ABSTRACT
Circadian clocks regulate rhythmic gene expression levels by means of mRNA oscillations that are mainly driven by post-transcriptional regulation. We identified a new post-transcriptional mechanism, which involves nuclear bodies called paraspeckles. Major components of paraspeckles including the long noncoding RNA Neat1, which is the structural component, and its major protein partners, as well as the number of paraspeckles, follow a circadian pattern in pituitary cells. Paraspeckles are known to retain within the nucleus RNAs containing inverted repeats of Alu sequences. We showed that a reporter gene in which these RNA duplex elements were inserted in the 3’-UTR region displayed a circadian expression. Moreover, circadian endogenous mRNA associated with paraspeckles lost their circadian pattern when paraspeckles were disrupted. This work not only highlights a new paraspeckle-based post-transcriptional mechanism involved in circadian gene expression but also provides the list of all mRNA associated with paraspeckles in the nucleus of pituitary cells.
KEYWORDS: circadian oscillators, circadian rhythm, nuclear RNA retention, paraspeckle, post-transcriptional mechanism
Most organisms have built-on time-measuring devices that are commonly known as circadian clocks. These circadian clocks allow them to anticipate the time of day and hence to temporally organize behavior as well as physiologic and biochemical processes. They therefore play a key role as adaptive mechanisms to permanent changes in the environment necessary for the individual survival and the sustainability of the species. In living systems ranging from bacteria to humans,1,2 circadian rhythms are generated endogenously through genetic control3 and regulate vital aspects of the organism physiology, from sleeping and waking to neurotransmitter secretion and cellular metabolism. At the center of these rhythms is the circadian clock machinery that consists in a transcription-translation feedback system regulated by a group of genes that oscillate in a circadian manner, the so-called clock genes. In mammals, the circadian system is hierarchically organized. Indeed, while molecular oscillations occur in most cells and tissues of the body, a central structure, the suprachiasmatic nucleus (SCN) of the hypothalamus, functions as the master regulator to synchronize the phase of the other slave oscillating tissues.4,5
Briefly, the molecular mechanism that generates circadian rhythms involves the interacting positive and negative feedback loops of transcriptional or translational processes of clock genes (Fig. 1).1,6 In mammals, 2 basic helix-loop-helix transcription factors, Circadian Locomotor Output Cycles Kaput (CLOCK) and Brain and Muscle Aryl Hydrocarbon Receptor Nuclear Translocator-Like Protein 1 (ARNTL also named BMAL1), heterodimerize and subsequently bind to conserved E-box sequences in target gene promoters. In this manner, this complex activates the transcription of mammalian Period (Per1, Per2, Per3) and Cryptochrome (Cry1, Cry2) genes.6 The PERs and CRYs proteins are expressed, post-translationally modified, feedback to inhibit their own transcription and are then degraded to lead to a new round of BMAL1:CLOCK mediated transcription (For a review see ref.7). Another regulatory loop is mediated by the orphan nuclear receptors, the Retinoic Acid Receptor-Related Orphan Receptor α/β/γ (ROR α/β/γ) and the Reverse Erb α/β (Rev-erb α/β), that activate and inhibit, respectively, transcription of Bmal1 through the retinoic acid Receptor Response Element (RRE) in its promoter, leading it to oscillate in a circadian manner (Fig. 1).
Figure 1.
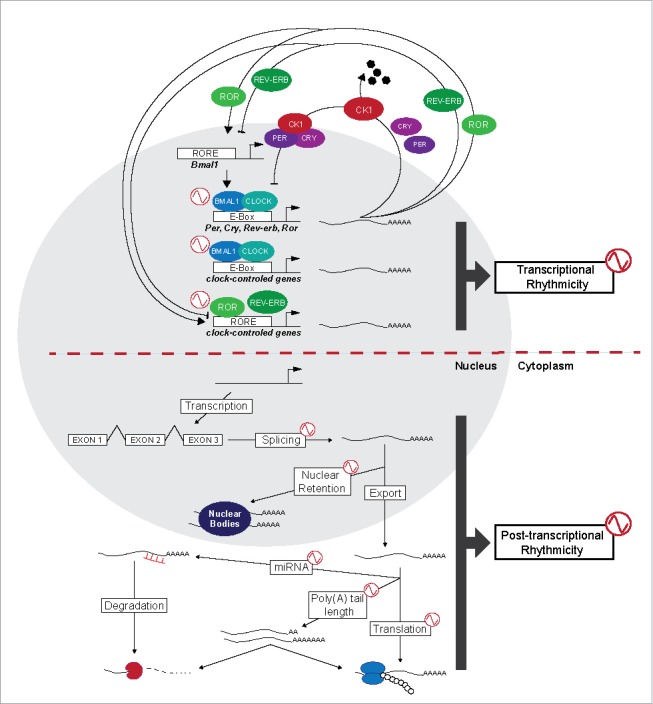
Molecular mechanisms of the circadian clockwork in mammals. In mammals, rhythmically transcribed BMAL1 heterodimerizes with CLOCK, and together they bind to target E-boxes in the promoters of Per, Cry, Rev-erb, and Ror. PER and CRY proteins are synthesized in the cytoplasm and may be phosphorylated by CK1 kinases including CK1ε/δ. PER/CRY heterodimers translocate to the nucleus where they repress their own transcription, generating a near 24-h feedback loop. A second feedback loop represses or activates the transcription of Bmal1 through the actions of REV-ERB or ROR, respectively. BMAL1/CLOCK heterodimer drives oscillating expression of clock-controlled genes with E-box or RORE containing promoters. Post-transcriptional events in the life cycle of a (pre-) mRNA that have been reported to directly influence the circadian clock and/or to be controlled in a circadian manner include splicing, nuclear retention and cytoplasmic export, regulation by miRNA and polyadenylation at the 3’ end. Translation and degradation of the mature mRNA are rhythmic processes as well.
In addition to the core regulation at the level of transcription or translation, circadian clock proteins are also subjected to extensive post-translational modifications that appear to control their cellular localization, protein stability, and activity. For example, Casein Kinase Iε and Iδ (CKIε/δ) are known to be critical factors that regulate the turnover of PERs and CRYs in mammals;8-10 however, kinase CKIε also activates BMAL1-mediated transcription9 (Fig. 1).
Importantly, circadian transcription factors not only regulate their own transcription but also regulate the expression of numerous other clock-controlled genes6 [CCGs; (Fig. 1)]. Over the past decade, clock gene transcriptional regulation has been described in many species and tissues, where it drives rhythmic mRNA expression. By use of techniques such as microarrays,11-13 a large fraction of the mRNA population (up to 10-15% of all mRNAs in a single mammalian tissue14) has been shown to display a rhythmic expression that has been initially assumed to result from temporal changes in transcription. However, data from mouse liver demonstrate poor correlation between the activation of a promoter and the amount of the corresponding transcript for genes that are rhythmic at the steady-state level.15 Actually, with the development of high-throughput sequencing, results obtained in the last years indicate that approximately 43% of the mammalian genome is rhythmic and analysis of circadian nascent RNA has allowed to show that less than 30 % of circadian mRNA are regulated by de novo transcription, suggesting that post-transcriptional regulation contributes mostly to rhythmic mRNA expression (Fig. 1).15-19 Much of what we initially knew about post-transcriptional regulation came from studies of fungi, plants and flies (For a review see20), but circadian post-transcriptional mechanisms involved in rhythmic control of mRNA expression have now also been reported in mammals at many different levels (For a review see ref. 21), such as RNA splicing, poly-adenylation, mRNA stability, mRNA cytoplasmic export and RNA nuclear retention (Fig. 1).
RNAs can be retained in the nucleus by particular bodies called paraspeckles. These nuclear bodies are found in almost all of the cultured cell lines and primary cultures from tissues,22 except for embryonic stem cells.23 Paraspeckles are detected as discrete dots found in inter-chromatin space, close to nuclear speckles.22 A long noncoding RNA, nuclear-enriched abundant transcript one (Neat1) is the structural component (Fig. 2).23-26 While a short and a long transcript previously identified as MENε (Neat1–1) and MENβ (Neat1–2), respectively.25,27 are generated from the same promoter, Neat1–1 alone cannot induce paraspeckle formation since specific depletion of Neat1–2 leads to disruption of paraspeckles.25 While paraspeckles detected by RNA FISH of Neat1 appeared as round foci when visualized under a confocal microscope, we showed that they appeared more likely as oblong structures with smaller dimensions after use of a combination of Neat1 RNA FISH and Super Resolution STORM analysis (as designed in Fig. 2). Paraspeckles have been shown to retain in the nucleus RNAs containing duplex structures.23 This has been shown for the mouse cationic amino acid transporter 2 (Cat2) transcribed nuclear RNA, Ctn-RNA, an alternatively spliced form of the Cat2 mRNA, which contains a dsRNA structure resulting from inverted short inter-spersed nuclear elements (SINEs) in its 3’-UTR.28 In primate cells, the most common inverted repeated SINEs are Alu elements. Alu elements are unique to primates and account for almost all of the human SINEs and for more than 10% of the genome and inverted repeat structures (inverted repeated Alu elements [IRAlus]) occur frequently in gene regions.29 Paraspeckles have been shown to retain in the nucleus mRNAs containing IRAlus in their 3’-UTRs like Nicolin 1 (NICN1) or Lin 28.23,29
Figure 2.
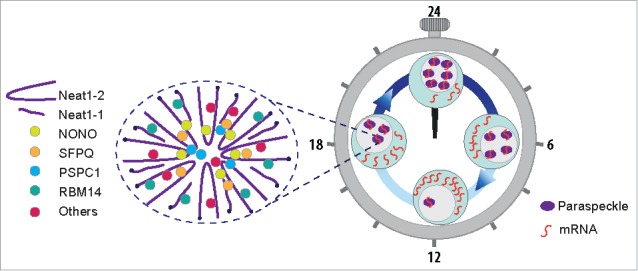
Structure and functional implication of paraspeckles in the rhythmic expression of mRNA. Paraspeckles are schematically drawn as oblong structures organized around the short (Neat1–1) and long (Neat1–2) transcripts of the long non-coding Neat1 RNA. Major protein entities of paraspeckles are also shown in the scheme. The roles of paraspeckles in circadian gene expression is schematically represented. The rhythmic number of paraspeckles inside the cells drives a circadian nuclear retention of paraspeckles-associated mRNA and thus leads to a circadian gene expression.
The nuclear retention of mRNAs containing IRAlus by paraspeckles can be regulated by a methyl-transferase, CARM1 (coactivator-associated arginine methyl-transferase1), which control both the binding capacity of some paraspeckle proteins to mRNAs containing IRAlus as well as NEAT1 transcription and then paraspeckle formation.30 Among the 40 paraspeckle proteins identified thus far,31 researchers classified 4 RNA-binding proteins, including 3 members of the Drosophila melanogaster behavior human splicing (DBHS) family proteins (NONO, PSPC1 and SFPQ) and RNA-binding motif protein 14 (RBM14) as major paraspeckle protein components (Fig. 2).22,28,32 We had previously reported that 2 of these major protein components of paraspeckles, namely NONO and SFPQ, display a circadian expression pattern in primary cultures of pituitary cells as well as in a rat pituitary cell line, the GH4C1 cells.33,34 In this latter cell line, we found thereafter that 2 other major paraspeckle proteins, PSPC1 and RBM14, display also a circadian pattern. All 4 proteins further bind rhythmically to Neat1 and Neat1 itself displays a circadian expression pattern. The expression of the long form of Neat1 RNA, Neat1–2, that is known to be sufficient for the formation of paraspeckle displays also a circadian pattern. In addition we showed that circadian expression of these different components leads to rhythmic variations in paraspeckle number within the cells.35 Thanks to their circadian expression pattern and given their presumed functions in gene expression through corresponding mRNA nuclear retention, we asked whether paraspeckle bodies can rhythmically retain RNAs in the nucleus leading to a rhythmic expression of the corresponding gene. This hypothesis was first tested with a reporter gene. Indeed, by using a construct of the EGFP reporter gene fused to an IRAlu and by transfecting the construct into GH4C1 cells, we obtained evidence that IRAlu elements inserted in 3’-UTR of egfp reporter mRNA allow for its circadian retention within the nucleus This rhythmic nuclear retention is abolished after disruption of paraspeckles by siRNA or Neat1 antisens oligonucleotides. Using real-time video microscopy, IRAlu elements inserted in 3’-UTR of egfp reporter mRNA was shown to cause rhythmic cytoplasmic expression of the EGFP protein. Paraspeckles through their circadian expression, could then control circadian expression pattern of a reporter gene containing IRAlu elements in 3’-UTR.35 We then asked whether this circadian post-transcriptional regulation exerted by paraspeckle bodies applies to endogenous genes. To address this issue, it was necessary to determine which mRNA are associated with paraspeckle bodies in the nucleus of GH4C1 cells. To this end, we developed a hybridization-based strategy that uses complementary oligonucleotides to purify Neat1 RNA together with its RNA targets from reversibly cross-linked extracts. Two antisense biotinylated oligonucleotide probes that target accessible regions of Neat1 RNA, as predicted by modeling its secondary structure by bioinformatics, were designed and used for Neat1 RNA specific pull-down whereas one biotinylated irrelevant probe was used for Neat1 RNA non-specific pull-down. Genes which were considered specifically associated with paraspeckles exhibited values of fragment per kilobase per million of mapped reads (FPKM) higher than 1 and were common to the lists obtained with the 2 specific probes. Overlapping list represented 65% of the list obtained with one probe and 83% of the list obtained with the other one. By comparing this gene list to available data in the literature such as the list of rhythmic mRNA in the mouse pituitary13 or the mRNA whose rhythm is post-transcriptionally controlled in the liver of mice,15 we achieved significant recovery rates of 18 and 27%, respectively. These rates are probably underestimated since they come from the comparison of gene lists obtained in different structures and species.
By selecting a few genes from our list, we showed that the mRNA of these genes exhibit a circadian rhythm of their nuclear retention and that this rhythm is abolished when paraspeckles are disrupted.35 These results obtained on endogenous mRNA, together with results obtained with a reporter gene, allow to conclude that paraspeckles participate in the post-transcriptional control of rhythmic gene expression (Fig. 2). Besides deciphering a post-transcriptional mechanism involved in the circadian regulation of gene expression, a problematic anchored in the field of chronobiology, our study from an endocrine point of view allows to assign a role in the physiology of the pituitary gland to the long non-coding RNA Neat1 whose functions remain enigmatic.
Although it has been shown that paraspeckles can retain in the nucleus genes which have sequences of IRAlu type in their 3′-UTR,23,28 endogenous mRNA we found associated with Neat1, actually do not contain such sequences or equivalent IR-SINE sequences in our model of rat pituitary cells. This suggests either that paraspeckles can recognize IR-sequences localized anywhere along the mRNA or that double-stranded RNA structures with such IR-sequences are not the only one recognized by paraspeckles. The challenge will be now to elicite the nature of the sequences together with their position along the mRNAs that are involved in the binding of paraspeckles and therefore form the basis of circadian expression.
In conclusion, circadian regulation has been investigated mainly at the transcriptional or the post-translational level, and RNA-based mechanisms contributing to this regulation are only beginning to emerge. Therefore, the impact of such mechanisms on circadian biology remains to be evaluated. However, given the finding that the majority of circadian mRNAs is not regulated by de novo transcription and given from the study presented here, the high number of mRNA that are potentially regulated by the new post-transcriptional mechanism we described, we predict all these post-transcriptional mechanisms will be shown to play extensive and widespread roles in circadian biology. It is also tempting to imagine that after the identification of mechanisms involved in post-transcriptional control of the circadian clock, connections to diseases will follow. Actually presumably because circadian rhythms play a key role as adaptive mechanisms to permanent changes in the environment, their deregulation is associated with many diseases the best known of which are neuropsychiatric, metabolic, cardiovascular disorders and cancers. A better understanding of the molecular mechanisms that govern the mammalian circadian clock is thus required to first identify connections to diseases, which may then give opportunities for new therapeutic concepts. Therapeutic targeting of paraspeckles could be for instance the basis for new strategies to control circadian rhythms, with the aim of improving human disease associated with rhythmicity dysfunction, in particular rhythmic hormonal dysfunctioning.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by Pfizer-SFE award - Research price Anne-Marie François-Bellan 2014.
References
- [1].Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Ann Rev Cell Dev Biol 2001; 17:215-253; PMID:11687489 [DOI] [PubMed] [Google Scholar]
- [2].Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thoma TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 2005; 6:544-556; PMID:15951747; https://doi.org/ 10.1038/nrg1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Ann Rev Neurosci 2000; 23:713-742; PMID:10845079 [DOI] [PubMed] [Google Scholar]
- [4].Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell 2002; 111:919-922; PMID:12507418; https://doi.org/ [DOI] [PubMed] [Google Scholar]
- [5].Hastings MH, Maywood ES, Reddy AB. Two decades of circadian time. J Neuroendocrinol 2008; 20:812-819; PMID:18601704 [DOI] [PubMed] [Google Scholar]
- [6].Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Ann Rev Physiol 2001; 63:647-676; PMID:11181971 [DOI] [PubMed] [Google Scholar]
- [7].Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet 2006; 15:R271-R277; https://doi.org/ 10.1093/hmg/ddl207 [DOI] [PubMed] [Google Scholar]
- [8].Akashi M, Tsuchiya Y, Yoshino T, Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol 2002; 22:1693-1703; PMID:11865049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J Biol Chem 2002; 277:17248-17254; PMID:11875063; https://doi.org/ 10.1074/jbc.M111466200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 2007; 8:139-148; PMID:17245414; https://doi.org/ 10.1038/nrm2106 [DOI] [PubMed] [Google Scholar]
- [11].Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002; 109:307-320; PMID:12015981 [DOI] [PubMed] [Google Scholar]
- [12].Storch K-F, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature 2002; 417:78-83; https://doi.org/ 10.1038/nature744; PMID:11967526 [DOI] [PubMed] [Google Scholar]
- [13].Hughes M, Deharo L, Pulivarthy SR, Gu J, Hayes K, Panda S, Hogenesch JB. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Symp Quant Biol 2007; 72:381-386; PMID:18419295; https://doi.org/ 10.1101/sqb.2007.72.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 2004; 5:407-441; PMID:15485355; https://doi.org/ 10.1146/annurev.genom.5.061903.175925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife 2012; 1:e00011; PMID:23150795; https://doi.org/ 10.7554/eLife.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Partch CL, Green CB, Takahashi JS. Molecular architec-ture of the mammalian circadian clock. Trends Cell Biol 2013; 24:90-9 PMID:23916625; https://doi.org/27498225 10.1016/j.tcb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hurley JM, Loros JJ, Dunlap JC. Circadian oscillators: Around the transcription-translation feedback loop and on to output. Trends Biochem Sci. 2016; 41(10):834-46; PMID:27498225; https://doi.org/ 10.1016/j.tibs.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012; 338:349-354; PMID:22936566; https://doi.org/ 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li J, Grant GR, Hogenesch JB, Hughes ME. Considerations for RNA-seq analysis of circadian rhythms. Methods Enzymol 2015; 551:349-367; https://doi.org/ 10.1016/bs.mie.2014.10.020 [DOI] [PubMed] [Google Scholar]
- [20].Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci 2011; 124:311-320; PMID:21242310; https://doi.org/ 10.1242/jcs.065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Preußner M, Heyd F. Post-transcriptional control of the mammalian circadian clock: implications for health and disease. Pflugers Arch 2016; 468(6):983-91; https://doi.org/ 10.1007/s00424-016-1820-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 2009; 186:637; PMID:19720872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 2009; 35:467-478; PMID:19716791; https://doi.org/ 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33:717-726; PMID:19217333; https://doi.org/ 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sasaki YT, Hirose T. How to build a paraspeckle. Genome Biol 2009; 10:227; PMID:19664169; https://doi.org/ 10.1186/gb-2009-10-7-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 2009; 19:347-359; PMID:19106332; https://doi.org/ 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Guru SC, Agarwal SK, Manickam P, Olufemi SE, Crabtree JS, Weisemann JM, Kester MB, Kim YS, Wang Y, Emmert-Buck MR. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res 1997; 7:725-735; PMID:9253601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell 2005; 123:249-263; PMID:16239143; https://doi.org/ 10.1016/j.cell.2005.08.033 [DOI] [PubMed] [Google Scholar]
- [29].Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle 2008; 7:3294-3301; PMID:18948735; https://doi.org/ [DOI] [PubMed] [Google Scholar]
- [30].Hu SB, Xiang JF, Li X, Xu Y, Xue W, Huang M, Wong CC, Sagum CA, Bedford MT, et al.. Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev 2015; 29:630-645; PMID:25792598; https://doi.org/ 10.1101/gad.257048.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long noncoding RNAs. RNA Biol 2013; 10:456-61; PMID:23324609; https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nakagawa S, Hirose T. Paraspeckle nuclear bodies-useful uselessness? Cell Mol Life Sci 2012; 69:3027-3036; PMID:22476590; https://doi.org/ 10.1007/s00018-012-0973-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Guillaumond F, Boyer B, Becquet D, Guillen S, Kuhn L, Garin J, Belghazi M, Bosler O, Franc JL, François-Bellan AM. Chromatin remodeling as a mechanism for circadian prolactin transcription: rhythmic NONO and SFPQ recruitment to HLTF. FASEB J. 2011; 25:2740-2756; PMID:21507896; https://doi.org/ 10.1096/fj.10-178616 [DOI] [PubMed] [Google Scholar]
- [34].Becquet D, Boyer B, Rasolonjanahary R, Brue T, Guillen S, Moreno M, Franc JL, François-Bellan AM. Evidence for an internal and functional circadian clock in rat pituitary cells. Mol Cell Endocrinol 2014; 382:888-898; PMID:24239982; https://doi.org/ 10.1016/j.mce.2013.11.004 [DOI] [PubMed] [Google Scholar]
- [35].Torres M, Becquet D, Blanchard MP, Guillen S, Boyer B, Moreno M, Franc JL, Fran , c ois-Bellan AM. Circadian RNA expression elicited by 3 0 -UTR IRAlu-paraspeckle associated elements. Elife 2016;5:e14837 https://doi.org/ 10.7554/eLife.14837 [DOI] [PMC free article] [PubMed] [Google Scholar]


