ABSTRACT
Contrast in electron cryo-microscopy (cryo-EM) is limited by the weak phase and radiation sensitive nature of biologic samples embedded in vitrified ice. We have recently shown that a new contrast enhancement technique utilizing the Volta phase plate can be combined with single particle analysis to determine the structure of a small chromatin complex, the nucleosome core particle, at near-atomic resolution. Here, we discuss advantages and limitations of the technique in terms of data collection, particle detection, and visualization of individual DNA molecules and higher-order chromatin structure.
KEYWORDS: chromatin, cryo-EM, DNA, nucleosome, Volta phase plate
The “phase-strip method for observing phase objects in good contrast” was discovered by Fritz Zernike in the 1930s.1 By introducing a 90° phase shift between the direct and diffracted light, Zernike's phase contrast microscope enabled scientists to image live cells and their internal structures with enhanced contrast without the need for cytotoxic stains.2 The idea to use phase plates in transmission electron microscopy was first proposed in 1947 by Hans Boersch.3 This was followed in 1958 by the construction of the first Zernike phase plate (ZPP), a thin carbon film with a central hole,4 and the first wire-type electrostatic phase plate constructed from spider thread in 1970.5,6 In 2001 Danev and Nagayama applied the ZPP to visualize negatively stained ferritin particles.7 The method was subsequently applied to single particle cryo-EM, electron diffraction, and electron tomography.8,9 However, the resolution was limited to 10 Å and collection of high quality images was challenging due to charging, contamination, and alignment problems10 (an overview of EM phase plate development and designs is reviewed in ref. 11).
The development of the Volta phase plate (VPP) in 2014 solved many of the practical challenges of the ZPP.12 Danev et al. discovered that pre-irradiating a thin continuous film of amorphous carbon with the electron beam caused the build-up of a local Volta potential. This potential would introduce a phase shift between the central and diffracted electrons, thereby generating phase contrast.12 Furthermore, beam-induced contamination could be reduced by heating the plate to ∼200°C. The ability to create phase shifts “on the fly” could then be combined with semi-automated data collection routines for single particle analysis. This was applied successfully to obtain a 4.4 Å reconstruction of the 257 kDa peroxiredoxin-3 protein complex13 and a 3.2 Å reconstruction of the 700 kDa 20S proteasome.14
We were excited by the potential of the VPP for imaging challenging protein-DNA complexes, such as small particles (100–200 kDa) that have low contrast in ice, as well as structurally heterogeneous macromolecular assemblies. We first tested the VPP to image an object of known structure, the 200 kDa nucleosome core particle (NCP). We found that one important advantage of using the VPP was that low contrast views (disk views) of the NCP could easily be detected. Fig. 1A shows the angular distribution of NCP views in micrographs recorded with and without the VPP on a 200 kV FEI Tecnai Arctica. A more isotropic reconstruction was obtained from micrographs recorded with the VPP. Particles were picked in EMAN215 and processed in RELION 1.3.16 Next we recorded a high-resolution in-focus VPP data set on a 300 kV FEI Titan Krios, in counting mode on a K2 detector (Fig. 1B). From only 26,060 particles and after applying C2 symmetry, we obtained an electron density map at a resolution of 3.9.17 Consistent with the attained resolution, the electron density map clearly showed amino acid side chains and DNA phosphate groups. Individual base pairs, however, could not be resolved (Fig. 1C). Moreover, we observed slightly lower local resolution at the DNA termini compared with the rest of the NCP. We interpreted this to be an indicator of DNA breathing. Förster resonance energy transfer studies have shown that at a given point in time, 3% of NCPs undergo transient DNA unwrapping at the termini, with a lifetime of 10–120 ms.18,19 Extrapolating this to our 26,060 particles, ∼780 particles would be in an unwrapped state. However, we did not observe 2D classes of NCPs with unwrapped DNA. This might be due to the small number of particles analyzed and/or the low ionic strength buffer (no salt) used. We hypothesize that at least a 10x increase in the number of particles and ∼50 mM NaCl in the buffer would be required to observe classes with DNA breathing at the termini.
Figure 1.
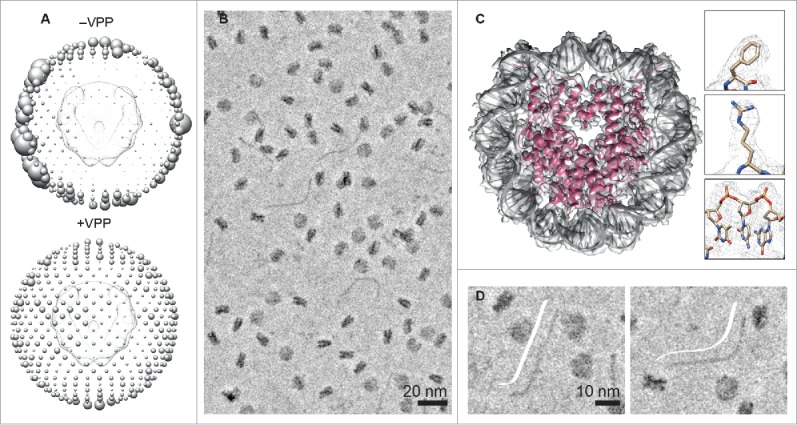
Phase plate imaging and analysis of nucleosome core particles and DNA in ice. (A) Angular distribution of views in micrographs recorded without (-VPP) and with (+VPP) the Volta phase plate. The size of each sphere is proportional to the number of particles per view. (B) Phase plate cryo-EM image of nucleosome core particles (NCPs) and excess DNA in ice. (C) 3.9 Å phase plate structure of the NCP. (D) Representative phase plate images of straight (left) and bent DNA (right). DNA trace (cartoon in white).
To prevent chromatin precipitation, we used excess DNA in our NCP reconstitution. As shown in Fig. 1D, individual DNA molecules can be clearly seen in micrographs recorded with the VPP. Measuring the DNA dimensions, we obtained a maximum DNA length of 480 Å, and a diameter of about 20 Å, which is expected for 145 base pairs of B-form DNA. Shorter DNA lengths were also observed, which we interpret as DNA molecules that are pointing slightly downwards in ice (not parallel to the image plane). We also observed a variety of DNA conformations that we classify as being straight, bent, or wavy. This demonstrates that cryo-EM imaging with the VPP offers a complementary technique to AFM and rotary-shadowing EM to visualize the solution structure of DNA. A clear advantage of cryo-EM is that the sample does not have to be immobilized on a substrate such as mica. We note that DNA can also be visualized in cryo-EM without the VPP, by applying a defocus that enhances the 20 Å frequency range. However, at this particular defocus value and acceleration voltage, it may be difficult to detect a small protein bound to DNA. In summary, we found that imaging of protein-DNA complexes in ice with the VPP enables direct interpretation of structure and composition without the need to apply CTF (contrast transfer function) correction or 2D class averaging.
The VPP has recently been used to reconstruct a 3.2 Å structure of hemoglobin,20 which at 64 kDa is thus far the smallest molecule to be successfully reconstructed to near-atomic resolution. Defocused data collection with the VPP followed by CTF correction has now been implemented, removing the requirement for highly accurate focusing and enabling reconstructions to 2.2 Å.21 We note that the VPP is not required to obtain high-resolution structures by single particle cryo-EM. Defocus cryo-EM has been successfully applied to relatively small particles, such as the 2.8 Å and 3.8 Å reconstructions of the 145 kDa lactate dehydrogenase and 98 kDa isocitrate dehydrogenase enzymes.22 Furthermore, for the proteasome, phase plate cryo-EM only marginally improved resolution compared with defocus cryo-EM.14 However, we believe that phase plate cryo-EM is of great value, as it allows the user to directly interpret raw micrographs and hence more accurately determine the identity of particles. This is particularly helpful for imaging new proteins, small and low contrast molecules, samples that are flexible or heterogeneous in structure and composition, as well as those that require embedding in thick ice.
A current disadvantage of data collection with the VPP is its lack of automation; several steps of the data collection process still require manual adjustments. This includes the change of phase plate position, activation (conditioning) of the phase plate, and correction of astigmatism at each phase plate position used. Furthermore, the performance of the VPP can vary markedly on different microscopes, and even on a given microscope different phase plate positions can have slightly different properties. This may be due to contamination build-up over time and/or the quality of the carbon film. Complete automation of the data collection process will be required to render the technique more user-friendly.
The increase in contrast from using the VPP has also been shown to be useful for electron cryo-tomography of macromolecules and cellular structures in situ.23-25 Imaging of nuclear pore complexes in situ by VPP-cryo-ET25 demonstrated that the technique can be applied in the future to visualize nucleosome arrays and chromatin inside the nucleus. We expect, therefore, that phase plate cryo-EM will continue to drive the resolution revolution in cryo-EM for structural analyses of small, flexible and challenging protein and chromatin complexes, and for visualizing native cellular structures in situ.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Singapore Ministry of Education under grant MOE2012-T3–1–001; NTU Singapore under a start-up grant to S.S.; and the NTU Institute of Structural Biology.
References
- [1].Zernike F. How I discovered phase contrast. Science 1955; 121:345-9; PMID:13237991; https://doi.org/ 10.1126/science.121.3141.345 [DOI] [PubMed] [Google Scholar]
- [2].Zernike F. Phase contrast, a new method for the microscopic observation of transparent objects. Physica 1942; 9:686-98; https://doi.org/ 10.1016/S0031-8914(42)80035-X [DOI] [Google Scholar]
- [3].Boersch H. Über die Kontraste von Atomen im Elektronenmikroskop. Zeitschrift für Naturforsch A 1947; 2:615-33. [Google Scholar]
- [4].Kanaya K, Kawakatsu H, Itō K, Yotsumoto H. Experiment on the Electron Phase Microscope. J Appl Phys 1958; 29:1046-9; https://doi.org/ 10.1063/1.1723360 [DOI] [Google Scholar]
- [5].Unwin PNT. An Electrostatic Phase Plate for the Electron Microscope. Berichte der Bunsengesellschaft für Phys Chemie 1970; 74:1137-41. [Google Scholar]
- [6].Unwin PNT. Phase Contrast and Interference Microscopy with the Electron Microscope. Philos Trans R Soc B Biol Sci 1971; 261:95-104; https://doi.org/ 10.1098/rstb.1971.0039 [DOI] [PubMed] [Google Scholar]
- [7].Danev R, Nagayama K. Transmission electron microscopy with Zernike phase plate. Ultramicroscopy 2001; 88:243-52; PMID:11545320; https://doi.org/ 10.1016/S0304-3991(01)00088-2 [DOI] [PubMed] [Google Scholar]
- [8].Danev R, Nagayama K. Single particle analysis based on Zernike phase contrast transmission electron microscopy. J Struct Biol 2008; 161:211-8; PMID:18082423; https://doi.org/ 10.1016/j.jsb.2007.10.015 [DOI] [PubMed] [Google Scholar]
- [9].Murata K, Liu X, Danev R, Jakana J, Schmid MF, King J, Nagayama K, Chiu W. Zernike Phase Contrast Cryo-Electron Microscopy and Tomography for Structure Determination at Nanometer and Subnanometer Resolutions. Structure 2010; 18:903-12; PMID:20696391; https://doi.org/ 10.1016/j.str.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Danev R, Glaeser RM, Nagayama K. Practical factors affecting the performance of a thin-film phase plate for transmission electron microscopy. Ultramicroscopy 2009; 109:312-25; PMID:19157711; https://doi.org/ 10.1016/j.ultramic.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nagayama K. Another 60 years in electron microscopy: development of phase-plate electron microscopy and biological applications. Microscopy 2011; 60, S43-62; https://doi.org/ 10.1093/jmicro/dfr037 [DOI] [PubMed] [Google Scholar]
- [12].Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci 2014; 111:15635-40; PMID:25331897; https://doi.org/ 10.1073/pnas.1418377111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Khoshouei M, Radjainia M, Phillips AJ, Gerrard JA, Mitra AK, Plitzko JM, Baumeister W, Danev R. Volta phase plate cryo-EM of the small protein complex Prx3. Nat Commun 2016; 7:10534; PMID:26817416; https://doi.org/ 10.1038/ncomms10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Danev R, Baumeister W. Cryo-EM single particle analysis with the Volta phase plate. eLife 2016; 5:e13046; PMID:26949259; https://doi.org/16859925 10.7554/eLife.13046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol 2007; 157:38-46; PMID:16859925; https://doi.org/ 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- [16].Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519-30; PMID:23000701; https://doi.org/ 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chua EYD, Vogirala VK, Inian O, Wong AS, Nordenskiöld L, Plitzko JM, Danev R, Sandin S. 3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM. Nucleic Acids Res 2016; 44:8013-9; PMID:27563056; https://doi.org/ 10.1093/nar/gkw708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol 2005; 12:46-53; PMID:15580276; https://doi.org/ 10.1038/nsmb869 [DOI] [PubMed] [Google Scholar]
- [19].Koopmans WJA, Brehm A, Logie C, Schmidt T, van Noort J. Single-Pair FRET Microscopy Reveals Mononucleosome Dynamics. J Fluoresc 2007; 17:785-95; PMID:17609864; https://doi.org/27238019 10.1007/s10895-007-0218-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Khoshouei M, Radjainia M, Baumeister W, Danev R. Cryo-EM structure of haemoglobin at 3.2 A determined with the Volta phase plate. bioRxiv 087841; https://doi.org/ 10.1101/087841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Danev R, Tegunov D, Baumeister W. Using the Volta phase plate with defocus for cryo-EM single particle analysis. eLife 2017; 6:e23006; https://doi.org/ 10.7554/eLife.23006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JL, et al.. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell 2016; 165:1698-707; PMID:27238019; https://doi.org/ 10.1016/j.cell.2016.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Asano S, Fukuda Y, Beck F, Aufderheide A, Förster F, Danev R, Baumeister W. A molecular census of 26S proteasomes in intact neurons. Science 2015; 347:439-42; https://doi.org/ 10.1126/science.1261197 [DOI] [PubMed] [Google Scholar]
- [24].Fukuda Y, Laugks U, Lučić V, Baumeister W, Danev R. Electron cryotomography of vitrified cells with a Volta phase plate. J Struct Biol 2015; 190:143-54; PMID:25770733; https://doi.org/ 10.1016/j.jsb.2015.03.004 [DOI] [PubMed] [Google Scholar]
- [25].Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, Baumeister W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 2016; 351:969-72; https://doi.org/ 10.1126/science.aad8857 [DOI] [PubMed] [Google Scholar]


