Abstract
The dynamics of cell morphology in eukaryotes is largely controlled by small GTPases of the Rho family. Rho GTPases are activated by guanine nucleotide exchange factors (RhoGEFs), of which diffuse B-cell lymphoma (Dbl)-like members form the largest family. Here, we surveyed Dbl-like sequences from 175 eukaryotic genomes and illuminate how the Dbl family evolved in all eukaryotic supergroups. By combining probabilistic phylogenetic approaches and functional domain analysis, we show that the human Dbl-like family is made of 71 members, structured into 20 subfamilies. The 71 members were already present in ancestral jawed vertebrates, but several members were subsequently lost in specific clades, up to 12% in birds. The jawed vertebrate repertoire was established from two rounds of duplications that occurred between tunicates, cyclostomes, and jawed vertebrates. Duplicated members showed distinct tissue distributions, conserved at least in Amniotes. All 20 subfamilies have members in Deuterostomes and Protostomes. Nineteen subfamilies are present in Porifera, the first phylum that diverged in Metazoa, 14 in Choanoflagellida and Filasterea, single-celled organisms closely related to Metazoa and three in Fungi, the sister clade to Metazoa. Other eukaryotic supergroups show an extraordinary variability of Dbl-like repertoires as a result of repeated and independent gain and loss events. Last, we observed that in Metazoa, the number of Dbl-like RhoGEFs varies in proportion of cell signaling complexity. Overall, our analysis supports the conclusion that Dbl-like RhoGEFs were present at the origin of eukaryotes and evolved as highly adaptive cell signaling mediators.
Keywords: Dbl, guanine nucleotide exchange factors, Rho GTPases, cell signaling
Introduction
The Ras-like superfamily is made of 167 proteins in human, which are distributed into five major families (Arf/Sar, Rab, Ran, Ras, Rho) and are highly conserved across evolution. Orthologs generally share 65–85% amino-acid sequence similarity, even between distantly related clades (Rojas et al. 2012). The extreme conservation is in keeping with their roles in basic cellular functions such as endo/exocytosis, F-actin dynamics, vesicular and nucleo-cytoplasmic trafficking. Ras-like proteins biochemically act as binary signaling switches that rely on structural changes between their GDP-bound and GTP-bound conformations (Bosco et al. 2009; Raimondi et al. 2011). Ras-like GTPases undergo activation and inactivation steps. Activation is promoted by guanine nucleotide exchange factors (GEFs), which reduce affinity of the GTPase for nucleotides thereby allowing entry of GTP, which is more abundant than GDP in the cytosol. When bound to GTP, Ras-like proteins activate a set of downstream effector proteins that mediate their cellular effects. Inactivation of Ras-like GTPases is controlled by GTPase activating proteins (GAPs), which stimulate intrinsic GTPase activity and thus favor the inactive GDP-bound form.
Members of the Rho family control F-actin dependent reorganization of the cell membrane and associated intracellular macromolecular scaffolds. Consequently, they play major roles in cell adhesion, polarity and locomotion processes. The human genome encodes 20 Rho GTPases, including the well-studied Rac1, Cdc42, and RhoA (Boureux et al. 2007), as well as 82 RhoGEFs. The RhoGEFs can be divided into two families. There are 11 DOCK (Dedicator Of CytoKinesis)-related proteins (Meller et al. 2005) and the remaining 71 form the Dbl-like family, due to their similarity to the Dbl (diffuse B-cell lymphoma) protein, which is an oncogenic protein that activates the Cdc42 Rho GTPase (Eva and Aaronson 1985; Hart et al. 1991). Dbl-like RhoGEFs all share a 170–190 amino acid Dbl Homology (DH) domain, which is responsible for the guanine nucleotide exchange activity on Rho GTPases (Jaiswal et al. 2013; Rossman et al. 2005). Humans also have ∼70 RhoGAPs (Amin et al. 2016; Tcherkezian and Lamarche-Vane 2007) and over 70 effectors (Bustelo et al. 2007). The Rho signaling module is thus a complex regulatory network that includes over 240 proteins.
Only a few Dbl-like members have been functionally studied in model organisms like Drosophila (Sone et al. 1997), Caenorhabditiselegans (Steven et al. 2005), yeast (Hart et al. 1991) and Dictyostelium discoideum (Vlahou and Rivero 2006) since the late 1990s. Most Dbl-like RhoGEFs have a high diversity of functional domains, in contrast with DOCK RhoGEFs that have a conserved core organization, made of DHR1/C2 and DHR2 domains, either alone or associated with single SH3 or PH domains (Meller et al. 2005). In addition to the DH domain, Dbl RhoGEFs contain domains that either mediate interaction with membranes, proteins or phosphorylated amino acids (e.g., C1, SH2, SH3, PDZ, PH domains) or that have diverse enzymatic activities (e.g., kinases, phosphatases, GEF or GAP).
The physiological importance of cell adhesion and locomotion, combined with the complexity of the Rho signaling network, raise the issue of when this network emerged and how it evolved in eukaryotic taxa, in particular in relation with multi-cellularity. We previously reported that the Rho GTPase family was already present as Rac-like proteins in the Last Eukaryotic Common Ancestor (LECA; Boureux et al. 2007), that is, 1.7–2.3 billion years ago (Hedges et al. 2004; Parfrey et al. 2011). The complexity of the Rho family remained at a low level in unicellular eukaryotes, fungi and ancestral metazoa, which have a minimal Rho family repertoire, comprising just Rac, Cdc42, and RhoA. The Rho family expanded in Metazoa (i.e., around 700 million years ago [Ma]), as a consequence of duplications and lateral gene transfers (LGTs). In contrast, little is known about the evolutionary history of the Dbl-like RhoGEF family, in particular how members are related to each other within and between eukaryotic clades and how the family evolved in terms of diversity, gain/loss events, and domain organization.
Here we performed a comprehensive analysis of Dbl-like RhoGEF sequences from all eukaryotic supergroups. In most eukaryotic clades, several species were examined, thus reducing the impact of incomplete assemblies on RhoGEF identification. Using annotation and phylogenetic tools, we trace the history of Dbl-like RhoGEF proteins back from extant species to the LECA and reveal a much higher plasticity of Dbl-like repertoires compared with Rho families.
Materials and Methods
Genomes and Annotated Sequences
Most sequences were retrieved from the NCBI annotated databases (nr and EST, http://www.ncbi.nlm.nih.gov), using NCBI PHI-BLAST as well as BLAST and Annotation search tools available in the Geneious 9.1.6 software package (Biomatters, http://www.geneious.com/). For specific searches, additional genome browsers were used (see supplementary table S6, Supplementary Material online). Protein sequences derived from genomes lacking annotations were annotated by searching Pfam, CDD or SMART domain databases using the InterProScan tool, integrated in the Geneious software. The InterProScan tool is freely available on the InterPro resource (http://www.ebi.ac.uk/interpro/).
Sequence Alignments
Amino acid sequences were aligned using MAFFT v7.017, available in the Geneious 9.1.6 package (Katoh et al. 2002). For human Dbl RhoGEFs, alignments were confirmed by using MUSCLE (Edgar 2004) and Promals3D (Pei et al. 2008), (http://prodata.swmed.edu/promals3d/promals3d.php), using the ARHGEF7 (PDB: 1by1) and ARHGEF3 (PDB:2z0q) X-ray 3D structures as guides. MSAs were manually edited for minor corrections and processed by BMGE (Block Mapping and Gathering with Entropy, Criscuolo and Gribaldo 2010) with a 0.6 cut-off value.
Phylogenetic Analyses
Phylogenetic trees were estimated by two probabilistic methods, that is, ML PhyML (Guindon and Gascuel 2003) and Bayesian analysis MrBayes (Ronquist et al. 2012), as implemented in Geneious. ML returns the topology that maximizes the likelihood function and estimates node support (i.e., robustness of the topology) by nonparametric bootstrapping. MrBayes samples trees according to their PP and directly measures node support. Models for amino acid substitution were chosen using ProtTest (Abascal et al. 2005). In most analyses, the best-fitting model was LG + I+G. PhyML was set-up using the gamma shape and proportion of invariable site parameters produced by ProtTest. ML trees were optimized for topology, length and rate and were generated using the best of nearest-neighbor interchange and subtree-pruning-regrafting tree search algorithms, with 100 bootstrap replicates.
MrBayes consensus trees were generated after two independent runs of four Markov chains for 1,100,000 generations sampled every 200 generations, with sampled trees from the first 100,000 generations discarded as “burn-in”. At the end of each run, average standard deviations of split frequencies were below 0.01 and the estimated sample sizes (ESS) were above 200 for all sampled parameters. Minimum ESS values were 787.01 (fig. 1A), 247.52 (fig. 3A), 313.87 (fig. 3B), 298.59 (fig. 3C), 347.39 (fig. 3D), 428.16 (fig. 6C left panel), and 591.28 (fig. 6C right panel). For each analysis, 50% majority-rule consensus trees and associated clade PPs were computed from sample trees. Trees were visualized and exported as PDF files with FigTree (v1.4.2, http://tree.bio.ed.ac.uk/software/figtree/) then assembled in Adobe Illustrator. PP values below 0.95 are considered unreliable for topological reconstruction (Erixon et al. 2003). Divergence times between taxa were collected from the TimeTree database (Hedges et al. 2006) (http://www.timetree.org/).
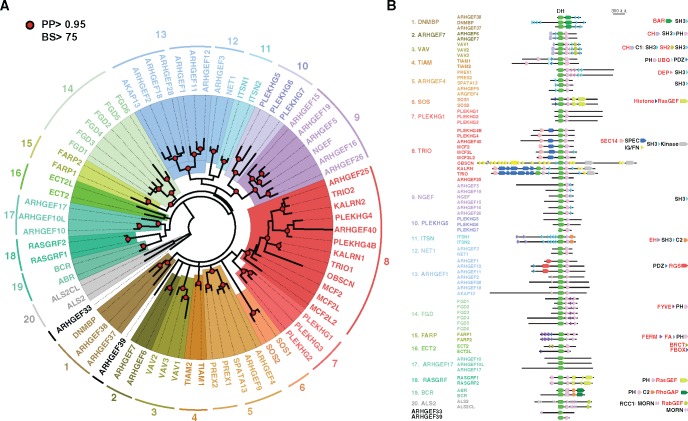
Fig. 1.
—The 71 human Dbl-like RhoGEFs cluster into 20 structurally related subfamilies. (A) Phylogenetic cladogram of DH domains. The tree was deduced from multiple sequence alignment and processed by PhyML and MrBayes analysis. RhoGEFs subfamilies were delineated from node supports (PP: Posterior probability, BS: Bootstrap proportion). Only highly supported nodes (PP > 0.95 and BS > 75) are indicated (red circle). Numbers indicate subfamilies. (B) Subfamilies cluster members with similar functional domain organization. For each subfamily, the structural domains associated with the catalytic DH domain are indicated. All proteins are drawn at the same scale, except OBSCN. Domains typical of subfamilies are in red. BAR: Bin, Amphiphysin, Rvs; SH3: Src homology 3; CH: Calponin homology; PH: Pleckstrin homology; C1: N-terminal region of PKC; SH2: Src homology 2; UBQ: Ubiquitin; DEP: SEC14: Dishevelled, Egl10, Pleckstrin; SPEC: Spectrin homology; IG: Immunoglobulin-like; FN: Fibronectin-like; EH: Eps15 homology; C2: Ca2+ binding domain of PKC; PDZ: PSD95, Dlg1, Zo-1; RGS: Regulator of G protein Signaling; FYVE: Fab1, YOTB, ZK632.12, Vac1, EEA1; FERM: Four-point-one, Ezrin, Radixin, Moesin; FA: FERM Adjacent; BRCT: BRCA1 C-Terminus; RCC1: Regulator of Chromosome Condensation 1; MORN: Membrane Occupation and Recognition Nexus.
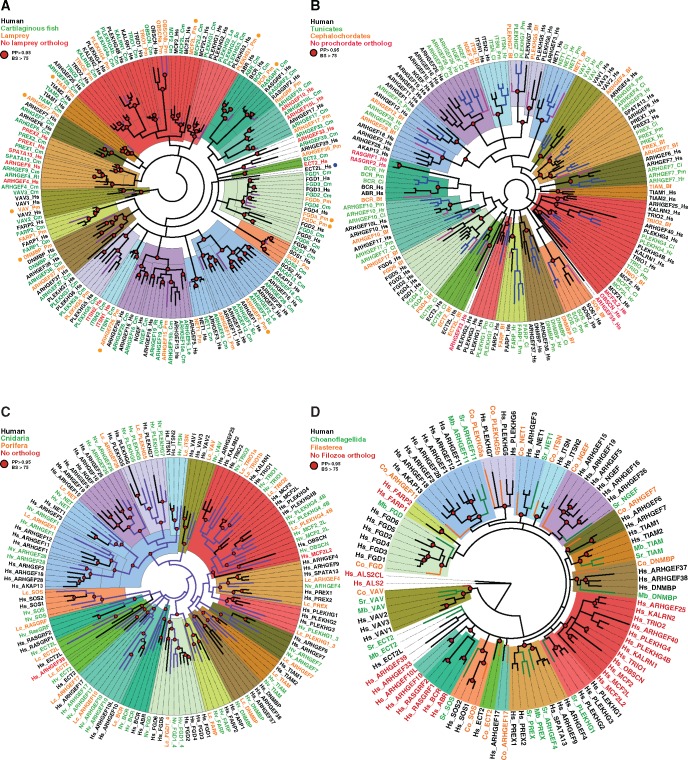
Fig. 3.
—Phylogenetic analyses of Dbl-like RhoGEFs in Metazoa. (A) Clustering of human and early vertebrate RhoGEFs. Hs: Homo sapiens; Cartilaginous fishes: Cm: Callorhinchus milii; Le: Leucoraja erinacea; Rt: Rhincodon typus; Lamprey: Pm: Petromyzon marinus. Lamprey members (names in orange and colored branches) at the roots of subfamilies or clusters are figured by an orange dot. No ortholog to ECT2L (blue dot) was found in Cm, Le, Rt or Pm genomes. (B) Clustering of human and prochordate RhoGEFs. Tunicates (names in green, blue branches): Ci: Ciona intestinalis; Hr: Halocynthia roretzi; Pm: Phallusia mammillata; Cephalochordate (names in orange, red branches) Bf: Branchiostoma floridae. (C) Clustering of human and early metazoan RhoGEFs. Cnidaria (green): Hv: Hydra vulgaris; Nv: Nematostella vectensis. Porifera (orange): Lc: Leucosolenia complicata. (D) Clustering of human and nonmetazoan Filozoa RhoGEFs. Choanoflagellida (green): Mb: Monosiga brevicollis; Sr: Salpingoeca rosetta. Filasterea (orange): Co: Capsaspora owczarzaki. DH domain amino acid sequences were aligned and analyzed by PhyML and MrBayes methods. Highly supported nodes are figured by a red circle. Names and accessions are listed in supplementary tables S1 and S3, Supplementary Material online. Color codes for RhoGEF subfamilies are as in figure 1A.
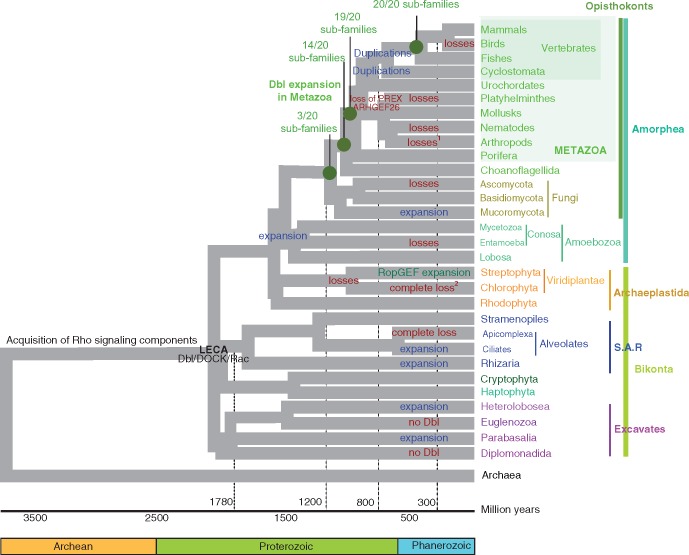
Fig. 6.
—Summary of Dbl-like RhoGEF gain and loss events across eukaryote supergroups. A global view of the phylogeny of supergroups and taxa examined is presented, with the timeline of eukaryote emergence. Gain (in blue) and loss (in red) events that have built the repertoires of extant species are indicated. 1: In Holometabola and Paraneoptera, and 2: In Chlamydomonales. The number of subfamilies, as defined in figure 1, is indicated for Metazoa. Green dots indicate nodes important for inferring the expansion of the Dbl family in Metazoa.
Species and Tissue-Wise Gene Expression Analysis
We used mRNA-seq data sets, generated from tissues of various species, as described in (Brawand et al. 2011) and (Barbosa-Morais et al. 2012). As comparison of gene expression levels between species relied on orthology relationships, we established a RhoGEF orthology table (see supplementary table S5, Supplementary Material online) by using Ensembl orthologous gene data (Flicek et al. 2014), which we further refined by phylogenetic analysis. For each species, RhoGEF data were retrieved from global mRNA-seq data and expressed as Reads Per Kilobase of transcript per Million reads mapped (RPKM). RhoGEF mRNA-seq data were then clustered using Cluster 3.0 (http://bonsai.hgc.jp/∼mdehoon/software/cluster/; de Hoon et al. 2004). Mean centered log10 (RPKM) values were normalized and hierarchically clustered (Euclidian distance, complete method). Cluster heatmaps were created with Java Treeview 1.1.6r4 (Saldanha 2004). For analysis of tissue-specificity RhoGEF expression across species, the Spearman correlation was applied to log2 (RPKM) in the nine species studied. Correlation heatmaps were drawn with Excel 14.0.0.
Results
The Human Dbl RhoGEF Repertoire Was Already Set Up at the Onset of Vertebrates
We mined the available Dbl-like RhoGEF sequences in the human genome and found the whole family includes 71 members (see supplementary table S1, Supplementary Material online). This represents 73 DH domains, since Trio and Kalirin each have two domains arranged in tandem. The amino acid sequences of the 73 DH domains are significantly divergent, sharing only 25 ± 8% identity, suggesting an ancient origin and/or a rapid evolution rate. To gain further insight into Dbl-like RhoGEF ontogeny, we deduced phylogenies by combining MrBayes and maximum-likelihood, two site-based phylogenetic approaches, on a multiple sequence alignment (MSA) of the 73 DH domains. The resulting tree topology has internal branches that are strongly supported by both analytic methods (fig. 1A). Globally, DH domains appear structured into 20 clusters or subfamilies, with just two isolated members (ARHGEF33 and ARHGEF39). Most of the shallow branches (close to the periphery of the tree) contain two members, as a likely result of the whole genome duplication that occurred at the onset of vertebrate radiation. Deeper branches connect more than two RhoGEFs (ten for TRIO, seven for ARHGEF28, six for NGEF and FGD), indicating that these groups resulted from more ancient duplications. The clustering based on DH domain sequence is further supported by the similar functional domain organization shared by members of each cluster (fig. 1B). Ontogeny of the TRIO cluster is complex, because OBSCN, TRIO and KALRN have gained kinases and immunoglobulin (IG)/fibronectin (FN) domains by recombination (see supplementary fig. S1, Supplementary Material online). Except ARHGEF33 and members of clusters 1 and 17, all Dbl-like RhoGEFs have a PH domain adjacent to the DH domain, suggesting that the two domains represent the ancestral architecture. In support of this, tree topology deduced from phylogenetic analysis of DH/PH domains (see supplementary fig. S2A, Supplementary Material online) and DH domain (fig. 1A) are highly similar and identify same clusters. PH domains appear less conserved, because PH-only based topology, although similar, has lower internal nodes supports for clusters 8, 14–15, and 9–12, and does not group cluster 13 with clusters 9–12 (see supplementary fig. S2B, Supplementary Material online). Note also that ECT2 and ECT2-like, which form a DH-based cluster with low support, failed to group when PH sequences are included in the analysis.
To gain insight in the timing of duplications detected by phylogenetic analyses, we looked for orthologs of human Dbl-like RhoGEFs across Vertebrates (fig. 2). Orthology was calculated by reciprocal BLAST analysis and comparison of structural domains. In all cases, orthologous members had unambiguous BLAST E-values, making it unnecessary to perform phylogenetic analysis. In Primates and Glires (Rodents and Lagomorpha) we identified the full set of 71 human orthologs (fig. 2 and see supplementary table S1, Supplementary Material online). However, all rodent species examined lacked PLEKHG4B and several rodent species like mice lacked MCF2L2 and PLEKHG7 (see supplementary table S2, Supplementary Material online). Very little is known about the physiological functions associated with these genes, except a moderate association of PLEKHG4B and MCF2L2 polymorphisms with type 2 diabetes and associated comorbidity (Raffield et al. 2015; Zheng et al. 2009).
Fig. 2.
—Conservation of the human Dbl-like RhoGEF repertoire across Metazoa. Human RhoGEF orthologs were searched in genomes of species covering the major Metazoa clades, as indicated on the top. In most clades, three or more species were examined and orthology was deduced from reciprocal BLAST scores (“Vertebrates”, from mammals to bony fish) and by phylogenetic analysis (shark, lampreys, Ambulacraria, Cnidaria, Porifera, and nonmetazoan Filozoa). Members that were not found are indicated by an x box. Vertical bars show duplications that were deduced from phylogenetic analyses. Dashed vertical bars indicate duplications that cannot be precisely dated. The color code for Dbl-like subfamilies is the same as in figure 1A.
The mammalian Laurasiatheria and Afrotheria share the same RhoGEF repertoire as Primates and Glires (fig. 2, see supplementary table S1, Supplementary Material online). Analysis of two Xenarthra genomes (armadillo and sloth) revealed a nearly complete set of RhoGEFs, missing only PLEKHG4B. Finally, merging analyses of the gray short-tailed opossum and Tasmanian devil genomes (Marsupials) recapitulated the 71 RhoGEF set, whereas the platypus genome (Monotremata) only lacks ALS2CL. We also identified a RhoGEF that is closely related to the drosophila GEF64C in marsupials (Bashaw et al. 2001; see supplementary table S1, Supplementary Material online). This RhoGEF was not found in other mammals or in platypus.
The RhoGEF repertoire has thus remained stable from the onset of Mammals, that is, 160–290 Ma (dos Reis et al. 2012). Such long-term stability is indicative of strong positive selection of all members. Of the 71 widely shared RhoGEFS, three were lost in several rodent species of the Murinae sub-family, suggesting they became neutral in this clade. Note that Murinae have gained a competitive advantage during middle Miocene and currently constitutes the largest extant mammalian subfamily (Tiphaine et al. 2013).
In nonmammalian Amniotes, there have been greater RhoGEF losses. The chicken genome (Sauria/Archelosauria/Dinosauria) lacked nine RhoGEFs (i.e., 12% of the repertoire; see supplementary table S2, Supplementary Material online). All nine were absent in all the 48 bird genomes available. Three of the missing RhoGEFs (ARHGEF15, PLEKHG6, and FGD1) were also missing in Crocodylia, the sister taxon of birds, while turtles (Testudines) only lacked PLEKHG6. As bony fishes and coelacanth have 72 RhoGEFs (the canonical 71 RhoGEFs plus GEF64C), losses detected in Sauria are specific to this clade and have occurred sequentially (see supplementary table S2, Supplementary Material online). Amphibia lack FGD3 and PLEKHG6 and cartilaginous fishes lack ARHGEF40 and ARHGEF11 (fig. 2, see supplementary table S2, Supplementary Material online). The absence of GEF64C in placentals can be considered as a specific loss as it is present in other vertebrate clades.
We next analyzed the Dbl-like RhoGEF repertoire in two jawless fish genomes (Lampreys, Agnatha). Merging member lists from the two genomes produced a set of 31 RhoGEF proteins. These 31 RhoGEFs include members of all the 20 mammalian RhoGEF subfamilies except ITSN (figs. 2 and 3A). Our phylogenetic analysis identified 21 vertebrate orthologs and 10 cases in which lamprey sequences branched at the roots of vertebrate clusters (orange dots in fig. 3A). This indicates that duplications took place between Agnatha and Gnathostomata, increasing the copy number from 10 to 25. The 31 RhoGEFs in extant Agnatha thus covers homologs to 46 vertebrate RhoGEFs. Since the Vertebrate repertoire is 72, including GEF64C, at most 26 RhoGEFs can be considered as missing or lost in lamprey genomes. This implies that ancestral Agnatha might have had at most 57 Dbl-like members.
We extended Dbl-like RhoGEF analysis to Tunicates, the sister clade to Vertebrates, and to Cephalochordates (lancelet). Tunicates and Cephalochordates (Prochordates) have a bilateral body plan, a notochord, a dorsal neural tube, pharyngeal slits, a postanal tail and an endostyle (Holland 2005). By combining four tunicate species, we identified 25 RhoGEFs, including members of most of the 20 mammalian subfamilies (figs. 2 and 3B, see supplementary table S1, Supplementary Material online). All species lacked ALS2, ARHGEF33, ARHGEF39, OBSCN, and RASGRF. Phylogenetic analysis showed that the vast majority of Tunicate and Cephalochordate DH domains branched at the roots of mammalian shallow clusters (fig. 3B). This indicates that widespread RhoGEF duplications took place between Tunicates and Vertebrates as a result of whole genome duplications that occurred between Tunicates and Agnatha (Smith et al. 2013). The timing of duplications cannot be precisely determined for members of the ARHGEF4, ITSN, ARHGEF1, FGD, and ARHGEF17 subfamilies, since they are absent in the two jawless fish genomes (dotted lines, fig. 2).
Thus, the ancestral vertebrate repertoire was established from two rounds of duplications: One that occurred between Tunicates and Agnatha, that is > 547 Ma, which produced 57 RhoGEFs, the second, between Agnatha and bony fishes, that is, 485 Ma, produced 15 additional RhoGEFs.
Inferring Subfunctionalization from Tissue Distribution
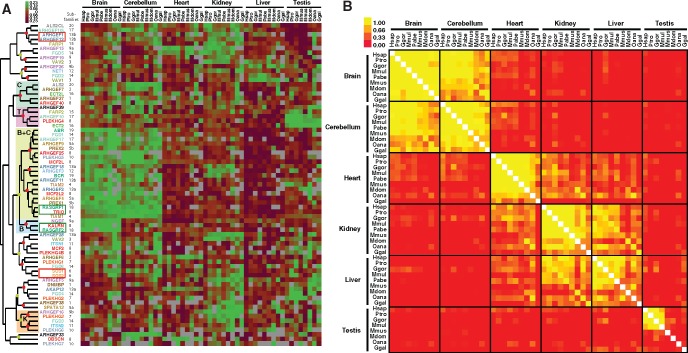
We next examined how tissue distribution of Dbl-like RhoGEF expression correlates with their ontogeny. We analyzed RhoGEF expression in high-throughput RNA sequencing (RNA-Seq) data, from brain, cerebellum, heart, kidney, liver and testes, of man, chimp, gorilla, orangutan, macaque, mouse, opossum, platypus, and chicken (Barbosa-Morais et al. 2012; Brawand et al. 2011). Hierarchical clustering of expression values shows that most RhoGEFs are differentially expressed (fig. 4). RhoGEFs grouped into a small number of well-supported coregulated clusters, based on their distribution across tissues and species. The three brain + cerebellum (B + C) sub-clusters have Pearson correlations ranging from 0.72 to 0.89. In the brain cluster (B), brain-specific expression of NGEF, KALRN and RASGRF2 is highly correlated (Pearson’s r = 0.7) and conserved from primates to chicken.
Fig. 4.
—Conservation of tissue specific expression of Dbl-like RhoGEFs in vertebrates. (A) Heatmap of RhoGEF mRNA expression in six tissues across nine vertebrate species. Orthologous RhoGEF values were extracted from global RNA-seq data. Mean centered log10(RPKM) values were normalized and hierarchically clustered (Euclidian distance, complete method). Colors of RhoGEF names and sub-families correspond to those delineated in figure 1A. Red and green frames illustrate closely related members expressed in same or distinct tissues, respectively. Red and green dots indicate clusters with Pearson correlations of > 0.7 and >0.5, respectively. B: Brain; C: Cerebellum; K: Kidney. Hsap: Homo sapiens; Ptro: Pan troglodytes; Ggor: Gorilla gorilla; Mmul: Macaca mulatta; Pabe: Pongo abelii; Mmus: Mus musculus; Mdom: Monodelphis domesticus; Oana: Ornithorhynchus anatinus; Ggal: Gallus gallus. (B) Symmetrical heat map of Pearson correlations from RhoGEF gene mRNA expression (log RPKM values) for the six tissues and nine species examined.
The tree topology of hierarchical clustering did not correlate with that of the phylogenetic analysis. This argues in favor of subfunctionalization (i.e., differential expression of the two copies), a process that promotes conservation of functional duplicate gene copies (Krakauer and Nowak 1999). Indeed, most coregulated clusters include RhoGEFs from different subfamilies, as shown in figure 4A. This is illustrated by KALRN and RASGRF2, which are mostly expressed in the brain, whereas their respective paralogs TRIO and RASGRF1 are highly expressed in the cerebellum. Conservation of RhoGEF expression across species and tissues was further estimated by Pearson correlation analysis from gene expression profiles (fig. 4B). In all species, correlation values were highly significant (>0.6), even for chicken, despite its higher divergence time (≃ 310 Myr). These data indicate that the last round of duplications within RhoGEF subfamilies was rapidly followed by subfunctionalization events and that tissue expression profiles of RhoGEFs were likely set up around 630 million years ago in early vertebrates.
Overall, this study shows that the ancestral vertebrate repertoire was established from two rounds of duplications that occurred between Tunicates and bony fishes, and since then, it has remained mostly unchanged. Since duplications were followed rapidly by subfunctionalization events, it is, therefore, implicit that major tissue-specific and Rho-controlled regulatory pathways were already set up in early Vertebrates. The Dbl-like repertoire is nearly conserved between mammals and fishes, two clades with widely different physiology. One may thus infer that the Dbl-like members have been strongly selected for their roles in basic vertebrate-specific functions. This may be more complex, as evolution of several vertebrate clades has been associated with Dbl-like RhoGEF losses: GEF64C was lost in placental mammals; PLEKHG4B and MCF2L2, two members of the TRIO subfamily, and PLEKHG7 were lost in Rodents; Nine Dbl-like RhoGEF members were lost in Birds. The losses of members that are otherwise highly conserved across Vertebrates indicate that the selective pressure exerted on tissue-specificity or expression of Rho-controlled pathways may considerably change during clade specialization.
Early Metazoans Had Most Vertebrate Dbl-like RhoGEF Sub Families
We next examined the RhoGEF repertoires in Ambulacraria (Echinoderms and Hemichordates) and Protostomia (fig. 2, see supplementary table S3, Supplementary Material online). Ambulacraria have 30 RhoGEFs (30 in Hemichordates, 28 in Echinoderms), including members of all the 20 vertebrate subfamilies except the PREX branch. Protostomes also have members of the 20 vertebrate subfamilies, but all lack the PREX and NGEF branches of the ARHGEF4 and NGEF subfamilies, respectively (fig. 2). However, Dbl-like repertoires greatly differ within Protostome clades. In Lophochotrozoa, Mollusks have the complete Protostome repertoire whereas Platyhelminthes (flatworms) lack six subfamilies (fig. 2, see supplementary table S3, Supplementary Material online). In Ecdysozoa, Crustaceans, and Hymenoptera (Insecta) lack ECT2L (fig. 2, see supplementary tables S2 and S3, Supplementary Material online). All other Insecta clades examined lack additional RhoGEFs; all lack DNMBP and NET1, while Diptera and Lepidoptera (Mecopterida) have also lost ITSN. According to holometabola phylogeny (Peters et al. 2014), DNMBP and NET1 were lost in Aparglossata (Coleoptera, Lepidoptera, and Diptera), then ITSN in Mecopterida (Lepitoptera and Diptera; see supplementary table S2, Supplementary Material online). The absence of DNMBP and NET1 in Hemiptera can be considered as independent losses. Last, Nematodes lack five Dbl-like proteins as compared with other Ecdysozoa: NET1, BCR, GEF64C, RasGRF, and ARHGEF10 (see supplementary table S2, Supplementary Material online).
Thus, the Dbl-like RhoGEF repertoire of ancestral Protostomia is very similar to that of Ambulacraria. However, with the exception of Mollusks, several RhoGEF subfamilies were lost in other Protostomia clades (up to five in Lepidoptera and Nematodes). In addition to vertebrate orthologs, three specific RhoGEFs were found in most Protostomia clades: PsGEF (Higuchi et al. 2009) and Tag-52, specific to Protostomia, and RhoGEF64C, conserved in Protostomia and Deuterostomia Vertebrates then lost in placentals (see supplementary tables S1 and S2, Supplementary Material online).
In non bilateria metazoans, we identified 28, 22, and 26 RhoGEF members in Cnidaria, Placozoa and Porifera genomes, respectively. Phylogenetic analyses placed Cnidaria and Porifera DH amino acid sequences at the roots of most vertebrate subfamilies. Only orthologs to PREX and ARHGEF28 were missing in Cnidaria and Porifera, respectively (fig. 3C). Since Porifera is a monophyletic group that diverged first in Metazoa, this suggests that ancestral Metazoa had at least 19 of the 20 vertebrate subfamilies (fig. 3C, see supplementary table S3, Supplementary Material online). This would imply that the metabolic pathways controlled by RhoGEFs subfamilies were already set up at the onset of Metazoa.
Evolution from single-celled ancestors to metazoa is hypothesized to have involved a colony-forming transition stage, formed by cells that were similar to the extant Choanoflagellates (Monosiga brevicollis, Salpingoeca rosetta; Nielsen 2008). Furthermore, Choanoflagellates and the Filasterea amoeba Capsaspora owczarzaki are closely related to metazoans (King et al. 2008; Suga et al. 2013). Together with Metazoans, these single-celled lineages formed the Filozoa group (Torruella et al. 2012). We identified 13 Dbl-like RhoGEFs in M. brevicollis and S. rosetta, and 12 in C. owczarzaki (fig. 2, see supplementary table S3, Supplementary Material online). Phylogenetic analyses of DH amino acid sequences showed that Choanoflagellates and Filasterea RhoGEFs clustered with Vertebrate subfamilies (fig. 3D). The clustering is further supported by the striking conservation of functional domains associated to the DH domains (see supplementary fig. S3, Supplementary Material online). Only six subfamilies have no homolog: TRIO, FARP, ALS2, RasGRF, BCR, and ARHGEF10. We also deduced that two ancestral duplications took place between Choanoflagellida and Porifera, which produced two branches in the ARHGEF1 and FGD subgroups in Porifera (fig. 2). Concerning the ARHGEF1/ARHGEF2 duplication, the ancestral architecture was likely (PDZ)/RGS/C1/DH/PH, as observed in C. owczarzaki. After duplication, one copy lost the N-terminal PDZ/RGS, likely by truncation (see supplementary fig. S3, Supplementary Material online).
Thus, a large fraction of the vertebrate RhoGEF repertoire is present in extant unicellular organisms that are closely related to metazoans, strongly suggesting it was also the case of the last common ancestor of Filozoa >1,000 Ma (Hedges et al. 2004; Parfrey et al. 2011). The transition to multi-cellularity has thus been associated with emergence of a restricted number of novel RhoGEFs with new functional domains, such as the SEC14 and spectrin domains in TRIO or the Ras guanine nucleotide exchange domain in RasGRF. The repertoire of RhoGEFs then remained fairly stable in metazoans until the vertebrate transition, at which time whole genome duplications generated a 2–3-fold increase in RhoGEF members and further sub functionalization.
The many expansion and loss events observed across and within Metazoan clades suggest that sizes of Dbl-like repertoires reflect the diversity of external stimuli cells respond to. We tested this hypothesis by considering the number of cell types in an organism as a proxy of its cell signaling complexity (Carroll 2001; McCarthy and Enquist 2005; Valentine et al. 1994; Hedges et al. 2004). The number of RhoGEFs varies linearly with the number of cell types, with a steep slope (0.38 ±0.05, P < 10−4; fig. 5). In contrast, the number of Rho GTPases activated by Dbl-like RhoGEFs varied with a much lower slope (0.035 ± 0.007, P = 8.10−4). This supports the notion that Dbl-like RhoGEF and not Rho repertoires vary in proportion of cell signaling complexity.
Fig. 5.
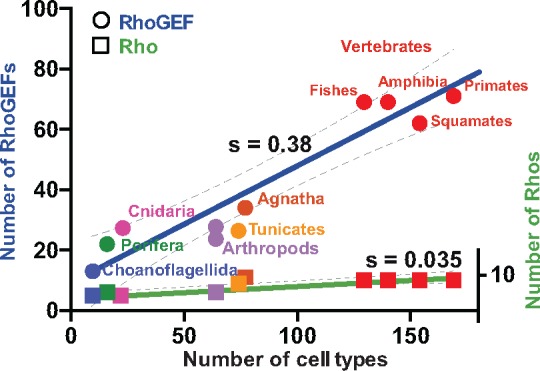
—Sizes of Dbl-like families in Metazoa correlate with numbers of cell types. Dot plot showing the relationship between the numbers of cell types in different metazoan species (Schad et al. 2011; Hedges et al. 2004; Valentine et al. 1994) and the numbers of RhoGEFs (circles) and their target Rho GTPases (squares) (calculated from Metazoa data in supplementary tables S1 and S3, Supplementary Material online). Data from the following species were used: Primates: H. sapiens; Squamates: A. carolinensis; Amphibia: X. tropicalis; Fishes: Danio rerio; Agnatha: P. marinus; Tunicates: C. intestinalis; Arthropods: D. melanogaster, A. gambiae; Cnidaria: H. magnipapillata; Porifera: A. queenslandica; Choanoflagellida: M. brevocollis. S indicates the slopes of regression lines, whose confidence intervals are indicated by dashed lines.
Multiple Independent Dbl-like Expansion and Reduction Events in Amorphea/Unikonta
The observation that 14 Dbl-like RhoGEF Vertebrate subfamilies were already present at the root of Metazoa prompted us to look at sister clades of Metazoa (fig. 6). Metazoa and Fungi form the monophyletic eukaryotic supergroup known as Opisthokonta (Baldauf and Palmer 1993), and they diverged around 1,300 Ma (Hedges et al. 2006; Parfrey et al. 2011). Opisthokonta, Breviatea, and Apusomonads form the supergroup Obazoa (Brown et al. 2013). Obazoa and its sister group Amoebozoa form the Amorphea supergroup (previously known as Unikonta; Adl et al. 2012), and they diverged 1,480 Ma (Hedges et al. 2006; Parfrey et al. 2011). A common feature of Amorphea is the presence of a single cilium or flagellum associated with a unique centriole, whereas the species of other eukaryotic clades have two centrioles and two flagella/cilia, that is, the ancestral state of all eukaryotes (Roger and Simpson 2009).
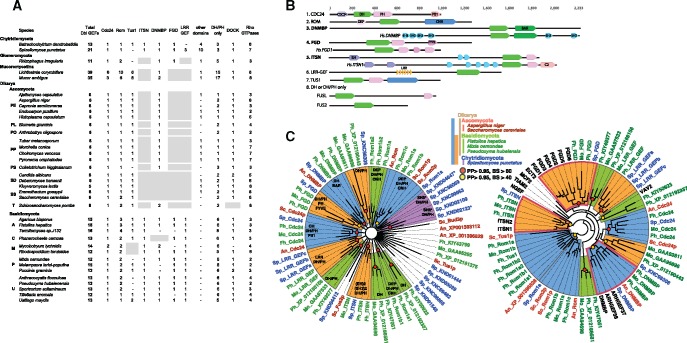
In Fungi, the number of extant species surpasses 100,000 and metagenomic data suggest there may actually be several million (O’Brien et al. 2005). Fungi are structured into a few phyla, among which Chytridiomycota, Mucoromycotina, and Glomeromycota diverged between 812 and 1,300 Ma whereas the Ascomycota and Basidiomycota (the two main phyla of Dikarya) diverged later (662–772 Ma; Floudas et al. 2012; Parfrey et al. 2011). We examined 37 fungal genomes from various clades and found a high variability in the number of Dbl-like members, ranging from 35 to 39 in Mucoromycotina, 11 to 21 in Chytridiomycota, Glomeromycota, and Basidiomycota, to only 5 to 8 in Ascomycota (fig. 7A, see supplementary table S4, Supplementary Material online). The Ascomycota Pezizomycetes morels and truffles (PP in fig. 7A), which develop fruiting bodies, have the same Dbl-like repertoire as yeasts (SS in fig. 7A), which do not. Thus, the higher Dbl-like content in Basidiomycota as compared with Ascomycota is probably not associated with the ability of Basidiomycota to develop fruiting bodies.
Fig. 7.
—The Dbl-like RhoGEF family in Fungi. (A) RhoGEFs were searched in genomes of species (in italics) distributed in the various Fungi phyla (in bold). RhoGEFs were classified according to their family (Dbl-like or DOCK) and their structural domain organization (see B). Ascomycota: Pezizomycotina: PE: Eurotiomycetes; PL: Leotiomycetes; PO: Orbiliomycetes; PP: Pezizomycetes; PS: Sordariomycetes. Ascomycota Saccharomycetales: SD: Debaryomycetaceae; SS: Saccharomycetaceae. T: Taphrinomycotina. Basisiomycota A: Agaricomycotina Agaricales; C: Agaricomycotina Corticiales; M: Pucciniomycotina Microbotryomycetes; P: Pucciniomycotina Pucciniomycetes; U: Ustilaginomycotina. (B) Eight types of RhoGEFs were identified in Fungi, based on the presence of functional domains. CH: Calponin homology, PB1: Phox/Bem1, DEP: Dishevelled/Egl10/Pleckstrin, CNH: Citron/Nik1 homology, BAR: Bin/Amphiphysin/Rvs, FYVE: Fab1/YOTB/ZK632.12/Vac1/EEA1, EH: Eps15 homology, SH3: Src homology 3, LRR: Leucine Rich Repeats. Three fungal RhoGEFs (DNMBP, FGD, ITSN) share similar functional domain organization with human RhoGEFs. (C) PhyML and MrBayes phylogenetic analysis of DH domains from Fungi of different clades, as indicated by the color code on the top left, excluding (left tree) or including (right tree) human DH sequences (in black). Nodes supported by posterior probabilities above 0.95 are indicated by red and yellow circles, with bootstrap BS values > 60 or >40, respectively. The domain organization of each cluster is shown.
In most fungal phyla and species examined, we identified eight types of Dbl-like proteins. Seven display specific structural domains associated with the DH (fig. 7B). These are Cdc24 (CH and PB1), Rom (DEP and CNH), DNMBP (BAR), Tus1 (CNH), FGD (FYVE), ITSN (EH, SH3), and LRR (Leucine Rich Repeats). Rom and Cdc24 are present in all examined phyla, DNMBP is only missing in Saccharomycotina (true yeasts) and FGD, ITSN, and LRR are missing in Ascomycota. In addition to these multi-domain Dbl-like proteins, all examined genomes encoded RhoGEFs with DH/PH or DH domains only, like Fusl in Agaricus bisporus and Fus2p in Saccharomycescerevisiae. Phylogenetic DH analysis clustered fungal Dbl-like proteins from multiple clades together and members of each cluster have the same functional domains (fig. 7C). Note that Mucoromycotina and Ascomycota have Dbl-like families with similar levels of diversity, although the former taxon encodes 6–8-fold more members than the latter (fig. 7A). This is consistent with multiploidy, which is suspected to have occurred in Mucoromycotina (Albertin and Marullo 2012).
Phylogenetic analysis of DH domains identified three clusters grouping fungal and metazoan RhoGEFs (FGD, ITSN, and DNMBP, fig. 7C, red frames), all of which are active on Cdc42 in mammals. The clustering is in agreement with the presence of the FYVE, BAR, EH, and SH3 domains (fig. 7B). All three groups are well supported by posterior probabilities (PPs) (>0.95) but have only moderate to low maximum likelihood (ML) bootstrap values, suggesting that fungal and vertebrate DH sequences have reached too high a proportion of saturated sites to be clustered with confident support.
In summary, Dbl-like repertoires in Fungi are highly heterogeneous, ranging from 5 to 39 in number, and this complexity appears unrelated to the ability to form fruiting bodies. Only three fungal Dbl-like RhoGEFs have metazoan orthologs (FGD, ITSN, and DNMBP), as supported both by the phylogenetic clustering of their DH domains and by the similar organization of structural domains. Nothing is known about the cellular roles of these RhoGEFs, due to their absence in yeast biological models.
Apusomonads are heterotrophic flagellate protozoa that live in soils, freshwater and marine habitats and which have an organic shell over the dorsal cell surface, called a theca. We identified 24 Dbl-like RhoGEFs in the genome of Thecamonas trahens, of which 10 contain only DH or DH/PH domains (see supplementary fig. S4A and table S4, Supplementary Material online). Other T. trahens RhoGEFs have DH/PH domains associated with additional domains, only two of which are classically found in Metazoa or Fungi (SH3 and MORN). The others domains were not observed in Fungi: The enzymatic Ras-like and ArfGAP, and the protein interacting motifs IQ (calmodulin-binding), LIM (Lin-11, Isl1, and Mec-3), SAM (Sterile Alpha Motif), ARM (Armadillo), and ANK (Ankyrin). Note also that the domains of the three RhoGEFs that are common to all Opisthokonts, (i.e., BAR for DNMBP, FYVE for FGD and EH for ITSN) are absent from T. trahens Dbl-like RhoGEFs (see supplementary fig. S4, Supplementary Material online).
Amoebozoa is the major protist phylum that regroups amoeba. Amoebozoa are defined as unicellular eukaryotes that move with highly dynamic pseudopodia. Amoebozoa are sub-divided into Conosa, which have a complex microtubule network at flagellate stages, and Lobosa, which do not (Cavalier-Smith et al. 2015). Among Conosa, the Mycetozoa class contains the true slime-molds, which are social amoebae that develop a multicellular fruiting body upon starvation. Previous work reported the presence of 45 genes for Dbl-like RhoGEFs in the slime mold D.discoideum (Mycetozoa/Conosa; Vlahou and Rivero 2006). To get a more robust view of Dbl-like repertoires in Mycetozoa, we examined four additional Conosa species, namely Dictyosteliumpurpureum, Dictyosteliumfasciculatum, Acytostelium subglobosum and Polysphondylium pallidum, which encoded respectively 45, 44, 64, and 44 Dbl-like RhoGEFs (see supplementary table S4, Supplementary Material online). Phylogenetic analysis of their DH domains identified 44 robust clusters from each species (see supplementary fig. S5A, Supplementary Material online). This indicates that the Dbl-like repertoire in Mycetozoa has remained stable for the last 600 Myr (Fiz-Palacios et al. 2013). All Dbl-like RhoGEFs that regrouped in clusters had same functional domains associated. Several of these domains were also found in Opisthokont RhoGEFs, like the BAR and CH domains (DNMBP, VAV, and ARHGEF6, fig. 1B) or LRR in Fungi (fig. 7B). Several domains that are not found in Opisthokonts, like ANK, ARM, ArfGAP and IQ, are present in T. trahens RhoGEFs (see supplementary fig. S4, Supplementary Material online). Note that all D. discoideum DH domains are equally distantly related to metazoan DH, which did not allow to detect any specific orthology.
We next identified the Dbl-like members in four Entamoeba species (E. histolytica, E. invadens, E. dispar, and E. nuttalli). Entamoeba also belong to Conosa and diverged from Mycetozoa around 1,500 Ma (Parfrey et al. 2011). The Entamoeba genus is made of amitochondriate and morphologically similar species, most of which are intestinal parasites (Stensvold et al. 2011). We identified 62 Dbl-like proteins in E. his, 95 in E. inv, 63 in E. dis and 60 in E. nut (see supplementary table S4, Supplementary Material online). Interestingly, although the numbers of Dbl-like members were higher in Entamoeba than in Mycetozoa, the structural diversity associated with their DH domains was much lower: Entamoeba Dbl-like RhoGEFs lack the ARM, BAR, and IQ domains (see supplementary fig. S4, Supplementary Material online). Phylogenetic analysis of the DH domains distributed them into 26 clusters, in which proteins have a similar domain organization (see supplementary fig. S5B, Supplementary Material online). It is probable that multiple Dbl-like members were lost in ancestral Entamoeba after the split with Mycetozoa, reducing their repertoire from 45 down to 26. This was followed by duplications leading to over 60 members. The four species examined diverged after the duplications.
Finally we examined the genome of Acanthamoeba castellanii, which belongs to Lobosa (Clarke et al. 2013), and identified 108 Dbl-like RhoGEFs (see supplementary fig. S4A, Supplementary Material online). Phylogenetic analysis showed that the 108 DH domains distributed into 22 clusters and 33 single sequences (see supplementary fig. S5C, Supplementary Material online). Although analysis of additional species is needed to get a more comprehensive view of the Dbl-like family in Lobosa, this nevertheless suggests that the number of independent members is of the same order in Lobosa and Mycetozoa. This implies that the number of Dbl-like members in ancestral Amoebozoa were in the same range. 74 of the 108 A.castellanii Dbl-like RhoGEFs have only DH or DH/PH domains (see supplementary fig. S4A, Supplementary Material online). Among the domains associated with DH/PH in the 34 other RhoGEFs, most are classically found in Metazoan RhoGEFs (CH, BAR, FYVE, RhoGAP, SH3, C1, C2, F-BOX, or MORN). However, A. castellanii Dbl-like members do not contain any of the kinase, RasGAP, LRR, and Myosin domains, found in Mycetozoa. Neither do they contain either the ArfGAP or RasGEF domains, which are found in of Entamoeba and Mycetozoa (see supplementary fig. S4, Supplementary Material online).
Thus, the repertoires of Dbl-like RhoGEFs in Amorphea clades are highly variable. Their numbers range from 15 to 72 in Metazoa, 5 to 35 in Fungi and from 46 to 108 in Amoebozoa. In the three clades, we observed multiple and independent loss and expansion events (fig. 6), which may be directly linked to complexity of cell signaling. In addition, their DH-associated domains are mostly different. The only domains common to Amorphea clades are CH and SH3, two protein–protein interaction domains, FYVE/PHD, which targets cell membranes, and RhoGAP, a negative regulator of Rho signaling. This suggests that they were likely present in ancestral Amorphea and have prominent roles in basal Rho signaling.
Contrasting Evolutionary Repertoires of RhoGEF Families in Bikonta
We next examined the presence of Dbl RhoGEFs in Bikonta eukaryotes (fig. 6). Bikonta are clustered into three major supergroups whose relative positions are still debated (Derelle et al. 2015; Adl et al. 2012; He et al. 2014). The Bikonta are divided into Archaeplastida, SAR and Excavates. The Archaeplastida are further divided into two supergroups, the Viridiplantae (land plants and green algae) and Rhodophyta (red algae). The SAR supergroup is divided into Stramenopiles (e.g., Phytophtora infestans, that causes the potato blight), Alveolates (e.g., the apicomplex Plasmodium falciparum or the ciliate Paramecium tetraurelia), and Rhizaria (e.g., the amoeba-like Reticulomyxa filosa, a model system for motility and organelle transport analysis, Ashkin et al. 1990). The Excavates supergroup is divided into Euglens (e.g., the parasites Trypanosoma or Leishmania), Diplomonads (e.g., the intestinal parasite Giardia intestinalis), Heterolobosea (Lee 2010), and Parabasalids (e.g., the sexually transmitted infection Trichomonas vaginalis). Haptophyta and Cryptophta are phylogenetically incertae sedis groups, although they have been proposed to be related to SAR and Rhodophyceae (Parfrey et al. 2011; Reeb et al. 2009). To gain a deeper insight in the Bikonta, we also looked, in each species, at the presence of Rho and DOCK proteins, as well as RopGEF, exchange factors that are active on Rac-like proteins in plants and characterized by a PRONE domain (Berken et al. 2005).
We identified Dbl RhoGEFs in all Bikonta supergroups (see supplementary fig. S6 and table S4, Supplementary Material online). However, the presence of Dbl-like proteins is variable within each supergroup and between phyla. Two clades have no Rho or RhoGEFs at all (i.e., Chlamydomonadales in Chlorophyta or Apicomplexa in Alveolates). The absence of Rho signaling may be associated with the fact that Chlamydomonadales and Apicomplexa use gliding as a particular mode of locomotion, which depends on actin and myosin class XIV but does not require membrane dynamics like amoeboid motion does (Heintzelman 2006). In Archaeplastida, Viridiplantae do not encode Dbl-like proteins, although the exchange domain of SWAP70 orthologs has been proposed to be structurally related to DH (Yamaguchi et al. 2012). Viridiplantae encode Rac-like GTPases (Valster et al. 2000) and GEFs of the DOCK and RopGEF families (see supplementary fig. S6, Supplementary Material online). In Rhodophyta, the sister clade of Viridiplantae, the Florideaphyceae Chondrus crispus (Irish moss, a multicellular red alga living in North Atlantic) encodes a Rho signaling set similar to other Viridiplantae. In contrast, the Cyanidiophyceae Galdieria sulphuraria and Cyanidioschyzon merolae, unicellular and extremophilic red algae living in acidic hot sulfur springs (Rothschild and Mancinelli 2001), encode Dbl-like proteins but no DOCKs. These two unicellular red algae species thus express both Dbl-RhoGEFs and RopGEFs. This is also the case of the Haptophyta Emiliania huxleyi, a phytoplanktonic coccolithophore that has two Rac GTPases, four Dbl-like members and one RopGEF but has no DOCK protein. In the current phylogenetic view, Dbl-like genes were lost twice in Archaeplastida, once in the Rhodophyta clades (but not in Cyanidiophyceae, the first clade that diverged from Archaeplastida, Verbruggen et al. 2010) and once at the onset of Viridiplantae. Alternately, a single Dbl-like loss may be invoked if Cyanidiophyceae had gained Dbl-like genes by LGT, which they are prone to (Qiu et al. 2013).
In the Excavates and SAR supergroups, the presence of genes for Dbl-like RhoGEFs is heterogeneous. A high copy number of Dbl, DOCK and Rho GTPases are found in Heterolobosea (Naegleria gruberi), Parabasalia (T.vaginalis) and Rhizaria (R.filosa and Plasmodiophora brassicae). These protists are not Amoebozoa yet they adopt a dynamic amoeba-like morphology, suggesting that independent amplification of Rho components has enabled the acquisition of the amoeboid phenotype.
In contrast to these amoeboid protists, Euglenozoa and Diplomonadida do not encode Dbl-like proteins but they do encode unique DOCK proteins. Unexpectedly, we even detected a DOCK protein in ten species that do not encode Rac proteins (boxed in supplementary fig. S6, Supplementary Material online). DOCK proteins of the ten species have a canonical domain structure (N-terminal lipid binding C2 and C-terminal catalytic DHR2 domains, see supplementary fig. S7A, Supplementary Material online) and are highly conserved at the amino acid level (70% overall similarity between Trypanosoma and Leishmania, 53% between Bonodidae and Trypanosomatidae). DOCK protein catalytic DHR2 domains are 40–45% similar between human and euglenozoan sequences, irrespective to the presence or absence of Rac proteins (see supplementary fig. S7B, Supplementary Material online). Although some cellular functions of DOCK do not require its RacGEF activity (Ogawa et al. 2014), the presence of an apparently normal DHR2 domain in organisms that do not encode any Rac protein suggests that DHR2 domains may have additional activities in addition to regulating Rac.
In summary, most Bikonta clades encode RhoGEFs of the Dbl and DOCK families, indicating that, together with Rac, they were part of the basic Rho signaling module in LECA. However, this module experienced independent loss or expansion events between and within taxa, varying from a total loss in Apicomplexa to acquisition of over 30 Dbl-like and Rac members in amoeba-like protists.
Discussion
By examining the genomes of 175 species covering all eukaryotic supergroups and spanning over 1.7 billion years of evolution, we show here that the Dbl RhoGEF family is present in all eukaryotic supergroups, implying it was already present in the LECA. However, this family has experienced many independent expansion or loss episodes in branches of the same clades (fig. 6). This plasticity suggests that most of RhoGEF diversity is not related to basic cellular metabolism but may rather reflect the diversity of external stimuli cells respond to. This hypothesis is supported by the tissue-specificity of RhoGEF mRNA expression in Vertebrates (fig. 4) and by the steepness of the linear relationship between the numbers of RhoGEFs and cell types in Metazoa (fig. 5). This is not the case for Rho GTPases activated by Dbl RhoGEFs (Jaiswal et al. 2013), as their expression is ubiquitous in human tissues (Boureux et al. 2007) and their numbers vary in proportion to cell types in Metazoa with a much lower slope than RhoGEFs (fig. 5). On the one hand, the remarkable conservation of 14 of the 20 vertebrate Dbl subfamilies as far away as in Choanoflagellida/Filasterea unicellular animals might thus have enabled the early emergence of the basic repertoire of receptors and signaling in eukaryotes. On the other hand, the many RhoGEF loss events observed in subclades may reflect a decreased diversity of signaling after their adaptation to specific ecological niches. For example, birds lost 12% of the vertebrate Dbl-like repertoire and this may be directly or indirectly associated with establishment of particular features, such as exceptionally enlarged and diversified muscles or atypical thermogenesis. Such features are thought to be driven by gene loss rather than gain (Newman et al. 2013). The loss of ARHGEF19 and ARHGEF25 in birds may have been instrumental, as these genes control myogenesis and adipogenesis in mice (Horii et al. 2009; Bryan et al. 2005). The loss of PLEKHG2 might also have been instrumental in the evolution of bird-specific features, since PLEKHG2 is involved in insulin-stimulated GLUT4-mediated glucose uptake in L6 rat myoblasts (Sato et al. 2014), and this process no longer occurs in birds (Seki et al. 2003). Establishment of particular features due to Dbl-like RhoGEF losses may concern other Metazoan sub-clades, like mice in mammals, nematodes and flies in Ecdyzsozoa or yeasts in Fungi, which all had faster evolution rates. The finding that these organisms lost several RhoGEFs implies that their respective physiology might be adapted to a reduced signaling diversity. This must be taken into account when addressing functional roles of RhoGEFs in these organisms, widely used as biological models.
Decreased signaling diversity is also consistent with the Dbl-like repertoire being smaller in Entamoeba than in Mycetozoa, since, as parasites of the intestines, Entamoeba inhabit a more constant environment than the free-living Mycetozoa. Besides, high numbers of Dbl-like RhoGEFs are observed in species of distinct clades but sharing all the amoeboid phenotype, that is, Amoebozoa, Heterolobosea, Parabasalia, and Rhizaria (see supplementary figs. S4 and S6, Supplementary Material online). Independent Dbl-like expansion events thus occurred at least four times in amoeboid protists, which supports the notion that Dbl-like RhoGEFs are functionally involved in acquisition of the amoeboid phenotype. Amoebae produce different types of pseudopodia, which they use to move and to feed on bacteria. Amoebae express different suites of genes when they encounter different bacterial species (Nasser et al. 2013), suggesting that distinct types of membrane receptors are involved and such receptor diversity in amoebae may have been enabled by the high number of Dbl-like proteins. Thus, despite their ancient origin in Eukaryotes and their overall conservation in Metazoa, Dbl RhoGEFs appear as a dynamic family, which can adapt its size to the level of cell signaling diversity.
We also show here that Dbl-like and DOCK RhoGEF families were both present in the LECA. This implies that these two families, which are both active on the same Rac-like proteins, have distinct properties. One straightforward difference is that DOCKs have additional functions as well as just activating Rac GTPases (Ogawa et al. 2014). This may explain the unexpected finding that several Euglenozoa and Stramenopiles species encode DOCK proteins but no GTPase of the Rho family. However, another striking difference between the Dbl-like and DOCK RhoGEFs concerns the high diversity of domains associated with the tandem DH/PH in Dbl-like RhoGEFs; DOCK proteins contain a C2 calcium-binding domain and a DHR2 catalytic domain, either alone or associated with single SH3 or PH domains (Meller et al. 2005). In this respect, Dbl-like RhoGEFs have a much higher capacity to evolve, by reshuffling various functional domains with the catalytic DH domain. Although a few domain combinations appeared conserved in the various clades studied (e.g., FGD, DNMBP or ITSN in Amorphea), the general trend is a high heterogeneity between eukaryotic clades, and even within clades. Domain reshuffling is also supported by phylogenetic analyses, in which Dbl-like RhoGEFs with similar DH domains have different auxiliary domains, like AKAP13, ARHGEF1, ITSN, PLEKHG5, or NGEF. Domain reshuffling has also occurred in the TRIO family, in which OBSCN, TRIO, and KALRN have gained kinase and IG/FN domains from SPEG kinases and Titin. Proteins with same domain organization as RhoGEFs but lacking the DH domains also add an indirect support to reshuffling.
In conclusion, this study establishes that the family of Dbl-like RhoGEFs had a highly complex pattern of evolution and underwent repeated expansion and reduction events. Given that Dbl-like family complexity reflects the diversity of cell signaling, this family of Rho regulators constitutes an adaptive toolbox whose requirement in eukaryotic cell physiology has greatly varied depending on species biology.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank J. Piette and P. Lemaire for giving access to tunicate transcriptomic data and P. Labbé for help in R scripts. We also thank CRBM members for fruitful discussions and Julian Venables for help in writing the manuscript. This work was supported by institutional grants from the Centre National de la Recherche Scientifique and from Montpellier Université.
Literature Cited
- Abascal F, Zardoya R, Posada D.. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Adl SM, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59:429–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin W, Marullo P.. 2012. Polyploidy in fungi: evolution after whole-genome duplication. Proc Biol Sci. 279:2497–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin E, et al. 2016. Deciphering the molecular and functional basis of RHOGAP family proteins: a systematic approach toward selective inactivation of RHO family proteins. J Biol Chem. 291:20353–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A, Schütze K, Dziedzic JM, Euteneuer U, Schliwa M.. 1990. Force generation of organelle transport measured in vivo by an infrared laser trap. Nature. 348:346–348. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Palmer JD.. 1993. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci U S A. 90:11558.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338:1587–1593. [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Hu H, Nobes CD, Goodman CS.. 2001. A novel Dbl family RhoGEF promotes Rho-dependent axon attraction to the central nervous system midline in Drosophila and overcomes Robo repulsion. J Cell Biol. 155:1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berken A, Thomas C, Wittinghofer A.. 2005. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436:1176–1180. [DOI] [PubMed] [Google Scholar]
- Bosco EE, Mulloy JC, Zheng Y.. 2009. Rac1 GTPase: a ‘Rac’ of all trades. Cell Mol Life Sci. 66:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureux A, Vignal E, Faure S, Fort P.. 2007. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 24:203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478:343–348. [DOI] [PubMed] [Google Scholar]
- Brown MW, et al. 2013. Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proc R Soc Lond B Biol Sci. 280:20131755.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan BA, et al. 2005. Modulation of muscle regeneration, myogenesis, and adipogenesis by the Rho family guanine nucleotide exchange factor GEFT. Mol Cell Biol. 25:11089–11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo XR, Sauzeau V, Berenjeno IM.. 2007. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. BioEssays News Rev Mol Cell Dev Biol. 29:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2001. Chance and necessity: the evolution of morphological complexity and diversity. Nature 409:1102–1109. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, et al. 2015. Multigene phylogeny resolves deep branching of Amoebozoa. Mol Phylogenet Evol. 83:293–304. [DOI] [PubMed] [Google Scholar]
- Clarke M, et al. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 14:R11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, et al. 2015. Bacterial proteins pinpoint a single eukaryotic root. Proc Natl Acad Sci U S A. 112:E693–E699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S.. 2004. Open source clustering software. Bioinformatics 20:1453–1454. [DOI] [PubMed] [Google Scholar]
- dos Reis M, et al. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc Biol Sci. 279:3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 5:113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erixon P, Svennblad B, Britton T, Oxelman B.. 2003. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol. 52:665–673. [DOI] [PubMed] [Google Scholar]
- Eva A, Aaronson SA.. 1985. Isolation of a new human oncogene from a diffuse B-cell lymphoma. Nature 316:273–275. [DOI] [PubMed] [Google Scholar]
- Fiz-Palacios O, et al. 2013. Did terrestrial diversification of amoebas (Amoebozoa) occur in synchrony with land plants? PLoS ONE. 8:e74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. 2014. Ensembl 2014. Nucleic Acids Res. 42:D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floudas D, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Hart MJ, Eva A, Evans T, Aaronson SA, Cerione RA.. 1991. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature 354:311–314. [DOI] [PubMed] [Google Scholar]
- He D, et al. 2014. An alternative root for the eukaryote tree of life. Curr Biol. 24:465–470. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL.. 2004. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S.. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22:2971–2972. [DOI] [PubMed] [Google Scholar]
- Heintzelman MB. 2006. Cellular and molecular mechanics of gliding locomotion in eukaryotes. In: Kwang, J, editor. International review of cytology. International review of cytology. Vol. 251. Academic Press. pp. 79–129. [accessed 2016 Sep 6]. Available from: http://www.sciencedirect.com/science/article/pii/S0074769606510034. [DOI] [PubMed]
- Higuchi N, Kohno K, Kadowaki T.. 2009. Specific retention of the protostome-specific PsGEF may parallel with the evolution of mushroom bodies in insect and lophotrochozoan brains. BMC Biol. 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland ND. 2005. Chordates. Curr Biol CB. 15:R911–R914. [DOI] [PubMed] [Google Scholar]
- Horii T, Morita S, Kimura M, Hatada I.. 2009. Epigenetic regulation of adipocyte differentiation by a Rho guanine nucleotide exchange factor, WGEF. PLoS ONE. 4:e5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal M, Dvorsky R, Ahmadian MR.. 2013. Deciphering the molecular and functional basis of Dbl family proteins: a novel systematic approach toward classification of selective activation of the Rho family proteins. J Biol Chem. 288:4486–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451:783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer DC, Nowak MA.. 1999. Evolutionary preservation of redundant duplicated genes. Semin Cell Dev Biol. 10:555–559. [DOI] [PubMed] [Google Scholar]
- Lee J. 2010. De novo formation of basal bodies during cellular differentiation of Naegleria gruberi: progress and hypotheses. Semin Cell Dev Biol. 21:156–162. [DOI] [PubMed] [Google Scholar]
- McCarthy MC, Enquist BJ.. 2005. Organismal size, metabolism and the evolution of complexity in metazoans. Evol Ecol Res. 7:681–696. [Google Scholar]
- Meller N, Merlot S, Guda C.. 2005. CZH proteins: a new family of Rho-GEFs. J Cell Sci. 118:4937–4946. [DOI] [PubMed] [Google Scholar]
- Nasser W, et al. 2013. Bacterial discrimination by Dictyostelid amoebae reveals the complexity of ancient interspecies interactions. Curr Biol CB. 23:862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SA, Mezentseva NV, Badyaev AV.. 2013. Gene loss, thermogenesis, and the origin of birds. Ann N Y Acad Sci. 1289:36–47. [DOI] [PubMed] [Google Scholar]
- Nielsen C. 2008. Six major steps in animal evolution: are we derived sponge larvae? Evol Dev. 10:241–257. [DOI] [PubMed] [Google Scholar]
- O’Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R.. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol. 71:5544–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, et al. 2014. DOCK5 functions as a key signaling adaptor that links FcεRI signals to microtubule dynamics during mast cell degranulation. J Exp Med. 211:1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA.. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 108:13624–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV.. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RS, et al. 2014. The evolutionary history of holometabolous insects inferred from transcriptome-based phylogeny and comprehensive morphological data. BMC Evol Biol. 14:52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Yoon HS, Bhattacharya D.. 2013. Algal endosymbionts as vectors of horizontal gene transfer in photosynthetic eukaryotes. Front Plant Sci. 4:366.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffield LM, et al. 2015. Heritability and genetic association analysis of neuroimaging measures in the Diabetes Heart Study. Neurobiol Aging. 36:1602.e7–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi F, Portella G, Orozco M, Fanelli F.. 2011. Nucleotide binding switches the information flow in ras GTPases. PLoS Comput Biol. 7:e1001098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeb VC, et al. 2009. Interrelationships of chromalveolates within a broadly sampled tree of photosynthetic protists. Mol Phylogenet Evol. 53:202–211. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Simpson AGB.. 2009. Evolution: revisiting the root of the eukaryote tree. Curr Biol CB. 19:R165–16R167. [DOI] [PubMed] [Google Scholar]
- Rojas AM, Fuentes G, Rausell A, Valencia A.. 2012. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol. 196:189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J.. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 6:167–180. [DOI] [PubMed] [Google Scholar]
- Rothschild LJ, Mancinelli RL.. 2001. Life in extreme environments. Nature 409:1092–1101. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview – extensible visualization of microarray data. Bioinformatics 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- Sato K, et al. 2014. PLEKHG2/FLJ00018, a Rho family-specific guanine nucleotide exchange factor, is tyrosine phosphorylated via the EphB2/cSrc signaling pathway. Cell Signal. 26:691–696. [DOI] [PubMed] [Google Scholar]
- Schad E, Tompa P, Hegyi H.. 2011. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 12:R120.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Sato K, Kono T, Abe H, Akiba Y.. 2003. Broiler chickens (Ross strain) lack insulin-responsive glucose transporter GLUT4 and have GLUT8 cDNA. Gen Comp Endocrinol. 133:80–87. [DOI] [PubMed] [Google Scholar]
- Smith JJ, et al. 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 45:415–421, 421. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M, et al. 1997. Still life, a protein in synaptic terminals of drosophila homologous to GDP-GTP exchangers. Science 275:543–547. [DOI] [PubMed] [Google Scholar]
- Stensvold CR, et al. 2011. Increased sampling reveals novel lineages of Entamoeba: consequences of genetic diversity and host specificity for taxonomy and molecular detection. Protist 162:525–541. [DOI] [PubMed] [Google Scholar]
- Steven R, Zhang L, Culotti J, Pawson T.. 2005. The UNC-73/Trio RhoGEF-2 domain is required in separate isoforms for the regulation of pharynx pumping and normal neurotransmission in C. elegans. Genes Dev. 19:2016–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, et al. 2013. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 4:2325.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Lamarche-Vane N.. 2007. Current knowledge of the large RhoGAP family of proteins. Biol Cell Auspices Eur Cell Biol Organ. 99:67–86. [DOI] [PubMed] [Google Scholar]
- Tiphaine C, et al. 2013. Correlated changes in occlusal pattern and diet in stem Murinae during the onset of the radiation of Old World rats and mice. Evol Int J Org Evol. 67:3323–3338. [DOI] [PubMed] [Google Scholar]
- Torruella G, et al. 2012. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 29:531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine JW, Collins AG, Meyer CP.. 1994. Morphological complexity increase in metazoans. Paleobiology 20:131–142. [Google Scholar]
- Valster AH, Hepler PK, Chernoff J.. 2000. Plant GTPases: the Rhos in bloom. Trends Cell Biol. 10:141–146. [DOI] [PubMed] [Google Scholar]
- Verbruggen H, et al. 2010. Data mining approach identifies research priorities and data requirements for resolving the red algal tree of life. BMC Evol Biol. 10:16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahou G, Rivero F.. 2006. Rho GTPase signaling in Dictyostelium discoideum: insights from the genome. Eur J Cell Biol. 85:947–959. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, et al. 2012. SWAP70 functions as a Rac/Rop guanine nucleotide-exchange factor in rice. Plant J. 70:389–397. [DOI] [PubMed] [Google Scholar]
- Zheng Q, et al. 2009. Family-based association study of the MCF2L2 gene and polycystic ovary syndrome. Gynecol Obstet Invest. 68:171–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.