Abstract
RALY is a member of the heterogeneous nuclear ribonucleoprotein family (hnRNP), a large family of RNA-binding proteins involved in many aspects of RNA metabolism. Although RALY interactome has been recently characterized, a comprehensive global analysis of RALY-associated RNAs is lacking and the biological function of RALY remains elusive. Here, we performed RIP-seq analysis to identify RALY interacting RNAs and assessed the role of RALY in gene expression. We demonstrate that RALY binds specific coding and non-coding RNAs and associates with translating mRNAs of mammalian cells. Among the identified transcripts, we focused on ANXA1 and H1FX mRNAs, encoding for Annexin A1 and for the linker variant of the histone H1X, respectively. Both proteins are differentially expressed by proliferating cells and are considered as markers for tumorigenesis. We demonstrate that cells lacking RALY expression exhibit changes in the levels of H1FX and ANXA1 mRNAs and proteins in an opposite manner. We also provide evidence for a direct binding of RALY to the U-rich elements present within the 3΄UTR of both transcripts. Thus, our results identify RALY as a poly-U binding protein and as a regulator of H1FX and ANXA1 in mammalian cells.
INTRODUCTION
Transcriptional regulation and post-transcriptional control of mRNA translation, mRNA turnover and post-translational regulation of proteins are mechanisms used by eukaryotic cells to control gene expression. Moreover, transport, subsequent subcellular localization and local translational control of mRNAs are additional mechanisms to target proteins to specific intracellular regions (1,2). In this context, RNA-binding proteins (RBPs) play a wide range of functions and are heterogeneous in terms of structure. The heterogeneous nuclear ribonucleoproteins (hnRNPs), for example, are a family of more than 40 RNA-binding proteins which exert several roles in RNA metabolism, such as splicing, mRNA stability, and nuclear export in many different cell types (3–7). Moreover, some hnRNPs might also recruit regulatory proteins associated with DNA-related processes such as double-stranded DNA break repair (8). HnRNPs are characterized by the presence of one or two RNA-recognition motifs (RRMs) whose sequence can vary among the members of the family (4,9). Moreover, sequence motifs rich in arginine and glycine are also present whose function, albeit still unclear, seems to mediate the intracellular trafficking of the hnRNPs (10–12).
Given its high similarity in amino acid sequence with the hnRNP-C (13,14), a putative member of the hnRNP family is RALY (RBP associated with lethal yellow mutation), also known as HNRPCL2 and P542. RALY was originally identified as an autoantigen cross-reacting with the Epstein-Barr nuclear antigen 1 (EBNA1), a viral protein associated with Epstein-Barr virus (15). Subsequent studies associated a deletion of the murine Raly gene with the lethal yellow mutation. This deletion of 170 kb is localized upstream the agouti allele, and it encompasses part of the coding region for Raly and the entire coding region of the eukaryotic initiation factor 2B (eIF2B), with the consequence that the agouti gene passes under control of Raly's promoter (14). Although RALY protein was found to be a component of the exon–junction complex, its down regulation did not impair splicing. Thus, the role of RALY in this process remains unclear (16–18). Interestingly, RALY protein was found to be upregulated in adenocarcinoma cell lines (19). In these cells, RALY together with NONO/p54nrb was identified as an interactor of YB-1, an RNA-binding protein involved in splicing, transcription and translational regulation of specific mRNAs (20,21). NONO/p54nrb is a DNA- and RNA-binding protein involved in several nuclear processes, including pre-mRNA splicing and double-strand DNA break repair (22,23). YB-1 mediates pre-mRNA alternative splicing regulation, controls the transcription of numerous genes, and can play a role in repairing nicks and/or breaks into double-stranded DNA (21,24). YB-1 over-expression in different tumors has been associated to an acquired resistance to specific drugs (25,26). Similarly, the levels of RALY mRNA are increased in different cancerous tissues such as ovarian, lung, bladder, brain and breast cancers as well as in multiple myelomas and melanomas with associated poor survival (19). These findings suggest a potential and still uninvestigated role of RALY in tumorigenesis.
Recently, we applied the iBioPQ approach to identify RALY-associated proteins and unravel the molecular mechanisms underlying the cellular function of RALY (27). Among the most abundant interacting proteins, we identified several factors involved in mRNA metabolism as well as translational control. Thus, although RALY might be involved in nuclear RNA processing, it can play still unknown roles in the cytoplasm. Moreover, RALY-associated RNAs are still unidentified. To fill this gap, we combined RNA-immunoprecipitation coupled to sequencing (RIP-seq) with transcriptome analysis of si-RALY cells to identify RALY-associated RNAs and to assess the possible role of the protein in RNA metabolism.
We identified 217 RNAs differentially expressed in RALY downregulated cells. Comparing the transcriptome findings with the RIP-seq results we identified 23 RNAs interacting with RALY, whose expression changed upon RALY silencing. Among the coding RNAs FOXI, ANXA1, CST6 mRNAs were upregulated and CXCR4, KNSTRN and H1FX mRNAs were downregulated in cells lacking RALY expression. We demonstrated that RALY specifically binds the uracil rich-regions present within the 3΄-UTRs of the transcripts. Moreover, we showed that RALY regulates the levels of ANXA1 and H1FX mRNAs, two transcripts encoding for Annexin A1 and for the linker variant of the histone H1X, respectively. In conclusion, we provide evidence of the function of RALY in RNA metabolism in mammal cells and of its possible role in tumorigenesis.
MATERIALS AND METHODS
Cell cultures and transient transfections
HEK293T, HeLa, Jurkat, MiaPaca, MCF7, Panc-1 and PK9 cells were grown in DMEM supplemented with 10% FCS as previously described (27). MCF-10A cells were cultured in DMEM/F12 Ham's Mixture supplemented with 5% Equine Serum, 20 ng/ml EGF (Sigma), 10 μg/ml insulin (Sigma), 0.5 mg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), 100 units/ml penicillin and 100 μg/ml streptomycin. HPNE cells were grown in 75% DMEM, 25% M3 Base, 5% FCS, 10 ng/ml human recombinant EGF and 750 ng/ml of Puromycin. OliNeu cells (28) were grown in SATO medium supplemented with 2% FCS. Murine primary cultures of hippocampal neurons were prepared as previously described (29). Cells were transfected using either the TransIT transfection reagent (Mirus, Bio LLC) or INTERFERin (Polyplus Transfection) according to the manufacturer's protocols.
Constructs
Reverse transcription PCR was performed on total RNA isolated from HeLa cells using the TRIzol reagent (Thermo Fisher Scientific). RALY cDNA was amplified with the Phusion High-Fidelity DNA polymerase (New England BioLabs) and cloned in frame with EGFP (pEGFP-N1, Clontech). To downregulate RALY, cells were transfected with ON-target plus SMART-pool (Dharmacon) using INTERFERin transfection reagent. All primers used in this study are listed in the Supplementary Table S5.
Microarray analysis
MCF7 cells were grown on 10 cm Petri dishes. Total RNA was extracted from four biological replicates using the Agilent Total RNA Isolation Mini kit (Agilent Technologies) according to the manufacturer's protocol. RNA was quantified using the NanoDrop spectrophotometer (Thermo Fisher) and the quality assessed by Agilent 2100 Bioanalyzer. Hybridization, blocking and washing were performed according to Agilent protocol ‘One-Color Microarray-Based Gene Expression Analysis (Quick Amp Labeling)’. Probes were hybridized on a chip of Agilent-039494 SurePrint G3 Human GE v2 8 × 60K Microarray. Hybridized microarray slides were then scanned with an Agilent DNA Microarray Scanner (G2505C) at 5-micron resolution with the manufacturer's software (Agilent ScanControl 8.1.3). The scanned TIFF images were analyzed numerically and the background was corrected using the Agilent Feature Extraction Software version 10.7.7.1 according to the Agilent standard protocol GE1_107_Sep09. The output of Feature Extraction was analysed with the R software environment for statistical computing (http://www.r-project.org/) and the Bioconductor library of biostatistical packages (http://www.bioconductor.org/). Low signal Agilent features, distinguished by a repeated ‘undetected’ flag across the majority of the arrays in every condition, were filtered out from the analysis, leaving features corresponding to 15 126 HGNC genes. Signal intensities across arrays were normalized with the quantile method. Differentially expressed genes (DEGs) were identified using linear models and moderated t-test as implemented in the Bioconductor Limma package, using a double threshold on the log2 fold change (absolute value > 0.75) and the correspondent statistical significance (P-value < 0.05).
Preparation of cell extracts and western blot
Cells were washed with PBS and lysed in lysis buffer (0.1% Triton X-100, 150 mM KCl, 50 mM HEPES, pH 7.4 and 1 mM DTT plus proteinase inhibitor mixture (Roche), including 1 mM phenylmethylsulfonyl fluoride (PMSF)). Equal amounts of proteins were separated on 12% SDS-PAGE and blotted onto nitrocellulose (Schleicher & Schuell). Western blots were probed with the following antibodies: rabbit polyclonal anti-RALY (Bethyl); mouse monoclonal anti-hnRNP A1 (Novus Biologicals); mouse monoclonal anti-Annexin A1 (Santa Cruz Biotechnology); rabbit polyclonal anti-Histone H1X (Abcam); rabbit polyclonal anti-PABP (Abcam); rabbit polyclonal anti-RPL26 (Abcam); mouse monoclonal anti-Tubulin (Santa Cruz Biotechnology); mouse monoclonal anti-Actinin (Santa Cruz Biotechnology). Horseradish peroxidase (HRP)-conjugated goat anti-mouse and anti-rabbit antibodies (Santa Cruz Biotechnology) were used as secondary antibodies.
Polyribosome profiling and pharmacological treatments
Polyribosome analysis was performed as described in (30,31). Briefly, MCF7 cells were grown on 10 cm Petri dishes with DMEM. When they reached 80% of confluence, cells were incubated in medium supplemented with cycloheximide (0.01 mg/ml) for 3 min. Then cells were washed three times with cold PBS plus cycloheximide (0.01 mg/ml) and lysed with the lysis buffer (10 mM NaCl, 10 mM MgCl2, 10 mM Tris–HCl pH 7.5, 1% Triton X-100, 100 U/ml of RNase inhibitors from human placenta (NEB), 1 mM DTT, 0.01 mg/ml cycloheximide, 0.1% NaDeoxycholate, proteinase inhibitor mixture (Roche)). Mitochondria and nuclei free lysates were loaded onto 15–50% (w/v) density gradient of sucrose in solution A (100 mM NaCl, 10 mM MgCl2, 30 mM Tris–HCl pH 7.5), and ultracentrifuged at 180 000 × g for 100 min at 4°C. For RNase treatment, the lysates were incubated with RNaseI for 20 min at room temperature then loaded on the sucrose gradient. The RNaseI units added to the lysate were calculated as follows: 3 U × A × lysate volume, where A = (260 nm Abs lysate – Abs buffer) × 10. EDTA was added at the final concentration of 10 mM and incubated for 10 min. The treatment with Puromycin (0.1 mg/ml) was performed for 3 h in the dishes. The sedimentation profiles were monitored by absorbance at 254 nm using an ISCO UA-6 UV detector, and 1 ml of each fraction was collected. For Western blot analysis, proteins precipitated with TCA and acetone and the pellets were solubilized directly in Laemmli buffer pH 8.
RNA immunoprecipitation (RIP)
RIP was performed as reported by Keene et al. with some modifications (32). In brief, 4 × 107 cells were lysed in lysis buffer (10 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.5% NP40, 1 mM DTT plus RNase and proteinase inhibitors) for 3 h at –80°C and centrifuged at 10 000 × g for 20 min at 4°C. The supernatants were incubated for 4 h at 4°C with protein A/G magnetic beads coated either with antibody anti-RALY (3 μg) or with normal rabbit IgG polyclonal antibody (Millipore). The beads were then washed three times with NT2 buffer [50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40, 0.5% urea] and RNA was isolated with TRIzol (Thermo Fisher Scientific) and processed for qRT-PCR analysis or RIP-seq analysis as described below. For UV-crosslinked RIP, 4 × 107 of cells grown in 15 cm-culture dishes were irradiated once with 150 mJ/cm2 at 254 nm using an UVLink UV-crosslinker (Uvitec Cambridge).
RIP-Seq analysis
RNA from three independent RALY-IP and controls in MCF7 cells was converted to cDNA libraries according to the Illumina TruSeq RNA Sample Prep Guide (version 2). Single-end reads (1 × 100) were generated from the RALY RIP-seq libraries using three lanes of the Illumina HiSeq2000 sequencer according to the standard Illumina protocol. Reads were aligned to the human genome (GRCh38.p2) with Tophat (version 2.0.14), using the GENCODE 22 transcript annotation as transcriptome guide. All programs were used with default settings unless otherwise specified. Mapped reads were subsequently assembled into transcripts guided by reference annotation (GENCODE 22) with Cufflinks (version 2.2.1). Expression levels were quantified as normalized FPKM (fragments per kilobase of exon per million mapped fragments) using Cufflinks. Enriched transcripts were detected with CuffDiff with a double threshold on the log2 fold change (>1) and the correspondent statistical significance (P-value < 0.05). Functional annotation enrichment analysis with Gene Ontology terms and KEGG pathways were performed using the clusterProfiler Bioconductor package. In order to perform motif analysis, transcript sequences were retrieved from GENCODE 22. Control sequences for motif analysis were generated by selecting non-enriched transcripts (18334 transcripts with RIP-seq log2 fold change < –1). The significance of motif enrichments was measured applying a proportion test implemented in the prop.test function in R (P-value < 0.05).
RALY Bind-n-Seq data were downloaded from ENCODE (https://www.encodeproject.org/experiments/ENCSR495OAI/).
RNA pull-down
Biotinylated RNA probes were purchased by IDT. The wild-type or mutant probes (50 pmol) were captured with 35 μl of streptavidin-coupled Dynabeads (Thermo Fisher) for 20 min at room temperature in RNA Capture Buffer [20 mM Tris (pH 7.5), 1 M NaCl, 1 mM EDTA]. MCF7 cells were washed with PBS and then lysed with the lysis buffer [20 mM Tris (pH 7.5), 50 mM NaCl, 2 mM MgCl2, 0.1% Tween-20]. The lysate (200 μg) was incubated with biotinylated RNA probe coupled to streptavidin Dynabeads for 1 h at 4°C under rotation. Dynabeads were then washed three times with washing solution [20 mM Tris (pH 7.5), 10 mM NaCl, 0.1% Tween-20], solubilized in Laemmli reducing buffer and boiled for Western blot analysis. For the competition experiments, 30 μg of cell lysate were incubated with 30 pmol of wild-type biotinylated RNA probes together with either 300 pmol (10×) or 1200 pmol (40×) of non-biotinylated RNA probes (wild-type or mutant).
Cell cycle assay
The Cycletest Plus DNA Reagent Kit (BD Bioscience, USA) was used to distinguish the distribution of cells between cell-cycle phases four days after the transfection of si-RALY in Panc-1 cells. The protocol recommended by BD Bioscience was followed and flow cytometry (FACSCanto II, BD Bioscience) was used to run samples. The results obtained were analyzed using the ModFit software.
Quantitative real-time PCR (qRT-PCR)
Total RNA was purified using TRIzol (Thermo Fisher) according to the manufacturer's protocol. The total RNA concentration and purity of samples were assessed using Nanodrop spectrophotometer (Thermo Fisher). Next, 1 μg of RNA was subjected to DNase (Thermo Fisher) treatment and retro-transcribed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher) in 20 μl-reaction, according to manufacturer's instructions. Then, the quantification of transcripts in the samples was performed by real time qRT-PCR analysis using TaqMan probes and it was carry out in CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). Briefly, the qPCR reaction (10 μl) contained: 1× KAPA PROBE FAST, 1 μM primers plus TaqMan probe and 2 μl of cDNA template obtained in the previous step, diluted 1:4. The qPCR assay was performed with the following amplification program: 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s, followed by a hold at 4°C. Primers and TaqMan probes for ACTB, ANXA1, CST6, CXCR4, GAPDH, H1FX, RALY and TMCO1 transcripts were purchased by IDT (TEMA Ricerca) (see Supplementary Table S5). The results were analyzed with the Bio-Rad CFX Manager version 2.1. The relative expression was calculated according to the 2−ΔΔCt method. Each reported experiments was performed, at least, in biological triplicates and technical duplicates.
Immunocytochemistry and fluorescence microscopy
Cells were washed in PBS and then fixed in 4% PFA for 15 min at room temperature. Immunofluorescence was carried out as previously described (27). The following antibodies were used: rabbit polyclonal anti-RALY (Bethyl Laboratories), mouse anti-betaIII tubulin (Sigma-Aldrich), Alexa 488-coupled goat anti-rabbit IgG (ThermoFisher Scientific), and Alexa 594-coupled goat anti-mouse IgG (ThermoFisher Scientific). Microscopy analysis was performed using the Zeiss Observer Z.1 microscope implemented with the Zeiss ApoTome device. Pictures were acquired using AxioVision imaging software package (Zeiss) and assembled with Adobe Photoshop CS6.
Immunogold staining and atomic force microscopy imaging
HeLa cells treated with siRNA for RALY and siRNA CTRL, as described in the previous paragraph, were employed to obtain polysomal lysates. Polysomal purification by sucrose gradient fractionation was performed according to (31). Briefly, after ultracentrifugation, sucrose gradients were fractionated in 1 ml volume fractions with continuously monitoring absorbance at 254 nm using an ISCO UA-6 UV detector. Aliquots (20 μl) of sucrose fractions corresponding to Medium Molecular Weight polysomes were stored at –80°C before use. For immunogold staining a primary antibody anti-RALY (1:100) was incubated with the aliquot containing polysomes for 1 h at 4°C. Then, the sample was absorbed on mica as described in (33). After 1 h and 30 min incubation on the mica, the samples were washed with cold AFM buffer (10 mM HEPES, 10 mM MgCl2, 0.1 mM cycloheximide, 3% sucrose, 100 mM NaCl (pH 7.4)) in RNase-free water to remove the excess of the primary antibody. Then the samples were covered with cold AFM buffer containing the secondary antibody conjugated to the gold beads (Abcam 41514, 1:200). After 1 h and 30 min incubation on ice, the sample was washed three times using the cold AFM solution. Finally, the sample was fixed using a mild fixing solution (1% PFA in cold AFM buffer) for 10 min. All incubations were performed at 4°C. Before imaging, the sample was washed with cold RNase-free water, 0.1 mM cycloheximide to remove salts according to (33). AFM images were acquired on dried samples at a temperature of 26°C using an Asylum Research Cypher (Oxford Instruments), equipped with an environmental scanner. AC200TS cantilevers (Olympus, nominal spring constant 9 N/m) were driven in AC mode at a typical frequency of 150 kHz. AFM images were leveled line-by-line and analysed using Gwyddion (34).
RESULTS
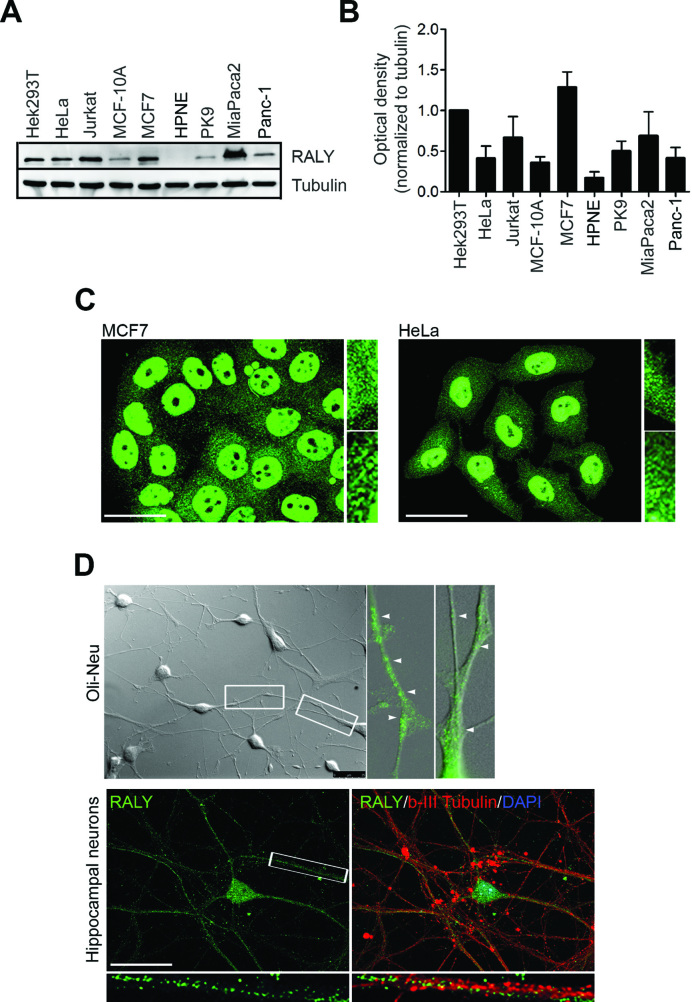
The human RNA-binding protein RALY consists of 306 amino acids with a predicted molecular mass of 36 kDa and shares 87.5% of identity with its murine homolog (35). Computational analysis of RALY protein predicted an RNA-recognition motif (RRM) at the N-terminal region followed by a consensus sequence for a bipartite nuclear localization signal (NLS1), a second NLS (NLS2), and a peculiar glycine-rich region (GRR) at the C-terminal region (27). We started to analyze the expression of RALY protein in several cell lines using a polyclonal antibody against the mammalian RALY (Figure 1A). The specificity of the antibody was verified by performing a competition assay (Supplementary Figure S1A).
Figure 1.
(A) Expression of RALY protein in different cell lines. Cell extracts were prepared from the indicated cell lines and analyzed by Western blot using anti-RALY and anti-Tubulin antibodies. The antibody anti-RALY recognizes a band at 36 kDa corresponding to the predicted full-length RALY protein. Tubulin served as a loading control. (B) Signals were quantified by band densitometry and normalized to tubulin signal (n = 3). (C) Representative fluorescent images showing RALY localization in MCF7 and HeLa cells. Scale bar = 25 μm. (D) Fluorescent images showing RALY localization in OliNeu cells, and hippocampal neurons. RALY forms discrete particles (arrowheads) distributed throughout the cytoplasm and at the periphery of the cells (insets magnified from boxed regions). Scale bar = 25 μm.
Compared to HEK293T cells, RALY was highly expressed by Jurkat, MCF7, and by the pancreatic carcinoma-derived cell line MiaPaca2 (Figure 1B). RALY was also expressed by the immortalized breast epithelial cell line MCF-10A and by Panc-1, a non-endocrine pancreatic cancer cell line. In contrast, low expression of RALY was observed in HPNE and PK9 cells, derived from the human pancreatic duct and from the pancreatic carcinoma, respectively (Figure 1A and B).
Immunostaining in both MCF7 and HeLa cells showed that RALY mainly localized in the nucleus with the exclusion of the nucleoli and also in the cytoplasm (Figure 1C) (27). In addition, we analyzed RALY expression in polarized cells such as Oli-Neu cells (36) and neurons. In these cells, RALY was detected as discrete granules in the cytoplasm as well as in distal processes (Figure 1D).
RALY associates with polysomes
The cytoplasmic localization of RALY prompted us to investigate its possible association to the translational machinery by studying the co-sedimentation profile of RALY with polysomes. MCF7 cells were lysed in the presence of cycloheximide and the cell extracts were fractionated on linear sucrose density gradients (37).
RALY co-sedimented with the 60S, 80S and polysome fractions in accordance with proteins associated with the translational machinery, namely the poly A-binding protein (PABP), the ribosomal protein L26 (RPL26) but not with hnRNP-A1, a nuclear hnRNP involved in the splicing of specific mRNAs (38,39) (Figure 2A).
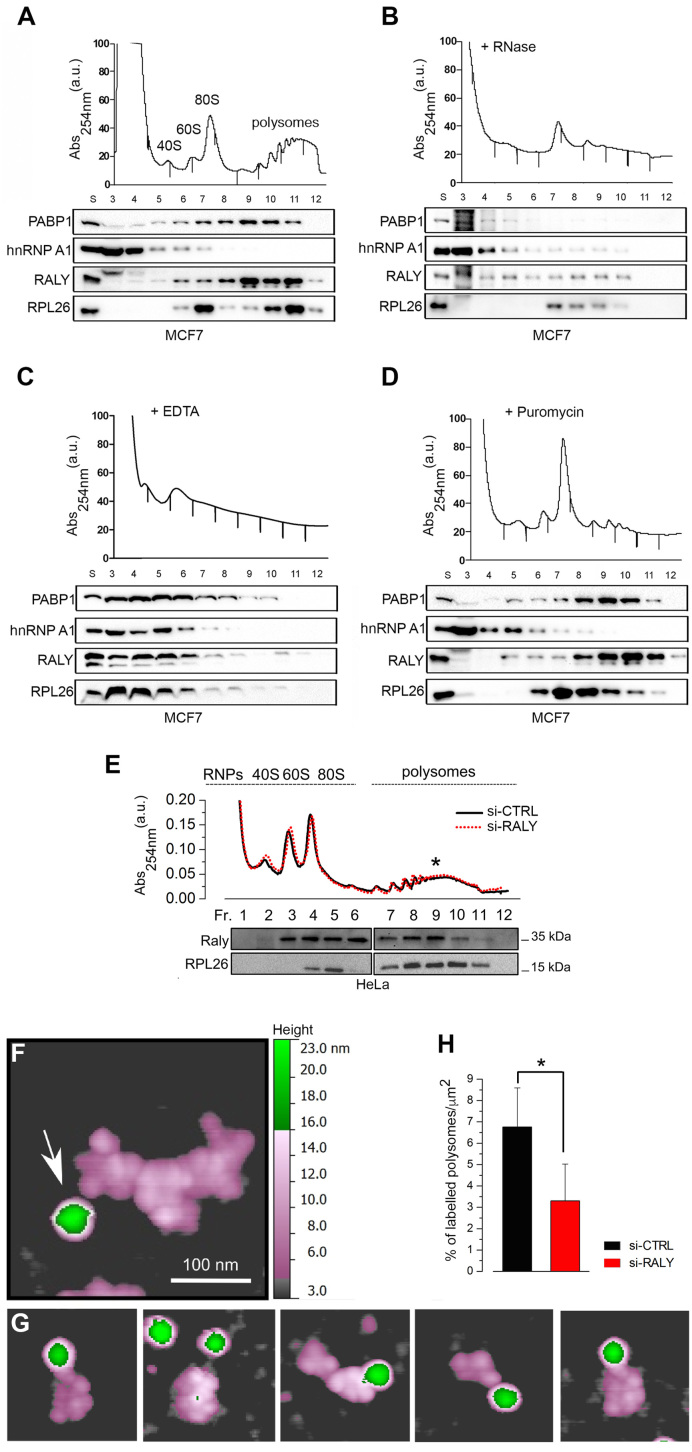
Figure 2.
RALY associates to a subset of polysomes. Absorbance profiles at 254 nm of sucrose gradient (15–50%) sedimentation of MCF7 cell extracts. The first peak contains free cytosolic light components (RNPs), the following peaks include ribosomal subunits (40S and 60S) and not translating monosomes (80S). The remaining peaks of the profile represent polysomes. Cellular extracts from MCF7 were either untreated (A) or treated with RNase (B), EDTA (C) or Puromycin (D). S, supernatant. Each gradient was fractionated and 10 fractions were collected, starting from the upper light fractions. The bottom panels show the Western blot analysis of RALY, PABP1, hnRNP A1 and RPL26 distribution in fractions 3–12. (E) Representative polysomal profiles of HeLa cells with and without RALY silencing (upper panel) after sucrose gradient fractionation. The star highlights the Medium Molecular Weight polysomes used for immunogold staining of RALY shown in panel F. The co-sedimentation profiles by Western blot of RALY and the ribosomal protein 26 (RPL26), used as control for co-sedimentation of the 60S and 80S, are shown in the lower panel. Each lane corresponds to a sucrose fraction and is located in correspondence to the polysomal profiles. The proteins extracted from sucrose fractions have been divided into two gels for technical reasons. The two blots were run in parallel and incubated together with the primary antibodies to avoid differences in the hybridization. The images were acquired at the same exposure time. (F) Example of a typical polysome after immunogold staining of RALY on Medium Molecular Weight (MMW) polysomes from HeLa cells. Purified polysomes were incubated with a primary antibody against RALY in combination with a secondary antibody conjugated with gold beads (25 nm diameter). Coloured height scale was used to highlight different height levels of objects: 0–3 nm gray background, 3–15 nm violet ribosomes; 15–23 nm green gold bead. The white arrow indicates a bead (in green corresponding to 25 nm in height) and the ribosomes in polysomes (Figure 2F and G) are in violet corresponding to 14 nm in height. (G) Gallery of MMW polysomes bearing a gold bead. (H) Percentage of positively labeled MMW polysomes in HeLa cells with and without RALY silencing. We identified and counted the number of polysomes per μm2 associated to the gold beads over the total number of polysomes observed per μm2. The total number of polysomes analysed was n = 416 (HeLa, si-control) and n = 324 (HeLa with si-RALY). A significant difference (P = 0.037) between the samples was assessed using two-tailed Student's t-test (*P < 0.05).
To determine if the association of RALY with polyribosomes was RNA-dependent, cell lysates were incubated with RNaseI before sucrose fractionation (Figure 2B). The treatment with RNaseI degrades single-stranded RNA preserving ribosome-protected fragments and inducing the disassembly of polysomes into single ribosomes (80S). Under these conditions, the co-sedimentation profile of RALY partially shifted to lighter fractions along the sucrose gradient, similarly to PABP (Figure 2B). The vast majority of RALY remained associated to 80S and polysomes, suggesting that its recruitment to the translational machinery may occur through both RNA and protein–protein interactions.
To further investigate the association of RALY with the polysomal fractions, we treated the cell lysate with EDTA before fractionation. This treatment disrupts the 40S–60S interaction of any ribosome. Indeed, the polysomes and 80S peaks disappeared, while 40S and 60S were maintained (Figure 2C). Under these conditions, the co-sedimentation profile of RALY shifted to lighter fractions, similarly to PABP and RPL26 (Figure 2C).
To test whether RALY was associated to actively translating polysomes, MCF7 cells were treated with the translation inhibitor Puromycin for 3 h. Puromycin is known to block the elongation step of translation causing a premature release of the growing polypeptide. Under these conditions, the sedimentation profile of RALY did not change (see Figure 2A) and RALY remained associated with the so-called Puromycin-insensitive polysomes (Figure 2D). The same pattern was observed for PABP, while RPL26 shifted to monosomes-containing fractions (Figure 2D). As expected, the sedimentation profile of hnRNP-A1 was not modified by any treatment. Since Puromycin-insensitive polysomes are considered as translationally inactive (40,41), it is tempting to speculate that RALY might have a potential role in the translational control of specific transcripts that will be a matter of future investigation.
To assess and quantify the association of RALY on the translation machinery, we took advantage of atomic force microscopy (AFM) imaging of polysomes (31,33) combined with immunogold labeling in HeLa cells (42–44). We analysed the localization of RALY within polysomes utilizing a secondary antibody conjugated to gold nanoparticles of 25 nm diameter. The height of the bead in AFM was 10 nm larger than that of mammalian ribosomes in polysomes observed in air-dried samples (around 14 nm) (31,33). This difference in height, allows to easily identifying the beads and distinguishing them from the ribosomes. The sucrose fraction containing the highest amount of RALY by western blotting (Figure 2E, lower panel) and corresponding to medium molecular weight polysomes (Figure 2E, star) were incubated with a primary antibody against RALY in combination with a secondary antibody conjugated with gold beads. Using a colored height scale, we identified the beads associated to the anti-RALY antibody (Figure 2F, white arrow) and the ribosomes in polysomes (Figure 2F and G). Then, we calculated the percentage of RALY-positive polysomes by counting the number of polysomes per μm2 associated to the gold beads over the total number of polysomes observed per μm2. We observed that 7 ± 2% of MMW was positively stained versus 3 ± 2% in si-RALY conditions (Figure 2H). Taken together, these results suggest that RALY is bound to a sub-population of polysomes and that the protein may be associated with specific, and not all, transcripts engaged with the translational machinery.
Identification of RALY-associated transcripts
We comprehensively identified the RALY-associated transcriptome by sequencing after RNA immunoprecipitation (RIP-seq) of RALY-complexes and assessed possible variations in the global transcriptome of RALY downregulated cells by microarray analysis. Both analyses were performed in MCF7 cells given their high levels of endogenous RALY.
Before performing the RIP-seq, we verified whether RNA could be purified from RALY-RNPs by performing RNA immunoprecipitation. After immunoprecipitation with either anti-RALY antibody or anti-rabbit IgG (Supplementary Figure S2A), RNA was extracted and run on a denaturing polyacrylamide-urea gel (Supplementary Figure S2B). We observed that RNA was immunoprecipitated. To confirm the nature of nucleic acids co-immunoprecipitated with RALY, we treated the samples with RNaseA and observed the disappearance of the signal. As expected, no RNA was immunoprecipitated when rabbit IgG was used as a negative control (Supplementary Figure S2B).
We then transfected MCF7 cells with either a myc-tagged wild-type RALY or a RRM-deficient mutant and extracted the RNA after the immunoprecipitation. We detected the presence of RNA only in the immunoprecipitated wild-type RALY, confirming the ability of RALY to bind RNA through its RRM (Supplementary Figure S2C) (27). These results demonstrate that endogenous RALY-RNP complexes can be efficiently immunoprecipitated together with the associated RNAs.
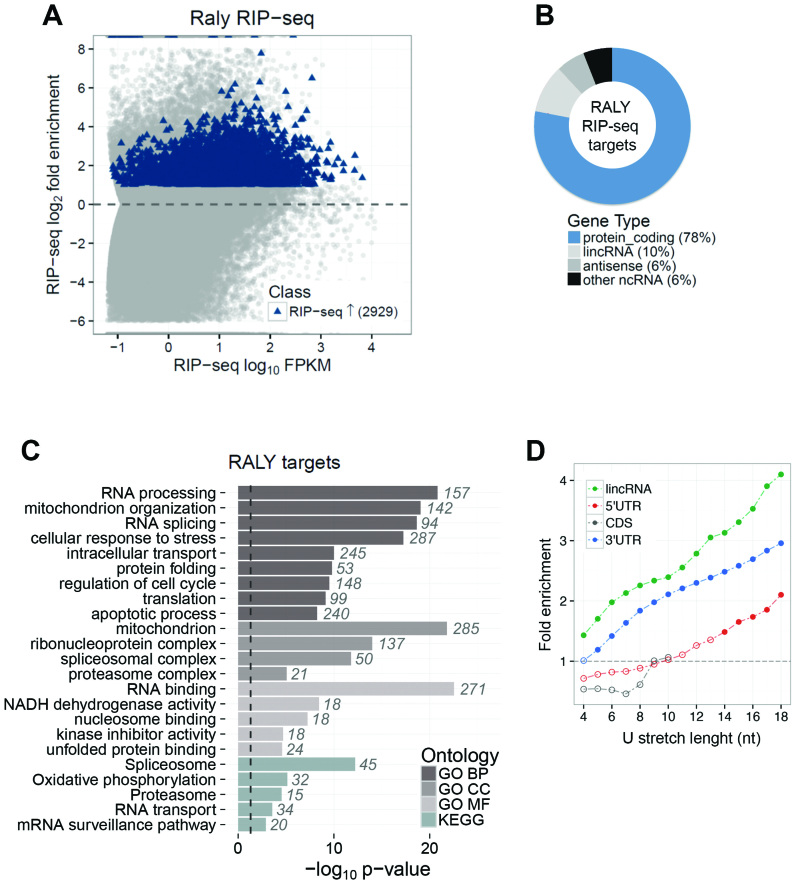
By sequencing the RNAs associated with RALY we found that 2929 transcripts were significantly enriched in the immunoprecipitated RALY-RNPs (Figure 3A and Supplementary Table S1). Most of the identified RNAs were protein-coding transcripts (78%), while long intervening noncoding RNAs (lincRNAs) and antisense RNAs were the most represented classes of non-coding RNAs (10% and 6% respectively) (Figure 3B). Gene ontology and pathway annotation enrichment analyses revealed that proteins coded by RNAs interacting with RALY were significantly associated with different RNA metabolism related processes such as RNA processing, splicing, translation and RNA transport (Figure 3C). Consistently, transcripts associated with RALY were enriched in RNA-binding proteins that are components of RNP complexes. Interestingly, other biological processes such as regulation of cell cycle and apoptosis were also significantly enriched (Supplementary Table S2).
Figure 3.
Identification of RALY-associated RIP-seq analysis performed in MCF7 cells. The total and the immunoprecipitated RNA were fragmented, retro-transcribed to cDNA, ligated with adapters, amplified and sequenced. Three independent experiments were carried out using an anti-RALY polyclonal antibody to immunoprecipitate endogenous RALY-containing RNPs. (A) MA plot of RALY RIP-seq. For each transcript, the average signal (measured as log10 FPKM) against the RIP-seq log2 Fold Enrichment (RIP versus INPUT) was plotted. Significantly enriched targets are highlighted in blue. (B) Classes of RALY-associated RNAs. The majority (78%) of identified targets are coding protein genes, but long intervening noncoding RNAs (lincRNAs) and antisense RNAs were also present (10% and 6%, respectively). (C) Functional annotation enrichment of RALY target genes. The barplot displays enriched classes from Gene Ontology terms (BP, biological process; CC, cellular component; MF, molecular function) and KEGG pathways. The length of each bar is proportional to the statistical significance of the enrichment. The number of RALY targets falling in each category is displayed beside the corresponding bar. (D) Enrichment of poly-U stretches of varying length (from 4 to 18 nucleotides) in the sequence of RALY targets compared to non-target sequences. The 5΄UTR, coding sequences (CDS), 3΄UTR sequences of RALY protein coding targets as well as lincRNA targets were analyzed separately. CDS regions do not have occurrences of poly-U stretches longer than 10 nucleotides. Filled dots highlight significant enrichments.
It has been reported that in vitro RALY might bind poly-U stretches with as high affinity as hnRNP-C (45). This affinity is also supported by the ENCODE data on RALY examined by RNA Bind-n-Seq (46). The analysis of these data showed that RALY unequivocally favors poly-U stretches in vitro (Supplementary Figure S3). Then, we performed motif enrichment analysis on the sequences of RALY interacting RNAs, comparing the occurrences of poly-U elements characterized by varying lengths with respect to non-interacting RNAs. This analysis revealed a significant enrichment of poly-U elements in the 3΄ UTRs as well as in the target lincRNAs. Enrichment in 5΄ UTR was significant only for longer poly-U stretches (>13 nt). On the contrary, the coding sequences of mRNAs interacting with RALY were not enriched in poly-U elements, suggesting a preferential binding of RALY to the 3΄ UTRs (Figure 3D).
RALY silencing affects gene expression
We then analyzed the impact of RALY downregulation on global gene expression. MCF7 cells were transfected either with siRNA targeting RALY or mismatch siRNA as negative control and the total RNA was isolated and analyzed using a whole human genome microarray. Compared to siRNA mismatch-treated cells, Western blot analysis confirmed that RALY was efficiently downregulated by 90% after 72 h (see Figure 6A). In contrast, the levels of hnRNP-C were not affected (Supplementary Figure S1B).
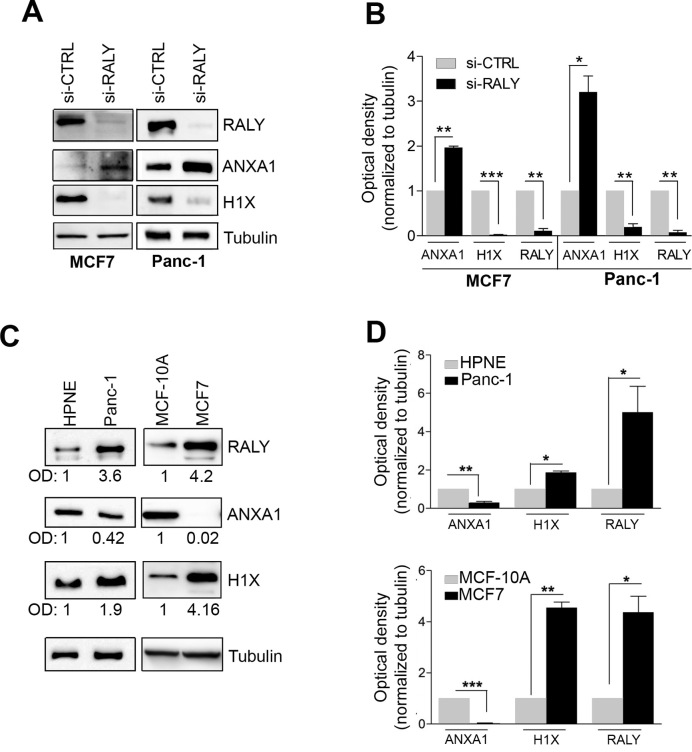
Figure 6.
RALY regulates ANXA1 and H1X protein levels. (A) Protein expression of RALY, ANXA1 and H1X in MCF7 and Panc-1 cell lines transfected with siRNA against RALY (si-RALY) and siRNA mismatch (si-CTRL). The endogenous levels of the indicated proteins were analysed by Western blotting using specific antibodies. In both cells lines, the downregulation of RALY caused a significant upregulation of ANXA1 protein and a reduction of H1X compared to controls. (B) The levels of ANXA1 and H1X were quantified by band densitometry analysis. The barplot shows the mean values of three independent experiments after normalization to Tubulin. Bars represent mean ± S.E.M. P-value was calculated using unpaired two-tailed t-test. *P < 0.05; **P < 0.01; ***P < 0.001 (C) Comparison of the expression of the endogenous RALY, ANXA1 and H1X proteins. Western blot analysis showed that in cancer cell lines (MCF7 and Panc-1) higher expression level of RALY is associated to a higher H1X and lower ANXA1 levels compared to non-tumorigenic cell lines (MCF10A and HPNE, respectively). The optical densitometry analysis (O.D.) normalized to tubulin is reported for each panel. (D) The graphs show the mean values of three independent experiments after normalization to Tubulin. Bars represent mean ± S.E.M. P-value was calculated using unpaired two-tailed t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
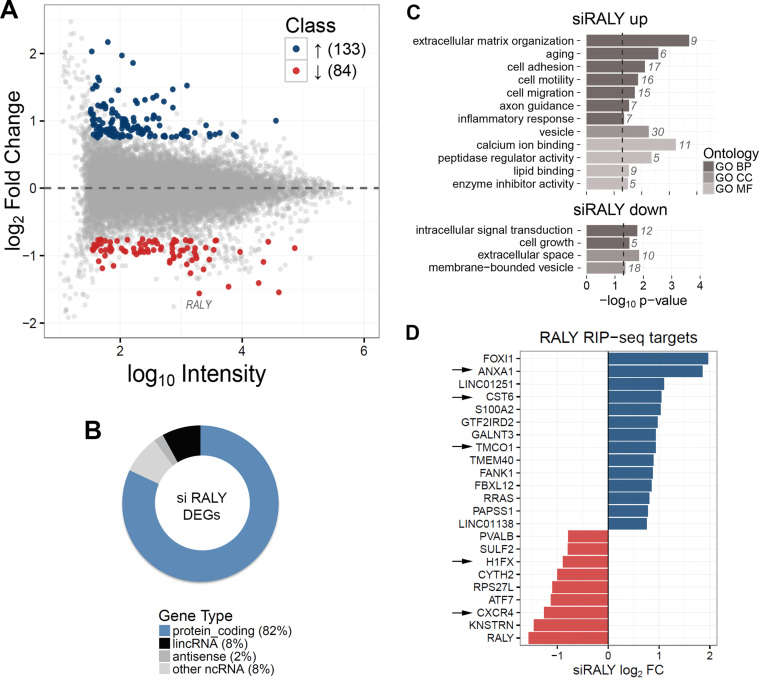
Out of 217 differentially expressed genes after RALY silencing (Supplementary Table S3), 133 were upregulated and 84 were downregulated (Figure 4A). As for the RIP-seq results, the majority of the identified RNAs was protein-coding genes, 8% was lincRNAs, 2% antisense RNAs and 8% other non-coding RNAs (Figure 4B). Gene Ontology enrichment analysis identified extracellular matrix organization, cell adhesion and cell motility as biological processes significantly associated with upregulated genes, while cell growth and intracellular signal transduction were significantly associated with downregulated genes (Figure 4C and Supplementary Table S4).
Figure 4.
Identification of up and downregulated genes upon RALY silencing in MCF7 cells. Four independent microarray experiments were performed using RNA preparations in four independent biological replicates. (A) MA plot of transcriptome profiling in RALY silenced MCF7. For each gene, the average log10 signal against the RALY silencing log2 Fold Change (siRALY versus control) is plotted. Genes significantly up- (blue) or down- (red) regulated upon RALY silencing are highlighted. (B) Classification of differentially expressed genes (DEGs) upon RALY silencing according to RNA classes. The majority (82%) of transcripts is protein-coding genes. LincRNAs (8%) and antisense RNAs (2%) are also present. (C) Functional annotation enrichment analysis of RALY upregulated and downregulated genes. The barplot displays enriched classes from Gene Ontology terms (BP, biological process; CC, cellular component; MF, molecular function). (D) Intersection of the list of RALY RIP-seq targets and the list of siRALY differentially expressed genes. Blue and red bars represent up and down DEGs, respectively. The arrows indicate the experimentally validated genes.
By intersecting the list of RNAs identified by RIP-seq with the list of differentially expressed genes upon RALY silencing, we obtained a set of 23 RNAs present in both lists (Figure 4D). Among these RNAs, we focused on the following transcripts: annexin A1 (ANXA1), the chemokine C-X-C motif receptor 4 (CXCR4), Cystatin-M (CST6), the transmembrane and coiled-coil domains 1 (TMCO1), and histone H1X (H1FX) (Figure 4D, arrows).
Validation of RALY-regulated transcripts
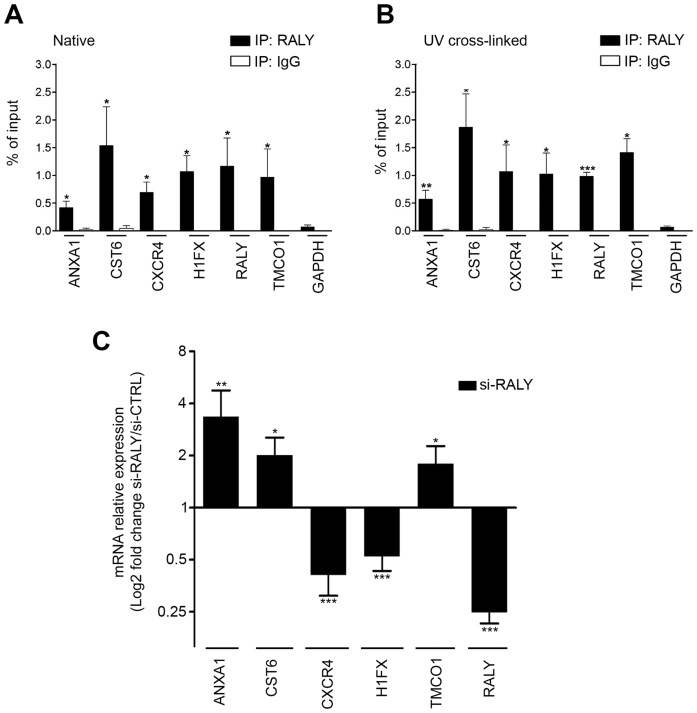
We validated the enrichment of the identified transcripts as components of RALY-RNPs by qRT-PCR after RALY immunoprecipitation. As expected, all the above-mentioned transcripts were significantly enriched in RALY immunoprecipitates compared to rabbit IgG. Since it was not identified by RIP-seq, we also analyzed GAPDH mRNA as negative control (Figure 5A). In particular, we observed that H1FX, CST6 and TMCO1 mRNAs were highly represented in RALY-RNP. Interestingly, RALY mRNA was also found in the immunoprecipitated complexes suggesting the potential interaction of RALY protein with its own mRNA (Figure 5A). The same mRNAs were enriched when the RIPs were performed after UV cross-linking (Figure 5B).
Figure 5.
Validation of the identified RALY-associated mRNAs (A) qRT-PCR was used to compare the indicated mRNAs isolated upon RALY immunoprecipitation with RNA recovered after immunoprecipitation with IgG. All the relative abundances were compared to 10% of input. The experiments were performed at least three times. Bars represent means ± S.E.M. P-value was calculated comparing the amount of each sample with the amount of GAPDH using an unpaired two tailed t-test (*P < 0.05; **P < 0.01; ***P < 0.001). (B) qRT-PCR was used to compare the indicated mRNAs isolated upon UV-crosslinked RALY immunoprecipitation with RNA recovered after immunoprecipitation with IgG. The experiments were performed at least three times. Bars represent means ± S.E.M. P-value was calculated comparing the amount of each sample with the amount of GAPDH using an unpaired two tailed t-test (*P < 0.05; **P < 0.01; ***P < 0.001). (C) qRT-PCR was performed to assess the levels of the indicated transcripts after silencing of RALY in MCF7 cells. Results are presented in terms of change after normalizing to GAPDH mRNA. Each value represents the mean of at least three independent experiments ± S.E.M. P-value was calculated using an unpaired two-tailed t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
We confirmed the specificity of these results by performing UV cross-linked RIP from MCF7 cells expressing myc-tagged RALY (Supplementary Figure S2D). In contrast to wild-type RALY, none of the analyzed mRNAs was enriched with RALY lacking the RRM domain (Supplementary Figure S2D).
Then, we validated the potential misregulation of the above mRNAs in the absence of RALY by qRT-PCR. The results confirmed the upregulation of ANXA1, CST6 and TMCO1 and the downregulation of CXCR4 and H1FX in RALY deficient cells (Figure 5C). Taken together, these results identified a specific set of mRNAs able to interact with RALY-RNPs and whose cellular levels are regulated by the expression of RALY protein.
RALY modulates the expression of ANXA1 and H1FX
Among the six validated mRNAs, we decided to investigate how RALY affected the amount of ANXA1 and H1FX mRNAs. Annexin A1 (ANXA1) is a 37 kDa calcium and phospholipid-binding protein that is involved in different biological processes, such as regulation of cell proliferation, cell death signaling, and apoptosis (47,48). The histone H1X (H1X) is a member of the ubiquitously expressed H1 histone family, composed of 11 subtypes of linker histones which bind the linker DNA between the nucleosome cores, and stabilizes compact, higher order structures of chromatin (49–51).
Since the levels of ANXA1 and H1FX mRNAs were regulated by RALY expression, we checked the protein level of ANXA1 and H1X in MCF7 and Panc-1 cells transfected with either control siRNA or siRNA against RALY (Figure 6A and B). In both cell lines, we observed a significant increase of ANXA1 when RALY was downregulated compared to control siRNA-transfected cells. In contrast, a significant decrease of H1X was observed. No change was detected for tubulin that was used as a negative control (Figure 6A and B).
To assess if RALY downregulation could change the nuclear versus cytoplasmic localization of H1X and ANXA1 proteins, we performed cell fractionation. No changes in the subcellular distribution was observed suggesting that the downregulation of RALY did not affect the localization of the H1X and ANXA1 proteins (Supplementary Figure S4A).
A further proof that RALY contributed to regulate ANXA1 and H1X levels came from experiments in which RALY was knocked out using CRISPR/Cas9 technology. Consistently with previous results, ANXA1 and H1X were altered at both transcriptional and protein levels (Supplementary Figure S4B and S4C and data not shown). In addition, when we overexpressed RALY in both HEK293T and HPNE cells, we observed an upregulation of H1X and a downregulation of ANXA1 proteins (Supplementary Figure S4D).
Based on these results we speculated that cells expressing a different amount of endogenous RALY might show variations in both ANXA1 and H1X expression. Thus, we verified this hypothesis by checking the expression of ANXA1 and H1X in HPNE and Panc-1 pancreatic cell lines as well as in MCF10A and MCF7 breast cell lines (Figure 6C and D). We observed that the expression of ANXA1 protein was lower in Panc-1 compared to HPNE, while H1X protein level was higher. The same results were found when comparing MCF7 with MCF10A cells, confirming that the effects on ANXA1 and H1X expression levels associated with the expression of the endogenous RALY.
RALY binds the poly-U elements of ANXA1 and H1FX 3΄UTRs
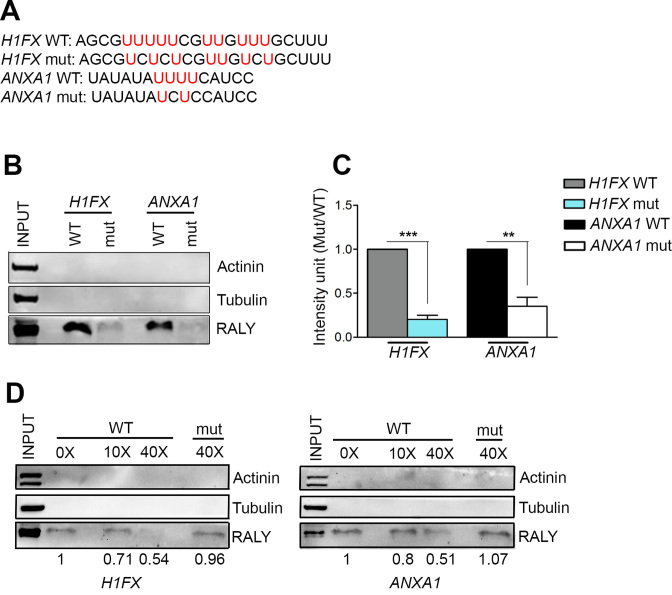
Our RIP-seq analysis showed that RALY could potentially recognize poly-U-rich stretches within the 3΄UTRs (Figure 3D). We checked for poly-U-rich elements within the 3΄ UTR of ANXA1, H1FX and RALY itself. We identified one poly-U-rich element in the 3΄UTR of RALY, seven in H1FX and four in ANXA1 (Supplementary Figure S5A).
To confirm that RALY binds ANXA1 and H1FX mRNAs in correspondence to poly-U we used a pull-down assay. We incubated biotinylated synthetic probes containing either wild-type or mutant poly-U-rich elements with MCF7 cell lysate, and assessed RALY association by Western blot (Figure 7A). As expected, we detected high signals for wild-type probes of H1FX and ANXA1 indicating a robust interaction. In contrast, a reduced association was observed with both mutated probes (Figure 7B and C). Similar results were obtained for CXCR4, another RALY mRNA target that contains 12 stretches of poly-U within the 3΄UTR (Supplementary Figure S5A and S5B).
Figure 7.
RALY binds the poly-U regions. (A) Sequences of biotinylated wild-type and mutant H1FX and ANXA1 3΄UTR probes. (B) MCF7 cells total protein extract was incubated with either wild-type or mutant biotinylated RNA probes (50 pmol) and captured by streptavidin beads. Western blot of RNA-bound protein fraction shows the positive interaction in vitro of RALY with the poly-U wild-type sequences, but not with mutant probes. Immunoblotting with anti-Actinin and anti-Tubulin served as a negative control. (B) The graphs show the mean values of three independent experiments. Bars represent mean ± S.E.M. P-value was calculated by unpaired two-tailed t-test (*P < 0.05; **P < 0.01; ***P < 0.001). (C) RNA pull-down competition assay was performed to assess the RNA-binding specificity of RALY. 30 pmol of either H1FX or ANXA1 wild-type probes were incubated with increasing amounts (10× and 40×, respectively) of non-biotinylated wild-type or 40× mutant probes and successively mixed with 30 μg of MCF7 cells extract. Western blot shows the amount of RALY bound to the probes in the different competitive conditions and quantified by band densitometry analysis. Immunoblotting with anti-Actinin and anti-Tubulin served as a negative control.
Three different biotinylated probes designed within the 3΄UTR were tested by RNA-pulldown assay, finding the strongest signal associated with the probe containing 8 poly-U stretches. In addition, the specificity of RALY interaction with wild-type H1FX and ANXA1 probes was confirmed by a competition assay with increasing amount of either wild-type or mutated non-biotinylated probes with wild-type biotinylated probes (Figure 7D). Taken together, these data confirmed that RALY binds to the poly-U elements of ANXA1 and H1FX mRNAs.
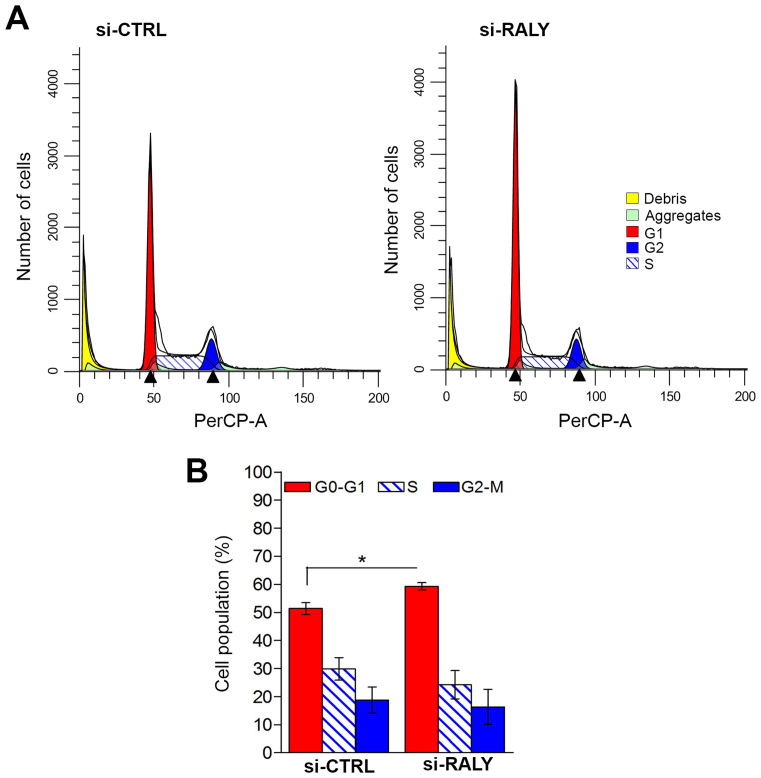
RALY downregulation affects cell cycle progression
It has been reported that the upregulation of ANXA1 suppresses cell proliferation, invasion and migration (52–54). These evidences, together with the observation that Panc-1 cells lacking RALY halt the proliferation six days after transfection, prompted us to check whether RALY downregulation affected cell proliferation. We silenced RALY in Panc-1 cells and performed cell cycle analyses measuring cellular DNA content by staining with propidium. FACS analysis revealed that a statistically significant population of Panc-1 cells stopped at the G1 phase of their cell cycle (Figure 8A and B). These results were consistent with a reduced proportion of cells in S and G2-M phases (5.6% and 2.5% of cells, respectively). In summary, the silencing of RALY contributes to the alteration of cell progression with a partial arrest of cells in G0–G1 phase.
Figure 8.
Down-regulation of RALY increases the number of Panc-1 cells in G1 phase of the cell cycle. (A) Control and si-RALY Panc-1 cells were fixed and stained with propidium iodide (PI). DNA content was analyzed by flow cytometry. Charts show a flow cytometric analysis of si-CTRL (left) and si-RALY (right) transfected Panc-1 cells in a representative experiment. 30,000 events were analyzed in each experiments. (B) The percent of cells in each phase was analyzed by ModFit LT (Verity Software). The graph represents the percentage of cell distribution and the mean of three independent experiments. Error bars, S.D; P = 0.0049 was calculated with unpaired two-tailed t-test. si-CTRL G0-G1 = 51.35 ± 1.87; si-RALY G0–G1 = 59.26 ± 0.75.
DISCUSSION
This study provides a characterization of RALY, a protein that belongs to the class of the RNA-binding proteins hnRNPs (55–59). RALY, previously known as hnRNP-C like-protein 2, shares common features with the majority of the hnRNPs: it contains a single RRM at the N-terminal region sharing 43% homology with hnRNP C, a glycine-rich region at the C-terminus and two uncharacterized NLS that have been predicted by computational analysis (27).
Although the hnRNPs are the most abundant nuclear proteins, some of them may form RNPs complexes shuttling from the nucleus into the cytoplasm. Here, they can be transported to distinct intracellular compartments together with their mRNA cargo (59). RALY was identified as a common component of Staufen2 and Barentsz transport RNPs (60). These proteins are two RBPs well characterized in the context of RNA transport, translational control and non-sense mediated decay in nerve cells and many other cell types (61–67).
Using biochemical gradient fractionation, we demonstrated that RALY localizes in the cytoplasmic compartment where it is able to co-sediment with ribosomes and polysomes (Figure 2). To test whether RALY was associated to either actively translating or stalled polyribosomes, we studied the changes of RALY co-sedimentation profile along the polysomal fractions upon treatment of MCF7 cells with the translation inhibitor Puromycin. Since this inhibitor is known to interfere with the elongation step of translation, all polysomes that remain insensitive to the treatment are stalled (40). Therefore, the co-sedimentation profile of the protein after Puromycin treatment is an indication of an activating or repressive function in translation (37,41,68). Under our conditions, Puromycin mildly disrupted RALY-polyribosomes associated complexes (Figure 2D), suggesting that RALY might have a potential role in the translational control of specific transcripts as FMRP does (41).
These findings confirm our previous study reporting the interactions of RALY with ribosomal proteins and with proteins involved in translation control (27). Since RALY is detected in distal processes of polarized cells such as oligodendrocytes and neurons, it would be interesting to investigate if RALY is involved in the transport and/or translational control of specific mRNAs in these compartments.
The unknown identities of RALY associated RNAs prompted us to identify the RALY–RNA interactome and to study its RNA-binding properties in mammalian cells taking advantage of two high-throughput approaches, the RIP-seq and microarray analyses (Figures 3 and 4). RIP-seq experiments identified 2929 RNA targets and the major class comprised protein-coding RNAs. Microarray experiments detected 217 RNAs whose expression is altered upon RALY silencing.
Among the jointly identified transcripts, we focused on ANXA1 and HF1X. Annexin A1 (ANXA1) is a well-studied calcium and phospholipid-binding protein that is involved in different biological processes: regulation of cell proliferation, cell death signalling and apoptosis (45,47). Although ANXA1 has been implicated in the biology of various tumors, the molecular mechanisms by which ANXA1 participates to carcinogenesis and tumor progression remains unclear and rather controversial (69).
The histone H1X is a member of the H1 histone family that binds the linker DNA between two nucleosomes contributing to stabilize the chromatin fibers (70,71). Despite previous convincement, it has become clear that linker histones are not merely static and structural components of chromatin but rather dynamic elements with important regulatory proprieties (72–74). For example, H1X shows a role in embryonic differentiation processes by regulating the expression of Nanog, a gene marker of stemness that is significantly suppressed in differentiated cells (75). Recent findings demonstrate an emerging role of H1X in cancer development and it has been proposed as a prognostic biomarker (76). Although some drugs have been used to modify the expression of H1X, the intracellular mechanisms involved in its regulation are still poorly characterized (75).
By computational analysis and biochemical RNA-pull down experiments, we discovered that RALY preferentially associated with the 3΄UTR of poly-U containing RNAs (Figures 3 and 7). We showed that RALY binds the poly-U stretches within the 3΄UTRs of these genes, and its downregulation could regulate both ANXA1 and H1X at mRNA as well as at protein levels (Figures 5–7). In addition, our data show that ANXA1 and H1X expression is correlated with the intracellular levels of RALY (Figure 6 and Supplementary Figure S4).
According to these findings, another member of the hnRNP family, the hnRNP-C, was originally identified as a poly-U binding protein (77). More recently, Ray and colleagues identified several RNA motifs, including poly-U stretches, which can be recognized by different RNA-binding proteins (44). Poly-U motifs are common sequences also within Alu elements, the most abundant class of transposable elements in primates (78). Interestingly, the presence of Alu elements can influence splicing. Splicing factors such as U2AF65 and TIA1 can recognize the poly-U tracts within Alu elements and create cryptic or alternative Alu exons (78). The binding of hnRNP-C to these elements can compete with the molecular components of splicing machinery preventing the formation of aberrant splicing events (79,80).
Our data show that RALY silencing alters H1FX and ANXA1 expression levels, but not their translational efficiency (data not shown). In addition, we blocked de novo transcription with Actinomycin D and analysed if RALY could affect ANXA1 and H1FX RNA-stability, in MCF7 cells, by comparing the half-lives of both transcripts. Surprisingly, we found no significant differences in the stability in the absence of RALY compared to control (data not shown). Hence, we hypothesize that RALY exerts a transcriptional control of these targets. Since the 3΄UTRs act as a scaffold where many proteins and microRNAs form complexes for post-transcriptional and/or translational regulation (81,82), RALY might compete or cooperate with one or more of these factors in regulating the level of these specific transcripts. RNA binding proteins can be recruited to promoter regions during transcription and they can interact with RNA polymerase II as well as with transcription factors and then bind to the nascent RNA (83). The lack of RNA-binding proteins, showing also DNA binding properties, can affect RNA metabolism at different levels (84,85).
Non-coding RNAs can modulate the activities of classes of RNA binding proteins in response to specific signals (86). While this manuscript was in preparation, a work reported that RALY can act as a transcriptional cofactor for genes involved in cholesterol biosynthesis (87). In particular, RALY was found associated with gene promoters and the long non-coding RNA LeXis inhibited this promoter-binding activity. Although LeXis was not in our list, our RIP-seq revealed that more than 20% of transcripts in RALY immunoprecipitates comprised lincRNA, antisense and ncRNAs (Figure 3). Interestingly, their sequences are also enriched in stretches of poly-U. Taken together, we hypothesize that RALY exerts a dual control, one at the transcriptional level that is mediated by ncRNAs and one posttranscriptionally mediated by its binding to the poly-U within the 3΄ UTRs of coding RNAs.
The involvement of hnRNPs in pathologies is also well documented (88–90). For example, several hnRNPs have been implicated in cancer development and aberrant expression of hnRNPs is characteristic of various types of cancer (3,91). RALY is upregulated in metastatic nasopharyngeal carcinoma cell lines compared to normal cells (92). Moreover, cells with upregulated levels of RALY and NONO become more resistant to the effects of the drug oxaliplatin (19). In contrast, the depletion of RALY expression by RNAi sensitized colorectal cancer cell lines treated with the oxaliplatin without affecting the cell growth rate (19).
We observed that RALY is expressed more in tumor cell lines such as MCF7 and PK9, MiaPaca2 and Panc-1 compared to non-tumorigenic cell lines such as MCF10A and HPNE (Figure 1A and B). Studying the role of hnRNPs in cell cycle regulation and cell differentiation has recently opened a new exciting area of investigation (93). Our data together with data present in public data bank (e.g. The Human Protein Atlas) suggest a possible functional link between RALY expression and cell proliferation. The knockdown of RALY in Panc-1 cells causes a moderate but significant arrest of the cell cycle with a discrete number of cells blocked in G0/G1 phase (Figure 8). This finding might be the result of the altered expression of ANXA1 and/or H1X but we cannot exclude that it might be due to alteration of other RALY RNA targets involved in controlling cell cycle progression. For example, FBXL12 transcript was identified as one RNA bound by RALY in our RIP-seq and it was found to be upregulated upon RALY downregulation (Figure 4D). In lung epithelia, FBXL12 protein induces G1 cell cycle arrest by promoting the proteasomal degradation of CAMK1 that triggers the disruption of the cyclin D1/CDK4 complex (94). An overexpression of FBXL12 due to RALY downregulation might result in an inhibition of cell cycle progression.
Taken together, we suggest a role of RALY as a possible oncogene during tumorigenesis. Further work will now be undertaken to define the precise mechanism of action.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Veronica De Sanctis and Dr Roberto Bertorelli (NGS Facility at the Centre for Integrative Biology and LaBSSAH, University of Trento) for NGS sequencing and helpful discussions. We are grateful to Dr Valentina Adami and Dr Michael Pancher (CIBIO-High Throughput Screening Core Facility) for the technical assistance during the microarray analysis. We also thank Dr Isabella Pesce (CIBIO-Cell Analysis and Separation Core) for the technical assistance during FACS analysis.
We acknowledge Dr Paola Zuccotti for the assistance in polysomal profiling. We are grateful to Dr Alessandro Provenzani, Bouchra Khalaf and Martin Hanczyc for their comments to the manuscript.
Footnotes
Present address: Albertomaria Moro, Yale Cardiovascular Research Center, Internal Medicine, Yale University, New Haven, CT 06511, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Trento – Progetto Biotecnologie (to P.M.); Autonomous Province of Trento through Madelena project (to P.M.); Axonomix project (to A.Q., L.L and G.V). Funding for open access charge: Madelena Project.
Conflict of interest statement. None declared.
REFERENCES
- 1. Huang Y., Carmichael G.G.. Nucleocytoplasmic mRNA transport. Results Probl. Cell Differ. 2001; 34:139–155. [DOI] [PubMed] [Google Scholar]
- 2. Palacios I.M., St Johnston D.. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 2001; 17:569–614. [DOI] [PubMed] [Google Scholar]
- 3. Carpenter B., MacKay C., Alnabulsi A., MacKay M., Telfer C., Melvin W.T., Murray G.I.. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim. Biophys. Acta. 2006; 1765:85–100. [DOI] [PubMed] [Google Scholar]
- 4. Han S.P., Tang Y.H., Smith R.. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 430:379–392. [DOI] [PubMed] [Google Scholar]
- 5. Krecic A.M., Swanson M.S.. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999; 11:363–371. [DOI] [PubMed] [Google Scholar]
- 6. Ostareck-Lederer A., Ostareck D.H.. Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol. Cell. 2004; 96:407–411. [DOI] [PubMed] [Google Scholar]
- 7. Weighardt F., Biamonti G., Riva S.. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996; 18:747–756. [DOI] [PubMed] [Google Scholar]
- 8. He Y., Smith R.. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell. Mol. Life Sci. 2009; 66:1239–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valverde R., Edwards L., Regan L.. Structure and function of KH domains. FEBS J. 2008; 275:2712–2726. [DOI] [PubMed] [Google Scholar]
- 10. Sun K.H., Tang S.J., Wang Y.S., Lin W.J., You R.I.. Autoantibodies to dsDNA cross-react with the arginine-glycine-rich domain of heterogeneous nuclear ribonucleoprotein A2 (hnRNP A2) and promote methylation of hnRNP A2. Rheumatology (Oxford). 2003; 42:154–161. [DOI] [PubMed] [Google Scholar]
- 11. Van Dusen C.M., Yee L., McNally L.M., McNally M.T.. A glycine-rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Mol. Cell Biol. 30:2552–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pesiridis G.S., Lee V.M., Trojanowski J.Q.. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum. Mol. Genet. 2009; 18:R156–R162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duhl D.M., Stevens M.E., Vrieling H., Saxon P.J., Miller M.W., Epstein C.J., Barsh G.S.. Pleiotropic effects of the mouse lethal yellow (Ay) mutation explained by deletion of a maternally expressed gene and the simultaneous production of agouti fusion RNAs. Development. 1994; 120:1695–1708. [DOI] [PubMed] [Google Scholar]
- 14. Michaud E.J., Bultman S.J., Klebig M.L., van Vugt M.J., Stubbs L.J., Russell L.B., Woychik R.P.. A molecular model for the genetic and phenotypic characteristics of the mouse lethal yellow (Ay) mutation. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:2562–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rhodes G.H., Valbracht J.R., Nguyen M.D., Vaughan J.H.. The p542 gene encodes an autoantigen that cross-reacts with EBNA-1 of the Epstein Barr virus and which may be a heterogeneous nuclear ribonucleoprotein. J. Autoimmun. 1997; 10:447–454. [DOI] [PubMed] [Google Scholar]
- 16. Jurica M.S., Licklider L.J., Gygi S.R., Grigorieff N., Moore M.J.. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002; 8:426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh G., Kucukural A., Cenik C., Leszyk J.D., Shaffer S.A., Weng Z., Moore M.J.. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012; 151:750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun S., Zhang Z., Fregoso O., Krainer A.R.. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA. 18:274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsofack S.P., Garand C., Sereduk C., Chow D., Aziz M., Guay D., Yin H.H., Lebel M.. NONO and RALY proteins are required for YB-1 oxaliplatin induced resistance in colon adenocarcinoma cell lines. Mol. Cancer. 10:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen C.Y., Gherzi R., Andersen J.S., Gaietta G., Jurchott K., Royer H.D., Mann M., Karin M.. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000; 14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 21. Raffetseder U., Frye B., Rauen T., Jurchott K., Royer H.D., Jansen P.L., Mertens P.R.. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J. Biol. Chem. 2003; 278:18241–18248. [DOI] [PubMed] [Google Scholar]
- 22. Bladen C.L., Udayakumar D., Takeda Y., Dynan W.S.. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005; 280:5205–5210. [DOI] [PubMed] [Google Scholar]
- 23. Sewer M.B., Nguyen V.Q., Huang C.J., Tucker P.W., Kagawa N., Waterman M.R.. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54(nrb)/NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology. 2002; 143:1280–1290. [DOI] [PubMed] [Google Scholar]
- 24. Gaudreault I., Guay D., Lebel M.. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 2004; 32:316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohga T., Uchiumi T., Makino Y., Koike K., Wada M., Kuwano M., Kohno K.. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J. Biol. Chem. 1998; 273:5997–6000. [DOI] [PubMed] [Google Scholar]
- 26. Schittek B., Psenner K., Sauer B., Meier F., Iftner T., Garbe C.. The increased expression of Y box-binding protein 1 in melanoma stimulates proliferation and tumor invasion, antagonizes apoptosis and enhances chemoresistance. Int. J. Cancer. 2007; 120:2110–2118. [DOI] [PubMed] [Google Scholar]
- 27. Tenzer S., Moro A., Kuharev J., Francis A.C., Vidalino L., Provenzani A., Macchi P.. Proteome-wide characterization of the RNA-binding protein RALY-interactome using the in vivo-biotinylation-pulldown-quant (iBioPQ) approach. J. Proteome Res. 2013; 12:2869–2884. [DOI] [PubMed] [Google Scholar]
- 28. Jung M., Kramer E., Grzenkowski M., Tang K., Blakemore W., Aguzzi A., Khazaie K., Chlichlia K., von Blankenfeld G., Kettenmann H. et al. . Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur. J. Neurosci. 1995; 7:1245–1265. [DOI] [PubMed] [Google Scholar]
- 29. Vessey J.P., Macchi P., Stein J.M., Mikl M., Hawker K.N., Vogelsang P., Wieczorek K., Vendra G., Riefler J., Tubing F. et al. . A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:16374–16379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Provenzani A., Fronza R., Loreni F., Pascale A., Amadio M., Quattrone A.. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis. 2006; 27:1323–1333. [DOI] [PubMed] [Google Scholar]
- 31. Viero G., Lunelli L., Passerini A., Bianchini P., Gilbert R.J., Bernabo P., Tebaldi T., Diaspro A., Pederzolli C., Quattrone A.. Three distinct ribosome assemblies modulated by translation are the building blocks of polysomes. J. Cell Biol. 2015; 208:581–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keene J.D., Komisarow J.M., Friedersdorf M.B.. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006; 1:302–307. [DOI] [PubMed] [Google Scholar]
- 33. Lunelli L., Bernabo P., Bolner A., Vaghi V., Marchioretto M., Viero G.. Peering at brain polysomes with atomic force microscopy. J. Visual. Exp.: JoVE. 2016; 109, doi:10.3791/53851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Necas D., Klapetek P.. Gwyddion: an open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012; 10:181–188. [Google Scholar]
- 35. Khrebtukova I., Kuklin A., Woychik R.P., Michaud E.J.. Alternative processing of the human and mouse raly genes(1). Biochim. Biophys. Acta. 1999; 1447:107–112. [DOI] [PubMed] [Google Scholar]
- 36. White R., Gonsior C., Bauer N.M., Kramer-Albers E.M., Luhmann H.J., Trotter J.. Heterogeneous nuclear ribonucleoprotein (hnRNP) F is a novel component of oligodendroglial RNA transport granules contributing to regulation of myelin basic protein (MBP) synthesis. J. Biol. Chem. 2012; 287:1742–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stefani G., Fraser C.E., Darnell J.C., Darnell R.B.. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 2004; 24:7272–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Izaurralde E., Jarmolowski A., Beisel C., Mattaj I.W., Dreyfuss G., Fischer U.. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J. Cell Biol. 1997; 137:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kashima T., Rao N., David C.J., Manley J.L.. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum. Mol. Genet. 2007; 16:3149–3159. [DOI] [PubMed] [Google Scholar]
- 40. Nilsson L., Nygård O.. Reduced puromycin sensitivity of translocated polysomes after the addition of elongation factor 2 and non-hydrolysable GTP analogues. FEBS Lett. 1992; 309:89–91. [DOI] [PubMed] [Google Scholar]
- 41. Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E. et al. . FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011; 146:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fritz J., Anselmetti D., Jarchow J., Fernandez-Busquets X.. Probing single biomolecules with atomic force microscopy. J. Struct. Biol. 1997; 119:165–171. [DOI] [PubMed] [Google Scholar]
- 43. Kang I., Raghavachari M., Hofmann C.M., Marchant R.E.. Surface-dependent expression in the platelet GPIb binding domain within human von Willebrand factor studied by atomic force microscopy. Thromb. Res. 2007; 119:731–740. [DOI] [PubMed] [Google Scholar]
- 44. Sharma S., Rasool H.I., Palanisamy V., Mathisen C., Schmidt M., Wong D.T., Gimzewski J.K.. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010; 4:1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. et al. . A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lim L.H., Pervaiz S.. Annexin 1: the new face of an old molecule. FASEB J. 2007; 21:968–975. [DOI] [PubMed] [Google Scholar]
- 48. Rescher U., Gerke V.. Annexins—unique membrane binding proteins with diverse functions. J. Cell Sci. 2004; 117:2631–2639. [DOI] [PubMed] [Google Scholar]
- 49. Happel N., Schulze E., Doenecke D.. Characterisation of human histone H1x. Biol. Chem. 2005; 386:541–551. [DOI] [PubMed] [Google Scholar]
- 50. Happel N., Warneboldt J., Hanecke K., Haller F., Doenecke D.. H1 subtype expression during cell proliferation and growth arrest. Cell Cycle. 2009; 8:2226–2232. [DOI] [PubMed] [Google Scholar]
- 51. Medrzycki M., Zhang Y., Cao K., Fan Y.. Expression analysis of mammalian linker-histone subtypes. J. Visual. Exp.: JoVE. 2012; 61, doi:10.3791/3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alldridge L.C., Bryant C.E.. Annexin 1 regulates cell proliferation by disruption of cell morphology and inhibition of cyclin D1 expression through sustained activation of the ERK1/2 MAPK signal. Exp. Cell Res. 2003; 290:93–107. [DOI] [PubMed] [Google Scholar]
- 53. Maschler S., Gebeshuber C.A., Wiedemann E.M., Alacakaptan M., Schreiber M., Custic I., Beug H.. Annexin A1 attenuates EMT and metastatic potential in breast cancer. EMBO Mol. Med. 2010; 2:401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ang E.Z., Nguyen H.T., Sim H.L., Putti T.C., Lim L.H.. Annexin-1 regulates growth arrest induced by high levels of estrogen in MCF-7 breast cancer cells. Mol. Cancer Res.: MCR. 2009; 7:266–274. [DOI] [PubMed] [Google Scholar]
- 55. Bailey-Serres J., Sorenson R., Juntawong P.. Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci. 2009; 14:443–453. [DOI] [PubMed] [Google Scholar]
- 56. Giorgi C., Moore M.J.. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Dev. Biol. 2007; 18:186–193. [DOI] [PubMed] [Google Scholar]
- 57. Hirokawa N. mRNA transport in dendrites: RNA granules, motors, and tracks. J. Neurosci. 2006; 26:7139–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kiebler M.A., Bassell G.J.. Neuronal RNA granules: movers and makers. Neuron. 2006; 51:685–690. [DOI] [PubMed] [Google Scholar]
- 59. Percipalle P., Raju C.S., Fukuda N.. Actin-associated hnRNP proteins as transacting factors in the control of mRNA transport and localization. RNA Biol. 2009; 6:171–174. [DOI] [PubMed] [Google Scholar]
- 60. Fritzsche R., Karra D., Bennett K.L., Ang F.Y., Heraud-Farlow J.E., Tolino M., Doyle M., Bauer K.E., Thomas S., Planyavsky M. et al. . Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013; 5:1749–1762. [DOI] [PubMed] [Google Scholar]
- 61. Buchwald G., Ebert J., Basquin C., Sauliere J., Jayachandran U., Bono F., Le Hir H., Conti E.. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:10050–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Macchi P., Kroening S., Palacios I.M., Baldassa S., Grunewald B., Ambrosino C., Goetze B., Lupas A., St Johnston D., Kiebler M.. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J. Neurosci. 2003; 23:5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Palacios I.M., Gatfield D., St Johnston D., Izaurralde E.. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004; 427:753–757. [DOI] [PubMed] [Google Scholar]
- 64. Park E., Gleghorn M.L., Maquat L.E.. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Heraud-Farlow J.E., Sharangdhar T., Li X., Pfeifer P., Tauber S., Orozco D., Hormann A., Thomas S., Bakosova A., Farlow A.R. et al. . Staufen2 regulates neuronal target RNAs. Cell Rep. 2013; 5:1511–1518. [DOI] [PubMed] [Google Scholar]
- 66. Miki T., Takano K., Yoneda Y.. The role of mammalian Staufen on mRNA traffic: a view from its nucleocytoplasmic shuttling function. Cell Struct. Funct. 2005; 30:51–56. [DOI] [PubMed] [Google Scholar]
- 67. Tang S.J., Meulemans D., Vazquez L., Colaco N., Schuman E.. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001; 32:463–475. [DOI] [PubMed] [Google Scholar]
- 68. Sanchez G., Dury A.Y., Murray L.M., Biondi O., Tadesse H., El Fatimy R., Kothary R., Charbonnier F., Khandjian E.W., Côté J.. A novel function for the survival motoneuron protein as a translational regulator. Hum. Mol. Genet. 2013; 22:668–84. [DOI] [PubMed] [Google Scholar]
- 69. Biaoxue R., Xiguang C., Shuanying Y.. Annexin A1 in malignant tumors: current opinions and controversies. Int. J. Biol. Markers. 2014; 29:e8–20. [DOI] [PubMed] [Google Scholar]
- 70. Harvey A.C., Downs J.A.. What functions do linker histones provide. Mol. Microbiol. 2004; 53:771–775. [DOI] [PubMed] [Google Scholar]
- 71. McBryant S.J., Lu X., Hansen J.C.. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell Res. 2010; 20:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lever M.A., Th’ng J.P., Sun X., Hendzel M.J.. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000; 408:873–876. [DOI] [PubMed] [Google Scholar]
- 73. Misteli T., Gunjan A., Hock R., Bustin M., Brown D.T.. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000; 408:877–881. [DOI] [PubMed] [Google Scholar]
- 74. Catez F., Ueda T., Bustin M.. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 2006; 13:305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shahhoseini M., Favaedi R., Baharvand H., Sharma V., Stunnenberg H.G.. Evidence for a dynamic role of the linker histone variant H1x during retinoic acid-induced differentiation of NT2 cells. FEBS Lett. 2010; 584:4661–4664. [DOI] [PubMed] [Google Scholar]
- 76. Sepsa A., Levidou G., Gargalionis A., Adamopoulos C., Spyropoulou A., Dalagiorgou G., Thymara I., Boviatsis E., Themistocleous M.S., Petraki K. et al. . Emerging role of linker histone variant H1x as a biomarker with prognostic value in astrocytic gliomas. A multivariate analysis including trimethylation of H3K9 and H4K20. PLoS One. 2015; 10:e0115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Swanson M.S., Dreyfuss G.. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 1988; 8:2237–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sibley C.R., Blazquez L., Ule J.. Lessons from non-canonical splicing. Nat. Rev. Genet. 2016; 17:407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tajnik M., Vigilante A., Braun S., Hanel H., Luscombe N.M., Ule J., Zarnack K., Konig J.. Intergenic Alu exonisation facilitates the evolution of tissue-specific transcript ends. Nucleic Acids Res. 2015; 43:10492–10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zarnack K., Konig J., Tajnik M., Martincorena I., Eustermann S., Stevant I., Reyes A., Anders S., Luscombe N.M., Ule J.. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013; 152:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Eliseeva I.A., Ovchinnikov L.P., Lyabin D.N.. Specific PABP effect on translation of YB-1 mRNA is neutralized by polyadenylation through a ‘mini-loop’ at 3΄ UTR. RNA Biol. 2012; 9:1473–1487. [DOI] [PubMed] [Google Scholar]
- 82. Lyabin D.N., Eliseeva I.A., Skabkina O.V., Ovchinnikov L.P.. Interplay between Y-box-binding protein 1 (YB-1) and poly(A) binding protein (PABP) in specific regulation of YB-1 mRNA translation. RNA Biol. 2011; 8:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Trcek T., Larson D.R., Moldon A., Query C.C., Singer R.H.. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell. 2011; 147:1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.C., Liang T.Y., Mazur C. et al. . Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 2012; 15:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tan A.Y., Riley T.R., Coady T., Bussemaker H.J., Manley J.L.. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R.. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008; 454:126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sallam T., Jones M.C., Gilliland T., Zhang L., Wu X., Eskin A., Sandhu J., Casero D., Vallim T.Q., Hong C. et al. . Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016; 534:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Geuens T., Bouhy D., Timmerman V.. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016; 135:851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Han N., Li W., Zhang M.. The function of the RNA-binding protein hnRNP in cancer metastasis. J. Cancer Res. Ther. 2013; 9(Suppl):S129–134. [DOI] [PubMed] [Google Scholar]
- 90. Piccolo L.L., Corona D., Onorati M.C.. Emerging roles for hnRNPs in post-transcriptional regulation: what can we learn from flies?. Chromosoma. 2014; 123:515–527. [DOI] [PubMed] [Google Scholar]
- 91. Shin K.H., Kang M.K., Kim R.H., Christensen R., Park N.H.. Heterogeneous nuclear ribonucleoprotein G shows tumor suppressive effect against oral squamous cell carcinoma cells. Clin. Cancer Res. 2006; 12:3222–3228. [DOI] [PubMed] [Google Scholar]
- 92. Yang X.Y., Ren C.P., Wang L., Li H., Jiang C.J., Zhang H.B., Zhao M., Yao K.T.. Identification of differentially expressed genes in metastatic and non-metastatic nasopharyngeal carcinoma cells by suppression subtractive hybridization. Cell Oncol. 2005; 27:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen Q., Jin M., Zhu J., Xiao Q., Zhang L.. Functions of heterogeneous nuclear ribonucleoproteins in stem cell potency and differentiation. BioMed Res. Int. 2013; 2013:623978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mallampalli R.K., Kaercher L., Snavely C., Pulijala R., Chen B.B., Coon T., Zhao J., Agassandian M.. Fbxl12 triggers G1 arrest by mediating degradation of calmodulin kinase I. Cell. Signal. 2013; 25:2047–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.