ABSTRACT
In the cell nucleus, DNA repair machineries operate on a chromatin substrate, whose integrity is key for preserving cell functions and identity. Yet, it is still unclear how the epigenetic information conveyed by chromatin is maintained during the DNA repair process. We recently characterized the dynamics of parental histones coupled to UV-C damage repair in human cells, providing insights into how the pre-damage chromatin state may be restored. Here, we summarize our main findings and discuss them in the context of epigenome maintenance following DNA damage. We further address the mechanistic aspects of repair-coupled histone dynamics and develop working hypotheses regarding their functional relevance in the cellular response to genotoxic stress.
KEYWORDS: chromatin rearrangements, DNA damage binding protein 2, DNA damage repair, epigenome integrity, histone dynamics
Introduction
In eukaryotes, the genetic material is packaged into the cell nucleus in the form chromatin, the basic unit of which is the nucleosome where DNA wraps around histone proteins.1 Beyond genome packaging, chromatin also conveys epigenetic information through the existence of histone variants2 and their post-translational modifications,3 the association of non-histone chromatin components and higher-order folding of the chromatin fiber into nuclear domains.4 Collectively, these chromatin features govern gene expression and therefore cell identity.5 Preserving chromatin integrity is thus of fundamental importance. However, the integrity of chromatin organization is challenged during DNA metabolic processes, including DNA damage, which can arise from multiple sources at any time. The cellular protection against DNA insults involves a broad range of repair factors and pathways, altogether building an orchestrated response to safeguard genome integrity.6-8 Conjointly with these genomic changes, chromatin rearrangements arise during the repair response, as described in the Access-Repair-Restore (ARR) model.9,10 According to this model, chromatin is transiently disorganized to facilitate access of repair factors to DNA lesions, followed by restoration of chromatin structure. Although critical for preserving chromatin functions, how this restoration process operates is not entirely clear. Studies in human cells showed that newly synthesized histone variants H2A, H3.1 and H3.3 were deposited in damaged chromatin.11-14 This raised the possibility that new histones were replacing parental histones, which carry the original epigenetic information. How the pre-damage chromatin state could be preserved in this context was elusive.15 We addressed this question in our recent study, by combining real-time tracking of parental histones with local induction of DNA damage by a UV-C laser.16 This provided important novel insights into parental histone mobilization following DNA damage, an understanding of the underlying mechanisms and of the contribution of parental histones to chromatin repair.
Revisiting the Access-Repair-Restore model: Conservative redistribution of parental histones to the periphery of UV-C damaged sites
Accessing DNA lesions within chromatin is thought to involve histone eviction from nucleosomes. Lending support to this hypothesis, several reports describe nucleosome disruption in response to DNA Double-Strand-Breaks (DSBs) in human cells,17,18 and even degradation of acetylated histones in response to alkylating damage or ionizing radiations in yeast and mouse cells.19 Such nucleosome disassembly may entail a loss of epigenetic information from chromatin through the loss of histone variants and their associated marks. To address how the original information can be preserved in this context, we recently developed two complementary imaging approaches, SNAP-tag- or photoactivation-based, for real-time tracking of parental histones following UV-C damage in human cells. Thus, we observed a local loss of density of parental histones H3 and H4 at UV-C-damaged sites, detectable within minutes after damage infliction. Remarkably, quantification of the signal associated with the H3.3 histone variant in damaged cell nuclei showed that parental histones were neither massively degraded nor evicted from chromatin following local UV-C irradiation. Instead, they redistributed to the periphery of the damaged chromatin region in a conservative manner, as revealed by analyzing their distribution in concentric regions around the point of UV-C laser impact over time. This parental histone redistribution was progressive and accompanied by an expansion of the damaged zone, which is indicative of an outward motion of chromatin.
Based on measurements of histone and DNA densities, we surmised that parental histone redistribution around UV-C damaged sites was driven by two mechanisms: chromatin decompaction, leading to an expansion of the damaged area, and histone mobilization on chromatin, potentially by nucleosome sliding. This scenario corroborates previous studies showing local chromatin decondensation upon UV-C damage detection by the repair machinery in human cells,20 and nucleosome unwrapping on a UV-damaged template in vitro.21 Contrary to chromatin decompaction, which is the major contributor to the observed reduction in histone density at UV-damaged sites, nucleosome sliding is unlikely to occur over very large distances. We rather favor the idea that all damaged nucleosomes can slide over short distances within the irradiated chromatin region, and only nucleosome sliding at the edge of that region would reduce histone density in the damaged area.
From these findings, it is tempting to speculate that parental histone redistribution could involve a decompaction of damaged chromatin accompanied by the displacement of non-damaged chromatin fibers toward the periphery of the damaged site, therefore creating a local environment suitable for DNA repair. It is important to consider also that these local chromatin alterations could have profound effects on nuclear organization, by iterative rearrangements or by disrupting long-range interactions between chromatin domains. This could be addressed by chromosome conformation capture approaches.22 It remains to be determined whether parental histones are mobilized in a nucleosomal form, as there is still no evidence for parental H2A-H2B redistribution jointly with H3-H4. However, a reduction in H2A density and an enhanced turnover of H2A-H2B have been observed at sites of UV-C damage in human cells.20,11 Modeling parental histone dynamics in response to DNA damage would be useful to get mechanistic insights into these chromatin rearrangements and to assess their long-range effects in the nucleus.
Altogether, our recent data support the view that chromatin is pushed away from the damage site to allow access to the repair machinery. In addition, we bring another piece to the puzzle by demonstrating that parental histones are kept in the vicinity of the damaged region, and thus potentially available to take part in the restoration of chromatin.
Recovery of parental histones in repairing chromatin
As parental H3.3 histones were retained proximal to the damage region, we examined their long-term fate and their contribution to repaired chromatin by live cell imaging during repair progression. Thus, we unveiled that chromatin restoration was a two-step process with new histone deposition occurring first, followed by parental H3.3 histone recovery. The recovery of parental histone signal within chromatin regions undergoing repair was almost complete, underlining the major contribution of parental histones to repaired chromatin. Furthermore, the recovery proceeded radially inwards, suggesting that the parental histones that recovered were the ones that originally redistributed away, although this still needs to be formally demonstrated.
Given the differences in the early chromatin response to DNA damage among the various types of DNA lesions, the mechanisms that we have uncovered following UV damage cannot be directly transposed to other damage responses. Nevertheless, two recent studies showed that nucleosome occupancy is restored after DSB repair both in yeast and human cells,18,23 most likely by nucleosome re-assembly. It will be important to determine whether and to which extent parental histones are recycled during the nucleosome re-assembly process at repaired DSBs.
Strikingly, in chromatin undergoing UV damage repair, newly deposited histones persist after the recovery of parental histones. This is a puzzling observation considering that parental histones almost completely recover. Assessing the relative proportions of new and parental histones in repaired chromatin will be critical to determine if new histones replace the small fraction of parental histones that do not recover. Nevertheless, the persistence of new histones suggests that they are not merely a by-product of the DNA damage response and raises questions about their function during or after repair. Considering the timing of new histone deposition, they could be part of a damage signaling process, and/or protect damaged chromatin from a second genotoxic insult. If new histones are maintained in chromatin after repair completion, they could even serve as a memory of damage infliction, thereby potentiating a second response to genotoxic stress. Noteworthy, newly synthesized histones H3 undergo a delay in mitotic phosphorylations associated with chromosome condensation, as shown in Drosophila and human cells.24,25 We could then envision that the incorporation of new histones during DNA damage repair could interfere with chromosome condensation in early mitosis and thus serve as a “chromatin checkpoint” mechanism ensuring that incompletely repaired material cannot be transmitted to the next cell generation.
Whatever the function of new histones in repaired chromatin, their maintenance along with the recovery of parental histones may end up in a higher histone density with a different nucleosomal/chromatin structure at repair sites. In agreement with this hypothesis, a reduction in chromatin accessibility was reported following UV irradiation in mouse cells,26 and chromatin compaction was observed in response to DSBs in human cells.27 Further investigations using super-resolution microscopy for instance could help decipher the relative distribution of parental and new histones and the overall chromatin structure at repair sites.
Coordination of parental histone dynamics with repair progression
To gain a better understanding of the mechanisms driving chromatin rearrangements during UV-C damage repair, we investigated the molecular interplay between histone dynamics and the DNA repair machinery. Using siRNA-mediated downregulation of repair factors, we determined that the UV damage sensor DNA Damage Binding protein 2 (DDB2)28 was the main regulator of parental histone dynamics in response to UV-C damage. DDB2 knockdown greatly impaired parental histone redistribution early after damage induction, and this effect was mostly independent of the ubiquitylation activity of the DDB2 complex, which is fully consistent with previous data.20 Furthermore, the recovery of parental histones was dependent on repair progression and correlated with DDB2 release from chromatin. The role of DDB2 in these two processes was further assessed by taking advantage of the LacO-LacR system, in which one can artificially tether/release a factor to/from chromatin in the absence of damage. This assay was already used in an elegant study,20 in which the authors established that DDB2 tethering to chromatin triggered chromatin unfolding at the LacO array. We further demonstrated that DDB2 was actually sufficient to govern parental histone dynamics, both in terms of displacement and recovery.
Mechanistically, we showed that DDB2 was promoting parental histone redistribution mostly by affecting chromatin compaction, in line with previous results.20 How DDB2 fulfills this activity is not yet clear and additional players that could account for histone mobilization on chromatin are still to be characterized. We may envision that the direct binding of DDB2 to DNA lesions could push away surrounding chromatin fibers by steric hindrance, and this would be sufficient to grant access to later repair factors. DDB2 could also promote chromatin decompaction by acting directly or indirectly on cohesins that were recently proposed to control chromatin compaction in interphase by promoting loop extrusion.29 DDB2 is able to bind DNA lesions within a nucleosomal structure30 and is unlikely to possess any nucleosome remodeling activity on its own. It is thus tempting to speculate that another factor, either a chromatin-remodeling factor or a histone chaperone, could assist DDB2 to provide such an activity. Interesting candidates are chromatin remodelers that are known to be involved in the DNA damage response.31 Identifying DDB2-associated factors after UV-C damage by proteomic approaches would also be helpful to uncover potential candidates for mediating DDB2 effect on parental histone dynamics.
Based on our findings, it appears that DDB2, by mediating both UV-C-damage sensing and parental histone dynamics, may be key for ensuring the coordination between genome and epigenome maintenance during UV-C-damage repair. It will be interesting to investigate whether similar repair-coupled mechanisms ensure the maintenance of higher-order chromatin domains. One could also wonder if other DNA damage sensors fulfill a function comparable to that of DDB2 in distinct repair contexts.
Contribution of parental histone dynamics to epigenome maintenance?
The functional relevance of parental histone dynamics in response to DNA damage is still a matter of debate. Indeed, it is not yet possible to uncouple histone dynamics from genome maintenance, as the only factors identified so far that drive parental histone dynamics are also involved in DNA repair. Thus, we can only speculate that parental histone redistribution may facilitate DNA repair and that their recovery may help preserve epigenome integrity. However, much remains to be investigated in this respect.
For example, we still ignore if parental histones retrieve their original positions on the DNA sequence when they recover. Solving this issue would require higher resolution approaches, combining the induction of sequence-specific damage and nucleosome profiling. Moreover, it is still unclear if parental histones come back with their original post-translational modifications (PTMs), and if these marks are eventually transferred onto newly incorporated histones, which could be part of a chromatin maturation process after DNA damage repair. Combining differential labeling of old and new histones with histone PTM detection, as it has been done at the replication fork,32,33 would be a powerful technique to investigate this issue. Nevertheless, one could wonder which level of precision is required in terms of PTM restoration. Indeed, a recent perspective on epigenetic inheritance suggests that, at least for repressive marks, a certain flexibility in histone PTM inheritance is tolerated without significant effects on the transcriptional outcome.34 It is also conceivable that new histone deposition coupled to repair of DNA damage may provide a window of opportunity for epigenome plasticity.
Conclusion
It becomes more and more evident that chromatin dynamics are an integral part of the DNA damage response.15 In this context, our recent study gives new insights into the coordinated maintenance of genome and epigenome integrity and leads to a revised version of the ARR model (Fig. 1), but also opens up several unresolved issues regarding the contribution of histone dynamics to epigenome maintenance. It will be of major interest to investigate the relevance of repair-coupled histone dynamics in preserving cellular homeostasis and in protecting cells against pathological conditions.
Figure 1.
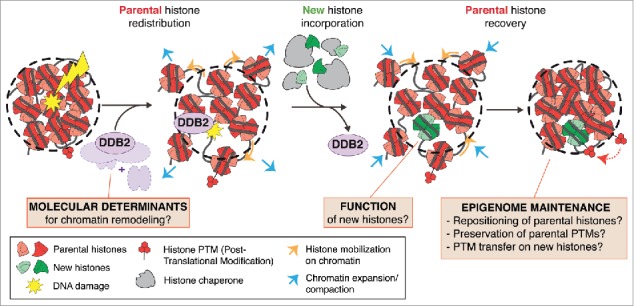
Model for histone dynamics in response to UV-C damage. Following local UV-C irradiation, parental histones (red) redistribute to the periphery of the damaged region by two mechanisms: chromatin expansion (light blue) and histone mobilization on chromatin (light orange). Newly synthesized histones (green) are incorporated into chromatin undergoing UV-C damage repair, followed by the recovery of parental histones. The redistribution and recovery of parental histones are coordinated with repair progression through binding and release of the UV damage sensor DDB2 (purple). This model raises questions regarding the molecular determinants of these chromatin rearrangements, their function and their impact on epigenome maintenance following DNA damage (orange boxes). Adapted from ref. 16.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Salomé Adam for critical reading of the manuscript.
Funding
Work in the S.E.P. Laboratory is supported by the European Research Council under Grant ERC-2013-StG-336427 “EpIn;” “Who am I?” laboratory of excellence under Grant ANR-11-LABX-0071. J.D. is the recipient of a PhD fellowship from Paris Diderot University.
References
- [1].Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997; 389:251-60; PMID:9305837; https://doi.org/ 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- [2].Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet 2014; 15:259-71; PMID:24614311; https://doi.org/ 10.1038/nrg3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011; 21:381-95; PMID:21321607; https://doi.org/ 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Cell Biol 2011; 21:175-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 2009; 10:192-206; PMID:19234478; https://doi.org/ 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- [6].Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071-8; PMID:19847258; https://doi.org/ 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hoeijmakers JHJ. DNA Damage, Aging, and Cancer. N Engl J Med 2009; 361:1475-85; PMID:19812404; https://doi.org/ 10.1056/NEJMra0804615 [DOI] [PubMed] [Google Scholar]
- [8].Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40:179-204; PMID:20965415; https://doi.org/ 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Smerdon MJ. DNA repair and the role of chromatin structure. Curr Opin Cell Biol 1991; 3:422-8; PMID:1892653; https://doi.org/ 10.1016/0955-0674(91)90069-B [DOI] [PubMed] [Google Scholar]
- [10].Polo SE, Almouzni G. Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model. DNA Repair (Amst) 2015; 36:114-21; PMID:26429064; https://doi.org/ 10.1016/j.dnarep.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, et al.. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell 2013; 51:469-79; PMID:23973375; https://doi.org/ 10.1016/j.molcel.2013.08.007 [DOI] [PubMed] [Google Scholar]
- [12].Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell 2006; 127:481-93; PMID:17081972; https://doi.org/ 10.1016/j.cell.2006.08.049 [DOI] [PubMed] [Google Scholar]
- [13].Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 2013; 155:94-106; PMID:24074863; https://doi.org/ 10.1016/j.cell.2013.08.029 [DOI] [PubMed] [Google Scholar]
- [14].Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJ, Pines A, Vertegaal AC, Jacobs JJ, Shah GM, et al.. PARP1 Links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell 2016; 61:547-62; PMID:26895424;https://doi.org/ 10.1016/j.molcel.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dabin J, Fortuny A, Polo SE. Epigenome maintenance in response to DNA damage. Mol Cell 2016; 62:712-27; PMID:27259203;https://doi.org/ 10.1016/j.molcel.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Adam S, Dabin J, Chevallier O, Leroy O, Baldeyron C, Corpet A, Lomonte P, Renaud O, Almouzni G, Polo SE. Real-time tracking of parental histones reveals their contribution to chromatin integrity following DNA damage. Mol Cell 2016; 64:65-78; PMID:27642047; https://doi.org/ 10.1016/j.molcel.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc Natl Acad Sci USA 2013; 110:16874-9; PMID:24082117; https://doi.org/ 10.1073/pnas.1306160110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li X, Tyler JK. Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. Elife 2016; 5:e15129; PMID:27269284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y, et al.. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013; 153:1012-24; PMID:23706739; https://doi.org/ 10.1016/j.cell.2013.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, Mullenders LH, Dantuma NP. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J Cell Biol 2012; 197:267-81; PMID:22492724;https://doi.org/ 10.1083/jcb.201106074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Duan MR, Smerdon MJ. UV damage in DNA promotes nucleosome unwrapping. J Biol Chem 2010; 285:26295-303; PMID:20562439; https://doi.org/ 10.1074/jbc.M110.140087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sati S, Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma 2016; 125:1-12; PMID:25921218; https://doi.org/ 10.1007/s00412-015-0517-x [DOI] [PubMed] [Google Scholar]
- [23].Tsabar M, Hicks WM, Tsaponina O, Haber JE. Re-establishment of nucleosome occupancy during double-strand break repair in budding yeast. DNA Repair (Amst) 2016; 47:21-29; PMID:27720308; https://doi.org/ 10.1016/j.dnarep.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lin S, Yuan Z-F, Han Y, Marchione DM, Garcia BA. Preferential phosphorylation on old histones during early mitosis in human cells. J Biol Chem 2016; 291:15342-57; PMID:27226594;https://doi.org/ 10.1074/jbc.M116.726067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xie J, Wooten M, Tran V, Chen BC, Pozmanter C, Simbolon C, Betzig E, Chen X. Histone H3 threonine phosphorylation regulates asymmetric histone inheritance in the drosophila male germline. Cell 2015; 163:920-33; PMID:26522592;https://doi.org/ 10.1016/j.cell.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schick S, Fournier D, Thakurela S, Sahu SK, Garding A, Tiwari VK. Dynamics of chromatin accessibility and epigenetic state in response to UV damage. J Cell Sci 2015; 128:4380-94; PMID:26446258; https://doi.org/ 10.1242/jcs.173633 [DOI] [PubMed] [Google Scholar]
- [27].Strickfaden H, McDonald D, Kruhlak MJ, Haince JF, Th'ng JPH, Rouleau M, Ishibashi T, Corry GN, Ausio J, Underhill DA, et al.. Poly(ADP-ribosyl)ation-dependent transient chromatin decondensation and histone displacement following laser micro-irradiation. J Biol Chem 2015; 291:1789-804; PMID:26559976; https://doi.org/ 10.1074/jbc.M115.694992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sugasawa K. Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Rep (Amst) 2016; 44:110-7; PMID:27264556; https://doi.org/ 10.1016/j.dnarep.2016.05.015 [DOI] [PubMed] [Google Scholar]
- [29].Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep 2016; 15:2038-49; PMID:27210764; https://doi.org/ 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Osakabe A, Tachiwana H, Kagawa W, Horikoshi N, Matsumoto S, Hasegawa M, Matsumoto N, Toga T, Yamamoto J, Hanaoka F, et al.. Structural basis of pyrimidine-pyrimidone (6–4) photoproduct recognition by UV-DDB in the nucleosome. Sci Rep 2015; 5:16330; PMID:26573481; https://doi.org/ 10.1038/srep16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 2012; 5:4; PMID:22289628; https://doi.org/ 10.1186/1756-8935-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Annunziato AT. The Fork in the road: histone partitioning during DNA replication. Genes (Basel) 2015; 6:353-71; PMID:26110314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Alabert C, Barth TK, Reverón-Gómez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 2015; 29:585-90; PMID:25792596; https://doi.org/ 10.1101/gad.256354.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huang C, Xu M, Zhu B. Epigenetic inheritance mediated by histone lysine methylation: maintaining transcriptional states without the precise restoration of marks? Philos Trans R Soc Lond B Biol Sci 2013; 368:20110332; PMID:23166395; https://doi.org/ 10.1098/rstb.2011.0332 [DOI] [PMC free article] [PubMed] [Google Scholar]


