AbSTRACT
Dissecting the complex network of epigenetic modifications requires tools that combine precise recognition of DNA sequences with the capability to modify epigenetic marks. The CRISPR/Cas system has been proven to be a valuable addition to existing methodologies that fulfill these tasks. So far, sequence-specific editing of epigenetic modifications such as DNA methylation and histone posttranslational modifications relied on direct fusions of enzymatically inactivated Cas9 (dCas9) with epigenetic effectors. Here, we report a novel, modular system that facilitates the recruitment of any GFP-tagged protein to desired genomic loci. By fusing dCas9 to a GFP-binding nanobody (GBP) we demonstrate that prevalent epigenetic modifications at mouse major satellite repeats can be erased or set de novo by recruiting GFP-coupled catalytic domains of TET1 and DNMT3A, respectively. Furthermore, we construct an inducible expression system that enables a temporally controlled expression of both GBP-dCas9 and the effector protein. Thus, our approach further expands the CRISPR/Cas toolbox for site-specific manipulation of epigenetic modifications with a modular and easy-to-use system.
KEYWORDS: CRISPR/Cas, DNA de novo methylation, DNA demethylation, GBP, GFP, Nanobody
Introduction
Eukaryotic gene expression is controlled by a complex network of epigenetic mechanisms that include the posttranslational modification of histones as well as covalent DNA modifications.1 Dissection of this network using knockout or overexpression studies have greatly advanced our understanding of how epigenetic modifications contribute to transcriptional regulation. However, using traditional techniques, the complex relationships and feedback circuits that interconnect epigenetic pathways make it difficult to differentiate direct consequences of epigenetic modifications on transcription from secondary effects. Site-specific manipulation of epigenetic marks therefore represents a highly desirable tool to study and understand their direct functional relevance on gene expression and genome organization.
Previously, tools that direct the enzymatic activity of epigenetic effectors to specific loci were based on zinc finger proteins (ZFPs)2-6 or transcription activator-like effectors (TALEs).6-8 However, custom design and engineering of ZFPs and TALEs is based on the rearrangement of their modular DNA-binding domains, requiring elaborate cloning techniques and rigorous testing.9-12 In contrast, the RNA-guided endonuclease Cas9 of the type II CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated) system recognizes specific loci via Watson-Crick base pairing between a readily exchangeable 20 bp sequence of the single guide RNA (sgRNA) and the target DNA in the direct vicinity of a PAM (protospacer adjacent motif).13-15 Due to this ease of use, Cas9-based approaches have been rapidly adopted for genome engineering strategies in a wide variety of cell types and organisms.16-20
Importantly, engineering of a catalytically inactive variant of Cas9 (dCas9) facilitates RNA-guided genome targeting in a sequence specific manner, without cleaving the underlying DNA.13,21 We and others have previously demonstrated that this programmable DNA-binding platform can be harnessed for in vivo visualization of specific genomic regions22-24 as well as for determination of local chromatin composition.25-26 In addition, fusion of dCas9 with the methyltransferase DNMT3A enables targeted transcriptional repression by catalyzing de novo methylation at gene regulatory regions.27-28 Vice versa, dCas9 mediated targeting of ten-eleven translocation methylcytosine dioxygenase 1 (TET1) to regulatory elements results in upregulation of silenced genes via active DNA demethylation.29-31 Similarly, transcriptional modulation has also been reported by targeting histone acetylase32 and histone demethylase activities33 via dCas9, highlighting the versatility of this approach.
Here, we introduce a modular CRISPR/Cas9 system, which combines the sequence specificity of dCas9 with stringent recruitment of GFP-coupled epigenetic effectors via a GFP-binding nanobody (GBP).34 We show that this versatile setup can be exploited to control the levels of DNA modifications at target loci using GFP fusions of DNA methyltransferases and methylcytosine dioxygenases. Furthermore, using a bidirectional doxycycline-inducible promoter we develop a single vector system that allows the timed expression and targeted recruitment of GBP-Cas9 and GFP-fusion proteins.
Results and discussion
We first set out to assess, whether GFP can be recruited to a defined genomic locus via dCas9. To this end, we constructed a GBP-dCas9-mRFP construct, which enabled us to simultaneously visualize dCas9 and GFP localization (Fig. 1A). Since heterochromatic chromocenters (CCs) are distinct subnuclear regions, which can be readily distinguished and are characterized by well-defined epigenetic marks,35-36 we decided to tether GBP-dCas9-mRFP to these loci via a major satellite specific sgRNA (MaSgRNA).
Figure 1.
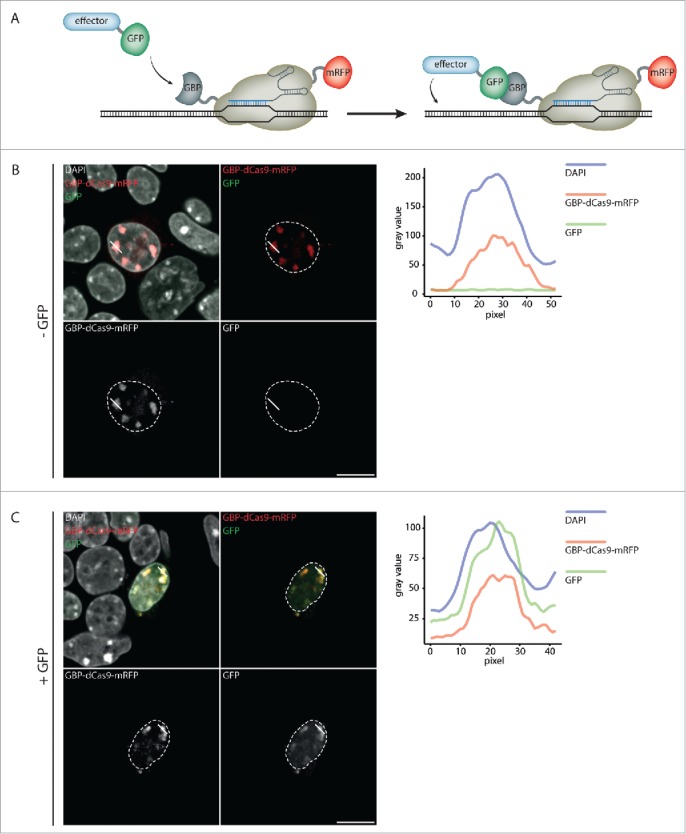
Targeted recruitment of GFP to major satellites. (A) Schematic outline of dCas9-mediated effector recruitment. GBP-dCas9-mRFP is guided to a desired locus by a sgRNA and interacts with a GFP-coupled epigenetic effector via GBP. Subsequently, the effector modifies the underlying DNA. (B-C) Representative confocal images of ESCs, co-transfected with GBP-dCas9-mRFP and major satellite specific sgRNA. GBP-dCas9-mRFP specifically localizes at CCs and recruits GFP, when it is additionally co-transfected (C). Line plots represent the signal intensity of the different channels along the indicated chromocenter (solid white line). White dashed lines indicate the nuclear border. Scale bar: 10 µm.
Transient co-transfection of mouse embryonic stem cells (ESCs wt J1) with GBP-dCas9-mRFP and MaSgRNA resulted in a specific enrichment of GBP-dCas9-mRFP at CCs. Importantly, when we additionally transfected a GFP-encoding plasmid, we observed co-localization of GFP with GBP-dCas9-mRFP at CCs (Fig. 1B and C). This experiment confirms the functionality of the GBP-dCas9-mRFP construct in facilitating the recruitment of GFP to target loci.
Next, we aimed to target GFP-tagged epigenetic effector proteins to chromocenters via GBP-dCas9-mRFP. To test the feasibility of such an approach we used the catalytic domains of the methylcytosine dioxygenase TET1 and the de novo methyltransferase DNMT3A coupled to GFP (GFP-TET1CD and GFP-DNMT3ACD, respectively). DNMT3A catalyzes the methylation of cytosine generating 5-methylcytosine (5mC), a repressive epigenetic mark enriched at CCs.36 In contrast, TET1 oxidizes 5mC to 5-hydroxymethylcytosine (5hmC), a DNA modification generally found in euchromatin and depleted at the heterochromatic CCs.37
Similar to GFP alone, GFP-TET1CD was successfully recruited to CCs in cells, which co-expressed GBP-dCas9-mRFP and MaSgRNA. Notably, TET1CD recruitment to the highly methylated CCs in wt ESCs, resulted in an ectopic enrichment of 5hmC at these sites (Fig. 2A). To test the feasibility of GFP-DNMT3ACD recruitment to CCs we used DNMT triple knockout (TKO) cells,38 which are virtually devoid of genomic DNA methylation. In TKO cells, which co-expressed GBP-dCas9-mRFP and MaSgRNA GFP-DNMT3ACD was successfully recruited to CCs, leading to a dramatic increase of 5mC at CCs (Fig. 2B). Taken together, these data show that GBP-dCas9-mRFP is capable of directing the enzymatic activity of epigenetic factors to targeted genomic regions.
Figure 2.
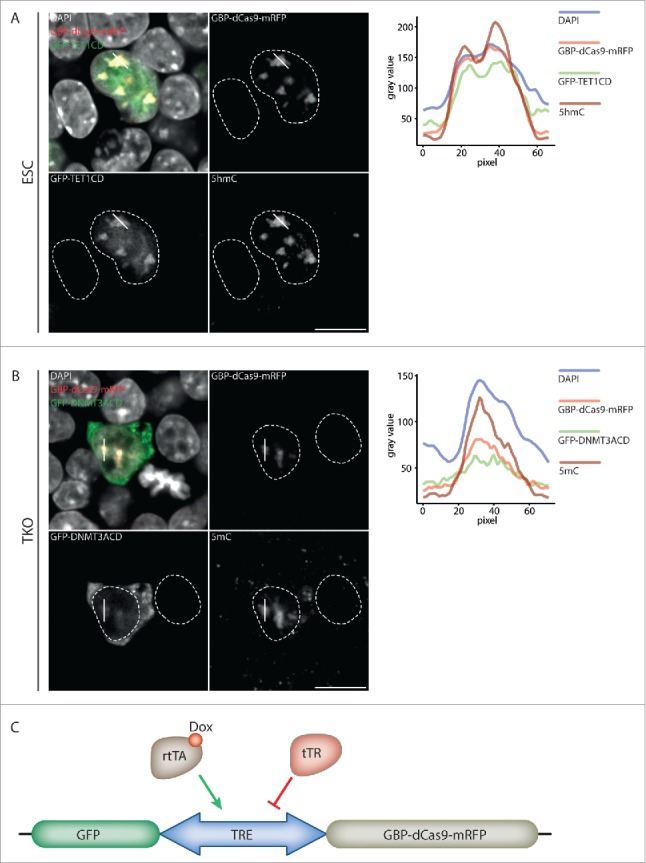
Targeted recruitment of GFP-tagged effector proteins. (A) GBP-dCas9-mRFP recruits GFP-TET1CD to chromocenters. Recruited GFP-TET1CD oxidizes 5mC to 5hmC at CCs in transfected ESCs. In untransfected cells, no 5hmC signal was detected. (B) When targeted to CCs, GFP-DNMT3ACD mediates de novo DNA methylation in TKO cells, which was not observed in untransfected control cells. Line plots represent the signal intensity of the different channels along the indicated chromocenter (solid white line). White dashed lines indicate the nuclear border. Scale bar: 10 µm. (C) Schematic representation of the inducible vector system. A bi-directional promoter drives the expression of GBP-dCas9-mRFP as well as GFP. The vector additionally encodes a transcriptional repressor (tTR) and a transcriptional activator (rtTA). In the absence of doxycycline (Dox), tTR binds to a tetracycline response element (TRE) within the promoter sequence and represses transcription. Upon addition of Dox to the culture medium, rtTA replaces tTR and induces gene transcription.
Besides targeted recruitment, control over the timing of site-specific epigenetic editing is crucial for the dissection of direct functional consequences resulting from local epigenetic perturbations. To this end, we constructed an inducible system for GBP-dCas9-mRFP and GFP/GFP-effector expression. We used an Epstein-Barr virus (EBV) derived episomal expression plasmid (pRTS), which harbors a doxycycline-inducible bi-directional promoter and additionally encodes a tet-transcriptional (tTR) repressor as well as a tet-transcriptional activator (rtTA).39-40 Conditional transcriptional activation is achieved in the presence of doxycycline (Dox) by the rtTA, whereas tTR mediates active repression in Dox-free conditions. This single vector system allows tight control over the timing and expression level of GBP-dCas9-mRFP as well as a GFP-fusion protein (pRTS-GBP-dCas9-mRFP; Fig. 2C). Moreover, using a bidirectional promoter ensures that both proteins are expressed at comparable levels reducing the amount of freely diffusing GFP-fusion protein, thereby minimizing off-target effects. We first tested the inducible system in mouse myoblast (C2C12) using a pRTS-GBP-dCas9-mRFP, additionally harboring either GFP alone or GFP-TET1CD. Both GFP and GFP-TET1CD were expressed and specifically recruited to CCs in a strictly sgRNA- and Dox-dependent manner (Fig. 3A and B; Figure S1A; Figure S2A and B). Similar to co-transfections performed in wt ESCs, GFP-TET1CD recruitment resulted in a drastic increase of hmC at myoblast CCs (Fig. 3B). Comparably, transfection of pRTS-GBP-dCas9-mRFP harboring GFP-DNMT3ACD into TKO cells led to a specific, Dox-dependent enrichment of 5mC at CCs (Fig. 3C; Figure S1B; Figure S2C).
Figure 3.
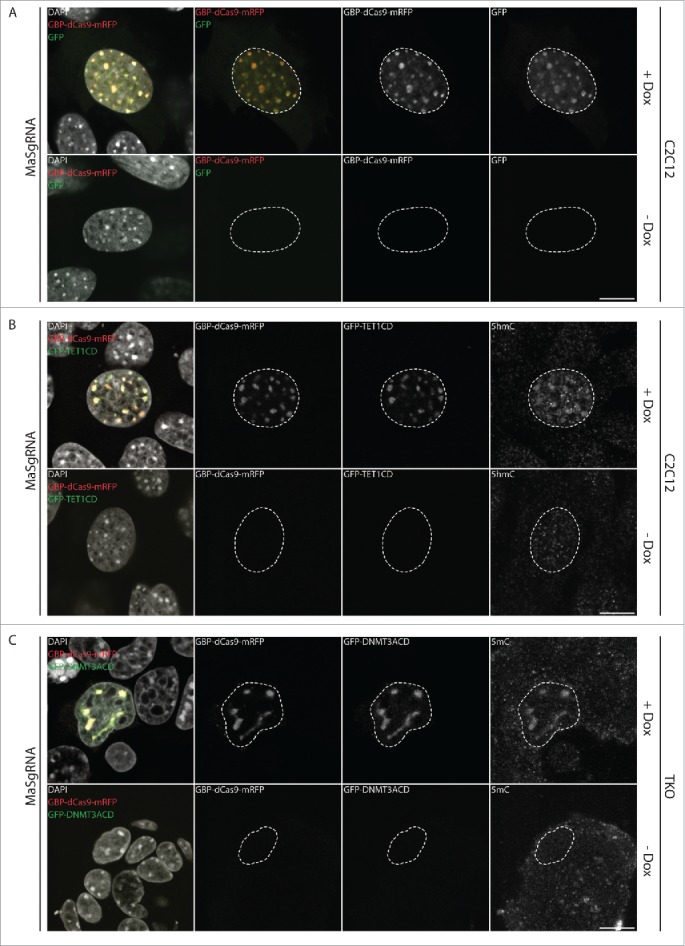
Doxycycline-dependent, coordinated expression of GBP-dCas9-mRFP and GFP-tagged effectors. Cells were co-transfected with MaSgRNA and pRTS-GBP-dCas9-mRFP, additionally encoding GFP (A), GFP-TET1CD (B) or GFP-DNMT3ACD (C). Upon induction with doxycycline both GBP-dCas9-mRFP and the corresponding GFP-tagged effector are expressed, resulting in the oxidation of 5mC (5 hmC; B) or de novo methylation of CCs (5 mC; C). White dashed lines indicate the nuclear border. Scale bar: 10 µm.
In summary, we demonstrated that our modular system can be used to edit prevalent epigenetic marks such as DNA methylation at heterochromatic chromocenters in a timely controlled manner. While we used repetitive target sequences, which allow a fast and simple readout of efficient effector recruitment by microscopy, it is in principle possible to apply our system for the epigenetic modification of single target loci. Using multiple gene/target specific sgRNAs in parallel will also enable the modification of multiple loci at once or the epigenetic editing of a single copy locus.22 The bidirectional, inducible system offers the added advantage to titrate of the amount of epigenetic modifier and Cas9 protein simultaneously, greatly reducing the possibility of off-target effects. Furthermore, our GBP-based approach for dCas9-assisted targeted recruitment can be combined with virtually any GFP-tagged protein. Considering the widespread use of GFP-fusions in cell lines and animal models, this system will help to facilitate the systematic dissection of biologic processes in basic and biomedical research.45
Material and methods
Cell culture and transient transfections
J1 ESCs and TKO cells were cultivated at 37°C and 5% CO2 on gelatin-coated dishes in Dulbecco´s modified Eagle´s medium (DMEM, Sigma), supplemented with 16% fetal bovine serum (FBS, Biochrom), 0.1 mM β-mercaptoethanol (Invitrogen), 2 mM L-glutamine, 1x MEM non-essential amino acids, 100 U/ml penicillin, 100 µg/ml streptomycin (PAA Laboratories GmbH), 1000 U/ml recombinant mouse LIF (Millipore), 1 µM PD032501 and 3 µM CHIR99021 (Axon Medchem). C2C12 cells were cultured at 37°C and 5% CO2 in DMEM, supplemented with 20% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. For conditional transcription activation, the culture medium was additionally supplemented with 1.5 µg/ml doxycycline for 24 hours. Transient transfections were performed using Lipofectamine® 3000 (Thermo Fisher Scientific) according to the manufacturer´s instructions and cells were analyzed 24 – 48 hrs post-transfection.
Plasmid generation
For generating the GBP-dCas9-mRFP expression plasmid, the GBP and mRFP coding sequences were amplified from pGFPbinderImR41 and cloned into pCAG-dCas923 via XbaI and AsiSI/NotI, respectively. The expression constructs for GFP-TET1CD and MaSgRNA were described previously.23,42 GFP-DNMT3ACD was constructed by amplifying the catalytic domain of DNMT3A from pCAG-GMT3a.43 Subsequently TET1CD was exchanged by DNMT3ACD using AsiSI and NotI restriction enzymes. The doxycycline-inducible expression system is based on the pRTS plasmid described previously.39 The GBP-dCas9-mRFP coding sequence was cloned into pRTS via SfiI, whereas TET1CD and DNMT3ACD, respectively, were inserted upstream of GFP via SmiI.
Constructs generated for this study are available via Addgene.
Immunofluorescence staining and microscopy
Immunofluorescence staining was performed as described previously.44 Briefly, cells were grown on coverslips (thickness 1.5H, 170 µm ± 5 µm; Marienfeld Superior) and transfected with the respective expression plasmids. 24 – 48 hours after transfection, cells were washed with phosphate buffered saline (PBS), fixed with 3.7% formaldehyde for 10 min and permeabilized with 0.5% Triton X-100 in PBS. For 5 mC and 5 hmC detection, DNA was first denatured with 1 N HCl for 15 min and then neutralized with 150 mM TRIS-HCl (pH 8.5). Subsequently, cells were transferred into blocking buffer (0.02% Tween and 2% bovine serum albumin in PBS) for 1 hour. Both primary and secondary antibodies were diluted in blocking buffer and cells were incubated in a dark, humidified chamber for 1 hour at room temperature. Nuclei were counterstained with 0.2 µg/ml DAPI in PBS or SiR-DNA (Spirochrome). Coverslips were mounted with antifade medium (Vectashield, Vector Laboratories) and sealed with colorless nail polish. Primary antibodies used in this study were: GFP- and RFP-booster conjugated to Atto 488 and Atto 593, respectively (1:200; Chromotek), anti-5hmC (1:250; Active Motif) and anti-5 mC (1:500, Diagenode). The secondary antibodies were: anti-rabbit IgG conjugated to Alexa 647, anti-mouse IgG conjugated to Alexa 647 (1:400; Thermo Fisher Scientific) and anti-mouse IgG conjugated to Alexa 405 (1:400; Invitrogen).
Confocal images were acquired with a Leica TCS SP5 microscope equipped with a Plan Apo 63x/1.4 NA oil immersion objective. Image processing and assembly of figures was performed with FIJI and Photoshop CS5.1 (Adobe), respectively.
Supplementary Material
Abbreviations
- CC
Chromocenter
- DNMT3A
DNA methyltransferase 3A
- GBP
GFP-binding protein.
- TET1
ten-eleven translocation methylcytosine dioxygenase 1
- 5hmC
5-hydroxymethylcytosine
- 5mC
5-methylcytosine
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
First and foremost, we would like to thank Heinrich Leonhardt (LMU Munich) for helpful discussions and advice on the manuscript. We further would like to thank Dirk Eick (Helmholtz Zentrum München) for providing the pRTS vector, Masaki Okano (RIKEN) for providing TKO cells and Kerry Tucker (Ruprecht-Karls-University, Heidelberg) for providing wt J1 ESCs. We also thank Sebastian Kaluscha, Tsotne Chitiashvili and Orsi Wade for help with cloning the pRTS constructs. T.A. thankfully acknowledges the Graduiertenkolleg GRK1721.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) under Grant SPP1623 and by the GRK1721.
References
- [1].Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016; 17:487-500; PMID:27346641; https://doi.org/ 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- [2].Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, Blancafort P. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 2012; 7:350-60; PMID:22419067; https://doi.org/ 10.4161/epi.19507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol 2013; 425:479-91; PMID:23220192; https://doi.org/ 10.1016/j.jmb.2012.11.038 [DOI] [PubMed] [Google Scholar]
- [4].Falahi F, Huisman C, Kazemier HG, van der Vlies P, Kok K, Hospers GA, Rots MG. Towards sustained silencing of HER2/neu in cancer by epigenetic editing. Mol Cancer Res 2013; 11:1029-39; PMID:23814024; https://doi.org/ 10.1158/1541-7786.MCR-12-0567 [DOI] [PubMed] [Google Scholar]
- [5].Chen H, Kazemier HG, de Groote ML, Ruiters MH, Xu GL, Rots MG. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res 2014; 42:1563-74; PMID:24194590; https://doi.org/ 10.1093/nar/gkt1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al.. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol 2013; 31:1137-42; PMID:24108092; https://doi.org/ 10.1038/nbt.2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol 2013; 31:1133-6; PMID:24013198; https://doi.org/ 10.1038/nbt.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature 2013; 500:472-6; PMID:23877069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].DeFrancesco L. Move over ZFNs. Nat Biotech 2011; 29:681-4; https://doi.org/ 10.1038/nbt.1935 [DOI] [PubMed] [Google Scholar]
- [10].Segal DJ, Dreier B, Beerli RR, Barbas CF 3rd.. Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci U S A 1999; 96:2758-63; PMID:10077584; https://doi.org/ 10.1073/pnas.96.6.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 2011; 39:e82; PMID:21493687; https://doi.org/ 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res 2011; 39:5790-9; PMID:21421566; https://doi.org/ 10.1093/nar/gkr151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337:816-21; PMID:22745249; https://doi.org/ 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al.. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819-23; PMID:23287718; https://doi.org/ 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339:823-6; PMID:23287722; https://doi.org/ 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31:227-9; PMID:23360964; https://doi.org/ 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 2013; 31:233-9; PMID:23360965; https://doi.org/ 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 2013; 23:720-3; PMID:23545779; https://doi.org/ 10.1038/cr.2013.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 2013; 10:741-3; PMID:23817069; https://doi.org/ 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31:230-2; PMID:23360966; https://doi.org/ 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- [21].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013; 152:1173-83; PMID:23452860; https://doi.org/ 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al.. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013; 155:1479-91; PMID:24360272; https://doi.org/ 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Anton T, Bultmann S, Leonhardt H, Markaki Y. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus 2014; 5:163-72; PMID:24637835; https://doi.org/ 10.4161/nucl.28488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A 2015; 112:3002-7; PMID:25713381; https://doi.org/ 10.1073/pnas.1420024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schmidtmann E, Anton T, Rombaut P, Herzog F, Leonhardt H. Determination of local chromatin composition by CasID. Nucleus 2016; 7:476-84; PMID:27676121; https://doi.org/ 10.1080/19491034.2016.1239000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun 2013; 439:132-6; PMID:23942116; https://doi.org/ 10.1016/j.bbrc.2013.08.013 [DOI] [PubMed] [Google Scholar]
- [27].McDonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R, Kramer A, Martens A, Edwards JR, Challen GA. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open 2016; 5:866-74; PMID:27170255; https://doi.org/ 10.1242/bio.019067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, Klasić M, Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 2016; 44:5615-28; PMID:26969735; https://doi.org/ 10.1093/nar/gkw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell 2016; 167:233-47 e17; PMID:27662091; https://doi.org/ 10.1016/j.cell.2016.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu GL, Hu R. A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2016; 2:16009; PMID:27462456; https://doi.org/ 10.1038/celldisc.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 2016; 7:46545-56; PMID:27356740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015; 33:510-7; PMID:25849900; https://doi.org/ 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 2015; 12:401-3; PMID:25775043; https://doi.org/ 10.1038/nmeth.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 2008; 7:282-9; PMID:17951627; https://doi.org/ 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- [35].Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 2004; 166:493-505; PMID:15302854; https://doi.org/ 10.1083/jcb.200403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gilbert N, Thomson I, Boyle S, Allan J, Ramsahoye B, Bickmore WA. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J Cell Biol 2007; 177:401-11; PMID:17485486; https://doi.org/ 10.1083/jcb.200607133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al.. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 2011; 42:451-64; PMID:21514197; https://doi.org/ 10.1016/j.molcel.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al.. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 2006; 11:805-14; PMID:16824199; https://doi.org/ 10.1111/j.1365-2443.2006.00984.x [DOI] [PubMed] [Google Scholar]
- [39].Bornkamm GW, Berens C, Kuklik-Roos C, Bechet JM, Laux G, Bachl J, Korndoerfer M, Schlee M, Hölzel M, Malamoussi A, et al.. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res 2005; 33:e137; PMID:16147984; https://doi.org/ 10.1093/nar/gni137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hölzel M, Rohrmoser M, Orban M, Homig C, Harasim T, Malamoussi A, Gruber-Eber A, Heissmeyer V, Bornkamm G, Eick D. Rapid conditional knock-down-knock-in system for mammalian cells. Nucleic Acids Res 2007; 35:e17; PMID:17169998; https://doi.org/ 10.1093/nar/gkl1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zolghadr K, Mortusewicz O, Rothbauer U, Kleinhans R, Goehler H, Wanker EE, Cardoso MC, Leonhardt H. A fluorescent two-hybrid assay for direct visualization of protein interactions in living cells. Mol Cell Proteomics 2008; 7:2279-87; PMID:18622019; https://doi.org/ 10.1074/mcp.M700548-MCP200 [DOI] [PubMed] [Google Scholar]
- [42].Müller U, Bauer C, Siegl M, Rottach A, Leonhardt H. TET-mediated oxidation of methylcytosine causes TDG or NEIL glycosylase dependent gene reactivation. Nucleic Acids Res 2014; 42:8592-604; PMID:24948610; https://doi.org/ 10.1093/nar/gku552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep 2009; 10:1259-64; PMID:19798101; https://doi.org/ 10.1038/embor.2009.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Solovei I, Cremer M. 3D-FISH on cultured cells combined with immunostaining. Methods Mol Biol 2010; 659:117-26; PMID:20809307 [DOI] [PubMed] [Google Scholar]
- [45].Casas-Delucchi CS, Becker A, Bolius JJ, Cardoso MC. Targeted manipulation of heterochromatin rescues MeCP2 Rett mutants and re-establishes higher order chromatin organization. Nucleic acids research 2012; 40:e176-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


