Abstract
The expression of CBF (C-repeat-binding factor) genes is required for freezing tolerance in Arabidopsis thaliana. CBFs are positively regulated by INDUCER OF CBF EXPRESSION1 (ICE1) and negatively regulated by MYB15. These transcription factors directly interact with specific elements in the CBF promoters. Mitogen-activated protein kinase (MAPK/MPK) cascades function upstream to regulate CBFs. However, the mechanism by which MPKs control CBF expression during cold stress signaling remains unknown. This study showed that the activity of MYB15, a transcriptional repressor of cold signaling, is regulated by MPK6-mediated phosphorylation. MYB15 specifically interacts with MPK6, and MPK6 phosphorylates MYB15 on Ser168. MPK6-induced phosphorylation reduced the affinity of MYB15 binding to the CBF3 promoter and mutation of its phosphorylation site (MYB15S168A) enhanced the transcriptional repression of CBF3 by MYB15. Furthermore, transgenic plants overexpressing MYB15S168A showed significantly reduced CBF transcript levels in response to cold stress, compared with plants overexpressing MYB15. The MYB15S168A-overexpressing plants were also more sensitive to freezing than MYB15-overexpressing plants. These results suggest that MPK6-mediated regulation of MYB15 plays an important role in cold stress signaling in Arabidopsis.
INTRODUCTION
Cold temperatures and freezing can adversely affect plant growth and development, thus limiting the geographical distribution of plants and decreasing agricultural yields. However, plants can acclimate to low temperatures via the reprogramming of gene expression, metabolism and cellular architecture (1,2). Most of our current knowledge on cold sensing and acclimation mechanisms comes from studies of the model plant Arabidopsis thaliana (3,4). Evidence suggests that cold stress is sensed through cold-induced changes in membrane fluidity and other signals, which induce fluxes in cytoplasmic Ca2+ currents. These Ca2+ fluxes activate kinases or kinase cascades, leading to the transcription of cold regulated (COR) genes, which are required for freezing tolerance.
Members of the C-repeat (CRT)-binding factor (CBF) family of transcription factors (also known as dehydration-responsive element-binding protein 1 family members, DREB1s) have central functions in the regulation of cold stress-induced transcription. The expression levels of CBFs determine the levels of COR gene expression and freezing tolerance (5,6). The transcription of CBFs is activated by INDUCER OF CBF EXPRESSION1 (ICE1) and ICE1-like transcription factors (MYC-family transcription factors) and repressed by MYB15 (an MYB family transcription factor), which bind to specific elements in the CBF promoters.
Forward and reverse genetic approaches have demonstrated that mitogen-activated protein kinase (MAPK/MPK) cascades regulate CBF gene expression. However, the molecular links connecting cold-responsive MPK cascades and CBF gene transcription remain to be identified. In eukaryotes, MPK cascades play essential roles in transmitting stimuli from mitogens, developmental cues and various environmental stresses (7,8). MPK cascades are major signaling pathways that function downstream of sensors/receptors that are positively or negatively regulated by cytosolic Ca2+ influx in a tissue-specific manner (9). MPK cascades consist of three sequentially acting protein kinases. Signal transduction via MPK cascades is initiated by the activation of MPK kinase kinase (MAPKKK/MEKK/MTK/MKKK). This enzyme phosphorylates and activates MPK kinase (MAPKK/MEK/MKK), which in turn phosphorylates and activates MPK. MKKKs are serine (Ser, S)/threonine (Thr, T) kinases that phosphorylate Ser/Thr on conserved S/T-X3–5-S/T motifs of the MKK activation loop (10). MKKs are dual-specificity kinases that doubly phosphorylate the T-X-Y motif in the activation loop of MPKs (10). The terminal kinases of the cascade, MPKs, are also Ser/Thr kinases. The substrates of MPKs include other kinases and various transcription factors.
The Arabidopsis thaliana genome encodes 20 predicted MPKs, which differ in the signature sequences of their activation loops (8,11). Of these, MPK3, MPK4 and MPK6, the most extensively studied MPKs, are activated by stress (pathogens, osmotic, cold and oxidative stress), developmental cues and auxin signaling (12–14). Each MPK is thought to participate in responses to a specific stress or a subset of stresses. The specificity of their activity appears to be determined by unique MKK-MPK combinations that participate in the responses to different stresses. For example, drought and wounding induce the MKK1-MPK4 module, abscisic acid activates MKK1-MPK3 and osmotic shock activates MKK1-MPK3/MPK6 (4,15–17). The MEKK1-MKK2-MPK4/MPK6 cascade plays an important role in the cold signal transduction pathway (4). MKK2 is specifically activated by cold stress in Arabidopsis protoplasts. Yeast two-hybrid analyses, as well as in vitro and in vivo protein kinase assays, showed that MKK2 phosphorylates MPK4 and MPK6. Transgenic plants overexpressing MKK2 exhibit constitutive MPK4 and MPK6 activity and improved freezing tolerance through increased expression of CBF genes (4).
The specificity of the response to a given stimulus could also be determined by the choice of MPK substrates. Several studies have attempted to identify the substrates and interaction partners of MPKs (18,19). Protein microarray analysis of 1690 unique Arabidopsis thaliana proteins identified 39 proteins as substrates of MPK6 and 48 proteins as substrates of MPK3 (20). Another protein microarray experiment identified the substrates of 10 different activated MPKs (21). More studies will be required to establish that these candidates truly function as MPK substrates. In addition, each candidate must still be connected to specific signaling outcomes.
Downstream components of the cold-responsive MPK cascade, such as MPK4 and MPK6 substrates, remain to be identified. The transcription factor MYB15 was revealed as an MPK6 interaction partner by yeast two-hybrid analysis of an Arabidopsis transcription factor library (22). Protein microarray analysis also predicted that MYB15 is phosphorylated by activated MPK6 and MPK10 (20). MYB15 represses CBF gene expression and freezing tolerance in Arabidopsis (23,24) and MYB15 is expressed in Arabidopsis in the absence of cold stress. Mutational inactivation of MYB15 results in enhanced expression of CBFs during cold acclimation and the freezing tolerance response, whereas overexpression of MYB15 has the opposite effect. MYB15 binds to the conserved MYB transcription factor recognition sequences in the promoters of CBFs and inhibits their transcription.
In this study, we obtained several lines of evidence that MYB15 is a substrate for MPK6 both in vitro and in vivo. MYB15 physically interacted with MPK6 in vitro (in a yeast two-hybrid assay) and in planta, and MYB15 was phosphorylated by both recombinant and native MPK6. We identified an MPK6 phosphorylation site in MYB15 and performed mutational analysis showing that the binding affinity of MYB15 to the CBF3 promoter fragment, as well as the effect of MYB15 on CBF3 gene expression, are regulated by the phosphorylation status of the MPK6-target phosphorylation site. Comparison of the freezing tolerance of transgenic plants overexpressing native MYB15 or MYB15S168A, a mutant MYB15 protein lacking the MPK6-phosphorylation site, demonstrated that an intact MPK6-target phosphorylation site is associated with freezing tolerance. The finding that transcriptional derepression of CBF genes and the freezing tolerance response are mediated by direct phosphorylation of MYB15 by MPK6 increases our understanding of the cold stress signaling pathway in Arabidopsis.
MATERIALS AND METHODS
Plant materials and growth conditions
The Arabidopsis thaliana lines used in this study were in the Columbia background. For surface sterilization, seeds were soaked for 1 min in 70% EtOH, followed by 10 min in 1/10-diluted commercial bleach (0.4% NaOCl) and three washes with sterile distilled water. Surface-sterilized seeds were sown on agar plates containing half-strength Murashige-Skoog (MS) salts and vitamins (25), 2.0% sucrose and 0.8% agar. The plates were incubated in the dark for 3 days at 4°C, followed by incubation at 22°C in a growth chamber under a 16 h light/8 h dark photoperiod. Ten- to twelve-day-old seedlings were transferred to soil and grown under the same conditions.
Nicotiana benthamiana seeds were sown in soil and grown in a growth chamber at 24°C under a 16 h light/8 h dark photoperiod. Seven-week-old N. benthamiana plants were used for Agrobacterium-mediated transient expression.
Plasmid construction, site-directed mutagenesis and expression of recombinant proteins
Full-length and partial open reading frames (ORFs) encoding the N-terminal (amino acids 1–172) and C-terminal (amino acids 173–285) regions of MYB15 were amplified by polymerase chain reaction (PCR) from a cDNA library of Arabidopsis seedlings using gene-specific primers (Supplementary Table S1). The amplicons were cloned into the pGEM-T Easy Vector, and their fidelity was verified by sequencing. To create in-frame N-terminal GST fusions, the inserts were excised with BamHI and EcoRI and cloned into the pGEX 4T-1 vector (Amersham Biosciences, USA). Single amino acid substitutions in full-length GST-MYB15 (T18A, S168A and T18A/S168A) were produced using the primers listed in Supplementary Table S2 using a QuikChange Site-directed Mutagenesis kit (Stratagene, USA). The mutations were confirmed by sequencing. The GST-fusion constructs were transformed into BL21 (DE3) Escherichia coli, and GST fusion proteins were expressed and purified using glutathione Sepharose-4B beads according to the manufacturer's instructions (GE Healthcare, USA). To express His- or GST-tag-fused MPKs in bacteria, full-length MPK3, MPK4 or MPK6 ORFs were amplified by PCR from a cDNA library of Arabidopsis seedlings using gene-specific primers (Supplementary Table S2) and cloned into the pGEM-T Easy Vector (Promega, USA). Verified inserts by sequencing were excised with BamHI and SalI, and subcloned into pQE30 or pGEX 4T-1 for His- or GST-tag-fused MPKs, respectively. The His-fusion constructs were transformed into E. coli (M15), and His-tag fusion proteins were expressed and purified using Ni-NTA agarose beads according to the manufacturer's instructions (Qiagen, Germany).
In vitro pull-down assay
Pull-down assays were carried out to examine the interaction of MYB15 with MPK3, MPK4 or MPK6. GST-MYB15 and GST were immobilized by incubating crude proteins (100 μg) with 10 μl of glutathione-Sepharose 4B beads in 1 ml buffer (50 mM Tris–HCl, pH 7.5, 200 mM NaCl, 1% Triton X-100, 0.1 mM ethylenediaminetetraacetic acid (EDTA) and 0.5 mM DTT) for 4 h at 4°C. Glutathione-Sepharose 4B beads containing immobilized proteins were collected by centrifugation and incubated overnight at 4°C with purified His-MPK3, His-MPK4, or His-MPK6 (10 μg) in 1 ml binding buffer (50 mM Tris–Cl pH 7.5, 200 mM NaCl, 0.1 mM EDTA and 0.5 mM DTT). Beads were collected by centrifugation and washed three times with binding buffer at 4°C. Proteins were eluted by boiling in sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, separated by 10% SDS-PAGE and transferred onto an Immobilon-P membrane for immunoblotting. His-tagged proteins were detected on the membrane with polyclonal anti-His antibodies (Abcam, UK) using the enhanced chemiluminescence (ECL) technique.
Yeast two-hybrid analysis
Full-length ORFS of MYB15, MPK3, MPK4 and MPK6 were amplified using gene-specific primers (Supplementary Tables S1 and 2) and cloned into pGAD424 (prey) and pAS2–1 (bait) vectors, respectively. Prey and bait plasmids were co-transformed into yeast strain pJ69–4A for interaction analysis, and colonies were selected after 2–3 days growth at 30°C on SD-Trp-Leu medium. The transformants were tested for positive bait–prey interactions by monitoring the activation of ADE2 and LacZ reporter genes as described.
Luciferase complementation imaging assay
The luciferase (LUC) complementation imaging assay was performed as described by Chen et al. (26). Full-length MPK6 and MYB15 ORFs were inserted into pCAMBIA1300-NLuc and pCAMBIA1300-CLuc, resulting in the production of CaMV35S: MPK6-NLuc and CaMV35S: CLuc-MYB15, respectively. The constructs were mobilized into Agrobacterium tumefaciens strain GV3101 (pMP90) (27). The Agrobacterium cells were grown in LB medium at 28°C overnight, collected by centrifugation, resuspended in induction medium at 0.3 OD600nm (28) and incubated for 8–12 h at 28°C with shaking. Bacteria were collected by centrifugation, washed once with infiltration buffer (10 mM MgCl2, 10 mM MES and 100 mM acetosyringone) and resuspended in infiltration buffer at 0.5 OD600nm. Bacterial suspensions were infiltrated into young, fully expanded leaves of 3-week-old N. benthamiana plants using a needleless syringe. After infiltration, the plants were immediately covered with plastic bags and incubated at 23°C for 3 days. The plastic bags were removed, and infiltrated leaves were sprayed with luciferin solution (100 μM D-luciferin and 0.01% Triton X-100) and incubated in the dark for 4 h to quench fluorescence. LUC activity was observed under a low-light EMCCD apparatus (AndoriXon; Andor, UK).
Mass spectrometric analysis of phosphopeptides using TiO2 microcolumns
GST-MYB15 was treated with His-MPK6 as described below for the in vitro kinase assays. Protein bands were excised after resolution by SDS-PAGE and in-gel digested with modified trypsin (Promega, USA) (29). The digested peptides were dissolved in loading buffer (80% acetonitrile and 5% trifluoroacetic acid) and passed through a TiO2 microcolumn (29). Phosphopeptides were eluted with NH4OH (pH 10.5), applied to a Poros Oligo R3 column (Applied Biosystems, USA) and eluted from the column using 2,5-dihydroxybenzoic acid (DHB; Fluka, USA) solution (20 mg/ml DHB, 50% acetonitrile, 0.1% trifluoroacetic acid and 1% ortho-phosphoric acid). MALDI-MS analysis was performed using a Voyager-DE STR mass spectrometer (PerSeptive Biosystems Inc., USA). Mass spectra were obtained in the reflectron/delayed extraction mode. Monoisotopic peptide masses were analyzed using MoverZ software (www.proteometric.com).
In vitro and in-gel kinase assays
In vitro kinase reactions were performed in kinase buffer (25 mM Tris–HCl, pH 7.5, 1 mM DTT, 20 mM MgCl2, 2 mM MnCl2, and 50 μM [γ-32P] ATP) containing GST-MPK6 or His-MPK6 (1 μg) and substrate in a total reaction volume of 20 μl. GST (1 μg; negative control), myelin basic protein MBP (0.5 μg; positive control) and GST-MYB15 variants (2 μg) were used as substrates. The reactions were initiated using 1 μCi [γ-32P] ATP and allowed to proceed at 30°C for 30 min. The kinase reactions were stopped by adding 6 μl of 4 × SDS sample buffer and boiling for 5 min. Reaction products were resolved by 10% SDS-PAGE. The gels were autoradiographed and stained with Coomassie Brilliant Blue (CBB) R-250, using pre-stained markers to estimate protein size.
The in-gel kinase assay was performed using extracts from 2- to 3-week-old Petri dish-grown wild-type (WT), mpk3 (Salk_127507) and mpk6 mutant (Salk_127507) seedlings treated with cold (4°C) for the indicated times. The in-gel kinase assay was performed as described previously with some modifications (30). In brief, cell-free extracts (30 μg total proteins) were incubated at 60°C for 10 min and separated by 10% SDS-PAGE on a separating gel embedded with 0.5 mg/ml purified GST-MYB15 as a kinase substrate. After electrophoresis, the gel was washed three times with washing buffer (25 mM Tris–HCl, pH 7.5, 0.5 mM DTT, 0.1 mM Na3VO4, 5 mM NaF, 0.5 mg/ml bovine serum albumin and 0.1% Triton X-100) to remove the SDS. The proteins were renatured by incubating the gel overnight at 4°C in renaturing buffer (25 mM Tris–HCl, pH 7.5, 1 mM DTT, 0.1 mM Na3VO4 and 5 mM NaF) with three changes of buffer. The gel was equilibrated in reaction buffer (25 mM Tris–HCl, pH 7.5, 2 mM EGTA, 12 mM MgCl2, 1 mM DTT and 0.1 mM Na3VO4) at room temperature for 30 min. The phosphorylation reaction was carried out by incubating the gel at room temperature for 1.5 h in 20 ml of reaction buffer containing 0.5 μM ATP and 50 μCi [γ-32P] ATP. The reaction was stopped by transferring the gel to stop solution (5% trichloroacetic acid and 1% disodium pyrophosphate). Gels were washed with stop solution for 5 h at room temperature with four changes of solution, dried on 3M paper and imaged using a Fuji Film FLA-5000 imaging system.
Electrophoretic mobility shift assay (EMSA)
The −984 to −785 bp region of the CBF promoter, which binds to MYB15 (23) and contains an MYB response element, was amplified by PCR from Arabidopsis genomic DNA, separated by agarose gel electrophoresis and recovered by elution using a gel purification kit (GeneAll, Korea). Eluted fragments were end-labeled with [γ-32P] ATP and T4 polynucleotide kinase. The DNA-binding reaction was performed at 25°C for 20 min in binding buffer (20 mM HEPES/KOH pH 7.9, 0.5 mM dithiothreitol, 0.1 mM EDTA, 50 mM KCl and 15% glycerol) containing 20 000 cpm of 32P-labeled DNA probe, 1 μg poly (dI-dC) and 0.5 μg bacterially produced GST-MYB15 variants that had been phosphorylated (or not) by MPK6. For competition, purified protein was incubated with unlabeled probes for 30 min at 25°C, before incubating with the labeled probe. The reaction mixture was subjected to electrophoresis on a 5% polyacrylamide gel in 0.5 × Tris-Borate-EDTA (TBE) buffer at 80 V for 3 h. The gel was dried and mounted for autoradiography at −70°C with an intensifying screen.
Transient expression assay
The reporter plasmid was a pUC19-derived plasmid containing the β-glucuronidase (GUS) reporter gene under the control of a chimeric promoter containing a CBF3 promoter region (−2000 to −1 bp) inserted in front of the CaMV35S minimal promoter (31). For effector plasmids, MYB15 or MYB15S168A ORFs were inserted into a plant expression vector (pHBT95) containing the 35S C4PPDK promoter and nos terminator (32). A construct carrying the 35S promoter fused to the LUC gene was used as an internal control to normalize the variation in cell numbers, transformation efficiency and cell viability of each transfection (33). Transient expression of these constructs was performed as described previously (34). Protoplasts (2 × 106) were co-transfected with 20 μg plasmid DNA consisting of a mixture of reporter, effector and internal control constructs. The transfected protoplasts were incubated for 16 h in the dark at 22°C before GUS and LUC activity measurements. In each sample, the GUS activity of the cell lysate was divided by the LUC activity, thereby normalizing the data to control for variations in transfection efficiency.
Construction of transgenic plants
The CaMV35S:3XFlag-MYB15 and CaMV35S:3XFlag-MYB15S168A constructs in the binary vector pCAMBIA 1300 were introduced into Agrobacterium strain GV3101 (pMP90) and used to transform Arabidopsis plants by the floral dip method (35). Transformants were selected on MS medium containing 40 μg/ml hygromycin. T3 homozygous progeny of transgenic plants expressing high levels of MYB15 were used for all experiments.
Extraction and immunoblot analysis of Arabidopsis proteins
Tissues were ground in liquid nitrogen and extracted in protein extraction buffer (50 mM HEPES, pH 7.5, 5 mM EDTA, 5 mM EGTA, 1 mM Na3VO4, 25 mM NaF, 50 mM-glycerophosphate, 2 mM DTT, 2 mM PMSF, 5% glycerol, 1% Triton X-100 and protease inhibitor). After two rounds of centrifugation at 12 000 × g for 10 min, the supernatants were transferred to clean tubes and stored at −80°C until use. Protein concentrations were determined using a Bio-Rad Protein Assay kit (Bio-Rad) with BSA as a standard. For immunoblot analysis, total protein was extracted from the leaves of 2-week-old plants and 30 μg protein samples were separated by 10% SDS-PAGE and transferred to PVDF membranes. Proteins were detected using mouse anti-Flag (1:5000; Sigma, USA) as a primary antibody and alkaline phosphate-conjugated anti-mouse as a secondary antibody (1:5000) and visualized using an ECL kit (Amersham Pharmacia Biotech, UK).
Quantitative RT-PCR
Total RNA was extracted using the LiCl/phenol method according to a published protocol (36), and 5 μg total RNA was reverse-transcribed in a 100-μl reaction volume using SuperScript II RNase-Reverse Transcriptase (Invitrogen, USA). Quantitative RT-PCR was performed in a 15 μl reaction volume containing 1 μl RT products, 10 pmole of gene-specific primers and 7.5 μl SsoFast EvaGreen Supermix (Bio-Rad, USA) using the CFX96 Real-Time System (Bio-Rad, USA). The reaction conditions included an initial 5 min pre-incubation at 94°C, 45 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 40 s followed by melting curves with 90 cycles at 55°C increasing 0.5°C/cycle and final cooling for 10 min at 72°C. Primers used for PCR are shown in Supplementary Table S2.
Ion leakage from leaves
Ion leakage tests were carried out as described by Ishitani et al. (37). Briefly, one excised leaf from a 3-week-old soil-grown plant was placed in a test tube containing 100 μl of deionized H2O, which was transferred to a circulating freezing bath set at 0°C. For each temperature treatment, three replicates were performed. The temperature of the bath was programmed to decrease to −10°C at a rate of 2°C per h. When the designated temperature was reached, the tubes were removed and placed immediately on ice to allow gradual thawing. The leaflets then were transferred carefully to another tube containing 20 ml of deionized water and shaken overnight, followed by conductivity measurements. The tubes containing leaves were then autoclaved. After cooling to room temperature for 4 h, the conductivity of the solution was again measured. The percentage of electrolyte leakage was calculated as the percentage of conductivity before autoclaving over that after autoclaving.
Plant freezing assay
Freezing stress was applied to 3-week-old plants grown in soil at 22°C under a long-day photoperiod by exposing them sequentially to 4°C for 1 h, followed by 0°C for 1 h and a programmed cycle of temperature reduction at a rate of 1°C per h to −8°C. After the freezing treatment, the plants were incubated at 4°C for 1 day, followed by a return to 22°C. Survival was evaluated 5 days later.
RESULTS
MYB15 interacts with MPK6
Separate high-throughput analyses initially identified MYB15 as a putative substrate for activated MPK6 and as an MPK6-interacting protein (4,20,22). Therefore, we re-examined the interaction between MYB15 and MPK6 using yeast two-hybrid analysis. As shown in Supplementary Figure S1, yeast cells co-expressing MYB15 and MPK6 grew on selective medium lacking adenine and acquired β-galactosidase activity, indicating a positive interaction between these proteins.
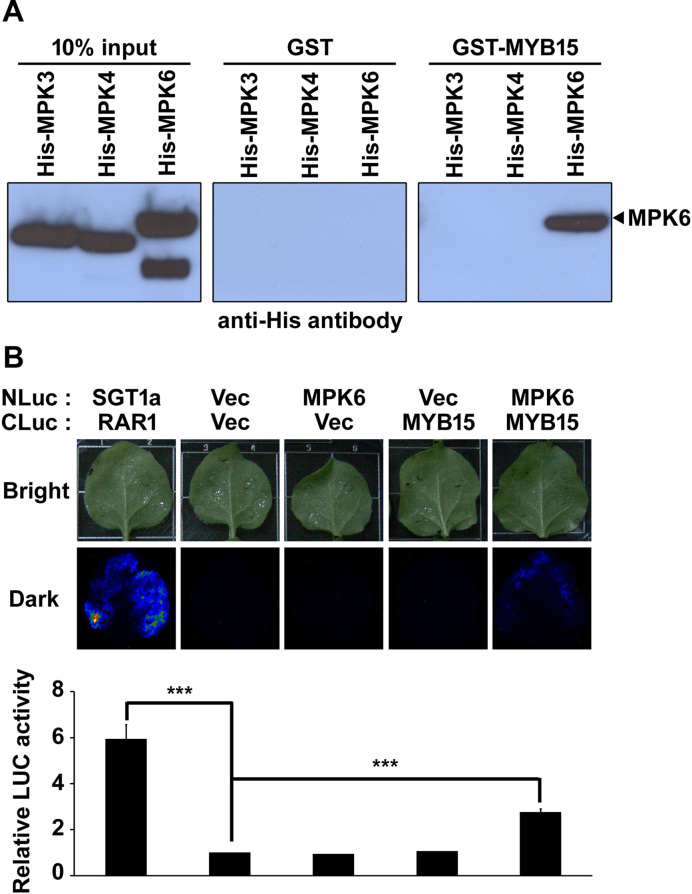
We then examined the specificity of the interaction of MYB15 with Arabidopsis MPKs using a pull-down assay (Figure 1A). As shown on the immunoblots, approximately equal amounts of the three MPKs were loaded onto the beads (left panel) and no His-MPKs were detected in eluates of immobilized GST-beads (middle panel). However, His-MPK6, but not His-MPK3 or His-MPK4, was detected in the eluates of immobilized GST-MYB15-beads (right panel). These results indicate that MYB15 interacts directly and specifically with MPK6 in vitro.
Figure 1.
MYB15 interacts with MPK6 in vitro and in planta. (A) Pull-down assay demonstrating the specificity of the interaction of MPK with MYB15. Shown is chemiluminescence detection of (His)6 on blots of SDS-polyacrylamide gels loaded with (left to right panels) input His-MPKs, GST and GST-MYB15 pull-down proteins. (B) LUC complementation imaging assay to detect the interaction of MPK6 with MYB15 in Nicotiana benthamiana leaves. Shown in the upper panel are images and luminescence of N. benthamiana leaves co-infiltrated with Agrobacterium strains containing the indicated combinations of NLuc- and CLuc-fusion constructs. The SGT1a-NLuc/CLuc-RAR1 combination was used as a positive control. Vec refers to NLuc and CLuc empty vectors. Leaves were photographed at 3 days after infiltration. Shown in the lower panel is the quantification of LUC activity in these leaves. The luminescence intensities are expressed relative to that of leaves infiltrated with the NLuc Vec/CLuc Vec combination. Data are presented as the mean ± SD of three independent experiments. P < 0.001 (***) indicate statistically significant changes.
We examined the interaction of MYB15 with MPK6 in planta in N. benthamiana leaves using an LUC complementation imaging assay. As shown in Figure 1B, strong LUC activity was detected at 3 days post-inoculation in positive control leaves co-infiltrated with Agrobacterium strains carrying CaMV35S: SGT1a-NLuc and CaMV35S: CLuc-RAR1 (26). No LUC activity was detected in leaves co-infiltrated with Agrobacterium strains carrying CLuc vector plus NLuc vector, CaMV35S: MPK6-NLuc plus CLuc vector or CaMV35S: CLuc-MYB15 plus NLuc vector combinations. However, LUC activity was detected in leaves co-infiltrated with Agrobacterium strains carrying CaMV35S: MPK6-NLuc plus CaMV35S: CLuc-MYB15 at levels ∼2- to 3-fold higher than that of control leaves co-infiltrated with empty vector(s). These results suggest that MYB15 directly interacts with MPK6 in planta.
MYB15 is phosphorylated by recombinant MPK6
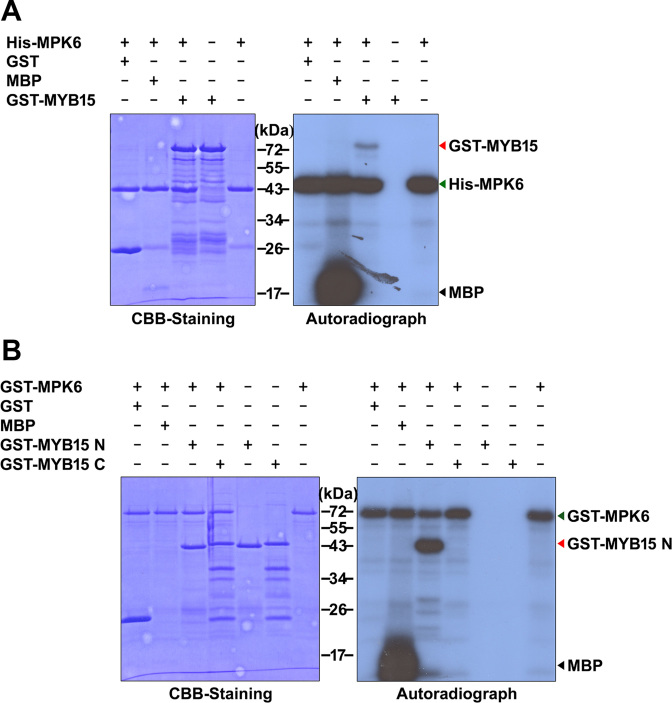
Having confirmed that MYB15 interacts directly with MPK6, we examined the ability of MPK6 to phosphorylate MYB15 using an in vitro kinase assay with E. coli-expressed purified proteins and [γ-32P] ATP. After separating the proteins by SDS-PAGE, we detected the phosphorylated proteins on the gel by autoradiography and visualized total proteins with CBB staining. As shown in Figure 2A, an autophosphorylation band of ∼48 kDa corresponding to His-MPK6 was observed in all samples. No phosphorylation band was observed corresponding to GST (∼25 kDa), the negative control substrate, whereas MBP (∼18.5 kDa; positive control) and GST-MYB15 (∼72 kDa) were phosphorylated by MPK6. Since MPKs usually phosphorylate Ser and Thr residues on S/T-P motifs, two MPK phosphorylation sites can be predicted in MYB15, on Thr18 and Ser168. To verify this prediction, we performed an in vitro kinase assay using purified GST-tagged MYB15 N-terminal fragment (GST- GST-MYB15N; amino acids 1–172, ∼45 kDa) and GST-tagged MYB15 C-terminal fragment (GST-MYB15C; amino acids 173–285, ∼45 kDa) as substrates for GST-MPK6 (∼72 kDa). GST-MPK6 autophosphorylation bands were detected in all samples (Figure 2B). A phosphorylation band was observed when we used GST-MYB15N as the substrate, but not with GST-MYB15C, suggesting that MYB15 is phosphorylated by MPK6 at the predicted sites.
Figure 2.
In vitro phosphorylation of MYB15 by recombinant MPK6. Coomassie Brilliant Blue (CBB) staining (A) and autoradiography (B) of an SDS-polyacrylamide gel with resolved kinase reactions containing the indicated combinations of purified Escherichia coli-expressed proteins and [γ-32P] ATP. Abbreviations: MBP, myelin basic protein; GST-MYB15, GST-tagged full-length MYB15; GST-MYB15N, GST-tagged MYB15 N-terminal fragment (amino acids 1–172) and GST-MYB15C, GST-tagged MYB15 C-terminal fragment (amino acids 173–285). MBP and GST were used as positive and negative control substrates, respectively.
Identification of a phosphorylation site in MYB15 by mass spectrometry
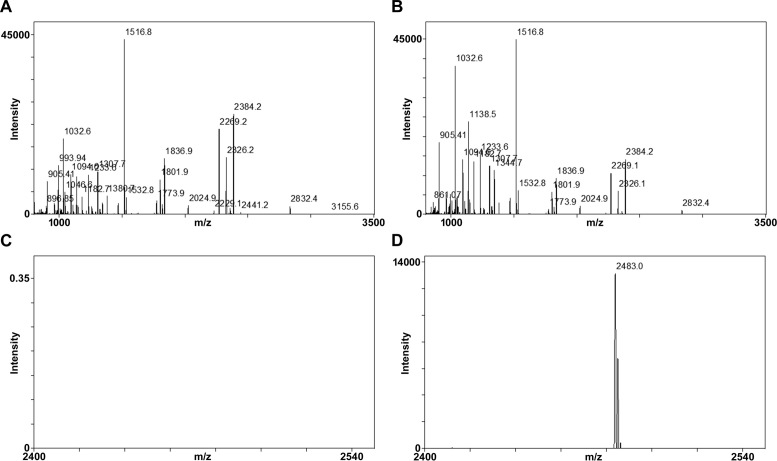
We investigated the MPK6-phosphorylation sites in MYB15 by mass spectrometry analysis after selectively enriching for phosphopeptides by TiO2 chromatography (Figure 3). A kinase reaction was performed with purified GST-MYB15N in the presence and absence of purified His-MPK6. After SDS-PAGE, we excised the bands corresponding to GST-MYB15N, digested them with trypsin and subjected the resulting peptides to MALDI-TOF MS. The mass spectrum of the phosphorylated sample prior to TiO2 chromatography identified the protein as GST and MYB15 with 56 and 34% coverage, respectively (Figure 3B). After TiO2 chromatography, one phosphopeptide was identified, as shown in Figure 3D and Supplementary Table S4. MPKs usually phosphorylate their substrates on Ser or Thr residues that are followed by a proline (Pro, P) (S/T-P motif). The phosphopeptide that was identified contained one putative MPK6 phosphorylation site, Ser168 (SESELADSSNPSGESLFSTS168PSTS). Therefore, Ser168 of MYB15 was identified as a putative MPK6 phosphorylation site.
Figure 3.
Identification of MPK6-phosphorylated peptides in GST-MYB15N using TiO2 chromatography and MALDI-TOF mass spectrometry. (A and B) Shown are the peptide mass fingerprint of unphosphorylated (A) or MPK6-phosphorylated (B) GST-MYB15 N-terminal fragment (amino acids 1–172) after trypsin digestion and without TiO2 purification. A database search of mass data identified GST and MYB15N in the sample with 62 and 38% coverage, respectively. (C and D) Shown are the mass fingerprints of phosphopeptides recovered after TiO2 enrichment from tryptic digests of unphosphorylated (C) or MPK6-phosphorylated (D) GST-MYB15N. One phosphopeptide peak (m/z 2482.9) originating from SESELADSSNPSGESLFSTSPSTS was detected from phosphorylated GST-MYB15N (D) but not from unphosphorylated GST-MYB15N (C).
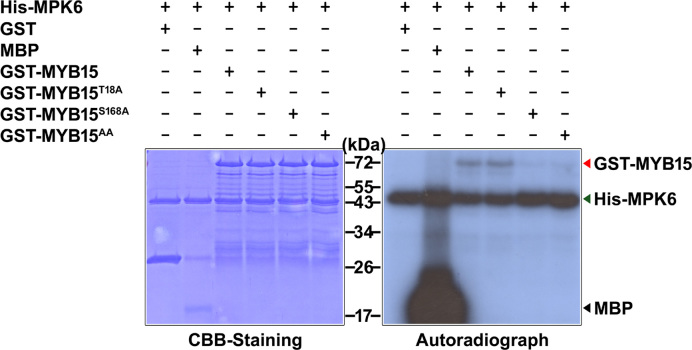
To confirm the phosphorylation site of MYB15, we performed in vitro kinase assays using point mutants of MYB15 as substrates. Specifically, purified GST-MYB15, GST-MYB15T18A, GST-MYB15S168A and GST-MYB15T18A/S168A (GST-MTB15AA) were used as substrates for His-MPK6. As shown in Figure 4, the intensity of the radiolabeled His-MPK6 band, indicating autophosphorylation, was roughly equal in all samples, confirming the presence of sufficient enzyme activity in all samples. Radiolabeled proteins of the expected size of GST-MYB15 (∼72 kDa) were detected in samples containing His-MPK6 along with GST-MYB15 or GST-MYB15T18A, indicating their phosphorylation. However, phosphorylation of GST-MYB15S168A and GST-MTB15AA by His-MPK6 almost disappeared compared to that seen in MYB15 or MYB15T18A proteins, even though faint bands were detected. Since mass spectrometry failed to identify any specific phosphorylation sites other than Ser168, these faint bands may indicate that the mutant proteins MYB15S168A and MTB15AA are non-specifically phosphorylated by MPK6 because of high protein levels in the assays (Figure 3). Thus, Ser168 residue is necessary and sufficient for MPK6-induced phosphorylation of MYB15.
Figure 4.
Identification of MPK6-phosphorylation sites on MYB15 by site-directed mutagenesis. Coomassie Brilliant Blue (CBB) staining and autoradiography of an SDS-polyacrylamide gel with resolved kinase reactions containing the indicated combinations of purified Escherichia coli-expressed proteins and [γ-32P] ATP. The mutant full-length MYB15 proteins MYB15T18A, MYB15S168A and MYB15T18A/S168A (MYB15AA) were generated by site-directed mutagenesis. Myelin basic protein (MBP) and GST were used as positive and negative controls, respectively.
MYB15 is phosphorylated by cold-activated plant MPK6
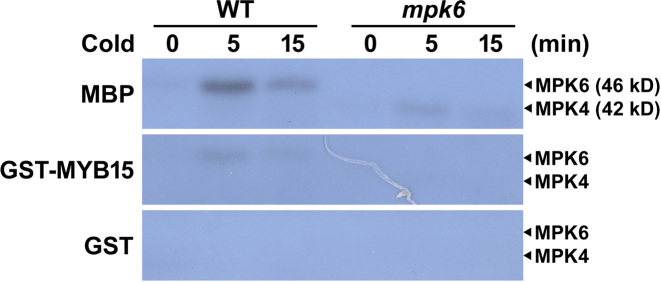
Immunocomplex kinase assays showed that cold stress activates endogenous MPK4 and MPK6 (4). To confirm that endogenous MPKs are activated by cold stress, we performed an in-gel kinase assay using MBP as substrate in cold stress-treated WT, mpk3 and mpk6 seedlings. MPK6 was mainly activated by cold stress (Supplementary Figure S2). To determine whether MYB15 is phosphorylated by native MPK6 and to associate MPK6-MYB15 signaling with cold stress, we performed an in-gel kinase assay using purified GST-MYB15, GST (negative control) and MBP (positive control) embedded in the gels and fractionated cell-free extracts prepared from 3-week-old WT and mpk6 seedlings subjected to cold stress for various time periods (Figure 5). No radiolabeled bands were detected in extracts from unstressed (0 min) WT, or mpk6 mutant leaves in gels embedded with GST-MYB15 (Figure 5). A radiolabeled phospho-MYB15 band at the expected location of MPK6 (∼46 kDa) was detected in the GST-MYB15-embedded gel containing resolved extracts of WT leaves, but not in the gel containing resolved extracts of mpk6 leaves from cold stress-treated plants. The phosphorylation of MYB15 was rapid and transient, as is expected if the phosphorylation requires activated endogenous MPK6. A similar pattern of phosphorylation was observed in the gel embedded with MBP, and no bands were observed in the gel embedded with GST (Figure 5). These observations suggest that MYB15 can be phosphorylated by native MPK6 and that the MPK6-MYB15 pathway participates in cold stress signaling.
Figure 5.
MYB15 is phosphorylated by cold-activated plant MPK6. Leaves of 3-week-old wild-type (WT) and mpk6 plants were subjected to cold (4°C) treatments for the indicated times to induce MPK activity. Shown are autoradiographs of SDS-polyacrylamide gels containing embedded MBP, GST-MYB15 and GST, depicting signals from an in-gel kinase assay on resolved proteins from extracts of treated leaves. The expected positions of MPK4 and MPK6 on the gels are indicated.
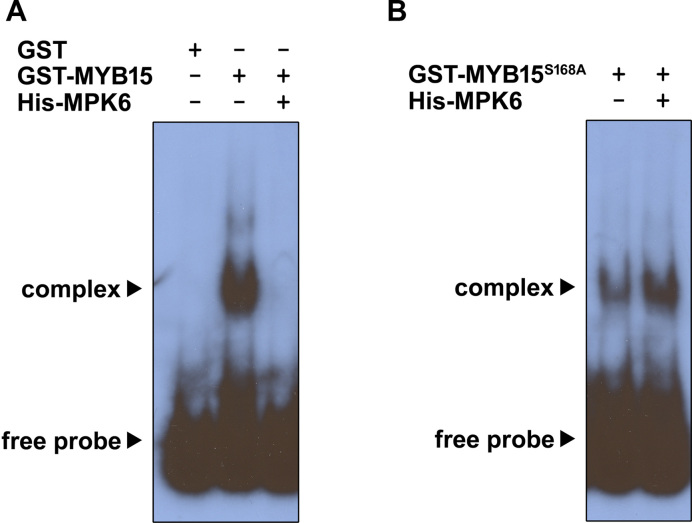
MPK6-induced phosphorylation of MYB15 affects its DNA-binding affinity
MYB15 binds to the promoters of CBFs and represses their transcription (23). Therefore, we hypothesized that the binding affinity of MYB15 to CBF promoters is regulated by MPK6-phosphorylation of MYB15. We tested this hypothesis by performing electrophoretic mobility shift assays (EMSAs) using purified proteins expressed in E. coli, with a radiolabeled CBF3 promoter fragment (−984 to −785 bp region) as a probe. This CBF3 promoter fragment binds strongly to MYB15 and contains two MYB recognition sequences (23). As expected (Figure 6), a shifted band was observed when the probe was incubated with GST-MYB15, but not with GST alone. The intensity of this shifted band decreased when a competitive unlabeled probe was added in a concentration-dependent manner (Supplementary Figure S3), indicating that MYB15 specifically interacts with MYB recognition sequences. No shifted band was detected when GST-MYB15 was incubated with His-MPK6 and ATP prior to the addition of the probe, suggesting that the phosphorylation of MYB15 by MPK6 diminishes its binding affinity (Figure 6A). The shifted band was observed with GST-MYB15S168A and when GST-MYB15S168A was pre-incubated with His-MPK6 and ATP (Figure 6B), suggesting that the loss of binding ability is mediated by phosphorylation of MYB15 by MPK6 at Ser168.
Figure 6.
Binding affinity of MYB15 to the CBF3 promoter fragment is reduced by MPK6-induced phosphorylation. Shown are the results of an EMSA performed using a 32P-labeled CBF3 promoter fragment (−984 to −785 bp region) as a probe. Equal amounts of purified Escherichia coli-expressed GST, GST-MYB15 (A) and GST-MYB15S168A (B) were incubated in the absence or presence of purified His-MPK6 at 30°C for 30 min before adding the probe.
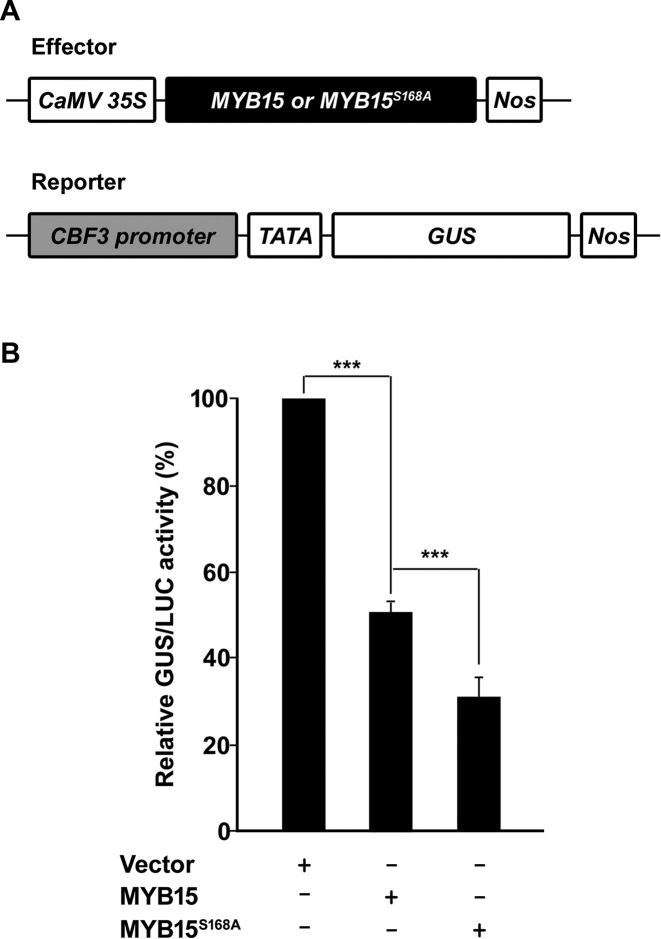
Phosphorylation of MYB15 is necessary for derepression of CBF3 gene expression
To investigate whether the transcriptional activity of MYB15 is regulated by its phosphorylation, we performed transient expression assays with Arabidopsis protoplasts (Figure 7) using the effector plasmids and reporter plasmids shown in Figure 7A. The relative GUS activity level was reduced (50.75 ± 2.84%) in protoplasts co-transfected with CaMV35S: MYB15 and reporter plasmid compared with protoplasts co-transfected with effector vector and reporter plasmid (100%); this was expected because MYB15 functions as a transcriptional repressor (Figure 7B). Interestingly, relative GUS activity was even lower (31.17 ± 6.55%) in protoplasts co-transfected with CaMV35S: MYB15S168A and reporter plasmid (Figure 7B). Protoplasts are more sensitive to subtle external stresses compared with normal plant cells; therefore, even in the absence of applied stress, MPKs can be somewhat activated in protoplasts (38). In fact, the activity of MPK6 extracted from protoplasts was increased compared with that of MPK6 from plant seedlings (0 min) (Supplementary Figure S4). Therefore, GUS activity was partly recovered in protoplasts transfected with MYB15 because some MYB15 proteins were phosphorylated by MPK6 and released from the promoter. In contrast, MYB15S168A proteins are unresponsive to MPK6 and are not released from the promoter at all. These results support that transcriptional activation of CBF3 is positively regulated by MPK6-induced phosphorylation of MYB15 at Ser168.
Figure 7.
Transcriptional repression activity of MYB15 is enhanced by a mutation in its phosphorylation site. (A) Schematic representation of reporter and effector constructs used in the transient expression assay. The effector construct consisted of the CaMV35S promoter fused to the full-length MYB15 and MYB15S168A ORFs. The CBF3 promoter (−2000 to −1 bp region) was fused to the GUS reporter gene containing a minimal CaMV35S promoter to generate the reporter construct. (B) Transient assay of CBF3 expression in Arabidopsis protoplasts. The GUS//LUC activities of Arabidopsis protoplasts that were co-transfected with the CBF3: GUS reporter plasmid, a CaMV35S: LUC internal control plasmid and the indicated effector plasmids were measured. Shown are relative GUS/LUC activities, which were calculated as a percentage of the GUS/LUC activities of samples transformed with the reporter construct plus empty effector vector. Data are presented as the mean ± SD of three independent experiments. P < 0.001 (***) indicate statistically significant changes.
Phosphorylation of MYB15 is required for the upregulation of CBF genes
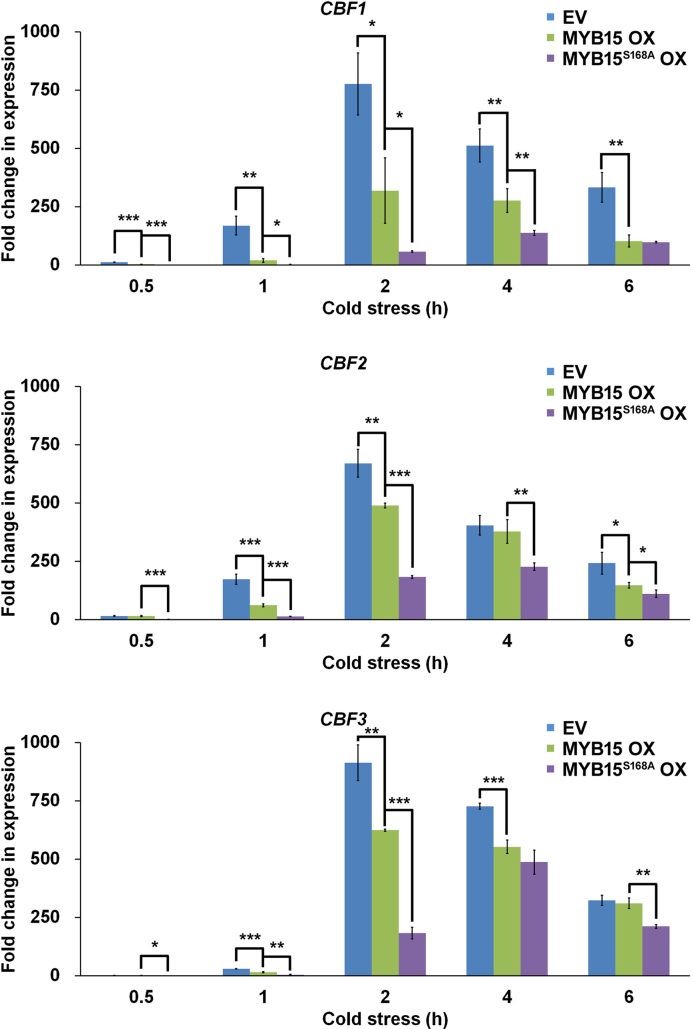
To examine the importance of an intact MPK6-phosphorylation site for the regulation of CBF genes and freezing tolerance by MYB15, we generated transgenic CaMV35S:3XFlag-MYB15 (MYB15 OX) and CaMV35S:3XFlag-MYB15S168A (MYB15S168A OX) lines via Agrobacterium-mediated transformation. We selected three independent MYB15 OX and MYB15S168A OX lines based on high levels of MYB15 expression (Supplementary Figure S5). We investigated the expression of CBF in each of these three lines subjected to cold stress for 3 h. Transcription of CBF decreased when MYB15 was overexpressed, and even more so when MYB15S168A was expressed (Supplementary Figure S6). Each of these three independent lines showed similar expression patterns and CBF levels. Therefore, we mixed seeds from these lines together and used these for further studies.
First, we compared the expression levels of CBF genes in seedlings harboring empty vector, MYB15 OX and MYB15S168A OX subjected to cold stress for various time periods (Figure 8). As expected (Agarwal et al., (23), the levels of CBF1, CBF2 and CBF3 transcripts increased upon cold treatment in all lines. The accumulation of all CBF transcripts peaked at 2 h of treatment in empty vector control and MYB15 OX plants but at 4 h in MYB15S168A OX. At 2 h of treatment, the transcript levels of CBF1, CBF2 and CBF3 were significantly lower in MYB15 OX seedlings than in the empty vector control and were lowest in MYB15S168A OX. The differences became less pronounced with increasing duration of cold treatment.
Figure 8.
Mutation of the MYB15 phosphorylation site increases its transcriptional repression of CBF genes in cold-stressed transgenic plants. CBF expression was measured in transgenic plants expressing empty vector (EV), MYB15 and MYB15S168A under cold stress. Quantitative RT-PCR analysis of CBF genes was performed using total RNA isolated from seedlings of the indicated lines subjected to cold (4°C) treatment for the indicated times. The cDNA was synthesized from 5 μg of total RNA and used as a template for RT-PCR. Data are presented as the mean ± SD of three independent experiments. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) indicate statistically significant changes.
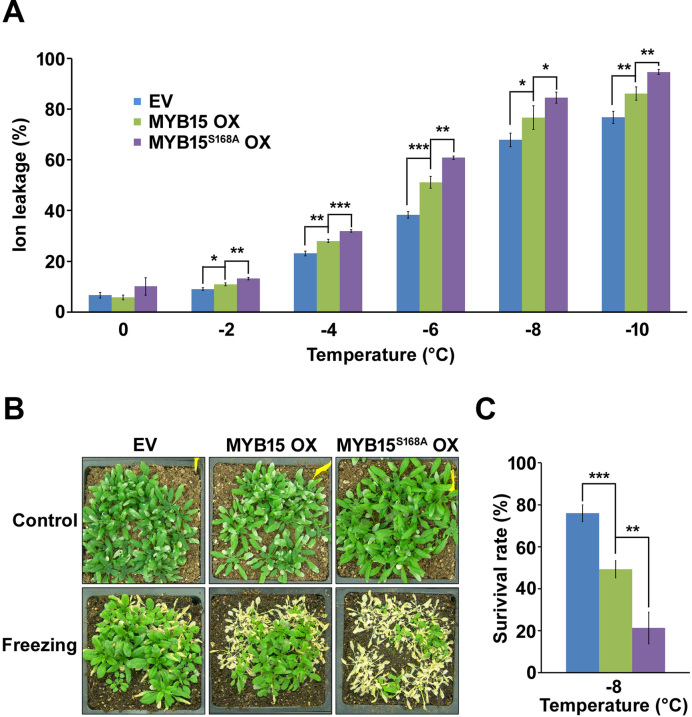
We then conducted an ion leakage test to evaluate the effect of MYB15 phosphorylation on freezing tolerance. Overexpression of MYB15 increased ion leakage in non-acclimated plants compared to the control, as previously reported (23), indicating increased freezing sensitivity (Figure 9A). The LT50 (the temperature of 50% ion leakage) values for empty vector control and MYB15 OX plants were −6.7°C and −5.7°C, respectively. The LT50 value for MYB15S168A OX plants was −5.4°C, that is, lower than that of MYB15 OX plants, indicating that these plants had the greatest freezing sensitivity among the lines tested. We also evaluated the role of MYB15 phosphorylation by performing a whole-plant freezing-survival test on 3-week-old soil-grown plants. As shown in Figure 9B and 9C, the lowest amount of tissue death was observed in leaves of empty-vector control plants and MYB15 OX plants were more sensitive to freezing than empty-vector plants but more tolerant than MYB15S168A OX plants. Therefore, the loss of the MPK6-phosphorylation site on MYB15 is associated with reduced freezing tolerance.
Figure 9.
Freezing tolerance is reduced by a mutation of MYB15 that prevents phosphorylation. (A) Ion-leakage test without cold acclimation. Empty vector (EV), MYB15 OX and MYB15S168A OX transgenic plants were used. The ion leakage experiment was repeated twice with three replicates per experiment. Shown are results from one representative experiment. Data are presented as the mean ± SD of three independent experiments. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) indicate statistically significant changes. (B and C) Freezing survival assay without acclimation. Three-week-old plants grown in soil at 22°C were incubated at 4°C for 1 h, followed by freezing treatment for 2 h at −8°C. After the freezing treatment, plants were incubated at 4°C for 1 day and returned to normal growing conditions for 5 days before photography (B) and survival rate measurements (C). Data are presented as the mean ± SD of three independent experiments. P < 0.01 (**) and P < 0.001 (***) indicate statistically significant changes.
DISCUSSION
MPK6 phosphorylates MYB15
MPK cascades are universal and highly conserved signal transduction modules that are present in all eukaryotes, including plants (10,39). These protein phosphorylation cascades link extracellular stimuli to a wide range of cellular responses. However, to date, only a limited number of MPK substrates have been found in plants. In this study, we provide experimental evidence that MYB15 is a substrate of MPK6. First, MYB15 specifically and directly interacted with MPK6 in vitro and in planta. Second, MYB15 was phosphorylated by recombinant MPK6 and cold-activated plant MPK6, as revealed by in vitro and in-gel kinase assays, respectively. Third, the site of MYB15 phosphorylation by MPK6 was identified by mass spectrometry and confirmed by site-directed mutagenesis. MYB15 is a member of the R2R3 family of MYB transcription factors, which control plant development, secondary metabolite biosynthesis and responses to environmental stresses (40,41). The activities of plant MYBs are regulated by phosphorylation. For example, NtMYBA2 is activated by the phosphorylation of a cyclin–CDK complex through the repression of its inhibitory domain (42,43). The DNA-binding ability of MYB41 is enhanced by the phosphorylation of MPK6, which is required for its physiological function in salt-stress tolerance (44). The phosphorylation of MYB44 by MPK3/MPK6 is required for the inhibition of seed germination (45). Also, phosphorylation is required for the activity of ZAT10, a positive regulator of osmotic stress tolerance in Arabidopsis (46). In addition, high-throughput protein microarray analysis in Arabidopsis revealed that many MYB proteins are potential targets of MPKs (20). In animal cells, murine c-Myb is phosphorylated and regulated by another MPK, p42mpk (47). The activity of B-Myb is also altered by the phosphorylation of the cyclin-dependent kinase (CDK) complex (48–51). Thus, the phosphorylation of MYB proteins by protein kinases appears to be an evolutionarily conserved regulatory mechanism.
The MPK cascade triggered by cold stress comprises MEKK1, MKK2, MPK4 and MPK6. Although MPK4 and MPK6 are activated by cold stress downstream of MEKK1–MKK2, we did not find experimental evidence for the direct regulation of MYB15 by MPK4, suggesting that MYB15 may not be a target of MPK4 during cold stress signaling. Therefore, MPK4 may be involved in cold stress signaling through a different pathway or a different substrate, which remains to be investigated.
DNA binding of MYB15 is reduced by its phosphorylation
The transcript and protein levels of plant MYB transcription factors are regulated by microRNAs and ubiquitin-mediated degradation, respectively (52). The activities of plant MYB proteins are thought to be governed by protein–protein interactions, redox control and phosphorylation. The phosphorylation of transcription factors can alter their subcellular location, stability, DNA-binding affinity and interactions with other regulatory proteins (53). As shown in Figures 6–8, the phosphorylation of MYB15 reduces its DNA-binding affinity to the promoters of CBF genes. In addition, a mutation in its phosphorylation site, MYB15S168A, disrupted its dissociation from the CBF3 promoter and the induction of cold-responsive genes such as CBF1, CBF2 and CBF3. Similarly, the binding of AmMYB340 to target DNA is inhibited by phosphorylation in Antirrhinum majus (54). Conversely, the transcriptional activities of PtMYB4 and AtMYB46 are positively regulated by phosphorylation in Pinus taeda and Arabidopsis thaliana, respectively (55). Ser168, the phosphorylation site of MYB15, is distantly located from an R2R3 DNA-binding domain corresponding to 11–116 amino acid residues. Therefore, it is unlikely that MYB15 phosphorylation directly changes the DNA binding ability of the R2R3 DNA-binding domain. Instead, it is more likely that the phosphorylation of MYB15 by MPKs is necessary for its conformational change, which induces its release from the promoters of cold-responsive genes. It is also possible that changes in hydrogen bonding capacity and electrostatic interactions caused by MYB15 phosphorylation affect the binding affinity between MYB15 and DNA. Therefore, structural comparisons of non-phosphorylated versus phosphorylated MYB15 would help clarify the molecular mechanism by which protein phosphorylation modulates MYB15 DNA-binding affinity.
Phosphorylation of MYB15 by MPK is required for freezing tolerance
Cold-mediated signaling during the freezing tolerance response requires fine-tuned transcriptional regulation. Cold temperatures trigger the transcription of CBF genes encoding transcription factors, which in turn activate the transcription of genes containing the DRE/CRT promoter element (1,56). The transcription of CBF genes is tightly controlled; uncontrolled expression of CBF genes may adversely affect growth and development (34,57,58). CBF genes are positively and negatively regulated by the transcription factors ICE and MYB15, respectively. ICE1 is a MYC-like bHLH transcription factor that binds to MYC elements of CBF gene promoters and activates their expression (3). Under normal conditions, ICE1 is ubiquitinated by the E3 ligase HOS1 and degraded by the 26S-proteasome pathway, whereas under cold conditions, ICE1 is stabilized by SIZ1-mediated sumoylation (59,60). MYB15 binds to MYB elements in the promoters of CBFs and negatively regulates their expression (23). However, the way(s) in which bound MYB15 is released from CBF promoters in response to cold stress has not yet been investigated.
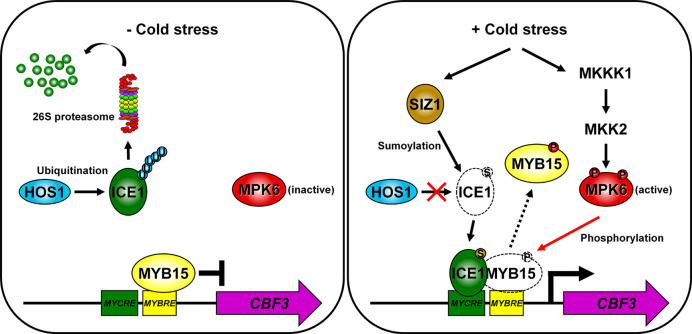
In the current study, we demonstrated that the DNA-binding ability of MYB15 is reduced by the phosphorylation activity of a cold-responsive MPK, releasing its repression of gene transcription. In our proposed model (Figure 10), under normal conditions, CBF transcription is repressed by the DNA-binding of unphosphorylated MYB15 to MYB recognition elements in their promoters and the absence of ICE1 activity. In response to cold stress, MYB15 is phosphorylated by cold-activated MPK6. The phosphorylated MYB15 dissociates from CBF promoters because of its reduced DNA-binding affinity. ICE1 that has been stabilized by SIZ1-mediated sumoylation binds to the MYC recognition elements in the CBF promoters and activates the expression of CBF genes, thereby conferring freezing tolerance to the plant. This suggests that release of MYB15 from the promoter is required for the activation of ICE1-mediated CBF genes because overexpression of MYB15S168A inhibits cold activation of CBF (Figure 8). It also suggests that MYB15S168A binds DNA in the presence of activated MPK6 (Figure 6B). To further elucidate the transcriptional regulation of CBF genes during the cold response, however, detailed studies should be performed on the release of MYB15 and the binding of ICE1 to the promoters of these genes.
Figure 10.
Model of the role of MYB15 in cold stress signaling. Under normal conditions, ICE1 is ubiquitinated by HOS1 and degraded by the 26S-proteasome pathway, whereas MYB15 binds to an MYBRE on the promoters of CBF genes to inhibit their transcription. In response to cold stress, MYB15 is phosphorylated by cold-activated MPK6 and released from the CBF gene promoters. ICE1 is stabilized by SIZ1-mediated sumoylation and sumoylated ICE1 binds to an MYCRE on the CBF promoters, leading to their increased expression and increased freezing tolerance. CBFs, C-repeat binding factors (an AP2-type transcription factors); MYBRE, MYB transcription factor recognition element; MYCRE, MYC transcription factor recognition element; P, phosphorylation; S, sumoylation; U, ubiquitination.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Dae-Jin Yun for critical comments of the article, and Dr Yong-Hwan Moon for helpful discussions and critique of the article.
Author contributions: S.H.K. and W.S.C. designed, planned and organized the experiments. S.H.K., S.B., and J.A. generated all Arabidopsis transgenic lines and all plant materials used in this study. S.H.K., H.S.K., S.B., J.A. and Y.Y. performed the research. S.H.K., J.Y.K. and W.S.C. analyzed the data. S.H.K. and W.S.C. wrote the article.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Next-Generation BioGreen 21 Program by the Rural Development Administration [#PJ01109101]; Basic Science Research Program through the National Research Foundation of Korea (NRF) by the Ministry of Education [2015R1D1A4A01020200]; National Research Foundation of Korea (NRF) grant by the Korean government (MSIP) [2016R1A2B4015859]. Funding for open access charge: Next-Generation BioGreen 21 Program by the Rural Development Administration [#PJ01109101].
Conflict of interest statement. None declared.
REFERENCES
- 1. Thomashow M.F. PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999; 50:571–599. [DOI] [PubMed] [Google Scholar]
- 2. Shinozaki K., Yamaguchi-Shinozaki K., Seki M.. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003; 6:410–417. [DOI] [PubMed] [Google Scholar]
- 3. Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K.. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003; 17:1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Teige M., Scheikl E., Eulgem T., Doczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H.. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 2004; 15:141–152. [DOI] [PubMed] [Google Scholar]
- 5. Stockinger E.J., Gilmour S.J., Thomashow M.F.. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Q., Sakuma Y., Abe H., Kasuga M., Miura S., Yamaguchi-Shinozaki K., Shinozaki K.. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998; 10:1391–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang S., Klessig D.F.. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001; 6:520–527. [DOI] [PubMed] [Google Scholar]
- 8. Jonak C., Ökrész L., Bögre L., Hirt H.. Complexity, crosstalk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002; 5:415–424. [DOI] [PubMed] [Google Scholar]
- 9. Tena G., Boudsocq M., Sheen J.. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 2011; 14:519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colcombet J., Hirt H.. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 2008; 413:217–226. [DOI] [PubMed] [Google Scholar]
- 11. Group MAPK. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002; 7:301–308. [DOI] [PubMed] [Google Scholar]
- 12. Ichimura K., Mizoguchi T., Yoshida R., Yuasa T., Shinozaki K.. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000; 24:655–665. [DOI] [PubMed] [Google Scholar]
- 13. Asai T., Tena G., Plotnikova J., Willman M.R., Chiu W.L., Gomez-Gomez L., Boller T., Asubel F.M., Sheen J.. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature. 2002; 415:977–983. [DOI] [PubMed] [Google Scholar]
- 14. Mishra N.S., Tuteja R., Tuteja N.. Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 2006; 452:55–68. [DOI] [PubMed] [Google Scholar]
- 15. Droillard M.J., Boudsoq M., Barbier-Brygoo H., Lauriere C.. Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett. 2002; 527:43–50. [DOI] [PubMed] [Google Scholar]
- 16. Gudesblat G.E., Iusem N.D., Morris P.C.. Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol. 2007; 173:713–721. [DOI] [PubMed] [Google Scholar]
- 17. Xing Y., Jia W., Zhang J.. AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J. Exp. Bot. 2007; 58:2969–2981. [DOI] [PubMed] [Google Scholar]
- 18. Link V., Sinha A.K., Vashista P., Hofmann M.G., Proels R.K., Ehness R., Roitsch T.. A heat-activated MAP kinase in tomato: a possible regulator of the heat stress response. FEBS Lett. 2002; 531:179–183. [DOI] [PubMed] [Google Scholar]
- 19. Kersten B., Agrawal G.K., Durek P., Neigenfind J., Schulze W., Walther D., Rakwal R.. Plant phosphoproteomics: an update. Proteomics. 2009; 9:964–988. [DOI] [PubMed] [Google Scholar]
- 20. Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P.. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009; 23:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Y., Zhang S.. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004; 16:3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nguyen X.C., Kim S.H., Lee K., Kim K.E., Liu X.M., Han H.J., Hoang M.H., Lee S.W., Hong J.C., Moon Y.H., Chung W.S.. Identification of a C2H2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Rep. 2012; 31:737–745. [DOI] [PubMed] [Google Scholar]
- 23. Agarwal M., Hao Y., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.K.. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006; 281:37636–37645. [DOI] [PubMed] [Google Scholar]
- 24. Jiang Y., Deyholos M.K.. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006; 6:6–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murashige T., Skoog F.. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 1962; 15:473–497. [Google Scholar]
- 26. Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M.. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008; 146:368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koncz C., Schell J.. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986; 204:383–396. [Google Scholar]
- 28. Bundock P., den Dulk-Ras A., Beijersbergen A., Hooykaas P.J.. Transkingdom T-DNA transfer from Agrobacterium tume faciens to Saccharomyces cerevisiae. EMBO J. 1995; 14:3206–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee K., Kye M., Jang J.S., Lee O.J., Kim T., Lim D.. Proteomic analysis revealed a strong association of a high level of alpha1-antitrypsin in gastric juice with gastric cancer. Proteomics. 2004; 4:3343–3352. [DOI] [PubMed] [Google Scholar]
- 30. Zhang S., Klessig D.F.. Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell. 1997; 9:809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D., Dean C., Ma H., Martienssen R.. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995; 9:1797–17810. [DOI] [PubMed] [Google Scholar]
- 32. Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science. 1996; 274:1900–1902. [DOI] [PubMed] [Google Scholar]
- 33. Kim H.S., Park B.O., Yoo J.H., Jung M.S., Lee S.M., Han H.J., Kim K.E., Kim S.H., Lim C.O., Yun D.J. et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 2007; 282:36292–36302. [DOI] [PubMed] [Google Scholar]
- 34. Raghothama K.G., Maggio A., Narasimhan M.L., Kononowicz A.K., Wang G., D’Urzo M.P., Hasegawa P.M., Bressan R.A.. Tissue-specific activation of the osmotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol. Biol. 1997; 32:393–404. [DOI] [PubMed] [Google Scholar]
- 35. Clough S.J., Bent A.F.. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 36. Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K.. Current Protocols in Molecular Biology. 1989; NY: Greene PublishingAssociates/Wiley Interscience. [Google Scholar]
- 37. Ishitani M., Xiong L., Le H., Stevenso B., Zhu J.K.. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell. 1998; 10:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mészáros T., Helfer A., Hatzimasoura E., Magyar Z., Serazetdinova L., Rios G., Bardóczy V., Teige M., Koncz C., Peck S. et al. The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 2006; 48:485–498. [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez M.C., Petersen M., Mundy J.. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2010; 61:621–649. [DOI] [PubMed] [Google Scholar]
- 40. Jin H., Martin C.. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999; 41:577–585. [DOI] [PubMed] [Google Scholar]
- 41. Stracke R., Werber M., Weisshaar B.. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001; 4:447–456. [DOI] [PubMed] [Google Scholar]
- 42. Araki S., Ito M., Soyano T., Nishihama R., Machida Y.. Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J. Biol. Chem. 2004; 279:32979–32988. [DOI] [PubMed] [Google Scholar]
- 43. Haga N., Kato K., Murase M., Araki S., Kubo M., Demura T., Suzuki K., Müller I., Voss U., Jürgens G. et al. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development. 2007; 134:1101–1110. [DOI] [PubMed] [Google Scholar]
- 44. Hoang M.H., Nguyen X.C., Lee K., Kwon Y.S., Pham H.T., Park H.C., Yun D.J., Lim C.O., Chung W.S.. Phosphorylation by AtMPK6 is required for the biological function of AtMYB41 in Arabidopsis. Biochem. Biophys. Res. Commun. 2012; 422:181–186. [DOI] [PubMed] [Google Scholar]
- 45. Nguyen X.C., Hoang M.H., Kim H.S., Lee K., Liu X.M., Kim S.H., Bahk S., Park H.C., Chung W.S.. Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochem. Biophys. Res. Commun. 2012; 423:703–708. [DOI] [PubMed] [Google Scholar]
- 46. Nguyen X.C., Kim S.H., Hussain S., An J., Yoo Y., Han H.J., Yoo J.S., Lim C.O., Yun D.J., Chung W.S.. A positive transcription factor in osmotic stress tolerance, ZAT10, is regulated by MAP kinases in Arabidopsis. J. Plant Biol. 2016; 59:55–61. [Google Scholar]
- 47. Miglarese M.R., Richardson A.F., Aziz N., Bender T.P.. Differential regulation of c-myb-induced transcription activation by a phosphorylation site in the negative regulatory domain. J. Biol. Chem. 1996; 271:22697–22705. [DOI] [PubMed] [Google Scholar]
- 48. Bessa M., Saville M.K., Watson R.J.. Inhibition of cyclin A/Cdk2 phosphorylation impairs B-Myb transactivation function without affecting interactions with DNA or the CBP coactivator. Oncogene. 2001; 20:3376–3386. [DOI] [PubMed] [Google Scholar]
- 49. Johnson L.R., Johnson T.K., Desler M., Luster T.A., Nowling T., Lewis R.E., Rizzino A.. Effects of B-Myb on gene ttranscription: PHSOPHORYLATION-DEPENDENT ACTIVITY AND ACETYLATION BY p300. J. Biol. Chem. 2002; 277:4088–4097. [DOI] [PubMed] [Google Scholar]
- 50. Li X., McDonnell D.P.. The transcription factor B-myb is maintained in an inhibited state in target cells through its interaction with the nuclear corepressors N-CoR and SMRT. Mol. Cell. Biol. 2002; 22:3663–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schubert S., Horstmann S., Bartusel T., Klempnauer K.H.. The cooperation of B-Myb with the coactivator p300 is orchestrated by cyclins A and D1. Oncogene. 2004; 23:1392–1404. [DOI] [PubMed] [Google Scholar]
- 52. Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L.. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010; 15:573–581. [DOI] [PubMed] [Google Scholar]
- 53. Yang S.H., Sharrocks A.D., Whitmarsh A.J.. Transcriptional regulation by the MAP kinase signaling cascades. Gene. 2003; 320:3–21. [DOI] [PubMed] [Google Scholar]
- 54. Moyano E., Martínez-Garcia J.F., Martin C.. Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in antirrhinum flowers. Plant Cell. 1996; 8:1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morse A.M., Whetten R.W., Dubos C., Campbell M.M.. Post-translational modification of an R2R3-MYB transcription factor by a MAP Kinase during xylem development. New Phytol. 2009; 183:1001–1013. [DOI] [PubMed] [Google Scholar]
- 56. Fowler S., Thomashow M.F.. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002; 14:1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.K.. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001; 15:912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gilmour S.J., Fowler S.G., Thomashow M.F.. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004; 54:767–781. [DOI] [PubMed] [Google Scholar]
- 59. Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K.. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M.. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007; 19:1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.