ABSTRACT
Cells undergoing meiosis produce haploid gametes through one round of DNA replication followed by 2 rounds of chromosome segregation. This requires that cohesin complexes, which establish sister chromatid cohesion during S phase, are removed in a stepwise manner. At meiosis I, the separase protease triggers the segregation of homologous chromosomes by cleaving cohesin's Rec8 subunit on chromosome arms. Cohesin persists at centromeres because the PP2A phosphatase, recruited by the shugoshin protein, dephosphorylates Rec8 and thereby protects it from cleavage. While chromatids disjoin upon cleavage of centromeric Rec8 at meiosis II, it was unclear how and when centromeric Rec8 is liberated from its protector PP2A. One proposal is that bipolar spindle forces separate PP2A from Rec8 as cells enter metaphase II. We show here that sister centromere biorientation is not sufficient to “deprotect” Rec8 at meiosis II in yeast. Instead, our data suggest that the ubiquitin-ligase APC/CCdc20 removes PP2A from centromeres by targeting for degradation the shugoshin Sgo1 and the kinase Mps1. This implies that Rec8 remains protected until entry into anaphase II when it is phosphorylated concurrently with the activation of separase. Here, we provide further support for this model and speculate on its relevance to mammalian oocytes.
KEYWORDS: APC/C-Cdc20, centromere, cohesin cleavage, Meiosis II, Mps1, PP2A, protection of centromeric cohesion, Rec8, separase, shugoshin
Introduction
Accurate chromosome segregation depends on cohesin complexes, which entrap sister chromatids within a proteinaceous ring from the time of DNA replication until anaphase when cohesin's kleisin subunit is cleaved by the separase protease.1-3 Due to cohesion, the kinetochores of chromosomes attached to microtubules from opposite spindle poles experience tension.4 This tension quenches an inhibitory signaling cascade, called the spindle assembly checkpoint (SAC), whereby unattached kinetochores prevent entry into anaphase.5 The target of the SAC is the ubiquitin-ligase APC/CCdc20, which induces anaphase by marking for degradation B-type cyclins and the separase inhibitor Pds1/securin.
Proliferating cells strictly alternate between establishing sister chromatid cohesion during S phase and removing cohesin along the entire length of chromosomes before nuclear division. While yeast uses separase to cleave all of cohesin at anaphase,6 vertebrates use a non-proteolytic mechanism to remove most of cohesin from chromosome arms during prophase, which is followed by separase-dependent cleavage of centromeric cohesin at entry into anaphase.7 In contrast to mitosis, sister chromatid cohesion established during S phase of meiosis mediates 2 rounds of chromosome segregation: disjunction of homologous chromosomes in meiosis I and segregation of chromatids in meiosis II.8 After DNA replication, homologous chromosomes pair and undergo reciprocal recombination between maternal and paternal chromatids. This creates bivalent chromosomes in which cohesin and crossovers hold together all 4 homologous chromatids. Attachment of maternal and paternal centromeres to microtubules from opposite spindle poles can now create the tension required for silencing the SAC.9 Meanwhile, biorientation of sister centromeres, which generates tension in mitosis and meiosis II, is suppressed by meiosis I-specific proteins, known as monopolins.10 Resolving bivalent chromosomes into their constituent chromatids requires 2 rounds of cohesin cleavage. The first wave of separase activity at the onset of anaphase I cleaves cohesin on chromosome arms, which gives rise to dyad chromosomes whose segregation to opposite poles of the meiosis I spindle halves the number of chromosomes.11-13 Dyads consist of 2 chromatids joined at their centromeres by cohesin, which enables sister centromeres to biorient on the meiosis II spindle. At the onset of anaphase II, centromeric cohesin is cleaved by a second wave of separase activity, leading to the disjunction of chromatids and the formation of the single-copy genome.14-16
The stepwise removal of meiotic cohesin depends on the exchange of cohesin's Scc1/Rad21 kleisin for a meiosis-specific paralogue, called Rec8.17-20 To be cleaved by separase, Rec8 has to be phosphorylated. In yeast, 2 kinases phosphorylate Rec8 to promote the cleavage of cohesin on chromosome arms at anaphase I: Hrr25, a conserved casein kinase 1δ, and the Dbf4-dependent Cdc7 kinase.21-23 Accordingly, inhibition of both kinases or mutations of Rec8 phospho-sites blocks Rec8 cleavage and homolog disjunction.22 Protection of centromeric Rec8 from cleavage by separase at anaphase I depends on orthologues of the Drosophila Mei-S332 protein, known as shugoshins (Sgo).24-28 These proteins accumulate around centromeres at least in part because they bind to histone H2A phosphorylated by the kinetochore-associated Bub1 kinase.29 Shugoshins recruit the phosphatase PP2A containing the regulatory B’ subunit (Rts1 in budding yeast), which dephosphorylates Rec8 and thereby protects it from cleavage by separase.30-32 In addition, shugoshins are involved in the stabilization of tension-generating microtubule-kinetochore attachments and, in vertebrate mitosis, protection of centromeric cohesin from removal by the prophase-pathway.33,34 Shugoshin functions are provided by a single protein in budding yeast and Drosophila, but by 2 paralogues in other organisms. While the protection of centromeric cohesin in meiosis I has been investigated extensively,35 less is known about the mechanism that leads to its “deprotection,” so that chromatids can segregate in meiosis II.
Models for the deprotection of centromeric cohesin in mammalian oocytes
At least in yeast, manipulating meiosis II without perturbing meiosis I is challenging because meiosis II occurs shortly (∼40 min) after meiosis I and the synchrony of meiotic cultures is poor. Furthermore, meiosis II shares key regulators with the first division. Thus, the first models for the deprotection of centromeric cohesin emerged from observations in mouse oocytes, which arrest at metaphase II until fertilization triggers entry into anaphase II. In these oocytes, the shugoshin SGOL2 co-localizes with, and protects centromeric Rec8 at anaphase I.36,37 At metaphase II, however, SGOL2 shifts toward the kinetochores and away from Rec8, which resides at the inner centromere. This observation led to the proposal of a deprotection-by-tension model in which bipolar spindle forces spatially separate SGOL2-PP2A from cohesin and thereby expose Rec8 to phosphorylation and subsequent cleavage by separase.37 However, this model remains to be tested in oocytes at meiosis II. The deprotection-by-tension model is not applicable to yeast, in which Sgo1-PP2A protects centromeric Rec8 even in mutants that biorient sister centromeres at meiosis I.38,39 Furthermore, fission yeast Sgo1 is capable of protecting cohesin at bioriented sister centromeres in mitotic cells provided that they express Rec8.26 A more recent oocyte study proposed that the conserved histone chaperone SET/TAF-1β inhibits PP2A activity specifically at centromeres in meiosis II.40,41 However, it remains unclear whether knockdown or overexpression of SET perturbs chromatid disjunction by an effect on PP2A's catalytic activity or the properties of centromeric nucleosomes.42 While both models are based on the assumption that PP2A protects centromeric cohesin by dephosphorylating Rec8, the relevant mammalian Rec8-kinases remain to be identified.
A model for APC/CCdc20-dependent deprotection of centromeric cohesin in yeast
To analyze chromatid segregation at meiosis II in budding yeast, we developed a synchronisation system, called CDC20-meiotic-arrest/release or CDC20-mAR, which is capable of resolving meiosis I and –II.43 With the help of a mitosis-specific promoter, cells induced to enter meiosis are depleted of the APC/C activator Cdc20, which leads to a uniform arrest at metaphase I. Expression of a second copy of CDC20 under the control of a copper-inducible promoter results in synchronous progression through both meiotic divisions and the formation of viable spores. This system enables us to study meiosis II following inactivation or expression of proteins of interest at mid-anaphase I.
To identify the kinase that phosphorylates centromeric Rec8 during meiosis II, we started with the finding that of the 2 kinases promoting cleavage of arm cohesin at meiosis I only one, namely Hrr25, is present in meiosis II.44 Thus, we introduced into CDC20-mAR cells the hrr25-as mutation, which renders Hrr25s activity sensitive to the cell-permeable inhibitor 1NM-PP1.38 Inhibition of Hrr25 activity at anaphase I showed that it is, indeed, required for the cleavage of centromeric Rec8, but not the activity of separase at anaphase II.43 Hrr25 phosphorylates Rec8 and simultaneously enables APC/CCdc20 to remove PP2ARts1 from centromeres through the degradation of Sgo1 and the Mps1 kinase at the onset of anaphase II. Whereas Mps1 is known as an upstream regulator of the SAC,5,45 we found that it is also required for Sgo1s binding to centromeres in meiosis II. Thus, APC/CCdc20 uses a dual and therefore robust mechanism to remove PP2ARts1 from centromeres.
The notion that Hrr25 rather than another kinase directly phosphorylates Rec8 is based on the finding that Hrr25 activity is essential for the cleavage of Rec8 even after Sgo1-PP2A has been experimentally removed from centromeres. Hrr25's second function, the removal of PP2ARts1 from centromeres, is similarly essential: the meiosis II division is blocked when PP2ARts1 is recruited directly (independently of Sgo1) to centromeric Rec8 with the help of an inducible protein interaction system. More importantly still, expression of non-degradable versions of Sgo1 and Mps1 in meiosis II retains PP2ARts1 at centromeres beyond anaphase II and blocks chromatid segregation in >50% of cells.43 This result seems incompatible with inhibition of PP2A's catalytic activity at centromeres in meiosis II, as suggested in mouse oocytes.40 Furthermore, this finding implies that deprotection results not from tension per se, but from the Hrr25- and APC/CCdc20-dependent removal of PP2ARts1 from centromeres. Since APC/CCdc20 also causes the degradation of Pds1, phosphorylation of Rec8 occurs concomitantly with the activation of separase. Consequently, the protection of centromeric Rec8 extends until the onset of anaphase II. Thus, our model for the deprotection of centromeric cohesin in budding yeast differs in fundamental aspects from the PP2A-inhibition and the deprotection-by-tension model. Herein, we provide further support for our model and speculate on the consequences of applying it to oocytes.
SET orthologues are dispensable for meiotic chromosome segregation in yeast
Our model suggests that in budding yeast, centromeric Rec8 is protected for as long as PP2ARts1 resides at centromeres. In other words, these cells seem to lack mechanisms that inhibit PP2A activity at centromeres in meiosis II. We asked, therefore, whether the 2 SET orthologues of budding yeast, called Nap1 and Vps75,46,47 play a role in chromatid segregation at meiosis II. To this end, we deleted both genes and followed progression through meiosis by live-cell imaging (Fig 1). Spindle formation, nuclear divisions, and the segregation of chromosome V sister sequences occur with similar kinetics in wild-type and nap1Δ vps75Δ double mutant cells. Furthermore, the double mutant produces tetrad spores with similar viability (96.5%) as the wild-type (97.2%, n = 144 spores per strain). We conclude that in budding yeast, histone chaperones related to SET are dispensable for accurate chromosome segregation in meiosis and play no detectable role in the deprotection of centromeric Rec8.
Figure 1.
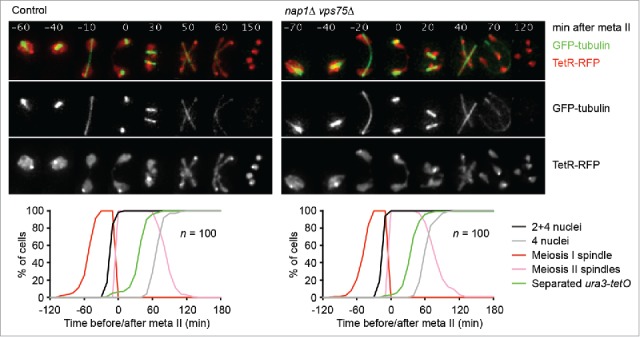
SET-related histone chaperones are dispensable for meiotic chromosome segregation in budding yeast. Spindles (GFP-tubulin), nuclei, and chromosome V sister sequences labeled with tet repressor (TetR) fused to the RFP tdTomato were imaged every 10 min in control (z21937) and nap1Δ vps75Δ (z21931) cells induced to enter meiosis. TetR-RFP bound to tet operators at the ura3 locus (ura3-tetO) on one copy of chromosome V produces one dot in meiosis I and 2 dots in meiosis II; free TetR-RFP illuminates the nucleoplasm.44 Top, representative time-lapse series. The width of one frame is 5 μm. Bottom, scoring of meiosis I and –II spindles, first (2 or 4 nuclei) and second (4 nuclei) division, and separated ura3-tetO sister sequences. Meiotic events were synchronised in silico by setting the time of meiosis II spindle formation (onset of metaphase II) to t = 0 in each cell.53
Sister centromere biorientation in meiosis II is not sufficient for the deprotection of centromeric cohesin
We found that sister centromere disjunction is blocked upon co-expression in meiosis II of Sgo1-mD and Mps1-mD, which resist APC/C-dependent proteolysis due to mutations in their destruction boxes.43 We ascribed this to prolonged protection of centromeric Rec8 by the persistence of PP2ARts1 at centromeres. However, an alternative explanation for the meiosis II division block might arise from the finding that Sgo1 and Mps1 are involved in the establishment of tension-generating microtubule-kinetochore attachments in both mitosis and meiosis.25,48-50 Therefore, we used live-cell imaging to investigate whether cells expressing Sgo1-mD and Mps1-mD put sister kinetochores under tension at meiosis II. CDC20-mAR strains containing RFP-tagged histone H2B and one copy of chromosome V marked with GFP at the centromere were released from the metaphase I arrest. In one of the strains, Sgo1-mD and Mps1-mD were expressed from an estradiol-inducible promoter (called PEST) at metaphase II (Fig 2). At anaphase I, cells show a single GFP dot as expected from the co-segregation of sister centromeres to the same spindle pole. In control cells, the GFP dot then splits into 2 closely spaced dots as sister centromeres biorient and come under tension at metaphase II. Consistent with the duration of metaphase II, split GFP dots can be observed in an undivided nucleus for a median time of 40 min, after which they abruptly segregate into 2 of the 4 nuclei at the onset of anaphase II. In control cells, the incidence of chromatid miss-segregation is very low: in less than 2% of tetranucleate cells, the GFP signals were found in the same nucleus. Co-expression of Sgo1-mD and Mps1-mD completely blocked the meiosis II division in 53% of cells. In these binucleate cells, the GFP-dots are split for a median time of 60 min before they coalesce into a single dot again. This is consistent with the notion that dot splitting in these cells is limited by the ∼60 min lifetime of meiosis II spindles.43 Cells that manage to undergo a meiosis II division in the presence of Sgo1-mD and Mps1-mD frequently produce nuclei of uneven size and show a high frequency of chromatid miss-segregation: in 20% of these tetranucleate cells, the GFP signals are found in only one of the 4 nuclei. We conclude that cells expressing Sgo1-mD and Mps1-mD at meiosis II fail to disjoin sister centromeres although sister kinetochores experience tension. Thus, in budding yeast, tension due to bipolar spindle forces is not sufficient to subject centromeric Rec8 to cleavage by separase, which differs from the deprotection-by-tension model proposed for oocytes.37
Figure 2.
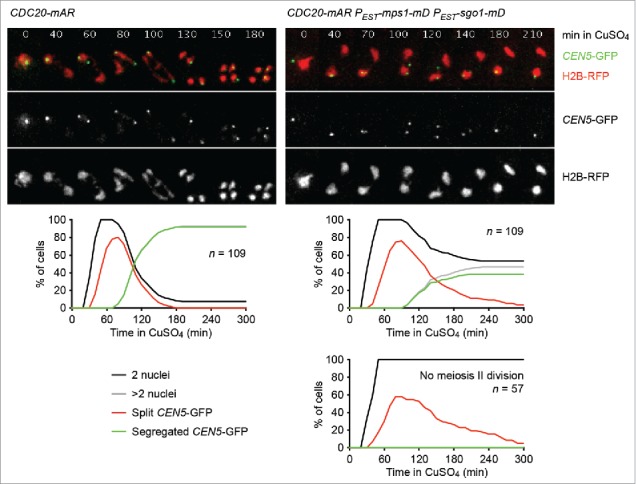
Sister centromeres come under tension but fail to disjoin upon co-expression of non-degradable versions of Sgo1 and Mps1 in meiosis II. Control (z26823) and PEST-sgo1-mD PEST-mps1-mD (z26825) cells containing the CDC20-mAR synchronisation system were released from the metaphase I arrest with CuSO4 (10 μM, t = 0). Estradiol (10 μM) was added at t = 15 min to induce PEST-sgo1-mD and PEST-mps1-mD.43 Nuclei containing histone H2B tagged with the RFP mCherry and the sister centromeres of one chromosome V copy labeled with the CEN5-tetO/TetR-GFP system (CEN5-GFP)19 were imaged every 10 min. Top, representative time-lapse series. The width of one frame is 5 μm. Middle, percentages of cells with one division (2 nuclei), 2 divisions (more than 2 nuclei), split GFP signals (2 GFP-dots in the same nucleus), and segregated GFP signals (GFP signals in separate nuclei). Bottom right, PEST-sgo1-mD PEST-mps1-mD cells that fail to undergo the meiosis II division are plotted separately.
Persistence of Sgo1 and Mps1 in meiosis II does not interfere with SAC silencing
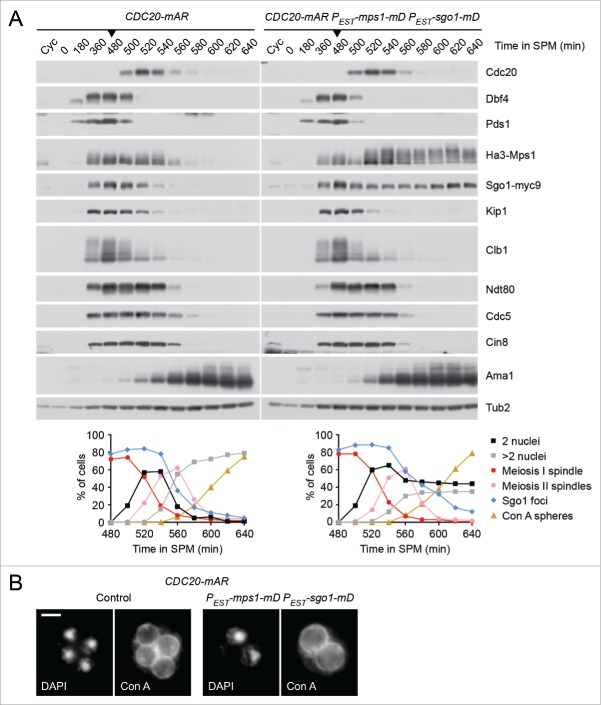
Sgo1 and Mps1 are also involved in the regulation of the SAC. Sgo1 is part of the tension-sensing arm,48 and Mps1 is a key upstream regulator whose overexpression activates the SAC even in the absence of spindle damage.51 Furthermore, APC/CCdc20-dependent degradation of these proteins has been proposed to facilitate silencing of the SAC at entry into anaphase.52 We asked, therefore, whether the presence of non-degradable Sgo1 and Mps1 at meiosis II interferes with silencing of the SAC, which would hinder chromatid segregation by delaying the activation of APC/CCdc20. To this end, we used the CDC20-mAR system to synchronize control cells and cells expressing Sgo1-mD and Mps1-mD at metaphase II. Immunoblot detection of proteins revealed that degradation of the APC/CCdc20 substrates Pds1, Kip1, and Clb1 is not affected by the persistence of Mps1-mD and Sgo1-mD (Fig 3A). Since the Cdk1-Clb1 kinase inhibits the meiosis-specific APC/C-activator Ama1, Clb1 degradation via APC/CCdc20 also induces proteolysis mediated by APC/CAma1.53 Accordingly, the APC/CAma1 substrates Cdc5 and Cin8 are degraded on time and meiosis II spindles disassemble normally in cells expressing Sgo1-mD and Mps1-mD. Furthermore, these cells initiate spore formation without delay (Fig 3A and B), but produce asci containing 2 diploid spores with high frequency (52%).43 We conclude that the persistence of Sgo1-mD and Mps1-mD at meiosis II hinders sister centromere disjunction but not SAC silencing, at least when the proteins are expressed from the estradiol-inducible promoter. These data strengthen our model that deprotection of centromeric Rec8 requires PP2A's removal from centromeres by Hrr25- and APC/CCdc20-dependent degradation of Sgo1 and Mps1.
Figure 3.
Co-expression of non-degradable Sgo1 and Mps1 in meiosis II does not interfere with silencing of the SAC. (A and B) Control (z26045) and PEST-sgo1-mD PEST-mps1-mD (z26015) cells containing the CDC20-mAR system were transferred to sporulation medium (SPM, t = 0) and released from the metaphase I arrest with CuSO4 at t = 480 min (arrow heads). Estradiol was added at t = 495 min. Note that endogenous and non-degradable Sgo1 carry Myc9 tags while both versions of Mps1 are tagged with Ha3.43 (A) Top, immunoblot detection of proteins in whole-cell extracts.44 Cyc, samples from proliferating cells. Bottom, percentages of cells with one (2 nuclei) or 2 divisions (more than 2 nuclei), meiosis I or meiosis II spindles, and spheres stained by FITC-concanavalin A (Con A spheres) were determined by immunofluorescence microscopy of fixed cells.14 Sgo1 foci were detected on chromatin spreads.44 100 cells were counted per time point. (B) Cells stained with FITC-concanavalin A and DAPI at t = 640 min. Spheres around the nuclei represent mannoproteins in the prospore membrane and the first (inner) spore wall layer.61 Scale bar = 2 μm.
Pds1 levels are lower at metaphase II than at metaphase I
Our results suggest that centromeric Rec8 remains protected until entry into anaphase II. What might be the advantage of protecting centromeric cohesin during metaphase II when separase is thought to be inhibited by the re-accumulation of Pds1? First, the amount of cohesin left to withstand bipolar spindle forces is relatively small. Cohesin persists at a few distinct sites within a 50 kb region around the “point” centromeres of budding yeast.54 Second, whereas metaphase I is preceded by a long prophase (∼3.5 hr) during which Pds1 can accumulate and “saturate” separase, there is much less time for the re-accumulation of Pds1 in meiosis II (∼40 min). Indeed, quantification of the nuclear Pds1-myc18 signal by immunofluorescence microscopy of cells from a conventional meiotic culture revealed that the levels of Pds1 at metaphase II are 3-fold lower compared with metaphase I (Fig 4A). By contrast, the levels at metaphase II of the M phase-cyclin Clb1-myc9 are comparable to those at metaphase I (Fig 4B), probably because Clb1 does not start to accumulate until the prophase-to-metaphase I transition. On the other hand, the cellular levels of the separase Esp1-myc18 remain constant during nuclear divisions. Indeed, separase/Esp1 is a very stable protein. Inhibition of protein synthesis with cycloheximide revealed a half-life of more than 2 hours irrespective of whether Esp1 is inactive (metaphase I, Fig. 5A) or active (anaphase I, Fig. 5B). It is possible, therefore, that inhibition of separase is less robust at metaphase II compared with metaphase I. Protection of centromeric Rec8 at metaphase II might help to safeguard the integrity of dyad chromosomes in organisms, such as budding yeast, in which centromeres are small and the time between meiosis I and –II is short.
Figure 4.
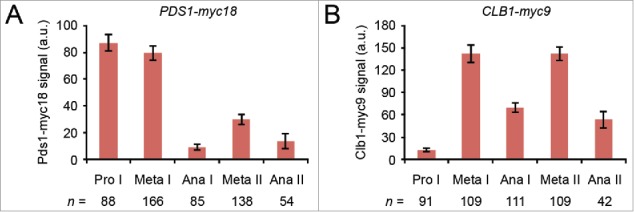
Levels of nuclear Pds1 and Clb1 at different stages of meiosis. (A and B) Formaldehyde-fixed cells from conventional meiotic cultures of untagged (z29971), PDS1-myc18 (z19647), and CLB1-myc9 (z29974) strains were stained with DAPI and antibodies to tubulin and Myc.14 The nuclear background signal generated by the α-Myc antibody was measured in 50–100 untagged cells at the indicated stage of meiosis, averaged, and the mean subtracted from the nuclear Myc signals of individual PDS1-myc18 (A) and CLB1-myc9 (B) cells at the corresponding meiotic stage. Columns indicate mean signal intensity in arbitrary units. Error bars show 95% confidence interval.
Figure 5.
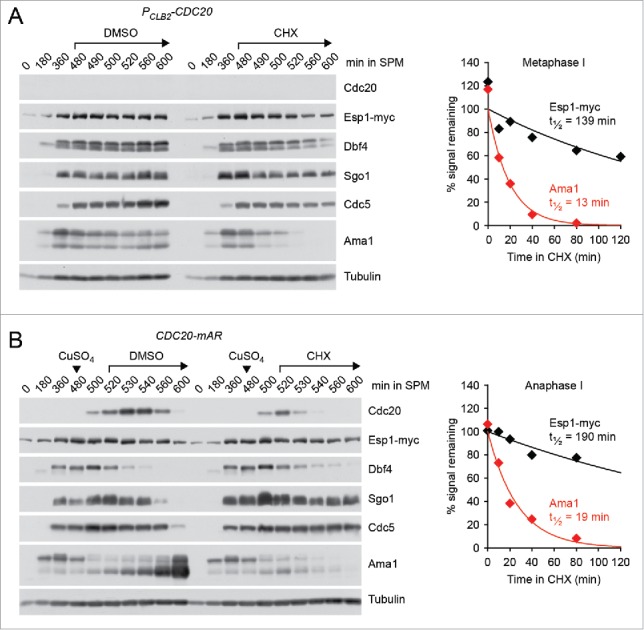
Levels and stability of separase/Esp1 in meiosis. (A and B) ESP1-myc18 strains were transferred to sporulation medium (SPM, t = 0) and treated with solvent (DMSO, 0.5%) or cycloheximide (CHX, 0.5 mg/ml) at the indicated times. Proteins were detected in whole-cell extracts by immunoblotting.44 Graphs show half-lives of Esp1-myc18 and Ama1 measured by scanning ECL signals on X-ray films. Data points are mean values from 2 gels. (A) Cells arresting at metaphase I due to CDC20 expression from the mitosis-specific CLB2 promoter (z30976) were treated with DMSO or CHX at t = 480 min. (B) CDC20-mAR cells (z29418) were released from the metaphase I arrest with CuSO4 at t = 480 min (arrowheads) and treated with DMSO or CHX at anaphase I (t = 520 min). Note that Dbf4 is degraded at anaphase I, Sgo1 at anaphase II, and Cdc5 at exit from meiosis II, whereas Ama1 is a constitutively unstable protein.
Is APC/CCdc20-dependent deprotection of centromeric Rec8 relevant to mammalian oocytes?
Mammalian oocytes arrest at metaphase II until fertilization initiates entry into anaphase II. This puts a higher demand on the function of centromeric cohesion than in spermatocytes, which usually progress through metaphase II without delay. While the density of cohesin appears higher around centromeres than on chromosome arms,13,55 studies in aged mouse oocytes revealed that sister centromere cohesion is particularly sensitive to reduced cohesin levels. As oocytes age, cohesin is gradually lost from chromosomes during the prolonged arrest in prophase of meiosis I.55,56 Although cohesin is lost globally, the incidence of single chromatids at metaphase II is much higher than that of unpaired dyads at meiosis I. The majority of observed aneuploidies is therefore best explained by premature loss of centromeric rather than arm cohesion. Interestingly, cohesin loss is accompanied by reduced levels of SGOL2,55,57 which might expose centromeric Rec8 to cleavage by separase at anaphase I or during metaphase II. Even in young oocytes, centromeric cohesin might be threatened by leaky inhibition of separase: Pds1/securin levels are much lower at metaphase II than at metaphase I.58 This potentially precarious situation could be ameliorated by additional mechanisms to inhibit separase during the metaphase II arrest. However, notwithstanding the finding that Cdk1-cyclin B can inhibit separase in vertebrates,59 separase inhibition at meiosis II in mouse oocytes seems to rely solely on Pds1/securin.58 An alternative solution to this problem might be to extend protection of centromeric Rec8 to metaphase II, as proposed in our yeast model. In this scenario, Rec8 would be deprotected not when sister centromeres come under tension and the SAC is silenced, but when APC/CCdc20 is activated at fertilisation. Whether SGOL2 is subject to APC/CCdc20-dependent degradation at anaphase II is not yet known. However, according to our model, even a stable SGOL2 would be removed from centromeres by APC/CCdc20-dependent degradation of Mps1, which is an APC/CCdc20 substrate also in mammalian cells.60
What protects the protector of centromeric cohesin at meiosis I?
While our model provides a mechanism for the deprotection of centromeric Rec8 in yeast, it cannot yet explain how this process is restricted to meiosis II. Two of its key components, APC/CCdc20 and Hrr25, are present not only in meiosis II but also in meiosis I. While degradation of Pds1 at meiosis I and –II requires Cdc20 but not Hrr25, the meiosis II-specific proteolysis of Sgo1 and Mps1 requires both Cdc20 and Hrr25. We speculate therefore that Hrr25 promotes ubiquitination of Sgo1 and Mps1 by APC/CCdc20 and that this activity is inhibited at meiosis I. Furthermore, we favor the idea that Hrr25 regulates the substrates Sgo1 and Mps1 rather than APC/CCdc20. Prime candidates for such regulators of Hrr25 activity are meiosis I-specific factors, such as monopolin or Spo13,10 which could coordinate the protection of centromeric Rec8 with the mono-orientation of sister kinetochores. Solving this problem would provide the first working model for the control of chromosome segregation during both meiotic divisions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Adam D. Rudner for antibodies to Sgo1 and Anthony A. Hyman for antibodies to Cin8 and Kip1.
Funding
This work was supported by the Max Planck Society and, in part, by the Deutsche Forschungsgemeinschaft (SPP1384 grant ZA519/1–1 to W.Z.).
References
- [1].Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol 2011; 13:1170-7; PMID:21968990; https://doi.org/ 10.1038/ncb2349 [DOI] [PubMed] [Google Scholar]
- [2].Haarhuis JHI, Elbatsh AMO, Rowland BD. Cohesin and Its Regulation: On the Logic of X-Shaped Chromosomes. Developmental Cell 2014; 31:7-18; PMID:25313959; https://doi.org/ 10.1016/j.devcel.2014.09.010 [DOI] [PubMed] [Google Scholar]
- [3].Gligoris T, Lowe J. Structural Insights into Ring Formation of Cohesin and Related Smc Complexes. Trends Cell Biol 2016; 26:680-93; PMID:27134029; https://doi.org/ 10.1016/j.tcb.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 2000; 101:763-75; PMID:10892747; https://doi.org/ 10.1016/S0092-8674(00)80888-0 [DOI] [PubMed] [Google Scholar]
- [5].Musacchio A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr Biol 2015; 25:R1002-18. [DOI] [PubMed] [Google Scholar]
- [6].Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 2000; 103:375-86; PMID:11081625; https://doi.org/ 10.1016/S0092-8674(00)00130-6 [DOI] [PubMed] [Google Scholar]
- [7].Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 2000; 103:399-410; PMID:11081627; https://doi.org/ 10.1016/S0092-8674(00)00132-X [DOI] [PubMed] [Google Scholar]
- [8].Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 2003; 112:423-40; PMID:12600308; https://doi.org/ 10.1016/S0092-8674(03)00083-7 [DOI] [PubMed] [Google Scholar]
- [9].Hauf S, Watanabe Y. Kinetochore orientation in mitosis and meiosis. Cell 2004; 119:317-27; PMID:15507205; https://doi.org/ 10.1016/j.cell.2004.10.014 [DOI] [PubMed] [Google Scholar]
- [10].Nasmyth K. A meiotic mystery: How sister kinetochores avoid being pulled in opposite directions during the first division. Bioessays 2015; 37:657-65; PMID:25874377; https://doi.org/ 10.1002/bies.201500006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 2000; 103:387-98; PMID:11081626; https://doi.org/ 10.1016/S0092-8674(00)00131-8 [DOI] [PubMed] [Google Scholar]
- [12].Terret ME, Wassmann K, Waizenegger I, Maro B, Peters JM, Verlhac MH. The meiosis I-to-meiosis II transition in mouse oocytes requires separase activity. Curr Biol 2003; 13:1797-802; PMID:14561405; https://doi.org/ 10.1016/j.cub.2003.09.032 [DOI] [PubMed] [Google Scholar]
- [13].Kudo NR, Wassmann K, Anger M, Schuh M, Wirth KG, Xu H, Helmhart W, Kudo H, McKay M, Maro B, et al.. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell 2006; 126:135-46; PMID:16839882; https://doi.org/ 10.1016/j.cell.2006.05.033 [DOI] [PubMed] [Google Scholar]
- [14].Salah SM, Nasmyth K. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma 2000; 109:27-34; PMID:10855492; https://doi.org/ 10.1007/s004120050409 [DOI] [PubMed] [Google Scholar]
- [15].Kitajima TS, Miyazaki Y, Yamamoto M, Watanabe Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. Embo J 2003; 22:5643-53; PMID:14532136; https://doi.org/ 10.1093/emboj/cdg527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lee J, Okada K, Ogushi S, Miyano T, Miyake M, Yamashita M. Loss of Rec8 from chromosome arm and centromere region is required for homologous chromosome separation and sister chromatid separation, respectively, in mammalian meiosis. Cell Cycle 2006; 5:1448-55; PMID:16855401; https://doi.org/ 10.4161/cc.5.13.2903 [DOI] [PubMed] [Google Scholar]
- [17].Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 1999; 98:91-103; PMID:10412984; https://doi.org/ 10.1016/S0092-8674(00)80609-1 [DOI] [PubMed] [Google Scholar]
- [18].Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 1999; 400:461-4; PMID:10440376; https://doi.org/ 10.1038/22774 [DOI] [PubMed] [Google Scholar]
- [19].Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 2000; 103:1155-68; PMID:11163190; https://doi.org/ 10.1016/S0092-8674(00)00217-8 [DOI] [PubMed] [Google Scholar]
- [20].Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 2001; 15:1349-60; PMID:11390355; https://doi.org/ 10.1101/gad.192701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol 2010; 12:500-6; PMID:20383139; https://doi.org/ 10.1038/ncb2052 [DOI] [PubMed] [Google Scholar]
- [22].Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell 2010; 18:397-409; PMID:20230747; https://doi.org/ 10.1016/j.devcel.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Le AH, Mastro TL, Forsburg SL. The C-terminus of S. pombe DDK subunit Dfp1 is required for meiosis-specific transcription and cohesin cleavage. Biol Open 2013; 2:728-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 1995; 83:247-56; PMID:7585942; https://doi.org/ 10.1016/0092-8674(95)90166-3 [DOI] [PubMed] [Google Scholar]
- [25].Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol 2004; 14:560-72; PMID:15062096; https://doi.org/ 10.1016/j.cub.2004.03.001 [DOI] [PubMed] [Google Scholar]
- [26].Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 2004; 427:510-7; PMID:14730319; https://doi.org/ 10.1038/nature02312 [DOI] [PubMed] [Google Scholar]
- [27].Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science 2004; 303:1367-70; PMID:14752166; https://doi.org/ 10.1126/science.1094220 [DOI] [PubMed] [Google Scholar]
- [28].Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol 2004; 14:287-301; PMID:14972679; https://doi.org/ 10.1016/j.cub.2004.01.051 [DOI] [PubMed] [Google Scholar]
- [29].Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 2010; 327:172-7; PMID:19965387; https://doi.org/ 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- [30].Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 2006; 441:46-52; PMID:16541025; https://doi.org/ 10.1038/nature04663 [DOI] [PubMed] [Google Scholar]
- [31].Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al.. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 2006; 441:53-61; PMID:16541024; https://doi.org/ 10.1038/nature04664 [DOI] [PubMed] [Google Scholar]
- [32].Xu Z, Cetin B, Anger M, Cho US, Helmhart W, Nasmyth K, Xu W. Structure and function of the PP2A-shugoshin interaction. Mol Cell 2009; 35:426-41; PMID:19716788; https://doi.org/ 10.1016/j.molcel.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gutierrez-Caballero C, Cebollero LR, Pendas AM. Shugoshins: from protectors of cohesion to versatile adaptors at the centromere. Trends Genet 2012; 28:351-60; PMID:22542109; https://doi.org/ 10.1016/j.tig.2012.03.003 [DOI] [PubMed] [Google Scholar]
- [34].Marston AL. Shugoshins: Tension-Sensitive Pericentromeric Adaptors Safeguarding Chromosome Segregation. Mol Cell Biol 2015; 35:634-48; PMID:25452306; https://doi.org/ 10.1128/MCB.01176-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Clift D, Marston AL. The role of shugoshin in meiotic chromosome segregation. Cytogenet Genome Res 2011; 133:234-42; PMID:21273764; https://doi.org/ 10.1159/000323793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Llano E, Herran Y, Garcia-Tunon I, Gutierrez-Caballero C, de Alava E, Barbero JL, Schimenti J, de Rooij DG, Sanchez-Martin M, Pendas AM. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J Cell Biol 2008; 197:877-85; https://doi.org/ 10.1083/jcb.201201100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol 2008; 10:42-52; PMID:18084284; https://doi.org/ 10.1038/ncb1667 [DOI] [PubMed] [Google Scholar]
- [38].Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell 2006; 126:1049-64; PMID:16990132; https://doi.org/ 10.1016/j.cell.2006.07.029 [DOI] [PubMed] [Google Scholar]
- [39].Hirose Y, Suzuki R, Ohba T, Hinohara Y, Matsuhara H, Yoshida M, Itabashi Y, Murakami H, Yamamoto A. Chiasmata promote monopolar attachment of sister chromatids and their co-segregation toward the proper pole during meiosis I. PLoS Genet 2011; 7:e1001329; PMID:21423721; https://doi.org/ 10.1371/journal.pgen.1001329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chambon JP, Touati SA, Berneau S, Cladiere D, Hebras C, Groeme R, McDougall A, Wassmann K. The PP2A inhibitor I2PP2A is essential for sister chromatid segregation in oocyte meiosis II. Curr Biol 2013; 23:485-90; PMID:23434280; https://doi.org/ 10.1016/j.cub.2013.02.004 [DOI] [PubMed] [Google Scholar]
- [41].Wassmann K. Sister chromatid segregation in meiosis II: deprotection through phosphorylation. Cell Cycle 2013; 12:1352-9; PMID:23574717; https://doi.org/ 10.4161/cc.24600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Higgins JM, Herbert M. Nucleosome assembly proteins get SET to defeat the guardian of chromosome cohesion. PLoS Genet 2013; 9:e1003829; PMID:24086159; https://doi.org/ 10.1371/journal.pgen.1003829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Arguello-Miranda O, Zagoriy I, Mengoli V, Rojas J, Jonak K, Oz T, Graf P, Zachariae W. Casein Kinase 1 Coordinates Cohesin Cleavage, Gametogenesis, and Exit from M Phase in Meiosis II. Dev Cell 2017; 40:37-52; PMID:28017619; https://doi.org/ 10.1016/j.devcel.2016.11.021 [DOI] [PubMed] [Google Scholar]
- [44].Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 2008; 135:662-78; PMID:19013276; https://doi.org/ 10.1016/j.cell.2008.10.026 [DOI] [PubMed] [Google Scholar]
- [45].Liu X, Winey M. The MPS1 family of protein kinases. Annu Rev Biochem 2012; 81:561-85; PMID:22482908; https://doi.org/ 10.1146/annurev-biochem-061611-090435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci U S A 2006; 103:1248-53; PMID:16432217; https://doi.org/ 10.1073/pnas.0508002103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tang Y, Meeth K, Jiang E, Luo C, Marmorstein R. Structure of Vps75 and implications for histone chaperone function. Proc Natl Acad Sci U S A 2008; 105:12206-11; PMID:18723682; https://doi.org/ 10.1073/pnas.0802393105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 2005; 307:130-3; PMID:15637284; https://doi.org/ 10.1126/science.1101366 [DOI] [PubMed] [Google Scholar]
- [49].Maure JF, Kitamura E, Tanaka TU. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr Biol 2007; 17:2175-82; PMID:18060784; https://doi.org/ 10.1016/j.cub.2007.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science 2013; 339:1071-4; PMID:23371552; https://doi.org/ 10.1126/science.1232518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 1996; 273:953-6; PMID:8688079; https://doi.org/ 10.1126/science.273.5277.953 [DOI] [PubMed] [Google Scholar]
- [52].Palframan WJ, Meehl JB, Jaspersen SL, Winey M, Murray AW. Anaphase inactivation of the spindle checkpoint. Science 2006; 313:680-4; PMID:16825537; https://doi.org/ 10.1126/science.1127205 [DOI] [PubMed] [Google Scholar]
- [53].Okaz E, Arguello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ, Markova Z, Zagoriy I, Novak B, Zachariae W. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell 2012; 151:603-18; PMID:PMID:23101628; https://doi.org/ 10.1016/j.cell.2012.08.044 [DOI] [PubMed] [Google Scholar]
- [54].Kiburz BM, Reynolds DB, Megee PC, Marston AL, Lee BH, Lee TI, Levine SS, Young RA, Amon A. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev 2005; 19:3017-30; PMID:16357219; https://doi.org/ 10.1101/gad.1373005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, et al.. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 2010; 20:1511-21; PMID:20817533; https://doi.org/ 10.1016/j.cub.2010.08.023 [DOI] [PubMed] [Google Scholar]
- [56].Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol 2010; 20:1522-8; PMID:20817534; https://doi.org/ 10.1016/j.cub.2010.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yun Y, Lane SI, Jones KT. Premature dyad separation in meiosis II is the major segregation error with maternal age in mouse oocytes. Development 2014; 141:199-208; PMID:24346700; https://doi.org/ 10.1242/dev.100206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nabti I, Reis A, Levasseur M, Stemmann O, Jones KT. Securin and not CDK1/cyclin B1 regulates sister chromatid disjunction during meiosis II in mouse eggs. Dev Biol 2008; 321:379-86; PMID:18639540; https://doi.org/ 10.1016/j.ydbio.2008.06.036 [DOI] [PubMed] [Google Scholar]
- [59].Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell 2001; 107:715-26; PMID:11747808; https://doi.org/ 10.1016/S0092-8674(01)00603-1 [DOI] [PubMed] [Google Scholar]
- [60].Cui Y, Cheng X, Zhang C, Zhang Y, Li S, Wang C, Guadagno TM. Degradation of the human mitotic checkpoint kinase Mps1 is cell cycle-regulated by APC-cCdc20 and APC-cCdh1 ubiquitin ligases. J Biol Chem 2010; 285:32988-98; PMID:20729194; https://doi.org/ 10.1074/jbc.M110.140905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tachikawa H, Bloecher A, Tatchell K, Neiman AM. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J Cell Biol 2001; 155:797-808; PMID:11724821; https://doi.org/ 10.1083/jcb.200107008 [DOI] [PMC free article] [PubMed] [Google Scholar]