Abstract
Purpose
Abnormal expression of miR-223 in cancerous tissue has confirmed it as an important player in tumorigenesis of cancers, such as hepatocellular carcinoma, colorectal carcinoma, osteosarcoma, gastric cancer, and chronic lymphocytic leukemia. The present meta-analysis aimed to explore the association between circulating miR-223 and prognosis of cancers.
Methods
The studies were accessed by an electronic search of multiple databases. RevMan5.3 and STATA14.0 were used to estimate the heterogeneity among studies, pooled effects, and publication bias.
Results
Ten studies with data of 1,002 patients with cancer were included in this meta-analysis. The risk of metastasis from stages 3 to 4 of TNM did not decrease when high versus low circulating expression of miR-223 were compared (pooled odds ratio =0.50, 95% CI: 0.24–1.03). In case of prognosis, the overall survival time was not significantly longer with high circulating miR-223 expression (pooled hazard ratio [HR] =0.64, 95% CI: 0.38–1.11) in all cancer types. However, the overall survival time of chronic lymphocytic leukemia (pooled HR =0.19, 95% CI: 0.07–0.54) increased in subgroup analysis. Moreover, the treatment-free survival of chronic lymphocytic leukemia was significantly increased with high circulating miR-223 expression (pooled HR =0.38, 95% CI: 0.23–0.64).
Conclusion
Circulating miR-223 was not an effective biomarker in prognosis surveillance in all cancers but in chronic lymphocytic leukemia.
Keywords: miR-223, carcinoma, metastasis, prognosis, meta-analysis
Background
MicroRNAs (miRNAs) are small noncoding RNA molecules (20–24 nucleotides) with negative regulation in gene expression at the posttranscriptional level.1 Experiments have shown that miRNA broadly impact hundreds of mRNA targets based on the ~7 nt complementary base-pairing to the “seed sequence” of an miRNA.2 The dysregulation of homeostatic control of miRNA biogenesis, such as miR-155, miR-17~92, miR-16, miR-10b, and miR-373, could be associated with multiple pathological cancers, especially tumors of the breast, lung, liver, pancreas, and bone marrow.3 Furthermore, the regulating miRNAs have shown aberrant expression in tumor tissues and patient serum and participate in the onset and progression of cancer.4,5 Because of the involvement of miRNAs in many cellular cancer pathways including development, cell proliferation, differentiation, and apoptosis,6–8 miRNAs were expected to play crucial roles in cancer diagnosis and therapy as well as in prognosis surveillance.
MiR-223, a common tumor-associated microRNA, has been reported to be dysregulated in various human tumors, such as hepatocellular carcinoma,9 colorectal carcinoma,10 osteosarcoma,11 gastric cancer,12 and chronic lymphocytic leukemia.13 Abnormal expression of miR-223 in cancerous tissue has confirmed it as an important player in tumorigenesis of cancers.14 Similar to the tissue, miR-223 could be steadily detected in patient’s circulating blood, and be used as a noninvasive biomarker for early cancer detection and diagnosis.9 Moreover, circulating miR-223 was found to be associated with metastasis and prognosis of cancers15–20 and it might be a diagnostic biomarker and prognostic factor. However, the effect of circulating miR-223 on cancer prognosis is controversial, and no meta-analysis has investigated the relationship between circulating miR-223 expression and prognosis.
Therefore, the present meta-analysis aimed to explore the association between circulating miR-223 level and clinical outcome of patients with cancer to further determine the biomarker role of circulating miR-223 in metastasis and prognosis of cancers.
Materials and methods
Literature search strategy
Reports of studies in Language of English or Chinese on the role of circulating miR-223 expression in the development of human cancer were searched in PubMed, EMBASE, the Cochrane Library, China National Knowledge Infrastructure, and Wanfang databases with the following keywords: (“cancer” or “tumor” or “neoplasm” or “malignant” or “carcinoma”) and (“prognos*” or “surviv*” or “follow-up” or “mortality” or “predict” or “outcome” or “metastas*”) and (“microRNA-223” or “miRNA-223”, or “miR-223”) and (“serum” or “plasma” or “blood” or “circulating”). The last search date was September 23, 2016. References of retrieved papers and conference reports were also searched to identify relevant studies.
Selection criteria
After removing the duplicate entries, titles and abstracts of articles were checked by 4 authors (YZ, JL, YC, YL). The full text of eligible articles was retrieved. The eligible articles had the following criteria: 1) the expression of circulating miR-223 was analyzed by metastasis or survival, 2) patients were divided by high and low expression of circulating miR-223, 3) odd ratios (ORs) for metastasis (tumor node metastasis [TNM] stage, international staging system) or hazard ratios (HRs) for survival (overall survival [OS], disease-free survival, event-free survival, progression-free survival, cause-specific survival, and treatment-free survival [TFS]) were provided or could be calculated from the available data; and 4) the expression of circulating miR-223 was tested in patients’ peripheral blood by real-time PCR or fluorescence in situ hybridization. Studies not fulfilling the criteria, reviews, and animal/cell-line studies were excluded. Furthermore, if more than 1 study of the same cohort was published, only the most recent English publication was included. Consensus on searching and exclusion was resolved by discussion and with 2 other investigators (TW, XC) if needed.
Data extraction and quality assessment
The general data was extracted by 4 authors (WH, WZ, DW, RM) using the following form: first author’s name, published year, region of cohort, sample size, cancer type, method to test miR-223, cases in each expression group (high/low), cases of metastasis (TNM/ISS) in each group, and survival results (OS, disease-free survival, event-free survival, progression-free survival, cause-specific survival, and TFS). Furthermore, the reference for all effects (ORs or HRs) was reformatted as low circulating miR-223 expression, and the multivariate analysis effects were used for pooled analysis. The quality of each eligible study was assessed by the Newcastle-Ottawa Scale, consisting of selection, outcome, and comparability, with scores from 0 to 9. A study with Newcastle-Ottawa Scale score ≥6 was considered at high quality.
Statistical methods
This meta-analysis involved the use of Review Manager 5.3 (Cochrane network) and STATA 14.0. When HRs and 95% CIs were not provided directly in some studies, Engauge Digitizer 4.1 was used to analyze them from Kaplan–Meier curve. The heterogeneity among eligible studies was tested by Inconsistency (I2) and Q tests (chi-squared test). If no statistical heterogeneity was found (PQ>0.05, I2<50%), a fixed-effects model was used to estimate the pooled OR and HR. Otherwise, a random-effects model was used. Moreover, Begg’s and Egger’s tests were used to assess publication bias. All tests were two-sided, and P<0.05 was considered statistically significant.
Results
Characteristics of eligible studies
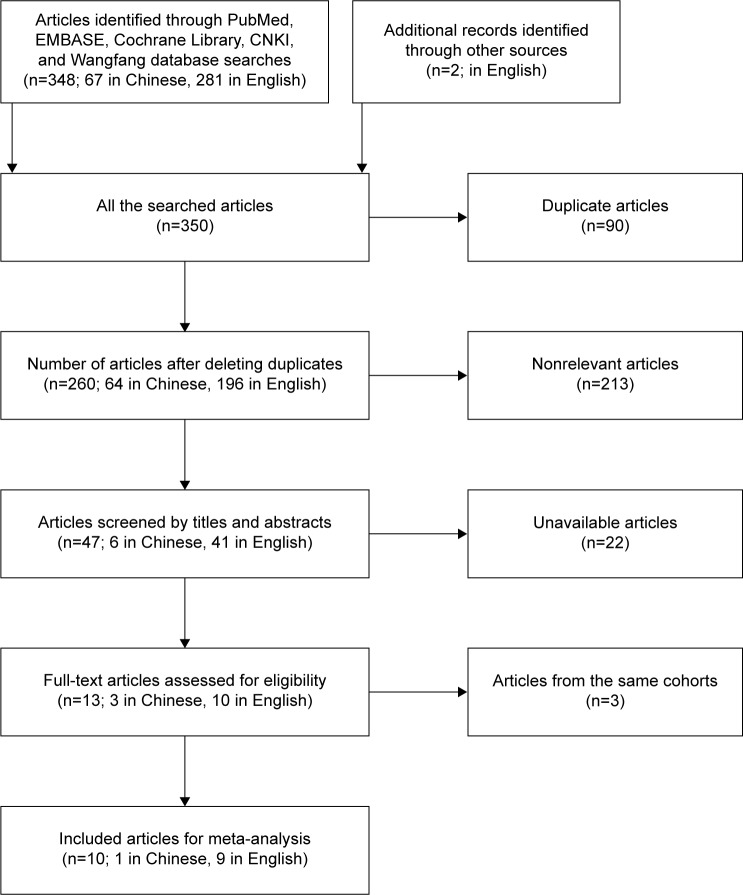
The literature search included 10 studies eligible in the meta-analysis (Figure 1), 6 from China13,17,18,21–23 and 1 each from France,24 United States,15 Japan,19 and Belgium.20 The studies involved a total of 1,002 patients with cancer, with mean sample size of 1,002 patients (range: 46–180). Six different types of cancers were evaluated: nonsmall cell lung carcinoma (n=3); esophageal squamous cell carcinoma and chronic lymphocytic leukemia (n=2 each); and pancreatic cancer, osteosarcoma, and multiple myeloma (n=1 each). The level of miR-223 was detected in patients’ plasma or serum by real-time PCR and the negative control was healthy plasma or serum. Furthermore, the group cutoff value determined by the original research depended on the median/mean value of miR-223 level receiver operating characteristic curve analysis. The main characteristics of each study are summarized in Table 1.
Figure 1.
Flow of the studies in the meta-analysis.
Table 1.
Basic data for all included studies in the meta-analysis
| References | Year | Country | Sample size | Cancer-type | Method | Follow-up (months) | Cutoff (high/low) | TNM stage
|
Survival
|
Quality score | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Method | Outcomes | |||||||||
| Sanfiorenzo et al24 | 2013 | France | 52 | NSCLC | qrt-PCR | 60 | 24/28 | NR | NR | Multivariate | DFS | 8 |
| Heegaard et al15 | 2012 | USA | 180 | NSCLC | qrt-PCR | 60 | 90/90 | NR | NR | Univariate | OS | 8 |
| Wu et al17,* | 2014 | China | 194 | ESCC | qrt-PCR | 60 | 113/81 | 1:3 | 1:5 | Multivariate | OS | 8 |
| 2:48 | 2:62 | |||||||||||
| 3:21 | 3:36 | |||||||||||
| 4:5 | 4:2 | |||||||||||
| Wu et al18 | 2014 | China | 63 | ESCC | qrt-PCR | 24 | NR | NR | NR | Univariate | OS | 8 |
| Komatsu et al19 | 2015 | Japan | 46 | PC | qrt-PCR | 50 | 25/20 | NR | NR | Univariate | CSS | 8 |
| Stamatopoulos et al20 | 2009 | Belgium | 110 | CLL | qrt-PCR | 300 | 37/73 | NR | NR | Univariate | OS, TFS | 8 |
| Zhou et al13 | 2012 | China | 53 | CLL | qrt-PCR | 90 | 31/22 | NR | NR | Univariate | OS, TFS | 8 |
| Dong et al21 | 2016 | China | 112 | OSC | qrt-PCR | 60 | 59/53 | 2a:22 | 2a:50 | Multivariate | OS | 8 |
| 2b/3:40 | 2b/3:10 | |||||||||||
| Wu et al22 | 2014 | China | 94 | NSCLC | qrt-PCR | 48 | 47/47 | 3:23 | 3:29 | Multivariate | OS | 8 |
| 4:24 | 4:18 | |||||||||||
| Qu23 | 2014 | China | 98 | MM | qrt-PCR | 48 | 49/49 | 1+2:22 | 1+2:30# | Univariate | OS, PFS | 8 |
| 3:27 | 3:19 | |||||||||||
Note:
ISS, international staging system.
Abbreviations: NSCLC, nonsmall cell lung carcinoma; ESCC, esophageal squamous cell carcinoma; PC, pancreatic cancer; CLL, chronic lymphocytic leukemia; OSC, osteosarcoma; OS, overall survival; MM, multiple myeloma; TNM, tumor node metastasis; qrt-PCR, quantitative real-time PCR; DFS, disease-free survival; TFS, treatment-free survival; CSS, cause-specific survival; PFS, progression-free survival; NR, not reported.
Association between circulating miR-223 and metastasis
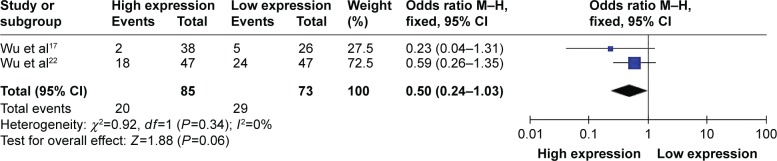
Although three studies reported the cases among TNM stage, only two in esophageal squamous cell carcinoma and nonsmall cell lung carcinoma had the same classification between high and low expression of circulating miR-223 with 158 cases in stages 3 and 4 of TNM. Because of severe heterogeneity with 2 studies (I2=0%, PQ=0.34), the fixed-effects model was used to calculate the pooled effect. High expression of circulating miR-223 did not significantly decrease the risk of stage 3 developing to 4, with pooled OR =0.50 (95% CI: 0.24–1.03) (Figure 2).
Figure 2.
Forest plot of association between circulating miR-223 expression and TNM.
Abbreviations: CI, confidence interval; M–H, Mantel–Haenszel test; TNM, tumor node metastasis.
Association between circulating miR-223 and OS
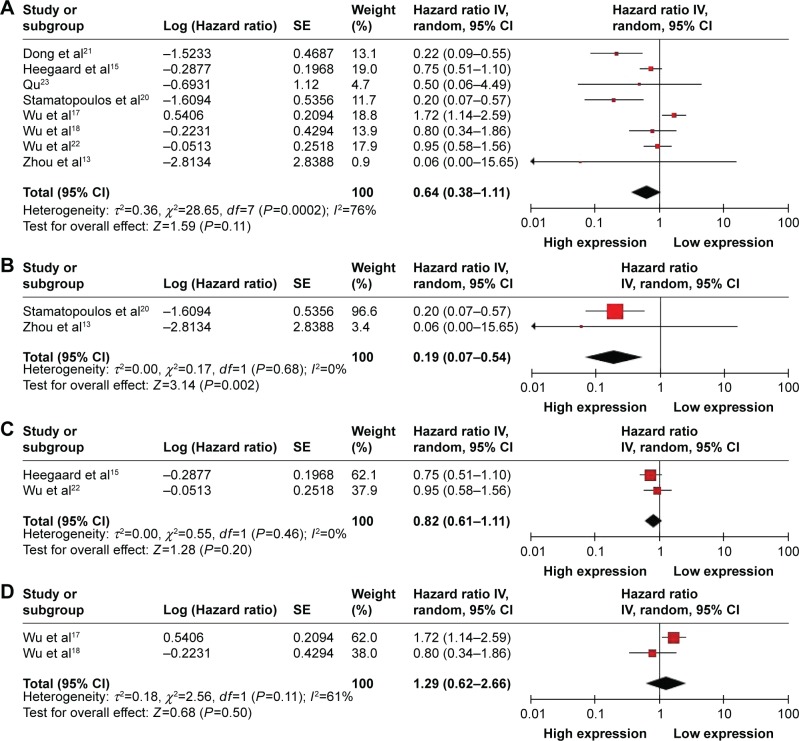
Eight studies showed data for OS in terms of circulating miR-223 level for 904 patients with cancer. Because of significant heterogeneity (I2=76%, PQ=0.0002), the random-effects model was used. The pooled HR of OS was 0.64 (95% CI: 0.38–1.11, P=0.11) for high versus low circulating miR-223 expression (Figure 3A); therefore, high miR-223 expression did not significantly increase the OS time. Because of the different types of cancer involved in this meta-analysis, we also calculated the pooled HR for OS by cancer-type. For studies of chronic lymphocytic leukemia, high miR-223 expression significantly increased the OS time with pooled HR of 0.19 (95% CI: 0.07–0.54) under the fixed-effects model (I2=0%, PQ=0.68) (Figure 3B). For studies of nonsmall cell lung carcinoma, the pooled HR was 0.82 (95% CI: 0.61–1.11) under the fixed-effects model (I2=0%, PQ=0.46) (Figure 3C) and for esophageal squamous cell carcinoma, it was 1.29 (95% CI: 0.62–2.66) under the random-effects model (I2=61%, PQ=0.11) (Figure 3D).
Figure 3.
Forest plot showing the association between circulating miR-223 expression and OS. (A) Pooled HR of circulating miR-223 on OS of all types of cancer under random-effects model. In subgroup analyses, (B) pooled HR of circulating miR-223 on OS of chronic lymphocytic leukemia under the fixed-effects model; (C) pooled HR of circulating miR-223 on OS of nonsmall cell lung carcinoma under fixed-effects model; (D) pooled HR of circulating miR-223 on OS of esophageal squamous cell carcinoma under the random-effects model. Data are HRs and 95% CIs.
Abbreviations: SE, standard error; IV, inverse variance; CI, confidence interval; OS, overall survival; HR, hazard ratio.
Association between circulating miR-223 and TFS
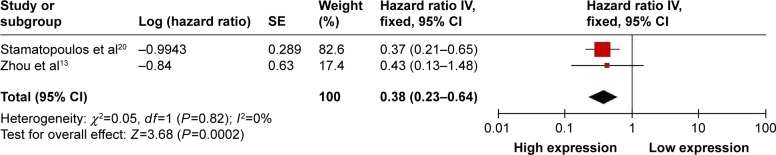
Two studies showed data for TFS with respect to circulating miR-223 level for 163 patients of chronic lymphocytic leukemia. These studies showed no significant heterogeneity (I2=0%, PQ=0.82) and therefore a fixed-effects model was used. High miR-223 expression significantly increased TFS time in patients with chronic lymphocytic leukemia (OR =0.38, 95% CI: 0.23–0.64) (Figure 4).
Figure 4.
Forest plot of association between circulating miR-223 expression and TFS.
Abbreviations: CI, confidence interval; IV, inverse variance; SE, standard error; TFS, treatment-free survival.
Sensitivity analysis
Sensitivity analysis was conducted to test the association between circulating miR-223 and OS. Each study was deleted in turn to examine the influence of the deleted data on the overall HR. Only exclusion of Wu et al’s data significantly changed the results of pooled HR of 0.54 (95% CI: 0.33–0.89) (Table 2), which suggested that the role of circulating miR-223 was differential or reverse in OS of esophageal squamous cell carcinoma. Therefore, we pooled the data on effect of circulating miR-223 on OS of all cancers except esophageal squamous cell carcinoma, which significantly increased the OS time under a random-effects model (HR =0.44, 95% CI: 0.24–0.82) (Figure S1).
Table 2.
Sensitivity analysis for the studies that resulted in loss of significance after omission
| Original summary effects
|
Omitted study | Resulting summary effects
|
||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | Nature of association | I2 | HR | 95% CI | Nature of association | I2 | |
| 0.64 | 0.38–1.11 | Not significantly decreased risk | 76% | Heegaard et al15 | 0.59 | 0.29–1.21 | Not significantly decreased risk | 78% |
| Wu et al17 | 0.54 | 0.33–0.89 | Significantly decreased risk | 57% | ||||
| Wu et al18 | 0.57 | 0.29–1.13 | Not significantly decreased risk | 79% | ||||
| Stamatopoulos et al20 | 0.77 | 0.46–1.29 | Not significantly decreased risk | 71% | ||||
| Zhou et al13 | 0.66 | 0.38–1.13 | Significantly increased risk | 78% | ||||
| Dong et al21 | 0.79 | 0.48–1.30 | Not significantly decreased risk | 69% | ||||
| Wu et al22 | 0.61 | 0.33–1.13 | Not significantly decreased risk | 79% | ||||
| Qu23 | 0.65 | 0.37–1.14 | Not significantly decreased risk | 79% | ||||
Abbreviations: CI, confidence interval; HR, hazard ratio.
Publication bias
Publication bias for the association between circulating miR-223 and OS was checked by a Begg’s funnel plot under the random-effects model (Figure 5). Begg’s test showed no significant rank correlation with Kendall score (Z=-1.24, Pr>|z| =0.216). Given this result, we performed Egger’s test where evidence of significance of publication bias was not found (r=−2.11, 95% CI: −5.09–0.87, P>|t| =0.133).
Figure 5.
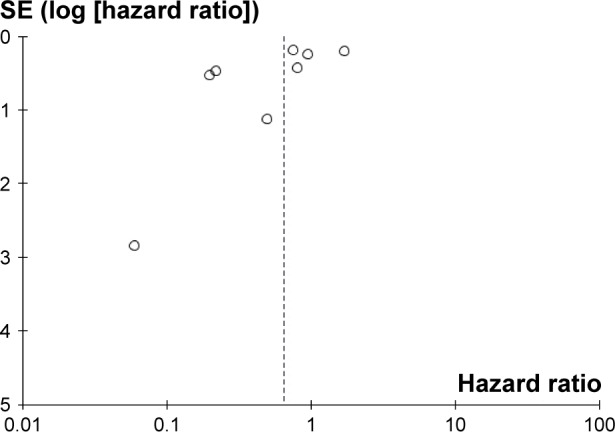
Begg’s funnel plot of the association between circulating miR-223 and OS in all cancer types under random-effects model.
Abbreviations: OS, overall survival; SE, standard error.
Discussion
This current study aimed to assess the pooled effect of circulating miR-223 expression on prognosis with cancer. High miR-223 expression was not significantly associated with decreased cancer metastasis and increased OS time of reported cancers. However, high miR-223 expression significantly increased the OS time as well as TFS time of patients with chronic lymphocytic leukemia.
MiR-223, initially identified as part of the hematopoietic system,25 has been widely reported to show association with different cancers. Research in the mechanism of miR-223 found that it could target C/EBPβ, E2F1, FOXO1, NFI-A, and MAFB to inhibit tumor cell proliferation, migration, and invasive capability in vitro;26–30 however, it could also deregulate the expression of tumor suppressors.31–33 As miR-223 affects multiple targets simultaneously that are involved in key processes,14 the role of miR-223 in cancer depends on the type of cancer and the total level in patient’s body. Although, circulating miR-223 was not always positively correlated with the level expressed in tumor tissue,19,34 the level of circulating miR-223 was proven as a high-quality biomarker for early cancer detection and diagnosis.9,35,36 Moreover, circulating miR-223 could contribute to the epithelial-to-mesenchymal transition, required for cancer metastasis and invasion,37 and could enhance response to preoperative chemo-radiotherapy in patients with cancer.38 Therefore, circulating miR-223 is an important cancer-associated miRNA and participates in tumor development and progression.
In our meta-analysis, although the risk of progression of stages 3 to 4 of TNM was not significantly decreased with high circulating miR-223 expression for the small sample size and few cancer types, it still suggested that high circulating miR-223 might be epidemiologically associated with decreased metastasis of cancers. For prognosis of patients, circulating miR-223 was not significantly associated with OS in all reported cancers; however, their association in subgroup analysis was not coincident. It epidemiologically supported the fact that the role of circulating miR-223 in cancers depends on the mechanism and character of cancer-type. It suggested that the circulating miR-223 could not be used as biomarker for prognosis surveillance of cancer but chronic lymphocytic leukemia.
Some meta-analyses focused on the association of miR-223 with cancer; all analyzed the role of miR-223 in the early detection and diagnosis of cancer.9,39–41 To search for an applicable biomarker for therapy, we focused on the association of circulating miR-223 on metastasis and prognosis. To the best of our knowledge, this is the first meta-analysis of the effect of circulating miR-223 on outcomes of patients with cancer.
Our study has some limitations. First, the studies of each cancer-type and respective patient samples were few in number. Second, because of heterogeneity of different cancer-type in the included studies, the results need to be further identified with more randomized clinical trials. Finally, because of using aggregated group data in the meta-analysis, some confounding factors could not be controlled.
Conclusion and recommendations
This meta-analysis is the first to demonstrate that high expression of circulating miR-223 is related to prognosis for patients with cancer. The expression of circulating miR-223 might be a biomarker for prognosis surveillance of some cancer types, especially chronic lymphocytic leukemia.
Supplementary material
Forest plot of the pooled HRs for OS with esophageal squamous cell carcinoma excluded under the random-effects model.
Abbreviations: HR, hazard ratio; OS, overall survival; IV, inverse variance; CI, confidence interval; SE, standard error.
References
- 1.Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J Bone Oncol. 2016;5(2):74–79. doi: 10.1016/j.jbo.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer. 2012;130(6):1378–1386. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu Y. The clinical significance of serum microRNA, long non-coding RNA and CRBN protein in multiple myeloma [master’s thesis] Shanghai: The Second Military Medical University; 2014. [Google Scholar]
- 4.Stamatopoulos B, Meuleman N, Haibe-Kains B, et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113(21):5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- 5.Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep. 2014;41(3):1257–1266. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 6.Zhou K, Yi S, Yu Z, et al. MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2012;53(6):1155–1161. doi: 10.3109/10428194.2011.642303. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This study was funded by the Shenzhen Technology Research and Development Funds (JCYJ20150403095530583 and 201604130086), and the Science & Technology Project of Shenzhen Longgang District (201406063001026 and 20160607153104624).
Footnotes
Author contributions
XLC conceived the study; YFZ, JBL, YC, and YL conducted the review and screened records for eligibility; WJH, WYZ, DLW, and RRM extracted data and conducted statistical analysis under the supervision of XLC and TQW. JBL prepared the initial report, which was read and edited by XLC. All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Shen J, Hung MC. Signaling-mediated regulation of MicroRNA processing. Cancer Res. 2015;75(5):783–791. doi: 10.1158/0008-5472.CAN-14-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JY, Ryu DS, Kim WJ, Kim SJ. Aberrantly expressed microRNAs in the context of bladder tumorigenesis. Investig Clin Urol. 2016;57(Suppl 1):S52–S59. doi: 10.4111/icu.2016.57.S1.S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med. 2016;14:143. doi: 10.1186/s12967-016-0893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pileczki V, Cojocneanu-Petric R, Maralani M, Neagoe IB, Sandulescu R. MicroRNAs as regulators of apoptosis mechanisms in cancer. Clujul Med. 2016;89(1):50–55. doi: 10.15386/cjmed-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwan JY, Psarianos P, Bruce JP, Yip KW, Liu FF. The complexity of microRNAs in human cancer. J Radiat Res. 2016;57(Suppl 1):i106–i111. doi: 10.1093/jrr/rrw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinweha P, Rattanapornsompong K, Charoensawan V, Jitrapakdee S. MicroRNAs and oncogenic transcriptional regulatory networks controlling metabolic reprogramming in cancers. Comput Struct Biotechnol J. 2016;14:223–233. doi: 10.1016/j.csbj.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, Shen Q, Li C, Li D, Chen J, He M. Identification of circulating MicroRNAs as novel potential biomarkers for hepatocellular carcinoma detection: a systematic review and meta-analysis. Clin Transl Oncol. 2015;17(9):684–693. doi: 10.1007/s12094-015-1294-y. [DOI] [PubMed] [Google Scholar]
- 10.Earle JS, Luthra R, Romans A, et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12(4):433–440. doi: 10.2353/jmoldx.2010.090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Yao Q, Hou Y, et al. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed Pharmacother. 2013;67(5):381–386. doi: 10.1016/j.biopha.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Shrestha S, Hsu SD, Huang WY, et al. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med. 2014;3(4):878–888. doi: 10.1002/cam4.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou K, Yi S, Yu Z, et al. MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2012;53(6):1155–1161. doi: 10.3109/10428194.2011.642303. [DOI] [PubMed] [Google Scholar]
- 14.Haneklaus M, Gerlic M, O’Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. 2013;274(3):215–226. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer. 2012;130(6):1378–1386. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joerger M, Baty F, Früh M, et al. Circulating microRNA profiling in patients with advanced non-squamous NSCLC receiving bevacizumab/erlotinib followed by platinum-based chemotherapy at progression (SAKK 19/05) Lung Cancer. 2014;85(2):306–313. doi: 10.1016/j.lungcan.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 17.Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep. 2014;41(3):1257–1266. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 18.Wu C, Wang C, Guan X, et al. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. 2014;9(3):e92292. doi: 10.1371/journal.pone.0092292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu S, Ichikawa D, Miyamae M, et al. Malignant potential in pancreatic neoplasm; new insights provided by circulating miR-223 in plasma. Expert Opin Biol Ther. 2015;15(6):773–785. doi: 10.1517/14712598.2015.1029914. [DOI] [PubMed] [Google Scholar]
- 20.Stamatopoulos B, Meuleman N, Haibe-Kains B, et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113(21):5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- 21.Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J Bone Oncol. 2016;5(2):74–79. doi: 10.1016/j.jbo.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C, Cao Y, He Z, et al. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med. 2014;232(2):85–95. doi: 10.1620/tjem.232.85. [DOI] [PubMed] [Google Scholar]
- 23.Qu Y. The clinical significance of serum microRNA, long non-coding RNA and CRBN protein in multiple myeloma [master’s thesis] Shanghai: The Second Military Medical University; 2014. [Google Scholar]
- 24.Sanfiorenzo C, Ilie MI, Belaid A, et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8(1):e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 26.Zhou H, Xiao J, Wu N, et al. MicroRNA-223 Regulates the differentiation and function of intestinal dendritic cells and macrophages by targeting C/EBPbeta. Cell Rep. 2015;13(6):1149–1160. doi: 10.1016/j.celrep.2015.09.073. [DOI] [PubMed] [Google Scholar]
- 27.Pulikkan JA, Dengler V, Peramangalam PS, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115(9):1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L, Li H, Jia CY, et al. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012;586(7):1038–1043. doi: 10.1016/j.febslet.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 29.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 30.Yang W, Lan X, Li D, Li T, Lu S. MiR-223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells. BMC Cancer. 2015;15:461. doi: 10.1186/s12885-015-1464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Zhang Y, Zhang H, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 32.Li ZW, Yang YM, Du LT, et al. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):256. doi: 10.1007/s12032-014-0256-5. [DOI] [PubMed] [Google Scholar]
- 33.Kurashige J, Watanabe M, Iwatsuki M, et al. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer. 2012;106(1):182–188. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cinpolat O, Unal ZN, Ismi O, Gorur A, Unal M. Comparison of microRNA profiles between benign and malignant salivary gland tumors in tissue, blood and saliva samples: a prospective, case-control study. Braz J Otorhinolaryngol. 2016 Apr 27; doi: 10.1016/j.bjorl.2016.03.013. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tachibana H, Sho R, Takeda Y, et al. Circulating miR-223 in oral cancer: its potential as a novel diagnostic biomarker and therapeutic target. PLoS One. 2016;11(7):e0159693. doi: 10.1371/journal.pone.0159693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li BS, Zhao YL, Guo G, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7(7):e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Y, Wang Y, Chen Q, Qiu N, Zhao Y, You X. MiR-223 inhibited cell metastasis of human cervical cancer by modulating epithelial-mesenchymal transition. Int J Clin Exp Pathol. 2015;8(9):11224–11229. [PMC free article] [PubMed] [Google Scholar]
- 38.Nakao T, Iwata T, Hotchi M, et al. Prediction of response to preoperative chemoradiotherapy and establishment of individualized therapy in advanced rectal cancer. Oncol Rep. 2015;34(4):1961–1967. doi: 10.3892/or.2015.4196. [DOI] [PubMed] [Google Scholar]
- 39.Fiorino S, Bacchi-Reggiani ML, Visani M, et al. MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis B- and C-related-hepatocellular-carcinoma. World J Gastroenterol. 2016;22(15):3907–3936. doi: 10.3748/wjg.v22.i15.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X, Ji G, Chen H, Jin W, Yin C, Zhang G. Clinical role of circulating miR-223 as a novel biomarker in early diagnosis of cancer patients. Int J Clin Exp Med. 2015;8(9):16890–16898. [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SY, Jeon TY, Choi CI, et al. Validation of circulating miRNA biomarkers for predicting lymph node metastasis in gastric cancer. J Mol Diagn. 2013;15(5):661–669. doi: 10.1016/j.jmoldx.2013.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plot of the pooled HRs for OS with esophageal squamous cell carcinoma excluded under the random-effects model.
Abbreviations: HR, hazard ratio; OS, overall survival; IV, inverse variance; CI, confidence interval; SE, standard error.