Abstract
Background:
The association of the WDR36 gene with glaucoma has been controversial in the literature. We therefore conducted a systematic review and meta-analysis to assess the association of all reported common polymorphisms in WDR36 with primary open angle glaucoma (POAG) and its subtypes: high tension glaucoma (HTG) and normal tension glaucoma (NTG).
Methods:
Publications in PUBMED and EMBASE databases up to March 9, 2016 were searched for case–control association studies of WDR36 with POAG, HTG, and/or NTG. Reported studies giving adequate genotype and/or allele information were included. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) of individual polymorphisms were estimated using the allelic model.
Results:
Our literature search yielded 122 records, among which 5 studies were eligible for meta-analysis, involving a total of 1352 POAG patients and 894 controls. Five WDR36 polymorphisms were meta-analyzed, rs11241095, rs10038177, rs17553936, rs13186912, and rs13153937. However, none of them was significantly associated with POAG, HTG, or NTG. The most-investigated polymorphisms, rs11241095 and rs10038177, had a pooled-OR of 1.09 (95% CI: 0.94–1.28, P = .25, I2 = 0) and 0.99 (95% CI: 0.71–1.39, P = .97, I2 = 77%), respectively, for POAG.
Conclusion:
The existing data in the literature do not support a significant role of WDR36 in the genetic susceptibility of POAG or its subtypes. Further replication studies in specific populations are warranted.
Keywords: genetics, glaucoma, meta-analysis, POAG, WDR36
1. Introduction
Glaucoma is a neurodegenerative disorder characterized by progressive damage of retinal ganglion cells, loss of optic nerve fibers, and visual field defect. It is a leading cause of irreversible blindness.[1] Primary open angle glaucoma (POAG) is the major form of primary glaucoma. According to the level of intraocular pressure (IOP), POAG can be classified into high tension glaucoma (HTG) and normal tension glaucoma (NTG).
POAG is a multifactorial disease involving both environmental and genetic factors.[2,3] To date, more than 20 loci have been linked to POAG, among which some causative genes were identified, including MYOC,[4]OPTN,[5] and WDR36.[6] In recent years, genome-wide association studies have led to the discovery of more susceptibility genes for POAG, such as CAV1/CAV2,[7]CDKN2B-AS1,[8] SIX1/SIX6,[9]ABCA1,[3,10]TXNRD2, ATXN2, and FOXC1.[11] The identification of these genes had very much enriched our understanding of the genetic architecture of POAG.
Among these genes, interestingly, the WDR36 gene has been reported as both causative and associated gene for POAG. The WDR36 gene was first identified as a causative gene for POAG at the GLC1G locus by using linkage analysis in 2 large Caucasian families.[6] Four mutations (N355S, A449T, R529Q, and D658G) in WDR36 were identified in approximately 6% of POAG patients.[6] However, conflict results were found in the studies on WDR36 mutations. The reported disease-causing variants were also detected in healthy subjects, which questioned the role of WDR36 mutations in POAG pathogenesis. Also, WDR36 variants showed inconsistent segregation with POAG in pedigrees, suggesting it may not be a disease-causing gene by itself.[12] Apart from mutations, common variants in the WDR36 gene have been assessed for association with POAG. Again, the results showed variable levels of involvement of WDR36 in POAG across different studies.[12–24] The previously reported major mutation D658G was found to be a neutral polymorphism and not associated with POAG in different Caucasian populations.[12–14] In contrast, several common variants were found to be associated with HTG in Asian populations.[16,19] Thus, the role of WDR36 variants in POAG remained inconclusive.
WDR36 is located on chromosomal region 5q22.1. It comprises 23 exons, encoding a protein of 951 amino acids. The WDR36 protein is a member of the WD repeat protein family, involving in a variety of cellular process including cell cycle progression, signal transduction, apoptosis, and gene regulation. WDR36 is ubiquitously expressed in different tissues, including a number of ocular tissues such as the lens, iris, sclera, ciliary body, trabecular meshwork, retina, and optic nerve.[6] After the identification of WDR36 as a disease gene for POAG, the pathogenesis of glaucoma due to WDR36 defects have been investigated. The glaucoma-associated WDR36 variants were found to encode functional defects in yeast model system. Variants of the stress inducible 1 conferred growth dysregulation in the context of mutations in the U 3 protein 21/WDR36 in yeast model.[25,26] A 3-bp amino-acid deletion at position 605–607 of mouse Wdr36, which corresponds to the location of the D658G mutation in human, led to the development of progressive retinal degeneration at the peripheral retina in mice with normal IOP, indicating that WDR36 mutations can cause retinal damage.[27] However, Gallenberger et al[28] found that heterozygote Wdr36-deficient mice model had no change in the number of optic nerve axons or susceptibility of retinal ganglion cells to excitotoxic damage, indicating that heterozygote Wdr36-deficient mice did not develop glaucoma, and that WDR36 might not play a causative role in the pathogenesis of POAG. Loss of Wdr36 function in mouse led to activation of the p53 stress–response pathway, suggesting that coinheritance of defects in the p53 pathway genes may influence the impact of Wdr36 variants.[29] Thus, WDR36 may act as a susceptibility gene for POAG, rather than a disease-causing gene.
Therefore, in order to assess the role of WDR36 as a susceptibility gene for POAG, we conducted a systematic review and meta-analysis to evaluate the associations of all reported WDR36 common single-nucleotide polymorphisms (SNPs) with POAG. Our results indicated that WDR36 does not play a major role in the genetic susceptibility of POAG.
2. Materials and methods
2.1. Literature search
A systematic literature search in the PubMed and Embase databases was conducted on March 9, 2016 to identify all published genetic studies on the association of WDR36 polymorphisms with POAG. We used MeSH terms and free words: (WD repeat domain 36 or WDR36) and (POAG or glaucoma, open angle or open angle glaucoma). All related articles were retrieved without language restriction.
2.2. Inclusion and exclusion criteria
A study was included if it fulfilled all of the following criteria: unrelated case–control study investigating the association between WDR36 and POAG; original research study, not a review, case report, editorial comment, or conference report; allele or genotype counts or frequencies of common SNPs (minor allele frequency >5%) in both the case and control groups being available from the articles; HTG being defined as reported IOP ≥ 21 mm Hg, NTG as reported IOP < 21 mm Hg, and POAG being defined as HTG and/or NTG; and unrelated control subjects being free of POAG. For studies that were published by the same group on the same gene and markers, only the one with the largest sample size was included.[30] Independent review was conducted by 2 reviewers (KL and WH). Resolution by a 3rd reviewer (JZ) was sought if there were any discrepancies.
2.3. Literature review and data extraction
Two reviewers (KL and WH) independently reviewed and extracted data from the retrieved articles. Any disagreement was resolved by a comprehensive reassessment until consensus was reached. The following data were extracted from each article: first author, year of publication, ethnicity of study subjects, study design, sample size, gender composition, mean age, disease subtype, allele counts of each SNP in the patients and controls, and Hardy–Weinberg equilibrium result in controls. The allelic counts, if not reported, were calculated from the genotype data or estimated by using the allelic frequencies and sample sizes if the genotype counts were not reported. The data were combined into 1 group as POAG if the allele and/or genotype data in HTG and NTG were reported separately.
2.4. Risk of bias assessment
The study quality was evaluated according to Newcastle Ottawa Scale (NOS) by 2 independent reviewers (KL and WH).[31] Further independent review and resolution by a 3rd researcher was sought for if discrepancies shown between the 2 reviewers. The NOS criteria included 3 categories: selection (from 0 to 4); comparability (from 0 to 2), and exposure (from 0 to 3). The total NOS scores ranged from 0 to 9. A study with less than 6 scores was considered as high risk in introducing bias.[32,33]
2.5. Data analysis
Meta-analysis for each polymorphism was performed if it has been reported in more than 2 studies. The genetic association was assessed using the allelic model. Based on the heterogeneity result tested by the I2 statistic,[34,35] the pooled odds ratio (OR) and 95% confidence interval (CI) of each SNP were estimated by using the fixed-effect or random-effect model. A fixed-effect model was adopted for the meta-analysis when I2 value ≤50%, which indicated low heterogeneity; while a random-effect model was adopted if the I2 value was >50%, which indicates a moderate to high heterogeneity.[36,37] Statistical analyses were performed by using the software Review Manager (RevMan, version 5.2, The Cochrane Collaboration, Copenhagen, Denmark). In the meta-analysis, an association was considered statistically significant if the summary P value was lower than 0.05. Sensitivity analysis was conducted to confirm the associations by sequentially removing the studies one at a time and recalculating the summary ORs.[38] Potential publication bias was evaluated by assessing the funnel plots in RevMan.
2.6. Ethical issues
Ethical approval is not required in this study because we collected and synthesized data from previous studies in which informed consent has already been obtained, and our study does not include confidential participant data and interventions.
3. Results
3.1. Eligible studies
A complete literature search was conducted and the flow of study inclusion was shown in Fig. 1. A total of 122 articles were identified by using the searching strategy, including 56 from PubMed and 66 from Embase. Among them, 94 were excluded because 52 were duplications, 25 were reviews, and 17 were about unrelated topics. We then retrieved the full texts of the remaining 28 records for review, and further excluded 23 articles, of which 7 were functional studies,[25,26,29,39–42] 7 were not case–control studies,[6,15,17,24,43–45] 4 did not provide allelic or genotype data,[12,13,18,23] 3 were animal studies,[27,28,46] and 2 were about rare variants with minor allele frequencies lower than 0.5%.[14,21] Finally, a total of 5 studies were included in the meta-analysis.[16,19,20,22,47]
Figure 1.
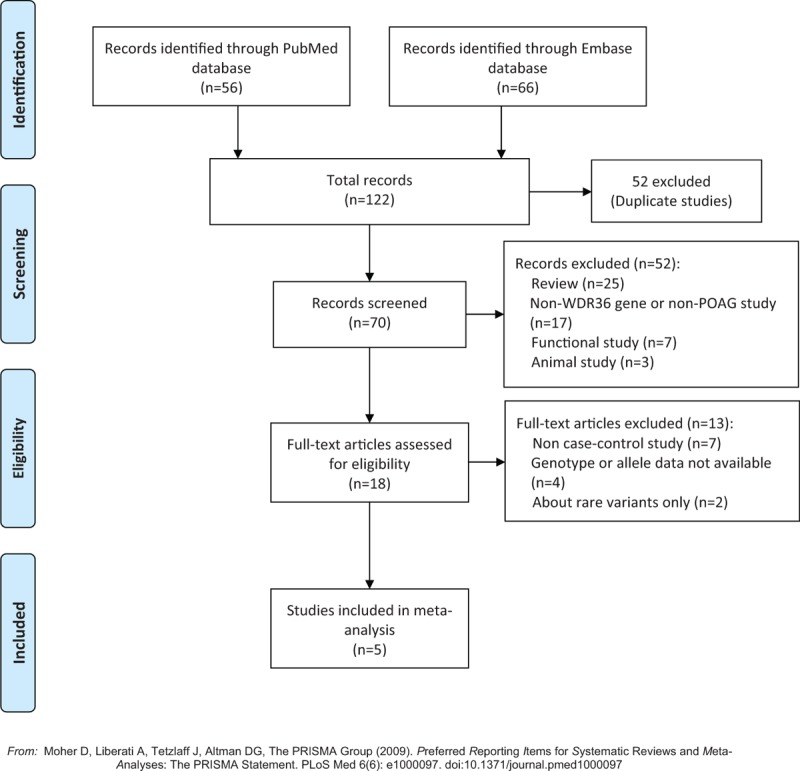
Flow diagram of literature search and study inclusion. The flow diagram depicts the screening process of retrieved articles, including the number and reason of exclusion. POAG = primary open angle glaucoma.
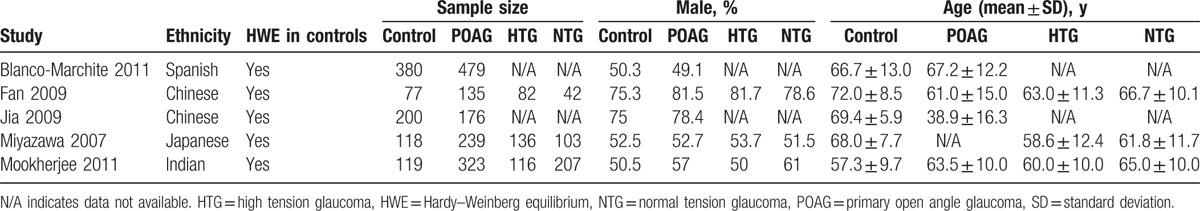
The main characteristics of included articles were showed in Table 1. This meta-analysis involved a total of 1352 POAG cases and 894 controls. The sample sizes varied from 135 to 479 among the patient groups and 77 to 380 among the control groups. The percentage of male ranged from 49.1% to 81.5% among the patient groups and from 50.3% to 75.3% among the control groups. The mean age varied from 38.9 to 67.2 years among the POAG groups and from 57.3 to 72 years among the control groups. Three studies were conducted in Asians,[16,19,20] 1 in East Indian,[22] and 1 in Caucasian.[47] Three studies included both HTG and NTG.[16,19,22] In the other 2 studies,[20,47] POAG was diagnosed with IOP higher than 21 mm Hg, so they were included in the HTG subgroup analysis. All of these 5 included studies were matched for ethnicity. The mean ages of the control groups were older than that of the POAG groups in 3 studies.[16,19,20] This is because older individuals were intentionally recruited as controls to reduce the chance of enrolling subjects with presymptomatic glaucoma. In the study of an East Indian cohort,[22] the mean age in the control group was younger than in the POAG group.
Table 1.
Characteristics of studies included in the meta-analysis.
3.2. Potential bias
None of the included studies reported deviation from Hardy–Weinberg equilibrium in the control subjects. Since controls in all the studies were recruited from those who attended the same clinic as the patients for other conditions, the NOS score achievable was 8 in 4 studies.[19,20,22,47] The lowest NOS score was 7 in 1 study,[16] because the same method of ascertainment for cases and controls was not reported. Except for rs10038177, there was no publication bias detected for the other 4 SNPs, with the funnel plots showing symmetry in comparisons. Since the sample size of Fan et al[19] was relatively small, the asymmetry in the funnel plot of rs10038177 suggested the possibility of publication bias or a systematic difference due to the small-study effect.
3.3. Meta-analysis of WDR36 polymorphisms in POAG
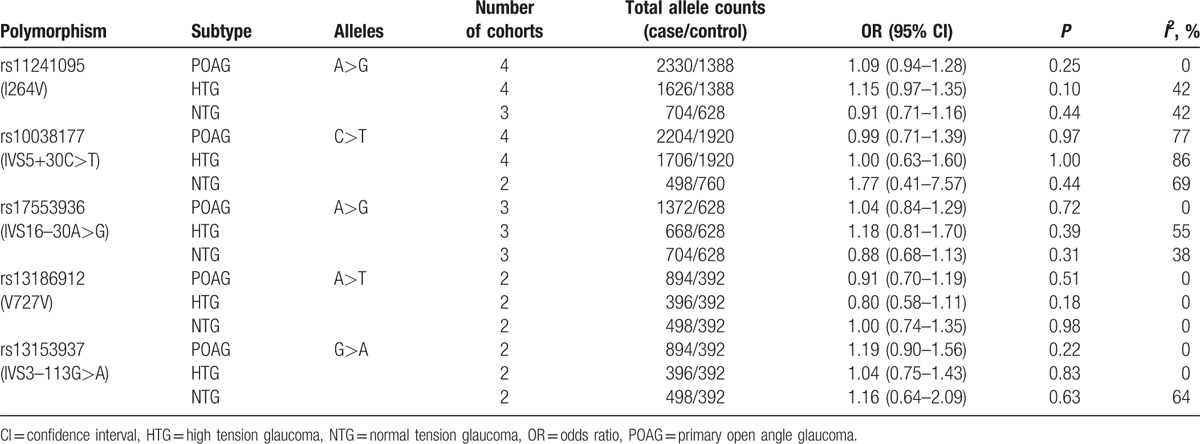
Five common SNPs in WDR36 (rs11241095, rs10038177, rs17553936, rs13186912, and rs13153937) have been reported in 2 or more studies and therefore eligible for the meta-analysis. The allelic associations of these 5 SNPs with POAG are summarized in Table 2. Among them, SNPs rs11241095 and rs10038177 were the most-frequently investigated.
Table 2.
Meta-analyses of allelic association of WDR36 polymorphisms with POAG, HTG, and NTG.
SNP rs11241095, a nonsynonymous variant – I264 V, has been reported in 4 studies involving a total of 1165 POAG cases and 694 controls.[16,19,22,47] The pooled results showed that there was no significant association between rs11241095 and POAG. The OR for the minor allele G in the allelic model was 1.09 (95% CI: 0.94–1.28, P = .25, I2 = 0, Fig. 2). In subgroup analyses, rs11241095 was reported in 4 studies with totally 813 HTG cases and 694 controls,[16,19,22,47] and in 3 studies involving totally 352 NTG cases and 314 controls.[16,19,22] The associations were not significant in HTG or NTG (P = .10, OR = 1.15, 95% CI: 0.97–1.35, I2 = 42% for HTG; and P = .44, OR = 0.91, 95% CI: 0.71–1.16, I2 = 42% for NTG; Fig. 2). Since the mean age of the controls was much younger than that of the cases in the East Indian cohort,[22] we excluded this study in the sensitivity analysis and found rs11241095 to be marginally associated with HTG (P = .03, OR = 1.22, 95% CI: 1.02–1.46, I2 = 31%), but not with POAG (P = .21, OR = 1.12, 95% CI: 0.94–1.34, I2 = 0) or NTG (P = .06, OR = 0.69, 95% CI: 0.46–1.02, I2 = 0).
Figure 2.
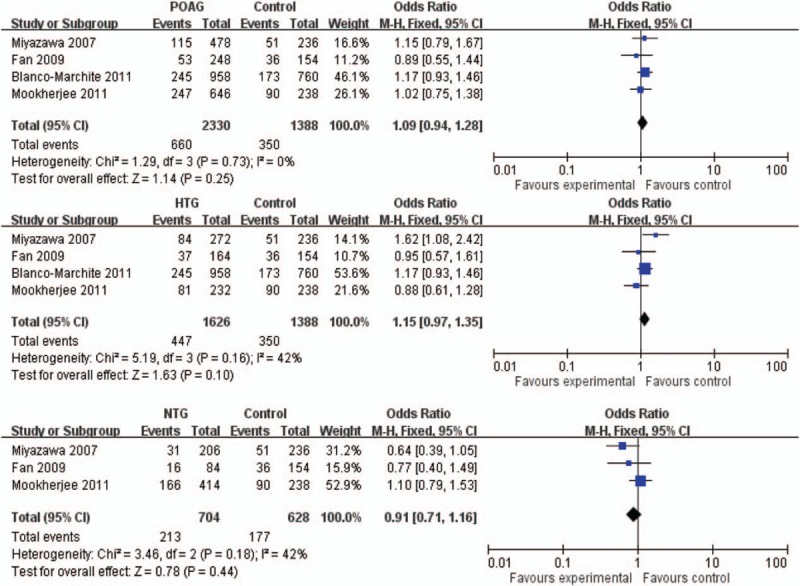
Forest plot of rs11241095 (G) in POAG, HTG, and NTG in allelic model. The bars with squares in the middle represent 95% CIs and ORs, respectively. The size of the square is proportional to the weight of the study. A diamond indicates the summary OR with its corresponding 95% CI. The central vertical solid line indicates the ORs for the null hypothesis. CI = confidence interval, HTG = high tension glaucoma, NTG = normal tension glaucoma, OR = odds ratio, POAG = primary open angle glaucoma.
SNP rs10038177 is located in the intronic region of WDR36. It was reported in 4 studies including a total of 1102 POAG cases and 960 controls.[19,20,22,47] This SNP was not associated with POAG (P = .97, OR = 0.99, 95% CI: 0.71–1.39, I2 = 77%; Fig. 3). In the subgroup analysis, rs10038177 was not associated with HTG or NTG (Fig. 3). In the sensitivity analysis, each of the studies was omitted 1 at a time and the pooled ORs were recalculated. Still no significant association was detected (data not shown).
Figure 3.
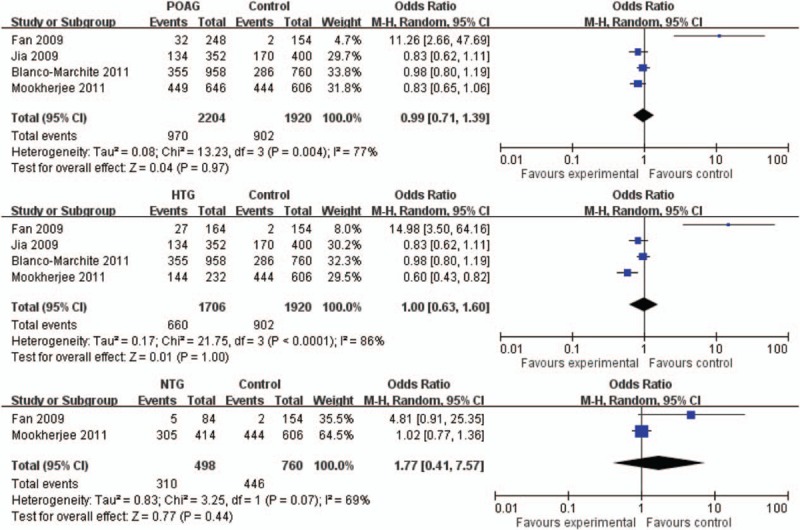
Forest plot of rs10038177 (T) in POAG, HTG, and NTG in allelic model. The bars with squares in the middle represent 95% CIs and ORs, respectively. The size of the square is proportional to the weight of the study. A diamond indicates the summary OR with its corresponding 95% CI. The central vertical solid line indicates the ORs for the null hypothesis. CI = confidence interval, HTG = high tension glaucoma, NTG = normal tension glaucoma, OR = odds ratio, POAG = primary open angle glaucoma.
Regarding the other 3 SNPs, rs17553936 (IVS16–30A>G), rs13153937 (IVS3–113G>A), and rs13186912 (V727V), which are intronic or synonymous polymorphisms, they were not significantly associated with POAG, HTG, or NTG (Table 2). Sensitivity analyses showed no significant change after excluding any study individually.
4. Discussion
In this study, we have conducted a systematic review and meta-analysis to summarize the genetic association of the WDR36 gene with POAG reported in candidate gene studies. We identified 5 eligible studies, in which the associations of 5 WDR36 SNPs (rs11241095, rs10038177, rs17553936, rs13186912, and rs13153937) with POAG, HTG, and NTG among different ethnic groups were assessed. We found no significant association between the reported WDR36 SNPs and POAG, HTG, or NTG.
The SNP rs11241095 (I264V) is a common variant in WDR36, conserved in 4 species. It is located in the 2nd G-beta WD40 repeat and the Cytochrome cd1-nitrite reductase-like, COOH-terminal haem d1 domain. Thus, this variant could be an important part of a WDR36 functional domain. In our meta-analysis, we found no significant association between rs11241095 and POAG. In a study in Japanese, rs11241095 was associated with HTG (OR = 1.92, 95% CI: 1.17–3.18 in the dominant model), but not with POAG or NTG.[16] This suggested that the association of rs11241095 with HTG may be subtype-specific or population-specific. However, as shown by the subgroup analysis in our meta-analysis, rs11241095 was not significantly associated with HTG or NTG, although there was a marginal association with HTG in the sensitivity analysis when the East Indian cohort was excluded (P = .03). Also, when we excluded both the Caucasian and Indian cohorts in meta-analysis to assess its effect in East Asians (including 1 Chinese cohort and 1 Japanese cohort), we found no significant association between rs11241095 and HTG (P = .08, OR = 1.33, 95% CI: 0.97–1.83). Of note, the ORs of the minor allele G were toward opposite directions between Chinese and Japanese (Fig. 2). Therefore, our findings did not support the role of the WDR36 SNP rs11241095 as a susceptibility marker for POAG, HTG, or NTG.
Reported associations of rs10038177 with POAG were controversial. In the study of Fan et al,[19] it was associated with HTG (P = 7.9 × 10–7, OR = 15) in a Taiwan Chinese cohort, with a higher frequency of the minor allele T in HTG (16.5%) than in controls (1.3%). However, this finding was not replicated in a northern Chinese cohort,[20] in which the minor allele frequency was higher (38.1% in HTG and 42.5% in controls), and the OR was toward the opposite direction. In HapMap, the frequency of rs10038177-T ranges from 37.5% to 76.8% among different populations (HapMap data release No.27), which is also higher than that of the control group in the study of Fan et al[19] (1.3%). Therefore, in view of the limited sample size (82 HTG patients and 77 controls) in the study of Fan et al,[19] the association should be replicated in larger Taiwan Chinese samples. Interestingly, a strong association of rs10038177 with HTG was also reported in an East Indian cohort.[22] However, the C allele was the minor allele, which conferred a risk effect on HTG (P = 1.4 × 10–4, OR = 2.19). Again, confirmation of this association is warranted in the East Indian population. Nevertheless, in our meta-analysis, where the effect of the T allele was summarized, we found no significant association between rs10038177 and POAG, HTG, or NTG. Of note, there was high interstudy heterogeneity. We therefore adopted a random-effect model to generate more conservative results, and conducted sensitivity analysis and subgroup analysis, but still we did not find any significant association. Such heterogeneity could be caused by the opposite effect in the Taiwan Chinese cohort[19] compared with other cohorts. Such drastic difference could be generated by chance due to small sample size. But it could also be driven by population-specific effect. For example, the effect size of SNP rs4236601 in the CAV1/CAV2 locus for POAG was much higher in Chinese (OR > 5) than in Caucasians (OR < 1.5).[7] Even within the Chinese population, the effects of this SNP were different.[48] Another example is about the SNP rs1061170 for age-related macular degeneration (AMD). In a Taiwan Chinese cohort, the frequency of the allele C of rs1061170 was 11.3% in AMD compared with 2.8% in controls (P < .00001).[49] In contrast, in a Hong Kong Chinese cohort the C allele presented in 5.8% of AMD and 3.9% of controls (P > .05).[50] Thus, replication studies are warranted to assess the SNP rs10038177 in HTG among Taiwan Chinese.
This systematic review and meta-analysis provides an overview of published candidate gene studies on WDR36 in POAG, HTG, and NTG. However, there are some limitations in this study. First, the number of eligible studies, and the study populations for certain SNPs and disease subtypes were limited. In particular, since the allelic data were available in only 1 Caucasian cohort, stratified analysis of different ethnicities was not performed. Therefore, the conclusions are powerful enough for specific populations. Notably, in the era of genomic study, genome-wide association studies, which involve thousands of samples, have provided a powerful platform for the gene discovery of POAG. Interestingly, the WDR36 gene was not pinpointed in any of the POAG GWAS. There are several possible explanations. First, in GWAS only the SNPs that had a P value passing a certain threshold (usually <10–5) would be subjected to replication, and the WDR36 SNPs might not among the top SNPs. In a recent large scale study by Bailey et al,[11] all variants with P < 1 × 10–5 in the NEIGHBORHOOD POAG cohort have been reported, but none of the WDR36 SNPs included in our meta-analysis was among the top SNPs. Second, comparing with GWAS, candidate gene studies usually included more stringently diagnosed patients and healthy controls, providing a relatively higher power to detect the association of candidate SNPs. This has been reflected in the NOS scores of the included studies (all >7), which assessed the ascertainment of subjects. Third, the positive findings in candidate gene studies of WDR36 could have been overestimated due to winner's curse when the statistical power of the original study is not sufficient. Therefore, meta-analysis of all reports on the same SNP(s) would be desirable to confirm the association by increasing the statistical power. Interestingly, by reviewing the POAG GWAS papers, including the supplementary materials, we found in the GWAS of Osman et al,[51] the associations of SNPs in MYOC, OPTN, and WDR36 were assessed, but no SNP revealed significant association with POAG (P > .05). This provides support to our claim that WDR36 does not play a major role in the genetic susceptibility of POAG. The 2nd limitation is that there were high interstudy heterogeneities of certain SNPs. This could be caused by the small sample sizes in some studies and/or different ethnicities being studied, which has been discussed in details above. Finally, only common SNPs were taken into account. The role of rare variants and disease-causing mutations is out of the scope of this study.
In summary, this systematic review and meta-analysis provided an overview of reported common WDR36 SNPs in POAG. Our results do not support WDR36 as a major susceptibility gene for POAG, HTG, or NTG. Further confirmation of the role of WDR36 in POAG among specific populations should be warranted.
Acknowledgments
The authors thank Shenzhen Science and Technology Innovation Committee, China (JZ; GJHZ20160229170608241), and the Medical Scientific and Technology Research Foundation of Guangdong Province, China (WH; A2014518) for the support.
Footnotes
Abbreviations: CI = confidence interval, HTG = high tension glaucoma, IOP = intraocular pressure, NOS = Newcastle Ottawa Scale, NTG = normal tension glaucoma, OR = odds ratio, POAG = primary open angle glaucoma, SNP = single-nucleotide polymorphism.
KL and WH are co-first authors.
Funding/support: This work was supported partially by the research grants from the Shenzhen Science and Technology Innovation Committee, China (JZ; GJHZ20160229170608241), and the Medical Scientific and Technology Research Foundation of Guangdong Province, China (WH; A2014518).
The authors have no conflicts of interest to disclose.
References
- [1].Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol 2006;90:262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li Z, Allingham RR, Nakano M, et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum Mol Genet 2015;24:3880–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen Y, Lin Y, Vithana EN, et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet 2014;46:1115–9. [DOI] [PubMed] [Google Scholar]
- [4].Stone EM, Fingert JH, Alward WL, et al. Identification of a gene that causes primary open angle glaucoma. Science 1997;275:668–70. [DOI] [PubMed] [Google Scholar]
- [5].Rezaie T, Child A, Hitchings R, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 2002;295:1077–9. [DOI] [PubMed] [Google Scholar]
- [6].Monemi S, Spaeth G, DaSilva A, et al. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet 2005;14:725–33. [DOI] [PubMed] [Google Scholar]
- [7].Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet 2010;42:906–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet 2011;43:574–8. [DOI] [PubMed] [Google Scholar]
- [9].Wiggs JL, Yaspan BL, Hauser MA, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet 2012;8:e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hysi PG, Cheng CY, Springelkamp H, et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet 2014;46:1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bailey JN, Loomis SJ, Kang JH, et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet 2016;48:189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hauser MA, Allingham RR, Linkroum K, et al. Distribution of WDR36 DNA sequence variants in patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci 2006;47:2542–6. [DOI] [PubMed] [Google Scholar]
- [13].Fingert JH, Alward WL, Kwon YH, et al. No association between variations in the WDR36 gene and primary open-angle glaucoma. Arch Ophthalmol 2007;125:434–6. [DOI] [PubMed] [Google Scholar]
- [14].Hewitt AW, Dimasi DP, Mackey DA, et al. A Glaucoma Case-control Study of the WDR36 Gene D658G sequence variant. Am J Ophthalmol 2006;142:324–5. [DOI] [PubMed] [Google Scholar]
- [15].Kramer PL, Samples JR, Monemi S, et al. The role of the WDR36 gene on chromosome 5q22.1 in a large family with primary open-angle glaucoma mapped to this region. Arch Ophthalmol 2006;124:1328–31. [DOI] [PubMed] [Google Scholar]
- [16].Miyazawa A, Fuse N, Mengkegale M, et al. Association between primary open-angle glaucoma and WDR36 DNA sequence variants in Japanese. Mol Vis 2007;13:1912–9. [PubMed] [Google Scholar]
- [17].Weisschuh N, Wolf C, Wissinger B, et al. Variations in the WDR36 gene in German patients with normal tension glaucoma. Mol Vis 2007;13:724–9. [PMC free article] [PubMed] [Google Scholar]
- [18].Pasutto F, Mardin CY, Michels-Rautenstrauss K, et al. Profiling of WDR36 missense variants in German patients with glaucoma. Invest Ophthalmol Vis Sci 2008;49:270–4. [DOI] [PubMed] [Google Scholar]
- [19].Fan BJ, Wang DY, Cheng CY, et al. Different WDR36 mutation pattern in Chinese patients with primary open-angle glaucoma. Mol Vis 2009;15:646–53. [PMC free article] [PubMed] [Google Scholar]
- [20].Jia LY, Tam PO, Chiang SW, et al. Multiple gene polymorphisms analysis revealed a different profile of genetic polymorphisms of primary open-angle glaucoma in northern Chinese. Mol Vis 2009;15:89–98. [PMC free article] [PubMed] [Google Scholar]
- [21].Ramdas WD, van Koolwijk LM, Cree AJ, et al. Clinical implications of old and new genes for open-angle glaucoma. Ophthalmology 2011;118:2389–97. [DOI] [PubMed] [Google Scholar]
- [22].Mookherjee S, Chakraborty S, Vishal M, et al. WDR36 variants in East Indian primary open-angle glaucoma patients. Mol Vis 2011;17:2618–27. [PMC free article] [PubMed] [Google Scholar]
- [23].Williams SE, Carmichael TR, Allingham RR, et al. The genetics of POAG in black South Africans: a candidate gene association study. Sci Rep 2015;5:8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Frezzotti P, Pescucci C, Papa FT, et al. Association between primary open-angle glaucoma (POAG) and WDR36 sequence variance in Italian families affected by POAG. Br J Ophthalmol 2011;95:624–6. [DOI] [PubMed] [Google Scholar]
- [25].Footz T, Dubois S, Sarfarazi M, et al. Co-variation of STI1 and WDR36/UTP21 alters cell proliferation in a glaucoma model. Mol Vis 2011;17:1957–69. [PMC free article] [PubMed] [Google Scholar]
- [26].Footz TK, Johnson JL, Dubois S, et al. Glaucoma-associated WDR36 variants encode functional defects in a yeast model system. Hum Mol Genet 2009;18:1276–87. [DOI] [PubMed] [Google Scholar]
- [27].Chi ZL, Yasumoto F, Sergeev Y, et al. Mutant WDR36 directly affects axon growth of retinal ganglion cells leading to progressive retinal degeneration in mice. Hum Mol Genet 2010;19:3806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gallenberger M, Kroeber M, Marz L, et al. Heterozygote Wdr36-deficient mice do not develop glaucoma. Exp Eye Res 2014;128:83–91. [DOI] [PubMed] [Google Scholar]
- [29].Skarie JM, Link BA. The primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Hum Mol Genet 2008;17:2474–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ma L, Tang FY, Chu WK, et al. Association of toll-like receptor 3 polymorphism rs3775291 with age-related macular degeneration: a systematic review and meta-analysis. Sci Rep 2016;6:19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [32].McPheeters ML, Kripalani S, Peterson NB, et al. Closing the quality gap: revisiting the state of the science (vol. 3: quality improvement interventions to address health disparities). Evid Rep Technol Assess (Full Rep) 2012;1–475. [PMC free article] [PubMed] [Google Scholar]
- [33].Rong SS, Tang FY, Chu WK, et al. Genetic associations of primary angle-closure disease: a systematic review and meta-analysis. Ophthalmology 2016;6:1211–21. [DOI] [PubMed] [Google Scholar]
- [34].Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med 1997;127:820–6. [DOI] [PubMed] [Google Scholar]
- [35].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [36].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ma L, Li Z, Liu K, et al. Association of genetic variants with polypoidal choroidal vasculopathy: a systematic review and updated meta-analysis. Ophthalmology 2015;122:1854–65. [DOI] [PubMed] [Google Scholar]
- [38].Thakkinstian A, McKay GJ, McEvoy M, et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol 2011;173:1365–79. [DOI] [PubMed] [Google Scholar]
- [39].Jia LY, Gong B, Pang CP, et al. Correction of the disease phenotype of myocilin-causing glaucoma by a natural osmolyte. Invest Ophthalmol Vis Sci 2009;50:3743–9. [DOI] [PubMed] [Google Scholar]
- [40].Rozsa FW, Scott K, Pawar H, et al. Effects of timolol on MYOC, OPTN, and WDR36 RNA levels. Arch Ophthalmol 2008;126:86–93. [DOI] [PubMed] [Google Scholar]
- [41].Gallenberger M, Meinel DM, Kroeber M, et al. Lack of WDR36 leads to preimplantation embryonic lethality in mice and delays the formation of small subunit ribosomal RNA in human cells in vitro. Hum Mol Genet 2011;20:422–35. [DOI] [PubMed] [Google Scholar]
- [42].Tenge VR, Knowles J, Johnson JL. The ribosomal biogenesis protein Utp21 interacts with Hsp90 and has differing requirements for Hsp90-associated proteins. PLoS One 2014;9:e92569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu T, He X. Mapping the disease-causative gene for a Chinese pedigree with primary open angle glaucoma. Chin Ophthalmic Res 2009;27:401–6. [Google Scholar]
- [44].Huang X, Li M, Guo X, et al. Mutation analysis of seven known glaucoma-associated genes in Chinese patients with glaucoma. Invest Ophthalmol Vis Sci 2014;55:3594–602. [DOI] [PubMed] [Google Scholar]
- [45].Patel HY, Richards AJ, De Karolyi B, et al. Screening glaucoma genes in adult glaucoma suggests a multiallelic contribution of CYP1B1 to open-angle glaucoma phenotypes. Clin Experiment Ophthalmol 2012;40:e208–17. [DOI] [PubMed] [Google Scholar]
- [46].Howell GR, Libby RT, John SW. Mouse genetic models: an ideal system for understanding glaucomatous neurodegeneration and neuroprotection. Prog Brain Res 2008;173:303–21. [DOI] [PubMed] [Google Scholar]
- [47].Blanco-Marchite C, Sanchez-Sanchez F, Lopez-Garrido MP, et al. WDR36 and P53 gene variants and susceptibility to primary open-angle glaucoma: analysis of gene-gene interactions. Invest Ophthalmol Vis Sci 2011;52:8467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rong SS, Chen LJ, Leung CK, et al. Ethnic specific association of the CAV1/CAV2 locus with primary open-angle glaucoma. Sci Rep 2016;6:27837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lau LI, Chen SJ, Cheng CY, et al. Association of the Y402H polymorphism in complement factor H gene and neovascular age-related macular degeneration in Chinese patients. Invest Ophthalmol Vis Sci 2006;47:3242–6. [DOI] [PubMed] [Google Scholar]
- [50].Chen LJ, Liu DT, Tam PO, et al. Association of complement factor H polymorphisms with exudative age-related macular degeneration. Mol Vis 2006;12:1536–42. [PubMed] [Google Scholar]
- [51].Osman W, Low SK, Takahashi A, et al. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum Mol Genet 2012;21:2836–42. [DOI] [PubMed] [Google Scholar]


