Abstract
Interstrand cross-links are exceptionally bioactive DNA lesions. Endogenous generation of interstrand cross-links in genomic DNA may contribute to aging, neurodegeneration, and cancer. Abasic (Ap) sites are common lesions in genomic DNA that readily undergo spontaneous and amine-catalyzed strand cleavage reactions that generate a 2,3-didehydro-2,3-dideoxyribose sugar remnant (3’ddR5p) at the 3’-terminus of the strand break. Interestingly, this strand scission process leaves an electrophilic α,β-unsaturated aldehyde residue embedded within the resulting nicked duplex. Here we present evidence that 3’ddR5p derivatives generated by spermine-catalyzed strand cleavage at Ap sites in duplex DNA can react with adenine residues on the opposing strand to generate a complex lesion consisting of an interstrand cross-link adjacent to a strand break. The cross-link blocks DNA replication by ϕ29 DNA polymerase, a highly processive polymerase enzyme that couples synthesis with strand displacement. This suggests that 3’ddR5p-derived cross-links have the potential to block critical cellular DNA transactions that require strand separation. LC-MS/MS methods developed herein provide powerful tools for studying the occurrence and properties of these cross-links in biochemical and biological systems.
INTRODUCTION
Damage to cellular DNA is unavoidable (1). A wide variety of chemical and enzymatic processes contribute to the degradation of cellular DNA, including oxidation, alkylation, misincorporation of ribonucleotides, hydrolytic deamination of the nucleobases, and hydrolysis of the glycosidic bonds to generate abasic (Ap) sites (1–8). DNA repair systems mitigate the effects of DNA damage (9–11), but some lesions inevitably evade repair with deleterious consequences. Endogenous DNA damage in mammalian cells may contribute to aging, neurodegeneration, mitochondrial dysfunction, mutagenesis and cancer (12–19).
Not all DNA lesions have equal bioactivity. Certain types of DNA damage such as double-strand breaks (20), interstrand cross-links (21–24), and clustered lesions (typically defined as more than one DNA lesion within a 10 base pair region) (25,26) have exceptionally potent biological effects even when generated at low levels in cellular DNA. To better understand the roles of endogenous DNA damage in human health and disease, it is critical to identify the exceptionally bioactive lesions that occur spontaneously in cellular DNA. Along these lines, we have identified a new, structurally complex, interstrand cross-link arising from strand breakage at Ap sites in duplex DNA.
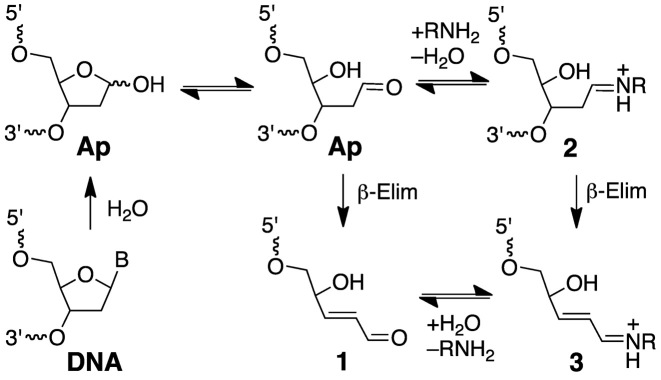
Ap sites are ubiquitous endogenous lesions in genomic DNA that are generated by spontaneous or enzyme-catalyzed hydrolysis of the glycosidic bonds linking the nucleobases to the sugar-phosphate backbone (1,27,28). Ap sites can be converted to strand breaks via β-elimination of the 3’-phosphoryl group (Scheme 1) (1,29–33). This reaction occurs spontaneously under physiological conditions and also can be catalyzed by amino groups in DNA repair enzymes, histones, small peptides and endogenous polyamines such as spermine (Scheme 1) (1,31–48). Interestingly, this strand scission process generates an electrophilic 2,3-didehydro-2,3-dideoxyribose derivative (3’ddR5p) embedded within the DNA duplex at the 3’-terminus of the strand break (Scheme 1) (30,44,49,50). Here, we present evidence that 3’ddR5p derivatives generated by spermine-catalyzed strand cleavage at Ap sites in duplex DNA can react with adenine residues on the opposing strand to generate a complex lesion consisting of an interstrand cross-link adjacent to a strand break.
Scheme 1.
Generation of strand breaks at Ap sites in DNA.
EXPERIMENTAL
Materials and methods
Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA), [γ-32P]-ATP (6000 Ci/mmol) was purchased from Perkin-Elmer, uracil DNA glycosylase (UDG) and ϕ29 DNA polymerase were from New England Biolabs (Ipswich, MA, USA), C-18 Sep-Pak cartridges were purchased from Waters (Milford, MA, USA), and BS Poly-prep columns were obtained from BioRad (Hercules, CA, USA). Acrylamide/bis-acrylamide 19:1 (40% solution, electrophoresis grade) was purchased from Fisher Scientific (Waltham, MA, USA), spermine and all other chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). A mixture of the four 2’-deoxynucleoside triphosphates (dNTPs) was purchased from Promega (Madison, WI, USA). Iron–EDTA–H2O2 footprinting (51–53), QTOF-MS (53–55), LC–MS/MS (53,55) and phi-29 (ϕ29) DNA polymerase primer extension reactions (56) were conducted as described in published procedures, with minor modifications. Detailed experimental protocols for these experiments are provided in the Supplementary Data.
Representative procedures for generation of the nicked duplex C and its conversion to the nicked cross-linked duplex D
In these experiments, 2’-deoxyuridine-containing oligonucleotides were used as precursors for the enzymatic generation of Ap-containing oligonucleotides (57,58). The single-stranded, uracil-containing 2’-deoxy-oligonucleotides were α-32P 5’-end-labeled using standard procedures (59), and then annealed with the complimentary strand. The labeled duplex DNA was treated with the UDG (200 units/ml, final concentration) in Tris–HCl buffer (20 mM, pH 8) containing DTT (1 mM), EDTA (1 mM) and spermine (1 mM). After incubation at 37°C for 45–60 min, the UDG enzyme was removed by phenol–chloroform extraction (59) and the DNA ethanol precipitated (59) to remove spermine and other low molecular weight compounds. The resulting nicked duplex was redissolved and incubated in a buffer composed of HEPES (50 mM, pH 7.4) and NaCl (100 mM) at 37°C for 72 h. The reaction mixture was then combined with formamide loading buffer (59) and the resulting sample loaded onto a 20% denaturing polyacrylamide gel (0.4 mm thick) and the gel electrophoresed for 6 h at 1600 V. A gel image and the amount of radiolabeled DNA in each band on the gel was obtained by phosphorimager analysis. The formation rate of cross-linked DNA was measured as described above except at specified time points, aliquots were removed and frozen at −20°C prior to gel electrophoretic analysis.
Reduction of the cross-link in duplex D by NaBH4
Duplex DNA containing 32P-labeled, nicked duplex D was incubated in a buffer composed of HEPES (50 mM, pH 7.4) and NaCl (100 mM) at 37°C for 72 h. NaBH4 was added (100 mM, final concentration), and the pH of the resulting solution adjusted by addition of aqueous NaOH to a final concentration of 0.01 N. The mixture was incubated at 37°C for another 4 h, followed by neutralization with HCl (0.1 N). The DNA was ethanol precipitated, resuspended in formamide loading buffer, loaded onto a 2 mm thick 20% denaturing polyacrylamide gel, and the cross-link isolated as described above.
RESULTS AND DISCUSSION
Generation of nicked DNA duplexes containing a 3’ddR5p sugar remnant and gel electrophoretic evidence for the formation of nicked cross-linked DNA duplexes
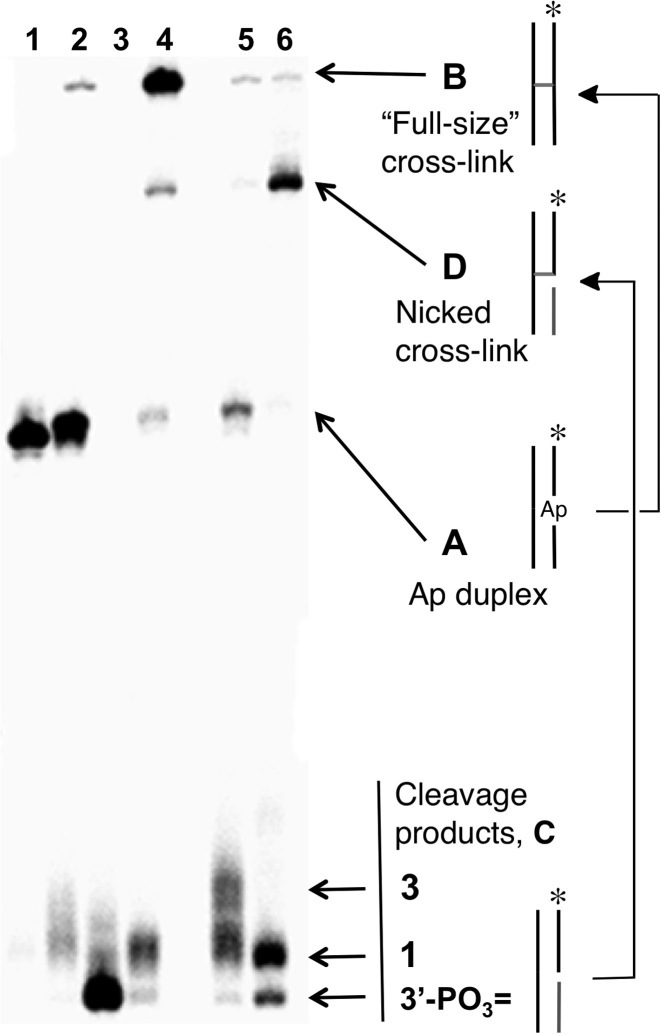
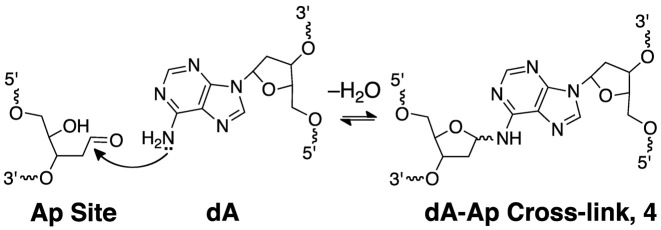
Nicked DNA duplexes containing the 3’ddR5p end group at the break were generated by reaction of the endogenous polyamine spermine (60) with the corresponding Ap duplexes (43,45,61). The 5’-32P-labeled, 35 base pair Ap-containing duplex A (Figure 1) was prepared from the corresponding 2’-deoxyuridine-containing duplex by treatment with uracil DNA glycosylase (UDG) as described previously (53–58,62–64). To confirm successful generation of the Ap site in duplex A, denaturing polyacrylamide gel electrophoretic analysis was used to show that treatment with piperidine (1 M, 95°C, 25 min) cleanly induced strand cleavage to give the expected (1,65) fast-migrating product with a 3’-phosphoryl terminus. In addition, incubation of duplex A, containing the intact Ap site, in HEPES buffer (50 mM, pH 7.4) containing NaCl (100 mM) at 37°C gave the expected slowly-migrating band (Figure 2, lane 4) corresponding to the previously characterized (55,63) full-length cross-linked duplex B (Figure 1, and structure 4 in Scheme 2).
Figure 1.
DNA duplexes used in this study.
Figure 2.
Formation of the nicked cross-linked duplex D by incubation of nicked duplex C in HEPES buffer (50 mM, pH 7.4) containing NaCl (100 mM) at 37°C. Lane 1, size-marker consisting of the 5’-32P labeled uracil-containing oligonucleotide precursor to duplex A; Lane 2, freshly-prepared Ap-containing duplex A; Lane 3, Ap-containing duplex A subjected to piperidine work-up (1 M, 95°C, 25 min) to induce strand cleavage; Lane 4, formation of full-length, cross-linked duplex B by incubation of duplex A in HEPES buffer (50 mM, pH 7.4) containing NaCl (100 mM) at 37°C for 120 h; Lane 5, freshly-prepared, nicked 3’ddR5p-containing duplex C; Lane 6, formation of the nicked cross-linked duplex D by incubation of the nicked duplex C for 72 h. The 5’-32P-labeled oligodeoxynucleotides in these reactions were resolved by electrophoresis on a 20% denaturing polyacrylamide gel and the radioactivity in each band quantitatively measured by phosphorimager analysis.
Scheme 2.
Generation of interstrand cross-links by reaction of an adenine residue with an intact Ap site in duplex DNA.
Nicked duplex C, containing the 3’ddR5p end group was generated by addition of spermine (1 mM), to the UDG reaction (43,45,61). Spermine is an endogenous polyamine that is present at millimolar concentrations in the nuclei of cells (60). Spermine and other polyamines associate with chromatin (66) and also serve as regulators of cell growth and apoptotic cell death (67). The cleavage of abasic sites by spermine and other low molecular weight polyamines has been described previously (43-45,61). Ethanol precipitation of the freshly prepared nicked duplex C, followed by electrophoretic analysis, revealed two cleavage products (Figure 2, lane 5) whose gel mobilities were consistent with the presence of the 3’-end groups 1 and 3 (Scheme 1) (42,44,68). Further incubation of the spermine-nicked duplex C in HEPES buffer (50 mM, pH 7.4) containing NaCl (100 mM) at 37°C led to the formation of a major slowly-migrating band whose mobility on the denaturing polyacrylamide gel—intermediate between the full-length single strand from duplex A and the full-length cross-linked duplex B—was consistent with that expected for the nicked cross-link (duplex D, lane 6).
A separate experiment provided evidence for the intermediacy of the spermine adducts 2 and 3 (Scheme 1). Specifically, treatment of the Ap-containing duplex A with various concentrations of spermine in the presence of NaCNBH3 (25 mM) for 1 h gave rise to two new bands (Supplementary Figure S1) with retarded gel mobility consistent with that expected for products arising from reduction of the spermine adducts 2 and 3 (44). The band arising from reduction of the full length spermine adduct 2 predominated (Supplementary Figure S1). This result is consistent with a system in which reduction of the intermediate Ap-derived imininium ion 2 by NaCNBH3 competes effectively against strand cleavage to generate 3.
The yield of the nicked cross-link formed following spermine-mediated generation of the nicked duplex C was ∼35%, with the half-maximal yield reached inside of 8 h at pH 7.4 and 37°C (Supplementary Figure S2). Easily detectable amounts of the cross-linked product were evident even at the earliest time points of our experiments, presumably formed during the generation of the nicked duplex C. It is noteworthy that a band consistent with the nicked cross-link was generated even in the absence of spermine (Figure 2, lane 3), albeit in lower yield, presumably arising via the relatively slow, spontaneous strand cleavage at the Ap site (31,38) to generate the 3’-ddR5p end product 1.
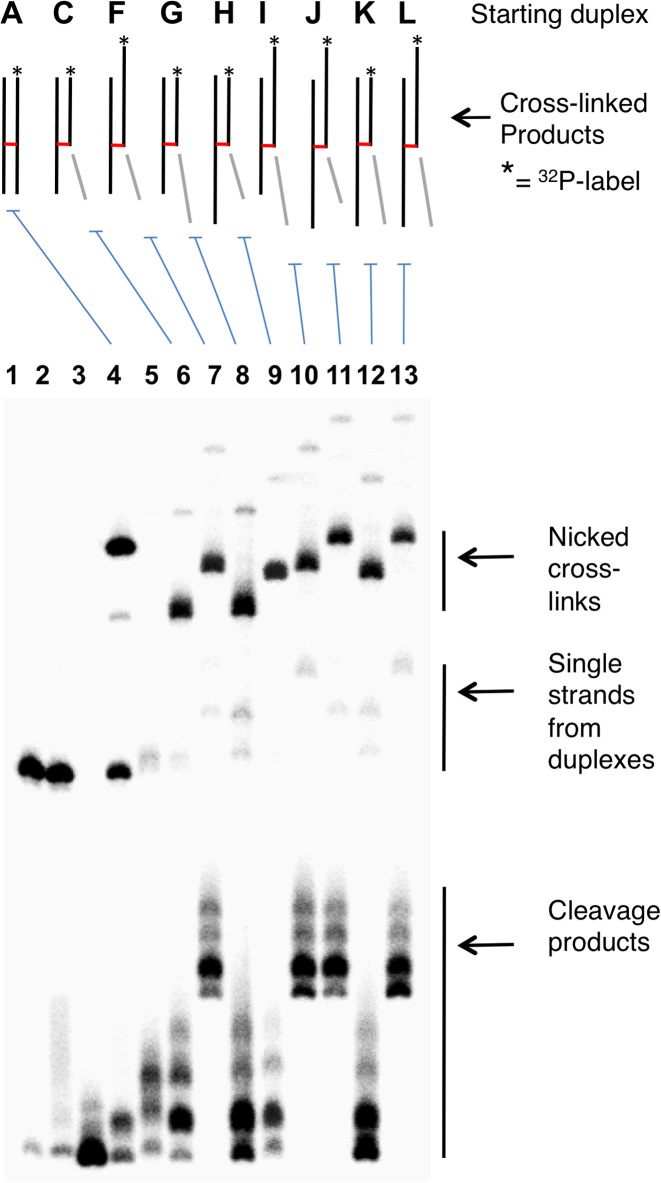
Gel shift analyses were used to provide evidence that the new, slowly-moving band generated from the nicked duplex C is a cross-linked species that contains three of the four ‘quadrants’ of the starting duplex A. We first showed that the new, slowly-migrating band has shed the oligonucleotide fragment on the 3’-side of the Ap site (light gray segments in Figures 1–3, and S3). We found that installation of a 5 nucleotide (nt) overhanging extension onto this end of the starting duplex caused no change in the mobility of the slowly-migrating band resulting from spermine-mediated strand cleavage (duplexes C and G, Figure 3). On the other hand, addition of 5 nt overhangs to the ends of the other four quadrants of the starting duplex caused retardation in the gel mobility of the slowly-migrating band resulting from spermine-nicked duplex (Figure 3). Overall, the gel electrophoretic results provided strong evidence that spermine-catalyzed cleavage of Ap sites in duplex DNA gave rise to a cross-linked species in which the 3’ddR5p sugar remnant is attached to the opposing strand of the duplex.
Figure 3.
Gel mobility studies provide evidence for the generation of nicked cross-linked DNA duplexes. Gel mobilities of cross-links generated from nicked duplexes G-M help define the size and structure of the species that gives the band of intermediate gel mobility between full length Ap-containing strand and the full-size cross-linked duplex B. Lane 1, size-marker consisting of the 5’-32P labeled uracil-containing oligonucleotide precursor to duplex A; Lane 2, freshly-prepared Ap-containing duplex A; Lane 3, Ap-containing duplex A subjected to piperidine work-up; Lane 4, formation of full-length cross-linked duplex B from duplex A; Lane 5, freshly-prepared, nicked 3’ddR5p-containing duplex C; Lane 6, formation of the nicked cross-linked duplex D from the nicked duplex C; Lanes 7–13, 3’ddR5p-containing DNA duplexes F-L were incubated in HEPES buffer (50 mM, pH 7.4) containing NaCl (100 mM) at 37°C to form the nicked, cross-linked duplexes. The 5’-32P-labeled oligodeoxynucleotides were resolved by electrophoresis on a 20% denaturing polyacrylamide gel, and the radioactivity in each band quantitatively measured by phosphorimager analysis. Sequences used in this experiment are shown in Supplementary Figure S3.
Iron–EDTA–H2O2 footprinting identifies the sites of cross-link attachment
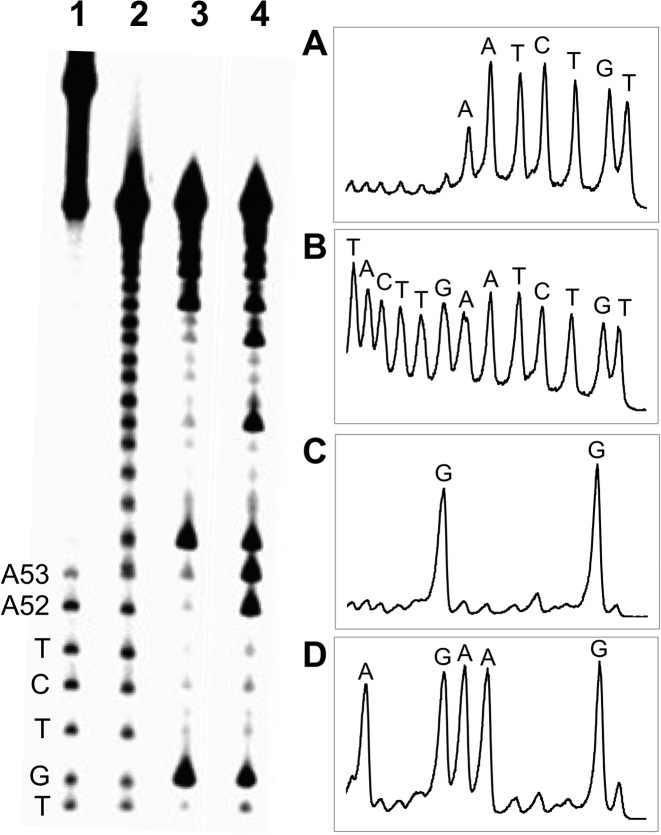
To identify the location of the cross-link attachment in duplex D, we conducted an iron–EDTA–H2O2 footprinting experiment (51,52). The strand complementary to the 3’ddR5p-containing oligonucleotide was 5’-32P-labeled and the gel-purified, nicked cross-link resulting from this duplex subjected to cleavage by a mixture of iron–EDTA–H2O2 (51–53). In this experiment, the site of cross-linking appears as an interruption in the ‘ladder’ of strand cleavage products generated by the iron–EDTA–H2O2 system, because cleavages beyond the cross-link yield large, slowly-migrating DNA fragments that are connected to the opposing strand (51,52). Footprinting of the cross-linked duplex D provided evidence that interstrand attachments were located at the adenine residues in positions 52 and 53 of duplex D, with adenine 52 being the predominant site of attachment (Figure 4).
Figure 4.
Iron–EDTA footprinting of the nicked cross-linked duplex D. The cross-linked duplex D, unlabeled on the 3’ddR5p-containing strand and 32P-labeled on the longer strand, was isolated from a denaturing gel and subjected to cleavage by a mixture of iron–EDTA–H2O2. The 32P-labeled oligodeoxynucleotides were resolved on a denaturing 20% polyacrylamide gel (left side of Figure) and the radioactivity in each band quantitatively measured by phosphorimager analysis. The right side of the Figure shows densitometry traces of: ane 1, Fe-EDTA footprinting reaction of the cross-linked duplex D (panel A); Lane 2, Fe-EDTA footprinting of the 5’-32P-labeled strand (panel B); Lane 3, Maxam–Gilbert G-specific sequencing reaction of the 32P-labeled strand (panel C); Lane 4 Maxam–Gilbert A+G sequencing reaction on the 32P-labeled strand (panel D).
Mass spectrometric analysis of nicked, cross-linked duplexes
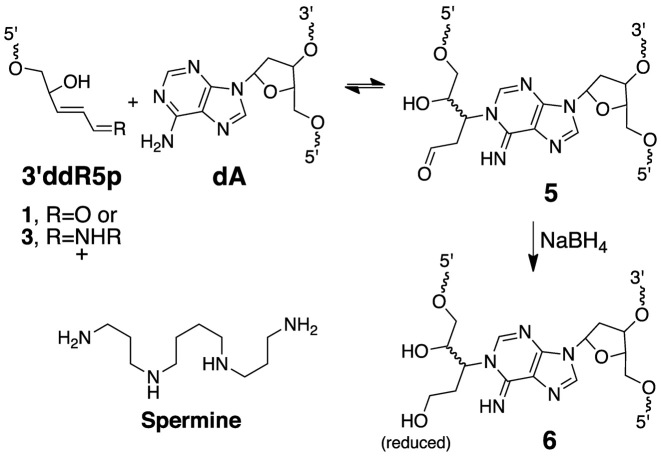
Analysis of the nicked cross-link generated in duplex E by nanospray QTOF-MS revealed a single major signal corresponding to an observed mass of 16115.65 amu (Supplementary Figure S4). This result combined with the data described above allowed us to suggest a reasonable mechanism for cross-link formation involving 1,4-addition of an adenine residue to the 3’ddR5p group (Scheme 3, calcd mass of 5 = 16115.44). Importantly, this proposed reaction is consistent with literature reports describing 1,4-conjugate addition of the N1-atom of the 2’-deoxyadenosine nucleoside to the low molecular weight α,β-unsaturated aldehyde, acrolein (69–71).
Scheme 3.
Formation of interstrand cross-links by reaction of an adenine residue with 3’ddR5p end groups arising from strand breakage at true Ap sites in duplex DNA.
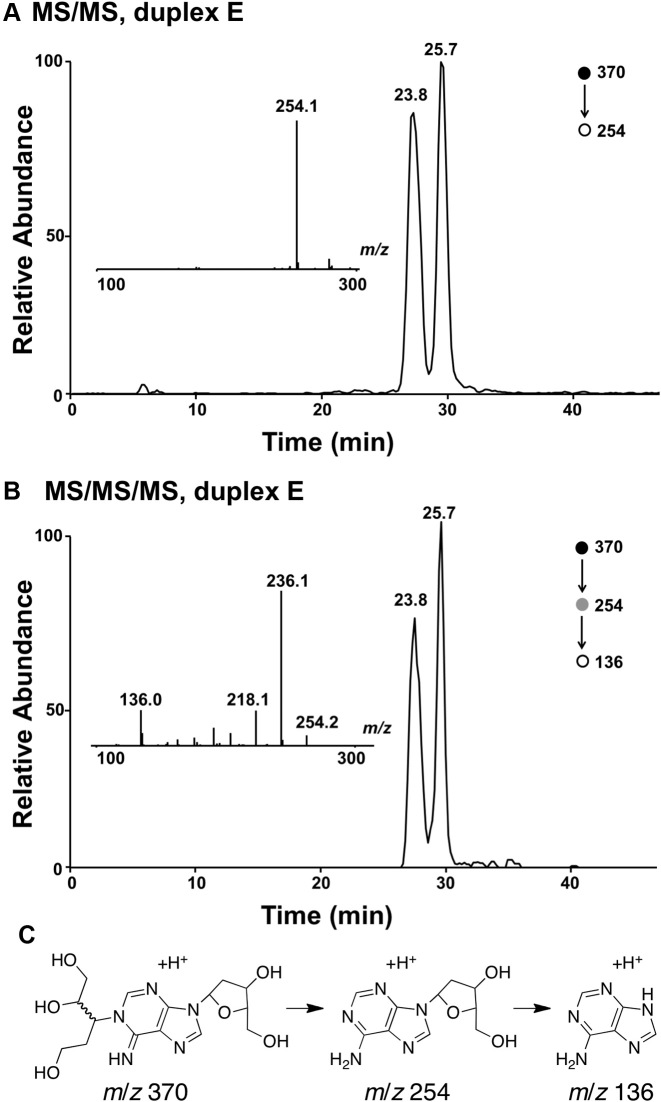
To further characterize the cross-linked DNA we undertook LC-MS analysis. In these experiments, we reasoned that reduction of the aldehyde group in the proposed adduct 5 would stabilize the cross-link against decomposition via a retro-Michael-type reaction (70) during sample handling and would also generate a diagnostic mass increase of 2 units. Toward this end, we added a solution of NaBH4 in NaOH to the cross-linking reaction after 72 h and incubated the mixture at 37°C for an additional 4 h, followed by neutralization with 0.1 N HCl, and isolation of the cross-linked DNA from a denaturing polyacrylamide gel. Digestion of the NaBH4-treated cross-linked DNA duplex E with a four-enzyme cocktail consisting of nuclease P1, alkaline phosphatase, and phosphodiesterases I and II, followed by LC-MS/MS analysis (63,72) revealed two peaks eluting at 23–26 min displaying the m/z 370→254 transition anticipated for the neutral loss of 2-deoxyribose (116 Da) from the reduced cross-link remnants 6 (Figures 5 and Supplementary Figure S5). The MS/MS/MS arising from the further cleavage of the ion of m/z 254 revealed fragment ions of m/z 236 and 218, which emanate from the neutral losses of one and two H2O molecules, respectively, along with the ion of m/z 136, which corresponds to the protonated adenine (Figures 5 and Supplementary Figure S5). The formation of these fragment ions was in line with the proposed structure for the cross-linked nucleobase remnant (Scheme 3). The presence of multiple peaks in the LC–MS/MS ion chromatogram was consistent with the anticipated diastereomeric nature of the nucleosidic cross-link remnant 6 (Scheme 3).
Figure 5.
LC–MS/MS and LC–MS/MS/MS analysis results of the nucleoside mixture arising from the enzymatic digestion of cross-linked duplex E. (A) The selected-ion chromatogram for the m/z 370→254 transition. (B) The selected-ion chromatogram for the m/z 370→254→136 transition. Shown in the insets are the MS/MS (A) and MS/MS/MS (B) averaged from the 23.8-min fraction. (C) Possible structures for the ions observed at m/z 370, 254 and 136.
Overall, the mass spectrometric data are consistent with formation of a nicked, cross-linked duplex arising from 3’ddR5p derivatives generated by strand scission at the Ap site. The data further indicates that cross-linking involves attachment to adenine on the opposing strand. Finally, the results are consistent with cross-link formation via 1,4-addition of adenine to the 3’ddR5p group.
The 3’ddR5p-derived cross-link blocks DNA replication by the strand-displacing ϕ29 DNA polymerase
The results described above indicate that the (unreduced) cross-link derived from the 3’ddR5p group is stable to sample handling and gel electrophoretic analysis (Figures 2 and 3). Nonetheless, we felt it was important to investigate whether the cross-linkage can withstand DNA processing enzymes that actively induce strand separation. To examine this issue, we used ϕ29 DNA polymerase, an enzyme that has been described as a hybrid helicase–polymerase (73). Bacteriophage ϕ29 DNA polymerase is a highly processive polymerase that couples synthesis and strand displacement to carry out DNA replication without assistance from helicases, clamp proteins, or other accessory factors (74).
We prepared a series of ϕ29 DNA polymerase substrates to examine whether the 3’ddR5p cross-link was able to block strand separation by the enzyme (Figure 6 and Supplementary Figure S6). We first carried out a series of control experiments showing that the enzyme, in combination with dNTPs fully extended a 15 nt 32P-labeled primer annealed to a 56 nt template strand (substrate M), a template strand with a 35 nt duplex region downstream of the primer (N), and a 56 nt template strand containing a 35 nt nicked duplex downstream of the primer (O, Figures 6 and Supplementary Figure S6). As expected based upon our previous work (56), extension of the primer in substrate R containing the dA-Ap cross-link 4 in the downstream duplex region was stalled immediately preceding the cross-link. Primer extension in substrate Q, containing the nicked 3’ddR5p cross-link in a downstream duplex region was almost completely stalled at last unmodified base preceding the cross-link. Extension of the primer in cross-linked substrate P, lacking the duplex region on the 3’-side of the cross-link was similarly stalled at the cross-link. Overall, the results provided evidence that the dA-3’ddR5p cross-link was stable against the powerful strand-separating properties (73) of the helicase-polymerase activity of the bacteriophage ϕ29 DNA polymerase.
Figure 6.
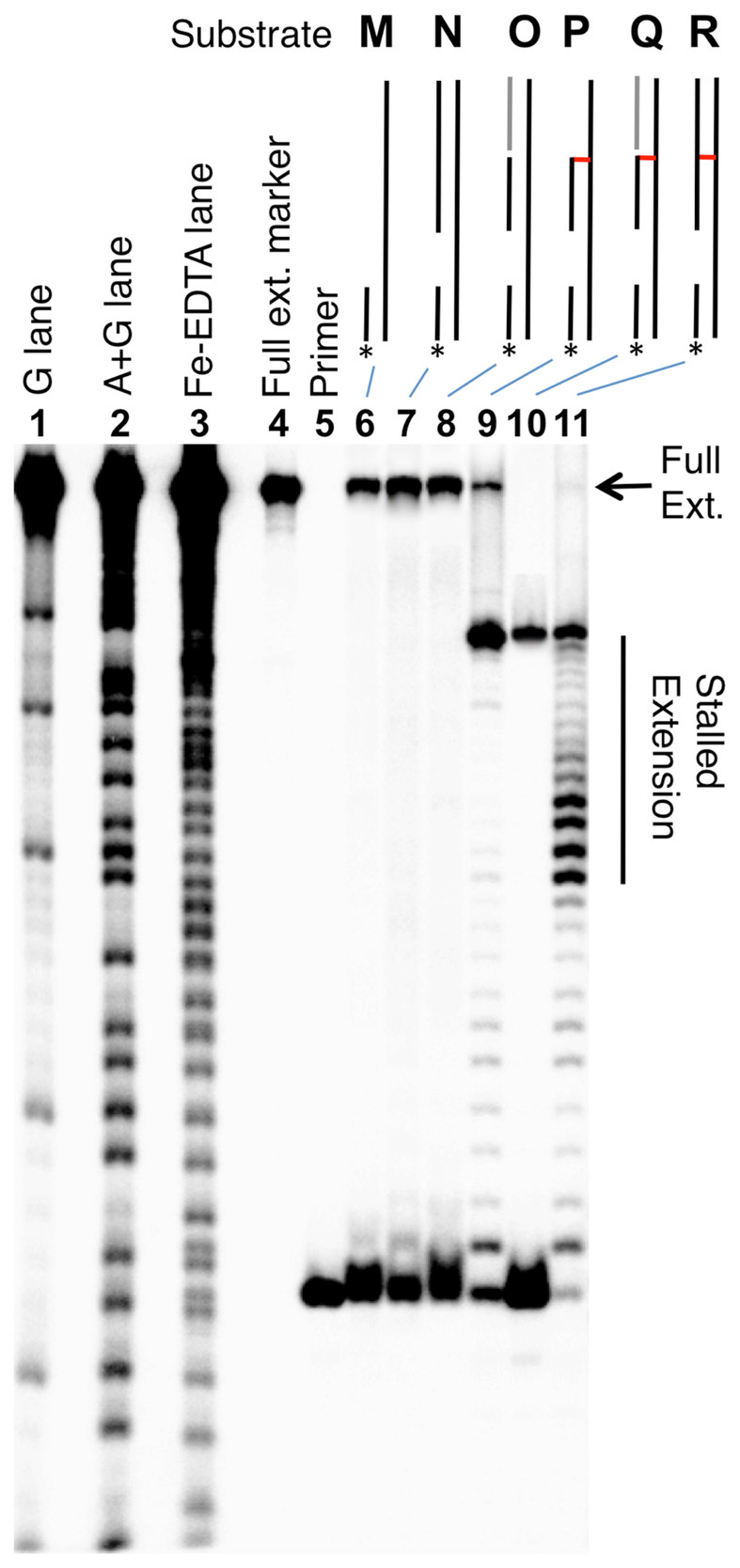
The dA-3’ddR5p cross-link blocks DNA replication by ϕ29 polymerase. The 15 nt 32P-labeled primers were incubated with the DNA substrates, the polymerase enzyme (10 U), and the four dNTPs (1 mM in each) in Tris–HCl (50 mM, pH 7.5), MgCl2 (10 mM), (NH4)2SO4 (10 mM), DTT (4 mM), and bovine serum albumin (0.1 mg/ml) for 30 min at 24°C. After reaction work-up, the primer extension products were analyzed on a 20% denaturing polyacrylamide gel. Lanes 1, 2 and 3 are Maxam–Gilbert G, A+G, and iron–EDTA–H2O2 cleavage reactions carried out on a 5’-32P-labeled standard of the full-length extension product (5’-32P-GAT CAC AGT GAG TAG AAT AGA ATA CCA GAT ACA TGA ACT TAG ACA TAT ACA CAG AT); lane 4 is the 5’-32P-labeled full-length extension product; lane 5 is the 15 nt, 5’-32P-labeled primer; lanes 6–11, primer extension on substrates M–R. Sequences of these substrates are shown in Supplementary Figure S6.
CONCLUSIONS
Strand breaks at Ap sites in cellular DNA are generated by spontaneous and enzymatic processes (1,31–48). Interestingly, this reaction generates an electrophilic 3’ddR5p derivative embedded within the nicked duplex (30,44,49,50). A few studies have shown that nucleophilic small molecules (34,37,75,76) and proteins (77–79) can react with the 3’ddR5p sugar remnant. However, our work provides the first characterization of interstrand cross-links resulting from the reaction of the 3’ddR5p group with a nucleobase on the opposing strand. Catalano et al. previously proposed a product of this type to explain a band observed in the denaturing gel analysis of Ap-containing DNA duplexes (80) but information related to the structure, origin and properties of this cross-link have been lacking until now. In addition, cross-links of this type were proposed to explain high molecular weight impurities generated in the manufacture of therapeutic oligonucleotides, but again the structure, origin and properties of these cross-linked species were not characterized (81).
Our results provide evidence that spermine-catalyzed strand cleavage at a true Ap site in duplex DNA can lead to generation of a complex lesion consisting of an interstrand cross-link to adenine adjacent to a strand break. This reaction is distinct from previous work involving strand cleavage at oxidized Ap sites (52,82–85). The exact chemical structure of the interstrand attachment remains to be defined, but our data combined with literature precedents (69,71) allow us to propose a cross-link structure 5 derived from 1,4-addition of the N1-atom of adenine to the 3’ddR5p residue (Scheme 3). Importantly, we provided evidence that this cross-link blocks DNA replication by ϕ29 DNA polymerase, a highly processive polymerase enzyme that couples synthesis and strand displacement (74). This forecasts the potential for the dA-3’ddR5p cross-link to block mammalian DNA replication and transcription. In addition, cross-links of this type may present special challenges to repair systems (52,83). Cross-links could contribute to the toxicity and generation of slow-moving chromosomal DNA intermediates caused by increased levels of 3’ddR5p strand breaks in Saccharomyces cerevisiae mutants with dysregulated base excision repair pathways (86–88). It will be interesting to explore how spermine and other polyamines in the nucleus modulate the fate of Ap sites in genomic DNA. The polyamine-derived imines 2 and 3 (Scheme 1) are expected to be much more reactive than the parent aldehydes Ap and 1 (89–91). Indeed, the spermine-mediated formation of the nicked cross-links described here is substantially faster than the formation of the full-length dA-Ap cross-link described previously (4, Scheme 2) (55).
Chemical precedents involving the reaction of nucleosides with low molecular weight compounds such as HNE and acrolein show that all four of the canonical DNA bases have the potential to react with α,β-unsaturated aldehydes (69,71,92–95). Thus, it seems likely that interstrand cross-links derived from the 3’ddR5p group in duplex DNA will prove to be a structurally diverse family of complex lesions that can form in a wide variety of sequence contexts. The LC–MS/MS methods developed here provide a powerful tool for studying the occurrence and properties of these cross-links in biochemical and biological systems (96,97).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Marc Greenberg for helpful conversations on this topic.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
National Institute of Environmental Health Science of the National Institutes of Health [ES021007 to K.S.G., Y.W.]; NRSA T32 institutional training grant [ES018827 to N.P.] (in part). Funding for open access charge: National Institute of Environmental Health Sciences [ES021007].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gates K.S. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol. 2009; 22:1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 3. Potenski C.J., Klein H.L.. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 2014; 42:10226–10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Bont R., van Larebeke N.. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004; 19:169–185. [DOI] [PubMed] [Google Scholar]
- 5. Barnes D.E., Lindahl T.. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004; 38:445–476. [DOI] [PubMed] [Google Scholar]
- 6. Pham P., Bransteitter R., Goodman M.F.. Reward versus risk: DNA cytidine deaminases triggering immunity and disease. Biochemistry. 2005; 44:2703–2715. [DOI] [PubMed] [Google Scholar]
- 7. Marnett L.J., Burcham P.C.. Endogenous DNA adducts: potential and paradox. Chem. Res. Toxicol. 1993; 6:771–785. [DOI] [PubMed] [Google Scholar]
- 8. Swenberg J.A., Moeller B.C., Gao L., Upton P.B., Nakamura J., Starr T.. Endogenous versus exogenous DNA adducts: their role in carginogenesis, epidemiology, and risk assessment. Toxicol. Sci. 2011; 120:S130–S145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindahl T., Wood R.D.. Quality control by DNA repair. Science. 1999; 286:1897–1905. [DOI] [PubMed] [Google Scholar]
- 10. Lindahl T., Barnes D.E.. Repair of endogenous DNA damage. Cold Spring Harb. Perspect. Biol. 2000; 65:127–134. [DOI] [PubMed] [Google Scholar]
- 11. Schärer O.D. Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. Eng. 2003; 42:2946–2974. [DOI] [PubMed] [Google Scholar]
- 12. McKinnon P.J. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 2009; 10:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoeijmakers J.H.J. DNA damage, aging, and cancer. J. Engl. J. Med. 2009; 361:1475–1485. [DOI] [PubMed] [Google Scholar]
- 14. Borgesius N.Z., de Waard M., van der Pluijm I., Omrani Z., Zondag G.C.M., ver der Horst G.T.J., Melton D.W., Hoeijmakers J.H.J., Jaarsma D., Elgersma Y.. Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. J. Neurosci. 2011; 31:12543–12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Menck C.F.M., Munford V.. DNA repair diseases: what do they tell us about cancer and aging?. Genet. Mol. Biol. 2014; 37:220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cline S.D. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim. Biophys. Acta. 2012; 1819:979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedberg E.C., McDaniel L.D., Schultz R.A.. The role of endogenous and exogenous DNA damage and mutagenesis. Curr. Opin. Genet. Dev. 2004; 14:5–10. [DOI] [PubMed] [Google Scholar]
- 18. Madabhushi R., Pan L., Tsai L.-H.. DNA damage and its links to neurodegeneration. Neuron. 2014; 83:266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schumacher B., Garinis G.A., Hoeijmakers J.H.J.. Age to survive: DNA damage and aging. Trends Genet. 2007; 24:77–85. [DOI] [PubMed] [Google Scholar]
- 20. Mehta A., Haber J.E.. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014; 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clauson C., Schärer O.D., Niedernhofer L.J.. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspect. Biol. 2013; 5:a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schärer O.D. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. ChemBioChem. 2005; 6:27–32. [DOI] [PubMed] [Google Scholar]
- 23. Lawley P.D., Phillips D.H.. DNA adducts from chemotherapeutic agents. Mutat. Res. 1996; 355:13–40. [DOI] [PubMed] [Google Scholar]
- 24. Grossman K.F., Ward A.M., Matkovic M.E., Folias A.E., Moses R.E.. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat. Res. 2001; 487:73–83. [DOI] [PubMed] [Google Scholar]
- 25. Eccles L.J., O’Neill P., Lomax M.E.. Delayed repair of radiation induced clustered DNA damage: friend or foe. Mutat. Res. 2011; 711:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Georgakilas A.G., O’Neill P., Stewart R.D.. Induction and repair of clustered DNA lesions: what do we know so far. Radiat. Res. 2013; 180:100–109. [DOI] [PubMed] [Google Scholar]
- 27. Nakamura J., Swenberg J.A.. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999; 59:2522–2526. [PubMed] [Google Scholar]
- 28. Guillet M., Bioteux S.. Origin of endogenous DNA abasic sites in Saccaromyces cerevisiae. Mol. Cell Biol. 2003; 23:8386–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tamm C., Shapiro H.S., Lipshitz R., Chargaff E.. Distribution density of nucleotides within a desoxyribonucleic acid chain. J. Biol. Chem. 1953; 199:673–688. [PubMed] [Google Scholar]
- 30. Bayley C.R., Brammer K.W., Jones A.S.. The nucleotide sequence in deoxyribonucleic acids. Part V. The alkaline degradation of apurinic sites. J. Chem. Soc. 1961; 1903–1907. [Google Scholar]
- 31. Lindahl T., Andersson A.. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972; 11:3618–3623. [DOI] [PubMed] [Google Scholar]
- 32. Crine P., Verly W.G.. A study of DNA spontaneous degradation. Biochim. Biophys. Acta. 1976; 442:50–57. [DOI] [PubMed] [Google Scholar]
- 33. Luke A.M., Chastain P.D., Pachkowski B.F., Afonin V., Takeda S., Kaufman D.G., Swenberg J.A., Nakamura J.. Accumulation of true single strand breaks and AP sites in base excision repair deficient cells. Mutat. Res. 2010; 694:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bailly V., Verly W.G.. Importance of thiols in the repair mechanisms of DNA containing AP (apurinic/apyrimidinic) sites. Nucleic Acids Res. 1988; 16:9489–9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Oliveira M.P., Constant J.-F., Peuchmaur M., Pitta I., Décout J.-L.. Antibiotic drugs aminoglycosides cleave DNA at abasic sites: shedding new light on their toxicity. Chem. Res. Toxicol. 2013; 26:1710–1719. [DOI] [PubMed] [Google Scholar]
- 36. Mirzabekov A.D., Shick V.V., Belyavsky A.V., Bavykin S.G.. Primary organization of nucleosome core particle of chromatin: Sequence of histone arrangement along DNA. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:4184–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vasseur J.-J., Rayner B., Imbach J.-L.. Apurinic DNA reactivity: modelisation of apurinic DNA breakage by phenylhydrazine and formation of pyrazole adduct. J. Heterocyclic Chem. 1988; 25:389–392. [Google Scholar]
- 38. Zhou C., Sczepanski J.T., Greenberg M.M.. Mechanistic studies on histone catalyzed cleavage of apyrimidinic/apurinic sites in nucleosome core particles. J. Am. Chem. Soc. 2012; 134:16734–16741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brooks S.C., Adhikary S., Rubinson E.H., Eichman B.F.. Recent advances in the structural mechanisms of DNA glycosylases. Biochim. Biophys. Acta. 2013; 1834:247–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. David S.S., Williams S.D.. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev. 1998; 98:1221–1261. [DOI] [PubMed] [Google Scholar]
- 41. Piersen C.E., McCullough A.K., Lloyd R.S.. AP lyases and dRPases: commonality of mechanism. Mutat. Res. 2000; 459:43–53. [DOI] [PubMed] [Google Scholar]
- 42. McHugh P.J., Knowland J.. Novel reagents for chemical cleavage at abasic sites and UV photoproducts in DNA. Nucleic Acids Res. 1995; 23:1664–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Male R., Fosse V.M., Kleppe K.. Polyamine-induced hydrolysis of apurinic sites in DNA and nucleosomes. Nucleic Acid Res. 1982; 10:6305–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kurtz A.J., Dodson M.L., Lloyd R.S.. Evidence for multiple imino intermediates and identification of reactive nucleophiles in peptide-catalyzed beta-elimination at abasic sites. Biochemistry. 2002; 41:7054–7064. [DOI] [PubMed] [Google Scholar]
- 45. Minko I.G., Jacobs A.C., de Leon A.R., Gruppi F., Donley N., Harris T.M., Rizzo C.J., McCullough A.K., Lloyd R.S.. Catalysts of DNA strand cleavage at apurinic/apyrmidinic sites. Sci. Rep. 2016; 6, doi:10.1038/srep28894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stande N.T., Carvajal J., Hallett R.A., Waters C.A., Roberts S.A., Strom C., Kuhlman B., Ramsden D.A.. Requirements for 5΄-dRP/AP lyase activity in Ku. Nucleic Acids Res. 2014; 42:11136–11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sczepanski J.T., Zhou C., Greenberg M.M.. Nucleosome core particle-catalyzed strand scission at abasic sites. Biochemistry. 2013; 52:2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sugiyama H., Fujiwara T., Ura A., Tashiro T., Yamamoto K., Kawanishi S., Saito I.. Chemistry of thermal degradation of abasic sites in DNA. mechanistic investigation on thermal DNA strand cleavage of alkylated DNA. Chem. Res. Toxicol. 1994; 7:673–683. [DOI] [PubMed] [Google Scholar]
- 49. Manoharan M., Ransom S.C., Mazumder A., Gerlt J.A., Wilde J.A., Withka J.A., Bolton P.H.. The characterization of abasic sites in DNA heteroduplexes by site specific labeling with carbon-13. J. Am. Chem. Soc. 1988; 110:1620–1622. [Google Scholar]
- 50. Bailly V., Verly W.G.. Possible roles of β-elimination and δ-elimination reactions in the repair of DNA containing AP (apurinic/apyrimidinic) sites in mammalian cells. Biochem. J. 1988; 253:553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luce R.A., Hopkins P.B.. Chemical cross-linking of drugs to DNA. Methods Enzymol. 2001; 340:396–412. [DOI] [PubMed] [Google Scholar]
- 52. Sczepanski J.T., Jacobs A.C., Majumdar A., Greenberg M.M.. Scope and mechanism of interstrand crosslink formation by the C4΄-oxidized abasic site. J. Am. Chem. Soc. 2009; 131:11132–11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Johnson K.M., Price N.E., Wang J., Fekry M.I., Dutta S., Seiner D.R., Wang Y., Gates K.S.. On the formation and properties of interstrand DNA-DNA cross-links forged by reaction of an abasic site with the opposing guanine residue of 5’-CAp sequences in duplex DNA. J. Am. Chem. Soc. 2013; 135:1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gamboa Varela J., Gates K.S.. A simple, high-yield synthesis of DNA duplexes containing a covalent, thermally-reversible interstrand cross-link at a defined location. Angew. Chem. Int. Ed. Engl. 2015; 54:7666–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Price N.E., Johnson K.M., Wang J., Fekry M. I., Wang Y., Gates K.S.. Interstrand DNA−DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J. Am. Chem. Soc. 2014; 136:3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang Z., Johnson K.M., Price N.E., Gates K.S.. Characterization of interstrand DNA-DNA cross-links derived from abasic sites using bacteriophage ϕ29 DNA polymerase. Biochemistry. 2015; 54:4259–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Varshney U., van de Sande J.H.. Specificities and kinetics of uracil excision from uracil-containing DNA oligomers by Escherichia coli uracil DNA glycosylase. Biochemistry. 1991; 30:4055–4061. [DOI] [PubMed] [Google Scholar]
- 58. Lindahl T., Ljunquist S., Siegert W., Nyberg B., Sperens B.. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977; 252:3286–3294. [PubMed] [Google Scholar]
- 59. Sambrook J., Fritsch E.F., Maniatis T.. Molecular Cloning: A Lab Manual. 1989; NY: Cold Spring Harbor Press. [Google Scholar]
- 60. Sarhan S., Seiler N.. On the subcellular localization of the polyamines. Biol. Chem. Hoppe Seyler. 1989; 370:1279–1284. [DOI] [PubMed] [Google Scholar]
- 61. Bailly V., Verly W.G.. Possible roles of beta-elimination and gamma-elimination reactions in the repair of DNA containing AP (apurinic/apyrimidinic) sites in mammalian cells. Biochem. J. 1988; 253:553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dutta S., Chowdhury G., Gates K.S.. Interstrand crosslinks generated by abasic sites in duplex DNA. J. Am. Chem. Soc. 2007; 129:1852–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Price N.E., Catalano M.J., Liu S., Wang Y., Gates K.S.. Chemical and structural characterization of interstrand cross-links formed between abasic sites and adenine residue in duplex DNA. Nucleic Acids Res. 2015; 43:3434–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gamboa Varela J., Gates K.S.. Simple, high-yield syntheses of DNA duplexes containing interstrand DNA-DNA cross-links between an N4-aminocytidine residue and an abasic site. Curr. Protoc. Nucleic Acid Chem. 2016; 65, doi:10.1002/cpnc.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Maxam A.M., Gilbert W.. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980; 65:499–560. [DOI] [PubMed] [Google Scholar]
- 66. Raspaud E., Chaperon I., Leforestier A., Livolant F.. Spermine-induced aggregation of DNA, nucleosome, and chromatin. Biophys. J. 1999; 77:1547–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wallace H.M., Fraser A.V., Hughes A.. A perspective of polyamine metabolism. Biochem. J. 2003; 376:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bailly V., Derydt M., Verly W.G.. δ-Elimination in the repair of AP (apurinic/apyrimidinic) sites in DNA. Biochem. J. 1989; 261: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pawlowicz A.J., Munter T., Zhao Y., Kronberg L.. Formation of acrolein adducts with 2΄-deoxyadenosine in calf thymus DNA. Chem. Res. Toxicol. 2006; 19:571–576. [DOI] [PubMed] [Google Scholar]
- 70. Chung F.-L., Nath R.G., Ocando J., Nishikawa A., Zhang L.. Deoxyguanosine adducts of t-4-hydroxy-2-nonenal are endogenous DNA lesions in rodents and humans: detection and potential sources. Cancer Res. 2000; 60:1507–1511. [PubMed] [Google Scholar]
- 71. Smith R.A., Williamson D.S., Cerny R.L., Cohen S.M.. Detection of 1,N6-propanodeoxyadenosine in acrolein-modified polydeoxyadenylic acid and DNA by 32P-postlabeleing. Cancer Res. 1990; 50:3005–3012. [PubMed] [Google Scholar]
- 72. Catalano M.J., Liu S., Andersen N., Yang Z., Johnson K.M., Price N.A., Wang Y., Gates K.S.. Chemical structure and properties of the interstrand cross-link formed by the reaction of guanine residues with abasic sites in duplex DNA. J. Am. Chem. Soc. 2015; 137:3933–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morin J.A., Cao F.J., Lázaro J.M., Arias-Gonzalaz R., Valpuesta J.M., Carrascosa J.L., Salas M., Ibarra B.. Active DNA unwinding dynamics during processive DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:8115–8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rodríguez I., Lázaro J.M., Blanco L., Kamtekar S., Berman A.J., Wang J., Steitz T.A., Salas M., de Vega M.. A specific subdomain in ϕ29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:6407–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shishkina I.G., Johnson F.. A new method for the postsynthetic generation of abasic sites in oligomeric DNA. Chem. Res. Toxicol. 2000; 13:907–912. [DOI] [PubMed] [Google Scholar]
- 76. Vasseur J.-J., Rayner B., Imbach J.-L., Verma S., McCloskey J.A., Lee M., Chang D.-K., Lown J.W.. Structure of the adduct formed between 3-aminocarbazole and the apurinic site oligonucleotide model d[Tp(Ap)pT]. J. Org. Chem. 1987; 52:4994–4998. [Google Scholar]
- 77. Sczepanski J.T., Wong R.S., McKnight J.N., Bowman G.D., Greenberg M.M.. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:22475–22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pruss D., Gravin I.M., Melnik S., Bavykin S.G.. DNA-protein cross-linking applications for chromatin studies in vitro and in vivo. Methods Enzymol. 1999; 304:516–533. [DOI] [PubMed] [Google Scholar]
- 79. Prasad R., Horton J.K., Chastain P.D. II, Gassman N.R., Freudenthal B.D., Hou E.W., Wilson S.H.. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic Acids Res. 2014; 42:6337–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Catalano M.J., Price N.E., Gates K.S.. Effective molarity in a nucleic acid controlled reaction. Bioorg. Med. Chem. Lett. 2016; 26:2627–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kurata C., Bradley K., Gaus H., Luu N., Cedillo I., Ravikumar V.T., Sooy K.V., McArdle J.V., Capaldi D.C.. Characterization of high molecular weight impurities in synthetic phosphorothioate oligonucleotides. Bioorg. Med. Chem. Lett. 2006; 16:607–614. [DOI] [PubMed] [Google Scholar]
- 82. Sczepanski J.T., Jacobs A.C., Greenberg M.M.. Self-promoted DNA interstrand cross-link formation by an abasic site. J. Am. Chem. Soc. 2008; 130:9646–9647. [DOI] [PubMed] [Google Scholar]
- 83. Ghosh S., Greenberg M.M.. Synthesis, of cross-linked DNA containing oxidized abasic site analogues. J. Org. Chem. 2014; 79:5948–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sun J., Tang X.. Photouncaged sequence-specific interstrand DNA cross-linking with photolabile 4-oxo-enal-modified oligonucleotides. Sci. Rep. 2015; 5, doi:10.1038/srep10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Regulus P., Duroux B., Bayle P.-A., Favier A., Cadet J., Ravanat J.-L.. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:14032–14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Guillet M., Bioteux S.. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2, and Rad1/Rad10 in Saccaromyces cerevisae. EMBO J. 2002; 21:2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ma W., Westmoreland J.W., Gordenin D.A., Resnick M.A.. Alkylation base damage is converted into repairable double-strand breaks and complex intermediates in G2 cells lacking AP endonuclease. PLoS Genet. 2011; 7:e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Boiteux S., Guillet M.. Abasic sites in DNA: repair and biological consequences in Saccaromyces cerevisiae. DNA Repair. 2004; 3:1–12. [DOI] [PubMed] [Google Scholar]
- 89. Bender M.L., Williams A.. Ketimine intermediates in amine-catalyzed enolization of acetone. J. Am. Chem. Soc. 1966; 88:2502–2508. [Google Scholar]
- 90. Erkkilä A., Majander I., Pihko P.M.. Iminium ion catalysis. Chem. Rev. 2007; 107:5416–5470. [DOI] [PubMed] [Google Scholar]
- 91. Appel R., Chelli S., Tokuyasu T., Troshin K., Mayr H.. Electrophilicities of benzaldehyde-derived iminium ions: quantification of the electrophilic activation of aldehydes by iminium formation. J. Am. Chem. Soc. 2013; 135:6579–6587. [DOI] [PubMed] [Google Scholar]
- 92. Pawlowicz A.J., Klika K.D., Kronberg L.. The structural identification and conformational analysis of the products from the reaction of acrolein with 2΄-deoxycytidine, 1-methylcytosine and calf thymus DNA. Eur. J. Org. Chem. 2007; 2007; 1429–1437. [Google Scholar]
- 93. Pawlowicz A.J., Kronberg L.. Characterization of adducts formed in the reactions of acrolein with thymidine and calf thymus DNA. Chem. Biodivers. 2008; 5:177–188. [DOI] [PubMed] [Google Scholar]
- 94. Chung F.-L., Young R., Hecht S.M.. Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 1984; 44:990–995. [PubMed] [Google Scholar]
- 95. Kowalczyk P., Ciesla J.M., Komisarski M., Kusmierek J., Tudek B.. Long-chain adducts of trans-4-hydroxy-2-nonenal to DNA bases cause recombination, base substitutions and frameshift mutations in M13 phage. Mutat. Res. 2004; 550:33–48. [DOI] [PubMed] [Google Scholar]
- 96. Liu S., Wang Y.. Mass spectrometry for the assessment of the occurrence and bioloigical consequences of DNA adducts. Chem. Soc. Rev. 2015; 44:7829–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. You C., Wang Y.. Mass spectrometry-based quantitative strategies for assessing the biological consequences and repair of DNA adducts. Acc. Chem. Res. 2016; 49:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.