Abstract
Regulation of gene expression at the level of cytoplasmic polyadenylation is important for many biological phenomena including cell cycle progression, mitochondrial respiration, and learning and memory. GLD4 is one of the non-canonical poly(A) polymerases that regulates cytoplasmic polyadenylation-induced translation, but its target mRNAs and role in cellular physiology is not well known. To assess the full panoply of mRNAs whose polyadenylation is controlled by GLD4, we performed an unbiased whole genome-wide screen using poy(U) chromatography and thermal elution. We identified hundreds of mRNAs regulated by GLD4, several of which are involved in carbohydrate metabolism including GLUT1, a major glucose transporter. Depletion of GLD4 not only reduced GLUT1 poly(A) tail length, but also GLUT1 protein. GLD4-mediated translational control of GLUT1 mRNA is dependent of an RNA binding protein, CPEB1, and its binding elements in the 3΄ UTR. Through regulating GLUT1 level, GLD4 affects glucose uptake into cells and lactate levels. Moreover, GLD4 depletion impairs glucose deprivation-induced GLUT1 up-regulation. In addition, we found that GLD4 affects glucose-dependent cellular phenotypes such as migration and invasion in glioblastoma cells. Our observations delineate a novel post-transcriptional regulatory network involving carbohydrate metabolism and glucose homeostasis mediated by GLD4.
INTRODUCTION
Dynamic and bidirectional regulation of poly(A) tail length in the cytoplasm often regulates mRNA stability and translation. In addition to canonical nuclear poly(A) polymerase (PAP), seven non-canonical PAPs catalyze the addition of polynucleotides (adenosine or uridine): PAPD1 (mitochondrial PAP), RBM21 (Star-PAP/PAPD2/TUT6), ZCCHC6 (TUT7), ZCCHC11 (TUT4), GLD2 (germline development 2, PAPD4/TUT2), GLD4 (PAPD5/TUT3/TRF4-2) and POLS (PAPD7/TUT5) (1). Some of these PAPs have specific subcellular localizations; for example, PAPD1 is predominantly mitochondrial (2) whereas RBM21 is found in nuclear speckles (3). Other PAPs such as ZCCHC6 and ZCCHC11 mainly reside in the cytoplasm where they catalyze terminal uridylation induced-mRNA degradation (4).
GLD2 is the most intensively studied member of this family, and is linked to multiple biological pathways in worms, flies, and mice (5–8); it lacks classical RNA binding motifs and requires association with RNA binding proteins to promote polyadenylation (9). GLD2 is bound to CPEB1 (cytoplasmic polyadenylation element binding protein 1), which associates with 3΄ UTR cytoplasmic polyadenylation elements (CPEs). To form the cytoplasmic polyadenylation ribonucleoprotein (RNP) complex, CPEB1 nucleates factors on mRNA such as GLD2 and PARN (poly(A) specific ribonuclease) (10). Polyadenylation is induced by signal-dependent phosphorylation of CPEB1, which results in the dissociation of PARN, thereby allowing for GLD2 to catalyze poly(A) addition (11) and resulting translational activation (11–13).
GLD4 and POLS are human homologues of yeast Trf4/Trf5, which are involved in quality control of RNA through polyadenylation and exosome-mediated degradation (14,15). Although the function of POLS is unknown, GLD4 has a role in processing rRNA precursors (16) and snoRNAs (17). It also regulates histone mRNA degradation in the cytoplasm (18), although another study observed no such effect (19). The C-terminus of GLD4 contains several basic amino acids that promote RNA binding indicating that GLD4 is active without an RNA binding protein cofactor (20). PAR-CLIP (Photoactivatable Ribonucleoside Enhanced Crosslinking and Immunoprecipitation) analysis of ectopically-expressed GLD4 identified rRNAs, but not mRNAs, as its main targets (20), suggesting cofactors may be required for binding to mRNAs. Indeed, GLD4 interacts with P53 mRNA in a CPEB1-dependent manner, and depletion of GLD4 or CPEB1 reduces P53 mRNA polyadenylation-induced translation and consequent bypass of cellular senescence (21,22).
Although the nuclear function of GLD4 has been explored (16,17), its role in the cytoplasm is largely unknown. The fly ortholog Trf4-1 is involved in cytoplasmic oligoadenylation-mediated exosomal mRNA degradation in Drosophila cells (23). In Caenorhabditis elegans, GLD4 (worm ortholog of human GLD4/PAPD5 and POLS/PAPD7) localizes in the cytoplasm where it forms a complex with gls-1 to promote translation of glp-1 mRNA and maintains germ cell proliferation (24). The molecular function of the mammalian orthologs of gls-1, ZCCHC14 and ZCCHC2, is unknown. A genome-wide polysome profiling analysis of gld-4(RNAi) animals revealed that GLD4 only mildly changes bulk mRNA poly(A) tail extension, but that it may actively promote general translational efficiency in C. elegans (25). With the exception of P53, molecular targets of GLD4-mediated cytoplasmic polyadenylation as well as the biological pathways GLD4 controls are largely unidentified in vertebrates. Based on previous studies (22), we hypothesized that GLD4 regulates poly(A) tail length of several mRNAs, and consequently sought to identify them in human cells as well as the cellular pathways they may control. In our analysis, we found hundreds of GLD4-regulated mRNAs including several in the carbohydrate metabolism pathway. One of these is the glucose transporter GLUT1 (SLC2A1), which we show is crucial for cellular glucose uptake and affects cellular migration. Moreover, we found that GLD4 regulates glucose deprivation-mediated GLUT1 expression. These observations suggest that GLD4 regulates glucose metabolism and homeostasis via regulating gene expression at the level of cytoplasmic polyadenylation-mediated translation.
MATERIALS AND METHODS
Cell culture
Human foreskin fibroblasts (Yale Skin Disease Research Center), U87MG human glioblastoma cells (a Gift of Dr Alonzo Ross, University of Massachusetts Medical School) and HEK293T human embryonic kidney cells (ATCC) were cultured in DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic compounds.
Thermal elution and microarray analysis
Total RNA from human foreskin fibroblasts was extracted with TRIzol reagent (Invitrogen). Polyadenylated RNAs were bound to poly(U) agarose beads (Sigma), washed at 50°C, and eluted at 65°C (8). Total RNA (input) as well as RNA from the thermally eluted fraction were precipitated and contaminants removed by an RNeasy MinElute purification kit (Qiagen). The purified RNAs were used as templates to synthesize biotin-labeled cRNAs that were annealed to the human HT-12 Expression BeadChip array (Illumina), which was performed at the Genomics core facility of the Pennsylvania State University. Differentially expressed RNAs were analyzed with Genespring Suite (Agilent Technologies) using unpaired asymptotic T-test (P-value < 0.05) and were deposited in Gene Expressing Omnibus (accession number: GSE77747).
PCR primers
All oligonucleotide sequences for PCR are provided in the Supplementary Table S1.
Lentivirus
shRNAs against GLD4 and GLUT1 were cloned into pLL3.7 (syn) lentivirus vector in which the CMV promoter is replaced with a synapsin promoter (8). Lentiviral shRNAs targeting GLD2 and CPEB1 are described elsewhere (8). Targeting sequences for shGLD4 and shGLUT1 are provided in the Supplementary Table S1.
Wound healing assay
Twenty thousand U87MG glioblastoma cells were plated in each well of a six-well plate, and were infected lentiviruses expressing shRNA after 1 day. After 3 days, the cultured cells were scratched with a 200 ul pipette tip and cell migration was first assessed and pictured after 12 hours (h), and then every 6 h thereafter. The wound area was measured using TScratch, a software tool to analyze wound healing assays developed by the CSElab. The details of the assay are in Supplementary Experimental Procedure.
Statistical analyses
Statistical analyses were performed with Student's t-test as indicated in the figure legends.
RESULTS
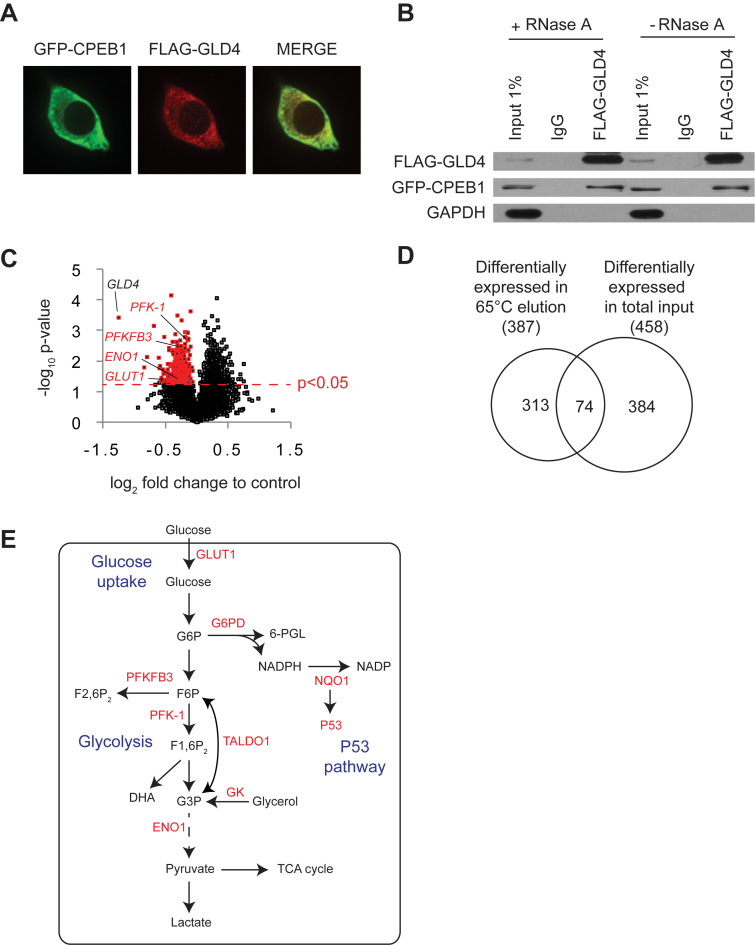
Some studies suggest that GLD4 is a component of the nuclear exosome complex (16,20,26); others, however, indicate that it has cytoplasmic functions (18,22–25). After unsuccessful attempts to detect endogenous GLD4 and CPEB1 by western blotting, we ectopically expressed tagged version of these proteins to determine their cellular localization. FLAG-GLD4 was nearly exclusively cytoplasmic as was GFP-CPEB1 (Figure 1A), which is a known cytoplasmic RNA-binding protein. Moreover, GLD4 was co-immunoprecipitated with CPEB1, but not the negative control GAPDH, in the presence of RNase A (Figure 1B) indicating GLD4 interacts with a cytoplasmic polyadenylation factor.
Figure 1.
Unbiased genome-wide screen identifies GLD4-regulated mRNAs. (A) Analysis of GLD4 protein localization. HEK 293T cells were transfected with GFP-CPEB1 and FLAG-GLD4, and immunostained with GFP (green) and FLAG (red) antibodies. (B) The same cells as in (A) were used for co-immunoprecipitation. FLAG-GLD4 was immunoprecipitated with IgG or FLAG antibody in the presence or absence of RNase A, and immunoblotted for FLAG, GFP and GAPDH. (C) Venn-diagram showing overlap of 74 mRNAs between the 387 that were differentially expressed in 65°C thermal elution fraction and the 458 that were differentially expressed in the total input fraction. (D) RNA from siNT or siGLD4-treated human primary foreskin fibroblasts was fractionated on poly(U) agarose, thermally eluted at 65°C and then subjected to microarray analysis. All 14666 detected RNAs were plotted as log2 fold change (abscissa) versus –log10P-value (ordinate) in GLD4 knockdown cells compared to control cells. 387 down-regulated RNAs in the 65°C fraction with P-value <0.05 are highlighted in red (see Supplementary Table S2). Four GLD4 target mRNAs (GLUT1, PFKFB3, PFK-1 and ENO1) are also labeled. (E) A schematic diagram showing glucose uptake, glycolysis and P53 pathways and GLD4 regulated RNAs (labeled in red; see main text for full name of the enzymes). G6P, glucose-6-phosphate; 6-PGL, 6-phosphoglconolactone; NADP, nicotinamide adenine dinucleotide phosphate; F6P, fructose-6-phosphate; F2,6P2, fructose-2,6-biphosphate; F1,6P2, fructose-1,6-biphosphate; G3P, glyceraldehyde-3-phosphate.
In human primary foreskin fibroblasts, GLD4 promotes polyadenylation and translation of P53 mRNA (22). However, other GLD4 target mRNAs are largely unknown. To identify mRNAs whose polyadenylation is controlled by GLD4, we employed poly(U) agarose chromatography, a procedure in which RNA bound to poly(U) beads is washed at 50°C and then collected at 65°C. Generally, mRNAs with relatively short poly(A) tails (∼50 nucleotides) elute at 50°C whereas RNAs with longer tails predominantly elute at a higher temperature (Supplementary Figure S1A). Primary human fibroblasts were transfected with non-targeting siRNA (siNT) or siRNA targeting GLD4 (siGLD4), which reduced GLD4 mRNA levels by ∼60% (Supplementary Figure S1B). Total RNA from three biological replicates were subjected to poly(U) chromatography and thermal elution followed by microarray analysis. Depletion of GLD4 would be expected to result in shortened poly(A) tails and hence the target RNAs would be reduced in the 65°C eluted fraction. Figure 1C demonstrates that many RNAs (highlighted in red) were significantly reduced in the 65°C eluate. Surprisingly, other RNAs were increased in that same RNA fraction. Here, we focused on 387 RNAs that were significantly down-regulated (P-value <0.05 by student's t-test) in the 65°C fraction (Supplementary Table S2). Of these 387 mRNAs, 74 were also reduced in the input RNA indicating not all the changes in the 65°C eluted fraction are due to shortened poly(A) (Figure 1D). The decreased expression levels of five selected mRNAs were confirmed by RT-qPCR (Supplementary Figure S1C). Gene ontology (GO) analysis of the down-regulated mRNAs identified carbohydrate metabolism as one of the enriched biological processes in this set of RNAs (Supplementary Figure S1D). Among these were a glucose transporter and many glycolytic enzymes (Figure 1E, labeled in red) such as GLUT1 (facilitated glucose transporter 1), G6PD (glucose-6-phosphate dehydrogenase), PFKFB3 (6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase), PFK-1 (phophofructokinase-1), GK (glycerol kinase), TALDO1 (transaldolase 1) and ENO1 (enolase 1). In addition, a known target mRNA of GLD4, P53 was also detected as a down-regulated RNA along with NQO1 (NAD(P)H dehydrogenase, quinone 1), which stabilizes P53 protein (27). These results suggest a role of GLD4 in carbohydrate metabolism by regulating 3΄ end modification of specific mRNAs.
GLD4 and CPEB1 control GLUT1 mRNA polyadenylation and translation
The central role of CPEB1 in cytoplasmic polyadenylation is to connect target RNAs with poly(A) polymerases. A specific RNA element in 3΄UTRs of mRNAs, the cytoplasmic polyadenylation element (CPE, consensus sequence UUUUAU or UUUUAAU) (10), is responsible for interaction with CPEB1. Many of the GLD4 targets in the carbohydrate metabolism have CPEs in their 3΄ (Supplementary Figure S2A). To test whether the identified GLD4 target mRNAs involved in glucose metabolism are associated with CPEB1, RNA-immunoprecipitation with FLAG-CPEB1 was performed (Supplementary Figure S2B). As expected, CPE-lacking RNAs did not interact with CPEB1 (G6PD, ENO1). Interestingly, some of the CPE-containing RNAs were associated with CPEB1 as expected (GLUT1, P53) while others were not (PFKFB3, PFK-1) (Supplementary Figure S2B) suggesting possible interactions with other RNA-binding proteins. We focused on GLD4 regulation of GLUT1 mRNA because GLUT1 is the starting point of glucose uptake and subsequent glycolytic processes and because the transcript interacts with CPEB1 (Figure 1E and Supplementary Figure S2B). GLUT1 is expressed in many cell types including erythrocytes, endothelial cells that form the blood-brain barrier, astrocytes, and several tumor types (28). Its major function is to maintain cellular glucose concentration, and mutations in this gene lead to a neurodevelopmental disorder, GLUT1 deficiency syndrome-1 (29), which is characterized by impaired brain function.
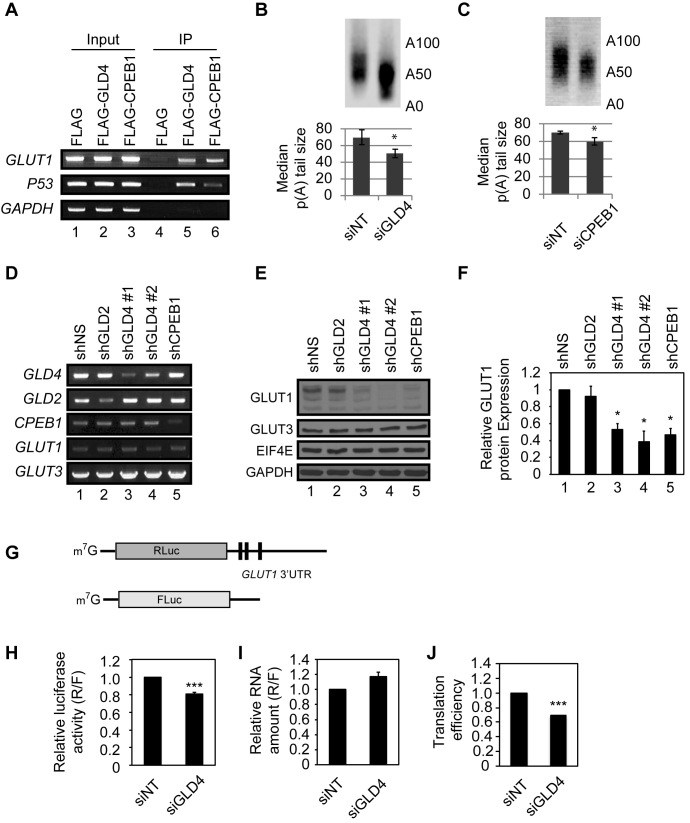
To investigate GLD4 and CPEB1 in GLUT1 mRNA regulation, we used U87MG astrocytoma cells, which have a defined glucose-sensitive metabolism (30). RNA-seq analysis of these cells showed that GLD4 RNA in U87MG cells is moderately abundant (14.14 TPM; transcripts per million) and ranked 7665th among 14473, and is comparable to other genes involved in cytoplasmic polyadenylation (GLD2: 24.36 TPM; CPEB1: 42.12 TPM) (Supplementary Figure S2C). To investigate GLD4 and CPEB1 interaction with GLUT1 mRNA, FLAG-GLD4 or FLAG-CPEB1 complexes were co-immunoprecipitated and the extracted RNA was analyzed for GLUT1, P53 (positive control), and GAPDH (negative control) sequences by RT-PCR. Both FLAG-GLD4 and FLAG-CPEB1 were co-immunoprecipitated with GLUT1 mRNA (Figure 2A). CPEB1 knockdown impaired the interaction between GLD4 and GLUT1 mRNA, indicating that GLUT1 mRNA is anchored to GLD4 in a CPEB1-dependent manner (Supplementary Figure S2D). We surmised that GLD4 binds to the GLUT1 mRNA by interacting with CPEB1 and the change in GLUT1 levels in the 65°C eluted fraction (but not in total input) was due to shortened poly(A) tail of the GLUT1 mRNA following knockdown of GLD4. To validate this hypothesis, we performed an RNA-ligation coupled, PCR-based poly(A) tail length (RL-PAT) assay to independently measure tail size of the GLUT1 mRNA. As expected, we observed a shortened tail length of the GLUT1 mRNA upon GLD4 or CPEB1 depletion (Figure 2B and C, Supplementary Figure S3A and S3B).
Figure 2.
GLD4 regulates GLUT1 poly(A) tail length and protein levels. (A) U87MG cells were transfected with plasmids carrying FLAG epitope only, FLAG-GLD4, or FLAG-CPEB1. After immunoprecipitation with FLAG antibody, extracted RNAs were subjected to RT-PCR for GLUT1 and P53 transcripts. GAPDH is a negative control. (B) U87MG cells were transfected with siNT and siGLD4 for 24 h, and they were subjected to RL-PAT assay to measure poly(A) tail size of GLUT1 mRNA. Relative signal intensity was quantified to estimate median poly(A) tail size (median ± SEM) (n = 3). (C) U87MG cells were transfected with siNT and siCPEB1 for 24 h, and were subjected to RL-PAT assay (median ± SEM) (n = 3). (D) U87MG cells were transduced with shNS, or shRNAs targeting GLD2, GLD4 and CPEB1. GLD4#1 and #2 indicate two separate shRNAs targeting different regions of GLD4. After 72 h after transduction, these cells were subjected to RT-PCR to measure knockdown efficiency of each shRNA and GLUT1 mRNA. GLUT3 mRNA was not changed and served as a control. (E) The same set of cells as in (D) was used for immunoblot analysis for GLUT1 and GLUT3 protein. eIF4E and GAPDH served as loading controls. (F) Quantification of relative GLUT1 protein with knockdown of GLD2, GLD4 and CPEB1 (mean ± SEM) (n = 3). (G) RNA reporter constructs used to measure translation efficiency. m7G denotes 7-methyl-guanosine cap structure. Solid line and black columns represents untranslated region (UTR) and CPE, respectively. (H) siNT and siGLD4 treated U87MG cells were transfected with Renilla luciferase RNA harboring GLUT1 3΄UTR and firefly luciferase as a transfection control. After 6 h, cells were harvested and measured for luciferase activity (R/F) (mean ± SEM) (n = 6). (I) The same set of cells as in (H) was used to assess the relative amount of RNA (R/F) (mean ± SEM) (n = 3) by real-time PCR. (J) Translation efficiency is calculated by dividing numbers from (H) with those from (I).* indicates P < 0.05, ** indicates P < 0.01 and *** indicates P < 0.001 by two-tailed Student's t-test.
Next we examined the consequence of GLUT1 tail length shortening following GLD4 or CPEB1 knockdown. Because elongated poly(A) tails are associated with elevated translation of various mRNAs (10), we speculated that GLD4 or CPEB1 knockdown would result in decreased production of GLUT1 protein. Western blot analysis of cells transduced with shRNA lentivirus targeting two different regions of GLD4 (Figure 2D, lanes 3 and 4) showed a ∼50% reduction of GLUT1 (Figure 2E, lanes 3 and 4; quantification in Figure 2F). Surprisingly, the level of GLUT3, whose mRNA polyadenylation was also reduced by GLD4 depletion (Supplementary Table S2), was unaffected by GLD4 knockdown, demonstrating that not all the RNAs identified in the screening have commensurate changes in protein levels (Figure 2E). Cells treated with shRNA lentivirus targeting CPEB1 also had a similar ∼50% reduction of GLUT1 (Figure 2E, lane 5; quantification in Figure 2F, lane 5). In contrast, knockdown of GLD2 had no effect on GLUT1 (Figure 2E and F, lane 2) showing specificity of target selection by PAPs. GLUT1 mRNA levels were not changed by GLD4 or CPEB1 depletion (Figure 2D), suggesting that the regulation of GLUT1 occurs at the translational level. In concordance to our data with shRNA, depletion of GLD4 with siRNAs also reduced GLUT1 levels (Supplementary Figure S3C–E). To support our model of GLD4-mediated translational control of GLUT1 mRNA, we employed a separate translational assay using a reporter RNA harboring the 3΄ UTR of GLUT1 mRNA (Figure 2G). The luciferase activity derived from the reporter was reduced 20% upon GLD4 depletion without affecting the luciferase RNA level, indicating translational control of GLUT1 mRNA by GLD4 (Figure 2H–J).
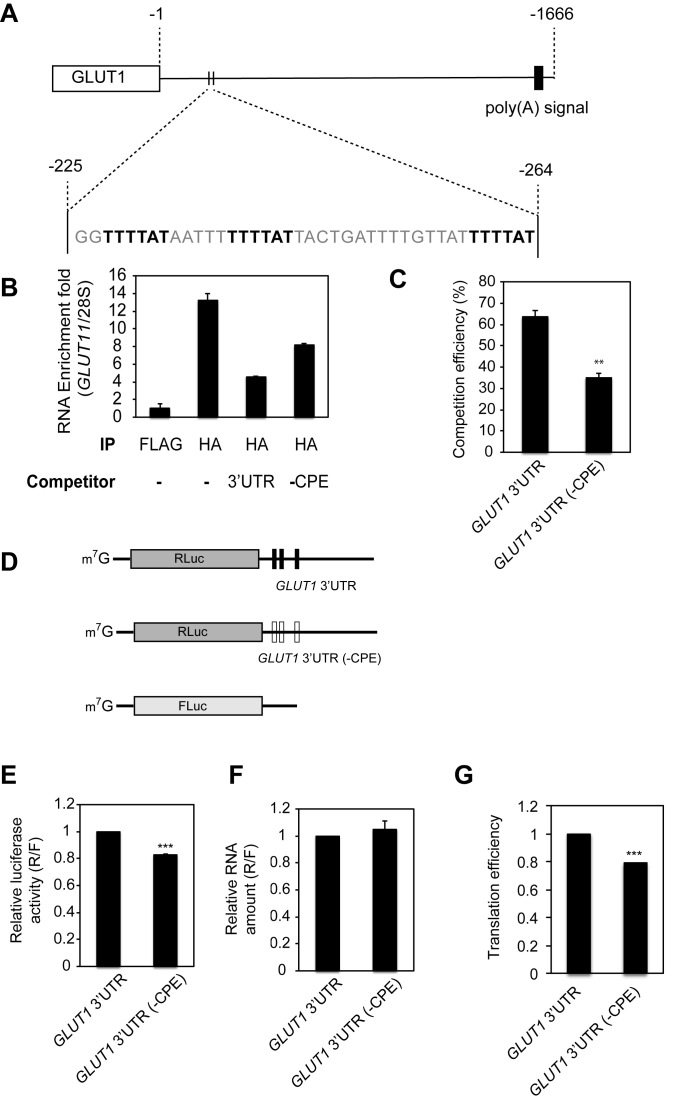
As indicated above, CPEB1-mediated cytoplasmic polyadenylation requires the 3΄ UTR cytoplasmic polyadenylation element (CPE, consensus sequence UUUUAU) (10). We found three CPEs in a 40 nucleotide window (Figure 3A) of the 1.7 kb GLUT1 3΄UTR, which are conserved among mammals (Supplementary Figure S4A). To determine whether these sequences are CPEB1 binding sites, we performed an RNA-IP competition assay using HA-CPEB1 expressed in U87MG cells with GLUT1 3΄ UTR containing or lacking three CPEs as competitors. Following immunoprecipitation of CPEB1-RNA complexes, the extracted RNA was analyzed for enrichment of endogenous GLUT1 mRNA relative to 28S rRNA (Figure 3B), and the relative competition efficiency by competitor RNA was calculated (Figure 3C). The exogenous GLUT1 3΄ UTR containing CPEs competed ∼65% of endogenous GLUT1 mRNA whereas GLUT1 3΄ UTR lacking CPE (-CPE) competed less efficiently (∼35%) (Figure 3C). To determine the importance of the CPEs in translational regulation of GLUT1 mRNA, we performed an assay using reporter RNAs of GLUT1 3΄ UTR containing or lacking CPEs (Figure 3D). As expected, a reporter RNA without CPEs binds to CPEB1 less efficiently than a reporter with CPEs (Supplementary Figure S4B). A reporter RNA with GLUT1 3΄ UTR lacking CPEs (-CPE) was translated 20% less efficiently than CPE-containing GLUT1 3΄ UTR without affecting the reporter RNA level (Figure 3E–G), indicating these 3 CPEs of GLUT1 3΄ UTR are important for translational regulation by CPEB1. Interestingly, the 40 nucleotides containing the three CPEs is sufficient to enhance translation as assessed by the use of a reporter RNA containing this region, which has a significantly higher translational efficiency than reporter that lacks this region (Supplementary Figure S5A-C). The reporter RNA containing the 40 nucleotide region with a mutant version of CPEs did not show the translational enhancement, indicating that the CPEs are important for the translation efficiency (Supplementary Figure S5C).
Figure 3.
CPEB1 regulates GLUT1 translation. (A) Schematic diagram for the 3΄ UTR of the GLUT1 mRNA. The rectangle indicates the open reading frame and the solid line indicates the 1.67 kb-long 3΄ UTR. Three CPEs within a 40nt window between positions 225 and 264 are bold highlighted. (B) U87MG cells were transduced with HA-CPEB1 expressing lentivirus. GLUT1 3΄ UTR RNA containing or lacking three CPEs were added to the lysate as competitors of RNA-IP reactions. After immnoprecipitation with HA antibody, RNAs were subjected to real-time PCR for the levels of GLUT1 RNA and normalized to 28S rRNA to quantify fold enrichment in each condition. The experiments were performed three times and one representative result is shown here. (C) The relative competition efficiency was calculated compared to the reaction without any competitor (mean ± SEM) (n = 3). (D) Schematic diagram of RNA reporter constructs. Three vertical black bars in GLUT1 3΄UTR denote CPEs, and white bars indicate lack of CPEs. (E) U87MG cells were transfected with a luciferase reporter harboring GLUT1 3΄ UTR containing or lacking three CPEs. After 6 h, relative Renilla luciferase (R) to firefly (F) activity was measured (R/F) (mean ± SEM) (n = 5). (F) The same set of cells as in (E) was used to assess the relative amount of RNA (R/F) (mean ± SEM) (n = 4) by real-time PCR. (G) Translation efficiency is calculated by dividing numbers from (E) with those from (F).* indicates P < 0.05, ** indicates P < 0.01 and *** indicates P < 0.001 by two-tailed Student's t-test.
GLD4-mediated polyadenylation regulates glucose homeostasis
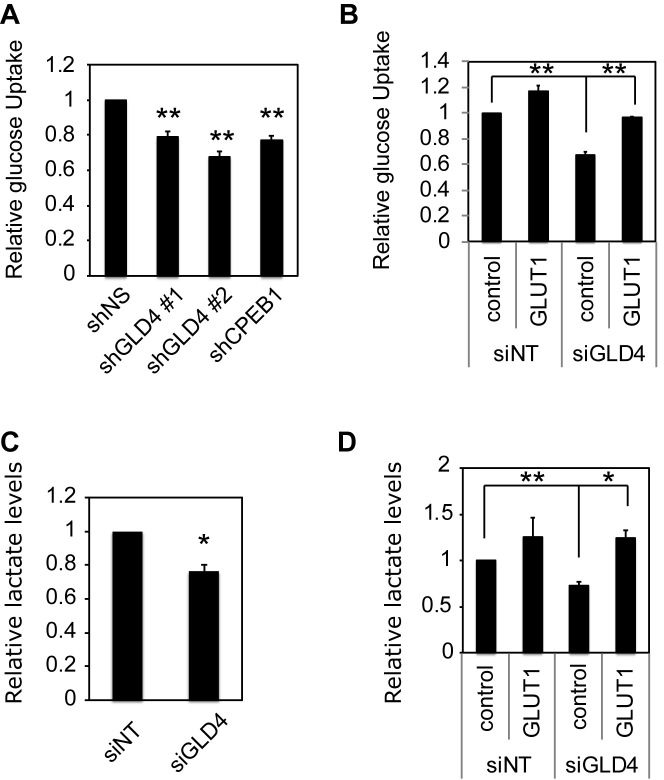
Because GLUT1 is a major glucose transporter and is responsible for the maintenance of basal glucose levels (28), we speculated that reduced GLUT1 resulting from GLD4 knockdown would diminish glucose uptake into cells. To assess this possibility, cells transduced with lentivirus expressing a non-specific (NS) shRNA or shRNAs against GLD4 or CPEB1 were starved for glucose for 2 h and then incubated with the glucose analog [3H]2-deoxyglucose. Uptake of the glucose analog was reduced by ∼25% following GLD4 or CPEB1 depletion (Figure 4A). Similar reductions of glucose uptake were observed when the cells were transfected with siGLD4 (Figure 4B, lanes 1 and 3, and Supplementary Figure S6). The reduced glucose uptake was rescued when GLUT1 was ectopically expressed in the GLD4-depleted cells (Figure 4B, lanes 3 and 4) indicating GLD4 control of glucose uptake is mediated by GLUT1. Because glucose is the major fuel source for energy through glycolysis that leads to production of other metabolites such as lactate (Figure 1E), we surmised that reduced glucose uptake following GLD4 knockdown would reduce lactate, which indeed occurred (Figure 4C). Reduced lactate was rescued by the ectopic expression of GLUT1 suggesting GLD4 regulation of lactate is mediated by GLUT1 (Figure 4D, lanes 3 and 4).
Figure 4.
GLD4 regulates glucose metabolism. (A) U87MG cells were transduced with shNS, or shRNAs targeting GLD4 or CPEB1 and measured for glucose uptake (mean ± SEM) (n = 4) after 72 h. (B) U87MG cells were transduced with lentivirus expressing empty vector (control) or a GLUT1 expressing vector and 24 h later the same cells were transfected with siNT or siGLD4. After 48 h, cells were measured for glucose uptake (mean ± SEM) (n = 3). (C) U87MG cells were transfected with siNT or siGLD4 and measured for lactate levels (mean ± SEM) (n = 4). (D) U87MG cells were transduced with lentivirus expressing empty vector (control) or a GLUT1 expressing vector and 24 h later the same cells were treated with siNT or GLD4. After 48 h, cells were measured for lactate levels (mean ± SEM) (n = 4). * indicates P < 0.05, ** indicates P < 0.01 and *** indicates P < 0.001 by two-tailed Student's t-test.
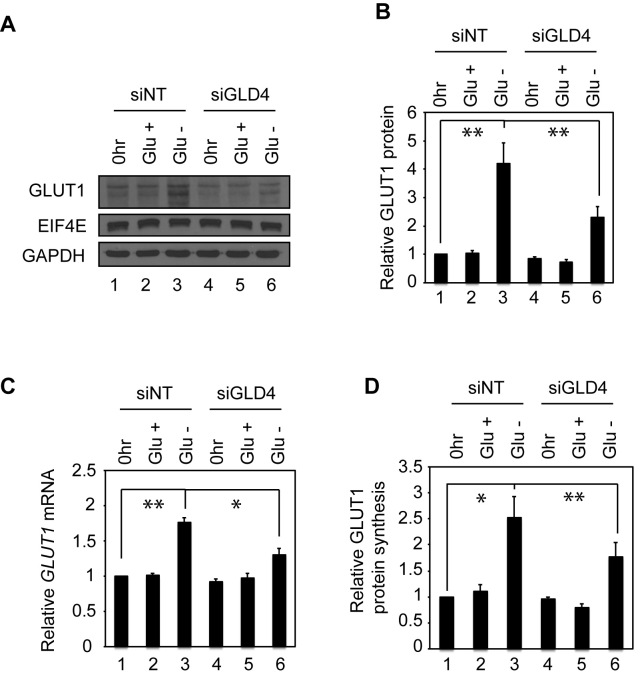
GLUT1 levels are dynamically regulated to maintain cellular glucose homeostasis (31). Previous studies indicate that increased GLUT1 mRNA stability upon glucose deprivation is mediated by the mRNA's 3΄ UTR, suggesting that poly(A) tail lengthening may be an underlying molecular mechanism (32). Indeed, glucose deprivation triggers an up-regulation of GLUT1 protein to facilitate the efficient uptake of glucose (Figure 5A, lanes 1–3), which confirms previous observations (31–33). In glucose-deprived cells, GLUT1 increased by ∼4-fold compared to cells maintained with constant high glucose (Figure 5A and B, lanes 1–3). In contrast, GLUT1 mRNA triggered by glucose deprivation changed <2-fold (Figure 5C lanes 1–3) indicating at least half of the up-regulation of GLUT1 occurred at the translational level (Figure 5D, lanes 1–3, GLUT1 protein to mRNA). The remaining up-regulation of GLUT1 is unlikely to be due to a transcriptional change but instead probably due to increased stability, which we infer from the observation that the GLUT1 pre-mRNA level was not changed (Supplementary Figure S7A). Interestingly, a subtle but significant increase of GLUT1 mRNA poly(A) tail length was observed upon glucose depletion, indicating that cytoplasmic polyadenylation is involved in the GLUT1 protein up-regulation (Supplementary Figure S7B, lanes 1 and 2). We next investigated whether GLD4 helps maintain glucose homeostasis by regulating GLUT1 following glucose depletion. When cells were transfected with siGLD4, the up-regulation of GLUT1 induced by glucose deprivation was suppressed ∼50% compared to siNT (Figure 5A and B, lanes 3 and 6). Moreover, depletion of GLD4 hampers increase of GLUT1 mRNA poly(A) tail under glucose deprivation (Supplementary Figure S7B). These data indicate that the GLD4-GLUT1 axis is a component of the glucose homeostasis mechanism.
Figure 5.
GLD4 regulates glucose deprivation-induced GLUT1 expression. (A) GLUT1 protein levels upon glucose deprivation (Glu–) in control (siNT) or GLD4 knockdown U87MG cells (siGLD4). 0 h rindicates pre-deprivation and Glu+ indicates a non-deprivation control. Glu– indicates glucose deprivation for 24 h. EIF4E and GAPDH were used as loading controls. (B) Relative GLUT1 protein levels were quantified compared to 0 h of the control cells (mean ± SEM) (n = 6). (C) The same set of cells as (B) was subjected to RT-qPCR to test GLUT1 mRNA levels. (D) Relative GLUT1 protein synthesis is calculated by dividing numbers from (B) with those from (C). * indicates P < 0.05, ** indicates P < 0.01 and *** indicates P < 0.001 by two-tailed Student's t-test.
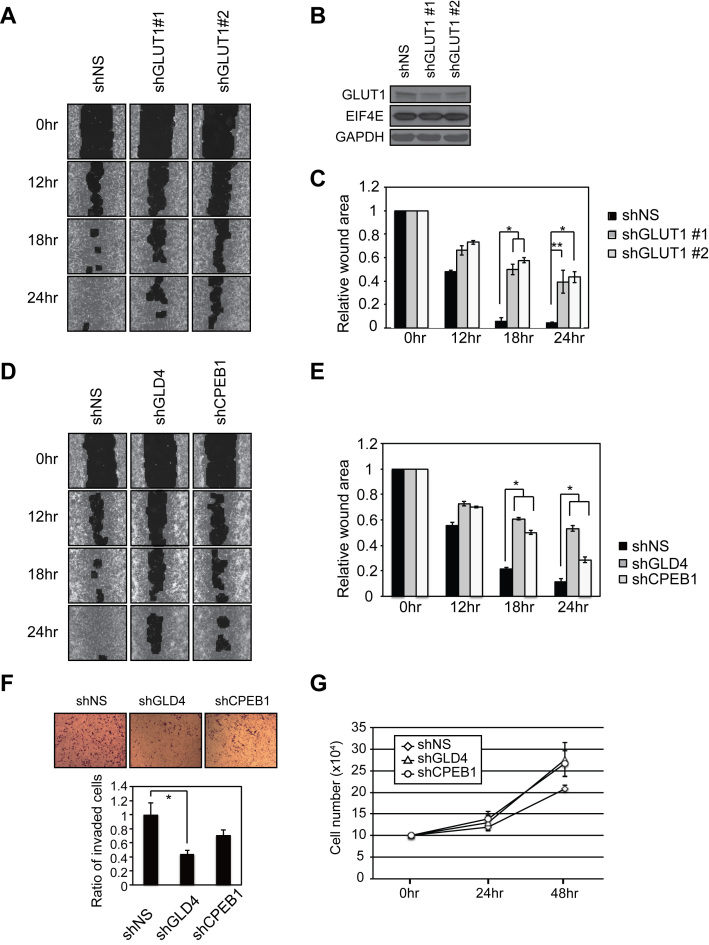
Given the importance of GLD4 and its interacting partner CPEB1 in GLUT1 regulation and glucose metabolism, we speculated that GLD4 and CPEB1 may regulate cellular physiologies that are sensitive to the glucose utilization such as cell migration (34,35). Consequently, we tested whether GLD4 or CPEB1 controls this process in a wound-healing assay that measures the rate of cell infiltration into a scratch made in confluent cells (36). To ensure U87MG cell migration is directly affected by GLUT1, we first tested cell migration of GLUT1-depleted cells. Following infection with lentivirus expressing a NS shRNA, or shRNAs against two different regions of GLUT1 (Figure 6B), confluent cells were scratched and the rate of infiltration into the wounded area was measured after 12, 18 and 24 h. At 18 and 24 h, depletion of GLUT1 significantly delayed mobility of cells into the wound (Figure 6A and C). Next, we tested effects of depletion of GLD4 or CPEB1 on cell migration. At 18 and 24 h, depletion of GLD4 or CPEB1 significantly delayed mobility of the cells into wound (Figure 6D and E). Moreover, depletion of GLD4 also elicited a decrease in transwell cell invasion through Matrigel, which is a surrogate basement membrane (Figure 6F). In contrast, neither GLD4 nor CPEB1 affected the rate of cell proliferation (Figure 6G). These data indicate that GLD4-mediated translational control of GLUT1 mRNA regulates not only cellular glucose metabolism, but also the glucose-related cell migration and invasion.
Figure 6.
GLD4 and GLUT1 regulate cell migration. (A) U87MG cells were infected with shNS or lentivirus expressing two different GLUT1 shRNAs. After 72 h, cells were subjected to wound healing assays. Images were captured 12, 18 and 24 h after the initial wound. Images from each time point are shown for graphical representations of wound healing ability. (B) Knockdown efficiency of two different shRNAs targeting GLUT1 was analyzed by western blot. eIF4E and GAPDH were negative controls. (C) Quantification of the relative wound area after 0, 12, 18 and 24 h after initial wound (mean ± SEM) (n = 6). (D) Wound healing assay as described in (A) was performed for shNS, shGLD4 or shCPEB1 infected cells. (E) Quantification of the relative wound area (mean ± SEM) (n = 6). (F) Matrigel invasion assays with U87MG cells infected with lentivirus expressing the indicated shRNAs (mean ± SEM) (n = 3). (G) Proliferation rate of the shRNA knockdown cells 24 and 48 h post-infection (mean ± SEM) (n = 4). * indicates P < 0.05, ** indicates P < 0.01 and *** indicates P < 0.001 by two-tailed Student's t-test.
DISCUSSION
The glucose transporter GLUT1 is widely expressed and is important for basal level glucose uptake. Aberrant expression of GLUT1 is detected in various disease conditions such as experimentally induced diabetes in animals, Alzheimer's disease and certain cancers where elevated GLUT1 is associated with a poor prognosis (37–41). Inhibition of GLUT1 expression as well as targeting of the enzyme by small molecules impairs growth of various cancers, indicating a pivotal role of GLUT1 in oncogenesis (42,43). Despite the fact that multiple glucose transporters are expressed in a variety of cell types, GLUT1 seems to have non-redundant functions in several of them (44,45). In addition, heterozygous mutations in this gene lead to a neurodevelopmental disease, GLUT1 deficiency syndrome-1, which is diagnosed by low glucose and lactate levels in cerebrospinal fluid (29).
Our data demonstrate that the CPEB1-GLD4 cytoplasmic polyadenylation complex binds to and facilitates translation of GLUT1 mRNA. Consequently, depletion of GLD4 reduces glucose uptake and lactate production. Our data also indicate that although GLD4 and CPEB1 knockdown had no effect on the proliferation/cell death of U87MG cells, it attenuated cell migration and invasion. These results agree with a previous observation that ectopic expression of a dominant negative CPEB1 reduces cell migration in rat glioblastoma cells (46). On the other hand, CPEB1 depletion promotes cell migration in breast cancer cells, suggesting that it can have opposite effects dependent on cell type (47). This duality is also illustrated by GLD4, which promotes translation of the major tumor suppressor P53 on one hand and translation of GLUT1 on the other, thereby simultaneously regulating seemingly opposing metabolic pathways. These observations suggest versatile regulation of biological pathways by cytoplasmic polyadenylation; that is, cells may be equipped with a multifaceted system that they can rapidly adapt to necessary changes by targeting multiple (sometimes contradictory) biological pathways depending on the biological context.
In addition to GLUT1, GLD4 regulates the poly(A) tails of a number of mRNAs that are involved in carbohydrate metabolism including G6PD, PFKFB3, PFK-1, GK, TALDO1 and ENO1. These genes encode essential glycolytic enzymes that convert glucose to pyruvate for energy production. The coordinated regulation of a cohort of enzymes in this pathway by GLD4 is reminiscent of the CPEB1-mediated regulation of multiple components in the insulin-signaling pathway (48). While not all of the mRNAs that we identified in this study are likely to be directly regulated by GLD4, it is remarkable that expression of several key enzymes in a single physiological pathway was affected by depletion of GLD4.
The changes in poly(A) tail length by GLD4 depletion were often <50%, which is of the same general magnitude as observed in neurons depleted of GLD2, another non-canonical PAP (8). The change in poly(A) tail size by GLD4 knockdown may be partly attributable to the fact that the cells we used were not stimulated. Indeed, GLD2- or CPEB1-mediated cytoplasmic polyadenylation is robust when cells respond to external signaling cues such as during development or upon synaptic activation, or stimulation of the acute inflammatory response (10). These observations suggest that a more substantial change in GLD4-mediated cytoplasmic polyadenylation may be associated with environmental cues such as hormone or stress-induced signaling. Therefore, investigation of GLD4-mediated cytoplasmic polyadenylation under various physiological contexts may identify different sets of GLD4-regulated mRNAs from those mRNAs identified in this study. GLD4 is widely expressed suggesting possible broad roles in various biological processes. The tail length changes we observe are consistent with studies from C. elegans where GLD4– compromised animals have slightly shorter poly(A) tails (25,49). Interestingly, these animals have profound defects in polysome assembly suggesting a role of GLD4 in promoting general protein synthesis (25).
It is evident that a number of mRNAs regulated by GLD4 at the level of cytoplasmic polyadenylation do not interact with CPEB1, suggesting the involvement of other RNA binding proteins in GLD4-mediated cytoplasmic polyadenylation (Supplementary Figure S2B). Bidirectional regulation of poly(A) tail length involves combinatorial interactions of RNA binding proteins, deadenylases, and poly(A) polymerases. Multiple RNA binding proteins are thought to drive cytoplasmic deadenylation (50) including the AU-rich element (ARE) binding protein, KSRP and TTP, PUF family members Pumilio and FBF, as well as a GU-rich sequence binding protein CUG-BP (51,52). Whether polyadenylation similarly involves multiple factors that differentiate cis-elements of mRNAs is less clear except for a few examples in C. elegans and Xenopus (53,54). Therefore, decoding the polyadenylation network woven by RNA binding proteins interacting with GLD4 is a necessary next step.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Anthony Carruthers for sharing GLUT1 antibody, Dr Alonzo Ross for U87MG cells, Dr Michael R. Green for pTRIPZ lentiviral vector, Drs Adilson Guilherme and Myoung Sook Han for the glucose uptake protocol, and Hui-Fang Hung for the Matrigel invasion analysis. We also thank Drs Andrea D’Ambrogio and Sharvari Gujja for the technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [GM46779 and NS079415 to J.D.R.]. Funding for open access charge: NIH [GM46779 and NS079415].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schmidt M.J., Norbury C.J.. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscipl. Rev. RNA. 2010; 1:142–151. [DOI] [PubMed] [Google Scholar]
- 2. Tomecki R., Dmochowska A., Gewartowski K., Dziembowski A., Stepien P.P.. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004; 32:6001–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mellman D.L., Gonzales M.L., Song C., Barlow C.A., Wang P., Kendziorski C., Anderson R.A.. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008; 451:1013–1017. [DOI] [PubMed] [Google Scholar]
- 4. Lim J., Ha M., Chang H., Kwon S.C., Simanshu D.K., Patel D.J., Kim V.N.. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014; 159:1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnard D.C., Ryan K., Manley J.L., Richter J.D.. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004; 119:641–651. [DOI] [PubMed] [Google Scholar]
- 6. Benoit P., Papin C., Kwak J.E., Wickens M., Simonelig M.. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008; 135:1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakanishi T., Kubota H., Ishibashi N., Kumagai S., Watanabe H., Yamashita M., Kashiwabara S., Miyado K., Baba T.. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev. Biol. 2006; 289:115–126. [DOI] [PubMed] [Google Scholar]
- 8. Udagawa T., Swanger S.A., Takeuchi K., Kim J.H., Nalavadi V., Shin J., Lorenz L.J., Zukin R.S., Bassell G.J., Richter J.D.. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol. Cell. 2012; 47:253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J.. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002; 419:312–316. [DOI] [PubMed] [Google Scholar]
- 10. Ivshina M., Lasko P., Richter J.D.. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 2014; 30:393–415. [DOI] [PubMed] [Google Scholar]
- 11. Kim J.H., Richter J.D.. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006; 24:173–183. [DOI] [PubMed] [Google Scholar]
- 12. Mendez R., Murthy K.G., Ryan K., Manley J.L., Richter J.D.. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell. 2000; 6:1253–1259. [DOI] [PubMed] [Google Scholar]
- 13. Sarkissian M., Mendez R., Richter J.D.. Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Genes Dev. 2004; 18:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin G., Keller W.. RNA-specific ribonucleotidyl transferases. RNA. 2007; 13:1834–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D.. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005; 121:713–724. [DOI] [PubMed] [Google Scholar]
- 16. Shcherbik N., Wang M., Lapik Y.R., Srivastava L., Pestov D.G.. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010; 11:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berndt H., Harnisch C., Rammelt C., Stohr N., Zirkel A., Dohm J.C., Himmelbauer H., Tavanez J.P., Huttelmaier S., Wahle E.. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012; 18:958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mullen T.E., Marzluff W.F.. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5΄ to 3΄ and 3΄ to 5΄. Genes Dev. 2008; 22:50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmidt M.J., West S., Norbury C.J.. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011; 17:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rammelt C., Bilen B., Zavolan M., Keller W.. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA. 2011; 17:1737–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burns D.M., Richter J.D.. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008; 22:3449–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burns D.M., D’Ambrogio A., Nottrott S., Richter J.D.. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011; 473:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harnisch C., Cuzic-Feltens S., Dohm J.C., Gotze M., Himmelbauer H., Wahle E.. Oligoadenylation of 3΄ decay intermediates promotes cytoplasmic mRNA degradation in Drosophila cells. RNA. 2016; 22:428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Millonigg S., Minasaki R., Nousch M., Eckmann C.R.. GLD-4-mediated translational activation regulates the size of the proliferative germ cell pool in the adult C. elegans germ line. PLoS Genet. 2014; 10:e1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nousch M., Yeroslaviz A., Habermann B., Eckmann C.R.. The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms. Nucleic Acids Res. 2014; 42:11622–11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., Jensen T.H.. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 2011; 43:624–637. [DOI] [PubMed] [Google Scholar]
- 27. Asher G., Lotem J., Kama R., Sachs L., Shaul Y.. NQO1 stabilizes p53 through a distinct pathway. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:3099–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carruthers A., DeZutter J., Ganguly A., Devaskar S.U.. Will the original glucose transporter isoform please stand up!. Am. J. Physiol. Endocrinol. Metabol. 2009; 297:E836–E848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rotstein M., Engelstad K., Yang H., Wang D., Levy B., Chung W.K., De Vivo D.C.. Glut1 deficiency: inheritance pattern determined by haploinsufficiency. Ann. Neurol. 2010; 68:955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jelluma N., Yang X., Stokoe D., Evan G.I., Dansen T.B., Haas-Kogan D.A.. Glucose withdrawal induces oxidative stress followed by apoptosis in glioblastoma cells but not in normal human astrocytes. Mol. Cancer Res.: MCR. 2006; 4:319–330. [DOI] [PubMed] [Google Scholar]
- 31. von der Crone S., Deppe C., Barthel A., Sasson S., Joost H.G., Schurmann A.. Glucose deprivation induces Akt-dependent synthesis and incorporation of GLUT1, but not of GLUT4, into the plasma membrane of 3T3-L1 adipocytes. Eur. J. Cell Biol. 2000; 79:943–949. [DOI] [PubMed] [Google Scholar]
- 32. Boado R.J., Pardridge W.M.. Glucose deprivation causes posttranscriptional enhancement of brain capillary endothelial glucose transporter gene expression via GLUT1 mRNA stabilization. J. Neurochem. 1993; 60:2290–2296. [DOI] [PubMed] [Google Scholar]
- 33. Bruckner B.A., Ammini C.V., Otal M.P., Raizada M.K., Stacpoole P.W.. Regulation of brain glucose transporters by glucose and oxygen deprivation. Metab.: Clin. Exp. 1999; 48:422–431. [DOI] [PubMed] [Google Scholar]
- 34. Takatani-Nakase T., Matsui C., Maeda S., Kawahara S., Takahashi K.. High glucose level promotes migration behavior of breast cancer cells through zinc and its transporters. PLoS One. 2014; 9:e90136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wahdan-Alaswad R., Fan Z., Edgerton S.M., Liu B., Deng X.S., Arnadottir S.S., Richer J.K., Anderson S.M., Thor A.D.. Glucose promotes breast cancer aggression and reduces metformin efficacy. Cell Cycle. 2013; 12:3759–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagaoka K., Udagawa T., Richter J.D.. CPEB-mediated ZO-1 mRNA localization is required for epithelial tight-junction assembly and cell polarity. Nat. Commun. 2012; 3:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Simpson I.A., Chundu K.R., Davies-Hill T., Honer W.G., Davies P.. Decreased concentrations of GLUT1 and GLUT3 glucose transporters in the brains of patients with Alzheimer's disease. Ann. Neurol. 1994; 35:546–551. [DOI] [PubMed] [Google Scholar]
- 38. Brown R.S., Wahl R.L.. Overexpression of Glut-1 glucose transporter in human breast cancer. An immunohistochemical study. Cancer. 1993; 72:2979–2985. [DOI] [PubMed] [Google Scholar]
- 39. Haber R.S., Rathan A., Weiser K.R., Pritsker A., Itzkowitz S.H., Bodian C., Slater G., Weiss A., Burstein D.E.. GLUT1 glucose transporter expression in colorectal carcinoma: a marker for poor prognosis. Cancer. 1998; 83:34–40. [DOI] [PubMed] [Google Scholar]
- 40. Pardridge W.M., Triguero D., Farrell C.R.. Downregulation of blood-brain barrier glucose transporter in experimental diabetes. Diabetes. 1990; 39:1040–1044. [DOI] [PubMed] [Google Scholar]
- 41. Amann T., Maegdefrau U., Hartmann A., Agaimy A., Marienhagen J., Weiss T.S., Stoeltzing O., Warnecke C., Scholmerich J., Oefner P.J. et al. . GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am. J. Pathol. 2009; 174:1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Y., Cao Y., Zhang W., Bergmeier S., Qian Y., Akbar H., Colvin R., Ding J., Tong L., Wu S. et al. . A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol. Cancer Ther. 2012; 11:1672–1682. [DOI] [PubMed] [Google Scholar]
- 43. Amann T., Hellerbrand C.. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin. Ther. Targets. 2009; 13:1411–1427. [DOI] [PubMed] [Google Scholar]
- 44. Macintyre A.N., Gerriets V.A., Nichols A.G., Michalek R.D., Rudolph M.C., Deoliveira D., Anderson S.M., Abel E.D., Chen B.J., Hale L.P. et al. . The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014; 20:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei J., Shimazu J., Makinistoglu M.P., Maurizi A., Kajimura D., Zong H., Takarada T., Iezaki T., Pessin J.E., Hinoi E. et al. . Glucose uptake and Runx2 synergize to orchestrate osteoblast differentiation and bone formation. Cell. 2015; 161:1576–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jones K.J., Korb E., Kundel M.A., Kochanek A.R., Kabraji S., McEvoy M., Shin C.Y., Wells D.G.. CPEB1 regulates beta-catenin mRNA translation and cell migration in astrocytes. Glia. 2008; 56:1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nagaoka K., Fujii K., Zhang H., Usuda K., Watanabe G., Ivshina M., Richter J.D.. CPEB1 mediates epithelial-to-mesenchyme transition and breast cancer metastasis. Oncogene. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alexandrov I.M., Ivshina M., Jung D.Y., Friedline R., Ko H.J., Xu M., O'Sullivan-Murphy B., Bortell R., Huang Y.T., Urano F. et al. . Cytoplasmic polyadenylation element binding protein deficiency stimulates PTEN and Stat3 mRNA translation and induces hepatic insulin resistance. PLoS Genet. 2012; 8:e1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Minasaki R., Rudel D., Eckmann C.R.. Increased sensitivity and accuracy of a single-stranded DNA splint-mediated ligation assay (sPAT) reveals poly(A) tail length dynamics of developmentally regulated mRNAs. RNA Biol. 2014; 11:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weill L., Belloc E., Bava F.A., Mendez R.. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 2012; 19:577–585. [DOI] [PubMed] [Google Scholar]
- 51. Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M.. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006; 13:533–539. [DOI] [PubMed] [Google Scholar]
- 52. Moraes K.C., Wilusz C.J., Wilusz J.. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006; 12:1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim K.W., Nykamp K., Suh N., Bachorik J.L., Wang L., Kimble J.. Antagonism between GLD-2 binding partners controls gamete sex. Dev. Cell. 2009; 16:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pique M., Lopez J.M., Foissac S., Guigo R., Mendez R.. A combinatorial code for CPE-mediated translational control. Cell. 2008; 132:434–448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.