Abstract
Protein ADP-ribosylation (ADPr), a biologically and clinically important post-translational modification, exerts its functions by targeting a variety of different amino acids. Its repertoire recently expanded to include serine ADPr, which is emerging as an important and widespread signal in the DNA damage response. Chemically, serine ADPr (and more generally o-glycosidic ADPr) is a form of o-glycosylation, and its extreme lability renders it practically invisible to standard mass spectrometry approaches, often leading to erroneous localizations. The knowledge from the mature field of o-glycosation and our own initial difficulties with mass spectrometric analyzes of serine ADPr suggest how to avoid these misidentifications and fully explore the scope of o-glycosidic ADPr in DNA damage response and beyond.
A variety of chemically diverse post-translational modifications (PTMs) regulate fundamental biological processes by dynamically changing the properties of their substrate proteins. PTMs can be based on small moieties, like methylation, acetylation and phosphorylation, or much larger ones, such as ubiquitin and ubiquitin-like proteins or long glycan chains. Nucleotides, the building blocks of nucleic acids, are emerging as the basis of important regulatory PTMs as well. Together with adenylylation (1) (also called AMPylation), ADP-ribosylation (ADPr) is the best-characterized nucleotide-based PTM and is especially known for its role in DNA repair signaling and as a target in cancer therapy (2–5). Though enzymatic conjugation of a PTM is usually limited to amino acids with similar functional groups (for example phosphorylation of hydroxyl groups on serine, threonine and tyrosine in eukaryotes), almost all chemically reactive amino acid side-chains have been reported to be ADP-ribosylated (6) with glutamate, aspartate, lysine and arginine historically considered as the main targets (7–14).
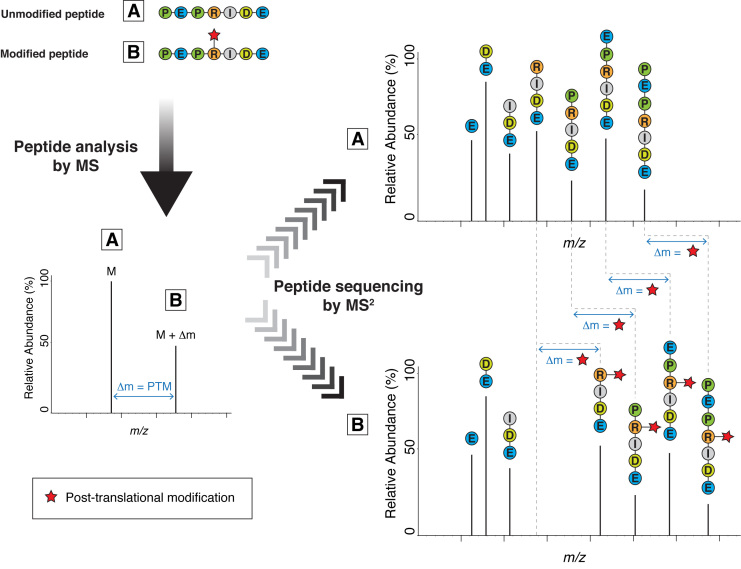
Mass spectrometry (MS) is an indispensable technology for analyzing PTMs via the identification of the modified peptides (15). When we analyze peptides by MS, we not only measure the molecular mass of the peptide molecules entering the spectrometer (the ‘precursors’), we also break these molecules into smaller pieces generating fragment ions, which cover different parts of the original peptide. From these ladders of fragment ions, we can ‘read off’ the amino acid sequence of the peptide; the masses of adjacent fragment ions differ by the masses of the respective amino acids. When an amino acid is modified by a PTM, an altered mass difference typically appears in the series (Figure 1). This is how standard modern MS technology can precisely map most PTMs to specific residues within target proteins. But the lability of some more challenging PTMs severely hampers their analysis by conventional MS at exactly this step. Fortunately, a better understanding of MS behavior often suggests a strategy to circumvent such technical limitations.
Figure 1.
Mass spectrometry (MS) for the analysis of post-translational modifications (PTMs) via the identification of the modified peptides. When we analyze peptides by mass spectrometry, we first acquire an MS spectrum (left-hand spectrum) to determine the molecular mass of the peptide molecules entering the spectrometer (the ‘precursors’). Next, each of these peptide species is selected and fragmented into smaller pieces, producing a set of fragment ions (right-hand spectra). From these fragment ions (MS2 peaks), we can extract information about the sequence of the peptide. The masses of adjacent fragment ions differ by the masses of the corresponding amino acids. The presence of a typical PTM changes the mass of the peptide, the modified amino acid residue and all fragment ions containing the modified residue by Δm.
Several different strategies for site-specific analysis of ADPr by MS have been considered over the last years (6,7,13,16–20). Excellent recent reviews (6,19) give an in-depth overview of the various proteomic approaches for the detection of ADP-ribosylated proteins and the specific ADPr sites. In this short commentary we would like to raise awareness of the hazards of ADPr misidentification by summarizing a few underlying concepts and illustrating the pitfalls of ADPr site mapping by describing our own initial mistakes.
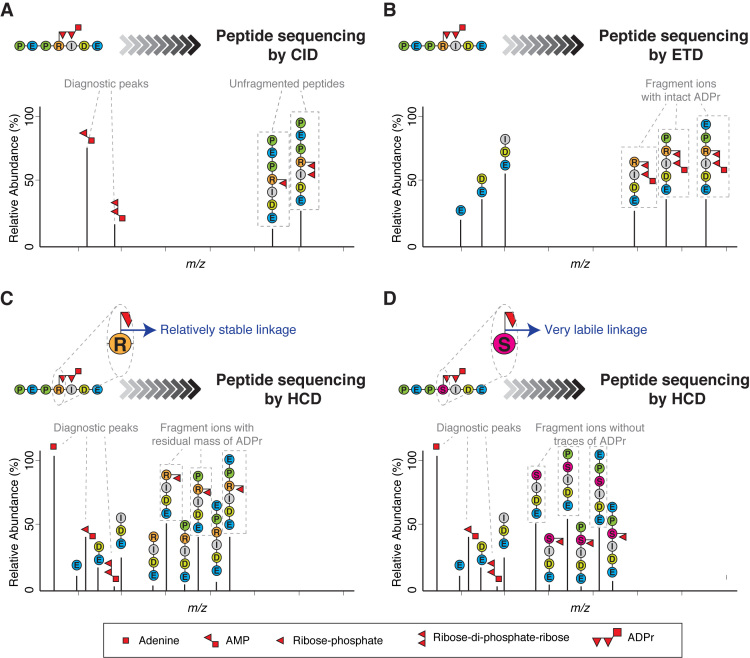
Early attempts to analyze ADPr by MS using collision-induced dissociation (CID), then the predominant peptide fragmentation technique, generally failed to identify the peptides modified by ADPr (21–25). This mode of fragmentation was simply unsuitable for this particular modification: CID essentially breaks one bond in each molecule, with the weakest chemical linkage breaking most frequently (26). In ADP-ribosylated peptides, this results in preferential breakage within ADP-ribose since this PTM is dramatically more fragile than the peptide itself. The predominance of this breakage means that the peptide itself seldom fragments, with consequently little information about the peptide sequence (21) (Figure 2A).
Figure 2.
Different fragmentation methods applied to ADP-ribosylated peptides. (A) Schematic representation of a typical collision-induced dissociation (CID) fragmentation spectrum from an ADP-ribosylated peptide. The CID spectrum contains more ions formed through fragmentation of the ADP-ribose itself than through fragmentation of the peptide backbone. As a consequence, it yields little or no information about the peptide sequence. However, CID can be used to confirm the presence of ADP-ribose on a precursor by generating modification-specific diagnostic peaks (e.g. AMP). (B) Schematic representation of a typical electron-transfer dissociation (ETD) fragmentation spectrum from an ADP-ribosylated peptide. The ETD spectrum is rich in information about the peptide sequence and the site of modification. A characteristic feature of ETD is the retention of the intact ADP-ribose. The masses of adjacent fragment ions differ by the masses of the corresponding amino acids. The presence of an ADP-ribose changes the mass of the modified amino acid residue and the ions containing the modified residue by 541.06 Da. (C) Schematic representation of a typical higher-energy collisional dissociation (HCD) fragmentation spectrum from an ADP-ribosylated peptide in which the ADP-ribose linkage to the modified amino acid is relatively stable under HCD fragmentation (e.g. ADPr on Arg). The HCD spectrum contains ions from fragmentation of both the peptide backbone and ADP-ribose. Usually, the HCD spectrum is dominated by one of the diagnostic ions generated by the internal fragmentation of ADP-ribose (adenine, at m/z 136.06), but additional fragments are also detected corresponding to the peptide with a residual part of ADP-ribose attached (typically ribose phosphate-H2O). These fragment ions provide the basis for determining the site of modification. (D) Schematic representation of a typical HCD spectrum from an ADP-ribosylated peptide in which the ADP-ribose linkage to the modified amino acid is very labile under HCD fragmentation (e.g. ADPr on Ser). The HCD spectrum contains ions from both the peptide backbone and from the ADP-ribose fragmentation. This spectrum can contain peptide fragment ions carrying part of ADP-ribose, but the strongest ion series is not that of a modified sequence, but of the native identified peptide. These de-modified fragment ions contain no information for determining the modification site. Since search algorithms score these de-modified ions as unmodified ions, conventional analysis of serine ADPr by HCD can lead to multiple, erroneous localizations arising from variations in spectrum quality. See Figure 3.
In contrast, fragmentation techniques that specifically target the bonds in the peptide backbone, like electron-capture dissociation (ECD) (27) or electron-transfer dissociation (ETD) (28), leave the ADP-ribose intact and yield spectra that are rich in information about the peptide sequence as well as the modification's location. Thus, data generated by ECD/ETD allow identification of ADP-ribosylated peptides and precise localization of the modification site (21) (Figure 2B).
The importance of this advantage of ETD diminished with the advent of higher-energy collisional dissociation (HCD) (29), which, unlike CID, fragments ADP-ribosylated peptides very well, since it is not limited to one fragmentation event per molecule. Compared to ETD, which has certain limitations with typical tryptic peptides, HCD allows for sequencing of a wider range of ADP-ribosylated peptides. Like CID, HCD confirms the presence of the modification of the peptide by ADP-ribose by generating modification-specific diagnostic peaks through breakage of the modifier (Figure 2C). Due to all these advantages, we, like others, quickly adopted high-resolution HCD as the technique of choice for the analysis of protein ADPr. HCD in combination with sophisticated biochemical and chemical approaches (6,13,19,30) has led to the identification of a large number of ADPr sites on glutamate, aspartate, lysine and arginine residues (10,11,13,17,30,31) and even allowed the discovery of cysteine ADPr (12).
But a troubling mystery took shape once we turned our attention to HCD data from our own ADP-ribosylated histone samples (32) and performed computational re-analyzes of large published HCD phosphoproteomics datasets (33,34) (in which ADP-ribosylated peptides are often co-enriched (18)). Although we confidently identified dozens of ADP-ribosylated peptides, the chemical bond through which ADP-ribose was linked to these peptides appeared to be extremely labile. Though the spectra displayed strong diagnostic ions for ADP-ribose, the fragment ion series from the peptide sequence bore little or no trace of the modification (Figure 2D), a phenomenon others have recently remarked upon (35). Following computational analysis of this HCD data, ADPr appeared to modify established target residues, such as lysine(14,36), but also virtually any residue we allowed, including clearly impossible ones such as alanine, that cannot be targeted by ADP-ribose as it lacks a chemically reactive side chain (Figure 3). For a while, we could not make sense of the mysterious form of ADPr that gave rise to these spectra and turned to ETD fragmentation for an answer.
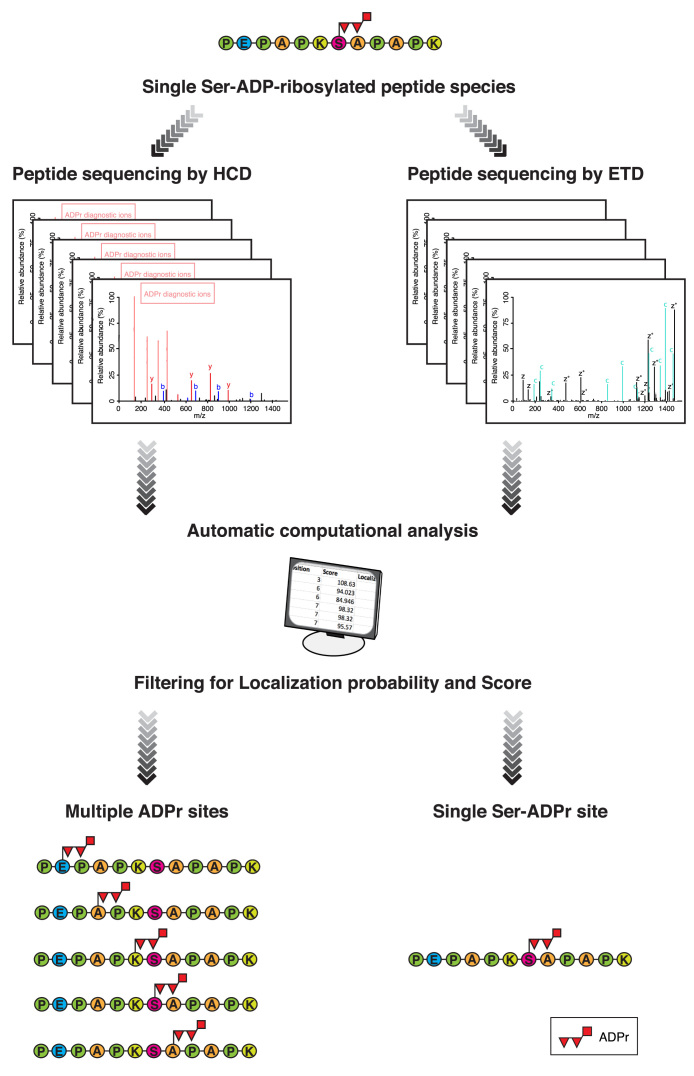
Figure 3.
Conventional analysis of serine ADPr by HCD can lead to multiple and erroneous ADPr site assignments. Schematic representation of a standard MS workflow illustrating the problematic behavior of Ser-ADP-ribosylated peptides under HCD fragmentation. In this representation, the same single Ser-ADP-ribosylated peptide species is subjected to MS analysis either using HCD or ETD fragmentation (left and right-hand respectively). The acquired mass spectra are searched against the appropriate database using any one of a number of database-search systems (e.g. Maxquant, Mascot, etc). The initial list of ADPr sites obtained after the automatic computational analysis is filtered by different criteria to eliminate unreliable (low scoring) or ambiguous (low localization score) results. The final filtered list of ADPr sites obtained from ETD fragmentation contains the same unique ADPr site as the single ADP-ribosylated peptide species initially subjected to MS analysis (bottom right-hand). On the other hand, the applying of the same filtering parameters to the sites found from HCD fragmentation data, reveals fictional ADPr sites in the final list, illustrating the fundamental weakness of this analysis approach (bottom left-hand). Readers can evaluate the misleading localization results provided by HCD MS spectra by re-analyzing publicly available data for themselves. A dataset containing HCD and ETD spectra from the same sample is recommended to demonstrate the fundamental weakness of HCD as well as the necessity of ETD for accurate localization of Ser-ADPr (31,37). Alternatively, large published HCD datasets from ADP-ribosylated peptides can also be re-analyzed permitting modification of chemically impossible ‘decoy’ sites (like alanine) to illustrate the weakness of automatic computational analysis of HCD data. This form of analysis illustrates the relative influence of data and prior assumptions on results.
With ETD, we could show that this extremely labile form of ADPr occurs on serine, a prominent target amino acid for cellular ADPr (32), which we have since shown is used in PARP-1/HPF1-mediated DNA repair signaling (37). In retrospect, the problematic behavior of our ADP-ribosylated peptides under HCD is no surprise. Although new to the ADPr field, the o-glycosidic bond that links the ribose of ADP-ribose and the hydroxyl of serine is well known to the many researchers using MS to study o-glycosylations (38) like O-GlcNAcylation (39). Fragmentation of the glycan-peptide linkage with HCD hampers the ability to localize the o-glycosylation site. For O-GlcNAc, it is well known that ‘ETD MS analysis of enriched peptides in many cases allowed assignment of the mass spectrum to a particular peptide sequence and unambiguous site localization of the modification (40–44). In HCD, the O-GlcNAc moiety usually dissociates (…), preventing the use of mass shifts in the peptide sequence ion series to establish the site(s) of this labile modification’ (45).
Since it had not previously been hypothesized that ADPr could target proteins via labile o-glycosydic linkages, this established knowledge about the MS behavior of o-glycosidic bonds was never applied to ADPr, and consequently o-glycosidic ADPr (on serine and possibly threonine and tyrosine) eluded discovery for decades. HCD works better with other less labile ADP-ribose-amino acid linkages, especially with arginine and cysteine ADPr (12). It is clear from our own experience, however, that by using HCD in combination with standard automatic computational analysis of ADP-ribosylated peptides, one runs the risk of erroneously assigning o-linked modifications to residues that are not the true attachment site (Figure 3). Misassignment by HCD is a particular concern whenever Ser-ADPr is strongly induced, namely DNA damage-treated cells and in vitro reactions in presence of HPF1. Erroneous localization of ADPr may also occur, although to a lesser extent, with other types of ADPr. Thus, we propose ETD as the method of choice for unambiguous ADPr site mapping.
By considering serine ADPr as a new type of o-glycosylation, we can take advantage of the experience and methodology amassed in the o-glycosylation field. This literature provides the guidelines we need for MS investigations of the puzzling amino acids specificities of ADPr. Thus: serine ADPr, welcome to the o-glycosylation club!
ACKNOWLEDGEMENTS
Many thanks to I. Atanassov (Max Planck Institute for Biology of Ageing) for comments on the manuscript.
FUNDING
Deutsche Forschungsgemeinschaft (Cluster of Excellence Cellular Stress Responses in Aging-Associated Diseases) [grant number EXC 229 to I.M.]; European Union's Horizon 2020 research and innovation program [Marie Skłodowska-Curie 657501 to J.J.B., I.M.]. Funding for open access charge: Deutsche Forschungsgemeinschaft [grant number EXC 229 to I.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hedberg C., Itzen A.. Molecular perspectives on protein adenylylation. Acs. Chem. Biol. 2015; 10:12–21. [DOI] [PubMed] [Google Scholar]
- 2. Feng F.Y., de Bono J.S., Rubin M.A., Knudsen K.E.. Chromatin to clinic: the molecular rationale for PARP1 inhibitor function. Mol. Cell. 2015; 58:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bock F.J., Chang P.. New directions in PARP biology. FEBS J. 2016; 283:4017–4031. [DOI] [PubMed] [Google Scholar]
- 4. Gupte R., Liu Z., Kraus W.L.. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017; 31:101–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martin-Hernandez K., Rodriguez-Vargas J.M., Schreiber V., Dantzer F.. Expanding functions of ADP-ribosylation in the maintenance of genome integrity. Semin. Cell Dev. Biol. 2017; 63:92–101. [DOI] [PubMed] [Google Scholar]
- 6. Daniels C.M., Ong S.E., Leung A.K.. The promise of proteomics for the study of ADP-Ribosylation. Mol. Cell. 2015; 58:911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gagne J.P., Ethier C., Defoy D., Bourassa S., Langelier M. F., Riccio A. A., Pascal J. M., Moon K. M., Foster L. J., Ning Z. et al. Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair (Amst). 2015; 30:68–79. [DOI] [PubMed] [Google Scholar]
- 8. Sharifi R., Morra R., Appel C. D., Tallis M., Chioza B., Jankevicius G., Simpson M. A., Matic I., Ozkan E., Golia B. et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013; 32:1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tao Z., Gao P., Liu H.W.. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 2009; 131:14258–14260. [DOI] [PubMed] [Google Scholar]
- 10. Rosenthal F., Nanni P., Barkow-Oesterreicher S., Hottiger M.O.. Optimization of LTQ-Orbitrap mass spectrometer parameters for the identification of ADP-ribosylation sites. J. Proteome Res. 2015; 14:4072–4079. [DOI] [PubMed] [Google Scholar]
- 11. Martello R., Leutert M., Jungmichel S., Bilan V., Larsen S. C., Young C., Hottiger M. O., Nielsen M. L.. Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun. 2016; 7:12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R. T., Ahel I., Chang P.. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014; 5:4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y., Wang J., Ding M., Yu Y.. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013; 10:981–984. [DOI] [PubMed] [Google Scholar]
- 14. Messner S., Altmeyer M., Zhao H., Pozivil A., Roschitzki B., Gehrig P., Rutishauser D., Huang D., Caflisch A., Hottiger M.O.. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010; 38:6350–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aebersold R., Mann M.. Mass-spectrometric exploration of proteome structure and function. Nature. 2016; 537:347–355. [DOI] [PubMed] [Google Scholar]
- 16. Palazzo L., Thomas B., Jemth A. S., Colby T., Leidecker O., Feijs K. L., Zaja R., Loseva O., Puigvert J.C., Matic I., Helleday T. et al. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem. J. 2015; 468:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daniels C.M., Ong S.E., Leung A.K.. Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl)ation sites from cells. J. Proteome Res. 2014; 13:3510–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matic I., Ahel I., Hay R.T.. Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat. Methods. 2012; 9:771–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vivelo C.A., Leung A.K.. Proteomics approaches to identify mono-(ADP-ribosyl)ated and poly(ADP-ribosyl)ated proteins. Proteomics. 2015; 15:203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daniels C.M., Thirawatananond P., Ong S.E., Gabelli S.B., Leung A.K.. Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci. Rep. 2015; 5:18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hengel S.M., Goodlett D.R.. A review of tandem mass spectrometry characterization of adenosine diphosphate-ribosylated peptides. Int. J. Mass Spectrom. 2012; 312:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hengel S.M., Shaffer S.A., Nunn B.L., Goodlett D.R.. Tandem mass spectrometry investigation of ADP-ribosylated kemptide. J. Am. Soc. Mass Spectrom. 2009; 20:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Margarit S.M., Davidson W., Frego L., Stebbins C.E.. A steric antagonism of actin polymerization by a salmonella virulence protein. Structure. 2006; 14:1219–1229. [DOI] [PubMed] [Google Scholar]
- 24. Osago H., Yamada K., Shibata T., Yoshino K., Hara N., Tsuchiya M.. Precursor ion scanning and sequencing of arginine-ADP-ribosylated peptide by mass spectrometry. Anal. Biochem. 2009; 393:248–254. [DOI] [PubMed] [Google Scholar]
- 25. Zee B.M., Garcia B.A.. Electron transfer dissociation facilitates sequencing of adenosine diphosphate-ribosylated peptides. Anal. Chem. 2010; 82:28–31. [DOI] [PubMed] [Google Scholar]
- 26. Dookeran N.N., Yalcin T., Harrison A.G.. Fragmentation reactions of protonated alpha-amino acids. J. Mass Spectrom. 1996; 31:500–508. [Google Scholar]
- 27. Zubarev R.A., Kelleher N.L., McLafferty F.W.. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998; 120:3265–3266. [Google Scholar]
- 28. Syka J.E., Coon J.J., Schroeder M.J., Shabanowitz J., Hunt D.F.. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olsen J.V., Macek B., Lange O., Makarov A., Horning S., Mann M.. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods. 2007; 4:709–712. [DOI] [PubMed] [Google Scholar]
- 30. Gibson B.A., Zhang Y., Jiang H., Hussey K.M., Shrimp J.H., Lin H., Schwede F., Yu Y., Kraus W.L.. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016; 353:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bilan V., Leutert M., Nanni P., Panse C., Hottiger M.O.. Combining higher-energy collision dissociation and electron-transfer/higher-energy collision dissociation fragmentation in a product-dependent manner confidently assigns proteomewide ADP-ribose acceptor sites. Anal. Chem. 2017; 89:1523–1530. [DOI] [PubMed] [Google Scholar]
- 32. Leidecker O., Bonfiglio J.J., Colby T., Zhang Q., Atanassov I., Zaja R., Palazzo L., Stockum A., Ahel I., Matic I.. Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol. 2016; 12:998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Phanstiel D.H., Brumbaugh J., Wenger C.D., Tian S., Probasco M.D., Bailey D.J., Swaney D.L., Tervo M.A., Bolin J.M., Ruotti V., Stewart R. et al. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods. 2011; 8:821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagaraj N., D'Souza R.C., Cox J., Olsen J.V., Mann M.. Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 2010; 9:6786–6794. [DOI] [PubMed] [Google Scholar]
- 35. Bilan V., Selevsek N., Kistemaker H.A.V., Abplanalp J., Feurer R., Filippov D.V., Hottiger M.O.. New quantitative mass spectrometry approaches reveal different ADP-ribosylation phases dependent on the levels of oxidative stress. Mol. Cell Proteomics. 2017; 7:44668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altmeyer M., Messner S., Hassa P.O., Fey M., Hottiger M.O.. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009; 37:3723–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bonfiglio J.J., Fontana P., Zhang Q., Colby T., Gibbs-Seymour I., Atanassov I., Bartlett E., Zaja R., Ahel I., Matic I.. Serine ADP-ribosylation depends on HPF1. Mol. Cell. 2017; 65:932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moremen K.W., Tiemeyer M., Nairn A.V.. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012; 13:448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hart G.W. Minireview series on the thirtieth anniversary of research on O-GlcNAcylation of nuclear and cytoplasmic proteins: Nutrient regulation of cellular metabolism and physiology by O-GlcNAcylation. J. Biol. Chem. 2014; 289:34422–34423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trinidad J.C., Barkan D.T., Gulledge B.F., Thalhammer A., Sali A., Schoepfer R., Burlingame A.L.. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell Proteomics. 2012; 11:215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Z., Udeshi N.D., O'Malley M., Shabanowitz J., Hunt D.F., Hart G.W.. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell Proteomics. 2010; 9:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khidekel N., Ficarro S.B., Clark P.M., Bryan M.C., Swaney D.L., Rexach J.E., Sun Y.E., Coon J.J., Peters E.C., Hsieh-Wilson L.C.. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 2007; 3:339–348. [DOI] [PubMed] [Google Scholar]
- 43. Alfaro J.F., Gong C.X., Monroe M.E., Aldrich J.T., Clauss T.R., Purvine S.O., Wang Z., Camp D.G. 2nd, Shabanowitz J., Stanley P., Hart G.W. et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:7280–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steentoft C., Vakhrushev S.Y., Vester-Christensen M.B., Schjoldager K.T., Kong Y., Bennett E.P., Mandel U., Wandall H., Levery S.B., Clausen H.. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered simplecell lines. Nat. Methods. 2011; 8:977–982. [DOI] [PubMed] [Google Scholar]
- 45. Xu S.L., Chalkley R.J., Maynard J.C., Wang W., Ni W., Jiang X., Shin K., Cheng L., Savage D., Huhmer A.F., Burlingame A.L. et al. Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E1536–E1543. [DOI] [PMC free article] [PubMed] [Google Scholar]