Abstract
CRISPR/Cas9-based gene knockout libraries have emerged as a powerful tool for functional screens. We present here a set of pre-designed human and mouse sgRNA sequences that are optimized for both high on-target potency and low off-target effect. To maximize the chance of target gene inactivation, sgRNAs were curated to target both 5΄ constitutive exons and exons that encode conserved protein domains. We describe here a robust and cost-effective method to construct multiple small sized CRISPR library from a single oligo pool generated by array synthesis using parallel oligonucleotide retrieval. Together, these resources provide a convenient means for individual labs to generate customized CRISPR libraries of variable size and coverage depth for functional genomics application.
INTRODUCTION
The discovery of the bacterial CRISPR/Cas9 endonuclease system and its subsequent adaptation for genome editing in mammalian cells has created new opportunities for high throughput, functional genomics screens (1). For loss-of-function CRISPR screens, the most widely used CRISPR/Cas9 system is the Streptococcus pyrogenes Cas9 (SpCas9) enzyme. SpCas9 can utilize a single guide RNA (sgRNA) that contains a 20-nt targeting sequence to introduce double-stranded DNA breaks in the genome in a sequence-specific manner. The SpCas9 target site must be upstream of a ‘NGG’ protospacer adjacent motif (PAM). Repair of the double-stranded DNA break generated by SpCas9 through non-homologous end joining introduces small indel mutations at the break site. If the break site is within the coding region of a gene, these indel mutations frequently result in frame-shift mutations that knock out gene expression (2). Thus, virtually any gene can be knocked out by co-expressing SpCas9 and a sgRNA in cells.
Large pooled lentiviral CRISPR libraries have been recently generated for loss of function screens with SpCas9. To construct these libraries, sgRNA sequences are first synthesized as a pool of DNA oligos using array-based oligo synthesis and then PCR cloned into lentiviral expression vectors (1). Currently, most ready-to-use CRISPR libraries are genome-wide libraries that contain tens of thousands of distinct sgRNAs in a single pool (3–8). Although useful for genome-wide screens, these libraries are technically more challenging to generate, maintain and screen due to their large size. In addition, sgRNA representation in these libraries can be adversely affected by their high degree of pool complexity. In many experimental settings, smaller, customized CRISPR libraries are desirable. For example, one might wish to screen a CRISPR library targeting only a specific gene family (e.g. kinases) or a subset of genes that constitute a gene expression or mutational signature. Small libraries might also be more desirable for their utility in in vivo screens. To carry out these ‘focused’ screens, a cost-effective, easy-to-implement method is necessary to enable individual labs to generate custom CRISPR libraries on demand.
In this study, we describe a flexible CRISPR library construction method to enable the generation of sub-genome scale, focused CRISPR sgRNA libraries of variable sizes. Our method uses parallel PCR retrieval to recover subsets of oligos from a master oligo pool generated by array-based synthesis. We demonstrate that sgRNA representations in these smaller libraries are highly consistent. We also generated a pre-designed database of sgRNA sequences targeting most human and mouse protein coding genes that can be used for selecting sgRNAs for custom library construction. These resources will enable easy creation of customized CRISPR libraries for genetic screens.
MATERIALS AND METHODS
Genome-wide sgRNA libraries targeting coding exons and protein domains for the human and mouse genomes were constructed following the steps described below. The approach is the same for both the human and mouse libraries. For the first step, we curated sgRNA sequences from all common exons and exon regions in protein coding genes. For each gene, all coding exons were identified using the NCBI Consensus Coding Sequence (CCDS) database (release# 106 for human, and release# 104 for mouse). For genes with multiple alternative transcripts, only exons or parts of exons (if the alternative splicing site is inside the exon) that existed in all transcripts were selected. For these common exons and exon regions, all S. pyogenes Cas9 sgRNA sequences in the form of (N)20NGG in both strands DNA were selected. This results in a database of 4.3 × 106 sgRNA sequences for human and 4.5 × 106 sgRNA sequences for mouse.
In the second step, we generated scores that predicts the potency and on-target effect of each sgRNA. The on-target potency score (range 0–1) of each sgRNA was calculated using a previously published algorithm (9). To minimize off-target effects, each sgRNA sequence was mapped to its respective reference genome (hg38 for human and mm10 for mouse) using the Bowtie short read aligner (10), allowing up to three base mismatches. An off-target score for each potential off-target site identified was calculated using a modified version of a previously published algorithm (11), where we only used the empirically-determined mismatch weight table to score the sgRNA's off-target effects. Summing the number of potential off-target sites and each site's off-target scores, we calculated separate guide scores (range 0–100) for off-target sites within only the coding region (guide_c score), for off-target sites within only the non-coding region (guide_nc score), and for off-target sites in the genome (guide_g score) for each candidate sgRNA to measure its uniqueness within the genome. A higher guide score predicts the candidate sgRNA to have less off-target activity (12).
To curate sgRNAs for the exon library, we started by only considering sgRNAs that have no perfect match off-target sites in the genome. Among these, we initially only considered those with potency scores ≥0.4, guide_c scores ≥50 and guide_nc scores ≥10. For each gene we chose up to six sgRNAs with the highest potency scores that also satisfy the following criteria: if a gene has only a single coding exon, all sgRNAs were chosen from that exon, if a gene has two coding exons, up to three sgRNAs were selected from each exon; and if a gene has ≥3 coding exons, we selected sgRNAs from the first five coding exons with no more than two sgRNAs from the same exon. In addition, no two sgRNAs in the library were allowed to have ≥15 nucleotides overlap with each other, and sgRNA sequences that contain the restriction site sequence for BsmBI were excluded. For the small number of fusion genes (read-throughs) and for family of genes that have the same annotated exon genomic locations in the CCDS database, only one sgRNA was chosen for each genomic location. For the majority of genes, we were able to curate 6 sgRNAs per gene. For some genes which are mostly small genes, fewer than 6 sgRNAs satisfied the above selection criteria. In these cases, we allowed the potency score cut-off to be first relaxed to ≥0.3 and then relaxed to ≥0.2 in order to curate additional sgRNAs. If after these two rounds of score relaxation we were still unable to curate six sgRNAs, we did not attempt to further relax the selection criteria. Thus, for a minor fraction of gene there were less than six sgRNAs/gene in the library.
To curate sgRNAs for the domain library, we first used the Pfam protein domains database (version 30) to define the regions in each protein that mapped to conserved domains. The genomic location of each domain was extracted from UCSC genomic databases (hg38 for human and mm10 for mouse) by using the ucscGenePfam and pfamDesc tables to map amino acid sequences to their corresponding exon regions. Intersection of these protein domain exon regions with the total sgRNA database from step 1 above allowed us to extract all sgRNAs mapped to Pfam protein domains in all genes. For each gene, we chose up to six sgRNAs from the set of domain-targeted sgRNAs that satisfied the following criteria. We only considered sgRNAs that have no perfect match off-target sites in the genome, and they must have guide_c scores ≥50 and guide_nc scores ≥10. Unlike the exon sgRNA library, we did not impose a potency score cut-off for the domain sgRNA library. We chose six sgRNAs with the highest potency score without considering from which domains they came or from which constitutive exon they came from. Other selection criteria were the same as the exon library above. Any sgRNAs that were already selected for the exon sgRNA library were excluded from the domain sgRNA library.
To curate a set of PCR primers that could work with the human and mouse sgRNAs for library construction, we first generated 1000 random GC-balanced 24-mers. These primer sequences were filtered to remove those with repetitive sequences, those with BsmBI sites, those with significant sequence similarity with any members of the sgRNA database, those with extreme Tm and those with intramolecular hairpin propensity. In total, we curated 59 primers and 40 primers that are compatible with the human and mouse CRISPR libraries, respectively. These will enable the parallel retrieval of 29 and 20 sub-pools of human and mouse CRISPR library from a single oligo array, respectively.
To compare sgRNA identify among the published genome-wide human and mouse CRISPR KO libraries, we first removed any duplication sgRNA sequences within each library and carried out pair-wise gene and sgRNA overlap analysis among them. Because each library contains a slightly different set of genes, only sgRNAs from the shared genes were used for the overlap analysis.
The sgRNA oligo design for array synthesis was illustrated in Supplementary Figure S2. Each oligo contains a 5΄ and a 3΄ PCR primer sequence. Inside the primer sequences were adaptor sequences containing BsmBI sites that are compatible for cloning into the LentiGuide-puro and the LentiCRISPR v2 expression vector (12). Inside the BsmBI adaptor sequences was the sgRNA 20-mer sequence. For parallel sub-pool retrieval, the sgRNAs were divided into groups of ∼1000 sgRNAs per group and each group was assigned a unique primer pair chosen from Supplementary Table S3. For each array experiment, 12.4k sgRNA oligos were synthesized on a single array (CustomArray Inc.) and the oligos were cleaved from the array as a single master oligo pool.
Individual groups of sgRNAs were PCR recovered from the master oligo pool using their corresponding primer pairs. The PCR components were as follows. Chip oligos 10 ng, 5΄ primer 2 μM, 3΄ primer 2 μM, Accuprime Pfx supermix (Life Technologies) 45 μl, total reaction volume 10 μl. The PCR condition was as follows. Hot start: 95°C for 5 min; PCR reaction: 95°C (15 s) –55°C (15 s) – 72°C (15 s) cycles with 15 s per step for 15 cycles; final extension: 72°C for 2 min. PCR product was gel extracted (Qiagen) and quantified using PicoGreen (Life Technologies). To clone PCR products into the lentiviral sgRNA expression vector LentiGuide-puro (12), the following Golden Gate reaction mixture was used. PCR Amplicon of sgRNA insert 4.3 ng, linearized LentiGuide-puro plasmid 150 ng, Esp3I 10 U (ThermoFisher), T7 DNA ligase 3000 U (New England Biolabs), ATP 10 mM, total reaction volume was 20 μl. Esp3I is an isoschizomer of BsmBI that has optimal activity at 37°C. The reaction condition was as follows. Pre-incubation: incubate the reaction mixture without T7 ligase at 37°C for 2 h; Golden Gate reaction: Add T7 ligase and incubate the mixture with 20°C (20 min) – 37°C (5 min) cycle for 10 cycles. For bacterial transformation, ∼30 ng of the Golden Gate reaction mixture was electroporated into 20 μl of ElectroMAX Stbl4 competent bacteria per manufacturer's instructions (Life Technologies). The electroporated bacterial was recovered in 500 μl of SPC media at 30°C for 90 min. A small amount of the bacterial culture was plated on 10 cm LB-agarose plates with carbenicillin in serial dilutions to estimate transformation efficiency. The rest of the bacterial culture was plated out on 24 cm square LB-agarose plates with carbenicillin and incubated at 30°C for ∼36 h. Bacteria were scrapped off the plate and transferred to 100 ml of pre-warmed liquid LB media containing carbenicillin. After 2 h of growth at 30°C with shaking, plasmids were extracted as maxi-preps (Qiagen). A detailed, step-by-step cloning protocol is included in the Supplemental Materials section.
To verify the cloning efficiency of sgRNAs, LentiGuide-puro plasmid sub-pools were combined and the sgRNA PCR was recovered using the following PCR primers. Forward primer with Illumina adaptor: aatgatacggcgaccaccgagatctgacgctcttccgatcttatcttgtggaaaggacgaaacaccg, reverse primer with Illumina adaptor: caagcagaagacggcatacgagatnnnnnnnngtgactggagttcagacgtgtgctcttccgatctctttagtttgtatgtctgttgctattatgtctactattctttcc (where ‘n’ denotes Illumina indexing nucleotides). The PCR mixture was as follows. Plasmid templates 100 ng, forward and reverse primers 0.2 μM each, hot start Taq 5 U (Takara), dNTP 250 μM each, total reaction volume 50 μl. The PCR condition was as follows. Hot start step: 95°C 5 min; PCR 1: 95°C (15 s) – 57°C (30 s) – 72°C (30 s) for 5 cycles; PCR 2: 95°C (15 s) – 62°C (30 s) – 72°C (30 s) for 5 cycles; final extension: 72°C for 10 min. PCR product was gel purified (Qiagen) and sequenced on the MiSeq platform (Illumina) and reads with perfect match to sgRNA 20-mers were counted. A detailed, step-by-step plasmid sequencing protocol is included in the Supplemental Materials section.
RESULTS
Bioinformatics design of human and mouse exon and domain CRISPR libraries
Recent studies have identified a number of rules that govern the potency and off-target effect of SpCas9 sgRNAs (9,11,13–16). Taking advantage of these findings, we utilized a bioinformatics approach that combined both sgRNA potency prediction and off-target prediction to generate a curated set of sgRNAs that target almost all human and mouse protein coding genes. To do so, we first generated databases of all sgRNA 20-mers (with the ‘NGG’ PAM context) that map to constitutive exons of human and mouse protein coding genes (Figure 1). Each sgRNA is therefore expected to knock out all splice variants of its target gene. To evaluate each sgRNA for its on-target potency, we used a published potency prediction algorithm (9) to assign each sgRNA a potency score that ranges between 0 and 1 (corresponding to the lowest to highest predicted potency, respectively). To identify sgRNAs with low potentials for off-target effect, we first removed any sgRNAs that perfectly matched any off-target sites in the genome beyond its intended on-target site. For the remaining sgRNAs, we used a modified version of a published algorithm (11) to assign a set of guide scores that ranges between 0 and 100 (corresponding to the most to least predicted off-target effects, respectively). The guide score for a sgRNA was calculated based on how many one, two and three mismatched off-target sites it has and the positions in the sgRNA 20-mer where the mismatches occur. Because off-target sites in coding regions are more likely to interfere with functional studies than off-target sites in non-coding regions, for each sgRNA we calculated its guide score separately using only coding regions (guide_c score), only non-coding regions (guide_nc scores), and the whole genome (guide_g score). We used both the guide_c scores and guide_nc scores as measures for a sgRNA's off-target effects.
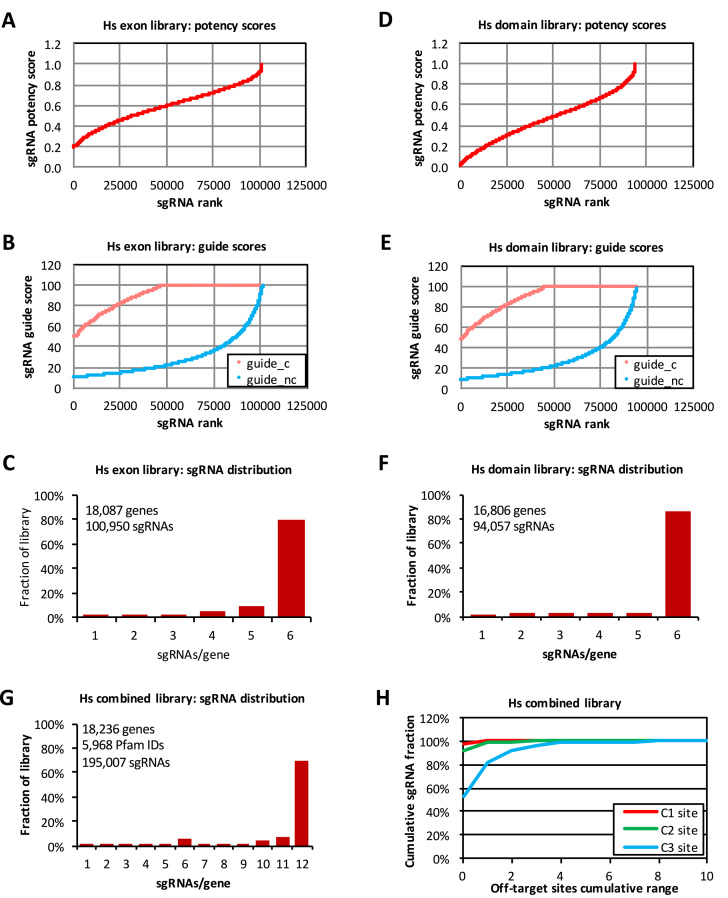
Figure 1.
Human CRISPR library characteristics. (A) Potency score distribution of the human (Hs) exon library. The potency score cut-off was 0.2. (B) Guide score distribution of the human exon library. Library sgRNAs were ranked separately for their guide score for coding regions (guide_c) and guide score for non-coding regions (guide_nc). The cut-off for guide_c score was 50 and for guide_nc was 10. (C) Distribution of sgRNA counts for all genes in the human exon library. (D) Potency score distribution of the human domain library (no cut-off was imposed for this library). (E) Guide score distribution of the human domain library. Library sgRNAs were ranked separately for their guide_c scores (cut-off at 50) and guide_nc scores (cut-off at 10). (F) Distribution of sgRNA counts for all genes in the human domain library. (G) Distribution of sgRNA counts for all genes in the combined human exon and domain library. (H) Distribution of off-target site counts for sgRNAs in the combined human exon and domain library. C1, C2 and C3 represent off-target sites in coding regions with up to one, two or three mis-matches, respectively.
To compile a CRISPR library that targets the constitutive exons of human genes, we selected sgRNAs using a set of criteria based on their potency score, guide score and exon location (Supplementary Figure S1). This library, which we referred to as the human exon library, consists of 100,950 sgRNAs targeting 18,087 human genes. All sgRNAs in this library have potency scores ≥0.2, with 83.3% of sgRNAs having potency scores ≥ 0.4, and 50.5% sgRNAs having potency scores ≥0.6 (Figure 1A). Thus, the majority of the sgRNAs in the library are expected to be highly effective at gene knockout (9). In addition, all sgRNAs in the exon library were selected to have guide_c scores ≥50 and guide_nc scores ≥10 (Figure 1B). Thus, the majority of the sgRNAs are expected to have low off-target effects (11). For each gene, we selected up to six sgRNAs that mapped to up to five of the 5΄ coding exons of the gene, with no more than two sgRNAs from the same exon. This increases the likelihood that at least some of sgRNAs will knock out the target gene's expression. For small genes and for genes with less than three exons, we allowed more than two sgRNAs to map to the same exon in order to curate six sgRNAs. For a small number of genes, fewer than six sgRNAs/gene satisfied our selection criteria. Nevertheless, in the human exon library 80.1% genes have six sgRNAs /gene, and 85.6% of the genes have at least four sgRNAs/gene (Figure 1C).
Recent studies suggest that sgRNAs targeting conserved domain regions of a protein could be particularly effective at disrupting the protein's function (5,17). We therefore further curated domain-targeted sgRNAs from our database to compile a library that we termed the human domain library. To do so, we first used the Pfam database to map regions in a human protein that belong to conserved domains. We then back-mapped the domains’ amino acid sequences to their corresponding exon regions in the genome and only selected sgRNAs that were located within these exon regions. We selected up to six sgRNAs per gene that target domains with the highest potency scores. Because 51% of sgRNAs in the exon library happened to map to protein domains fortuitously, any sgRNAs that were already present in the exon library were not selected again into the domain library to avoid sgRNA redundancy between the two libraries. The resulting domain library consists of 94,057 sgRNAs targeting 16,806 human genes. Among these sgRNAs, 84.8%, 59.6% and 28.5% have potency scores that are ≥0.2, ≥0.4 and ≥0.6, respectively (Figure 1D); and all sgRNAs have guide_c scores ≥50 and guice_nc scores ≥10 (Figure 1E). In the domain library, 86.2% of genes have six sgRNAs/gene and 92.0% of genes have at least four sgRNAs/gene (Figure 1F).
Because we designed the exon and domain libraries to have no overlapping sgRNA sequences, together the combined human exon and domain library consists of 195,007 sgRNAs targeting 18,236 genes. In the combined library, 81.5% of the genes having ≥10 sgRNAs/gene and 93.9% of the genes having ≥6 sgRNAs/gene, and a total of 5,968 Pfam domains were targeted (Figure 1G). A complete list of the human exon library and domain library sgRNAs is shown in Supplementary Table S1.
One of our goals in designing these CRISPR libraries is to balance the potency and off-target effects of a sgRNA. None of the sgRNAs in the library were allowed to have any perfectly matched off-target sites elsewhere in the genome. The majority of the sgRNAs also have very few predicted off-target sites in coding regions: in the combined human exon and domain library, 98.1%, 92.0% and 53.2% of the sgRNAs have zero off-target sites in coding regions with up to one, two and three mismatches, respectively; and 100%, 99.7% and 92.4% of the sgRNAs have ≤2 off-target sites in coding regions with up to one, two and three mismatches, respectively (Figure 1H). Thus, our library design should minimize off-target effects in a CRISPR KO screen.
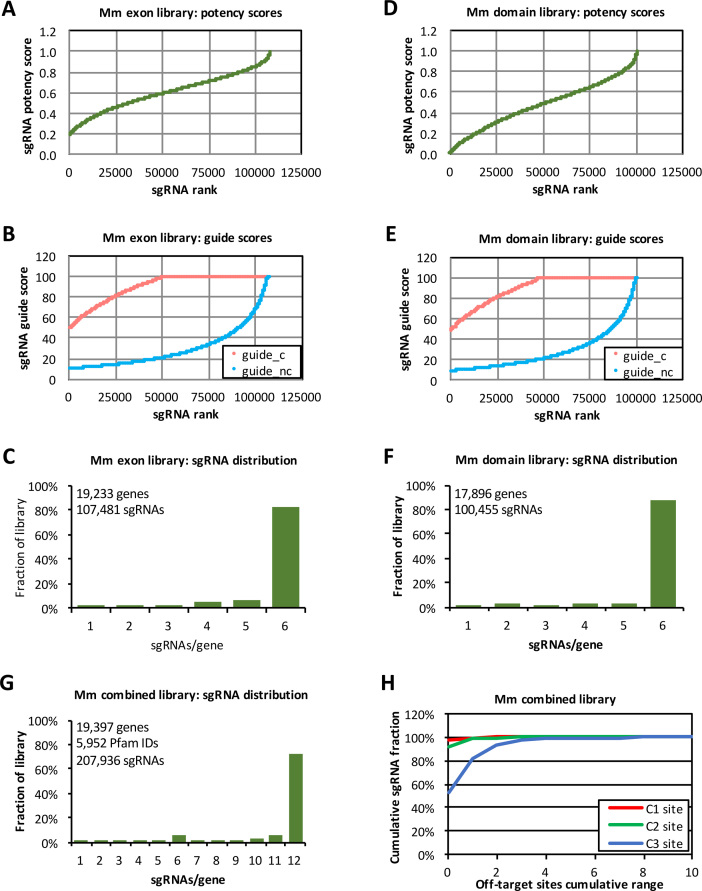
Using the same approach, we designed genome-wide exon and domain CRISPR KO libraries for the mouse genome. The overall characteristics of the mouse libraries are similar to those of the human libraries. In the mouse exon library, we curated 107,481 sgRNAs targeting 19,233 protein coding genes, with 82.1% of gene having six sgRNAs/gene and 93.7% of genes having ≥4 sgRNAs/gene (Figure 2A–C). In the mouse domain library, we curated 100,455 sgRNAs targeting 17,896 genes, with 87.0% of gene having six sgRNAs/gene and 92.3% of genes having ≥4 sgRNAs/gene (Figure 2D–F). In total, the combined mouse exon and domain libraries contain 207,936 sgRNAs (Supplementary Table S2) targeting 19,397 genes and 5,952 Pfam domains, with 93.5% of genes having ≥6 sgRNAs/gene (Figure 2G). Similar to the human libraries, most sgRNAs in the mouse libraries have very few off-target sites in coding regions. In the combined mouse exon and domain libraries, 97.5%, 91.2% and 53.4% of sgRNAs have zero off-target sites in coding regions with up to one, two and three mismatches, respectively; and 99.9%, 99.5% and 92.8% of sgRNAs have ≤2 off-target sites in coding regions with up to one, two and three mismatches, respectively (Figure 2H).
Figure 2.
Mouse CRISPR library characteristics. (A) Potency score distribution of the mouse (Mm) exon library. The potency score cut-off was 0.2. (B) Guide score distribution of the mouse exon library. Library sgRNAs were ranked separately for their guide score for coding regions (guide_c) and guide score for non-coding regions (guide_nc). The cut-off for guide_c score was 50 and for guide_nc was 10. (C) Distribution of sgRNA counts for all genes in the mouse exon library. (D) Potency score distribution of the mouse domain library (no cut-off was imposed for this library). (E) Guide score distribution of the mouse domain library. Library sgRNAs were ranked separately for their guide_c scores (cut-off at 50) and guide_nc scores (cut-off at 10). (F) Distribution of sgRNA counts for all genes in the mouse domain library. (G) Distribution of sgRNA counts for all genes in the combined mouse exon and domain library. (H) Distribution of off-target site counts for sgRNAs in the combined mouse exon and domain library. C1, C2 and C3 represent off-target sites in coding regions with up to one, two or three mismatches, respectively.
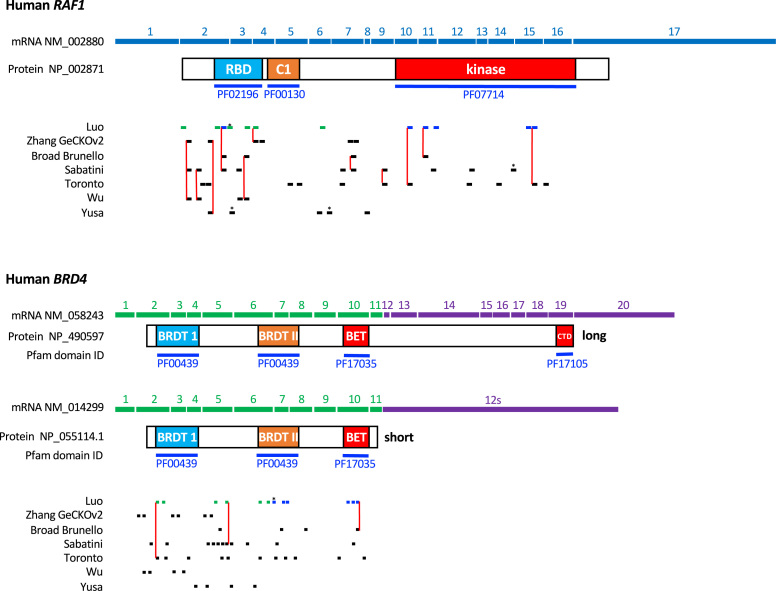
Several genome-wide CRISPR KO libraries have been generated using distinct guide RNA picking algorithms. We therefore compared the sgRNA composition of our human libraries with six published human CRISPR KO libraries: Zhang human GeCKOv2 KO library (12), Sabatini human KO library (18), Broad human Brunello KO library (16), Toronto human KO library (8), Wu human KO library (19), and Yusa human KO library (20). Each of these libraries target most of the protein coding genes in the human genome, although they vary in library coverage between ∼4 sgRNAs/gene to ∼11 sgRNAs/gene (Supplementary Table S4A). To illustrate the difference among these libraries, we mapped sgRNAs from each library to two representative genes. We chose the kinase RAF1, which has a single splice variant and the bromodomain protein BRD4, which has two splice variants that differ in their C-terminus (Figure 3). For each gene, some sgRNA sequences are present in multiple libraries, although the majority of sgRNAs are unique to each library. At the library level, the sgRNA overlap (i.e. number of identical sgRNAs) between any two libraries were relatively small, at only 5–20% of the sgRNAs (Supplementary Table S4B). This is not entirely surprising because each library only represents <5% of all 4.3 × 106 possible sgRNA sequences. Hence, minor differences in selection criteria could result in different sgRNAs being selected. Similarly, we carried out sgRNA overlap analysis with three published mouse CRISPR KO libraries: Zhang mouse GeCKOv2 KO library (12), Broad mouse brie KO library (16), and Yusa mouse KO library (20). We observed similar degree of sgRNA overlap (Supplementary Tables S5A and S5B).
Figure 3.
Location of sgRNAs from various CRISPR KO libraries in the human RAF1 and BRD4 genes. The positions of sgRNAs from our library and other published genome-wide libraries (8,12,16,18–20) in the gene are indicated as short lines. For our library, sgRNAs from the exon library are colored green and those from the domain library are colored blue. An * above the sgRNA indicates the target sequence spans an intron–exon junction. Identical sgRNA sequences from different libraries are connected by a vertical red line.
Parallel retrieval of library oligos for custom sgRNA pool construction
Pooled CRISPR library is a powerful tool for functional genomics screens. Typically, to construct a pooled CRISPR library, sgRNA sequences together with flanking PCR primers are synthesized on a high density oligo array, cleaved off, and PCR cloned into expression vectors as a mixture (3,4). Because oligo arrays have a pre-determined capacity format with a fixed number of oligo features (e.g. 12k, 90k or 1000k features per array), the size of a pooled CRISPR library often has to conform to the feature number on the oligo array. On the other hand, if smaller pools are desired, synthesizing each pool on a dedicated oligo array becomes costly as each oligo sequence will need to be duplicated to occupy multiple features on the array until all the features are filled. To circumvent this problem and enable the economical construction of CRISPR library pools of any size, we developed a parallel retrieval method to allow the construction of multiple CRISPR library pools from a single, high-capacity oligo array.
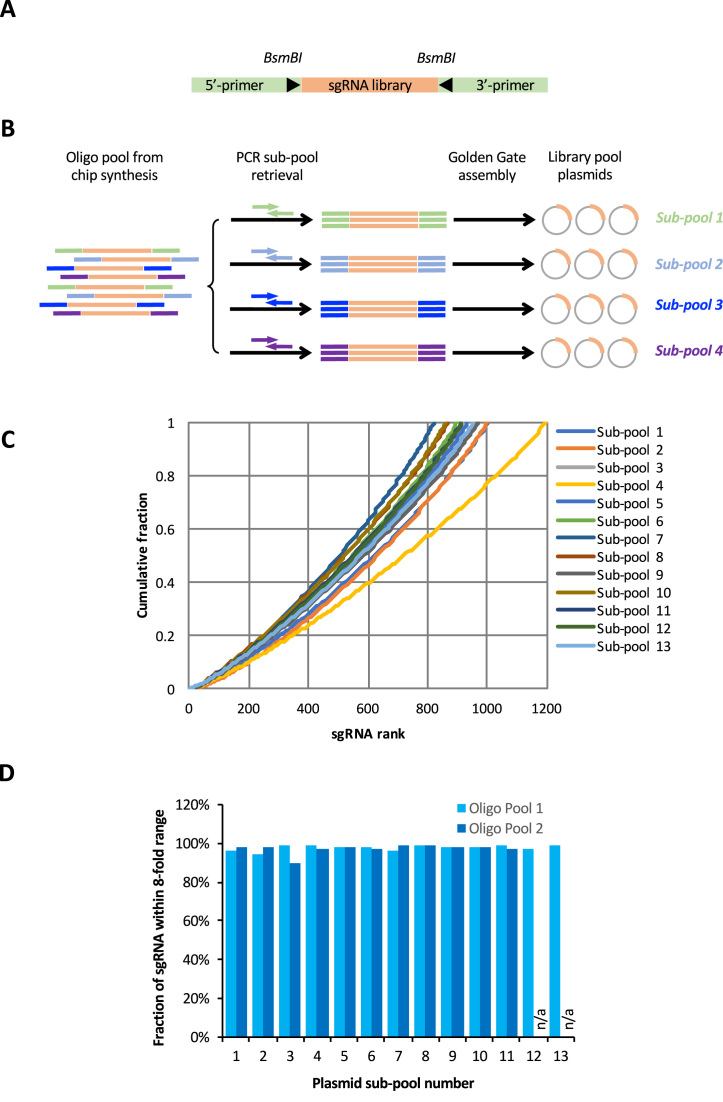
We designed our CRISPR library sequences for array synthesis as follows. Each sgRNA 20-mer was flanked by primer sequences and BsmBI sites for subsequent PCR cloning into expression vectors (Figure 4A). However, instead of using the same PCR primer pair for the entire array, we designed a set of PCR primers that can be used in multiplex format with our human and mouse sgRNA library sequences (Supplementary Table S3). These primers are GC-balanced and they do not have significant sequence similarity to library sgRNAs. Thus, the fixed number of oligo features on one array can be sub-divided into may sub-pools by using a different primer pair for each sub-pool (Figure 4B).
Figure 4.
Human CRISPR library construction and sgRNA read distribution. (A) Schematics of sgRNA oligo design. The pre-designed sgRNA 20-mers are flanked by 5΄- and 3΄-PCR primers and BsmBI sites. (B) Schematics of parallel retrieval of oligo pools. Library oligos with different flanking primer sequences were synthesized together as a single 12k oligo pool. Specific primer pairs were used to PCR amplify and retrieve subsets of oligos from the master pool. The PCR amplicons were cloned into lentiviral vectors using Golden Gate assembly. (C) Distribution of normalized sgRNA read frequency from 13 distinct plasmid sub-pools generated from a single oligo pool. (D) Percentage of sgRNA reads that are within an 8-fold range from 24 plasmid sub-pools generated from two master oligo pools.
To demonstrate that sub-pools can be reliably retrieved separately and cloned into plasmid vectors from a master oligo pool generated by array synthesis, we synthesized 12,251 sgRNA sequences using a 12k oligo array. These sgRNAs target 2,144 human kinase, phosphatase and transcription factor genes. The sgRNAs were bioinformatically divided into 13 sub-pools of ∼1,000 sgRNAs/sub-pool that map to ∼170 genes/sub-pool. Each sub-pool was given a unique pair of flanking primers for their selective retrieval from the master oligo pool. We used 13 parallel PCR reactions with low cycle numbers to retrieve each sub-pool separately from the master oligo pool, and we subsequently cloned each sub-pool into lentiviral expression plasmid using Golden Gate ligation (Supplementary Figure S2).
Illumina sequencing of the 13 plasmid pools showed that each sub-pool was retrieved from the master pool with comparable efficiency and consistency. Most of the designed sgRNA sequences were present in the plasmid pool after cloning, with only 0.2–1.2% of the sgRNAs failed to attain reads in the plasmid sub-pool at a sequencing coverage of ∼150. More importantly, the relative representation of each sgRNA within each plasmid sub-pool was highly consistent, with few sgRNAs that were either under- or over-represented in a given sub-pool (Figure 4C). We repeated this strategy with a second oligo array of 11,282 sgRNAs designed to target 1970 human metabolic enzyme genes. We bioinformatically divided the sgRNAs into 11 sub-pools for parallel PCR retrieval. Sequencing analysis of these two independent experiments, which involved the generation of 24 sub-pools from two independent oligo arrays, showed that our parallel retrieval strategy was highly consistent. In all 24 plasmid sub-pools, the within-pool raw sequencing reads for most sgRNAs fall in a narrow range (Supplementary Figure S3A and S3B), and > 90% of the sgRNAs were within an 8-fold range of representation to each other (Figure 4D).
We analyzed whether sgRNA representation in each plasmid sub-pool is influenced by the cloning representation during the bacterial transformation stage. The cloning representation (i.e. the fold-coverage of total number of bacterial transformants over pool complexity) for plasmid sub-pools varied between 62 and 410 in our experiments (Supplementary Figure S4). The variance of sgRNA sequence reads within each plasmid sub-pool was consistently between 0.05 and 0.15 for all but one sub-pool. Interestingly, the variance remained largely uncorrelated with the cloning representation of these sub-pools (Supplementary Figure S4). Thus, a cloning representation as low as ∼60 bacterial transform ants per sgRNA might be sufficient to yield a highly uniform plasmid library in this context. Taken together, these data indicate that our parallel retrieval method is highly robust at generating multiple customized CRISPR library pools using a single oligo pool from array synthesis.
DISCUSSION
In this study, we presented bioinformatics and technical resources that enable individual academic labs to create cost-effective custom CRISPR libraries for functional genomics screening. We generated SpCas9-compatible sgRNA databases that target most human and mouse protein coding genes. In designing the sgRNA sequences, we took several factors into consideration to balance sgRNAs features and achieve an optimal library design. The exon library and the domain library each has a coverage of ∼5.5 sgRNAs/gene and the combined library has a coverage of ∼11 sgRNAs/gene on average. This degree library coverage should be sufficiently deep for most screening purposes (7,18,21). Depending on the application and gene coverage depth desired, some or all of the sgRNAs for each gene can be chosen as part of a custom CRISPR library. In curating sgRNA sequences, we took particular care to balance both potency and off-target properties of sgRNAs. The majority of sgRNAs in our libraries have good scores with both of these measures. To further maximize the likelihood that these sgRNAs will effectively inactivate their target genes, we took two curation approaches such that for a given gene, half of its sgRNAs will target multiple 5΄ constitutive exons whereas the other half will target constitutive exons that encode conserved protein domains. The resulting human and mouse library, which constitute ∼200,000 sgRNAs each, represent ∼4.5% of the total candidate sgRNA sequences that are in the ‘NGG’ PAM context. Thus, we believe these curated sgRNA sequences will be useful for both custom library construction and for knocking out the expression of individual genes.
Prior to our library design, a number of genome-wide CRISPR KO libraries have been designed for the human and mouse genome (8,12,16,18–20). The relatively small sgRNA sequence overlap among these libraries, including ours, might be attributable to the fact that each library uses a distinct set of rules to pick <5% of all possible sgRNAs. Because sgRNA selection scores with any given algorithm is a near-continuous variable, a small difference in selection criteria could result in a fairly large difference in the sequence of the selected sgRNAs. Our library is the only one that balances sgRNA potency, off-target effects and domain targeting in a single design.
Our human and mouse CRISPR library design serve as a starting point for the construction of custom CRISPR libraries of virtually any size. Although a number of genome-wide CRISPR libraries have been made available (8,12,16,18–20), there is an increasing need among individual investigators to screen smaller, customized libraries that target a specific set of genes. This could be a functional category of genes such as kinases and metabolic enzymes, or a group of genes that represent either an over-expression signature or a mutational signature. Furthermore, with CRISPR screen moving towards in vivo applications (22), smaller libraries are likely to have better signal-to-noise ratio than genome-wide libraries in these settings because cell number, and hence library representation, could become a significant limiting factor in animal models (23,24). Our strategy thus enables the easy and cost-effective construction of sub-genome sized libraries to meet these special screening needs. We demonstrated that a dozen custom CRISPR library plasmid pools can be generated with a single master oligo pool from array synthesis. Although we chose to generate library pools of ∼1,000 sgRNAs, our method is scalable and should enable the generation of sub-genome scale CRISPR libraries of virtually any size.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Cancer Institute Intramural Grant [ZIA BC 011304 to J.L.]. Funding for open access charge: NIH Intramural Funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Shalem O., Sanjana N.E., Zhang F.. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015; 16:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Doudna J.A., Charpentier E.. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346:1258096. [DOI] [PubMed] [Google Scholar]
- 3. Wang T., Wei J.J., Sabatini D.M., Lander E.S.. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014; 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014; 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Munoz D.M., Cassiani P.J., Li L., Billy E., Korn J.M., Jones M.D., Golji J., Ruddy D.A., Yu K., McAllister G. et al. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov. 2016; 6:900–913. [DOI] [PubMed] [Google Scholar]
- 6. Aguirre A.J., Meyers R.M., Weir B.A., Vazquez F., Zhang C.-Z., Ben-David U., Cook A., Ha G., Harrington W.F., Doshi M.B. et al. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 2016; 6:914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morgens D.W., Deans R.M., Li A., Bassik M.C.. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016; 34:634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S. et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015; 163:1515–1526. [DOI] [PubMed] [Google Scholar]
- 9. Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E.. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014; 32:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanjana N.E., Shalem O., Zhang F.. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014; 11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heigwer F., Kerr G., Boutros M.. E-CRISP: fast CRISPR target site identification. Nat. Methods. 2014; 11:122–123. [DOI] [PubMed] [Google Scholar]
- 14. Liu H., Wei Z., Dominguez A., Li Y., Wang X., Qi L.S.. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics. 2015; 31:3676–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.-D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J.. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015; 12:982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016; 34:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi J., Wang E., Milazzo J.P., Wang Z., Kinney J.B., Vakoc C.R.. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 2015; doi:10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M.. Identification and characterization of essential genes in the human genome. Science. 2015; 350:1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma H., Dang Y., Wu Y., Jia G., Anaya E., Zhang J., Abraham S., Choi J.-G., Shi G., Qi L. et al. A CRISPR-based screen identifies genes essential for West-Nile-Virus-induced cell death. Cell Rep. 2015; 12:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tzelepis K., Koike-Yusa H., De Braekeleer E., Li Y., Metzakopian E., Dovey O.M., Mupo A., Grinkevich V., Li M., Mazan M. et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016; 17:1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo B., Cheung H., Subramanian A., Sharifnia T., Okamoto M., Yang X., Hinkle G., Boehm J., Beroukhim R., Weir B. et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2008; doi:10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen S., Sanjana N.E., Zheng K., Shalem O., Lee K., Shi X., Scott D.A., Song J., Pan J.Q., Weissleder R. et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015; 160:1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zender L., Xue W., Zuber J., Semighini C.P., Krasnitz A., Ma B., Zender P., Kubicka S., Luk J.M., Schirmacher P. et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008; 135:852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.-K., Jang H.G., Jha A.K. et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011; 476:346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.