Abstract
Pseudomonas aeruginosa has great intrinsic antimicrobial resistance limiting the number of effective antibiotics. Thus, other antimicrobial agents such as silver nanoparticles (AgNPs) are considered potential agents to help manage and prevent infections. AgNPs can be used in several applications against bacteria resistant to common antibiotics or even multi-resistant bacteria such as P. aeruginosa. This study assessed the antimicrobial activity of commercial 10 nm AgNPs on two hospital strains of P. aeruginosa resistant to a large number of antibiotics and a reference strain from a culture collection. All strains were susceptible to 5 µg/mL nanoparticles solution. Reference strains INCQS 0230 and P.a.1 were sensitive to AgNPs at concentrations of 1.25 and 0.156 µg/mL, respectively; however, this was not observed for hospital strain P.a.2, which was more resistant to all antibiotics and AgNPs tested. Cytotoxicity evaluation indicated that AgNPs, up to a concentration of 2.5 µg/mL, are very safe for all cell lines tested. At 5.0 µg/mL, AgNPs had a discrete cytotoxic effect on tumor cells HeLa and HepG2. Results showed the potential of using AgNPs as an alternative to conventional antimicrobial agents that are currently used, and a perspective for application of nanosilver with antibiotics to enhance antimicrobial activity.
Keywords: antimicrobial activity, bacterial resistance, Pseudomonas aeruginosa, silver nanoparticles, AgNPs
Introduction
Gram-negative bacteria are among the main agents in a nosocomial infection, contributing to longer hospital stay, mortality, and higher hospital costs. Among Gram-negative bacteria, Pseudomonas aeruginosa is highly susceptible to genetic modifications leading to resistance to antimicrobials and the consequent complications in impaired or immunocompromised patients. Due to its ability to survive in harsh environments, P. aeruginosa is known as one of the most important agents in hospital infections, and it is a suitable model to study control and combatting of nosocomial infection.1,2
More than 99% of bacteria exist in the ecosystems of biofilms attached to different surfaces. The formation of biofilms in implanted devices (eg, catheters, lenses, and artificial heart valves) is related to the development of chronic infections.3,4 These bacterial biofilms are frequently resistant to antibiotics. Therefore, actions to reduce or prevent increase in cases of hospital infections should also be directed to avoid and to control biofilm formation.
P. aeruginosa is associated with a strong increase in human infections.5 The most severe infections occur in health care units and include bacteremia, pneumonia, and postoperative urinary and gastrointestinal infections, besides endocarditis, osteomyelitis, and meningitis.5,6 They can cause opportunistic infections, especially in immunocompromised patients such as burn victims, patients with cancer, or those with cystic fibrosis. They grow easily even in conditions that are adverse for other organisms and have intrinsic and acquired resistance to common antimicrobials. Data related to reduction of P. aeruginosa susceptibility to antimicrobial agents have been described in Brazil,7–9 highlighting the decreased sensitivity to broad-spectrum antibiotics, such as carbapenems.10,11 Resistance to fluoroquinolones such as norfloxacin and ciprofloxacin has also been observed in Brazil.8,12
Moreover, epidemic reports of nosocomial infections caused by this bacterium are frequent, usually originating from hospital environments and cross-contamination that is often associated with the incorrect use of medical equipment,13,14 which could be minimized by the use of medical devices coated with antimicrobial agents, such as silver nanoparticles (AgNPs).
Studies in the medicine field have shown that silver is effective against more than 650 pathogens, having a broad spectrum of activity. Its use in the form of nanoparticles enhances this property, allowing its use in a wide range of applications.15,16 Therefore, nanosilver is now considered one of the most viable alternatives to antibiotics because it seems to have high potential to solve the problem of multidrug resistance, which is often observed in several bacterial strains.17–19
Nanoparticles are clusters of atoms, with sizes ranging between 1 and 100 nm, whereas a “nano” is used to indicate one billionth of a meter.20–22 Because of their size, AgNPs have different physical and chemical characteristics to that of metallic silver.23,24
Studies have demonstrated the effectiveness of AgNPs as dressings for covering burns to surgical devices and bone prostheses, and are incorporated into clothing – always with the aim of producing antimicrobial effect.25–28
This study was conducted to evaluate the antimicrobial activity of different concentrations of commercial AgNPs (10 nm) on two acquired nosocomial infectious strains of P. aeruginosa, resistant to a large number of antibiotics.
Materials and methods
Materials
Strains: P. aeruginosa INCQS 0230/ATCC 27853 and two strains of P. aeruginosa acquired from hospital infections – P.a.1 and P.a.2.
Silver nanoparticles 10 nm: AgNP 20 µg/mL solution (nanoparticles, 10 nm particle size [TEM], 0.02 mg/mL in aqueous buffer, contains sodium citrate as stabilizer – 730785, Sigma-Aldrich, St. Louis, MO, USA), tested at concentrations: 5.0, 1.25, and 0.156 µg/mL.
Antibiotics discs: ceftazidime (30 µg), meropenem (10 µg), amikacin (30 µg), ampicillin + sulbactam (20 µg), levofloxacin (5 µg), chloramphenicol (30 µg), vancomycin (30 µg), penicillin (10 µg), oxacillin (1 µg), cefoxitin (30 µg), and erythromycin (15 µg) (Laborclin, Sao Paulo, Brazil).
Cell lines: Mouse fibroblasts NCTC-929 were purchased from Adolfo Lutz Institute; HeLa and HepG2 were kindly provided by Hemocentro – USP, Ribeirao Preto; cell culture medium DME and trypsin-EDTA (Sigma-Aldrich; EUA) and fetal bovine serum (FBS, Invitrogen).
Methods
Inoculum of microorganisms
Fresh cultures with less than 24-hour incubation were prepared at a concentration of 0.5 McFarland Scale29 (1.5×108 CFU/mL) and used at different dilutions in the proposed evaluation tests.
Antimicrobial susceptibility testing with antibiotics
From the bacterial exponential growth (~16 hours), a cell suspension in saline was adjusted to 0.5 McFarland and inoculated in Muller Hinton Agar–MHA (Sigma-Aldrich) using agar diffusion method.30,31 After 15 minutes of being allowed to stand, discs recommended by the Clinical Laboratory Standards Institute (CLSI) were placed on the plate and incubated at 35°C (±2) for 16–18 hours. The antibiotics tested were: ceftazidime, meropenem, amikacin, ampicillin + sulbactan, levofloxacin, chloramphenicol, vancomycin, penicillin, oxacillin, cefoxitin, and erythromycin. Bacterial susceptibility to these antibiotics were verified by measuring the diameter of the inhibition zones formed and then interpreted according to values set by the CLSI.32
Antibacterial effect of AgNPs
To assess antimicrobial activity, 16 hours growth bacteria suspensions were incubated at 35°C (±2) and 150 rpm with different concentrations of 10 nm AgNPs (5.0, 1.25, and 0.156 µg/mL). Aliquots were taken every 1 hour for 12 hours to determine cell viability by the method of serial dilutions and counting of colonies on plates.
Cytotoxicity of AgNPs
Cytotoxicity evaluation was assessed according ISO 10993-533 by monitoring the neutral red uptake NRU assay using mouse fibroblast NCTC 929, tumoral HeLa, and HepG2 cells (100 µL; 1×105 cells/mL) seeded into 96-well microliter plates and left to adhere during 24 hours. Then, cells were exposed to the AgNPs previously dispersed and serially diluted in complete medium with 5% FBS (100 µL/mL) at the following concentrations: 10.0, 5.0, 2.5, 1.25, 0.625, 0.312, and 0.156 µg/mL. After 24-hour exposure, cells were incubated for a further 3 hours with neutral red dye (50 µg/mL) following dye extraction using ethanol/acetic acid/water (50%/1%/49%). The absorbance was measured at 540 nm on a spectrophotometer (LabSystemsTitertek Multiskan MCC/340 Plate Reader; Thermo Labsystems, Mountain View, CA, USA). Absorbance measurements of cells exposed only to medium were considered as 100% cell viability (ie, the negative control). The effect on cell growth was calculated from the relative absorbance of untreated control cells at 540 nm. The 3T3-Phototox© software was used for calculating the concentration inducing a 50% reduction of cell viability (IC50).
Results and discussion
Tests of susceptibility to antibiotics
P. aeruginosa is a ubiquitous bacterium with high incidence in nosocomial infections, and it is present in various environmental samples; moreover, it is frequently found to be related with biofilm formation, which is a critical factor for infections with this strain.
A significant and concerning feature of P. aeruginosa is the cross-resistance to antimicrobials, which results from the presence of multiple resistance mechanisms in a single host that lead to multidrug resistance.34,35
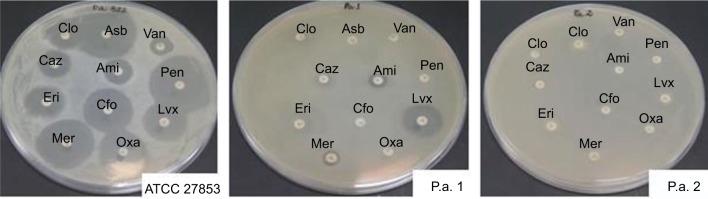
In this study, three strains of Pseudomonas were evaluated – one reference strain (INCQS 0230) and two strains from nosocomial infections. One of these (P.a.2) showed resistance to all 11 antibiotics tested (100% resistance), in contrast to the reference strain profile that showed sensitivity to almost all antibiotics tested, except for erythromycin (Figure 1 and Table 1).32,36
Figure 1.
Test of susceptibility to antimicrobials. Antibiotics tested: chloramphenicol (Clo), ampicillin + sulbactam (Asb), vancomycin (Van), ceftazidime (Caz), amikacin (Ami), penicillin (Pen), erythromycin (Eri), cefoxitin (Cfo), levofloxacin (Lvx), meropenem (Mer), and oxacillin (Oxa).
Table 1.
Antibiotic susceptibility test
| Strain | Antibiotics
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clo (mm) |
Asb (mm) |
Van (mm) |
Caz (mm) |
Ami (mm) |
Pen (mm) |
Eri (mm) |
Cfo (mm) |
Lvx (mm) |
Mer (mm) |
Oxa (mm) |
|
| INCQS 0230 | 26 S | 38 S | 14 S | 18 S | 21 S | 40 S | 19 I | 32 S | 24 S | 36 S | 30 S |
| P.a.1 | R | R | R | R | 12 R | R | R | R | 24 S | 19 R | R |
| P.a.2 | R | R | R | R | R | R | R | R | R | R | R |
Notes: The resistance or sensitivity of the samples to antimicrobials was determined by measuring the size (mm) of growth inhibition zones of each sample on the surface of the culture medium, where 108 CFU/mL of the test strain was inoculated. They were classified as follows according to the Clinical and Laboratory Standards Institute – CLSI: S – sensitive (inhibition zones ≥13 mm); R – resistant (inhibition zones ≤13 mm); I – intermediate (intermediate inhibition zones – sensitive/resistant).32
The resistance observed in P. aeruginosa against nosocomial infections may be due to mechanisms such as decreased permeability of the outer membrane due to loss or alterations in porin structure; activity of efflux pumps that promote the decrease of antimicrobial concentration within the bacterium; and/or by action of beta-lactamases (carbapenemases). Several acquired beta-lactamases belonging to class B of Ambler, and known as metallo-beta-lactamases (MBL), or to class D, also known as oxacillinases, have already been identified in these pathogens.34,37 These results were used as a reference for comparison with the results of antimicrobial susceptibility when using AgNPs.
Antibacterial effect of AgNPs
The broad antimicrobial effect of silver is well known, and it has been used in different fields in medicine for years – in wound healing or in biomaterials. The silver antimicrobial effect is dependent on superficial contact, in that silver can inhibit enzymatic systems of the respiratory chain and alter DNA synthesis.38 Due to their small size, nanoparticles have a large contact area compared to other salts and even the silver particulate, thus providing a better contact with the microorganisms by binding to the cell membrane and also penetrating inside.
The antimicrobial activity of AgNPs to the three P. aeruginosa strains (reference strain and two clinical strains), with resistance to some antibiotics, was evaluated using AgNPs at 5.0, 1.25, and 0.156 µg/mL concentrations. In these experiments, the CFU/mL of the three strains was determined every 1 hour in a 12-hour total assay to derive the behavior profile. Furthermore, all strains were also analyzed without AgNPs (control). The behavior profiles obtained are shown in Figures 2–4. In the control condition (without AgNPs), all strains showed similar profile, remaining at a concentration of approximately 1×108 CFU/mL, during the 12 hours of the experiment; this was as expected as the cultures were already in high cell concentration (1×108 CFU/mL), at the end of the exponential phase, and an expressive growth would not have been observed. However, with regard to bacterial behavior in the presence of AgNPs, the three strains showed different profiles, with reduction of viable cell concentration over time, indicating sensitivity to the AgNPs. The reference strain INCQS 0230 and P.a.1, showed bacterial reduction – at 1.25 and 0.156 µg/mL AgNP concentrations – which was not observed for strain P.a.2 at the same AgNPs concentrations. However, at the concentration of 5.0 µg/mL, all strains showed susceptibility to AgNPs. The reference strain showed a higher sensitivity to all tested AgNP concentrations as compared to hospital strains, corroborating the results of the antibiotic tests where the strain was sensitive to 10 of the 11 antibiotics tested. The results with the hospital strains with AgNPs are coherent and concur with the results obtained with antibiotic, showing the high resistance of the hospital strains. As shown earlier, the hospital strain P.a.2 was resistant to the 11 antibiotics tested.
Figure 2.
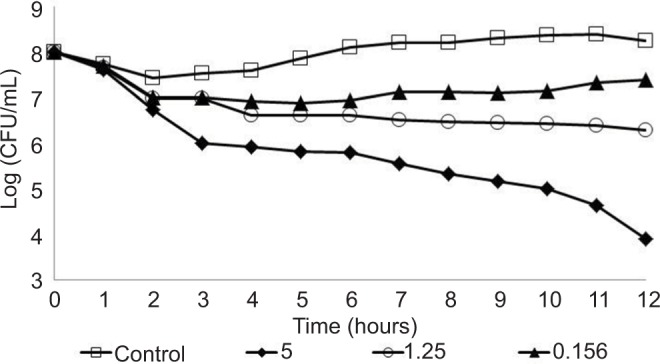
Behavior of the Pseudomonas aeruginosa INCQS 0230 strain tested at different concentrations of silver nanoparticles: 0.156, 1.25, and 5.0 µg/mL.
Figure 3.
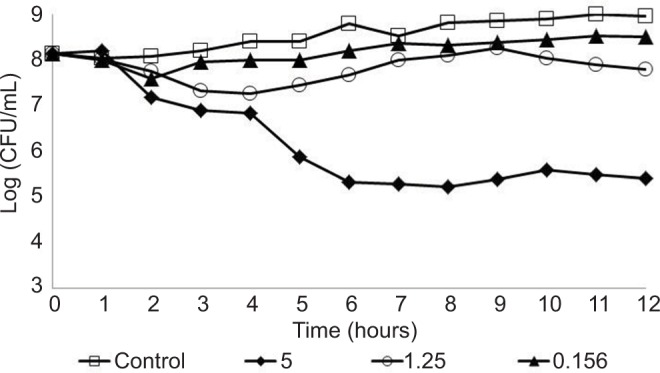
Behavior of the Pseudomonas aeruginosa P.a.1 strain tested at different concentrations of silver nanoparticles: 0.156, 1.25, and 5.0 µg/mL.
Figure 4.
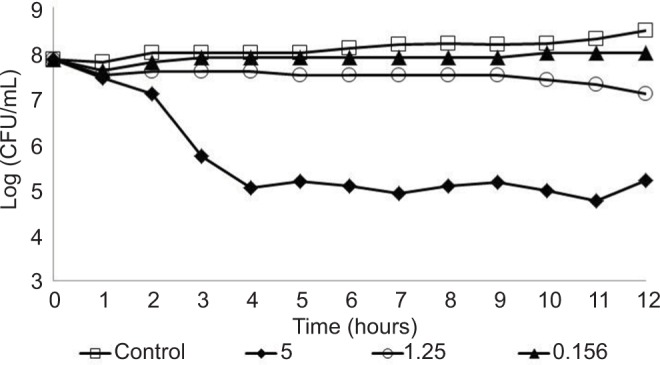
Behavior of the Pseudomonas aeruginosa P.a.2 strain tested at different concentrations of silver nanoparticles: 0.156, 1.25, and 5.0 µg/mL.
Hospital strains P.a.1 and P.a.2 showed some sensitivity against AgNP concentrations 1.25 and 0.156 µg/mL, with reduction of at least one log in the bacterial concentration, although the best effect was seen on the reference strain. After 12 hours of experiment, the highest concentration of AgNPs (5.0 µg/mL) showed excellent antibacterial activity with approximately 99.9% bacterial death, even when tested against hospital strains that presented high resistance to antibiotics, in contrast to the reference strain INCQS 0230 that was more sensitive during the whole experimental process.
Several authors have reported that the microbial activity of AgNPs measuring 20–80 nm was attributed to the release of silver ions, whereas 10 nm AgNPs proved more toxic to E. coli due to the smaller size of the nanoparticles. This result was justified by the fact that the contact cell-to-particle was more efficient with AgNPs measuring 10 nm than those measuring 20–80 nm, leading to a higher intracellular bioavailability of silver.39–41 Morones et al42 identified that AgNPs act primarily in three ways against Gram-negative bacteria: 1) nanoparticles mainly in the range of 1–10 nm attach to the surface of the cell membrane and drastically disturb its proper functioning, such as permeability and respiration; 2) they are able to penetrate inside the bacteria and cause further damage by possibly interacting with sulfur- and phosphorus-containing compounds such as DNA; 3) nanoparticles release silver ions, which will have an additional contribution to the bactericidal effect of the AgNPs.
Although it is well known that silver, whether in an ionic or nanoparticle form, it is highly toxic to microorganisms,43–45 the mechanism of its action has not been fully elucidated. In the case of Gram-negative bacteria, a recent report demonstrated that AgNPs induce breakages of the outer membrane, thereby affecting the permeability, and these disruptions have been termed “pits”.46
In a study by Lara et al,47 there was no significant difference in the antimicrobial activity of AgNPs on the different groups evaluated (Gram-positive versus Gram-negative and resistant to antibiotics versus susceptible), suggesting that AgNPs have a broad-spectrum bactericidal effect. One of the mechanisms suggested to explain the action of AgNPs on Gram-negative bacteria is in binding to their cell membranes and increased permeability due to structural changes that would result in cell lysis.48
The results of hospital strains corroborate the results of tests conducted using antibiotics, which show widespread resistance. The results of this work demonstrate the potential of using AgNPs as an alternative to conventional antimicrobial agents that are currently used. The use of nanosilver with antibiotics may enhance the antimicrobial action.
Further studies should investigate the combined action of AgNPs and antibiotics against resistant hospital strains as an alternative therapy option in controlling infections.
Cytotoxicity
There is growing concern about the biological impacts of large-scale use of AgNPs and possible risks to the environment and human health. The size of the nanoparticles allows them to easily enter the cells of living organisms, which can lead to several cell lesions.49,50 Concerns on the potential cytotoxicity and genotoxicity of nanoparticles has increased significantly; therefore, these have been intensively studied.51–53
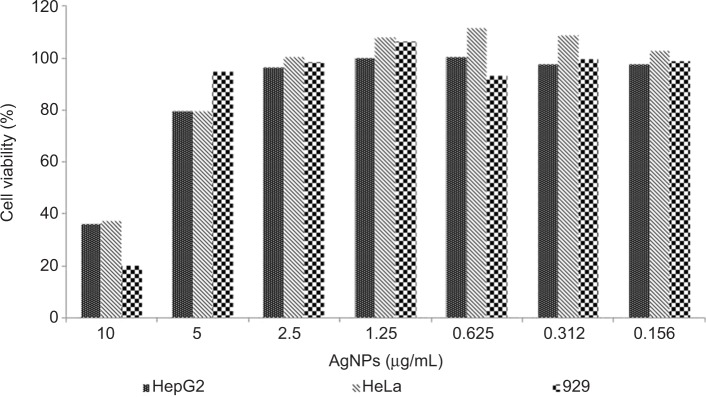
The absorbance values revealed after capture of the red dye and the respective concentrations of the AgNPs used in the cytotoxicity assays were treated by the 3T3 Phototox© program to determine the IC50 of the AgNP suspension in different cell lines. The results obtained showed that all cell lines presented similar IC50, approximately 7.0 µg/mL. These results indicate that the bactericidal AgNP concentrations were non-cytotoxic in NCTC 929, tumoral HepG2, and HeLa cells used in this study (Figure 5). A cytotoxic effect was demonstrated at 10 µg/mL AgNPs.
Figure 5.
Viability of NCTC 929, HeLa, and HepG2 cells after exposure to silver nanoparticles (AgNPs).
Notes: NCTC 929 (mouse fibroblasts), HeLa (human cervix adenocarcinoma), and HepG2 (human hepatocarcinoma) cells were pre-cultured for 24 hours, and then incubated at concentrations of 10, 5.0, 2.5, 1.25, 0.625, 0.312, and 0.156 µg/mL AgNPs (10 nm) for 24 hours. Cell viability was estimated by absorbance after uptake of neutral red.
Low toxicity to the tumoral cell lines HepG2 and HeLa was observed at 5.0 µg/mL – the concentration with the highest antimicrobial activity tested in this work. This result can indicate a certain degree of safety for lower concentrations. In the case of AgNP coating of medical devices, silver ions should be released slowly and the concentration of 5.0 µg/mL would probably never be reached.
Conclusion
The antimicrobial activity of silver is well known. In its nanometric form, this characteristic is accentuated. Due to their size, AgNPs can enter cells and inhibit enzymatic systems in the respiratory chain of some bacteria and thereby alter their DNA synthesis. In this work, three P. aeruginosa strains were evaluated against some antibiotics and 10 nm AgNPs at different concentrations. Different profiles of sensitivity to antibiotics and AgNPs were found among the strains tested. The suspension of AgNPs showed antimicrobial activity on the reference strain and against hospital strains that were resistant to the majority of antibiotics. These results support the importance of further studies on using nanosilver to control nosocomial infections caused by strains resistant to most antibiotics. Based on the results presented in this work concerning the action of commercial AgNPs, their use can be recommended as a good alternative for the control of microorganisms, with less risk of toxicity to mammalian cells. Further studies should investigate the combination of AgNPs and antibiotics against resistant hospital strains for the development of new materials and substances for medical application.
Acknowledgments
Financial support for this research was provided by the CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil) and the FIPT (Fundação do Instituto de Pesquisas Tecnológicas) by Novos Talentos scholarship. The authors thank the University of São Paulo – USP for academic support and the Institute for Technological Research – IPT for providing technical support.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bergogne-Bérézin E. Pseudomonas and miscellaneous Gram-negative bacilli. In: Cohen J, Powderly WG, editors. Infections Disease. 2nd ed. Philadelphia, PA: Mosby; 2004. pp. 2203–2217. [Google Scholar]
- 2.Pollack M. Pseudomonas aeruginosa. In: Mandell GL, Douglas RG Jr, Bernett JE, editors. Principles and Practice of Infectious Diseases. 5th ed. Philadelphia: Churchill Livingstone; 2000. pp. 2310–2317. [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Radzig MA, Nadtochenko VA, Koksharova OA, Kiwi J, Lipasova VA, Khmel IA. Antibacterial effects of silver nanoparticles on gram-negative bacteria: influence on the growth and biofilms formation, mechanisms of action. Colloids Surf B Biointerfaces. 2013;102:300–306. doi: 10.1016/j.colsurfb.2012.07.039. [DOI] [PubMed] [Google Scholar]
- 5.Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect. 2009;73(4):338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Mena KD, Gerba CP. Risk assessment of Pseudomonas aeruginosa in water. Rev Environ Contam Toxicol. 2009;201:71–115. doi: 10.1007/978-1-4419-0032-6_3. [DOI] [PubMed] [Google Scholar]
- 7.Iversen BG, Jacobsen T, Eriksen HM, et al. An outbreak of Pseudomonas aeruginosa infection caused by contaminated mouth swabs. Clin Infect Dis. 2007;44(6):794–801. doi: 10.1086/511644. [DOI] [PubMed] [Google Scholar]
- 8.Lanini S, D’Arezzo S, Puro V, et al. Molecular epidemiology of a Pseudomonas aeruginosa hospital outbreak driven by a contaminated disinfectant-soap dispenser. PLoS One. 2011;6(2):e17064. doi: 10.1371/journal.pone.0017064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiffer C, Hsiung A, Oplustil C, et al. MYSTIC Brazil Group Antimicrobial susceptibility of gram-negative bacteria in Brazilian hospitals: the MYSTYC Program Brazil 2003. Braz J Infect Dis. 2005;9(3):216–224. doi: 10.1590/s1413-86702005000300004. [DOI] [PubMed] [Google Scholar]
- 10.Andrade SS, Jones RN, Gales AC, Sader HS. Increasing prevalence of antimicrobial resistance among Pseudomonas aeruginosa isolates in Latin American medical centres: 5 year report of the SENTRY Antimicrobial Surveillance Program (1997–2001) J Antimicrob Chemother. 2003;52(1):140–141. doi: 10.1093/jac/dkg270. [DOI] [PubMed] [Google Scholar]
- 11.Zavascki AP, Goldani LZ, Gonçalves AL, Martins AF, Barth AL. High prevalence of metallo-beta-lactamase-mediated resistance challenging antimicrobial therapy against Pseudomonas aeruginosa in a Brazilian teaching hospital. Epidemiol Infect. 2007;135(2):343–345. doi: 10.1017/S0950268806006893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueiredo EA, Ramos H, Maciel MA, Vilar MC, Loureiro NG, Pereira RG. Pseudomonas aeruginosa: frequência de resistência a múltiplos fármacos e resistência cruzada entre antimicrobianos no Recife/PE [Pseudomonas aeruginosa: frequency of resistance to multiple drugs and cross resistance between antimicrobials in recife/PE] Rev Bras Ter Intensiva. 2007;19(4):421–427. Portuguese. [PubMed] [Google Scholar]
- 13.Gales AC, Torres PL, Vilarinho DS, Melo RS, Silva CF, Cereda RF. Carbapenem resistant Pseudomonas aeruginosa outbreak in an intensive care unit of a teaching hospital. Braz J Infect Dis. 2004;8(4):267–271. doi: 10.1590/s1413-86702004000400001. [DOI] [PubMed] [Google Scholar]
- 14.Menezes EA, Silveira LA, Cunha FA, et al. Perfil de resistência aos antimicrobianos de Pseudomonas isoladas no Hospital Geral de Fortaleza [Antimicrobials profile resistance from isolated pseudomonas at the Fortleza’s General Hospital] Rev Bras Anál Clín. 2003;35(4):177–180. Portuguese. [Google Scholar]
- 15.Dastjerdi R, Montazer M. A review on the application of inorganic nano-structured materials in the modification of textiles: focus on antimicrobial properties. Colloids Surf B Biointerfaces. 2010;79(1):5–18. doi: 10.1016/j.colsurfb.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 16.Yoon K, Hoon Byeon J, Park JH, Hwang J. Susceptibility constants of Escherichia coli and Bacillus subtilis to silver and copper nanoparticles. Sci Total Environ. 2007;373(2–3):572–575. doi: 10.1016/j.scitotenv.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Rai MK, Deshmukh SD, Ingle AP, Gade AK. Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. J Appl Microbiol. 2012;112(5):841–852. doi: 10.1111/j.1365-2672.2012.05253.x. [DOI] [PubMed] [Google Scholar]
- 18.Franci G, Falanga A, Galdiero S, et al. Silver nanoparticles as potential antibacterial agents. Molecules. 2015;20(5):8856–8874. doi: 10.3390/molecules20058856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salomoni R, Léo P, Rodrigues MFA. Antibacterial activity of silver nanoparticles (AgNPs) in Staphylococcus aureus and cytotoxicity effect in mammalian cells. Formatex Microbiol. 2015;5:851–857. [Google Scholar]
- 20.Brigger I, Dubernet C, Couvreur P. Nanoparticles in cancer therapy and diagnosis. Adv Drug Deliv Rev. 2002;54(5):631–651. doi: 10.1016/s0169-409x(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 21.Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 2009;27(1):76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Sudarenkov V. Nanotechnology: balancing benefits and risks to public health and the environment. Strasbourg: Council of Europe, Committee on Social Affairs, Health and Sustainable Development; 2013. [Google Scholar]
- 23.Nordberg GF, Fowler BA, Nordberg M, Friberg LT. Handbook on the Toxicology of Metals. 3rd ed. San Diego: Elsevier; 2007. [Google Scholar]
- 24.Marcone GP. Avaliação da ecotoxicidade de nanopartículas de dióxido de titânio e prata [Doutorado] [Assessment of ecotoxicity of nanoparticles of titatinium dioxide and silver] Campinas: UNICAMP; 2011. Portuguese. [Google Scholar]
- 25.Lansdown AB. Silver in health care: antimicrobial effects and safety in use. Curr Probl Dermatol. 2006;33:17–34. doi: 10.1159/000093928. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Han CM, Lin XW, Tang ZJ, Su SJ. Effect of silver nanoparticle dressing on second degree burn wound. Zhonghua Wai Ke Za Zhi. 2006;44(1):50–52. [PubMed] [Google Scholar]
- 27.Lee HY, Park HK, Lee YM, Kim K, Park SB. A practical procedure for producing silver nanocoated fabric and its antibacterial evaluation for biomedical applications. Chemic Communic (Camb) 2007;(28):2959–2961. doi: 10.1039/b703034g. [DOI] [PubMed] [Google Scholar]
- 28.Cohen MS, Stern JM, Vanni AJ, et al. In vitro analysis of a nanocrystalline silver-coated surgical mesh. Surg Infect (Larchmt) 2007;8(3):397–403. doi: 10.1089/sur.2006.032. [DOI] [PubMed] [Google Scholar]
- 29.McFarland J. The nephelometer: an instrument for media used for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. JAMA. 1907;49(14):1176–1178. [Google Scholar]
- 30.Bauer AW, Kirby EM, Sherris JC, Turck M. Antibiotic susceptibility testing by standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 31.Poletto KQ, Reis C. Antimicrobial susceptibility of the uropathogens in out patients in Goiania city, Goias state. Rev Soc Bras Med Trop. 2005;38(5):416–420. doi: 10.1590/s0037-86822005000500011. [DOI] [PubMed] [Google Scholar]
- 32.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing; 16th informational supplement. M100-S16. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- 33.International Organization for Standardization . ISO 10993-5, Biological evaluation of medical devices-part 5: tests for cytotoxicity: in vitro methods. Geneva: ISO; 1992. [Google Scholar]
- 34.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 2002;34(5):634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 35.McGowan JE., Jr Resistance in nonfermenting gram-negative bacteria: multidrug resistance to the maximum. Am J Infect Control. 2006;34(5 Suppl 1):S29–S37. doi: 10.1016/j.ajic.2006.05.226. discussion S64–S73. [DOI] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute . Publication M100-S21 Suggested Grouping of US-FDA Approved Antimicrobial Agents That Should Be Considered for Routine Testing and Reporting on Nonfastidious Organisms by Clinical Laboratories. 2011. [Google Scholar]
- 37.Perez F, Hujer MA, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brett DW. A discussion of silver as an antimicrobial agent: alleviating the confusion. Ostomy Wound Manage. 2006;52(1):34–41. [PubMed] [Google Scholar]
- 39.Hsiao IL, Hsieh YK, Wang CF, Chen IC, Huang YJ. Trojan-horse mechanism in the cellular uptake of silver nanoparticles verified by direct intra- and extracellular silver speciation analysis. Environ Sci Technol. 2015;49(6):3813–3821. doi: 10.1021/es504705p. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Sonshine DA, Shervani S, Hurt RH. Controlled release of biologically active silver from nanosilver surfaces. ACS Nano. 2010;4(11):6903–6913. doi: 10.1021/nn102272n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durán N, Durán M, de Jesus MB, Seabra AB, Fávaro WJ, Nakazato G. A new view on mechanistic aspects on antimicrobial activity. Nanomedicine. 2016;12(3):789–799. doi: 10.1016/j.nano.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Morones JR, Elechiguerra JL, Camacho A, et al. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16(10):2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 43.Schreurs WJ, Rosenberg H. Effect of silver ions on transport and retention of phosphate by Escherichia coli. J Bacteriol. 1982;152(1):7–13. doi: 10.1128/jb.152.1.7-13.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghandour W, Hubbard JA, Deistung J, Hughes MN, Poole RK. The uptake of silver ions by Escherichia coli K12: toxic effects and interaction with copper ion. Appl Microbiol Biotechnol. 1988;28(6):559–565. [Google Scholar]
- 45.Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J Biomed Mater Res. 2000;52(4):662–668. doi: 10.1002/1097-4636(20001215)52:4<662::aid-jbm10>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 46.Li WR, Xie XB, Shi QS, Zeng HY, OU-Yang YS, Chen YB. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl Microbiol Biotechnol. 2010;85(4):1115–1122. doi: 10.1007/s00253-009-2159-5. [DOI] [PubMed] [Google Scholar]
- 47.Lara HH, Ayala-Núñez NV, Turrent LD, Padilha CR. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J Microbiol Biotechnol. 2010;26(4):615–621. [Google Scholar]
- 48.Sondi I, Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J Coll Interf Sci. 2004;275(1):177–182. doi: 10.1016/j.jcis.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 49.Greulich C, Diendorf J, Simon T, Eggeler G, Epple M, Köller M. Uptake and intracellular distribution of silver nanoparticles in human mesenchymal stem cells. Acta Biomater. 2011;7(1):347–354. doi: 10.1016/j.actbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Liz R, Simard JC, Leonardi LB, Girard D. Silver nanoparticles rapidly induce atypical human neutrophil cell death by a process involving inflammatory caspases and reactive oxygen species and induce neutrophil extracellular traps release upon cell adhesion. Int Immunopharmacol. 2015;28(1):616–625. doi: 10.1016/j.intimp.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 51.Chairuangkitti P, Lawanprasert S, Roytrakul S, et al. Silver nanoparticles induce toxicity in A549 cells via ROS-dependent and ROS-independent pathways. Toxicol In Vitro. 2013;27(1):330–338. doi: 10.1016/j.tiv.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 52.Zhang T, Wang L, Chen Q, Chen C. Cytotoxic potential of silver nanoparticles. Yonsei Med J. 2014;55(2):283–291. doi: 10.3349/ymj.2014.55.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Składanowski M, Golinska P, Rudnicka K, Dahm H, Rai M. Evaluation of cytotoxicity, immune compatibility and antibacterial activity of biogenic silver nanoparticles. Med Microbiol Immunol. 2016;205(6):603–613. doi: 10.1007/s00430-016-0477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]