of either repressed or induced transcriptional activities termed TADs (Topologically Association Domains).5
The combination of cis and trans tethers during G1 that mediate regionalized chromatin condensation and nuclear architecture must remain plastic if cells are to respond appropriately to external cues. Numerous studies across several species document that the chromosome condensation state during interphase increases in response to heat-stress. For instance, hyperthermia induces premature chromosome condensation during S phase in CHO, HeLa and S3 cells and hypercondensation in Achlya ambisexualis hypha.6-12 Importantly, little is known regarding the mechanisms through which these induced and premature condensation reactions occur. Moreover, the extent to which cells respond to hyperthermia during mitosis, when chromosomes are already condensed, remains largely untested. Addressing these deficiencies becomes important since physiologic changes induced by heat stress are wide-ranging and include expression of heat shock proteins (HSPs), protein synthesis repression and altered rates of both transcription and mRNA turnover.13,14
Budding yeast remains a mainstay model organism from which numerous aspects of chromatin condensation have come to light. Common to most studies is the testing of temperature-dependent gene product inactivation on rDNA structure.15-27 This is because rDNA undergoes cell cycle-specific structural changes: forming diffuse puff-like structures during G1 and condensing through multiple stages that include cluster and line formations before coalescing into discrete loop-like structures during mitosis.16,17,21 When either SMC (structural maintenance of chromosomes) cohesin or condensin complexes are impaired (for instance through the use of conditional alleles), mitotic cells contain puff-like rDNA loci instead of tightly condensed rDNA loops.24,26-28 Since the rDNA locus is a highly-specialized domain comprising iterative repeats that are repressed for Homologous Recombination (HR) and under tight transcriptional regulation,29-31 parallel strategies were developed in yeast to assess changes in chromosome arm condensation.24,32 All of these studies operate under the assumption that mitotic chromatin structures in wildtype cells are refractory to the temperature shift required to inactivate conditional allele gene products. Conversely, a growing body of evidence suggests that DNA segment cis and trans tethers of interphase cells remain dynamic to respond to external cues.1-5,33 Here, we developed a streamlined rDNA condensation assay and report that wildtype budding yeast exhibit a mitotic chromatin hypercondensation activity that is induced at temperatures typically used to inactivate conditional alleles. Moreover, this hypercondensation activity predominantly targets the rDNA locus, revealing a heretofore unexplored and targeted physiologic response to hyperthermia.
Results
Temperature-dependent regulation of mitotic rDNA hypercondensation
Despite technical advances that include immunodetection, GFP tagging and Fluorescence in situ hybridization (FISH), barriers to detecting changes in chromatin architecture in yeast cells include small cell and nuclear sizes and limited longitudinal chromatin compaction relative to other eukaryotic cells.16,17,24-26,34,35 Here, we report on a streamlined procedure that provides for exquisite imaging of the yeast rDNA locus in the absence of GFP, stacked antibody complexes and hybridization of labeled nucleotide probes. To validate this procedure, we exploited a well-established observation that mutation of the cohesin subunit Mcd1/Scc1 results in condensation defects.17,19,24,26,36-38 Log phase wildtype and mcd1–1 mutant cells were synchronized in G1 (α factor) at 23°C and then released into 37°C (non-permissive temperature for mcdc1–1) fresh medium supplemented with nocodazole to arrest cells preanaphase. Cell cycle progression was confirmed by detection of DNA content using flow cytometry (Figure S1A). The resulting preanaphase wildtype cells contained predominantly loop-like rDNA structures while preanaphase mcd1–1 mutant cells failed to condense their chromatin such that the rDNA formed predominantly puff-like structures (Fig. 1A). We quantified these differences and found that our procedure detects similar levels of condensation defects in mcd1–1 mutant cells as those obtained using FISH and GFP-based methodologies (Fig. 1B).17,24,26,27
Figure 1.
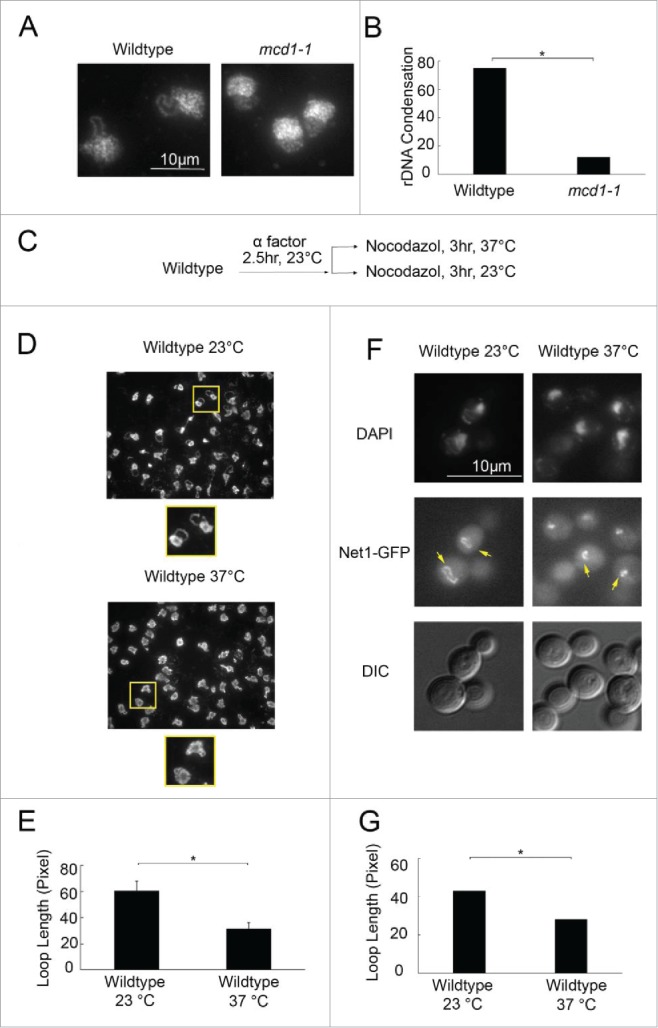
Temperature-dependent regulation of mitotic rDNA condensation. (A) Condensation assay validation. Micrographs of chromosomal mass and rDNA loop and puff structures detected using DAPI for wildtype (VG985) and mcd1–1 mutant cells. (B) Quantification of rDNA loops (condensation) in preanaphase wildtype and mcdc1–1 mutant strains (N = 100 cells for each strain). (C) Schematic of cell synchronization and experimental procedure performed on wildtype cells (YBS1019). (D) Micrographs of chromosome masses detected using DAPI. Regions demarcated by yellow squares equally magnified below. (E) Quantification and statistical analyses of rDNA loop lengths observed using our condensation assay in wildtype cells at 23°C and 37°C (3 biologic replicates with over 100 cells for each strain analyzed per replicate [N = 330 cells at 23°C and 352 cells at 37°C total]; p-value = 0.004). (F) Micrographs of preanaphase wildtype cells arrested at either 23°C or 37°C. Chromosome mass detected by DAPI, rDNA architecture detected using Net1GFP and cell morphology imaged using Differential Interference Contrast (DIC) microscopy. Yellow arrows indicate rDNA loops at 23°C and diminished loops at 37°C. (G) Quantification and statistical analyses of rDNA loop lengths measured using Net1-GFP in wildtype preanaphase cells arrested 23°C and then either maintained at 23°C or shifted to 37°C (N = 190 cells at 23°C and 157 cells at 37°C; P-value = 1.95E-28).
During this investigation, we discovered a robust temperature-dependent effect that occurs in wildtype cells, independent of gene mutation. We modified our prior experimental strategy to further analyze this hypercondensation activity by releasing G1 arrested cells into either 23°C or 37°C medium supplemented with nocodazole (Fig. 1C). Cell cycle progression was confirmed by detection of DNA content using flow cytometry (Figure S1B). Our condensation assay revealed that preanaphase wildtype cells condense rDNA loops to form tight and discrete loops at both 23°C or 37°C. Surprisingly, however, rDNA loops were condensed but appeared significantly shorter in wildtype cells shifted to 37°C upon release from G1, compared with loops in wildtype cells maintained at 23°C (Fig. 1D). We thus quantified the loop lengths in wildtype cells at both temperatures by tracing the entire length of the loop structures. The results clearly document that rDNA loop lengths are significantly shorter (herein termed hypercondensed) in cells that progress from G1 into mitosis at 37°C (Fig. 1E).
Our condensation assay provides for detailed images of rDNA structure, but we wondered if other procedures could detect temperature-sensitive hypercondensation of rDNA. Net1 is an rDNA binding protein and represents one current standard by which rDNA structure is assessed.24,26-28 Wildtype cells expressing GFP-tagged Net1 were synchronized in G1, released into either 23°C or 37°C rich medium supplemented with nocodazole, and the resulting preanaphase cells assessed for rDNA loop lengths. Cell cycle progression was confirmed by detection of DNA content using flow cytometry (Figure S1C). Wildtype cells that progressed from late G1 into mitosis at 23°C exhibited significant longer rDNA loop lengths than cells released from G1 into 37°C medium (Fig. 1F, G). In combination, these results reveal that rDNA condensation is exquisitely sensitive to temperature changes in wildtype cells with elevated temperatures leading to robust hypercondensation that is easily measurable using well-established procedures.
Temperature-induced hypercondensation is not based on increased condensation rates
What is the basis for this heretofore unreported temperature-dependent effect on rDNA condensation? We speculated that the decreased loop size that occurs in wildtype cells maintained at 37°C might reflect an accelerated rate of condensation, relative to that which occurs at 23°C. If this model is correct, then longer incubations at 23°C should ultimately lead to hypercondensed rDNA. To test this prediction, cultures released from G1 were maintained in fresh medium supplemented with nocodazole for either 2.5 or 5 hours at 23° and compared with cultures instead maintained for 2.5 or 5 hours at 37°C (Fig. 2A). Cell cycle progression was confirmed by detection of DNA content using flow cytometry (Figure S1D). The results reveal that rDNA loop lengths in cells maintained in preanaphase at 23°C for 5 hours failed to approach the loop lengths of cells maintained in preanaphase at 37°C for only 2.5 hours. Moreover, rDNA loop lengths in cells maintained in preanaphase at 23°C for 2.5 hours are identical to rDNA loop lengths in cells maintained in preanaphase at 23°C for 5 hours (Fig. 2B). The same is true for loop lengths in cells maintained at 37°C, regardless of incubation time. In all cases, rDNA loop lengths were significantly shorter in cells maintained at 37°C compared with cells maintained at 23°C (Fig. 2C). Thus, changes in rDNA architecture occur independent of condensation rates.
Figure 2.
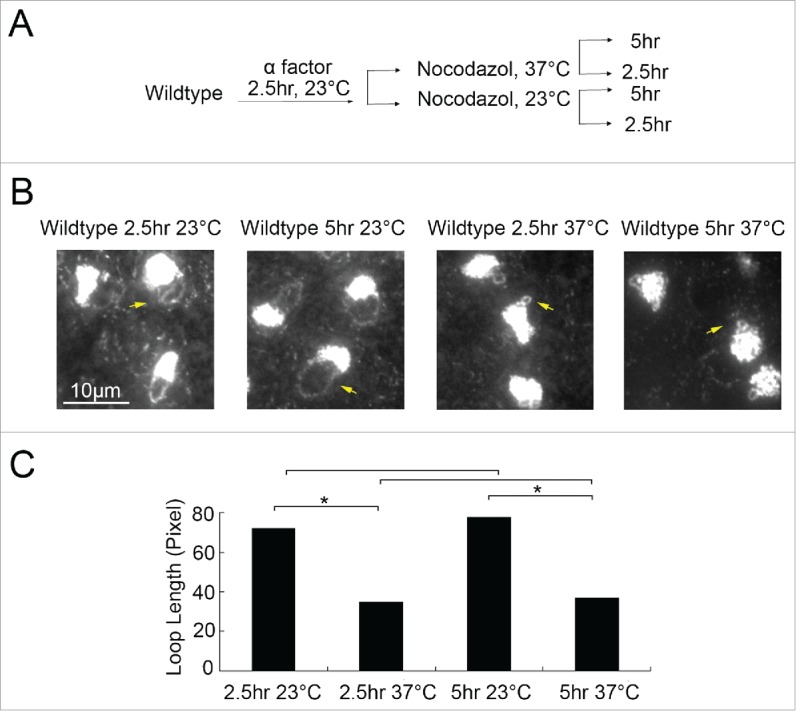
Temperature-induced hypercondensation is not based on increased condensation rates. (A) Schematic of synchronization and experimental procedure performed on wildtype cells (YBS1019). (B) Micrographs of chromosome masses and rDNA loops detected by DAPI staining. Yellow arrows point to rDNA loops at 23°C and diminished loops at 37°C. (C) Quantification and statistical analyses of rDNA loop lengths in each experimental group (N = 50 cells for each treatment, Pvalue = 0.149 for 23 °C 2.5 hr vs 5hr; P-value = 0.346 for 37 °C 2.5 hr vs 5hr; P-value = 5.54E-20 for 23 °C 2.5 hr vs 37 °C 2.5hr; P-value = 2.18E-20 for 23 °C 5 hr vs 37 °C 5hr; P-value = 5.90E-19 for 23 °C 2.5 hr vs 37 °C 5hr).
The rDNA hypercondensation machinery is active during mitosis
In most eukaryotes, chromosome condensation is a multi-step process that requires factors (such as core histones and cohesins) that are deposited during S phase and additional factors (such as linker histones and condensins) that associate or become activated later during the cell cycle.39,40 It thus became important to map the timing of this rDNA hypercondensation during the cell cycle. To test whether rDNA hypercondensation can be induced by elevated temperatures during M phase (after normal levels of condensation are already established), wildtype cells were synchronized in G1 at 23°C for 2.5 hours, then released at 23°C into fresh medium supplemented with nocodazole for 2.5 hours. Half of the resulting preanaphase cells were then shifted to 37°C for an additional hour and the other half maintained at 23°C for the same time period (Fig. 3A). Cell cycle progression was confirmed by changes in DNA content using flow cytometry (Figure S1E). As expected, rDNA in cells maintained at 23°C throughout the preanaphase arrest appeared as long loops that extend away from the DNA mass. In contrast, rDNA in cells arrested in mitosis at 23°C but then shifted to 37°C while maintaining the preanaphase arrest exhibited significantly hypercondensed rDNA loop lengths (Fig. 3B, C). Thus, cells exhibit an rDNA-directed hypercondensation activity during preanaphase that induces condensation beyond that which promotes entry into mitosis.
Figure 3.
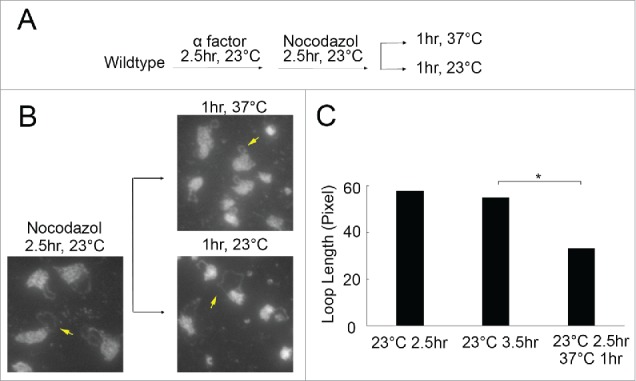
The rDNA hypercondensation machinery is active during mitosis. (A) Schematic of synchronization and experimental procedure performed on wildtype cells (YBS1019). (B) Micrographs of chromosome masses and rDNA loops detected by DAPI staining. Yellow arrows indicate loops at 23°C and diminished loops in response to a 1 hour shift to 37°C, but not 23°C, during preanaphase arrest. (C) Quantification and statistical analyses of the rDNA loop lengths in each experimental group (N = 138 cells at 23°C for 2.5 hr; N = 105 cells for 23°C for 3.5 hr; N = 115 cells for 23°C for 2.5 hrs and 37°C for 1 hr; P-value = 7.73E-17 for 23 °C 3.5 hr vs 23 °C 2.5 hr ->37 °C 1hr).
Temperature-induced hypercondensation is reversible and occurs independent of changes in rDNA repeat number
Budding yeast rDNA comprises approximately 150 repeats and is a hotspot for homologous recombination.41 This raised the possibility that the apparent hypercondensation is in reality a reduction in rDNA repeats through permanent recombination-based excisions and loss from the chromosome. We thus tested whether we could revert a hypercondensed rDNA loop back to an extended loop within a single preanaphase arrest. Wildtype cells were synchronized in G1 at 23°C, then released at 23°C into fresh medium supplemented with nocodazole. The resulting preanaphase arrested cells were then shifted to 37°C for 1 hour (which produces rDNA hypercondensation) before shifting back down to 23°C for an additional 1 hour (Fig. 4A). Cell cycle progression was confirmed by detection of DNA content using flow cytometry (Figure S1F). Similar to our earlier results, cells shifted up to 37°C during preanaphase contain hypercondensed rDNA (Fig. 4B). Importantly, when these cells were shifted down to 23°C for one hour, the rDNA recovered to a less condensed state – nearly matching the extended loop lengths of cells maintained at 23°C throughout the time course (Fig. 4B, C). Thus, rDNA hypercondensation is reversible and occurs independent of changes in the number of rDNA repeats – a revelation supported by the absence of Net1-GFP decorated excised rDNA circles under conditions that similarly produce short rDNA loops (Fig. 3C).
Figure 4.
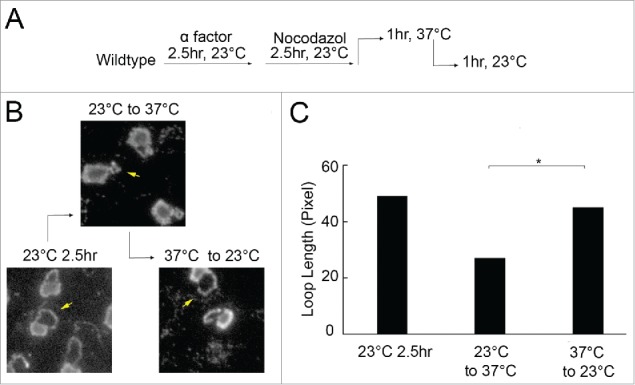
Temperature-induced hypercondensation occurs independent of changes in rDNA repeat number. (A) Schematic of synchronization and experimental procedure performed on wildtype cells (YBS1019). (B) Micrographs of chromosome mass and changes in rDNA loop lengths throughout the experiment detected by DAPI staining. (C) Quantification and statistical analyses of rDNA loop length in each experimental group (N = 130 cells at 23°C for 2.5 hr; N = 119 cells at 23°C for 2.5 hrs -> 37°C for 1 hr; N = 115 cells at 23°C for 2.5 hr -> 37°C for 1 hr -> 23°C for 1 hr; P-value = 2.53E-21 for shift from 23 °C to 37 °C vs shift from 37 °C to 23 °C).
Temperature-induced hypercondensation occurs independent of rDNA transcriptional inhibition
rDNA transcription, ribosome biogenesis and translational outputs are all upregulated during periods of accelerated growth. This increased rDNA transcription requires a relaxed DNA conformation and RNA polymerase accessibility.23,29,30,42-44 In contrast, yeast cell rDNA transcription levels decrease to basal line during mitotis that is coordinated with rDNA condensation.45,46 One prediction from these findings is that wildtype cells shifted to 37°C and that contain hypercondensed rDNA would exhibit slower growth kinetics (reduced rDNA transcription) compared with cells incubated at 23°C. To test this prediction, both S288C and W303 wildtype strains were sub-cultured over a 2 day period to ensure log phase growth before monitoring cell growth by spectroscopy. The results show that both yeast strains exhibit significantly increased growth rates at 37°C compared with 23°C (Fig. 5A, B). These findings negate the model that wildtype yeast cells exhibit hypercondensed rDNA due to slower growth at 37°C and instead are consistent with previous finding that wildtype yeast significantly increase ribosomes synthesis when shifted to 36°C from 23°C growth conditions. 47 The mechanism through which yeast cells exhibit increased growth rates that require elevated transcription from the rDNA locus, despite the hypercondensation that occurs at 37°C (Figs. 1–4), remains unknown.
Figure 5.
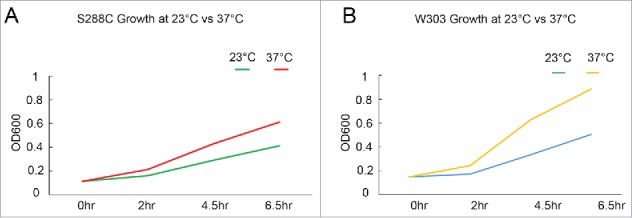
Temperature-induced hypercondensation occurs independent of rDNA transcription. (A) Growth curve of wildtype S288C background strain (YBS1019). (B) Growth curve of wildtype W303 background strain (YBS2020).
Temperature-induced hypercondensation is rDNA specific
rDNA is unique in terms of architecture, binding factors, transcription regulation, and altered levels of recombination, compared with the remainder of the genome.29,42,43 This raises the possibility that the temperature-dependent hypercondensation of rDNA is locus specific. To test this possibility, we obtained a chromosome arm condensation assay strain, kindly provided by Dr. Frank Uhlmann, that contains 2 lacO cassettes spaced 137 kb from each other on chromosome XII. Each LacO cassette is detected by lacR-GFP such that the inter-GFP distance allows for quantification of chromosome arm condensation.24 To validate this procedure, we placed through mating and dissection the LacO/lacR-GFP condensation cassette into mcd1–1 strains previously demonstrated as exhibiting condensation defects at both rDNA and chromosome arms.17 Log phase wildtype and mcd1–1 arm condensation assay strains were synchronized in G1 and released into either 23°C or 37° fresh medium supplemented with nocodazole to arrest cells preanaphase (Figure S2A). The resulting pre-anaphase synchronized cells were then fixed in paraformaldehyde and the disposition of arm condensation quantified by measuring the distance between GFP loci (Figure S2B). The results reveal a significant increase in the inter-GFP distance in mcd1–1 cells compared with wildtype cells (Figure S2C,D), confirming the efficacy of this arm condensation assay.
To test whether temperature-dependent hypercondensation extends to chromosome arm loci beyond that of rDNA, wildtype cells that harbor the arm condensation assay cassette were synchronized in G1 at 23°C and released into 23°C fresh medium supplemented with nocodazole for 2.5 hours. Half of the resulting preanaphase cells were shifted to 37°C for 1 hour while the other half was maintained at 23°C for 1 hour (Fig. 6A). Cell cycle progression and synchronization was confirmed by DNA content using flow cytometry (Figure S1G). The resulting preanaphase synchronized cells were then fixed and the distance between GFP loci quantified (Fig. 6B). The results reveal that preanaphase cells maintained at either 23°C or 37°C exhibit nearly identical inter-GFP distances (Fig. 6C, D). Thus, chromatin along the arm appears insensitive to temperature-induced hypercondensation that predominantly targets the rDNA locus.
Figure 6.
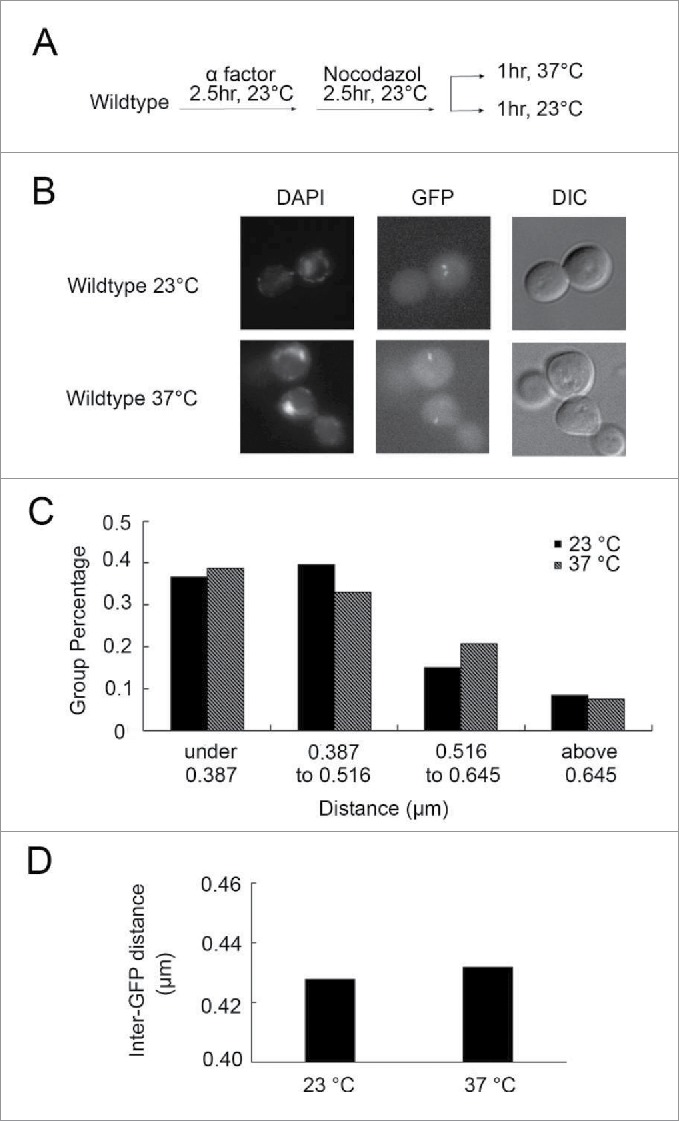
Temperature-induced hypercondensation is rDNA specific. (A) Schematic of synchronization and experimental procedure performed on wildtype cells (YBS2078). (B) Micrographs of DNA masses (DAPI), chromosome arm loci (GFP) and cell morphology (DIC) in preanaphase arrested cells at 23°C and 37°C. (C) Quantification of the distribution of measured distances between 2 arm loci GFP dots (N = 111 cells at 23°C; N = 106 cells at 37°C). (D) Quantification and statistical analyses of the GFP arm inter-loci distances in each experimental group (N = 111 cells at 23°C; N = 106 cells at 37°C; P-value = 0.812).
Discussion
Chromosome condensation ensures appropriate chromosome segregation but also influences transcriptional programs and nuclear architecture.48 Thus, it is not surprising that mutation of condensation pathways results in mitotic failure, aneuploidy and is further linked to developmental maladies such as microcephaly.49-53 A major revelation of the current study is that yeast cells contain hypercondensation machinery that is induced specifically during mitosis in response to elevated temperatures. This newly observed phenomenon is due neither to accelerated condensation reactions (hypercondensation at 37°C cannot be balanced by longer incubation time at 23°C), nor rDNA repeat reduction through recombination (hypercondensed rDNA loop lengths re-extend quickly upon shifting to 23°C and micrographs thus far fail to detect extrachromosomal rDNA signals in Net1-GFP cells that contain hypercondensed loops). Note that the homologous recombination machinery in general is suppressed during M phase such that changes in rDNA repeat numbers typically are monitored over several generations.30,54-56 In contrast, we observe inducible and reversible rDNA hypercondensation within a single M-phase arrest. Intriguingly, yeast hypercondense specific DNA loci in response to nutrient starvation,33 although the extent that these represent similar mechanisms in hypercondensation remains unknown.
What are the mechanisms through which rDNA hypercondensation at elevated temperature occurs? Early analyses of condensin mutants revealed reversibility in normal levels of chromosome condensation during mitosis: transient inactivation of temperature sensitive Brn1–9 condensin subunit during M phase resulted in rDNA decondensation that recondensed upon a shift back to permissive temperature. This reversibility of normal condensation levels during mitosis depends on both condensin and cohesin, although cohesin mutants thus far do not exhibit reversibility condensation - suggesting that cohesin promotes condensin activation.57 These observations raise the possibility that the reversible rDNA hypercondensation documented here requires similar molecular mechanisms that include cohesin and condensin - which are critical for mitotic chromosome condensation, stabilization of rDNA architecture and transcription regulation.57,62-65 Condensins in addition introduce positive supercoils into chromatin, promotes complementary single strand DNA reannealing and facilitates DNA catenation resolution.66 Cohesin-dependent tethering of DNA segments is also critical for transcription regulation, nuclear architecture and sister chromatid tethering.1 A final mechanism of hypercondensation could involve heat shock proteins,67 which may act in coordination or in parallel to the recruitment of condensins and cohesins (Fig. 7). The extent to which elevated temperature alters condensin, cohesin or heat shock protein deposition/activation onto rDNA remains an important goal of future research.
Figure 7.
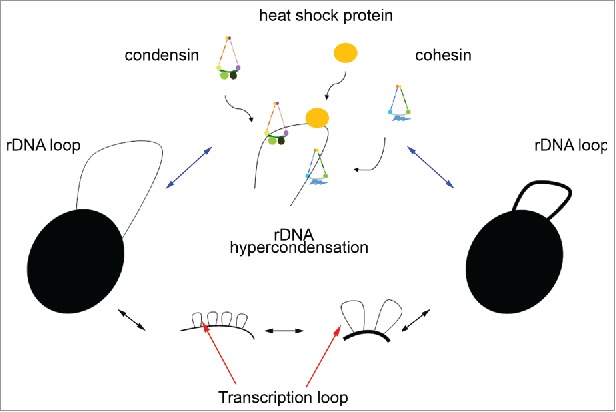
Model of rDNA transcription loop extension and possible mechanisms of hypercondensation. rDNA hypercondensation occurs in cells exposed to elevated temperature, potentially through transcription-dependent increase in axial loop extension (below). Candidates required to drive mitotic hypercondensation may include temperature-dependent enhanced activation of cohesin, condensin, or heat shock proteins (either directly or through condensation inhibitor inactivation) that specifically target the rDNA loci (above).
A second revelation of the current study is the apparent specificity of this hypercondensation activity. Using both our streamlined condensation assay and a well-established Net1-GFP strategy, we found that compaction along the longitudinal axis of rDNA is dramatically increased in response to elevated temperature. Conversely, genome-wide compaction along the chromosome arms is resistant to temperature-induced hypercondensation. In the context of cell physiology, mechanisms that selectively target rDNA are of critical interest. For instance, nutrient starvation induces rDNA hypercondensation and limits transcription factor accessibility which results in slow growth.33,58 On the other hand, increased cell growth kinetics correlate with elevated rates of rDNA transcription and ribosome assembly/maturation.58,59 In the current study, shifting wildtype cells to an elevated temperature (23°C to 37°C) produced an increased growth rate, consistent with previous findings that transcriptional silencing at rDNA loci is decreased and rRNA levels are increased at elevated temperatures.47,60,61 These observations raise a paradox: wildtype yeast at elevated temperatures exhibit both increased rDNA transcription and growth rates despite the hypercondensation of rDNA. Of the many possibilities, one model that resolves this apparent contradiction is that the shorter rDNA loop axis reflects significantly increased lateral loop extensions that accommodate elevated transcription during robust cell growth (Fig. 7). This transcriptional model is consistent with our results that exclude mechanisms of hypercondensation through accelerated condensation reactions and rDNA repeat reduction through recombination. Future efforts will be required to expose the mechanisms through which these potential longitudinal and lateral segment tetherings are regulated and document the extent to which transcriptionally active loops emerge laterally from the chromosomal axis.
The utilization of temperature-sensitive mutants remains a mainstay of yeast research. In particular, almost every condensation assay performed to date uses a temperature shift from permissive to non-permissive temperatures.16,17,19,24-27 Thus, our current study formally raises concerns regarding the extent through which defects in mutant strains (shifted to an elevated and non-permissive temperature) are predicated on comparisons to a hypercondensed rDNA locus. It thus becomes crucial to investigate the mechanism through which condensation is regulated by temperature, improve experimental strategies to better accommodate for those effects and then revisit results from prior studies predicated on conditional alleles and analysis of the rDNA locus. Moreover, our observations are likely to be of clinical interest, given the use of hyperthermia as a cancer treatment.68 For instance, the mechanism by which tumor cells become heat sensitive, compared with wildtype cells, is not fully understood. Our study provides a starting point through which locus-specific hypercondensation activity may promote hyperthermic resistance in wildtype cells and, in its absence, render tumor cells hyperthermically sensitive.
Materials and methods
Yeast strains and strain construction: Saccharomyces cerevisiae strains used in this study are listed in strain table (Table 1). GFP-tagging and deletion of genes was performed following published protocol.69
Table 1.
Yeast strains used in this study.
| Strain name | Genotype | Reference |
|---|---|---|
| VG955 | MATa mcd1–1 trp1 leu2 bar1 gal1 | 17 |
| VG982 | MATa trp1 ura3 bar1 gal1 | 17 |
| YBS1019 | MATa; S288C | 70 |
| YBS2020 | MATa; NET1:GFP:HIS3; w303 | For this study |
| YBS2078 | MATa; lacOs::YLR003c-1; lacOs::MMP1; LacI-GFP; w303 | 24 |
| YBS3017 | mcd1–1, MATa; lacOs::YLR003c-1; lacOs::MMP1; LacI-GFP; w303 | For this study |
rDNA condensation assay: Condensation assays were modified based on published FISH protocol.17 Briefly, synchronized cells were fixed by paraformaldehyde (100 μl 36% formaldehyde per 1 ml culture) for 2 hr at 23 °C. Cells were washed with distilled water 3 times and resuspended in spheroplast buffer (1M sorbitol, 20 mM KPO4, pH7.4), then spheroplasted by adding β-mercaptoethanol (1/50 volume) and Zymolyase T100 (1/100 volume) and incubating for 1 hour at 23°C. Resulting cells were pelleted and resuspended in 1.5 pellet volume of spheroplast buffer with 0.5% Triton X-100. 10 μl of the cell suspension were added to each well on poly-L-ysine coated slides, set at room temperature for 10 min, then removed the liquid gently by pipette. 20 μl of 0.5% SDS were added to each well, and set for 10 min, room temperature, then remove the liquid gently by pipette. Air dry the slides. Cells were then dehydrated by immersing the slides in fresh 3:1 methanol:acetic acid for 5 min at room temperature. Slides were stored at 4°C until completely dry, then cells were treated with RNase (100 μg/ml) in 2XSSC buffer (0.3M NaCl, 30 mM Sodium Citrate, pH7.0), incubate 1 hour at 37°C. Slides were washed 4 times in fresh 2XSSC (2 min/per wash), then went through a series of cold (−20°C) ethanol washes (start with 70%, followed by 80%, 95% ethanol washes, 2 min/per wash), then air dry. Slides were prewarmed to 37°C, then put into denaturing solution (70% formamide, 2X SSC) at 72°C for 2 min. Slides were immediately washed through a series of cold (−20°C) ethanol washes (start with 70%, followed by 80%, 90%, 100% ethanol washes, 1 min/per wash), then air dry. DNA mass were detected by DAPI (0.05ug/ml) staining and assayed under microscope. Cell cycle progression were confirmed by detection of DNA content using flow cytometry as described.27
Net1-GFP condensation assay: rDNA codensation assays were performed following similar strategy as previous publications.24,27 Briefly, cells were arrested at preanaphase and fixed by paraformaldehyde for 10 min at 30°C. GFP signal were then assayed under microscope. Cell cycle progression were confirmed by detection of DNA content using flow cytometry as described.27
Chromosome arm condensation assay: Chromosome arm condensation assays were performed as previous described.24 Briefly, cells were arrested at preanaphase and fixed by paraformaldehyde for 10 min at 30 °C. Distances between GFP dots were measured by microscopy. Cell cycle progression were confirmed by detection of DNA content using flow cytometry as described.27 To generate unbiased detection criteria of the GFP distance, cells contain a single GFP dot may reflect sister chromatids positioned vertical to the z-axial focal plane were thus excluded from analysis. In cases where GFP foci were planar to the field of view (eliminated z-axial contributions), we also excluded cells that contained 3 or 4 GFP loci, since the cohesion defect at one or both loci made it impossible to determine which GFP dot represented intra- or inter- sister chromatid loci. Thus, we focused our analysis on cells that contained 2 GFP dots resolvable within a single focal plane.
Statistical Analyses: F-Tests were used to assess the equality of 2 variances in chosen experimental groups, followed by Student's T-Tests to assess the statistical significance (P < 0.05).
Supplementary Material
Abbreviation
- (FISH)
Fluorescence in situ hybridization
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors thank Skibbens laboratory members, the Cassimeris laboratory and Cassbens members for helpful discussion throughout this process and Dr. Frank Uhlmann for kindly sharing yeast reagents.
Funding
RVS is supported by an award from the National Institutes of General Medicine (GM110631). Any opinions, findings, and conclusions or recommendations expressed in this study are those of the author(s) and do not necessarily reflect the views of the National Institutes of General Medicine.
References
- [1].Skibbens RV. Of rings and rods: Regulating cohesin entrapment of DNA to generate Intra- and intermolecular tethers. PLoS Genet 2016; 12(10):e1006337; PMID:27788133; https://doi.org/ 10.1371/journal.pgen.1006337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jeppsson K, Kanno T, Shirahige K, Sjögren C. The maintenance of chromosome structure: Positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol 2014; 15(9):601-14; PMID:25145851; https://doi.org/ 10.1038/nrm3857 [DOI] [PubMed] [Google Scholar]
- [3].Dorsett D. Cohesin: Genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 2011; 21(2):199-206; PMID:21324671; https://doi.org/ 10.1016/j.gde.2011.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Merkenschlager M, Nora EP. CTCF and cohesin in genome folding and transcriptional. Gene Regulation Annu Rev Genomics Hum Genet 2016; 17:8.1-8.27 [DOI] [PubMed] [Google Scholar]
- [5].Ciabrelli F, Cavalli G. Chromatin-driven behavior of topologically associating domains. J Mol Biol 2015; 427(3):608-25; PMID:25280896; https://doi.org/ 10.1016/j.jmb.2014.09.013 [DOI] [PubMed] [Google Scholar]
- [6].Pekkala D, Heath B, Silver JC. Changes in chromatin and the phosphorylation of nuclear proteins during heat shock of Achlya ambisexualis. Mol Cell Biol 1984; 4(7):1198-205; PMID:6504045; https://doi.org/ 10.1128/MCB.4.7.1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mackey MA, Morgan WF, Dewey WC. Nuclear fragmentation and premature chromosome condensation induced by heat shock in S-phase Chinese hamster ovary cells. Cancer Res 1988; 48(22):6478-83; PMID:3180064 [PubMed] [Google Scholar]
- [8].Flannery AV, Hill RS. The effect of heat shock on the morphology of amphibian lampbrush chromosomes. Exp Cell Res 1988; 177(1):9-18; PMID:3391242; https://doi.org/ 10.1016/0014-4827(88)90020-1 [DOI] [PubMed] [Google Scholar]
- [9].Roux C, Dadoune JP. Use of the acridine orange staining on smears of human spermatozoa after heat-treatment: Evaluation of the chromatin condensation. Andrologia 1988; 21(3):275-80; https://doi.org/ 10.1111/j.1439-0272.1989.tb02410.x [DOI] [PubMed] [Google Scholar]
- [10].Iliakis GE, Pantelias GE. Effects of hyperthermia on chromatin condensation and nucleoli disintegration as visualized by induction of premature chromosome condensation in interphase mammalian cells. Cancer Res 1989; 49(5):1254-60; PMID:2917355 [PubMed] [Google Scholar]
- [11].Rodriguez-Martin ML, Moreau N, Herberts C, Angelier N. Comparison between in vivo and in vitro heat-induced changes in amphibian lampbrush chromosomes. Chromosoma 1991; 100(2):79-86; PMID:2009818; https://doi.org/ 10.1007/BF00418240 [DOI] [PubMed] [Google Scholar]
- [12].Swanson PE, Carroll SB, Zhang XF, Mackey MA. Spontaneous premature chromosome condensation, micronucleus formation, and nonapoptotic cell death in heated HeLa S3 cells. Ultrastructural observations. Am J Pathol 1995; 146(4):963-71; PMID:7717463 [PMC free article] [PubMed] [Google Scholar]
- [13].Castells-Roca L, García-Martínez J, Moreno J, Herrero E, Bellí G, Pérez-Ortín JE. Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLoS ONE 2011; 6(2):e17272; PMID:21364882; https://doi.org/ 10.1371/journal.pone.0017272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Verghese J, Abrams J, Wang Y, Kevin A. Biology of the heat shock response and protein chaperones: Budding Yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev 2012; 76(2):115-58; PMID:22688810; https://doi.org/ 10.1128/MMBR.05018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Scherthan H, Loidl J, Schuster T, Schweizer D. Meiotic chromosome condensation and pairing in Saccharomyces cerevisiae studied by chromosome painting. Chromosoma 1992; 101(10):590-5; PMID:1424983; https://doi.org/ 10.1007/BF00360535 [DOI] [PubMed] [Google Scholar]
- [16].Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol 1994; 125(3):517-30 ; PMID:8175878; https://doi.org/ 10.1083/jcb.125.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 1997; 91(1):47-57; PMID:9335334; https://doi.org/ 10.1016/S0092-8674(01)80008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Castaño IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev 1996; 10(20):2564-76; PMID:8895658; https://doi.org/ 10.1101/gad.10.20.2564 [DOI] [PubMed] [Google Scholar]
- [19].Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev 1999; 13(3):307-19; PMID:9990855; https://doi.org/ 10.1101/gad.13.3.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol 2000; 149(4):811-24; PMID:10811823; https://doi.org/ 10.1083/jcb.149.4.811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev 2004; 18(1):76-87; PMID:14701879; https://doi.org/ 10.1101/gad.1150404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 2004; 117(4):471-82; PMID:15137940; https://doi.org/ 10.1016/S0092-8674(04)00415-5 [DOI] [PubMed] [Google Scholar]
- [23].Wang BD, Butylin P, Strunnikov A. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle 2006; 5(19):2260-7; PMID:16969110; https://doi.org/ 10.4161/cc.5.19.3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 2008; 22(16):2215-27; PMID:18708580; https://doi.org/ 10.1101/gad.1675708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, Fleharty B, Seidel C, Brickner JH, Gerton JL. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol 2009; 187(4):455-62; PMID:19948494; https://doi.org/ 10.1083/jcb.200906075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F. Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol 2013; 23(1):64-9; PMID:23219725; https://doi.org/ 10.1016/j.cub.2012.11.030 [DOI] [PubMed] [Google Scholar]
- [27].Tong K, Skibbens RV. Pds5 regulators segregate cohesion and condensation pathways in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 2015; 112(22):7021-6; PMID:25986377; https://doi.org/ 10.1073/pnas.1501369112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 1999; 97(2):245-56; PMID:10219245; https://doi.org/ 10.1016/S0092-8674(00)80734-5 [DOI] [PubMed] [Google Scholar]
- [29].Nomura M. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol 2001; 66:555-65; PMID:12762057; https://doi.org/ 10.1101/sqb.2001.66.555 [DOI] [PubMed] [Google Scholar]
- [30].Kobayashi T, Ganley AR. Recombination regulation by transcriptioninduced cohesin dissociation in rDNA repeats. Science 2005; 309(5740):1581-4; PMID:16141077; https://doi.org/ 10.1126/science.1116102 [DOI] [PubMed] [Google Scholar]
- [31].Huang J, Brito IL, Villén J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev 2006; 20(20):2887-901; PMID:17043313; https://doi.org/ 10.1101/gad.1472706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen T, Hiragami-Hamada K, Winkler M, Barral Y, Fischle W, Neumann H. A cascade of histone modifications induces chromatin condensation in mitosis. Science 2014; 343(6166):77-80; PMID:24385627; https://doi.org/ 10.1126/science.1244508 [DOI] [PubMed] [Google Scholar]
- [33].Tsang CK, Li H, Zheng XS. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J 2007; 26(2):448-58; PMID:17203076; https://doi.org/ 10.1038/sj.emboj.7601488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt EC. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J 1991; 10(3):573-83; PMID:1825809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Machín F, Torres-Rosell J, Jarmuz A, Aragón L. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J Cell Biol 2005; 168(2):209-19; PMID:15657393; https://doi.org/ 10.1083/jcb.200408087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol 2000; 151(3):613-26; PMID:11062262; https://doi.org/ 10.1083/jcb.151.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Woodman J, Hoffman M, Dzieciatkowska M, Hansen KC, Megee PC. Phosphorylation of the Scc2 cohesin deposition complex subunit regulates chromosome condensation through cohesin integrity. Mol Biol Cell 2015; 26(21):3754-67 ; PMID:26354421; https://doi.org/ 10.1091/mbc.E15-03-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Orgil O, Matityahu A, Eng T, Guacci V, Koshland D, Onn I. A conserved domain in the scc3 subunit of cohesin mediates the interaction with both mcd1 and the cohesin loader complex. PLoS Genet 2015; 11(3):e1005036; PMID:25748820; https://doi.org/ 10.1371/journal.pgen.1005036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Aragon L, Martinez-Perez E, Merkenschlager M. Condensin, cohesin and the control of chromatin states. Curr Opin Genet Dev 2013; 23(2):204-11; PMID:23312842; https://doi.org/ 10.1016/j.gde.2012.11.004 [DOI] [PubMed] [Google Scholar]
- [40].Hirano T. Condensin-Based chromosome organization from bacteria to vertebrates. Cell 2016; 164(5):847-57; PMID:26919425; https://doi.org/ 10.1016/j.cell.2016.01.033 [DOI] [PubMed] [Google Scholar]
- [41].Nomura M. Regulation of ribosome biosynthesis in escherichia coli and saccharomyces cerevisiae: Diversity and common principles. J Bacteriol 1999; 181(22):6857-64; PMID:10559149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Russell J, Zomerdijk JC. The RNA polymerase I transcription machinery. Biochem Soc Symp 2006; 73:203-16; PMID:16626300; https://doi.org/ 10.1042/bss0730203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Salminen A, Kaarniranta K. SIRT1 regulates the ribosomal DNA locus: Epigenetic candles twinkle longevity in the Christmas tree. Biochem Biophys Res Commun 2009; 378(1):6-9; PMID:19010308; https://doi.org/ 10.1016/j.bbrc.2008.11.023 [DOI] [PubMed] [Google Scholar]
- [44].Sáez-Vásquez J, Gadal O. Genome organization and function: A view from yeast and Arabidopsis. Mol Plant 2010; 3(4):678-90; PMID:20601371; https://doi.org/ 10.1093/mp/ssq034 [DOI] [PubMed] [Google Scholar]
- [45].Johzuka K, Horiuchi T. RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae. Genes Cells 2007; 12(6):759-71; PMID:17573776 [DOI] [PubMed] [Google Scholar]
- [46].Clemente-Blanco A, Mayán-Santo M., Schneider DA, Machín F, Jarmuz A, Tschochner H., Aragón L. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 2009; 458(7235):219-22; PMID:19158678; https://doi.org/ 10.1038/nature07652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Warner J, Udem S. Temperature sensitive mutations affecting ribosome synthesis in Saccharomyces cerevisiae. J Mol Biol 1972; 65:243-57; PMID:4557193; https://doi.org/ 10.1016/0022-2836(72)90280-X [DOI] [PubMed] [Google Scholar]
- [48].Cuylen S, Metz J, Hruby A, Haering CH. Entrapment of chromosomes by condensin rings prevents their breakage during cytokinesis. Dev Cell 2013; 27(4):469-78; PMID:24286828; https://doi.org/ 10.1016/j.devcel.2013.10.018 [DOI] [PubMed] [Google Scholar]
- [49].Martin CA, Murray JE, Carroll P, Leitch A, Mackenzie KJ, Halachev M, Fetit AE, Keith C, Bicknell LS, Fluteau A, et al.. Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev 2016; 30(19):2158-72; PMID:27737959; https://doi.org/ 10.1101/gad.286351.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kiraly G, Simonyi AS, Turani M, Juhasz I, Nagy G, Banfalvi G. Micronucleus formation during chromatin condensation and under apoptotic conditions. Apoptosis 2016; 22(2):207-219 [DOI] [PubMed] [Google Scholar]
- [51].Zhang C, Kuang M, Li M, Feng L, Zhang K, Cheng S. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Sci Rep 2016; 6:34508; PMID:27687868; https://doi.org/ 10.1038/srep34508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kagami Y, Yoshida K. The functional role for condensin in the regulation of chromosomal organization during the cell cycle. Cell Mol Life Sci 2016; 73(24):4591-98; PMID:27402120; https://doi.org/ 10.1007/s00018-016-2305-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Uhlmann F. SMC complexes: From DNA to chromosomes. Nat Rev Mol Cell Biol 2016; 17(7):399-412; PMID:27075410; https://doi.org/ 10.1038/nrm.2016.30 [DOI] [PubMed] [Google Scholar]
- [54].Houseley J, Tollervey D. Repeat expansion in the budding yeast ribosomal DNA can occur independently of the canonical homologous recombination machinery. Nucleic Acids Res 2011; 39(20):8778-91; PMID:21768125; https://doi.org/ 10.1093/nar/gkr589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lees-Miller SP. DNA double strand break repair in mitosis is suppressed by phosphorylation of XRCC4. PLoS Genet 2014; 10(8):e1004598; PMID:25165869; https://doi.org/ 10.1371/journal.pgen.1004598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mathiasen DP, Lisby M. Cell cycle regulation of homologous recombination in Saccharomyces cerevisiae. FEMS Microbiol Rev 2014; 38(2):172-84; PMID:24483249; https://doi.org/ 10.1111/1574-6976.12066 [DOI] [PubMed] [Google Scholar]
- [57].Lavoie BD, Hogan E, Koshland D. In vivo dissection of the chromosome condensation machinery reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol 2002; 156(5):805-15; PMID:11864994; https://doi.org/ 10.1083/jcb.200109056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Martin RM, Cardoso MC. Chromatin condensation modulates access and binding of nuclear proteins. FASEB J 2010; 24(4):1066-72; PMID:19897663; https://doi.org/ 10.1096/fj.08-128959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Moss T. At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev 2004; 14(2):210-7; PMID:15196469; https://doi.org/ 10.1016/j.gde.2004.02.005 [DOI] [PubMed] [Google Scholar]
- [60].Bi X, Yu Q, Sandmeier JJ, Elizondo S. Regulation of transcriptional silencing in yeast by growth temperature. J Mol Biol 2004; 344(4):893-905; PMID:15544800; https://doi.org/ 10.1016/j.jmb.2004.10.002 [DOI] [PubMed] [Google Scholar]
- [61].Hickman M, McCullough K, Woike A, Raducha-Grace L, Rozario T, Dula ML, Anderson E, Margalit D, Holmes SG. Isolation and characterization of conditional alleles of the yeast SIR2 gene. J Mol Biol 2007; 367(5):1246-57. Epub 2007January23; PMID:17316680; https://doi.org/ 10.1016/j.jmb.2007.01.044 [DOI] [PubMed] [Google Scholar]
- [62].Li P, Jin H, Yu HG. Condensin suppresses recombination and regulates double-strand break processing at the repetitive ribosomal DNA array to ensure proper chromosome segregation during meiosis in budding yeast. Mol Biol Cell 2014; 25(19):2934-47; PMID:25103240; https://doi.org/ 10.1091/mbc.E14-05-0957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Huang K, Jia J, Wu C, Yao M, Li M, Jin J, Jiang C, Cai Y, Pei D, Pan G, et al.. Ribosomal RNA gene transcription mediated by the master genome regulator protein CCCTC-binding Factor (CTCF) is negatively regulated by the condensin complex. J Biol Chem 2013; 288(36):26067-77; PMID:23884423; https://doi.org/ 10.1074/jbc.M113.486175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lu S, Lee KK, Harris B, Xiong B, Bose T, Saraf A, Hattem G, Florens L, Seidel C, Gerton J. The cohesin acetyltransferase Eco1 coordinates rDNA replication and transcription. EMBO Rep 2014; 15(5):609-17; PMID:24631914; https://doi.org/ 10.1002/embr.201337974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, Unruh J, Garrett A, McDowell W, Box A, et al.. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet 2012; 8(6):e1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hirano T. Condensins: Universal organizers of chromosomes with diverse functions. Genes Dev 2012; 26(15):1659-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Csermely P, Kajtár J, Hollósi M, Oikarinen J, Somogyi J. The 90 kDa heat shock protein (hsp90) induces the condensation of the chromatin structure. Biochem Biophys Res Commun 1994; 202(3):1657-63; PMID:8060353; https://doi.org/ 10.1006/bbrc.1994.2124 [DOI] [PubMed] [Google Scholar]
- [68].Asita E, Salehhuddin H. Heat sensitivity between human normal liver (WRL-68) and breast cancer (MDA-MB 231) cell lines. International Journal of Chemical, Environmental & Biological Sciences (IJCEBS) 2013; 1(1):191-195 [Google Scholar]
- [69].Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14(10):953-61; https://doi.org/ 10.1002/(SICI)1097-0061(199807)14:10%3c953::AID-YEA293%3e3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- [70].Rudra S, Skibbens RV. Chl1 DNA helicase regulates Scc2 deposition specifically during DNA-Replication in Saccharomyces cerevisiae. PLoS One 2013; 8(9):e75435; https://doi.org/ 10.1371/journal.pone.0075435 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


