Abstract
Genome rearrangements result in mutations that underlie many human diseases, and ongoing genome instability likely contributes to the development of many cancers. The tools for studying genome instability in mammalian cells are limited, whereas model organisms such as Saccharomyces cerevisiae are more amenable to these studies. Here, we discuss the many genetic assays developed to measure the rate of occurrence of Gross Chromosomal Rearrangements (called GCRs) in S. cerevisiae. These genetic assays have been used to identify many types of GCRs, including translocations, interstitial deletions, and broken chromosomes healed by de novo telomere addition, and have identified genes that act in the suppression and formation of GCRs. Insights from these studies have contributed to the understanding of pathways and mechanisms that suppress genome instability and how these pathways cooperate with each other. Integrated models for the formation and suppression of GCRs are discussed.
Keywords: DNA repair, DNA replication, genome rearrangements, telomerase, translocations
Gross Chromosomal Rearrangements (GCRs)
Genome stability is critical for cell survival and normal cell growth. Genomic rearrangements (herein called GCRs) include translocations, deletions, and amplifications. GCRs are associated with many human diseases including, but not limited to, cancers. The association of GCRs with different diseases has driven interest in how GCRs arise and are normally prevented. Remarkably, eukaryotic genomes are normally quite stable, despite the fact that they include many features that are at risk for causing the formation of GCRs, including duplicated sequences and double-strand break (DSB)-inducing sites (Gordenin and Resnick 1998; Lambert et al. 2005; Lemoine et al. 2005; Casper et al. 2009; Mizuno et al. 2009; Paek et al. 2009; Aksenova et al. 2013; Song et al. 2014).
A wide variety of GCRs have been observed in mammalian cancers (Inaki and Liu 2012; Janssen and Medema 2013; Macintyre et al. 2016). The most common cancers, excepting leukemias and lymphomas, often have large numbers of GCRs (Mitelman et al. 2006, 2007; Gordon et al. 2012; Cancer Genome Atlas Research Network et al. 2013) as well as ongoing genome instability (Nowell 1976; Campbell et al. 2010; Gundem et al. 2015; Gibson et al. 2016; Uchi et al. 2016). Many of the genes that are defective in inherited cancer susceptibility syndromes act in DNA damage response pathways (Friedberg et al. 2006; Ciccia and Elledge 2010), and these pathways suppress GCRs in the model organism Saccharomyces cerevisiae (Chen and Kolodner 1999; Myung et al. 2001a,b,c). Thus, both inherited and sporadic cancers may have genetic or epigenetic defects that destabilize their genomes.
Despite the considerable interest in studying genome instability in higher eukaryotes, the lack of facile genetic systems has limited progress in these organisms. In contrast, the conservation of DNA metabolism pathways has allowed experimental insights from more genetically tractable model systems to be applied to human diseases. Early S. cerevisiae studies identified rearrangements mediated by repetitive genomic features, including the ribosomal DNA array, CUP1 repeats, tRNA genes, Ty retrotransposon-related elements, and the 94 kb “Hawthorne” deletion between the homologous MATa and HMR loci (Hawthorne 1963; Rothstein 1979; Roeder and Fink 1980; Liebman et al. 1981; Rothstein et al. 1987; Christman et al. 1988; Keil and McWilliams 1993). At the same time, genome features designed to drive the formation of GCRs were engineered into normal S. cerevisiae chromosomes, demonstrating that GCRs could be observed (Mikus and Petes 1982; Sugawara and Szostak 1983; Haber and Thorburn 1984; Surosky and Tye 1985; Jinks-Robertson and Petes 1986; Kupiec and Petes 1988; Gordenin et al. 1993; Henderson and Petes 1993). In the last 15–20 years, considerable progress has been made in developing assays for detecting GCRs and structurally characterizing these GCRs, which has provided insights into both GCR-formation and GCR-suppression mechanisms. This article reviews our current understanding of GCRs in S. cerevisiae. We predict that the extensive knowledge that has accumulated in these areas should greatly facilitate the study of genome instability in higher eukaryotes.
How GCRs Arise
Based on the evidence described below, our current view is that GCRs are generated through normal DNA repair and homeostasis processes that act on some form of DNA damage but do so inappropriately. In these cases, the original sequence and structure of the genome are not restored. Importantly, GCRs are not damaged chromosomes themselves, but rather are the result of error-prone processing of damaged chromosomes. Most of the GCRs recovered in genetic assays appear to be stable, even when the GCR has undergone multiple rounds of rearrangement to reach its final structure. The stability of recovered GCRs is not surprising given that GCRs occur at low rates and are identified by plating cells on medium that selects for the presence of a GCR; in such selective medium, other rearranged chromosomes and chromosome fragments that are not under selection are likely lost due to segregation during the > 20 cell divisions required to form a S. cerevisiae colony from a single cell (Joseph and Hall 2004).
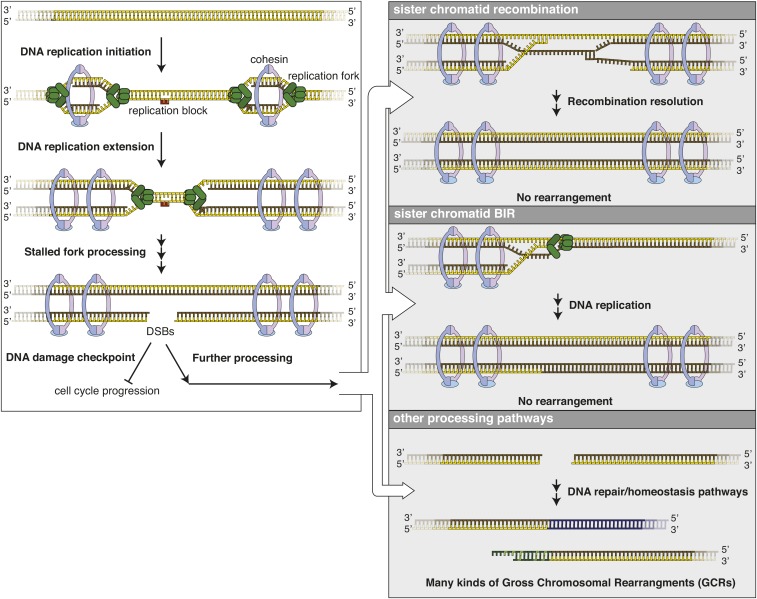
In most cases of spontaneous GCRs, the precise nature of the initiating damage is unknown; however, much of the genetic evidence described below strongly implicates DNA replication errors as an important but probably not the exclusive source of the broken chromosomes that result in GCRs (Figure 1). Replication errors could occur when replication encounters templates that are difficult to copy such as: (1) damaged DNA, including oxidatively damaged DNA; (2) difficult-to-replicate sequences, such as inverted repeats that can form a palindrome or interstitial telomere sequences; and (3) a block on the template, such as a bound protein or a transcriptional intermediate like a stable three-stranded RNA–DNA hybrid (R-loop) (Lambert et al. 2005; Lemoine et al. 2005; Casper et al. 2009; Mizuno et al. 2009; Paek et al. 2009; Aksenova et al. 2013; Song et al. 2014; Santos-Pereira and Aguilera 2015). These interactions potentially result in stalled replication forks, which are thought to be unstable, or structures like extruded palindromes that can be cleaved to generate DSBs. In some cases, regression of stalled forks may be linked to a restart mechanism involving template switching; these events likely prevent the formation of substrates that can lead to GCRs. In other cases, replication of nicked substrates or the action of nucleases and/or helicases might lead to replication fork collapse and the formation of DSBs (Figure 1) (Flores-Rozas and Kolodner 2000; Michel 2000). Replication can also misincorporate ribonucleotides that are then cleaved by topoisomerase I to produce aberrant DNA structures (Kim et al. 2011; Williams et al. 2013; Allen-Soltero et al. 2014). Other potential sources of damage include resection from deprotected telomeres and breakage of dicentric chromosomes formed by end-to-end fusion of chromosomes in strains with defects in telomere maintenance (Lydall and Weinert 1995; Craven et al. 2002; Maringele and Lydall 2002; Pennaneach and Kolodner 2004). Analysis of the structure of > 1000 GCR structures indicates that GCRs can be formed by mechanisms modeled on the assumption that the initiating damage is a DSB (Putnam et al. 2004, 2005, 2014; Pennaneach and Kolodner 2009; Chan and Kolodner 2012). Thus, for simplicity, we show DSBs as the initiating damage in this review, but we note that DSBs, if they are involved, are likely the result of processing more complicated forms of initiating DNA damage.
Figure 1.
A model for the formation of replication error-induced GCRs. Left: bidirectional DNA replication is initiated from origins, and sister chromatids are kept associated through the action of cohesins. DNA replication can be stalled by a variety of blocks on DNA, including DNA damage, DNA-binding proteins, transcriptional machinery, and R-loops. Processing of these stalled forks can give rise to DSBs. Right: at least some replication errors are probably repaired by recombination with the sister chromatid or BIR using the sister chromatid as template, and do not generate GCRs. Replication errors that are repaired by other DSB-processing pathways can give rise to a wide variety of GCRs depending on the nature of the damage, the genomic position of the damage, and the relative efficiency of competing DNA repair and homeostasis pathways. Note that other sources of DNA damage likely lead to the formation of GCRs, and mechanisms other than sister chromatid recombination can suppress the formation of GCRs. BIR, break-induced replication; DSBs, double-strand breaks; GCRs, Gross Chromosomal Rearrangements.
The most likely outcome for any DSB during the S- or G2-phases of the cell cycle is the initiation of homologous recombination (HR) with the sister chromatid (or a homologous chromosome in a diploid) to repair the DSB, or in some cases help rebuild replication forks (Figure 1). The result of this processing suppresses any genomic rearrangements and conserves the overall genome structure. Crucial players in this “conservative” repair reaction likely include proteins that mediate HR, sister chromatid cohesion, and DNA damage checkpoint signaling.
When sister chromatid recombination does not occur, then several “nonconservative” repair reactions can compete for these DSBs and lead to the formation of GCRs (Figure 1). DSBs can be repaired by different pathways, and in many cases, the intermediates formed in one repair pathway are substrates for other pathways. Thus, a single form of initiating damage can result in multiple types of GCRs (Figure 2), including terminal deletions healed by the addition of a de novo telomere, intra- or interchromosomal translocations, formation of hairpin-mediated inversions, and fusions to the telomeres of other chromosomes. The number of categories of GCRs shown in Figure 2 increases if additional factors are considered, including whether the initial rearrangement leads to chromosomes with one or two centromeres and whether additional rearrangements occur (described in detail below). Any observed GCR is the result of the individual steps that tend to be fastest during processing of the damage, given the nature of the damage, the chromosomal sequence context for the damage, the presence or absence of defects in specific DNA-processing pathways, and the idiosyncratic history of that particular event. Despite this complexity, the rates at which GCRs accumulate and the spectrum of GCR structures observed in the presence of different genetic defects have provided substantial insights into the mechanisms that form and prevent GCRs.
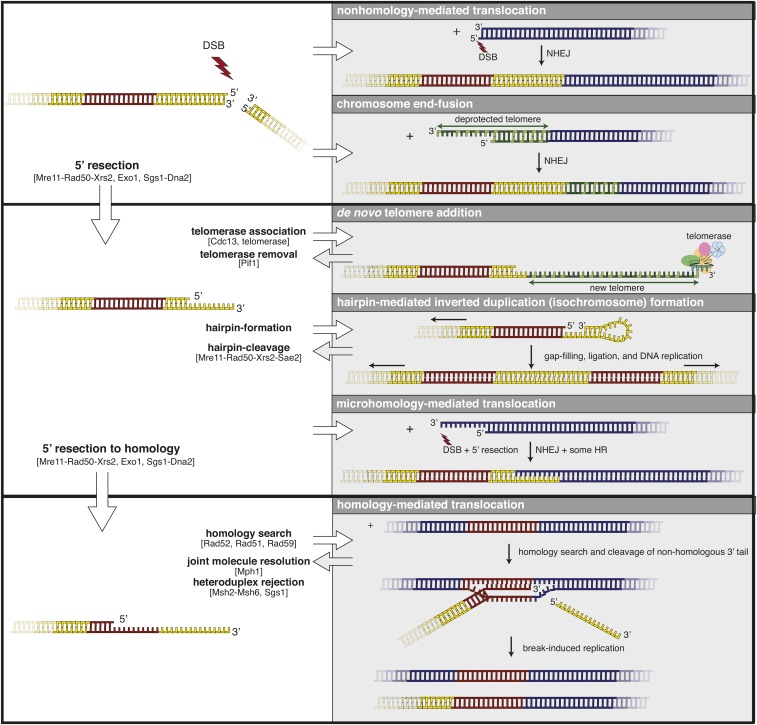
Figure 2.
DSBs can be acted on by a variety of DNA metabolic processes that can generate different kinds of GCRs. Top: DSBs lead to nonhomology-mediated translocations mediated by NHEJ and, in strains containing deprotected telomeres (green bases), can lead to dicentric chromosomes mediated by end-to-end fusions with other chromosomes (blue). Middle: 5′ resection of DSBs, which in mitotic cells is driven by a combination of Mre11-Rad50-Xrs2, Exo1, and Sgs1-Dna2, generates a 3′ overhang that is subject to several reactions. Cdc13 can recognize TG-rich ssDNA regions and promote the association of telomerase leading to synthesis of a de novo telomere; this association is antagonized by the activity of the Pif1 helicase. Overhang regions not bound by RPA can form hairpins. Upon gap filling by DNA polymerases, ligation, and a subsequent round of replication, these can give rise to dicentric (or centromere-less) inverted duplication chromosomes. These hairpins can also be cleaved through the action of Mre11-Rad50-Xrs2 in combination with Sae2. Resected ends can also lead to microhomology-mediated joining to other broken chromosomes resulting in translocations. Bottom: resection into sequences that have homology with other regions in the genome can lead to nonallelic HR, including BIR (data not shown) resulting in homology-mediated translocations. The Rad1-Rad10 nuclease has been implicated in the removal of the nonhomologous 3′-tail (yellow) during these types of events. BIR, break-induced replication; DSBs, double-strand breaks; GCRs, Gross Chromosomal Rearrangements; HR, homologous recombination; NHEJ, nonhomologous end joining; RPA, Replication Protein A.
Measuring Genome Instability
Overview
Methods for studying GCRs fall into two general categories. The first category, termed here “directed assays,” detects rearrangements mediated by specific sequence features. These assays usually probe high-frequency events, often mediated by a specific mechanism and often select for a specific rearrangement. For example, synchronous cleavage of an HO endonuclease site has been used to monitor DSB-mediated HR between specific target sequences (Connolly et al. 1988; Ira et al. 2003). Directed assays are useful for mechanistic studies (McEachern and Haber 2006; Mehta and Haber 2014) but may not necessarily detect the types of spontaneous GCRs that are associated with different diseases and arise due to DNA damage that occurs during normal cell growth. The second category, termed here “undirected assays,” detects GCRs that occur at low rates and targets native DNA sequences, DNA structures, and DNA damage but does not depend on specific engineered GCR-inducing structures or reflect the formation of a specific rearrangement. Undirected assays can probe the spectrum of spontaneous GCRs that grossly rearrange the genome and the broad diversity of pathways and mechanisms that impact the formation and suppression of GCRs. However, because the events occur at low frequencies, undirected assays usually provide more limited mechanistic insights than the directed assays. In this section, we will focus on many of the undirected assays used for studying GCRs.
The “classical” GCR assay
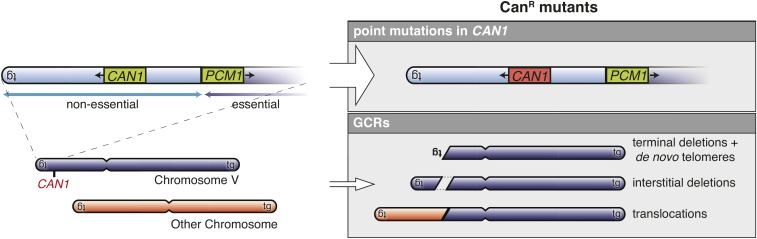
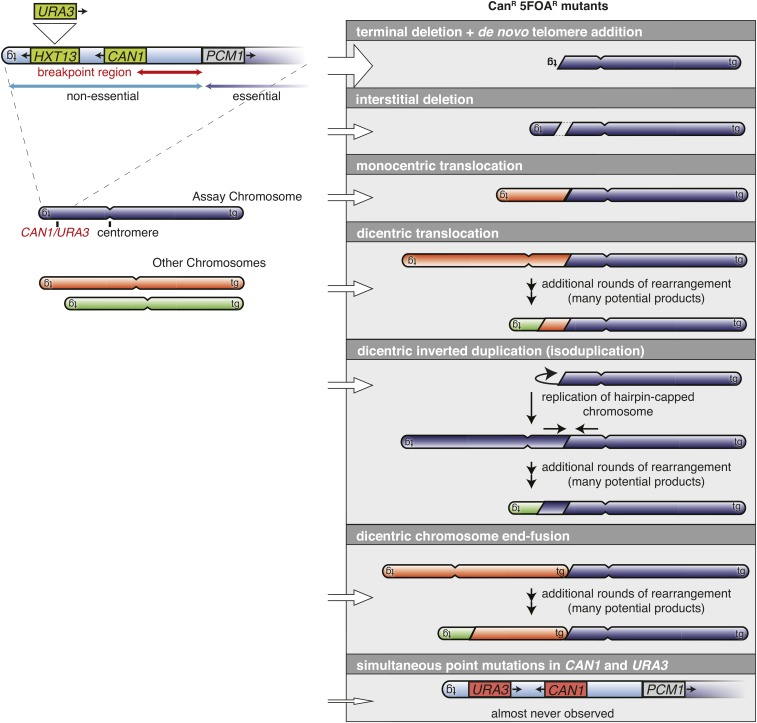
A key observation about the nature of some canavanine-resistant (Canr) S. cerevisiae mutations that proved to be GCRs led to the development of a series of undirected GCR assays (Figure 3). Most Canr mutations are point mutations in CAN1. However, some Canr mutations are GCRs causing a deletion of CAN1, which can form because CAN1 is on a terminal nonessential region of the left arm of chromosome V (Tishkoff et al. 1997; Chen et al. 1998). Modification of this chromosomal region by inserting a second marker, URA3, into the HXT13 gene generated an assay that selected for GCRs (Figure 4) (Chen and Kolodner 1999). This modified strain is sensitive to both Can and 5-fluoroorotic acid (5-FOA), and double-drug-resistant progeny arise at a rate that can be measured by fluctuation analysis [for a methods paper, see Schmidt et al. (2006a)]. Except in the case of a small number of mutants that accumulate point mutations at high rates or after treatment with some DNA-damaging agents (Myung et al. 2001b; Myung and Kolodner 2003), all of the double-drug-resistant progeny result from the formation of GCRs associated with the codeletion of CAN1 and URA3 (Figure 4). As a consequence, this assay, which we refer to as the “classical” GCR assay has been used to measure the rate of accumulating GCRs and, when coupled with detailed structural analysis of the GCRs that occur, can be used to determine the rate of accumulating specific types of GCRs.
Figure 3.
CanR progeny recovered from haploid S. cerevisiae strains. The CAN1 gene (green), which encodes a transporter that imports both arginine and the toxic arginine analog canavanine, is present on the terminal nonessential portion of the left arm of chromosome V, which is bounded by the TEL05 telomere (“tg”) and the essential PCM1 gene (green). Most CanR mutants isolated are due to point mutations in CAN1 indicated in red; however, some are GCRs that result in deletion of regions of the nonessential portion of chromosome V that includes the CAN1 gene. CanR, canavanine-resistant; GCRs, Gross Chromosomal Rearrangements.
Figure 4.
The “classical” GCR assay and products predicted from junction sequences from GCRs recovered in the assay. The classical GCR assay was created by inserting URA3 into the HXT13 gene telomeric to CAN1 (Chen and Kolodner 1999). The breakpoint region for this assay is between the first telomeric counterselectable gene (CAN1) and the most centromeric essential gene (PCM1). In this assay, the breakpoint region is comprised of single-copy sequences. The centromere is indicated by the invagination in the chromosome. The “tg” symbols indicate telomeric repeats at the chromosomal ends. Many GCRs selected in the classical GCR assay are terminal deletions healed by the addition of a de novo telomere. Other types of GCRs include interstitial deletions, inverted duplications, translocations, dicentric chromosome end-to-end fusions, and very rarely simultaneous point mutations in both URA3 and CAN1 indicated in red. In cases where dicentric products are initially formed, they undergo additional rearrangements to generate stable monocentric products. CanR, canavanine-resistant; GCRs, Gross Chromosomal Rearrangements.
Undirected loss GCR assays
The classical GCR assay (Figure 4 and Figure 5A) can be described as an “undirected loss GCR assay,” in which GCRs are selected on the basis of the loss of genetic markers in haploid strains. Undirected loss GCR assays share a key property: the placement of the selectable markers defines a “breakpoint region.” The breakpoint region is the portion of the chromosomal arm where one of the rearrangement breakpoints must occur; one end is the most centromeric counter-selectable marker, and the other end is the most telomeric essential gene (Figure 4 and Figure 5). Varying the chromosomal features in the breakpoint region has been a key feature of many next-generation assays described below and affects both the rearrangement rate and the sequences targeted at the other rearrangement breakpoint(s). This second rearrangement breakpoint, when there is one, can in principle be in any region of the genome provided that no essential genes are deleted by the formation of the GCR. Because of this latter requirement, virtually all translocations observed using undirected loss GCR assays in haploid strains are nonreciprocal; these translocations are associated with an intact copy of the target chromosome (Chen et al. 1998; Chen and Kolodner 1999; Pennaneach and Kolodner 2009; Putnam et al. 2009a, 2014). The accumulation of nonreciprocal translocations suggests that GCRs primarily form in S- or G2-phase after the donor chromosome is replicated or that their formation involves some type of copying mechanism like break-induced replication (BIR) (Bosco and Haber 1998; Flores-Rozas and Kolodner 2000).
Figure 5.
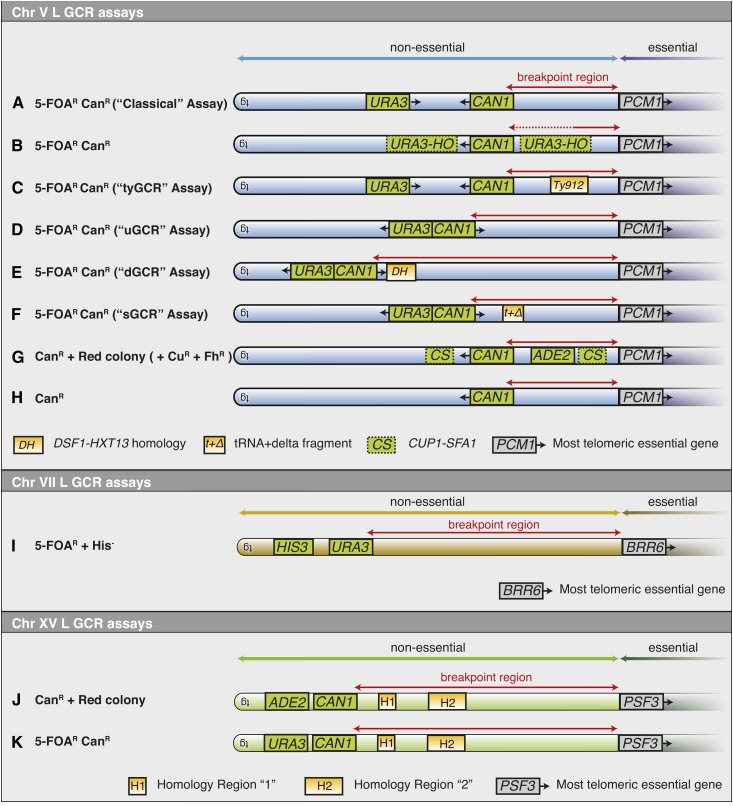
Comparison of a variety of haploid GCR assays that select for loss of markers. A number of variant GCR assays that select for loss of markers in haploid strains have been developed in S. cerevisiae utilizing chromosomes V (blue), VII (brown), and XV (green). For each assay, the nonessential (light color) and essential regions (dark color) of the chromosome arm are shown along with the relevant marker genes (red text), the breakpoint region (red horizontal line), and homologies to other regions of the genome (yellow boxes). The chromosome V L assays, which have a nonessential region telomeric to PCM1, are (A) the classical GCR assay selecting for loss of CAN1 and URA3 (Chen and Kolodner 1999), (B) assays introducing an HO-URA3 cassette either telomeric or centromeric to CAN1 indicated by one of the other of the dashed green boxes (Myung and Kolodner 2003), (C) the tyGCR assay in which Ty912 is inserted into the breakpoint region of the classical GCR assay (Chan and Kolodner 2011), (D) the “unique sequence” or uGCR assay (Putnam et al. 2009a), (E) the “duplication” or dGCR assay in which the DSF1-HXT13 segmental duplication is in the breakpoint region (Putnam et al. 2009a), (F) the “short duplication” or sGCR assay that includes the SUP53 tRNA gene and 100 bp of a Ty-related delta sequence in the breakpoint region (Putnam et al. 2016), (G) an assay that selects for loss of CAN1 and screens for loss of ADE2 by colony color and screens for amplification of a CUP1-SFA1 cassette (Indicated by one of the other of the dashed green boxes) by increased drug resistance (Narayanan et al. 2006), and (H) an assay that selects only for loss of CAN1 in strains with high GCR rates (Tishkoff et al. 1997; Chen et al. 1998; Craven et al. 2002). (I) The chromosome VII L assay, which has a nonessential region telomeric to BRR6, involves selection for loss of URA3 and screening for loss of HIS3 (Myung et al. 2001c). The chromosome XV L assays, which have a nonessential region telomeric to PSF3, include (J) an assay that has CAN1 and ADE2 telomeric to two homology regions (H1 and H2) and selects for loss of CAN1 followed by screening for loss of ADE2 by colony color (Hackett et al. 2001), and (K) a modified chromosome XV L assay that has CAN1 and URA3 telomeric to the homology regions (Kanellis et al. 2007). CanR, canavanine-resistant; GCRs, Gross Chromosomal Rearrangements.
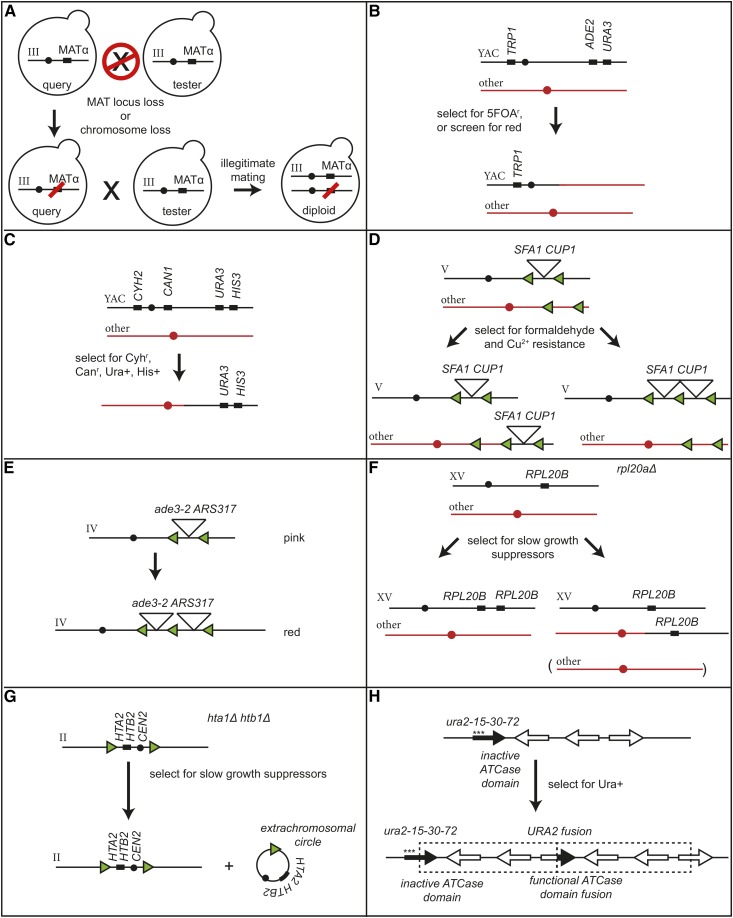
A number of next-generation undirected loss GCR assays have been devised in haploid strains. (1) Many assays have been developed in which potential at-risk sequences have been tested for their effects on the formation of GCRs. These at-risk sequences include HO endonuclease sites (Figure 5B) and a Ty1 element (Figure 5C), as well as trinucleotide repeat sequences, G-quadruplex motifs, GC-rich human minisatellite sequences, inverted Alu repeats, or inducible genes (Myung and Kolodner 2003; Sikdar et al. 2008; Kerrest et al. 2009; Chan and Kolodner 2011; Piazza et al. 2012; Y. Zhang et al. 2012, 2013; Paeschke et al. 2013). (2) A number of assays have utilized a CAN1-URA3 cassette that could be inserted at various locations to probe the effect of endogenous and engineered features such as single-copy sequences (Figure 5D), low-copy number repeat sequences (Figure 5E), or a short-homology region generated by ∼100 bp of a Ty-related δ sequence and a repetitive leucine tRNA (Figure 5F) (Putnam et al. 2009a, 2016). (3) A variant assay involving selection by ADE2 and CAN1 has been used to monitor terminal chromosome loss and SFA1-CUP1 cassette amplification due to rearrangements mediated by a LYS2 cassette containing an Alu sequence-derived direct or inverted repeat (Figure 5G) (Narayanan et al. 2006). (4) Selection against CAN1 alone can, in some genetic backgrounds, also identify chromosomal rearrangements similar to the observation of the first mutator mutants having high GCR rates (Figure 5H) (Chen et al. 1998; Craven et al. 2002). (5) The nonessential terminal region of chromosome VII L has also been engineered by insertion of URA3 and HIS3 markers to allow detection of GCRs, which demonstrated that features of the classical GCR assay could be generalized to another chromosome (Figure 5I) (Myung et al. 2001c). (6) The nonessential terminal region of chromosome XV L has been probed using CAN1 and ADE2 as markers (Figure 5J) (Hackett et al. 2001) and CAN1 and URA3 as markers (Figure 5K) (Kanellis et al. 2007). (7) A cassette bearing an intron-containing version of the URA3 gene, with or without intronic interstitial telomeric repeats, was inserted on chromosome III and used to demonstrate that these repeats increased the rate of loss of URA3 due to the formation of GCRs [assay not illustrated; Aksenova et al. (2013)]. In this assay, the URA3 insertion was within the essential region of chromosome III, and the observed rearrangements retained both fragments of the broken chromosome, including the retention of an 80-kb acentric minichromosome. (8) Haploid α cells can segregate variants that lose the MAT locus and can undergo illegitimate mating with other α strains (Figure 6A). This assay selects for both rearrangements of chromosome III as well as chromosome loss (Lemoine et al. 2005; Yuen et al. 2007). (9) In a yeast artificial chromosome (YAC)-based assay, GCRs that mediated the loss of URA3 and ADE2 but retained TRP1 were monitored (Figure 6B) (Huang and Koshland 2003; Wahba et al. 2013). Finally, (10) a system for monitoring translocations between a number of normal chromosomes and a YAC in HR-defective mutants was developed (Tennyson et al. 2002). In this assay, the centromere of the YAC was flanked by negative selection markers (CYH2 and CAN1), and the terminal region of the YAC contained positive selection markers (URA3 and HIS3) (Figure 6C). This assay is similar to the classical GCR assay, but it works by selecting for retention of a terminal region of the YAC and loss of the centromeric region of the YAC.
Figure 6.
Comparison of haploid GCR assays that select for amplification of markers or loss of internal markers. (A) The “a-like faker” mating assay that uses mating and selection to detect loss of information at the MAT locus (Lemoine et al. 2005; Yuen et al. 2007). (B) A YAC-based GCR assay that selects for loss of URA3 and ADE2 but retention of TRP1, which also detects translocations involving fragments of native S. cerevisiae chromosomes (Huang and Koshland 2003; Wahba et al. 2013). (C) A YAC-based GCR assay that selects for retention of the terminal YAC arm and loss of the remainder of the chromosome, and detects translocations involving fragments of native S. cerevisiae chromosomes (Tennyson et al. 2002). (D) Amplification of a SFA1-CUP1 cassette causes increased resistance to formaldehyde and copper ions that detects amplification resulting from HR-mediated unequal crossing over (Zhang et al. 2013a). (E) Amplification of ade3-2 resulting from HR-mediated unequal crossing over causes S. cerevisiae colonies to undergo a color change from pink to red (Green et al. 2010). (F) An assay for suppressors of the slow growth phenotype of an rpl20aΔ mutant strain selects for amplification of RPL20B (Koszul et al. 2004; Payen et al. 2008). (G) An assay for suppressors of the slow growth phenotype of an hta1Δ htb1Δ double-mutant strain selects for amplification of the centromere (black circle) proximal HTA2 and HTB2 resulting from the formation of a circular chromosome (Libuda and Winston 2006). (H) Reactivation of ura2-15-30-72, which has three nonsense mutations in the 5′-end of the gene (asterisks), by selecting for uracil prototrophy has identified the formation of large duplications in which the 3′-end of the ura2 allele is fused in-frame to another open reading frame resulting in expression of a functional Ura2 fusion protein (Schacherer et al. 2005). ATCase, aspartate carbamyltransferase; CanR, canavanine-resistant; GCRs, Gross Chromosomal Rearrangements.
Undirected gain GCR assays
Haploid strain-based assays that select for amplification of genetic markers that have dose-dependent effects are termed here “undirected gain GCR assays.” These assays are, in principle, less restrictive than undirected loss GCR assays because any chromosome can break at any site and the break healed by joining to a copy of a telomere-terminated fragment containing the selected genetic markers, provided that intact copies of the two chromosomes involved are maintained (Koszul et al. 2004; Libuda and Winston 2006; Payen et al. 2008; Green et al. 2010; H. Zhang et al. 2013). In practice, these assays mostly select for breakpoints in repeated sequences (H. Zhang et al. 2013) and are most useful for studying how cells maintain genome stability when repeated sequences are present (Deininger and Batzer 1999; Lobachev et al. 2000).
Multiple undirected gain GCR assays have been implemented in haploid cells. (1) Several assays have been devised that select for amplification of engineered markers that occurs when a broken chromosome is healed by joining to a telomere-terminated fragment containing the selected markers. The amplification markers include a SFA1-CUP1 cassette whose amplification causes increased resistance to formaldehyde and copper (Figure 6D) (H. Zhang et al. 2013), and the ade3-2 allele whose amplification causes strains to change color from pink to red (Figure 6E) (Green et al. 2010). (2) Other assays have taken advantage of the fact that deletion of one of the copies of a set of duplicated genes present in the S. cerevisiae genome can sometimes cause slow growth that can be suppressed by amplification of the remaining paralog. Such assays have utilized the RPL20A/RPL20B pair (Figure 6F) (Koszul et al. 2004; Payen et al. 2008) and the HTA1-HTB1/HTA2-HTB2 pair (Figure 6G) (Libuda and Winston 2006). (3) Selection of specific forms of gene duplication has been monitored by the reactivation of the ura2-15-30-72 allele. This allele has three nonsense mutations in the 5′-end of the gene and can be reactivated by gene duplication when the 3′-end of the URA2 is inserted in-frame into another open reading frame (Figure 6H) (Schacherer et al. 2005).
Diploid GCR assays
Diploid GCR assays are similar to undirected gain GCR assays in that they are not constrained by the loss of essential genes and, in principle, allow for a greater diversity of GCRs to occur (Hiraoka et al. 2000; Umezu et al. 2002; H. Zhang et al. 2013). This lack of constraint on where breakpoint junctions occur tends to lead to the formation of GCRs by HR between repetitive elements, which are distributed throughout the genome, especially in regions containing essential genes.
Only a relatively small number of diploid strain-based GCR assays have been constructed. (1) Several assays have monitored for loss of a single counterselectable marker, such as URA3 or CAN1, which can also measure chromosome loss (Hiraoka et al. 2000; Klein 2001; Umezu et al. 2002). Variants of these kinds of diploid assays have been performed in haploid cells that are disomic for a chromosome marked with CAN1 (Admire et al. 2006; Paek et al. 2009). (2) Assays that detect amplification of an SFA1-CUP1 cassette have been used to detect GCRs in diploid strains, which are mostly mediated by Ty × Ty HR (H. Zhang et al. 2013). Finally (3), amplification of the ura2-15-30-72 allele has also been studied in diploids, and selects for gene duplications and translocations as seen in the haploid assay (Schacherer et al. 2007).
Structural Analysis of GCRs
Methods for analyzing GCRs
Determining the structure of individual GCRs is important for understanding the mechanisms by which GCRs are formed. S. cerevisiae has advantages that facilitate the analysis of GCR structures, including the relatively small size of the genome, its organization into 16 chromosomes, the availability of the genome sequence, and the presence of a limited number of repeated sequences. Even with these advantages, elucidating the structure of individual GCRs including determining the connectivity of each segment at the DNA sequence level can be very difficult. A number of methods have been used to characterize GCRs; however, often no single method is sufficient to determine the complete GCR structure. Consequently, most studies have not determined complete structures but have inferred them from the limited available data.
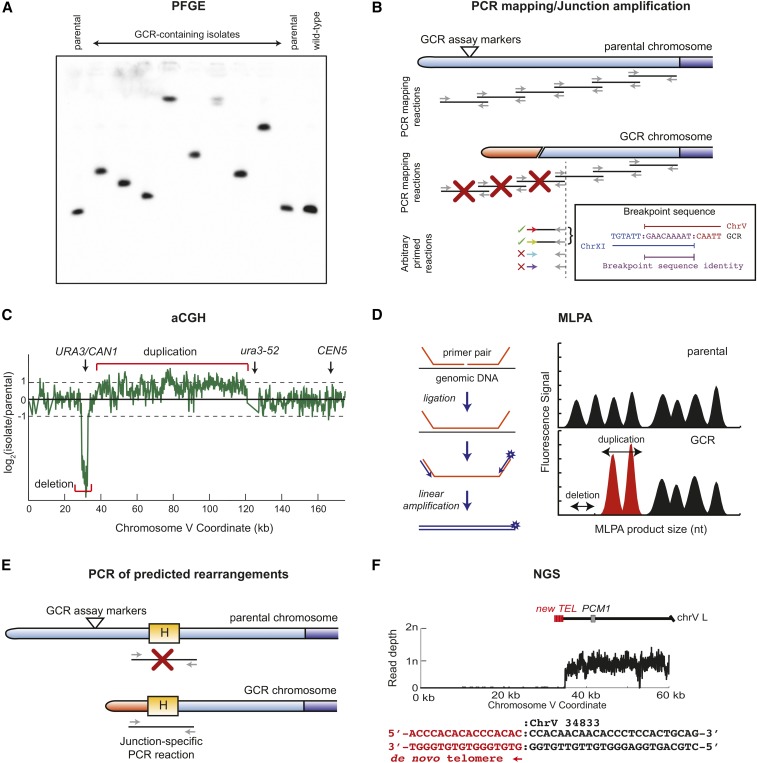
Methods that have been used to characterize GCRs include the following. (1) Pulsed-Field Gel Electrophoresis can detect aberrantly-sized chromosomes, and can provide information about the composition of these chromosomes by analyzing them by Southern blotting with appropriate hybridization probes (Figure 7A) (Chen et al. 1998; Narayanan et al. 2006; Pennaneach and Kolodner 2009; Chan and Kolodner 2011; Putnam et al. 2014; Serero et al. 2014; Deng et al. 2015). (2) PCR-based strategies for mapping, amplifying, and sequencing junction breakpoints have been used to characterize GCRs (Figure 7B) [for a methods paper see Schmidt et al. (2006a)]. Modifications of the original arbitrary-primed PCR strategies, involving use of telomere-specific primers and the ligation of linkers onto genomic DNA digested with restriction enzymes, have the potential to simplify the amplification of fragments containing junction sequences (Smith et al. 2004). The junction sequences determined by these methods provide insights into the structure of GCRs. (3) Array Comparative Genome Hybridization (aCGH) using densely tiled microarrays can provide a detailed map of the copy number changes (duplications and deletions) associated with a GCR (Figure 7C) (Lemoine et al. 2005, 2008; Pennaneach and Kolodner 2009; Putnam et al. 2009a; Zheng et al. 2016). However, aCGH provides no information about the connectivity of these changes, and any predicted GCR structure must be confirmed using secondary analyses. (4) Multiplex Ligation-mediated Probe Amplification (MLPA) (Schouten et al. 2002), which measures copy number changes at low resolution, has been used to rapidly and inexpensively identify duplication of chromosome arms associated with GCRs (Figure 7D) (Chan and Kolodner 2012). Like aCGH, MLPA requires secondary analysis to provide the connectivity information to confirm predicted GCR structures. (5) PCR amplification of the breakpoints predicted from the nature of the breakpoint region has been used for junction verification and, in some cases, was followed by DNA sequencing (Figure 7E) (Mieczkowski et al. 2003; Putnam et al. 2009a; Chan and Kolodner 2011, 2012). In cases where the breakpoints are difficult to amplify by PCR, such as those that occur between Ty elements, restriction mapping by Southern blotting with appropriate probes has been useful (Lemoine et al. 2005). (6) Given the relatively small size of the S. cerevisiae genome, whole-genome Next-Generation Sequencing of multiplexed libraries constructed from different individual GCR-containing isolates is a powerful and relatively inexpensive method for characterizing GCRs (Figure 7F) (Putnam et al. 2014; Serero et al. 2014; Zheng et al. 2016). However, use of this method for GCR analysis is relatively new. It has proven challenging to extract breakpoint sequences for breakpoint junctions mediated by repetitive regions, although variations between different repeated sequence elements can be exploited to detect rearrangement breakpoints (Putnam et al. 2009a); this approach has also been used to map crossovers (Smith et al. 2007; St Charles et al. 2012; Rosen et al. 2013; Song et al. 2014; Laureau et al. 2016; Zheng et al. 2016).
Figure 7.
Methods used to investigate the structures of Gross Chromosomal Rearrangements (GCRs). (A) Pulsed-field gel electrophoresis (PFGE) can separate individual S. cerevisiae chromosomes. Southern blotting with probes to a specific chromosome, in this case the GCR assay chromosome, reveals that all of the GCR-containing isolates have rearranged assay-containing chromosomes that are larger than the corresponding chromosome in a wild-type strain or the parental strain, which is the strain from which the GCR-containing isolates were derived. (B) PCR mapping and junction amplification can be used to map and sequence rearrangements in GCRs recovered from assays such as the classical GCR assay, where the general region in which one breakpoint must occur is known. A series of overlapping PCR products (primers are depicted as gray arrows, and PCR products are depicted as black lines) are generated in separate reactions that probe the breakpoint region. In the GCR-containing isolate, failure of some reactions (red crosses) allows approximate localization of one end of the rearranged assay chromosome. Primers within the last mapping region can be combined with a series of arbitrary primers containing randomized sequences (colored arrows). The products generated by successful PCR reactions (green checkmarks) can be sequenced to determine the breakpoint sequence. (C) Array comparative genomic hybridization (aCGH), in which genomic DNA from a wild-type or parental strain is labeled with one fluorophore and genomic DNA from a GCR-containing isolate is labeled with another fluorophore, and the DNAs are then competitively hybridized to chips containing immobilized oligonucleotides that sample regions across the entire genome. The log2 of the ratios of signals at each genomic position (green trace) reveal regions of copy number changes, including deleted and amplified regions. (D) Multiplexed ligation-mediated primer amplification (MLPA) probes the copy number at selected regions of the genome. For each location, a primer pair is annealed to single-stranded genomic DNA and ligated to form a ssDNA product of a unique size. Ligation products are linearly amplified with a primer pair that adds a fluorescent marker (star) and then separated and quantified on a DNA sequencer. Reductions in peak areas indicate deletions, and increases in peak areas indicate amplifications. (E) For GCR assays in which specific junctions can be predicted due to targeting of known homologies (yellow box labeled with the letter “H”), PCR reactions can be performed using primers specific to the two genomic regions that are joined together (gray arrows). In this case, a PCR product will be obtained from strains containing specific GCRs but will not be obtained from the wild-type or parental strains. (F) Whole-genome paired-end Next Generation Sequencing (NGS) can provide both copy number information via read depth at each base and some information regarding connectivity. Novel junction sequences can be identified using read pairs in which one read of the pair maps near a junction and the other read does not map to the reference genome because it spans the novel junction formed by the GCR. Depicted is a de novo telomere addition, which deletes ∼35 kb from the end of chromosome V L and whose junction sequence could be identified from the sequencing data.
Structures of GCRs selected in haploid strains
De novo telomere addition-mediated GCRs
The most prevalent type of GCR selected in the classical GCR assay in wild-type and some mutant strains is a terminally-deleted chromosome in which a de novo telomere is added at the broken end of the chromosome (Figure 2 and Figure 4) (Chen et al. 1998; Chen and Kolodner 1999; Myung et al. 2001a,c; Myung and Kolodner 2002). Telomerase and some but not all of the other telomere maintenance proteins are required to form these GCRs (Myung et al. 2001a). These GCRs form by telomerase targeting telomere-like TG sequences, which can be as short as two bases (Putnam et al. 2004). Analysis of the sequences of de novo telomeres provided insights into how the telomerase guide RNA is copied by telomerase (Putnam et al. 2004). Initially, the observation of de novo telomere additions seemed to contradict the specificity of telomerase for extending preexisting telomeres. However, telomerase preferentially extends extremely short telomeres, suggesting similar mechanisms for de novo telomere addition and extension of normal telomeres (Arneric and Lingner 2007; Chang et al. 2007; Sabourin et al. 2007). The high proportion of de novo telomere addition events obtained in GCR assays contrasts with the very low level of telomere additions targeted to HO endonuclease-induced DSBs that are not associated with telomere “seed” sequences (Schulz and Zakian 1994; Bosco and Haber 1998; Mangahas et al. 2001). One explanation for the difference may be that HO-induced DSBs do not provide a sequence or chromatin context that is amenable to de novo telomere addition.
GCRs with breakpoints at regions of short or no homology
Fusion of the broken assay chromosome to another chromosomal fragment can generate an interstitial deletion, if the terminal portion of the same chromosomal arm is captured, or a translocation, if a fragment of another chromosome is captured (Figure 2 and Figure 4) (Chen and Kolodner 1999; Myung et al. 2001a,c; Myung and Kolodner 2002; Pennaneach and Kolodner 2004). In GCR assays with only unique sequences in the breakpoint region, junctions typically form between sequences with little or no homology (Chen and Kolodner 1999; Putnam et al. 2005). The lengths of the sequence identities at the junctions were shorter when HR was defective (average length of 3.0 bases) and longer in when non-homologous end joining (NHEJ) was defective (average length of 6.1 bases), suggesting that both NHEJ and some type of HR can generate these GCRs (Putnam et al. 2005). In spite of the lack of homology at breakpoints, translocations with identical junction sequences have been recovered multiple times (Putnam et al. 2005), although the mechanisms and/or genomic features that underlie their formation have not yet been elucidated.
The junction sequences can be used to predict the structure of the rearranged chromosomes. In many cases, the junction sequences suggested the existence of monocentric products, including interstitial deletions, monocentric translocations, or, in the case of some GCRs identified in telomerase-defective strains, circular chromosomes (Chen and Kolodner 1999; Myung et al. 2001a,c; Putnam et al. 2005; Pennaneach and Kolodner 2009). In all cases where monocentric interstitial deletion and monocentric translocation GCRs were studied further, the structures predicted by the breakpoint junction sequences were confirmed (Pennaneach and Kolodner 2009; Putnam et al. 2009a, 2014).
In other cases, the junction sequences indicate the initial formation of three types of dicentric chromosomes (Figure 2 and Figure 4) (Myung et al. 2001c; Pennaneach and Kolodner 2004; Chan and Kolodner 2011; Putnam et al. 2014). (1) Dicentric translocations form when the broken assay chromosome is joined to a fragment of another chromosome such that the captured fragment contains a centromere and is terminated with a telomere. (2) Telomeric fusions form when the broken assay chromosome is fused to the telomere of another otherwise intact chromosome. (3) Inverted duplications (also called isoduplications due to their similarity to mammalian isochromosomes formed by centromere–centromere fusion) form when the broken assay chromosome is fused to a nearly identical copy of itself in the inverted orientation. Inverted duplications could form either by fusion to or invasion of a sister chromatid or, more likely, by formation of a hairpin-terminated chromosome that is then replicated (Pennaneach and Kolodner 2004, 2009; Narayanan et al. 2006; Putnam et al. 2014; Deng et al. 2015).
When predicted dicentric chromosomes have been further studied, evidence was found for secondary rearrangements that inactivated one of the two centromeres (Pennaneach and Kolodner 2009; Chan and Kolodner 2011, 2012; Putnam et al. 2014). Secondary rearrangements included: (1) dicentric chromosome breakage and healing of the DSB by de novo telomere addition, (2) dicentric chromosome breakage and formation of one or more secondary chromosomal fusions (typically but not exclusively by HR between repeated sequences such as Ty-related sequences) to generate a multipartite monocentric translocation, and (3) mutation or deletion of one of the centromeres. These additional rearrangements are consistent with early studies showing that dicentric chromosomes are prone to breakage when the two centromeres are pulled into different cells during mitosis (Scherer et al. 1982; Kramer et al. 1994; Thrower et al. 2003).
GCRs mediated by nonallelic recombination between large regions of homology
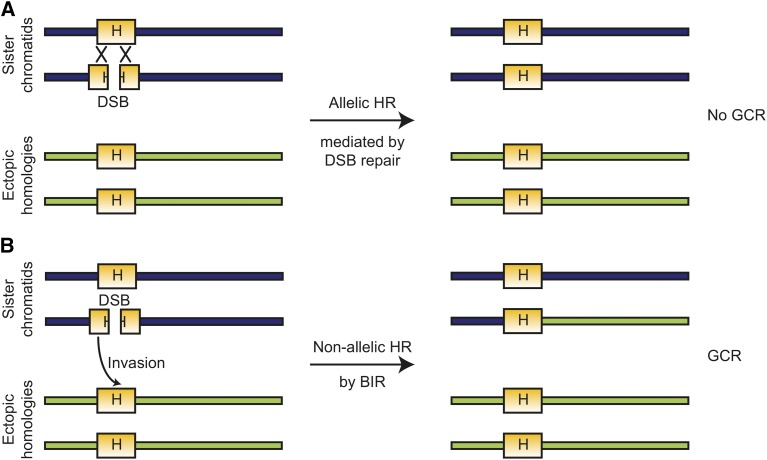
In GCR assays with a repeated sequence in the breakpoint region, the predominant types of GCRs recovered are translocations mediated by HR between the repeated sequence in the breakpoint region and a related sequence elsewhere in the genome (Figure 8) (Putnam et al. 2009a; Chan and Kolodner 2011, 2012). For example, in a duplication-mediated GCR assay (also called the dGCR or yel072w::CAN1/URA3 assay), which contains the chromosome V L HXT13-DSF1 sequences in the breakpoint region, the GCRs recovered were predominantly formed by HR with HXT13-DSF1-related sequences on chromosome IV L, X R, or XIV R (Putnam et al. 2009a). The formation of the duplication-mediated GCRs required DSB repair pathways, suggesting that they were formed by BIR, a half-crossover mechanism, or HR between more than one broken chromosomes (McEachern and Haber 2006; Deem et al. 2008).
Figure 8.
In the presence of homologies, HR can mediate both conservative repair as well as error-prone repair leading to the formation of a GCR. (A) After the formation of a DSB within a region of homology to different sites in the genome (or adjacent to the region of homology followed by resection into the homology), allelic HR targeted to the sister chromatid can repair the DSB such that the original structure of the chromosome is preserved and no GCR is formed. (B) If an ectopic homology is targeted, such as through BIR or a half-crossover mechanism, a GCR is formed and chromosomal regions lacking centromeres are lost. Note that the normal HR machinery is involved in both mechanisms that repair the initiating DSB. BIR, break-induced replication; DSBs, double-strand breaks; GCRs, Gross Chromosomal Rearrangements; HR, homologous recombination.
Other studies have analyzed GCRs whose formation is mediated by repeated Ty elements (Lemoine et al. 2005, 2008; Argueso et al. 2008; Chan and Kolodner 2011); in these cases, the GCRs detected appeared to be translocations or deletions mediated by HR between Ty elements. Insertion of a full length Ty1 element, Ty912, into the breakpoint region of the classical GCR assay chromosome resulted in an increased GCR rate (Chan and Kolodner 2011). The observed GCRs were mediated by HR between Ty912 and one of at least 254 known Ty or solo δ sequences in the S. cerevisiae genome (Chan and Kolodner 2011, 2012). The GCRs products were either monocentric translocations, if Ty912 and the recombination target had the same orientation relative to their respective centromeres, or dicentric translocations, if their orientations were the reverse of each other relative to their respective centromeres. As for dicentric translocations formed by nonrepetitive sequences (see section GCRs with breakpoints at regions of short or no homology), Ty-mediated dicentric chromosomes underwent one or more secondary rearrangements to yield stable monocentric translocations. Remarkably, rearrangements involving Ty912 preferentially targeted only a small subset of the Ty elements in the genome (six Ty elements accounted for 63% of the GCRs recovered) (Chan and Kolodner 2012), one of which had been previously been identified as a fragile site under low DNA polymerase α conditions (Lemoine et al. 2005).
An unusual type of GCR, observed in the classical GCR assay, involved multiple HR events between CAN1 and its distant homologs LYP1 and ALP1 (Schmidt et al. 2006b). These GCRs appeared to involve an initial crossover between CAN1 and LYP1, which would potentially give rise to a dicentric GCR, and a second crossover between LYP1 and ALP1, which is adjacent to LYP1 but in an inverted orientation, that potentially converted the predicted dicentric GCR to a monocentric GCR. Some examples of CAN1-LYP1-ALP1-LYP1-ALP1 translocations involving four crossovers were also observed (Schmidt et al. 2006b). These multipartite rearrangements appeared to reflect short sequence homology-mediated HR events that switch between CAN1, LYP1, and ALP1. These types of events were initially observed in strains with an sgs1 mutation and another mutation that increased the GCR rate, similar to the template switching observed in HXT13-DSF1 duplication-mediated GCRs in sgs1 single mutants (Putnam et al. 2009a), and were subsequently demonstrated with model HR substrates in wild-type strains (Smith et al. 2007; Anand et al. 2014). The observation of these unusual translocations demonstrates the promiscuity that HR can display during the processes that yield translocations.
Several studies have examined the formation of GCRs that create segmental duplications. In one study, duplication of the RPL20B region was selected for in haploid strains and resulted in tandem duplications or duplications mediated by the formation of nonreciprocal translocations (Koszul et al. 2004). Two types of breakpoints mediated these events: (1) short microhomologies like those seen in the classical GCR assay and (2) Ty element-related sequences. Another study selected for amplification of a SFA1-CUP1 cassette that was inserted between two Ty elements. In this study, the amplification events were mediated by unequal crossing over between Ty elements (H. Zhang et al. 2013).
Continuously shortening chromosomes
Analysis of GCR-containing strains has also revealed an unexpected type of fragmented chromosome lacking a telomere, which existed as a population of continually shortening chromosomes (Pennaneach and Kolodner 2009). These shortening chromosomes were seen in mutants defective for the checkpoints that cause cell cycle delay or arrest in response to DSBs. It appears that the loss of sequences from the broken end of these chromosomes during each round of replication may be slow enough that the cells containing such chromosomes can divide many times before growth terminates due to loss of essential genes and dilution of their encoded essential gene products. It is also possible that these chromosomes were stabilized by other mechanisms that can synthesize DNA, but not a telomere, onto the broken end of the fragmented chromosomes (Maringele and Lydall 2004).
Structures of GCRs selected in diploid strains
A limited number of studies have characterized in detail the GCRs selected in diploid strains. In one diploid assay, URA3 was inserted in different positions along one copy of chromosome III, and 5-FOA-resistant progeny were selected and analyzed (Umezu et al. 2002). URA3 was lost by multiple mechanisms, including: (1) chromosome loss; (2) mitotic HR between the two copies of chromosome III in the region between URA3 and the centromere combined with segregation of progeny lacking URA3 during cell division; (3) gene conversion of the inserted URA3 allele; (4) HR-mediated interstitial deletion of URA3; and (5) both interchromosomal and intrachromosomal translocations mediated by HR with a Ty element located between URA3 and the centromere. In the case of the interchromosomal translocations, the fate of the nonchromosome III target chromosome was not analyzed, so it is not known if the resulting translocations were reciprocal or nonreciprocal. In some cases, a region of chromosome III at the breakpoint junction was duplicated or triplicated (Umezu et al. 2002); studies of amplified regions associated with the formation of Ty element-mediated translocations selected in haploid GCR assays have provided structures for these types of amplified regions (Pennaneach and Kolodner 2009; Chan and Kolodner 2011, 2012).
In a second diploid assay, a SFA1-CUP1 cassette was inserted onto both copies of chromosome V R, followed by selection for amplification of the SFA1-CUP1 cassette (H. Zhang et al. 2013). The majority of events creating an extra copy of the SFA1-CUP1 cassette were nonreciprocal translocations mediated by Ty elements that fused an additional copy of the SFA1-CUP1-containing terminal segment of chromosome V R to the end of another chromosome. Other events included Ty element-mediated tandem duplication of the SFA1-CUP1 cassette-containing region, as observed in the haploid version of this assay (see sections Undirected gain GCR assays and GCRs mediated by nonallelic recombination between large regions of homology).
In a third assay, diploid cells were arrested in the G2-phase of the cell cycle, irradiated with x-rays to produce ∼250 DSBs/cell, and then plated onto growth media to select survivors, which were often found to contain one or more aberrant chromosomes (Argueso et al. 2008). The aberrant chromosomes either contained interstitial deletions or intrachromosomal or interchromosomal translocations, which were often nonreciprocal. The observed breakpoints involved Ty elements or related sequences (∼83%), repetitive gene families (∼10%), and single-copy sequences (∼7%), approximating the distribution that would be predicted from the rates that these types of events occur in wild-type haploid strains measured using assay systems that detect specific types of GCRs (Chen and Kolodner 1999; Putnam et al. 2009a; Chan and Kolodner 2011).
In a fourth and final series of assays, loss-of-heterozygosity (LOH) in diploid cells has been followed in a variety of studies. In some cases, loss of individual markers, such as ADE2 or SUP4-o, was followed and in other cases changes in single-nucleotide polymorphisms between sister chromosomes were followed (Barbera and Petes 2006; Smith et al. 2007; St Charles et al. 2012; Song et al. 2014; Zheng et al. 2016). For assays in which no markers were used (Song et al. 2014; Zheng et al. 2016), cells were grown under conditions of replication stress caused by reduced expression of DNA polymerase α or δ.
Genetic Analysis of Pathways that Suppress and Produce GCRs
A conceptual framework for understanding the pathways that suppress the formation of GCRs
A key challenge in studies of GCRs is how to use the GCR rate and structure data to understand the pathways that maintain genome stability. In some cases, the effects of individual mutations can be inferred from changes to the GCR rate and/or GCR spectrum. However, in most cases, detailed investigation of combinations of mutations is necessary to understand the role of genes of interest. This analysis can be challenging, particularly if different combinations of mutations alter growth rates and cell viability. In general, deciphering individual pathways often requires integration of data from other studies, such as identification of protein complexes, prior knowledge of genetic pathways, growth-based genetic interaction studies, and biochemical studies of the encoded proteins; many of these data are accessible through resources such as the Saccharomyces Genome Database and BioGRID (Cherry 2015; Oughtred et al. 2016).
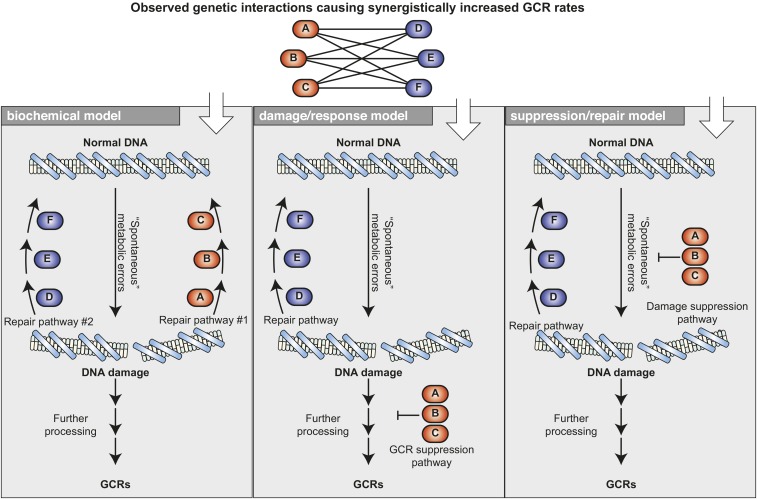
Models for interactions between mutations have been strongly influenced by the analysis of biochemical pathways. In these analyses, synergistic interactions between mutations are often thought to indicate interactions that inactivate independent pathways that perform the same function (Kaelin 2005; Ooi et al. 2006). Similarly, epistatic interactions between mutations are often though to indicate that these mutations affect the same pathway (Kaelin 2005; Ooi et al. 2006). This conceptual framework, termed here the “biochemical model,” is appropriate for understanding some GCR-suppressing interactions (Figure 9), such as the partial redundancy between the DNA damage checkpoint protein kinases Tel1 (homolog of human ATM) and Mec1 (homolog of human ATR) (Myung et al. 2001c; Craven et al. 2002; Mieczkowski et al. 2003). Deletion of the TEL1 gene causes no increase in the GCR rate relative to the wild-type strain (Myung et al. 2001c). In contrast, mec1 mutations cause a ∼200-fold increase, and the combination of mec1 and tel1 mutations causes a ∼13,000-fold increase in GCR rates (Myung et al. 2001c). Consistent with this synergistic increase in GCR rates, both protein kinases phosphorylate the same SQ and TQ sites in many common substrate proteins (Kim et al. 1999), and partial redundancy has also been observed for roles in telomere maintenance (Ritchie et al. 1999) and the DNA damage checkpoint response (Sanchez et al. 1996; Vialard et al. 1998). However, the extreme increase in GCR rate in the mec1tel1 double mutant likely reflects the deregulation of multiple processes that interact with each other including telomere maintenance and checkpoint responses (Pennaneach and Kolodner 2004). Another example consistent with the biochemical model is the observation of epistatic interactions within a pathway. Mutations in SGS1 or TOP3, which encode proteins that interact physically and biochemically (Gangloff et al. 1994; Ng et al. 1999), increase GCR rates in the classical GCR assay by ∼20-fold, and the sgs1top3 double mutant has a GCR rate similar to both of the single mutants (Myung et al. 2001b).
Figure 9.
Three genetic models can explain the same genetic interactions that cause synergistic increases in Gross Chromosomal Rearrangement (GCR) rates. Top: observed genetic interactions among deletions of genes A, B, and C and genes D, E, and F, causing synergistic increases in GCR rates can be explained by three distinct genetic models. Bottom: in the biochemical model, gene products A, B, and C function in one pathway and gene products D, E, and F function in another pathway that redundantly repairs the same type of DNA damage. In the damage/response model, one pathway suppresses the processing of DNA damage to GCRs, whereas the other pathway repairs the initiating DNA damage. In the suppression/repair model, one pathway suppresses the formation of DNA damage, whereas the other pathway promotes error-free repair of the damage.
For many other genetic interactions, the biochemical model is not appropriate, nor is the tendency to classify all synergistic genetic interactions as “buffering” interactions on the basis of this model (Hartman et al. 2001; Segre et al. 2005). In cases where mutations show interactions in GCR assays, and/or show growth-based synthetic interactions, the relevant genes can act within the same pathway as well as act in separate pathways that have different functions. In the case of the apparently paradoxical within-pathway interactions, an analysis by Heyer and colleagues has described mechanisms that may underlie these kinds of interactions (Zinovyev et al. 2013). When interacting genes function in nonredundant pathways, other models can sometimes explain these interactions.
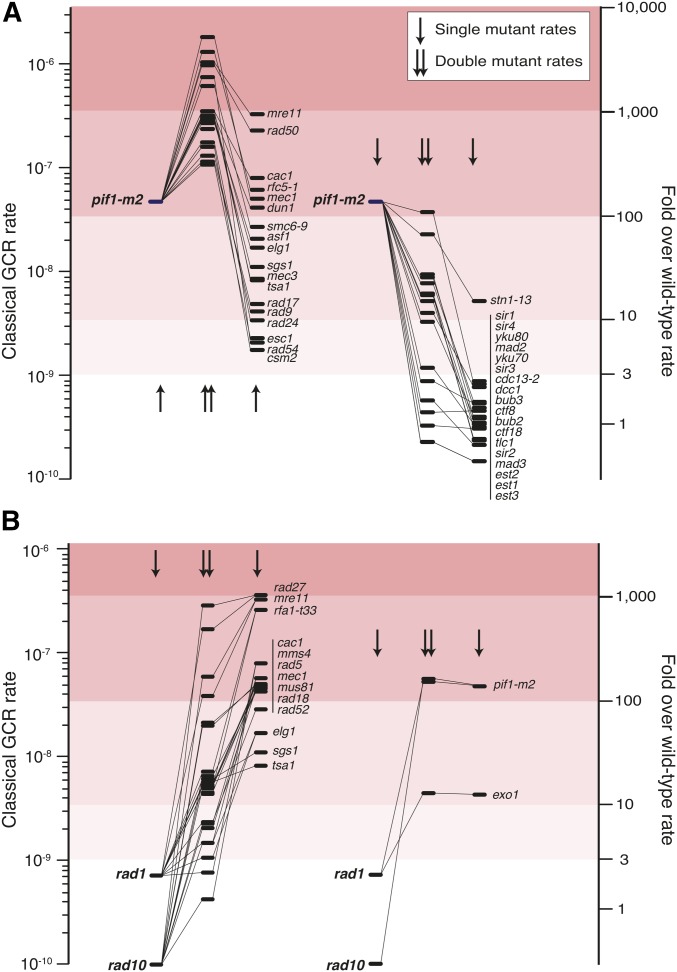
In the “damage/response model,” one gene plays a role in generally repairing DNA damage and the other plays a role in preventing DNA damage from generating GCRs (Figure 9). An example of a gene involved in these types of interactions is PIF1, which encodes a DNA helicase that plays a crucial role in suppressing de novo telomere additions by telomerase (Schulz and Zakian 1994; Zhou et al. 2000). Inactivation of the nuclear form of PIF1 with the pif1-m2 mutation causes an increase in GCR rates and synergistic increases in GCR rates when combined with a wide variety of mutations in genes that likely directly or indirectly suppress the accumulation of DNA damage (Figure 10A) (Myung et al. 2001a). The effect of PIF1-mediated suppression of telomerase is emphasized by the fact that deletion of PIF1 eliminates the duplication-mediated GCRs formed by HR typically seen in the duplication-mediated GCR assay, presumably by channeling damaged DNAs into de novo telomere addition reactions (Putnam et al. 2009a).
Figure 10.
Gross Chromosomal Rearrangement (GCR) rates of single and double mutants showing the effect of combining rad1, rad10, or pif1-m2 mutations with mutations affecting other pathways. (A) Combining a pif1-m2 mutation with many mutations affecting chromosome homeostasis causes a synergistic increase in GCR rates in the classical GCR assay (left), but is suppressed by mutations affecting specific pathways (right). GCR rates are displayed as horizontal bars, and related genotypes are connected with lines. Column 1: pif1-m2 single mutant. Column 2: double mutants with increased GCR rates. Column 3: single mutations that in combination with the pif1-m2 mutation increase the GCR rate. Column 4: pif1-m2 single mutant. Column 5: double mutants with decreased GCR rates. Column 6: single mutations that in combination with a pif1-m2 mutation decrease the GCR rate. The synergistically increased rates likely result from the mechanism depicted by the damage/response model, as PIF1 functions to suppress the formation of de novo telomere addition GCRs (Myung et al. 2001a), and many of the interacting genes are implicated in repairing or preventing the formation of DNA damage. Note that the increased GCR rate caused by a pif1-m2 mutation is suppressed by combining it with mutations affecting several pathways, including de novo telomere addition (est1, est2, est3, tlc1, yku70, yku80, cdc13-2, stn1-13, sir1, sir2, sir3, and sir4), the Ctf8-Ctf18-Dcc1 alternative RFC complex, and components of the spindle checkpoint (mad2, mad3, bub2, and bub3). Data are from a summary of the previous literature (Putnam et al. 2012). (B) Deletion of RAD1 or RAD10, which disrupts the Rad1-Rad10 endonuclease that targets nonhomologous ssDNA overhangs (Sugawara et al. 1997), suppresses the GCR rates caused by many (left), but not all (right), mutations that cause high GCR rates in the classical GCR assay (Hwang et al. 2005). Rates displayed as in panel A. Column 1: the rad1 and rad10 single mutants. Column 2: double mutants with decreased rates. Column 3: single mutations that cause increased GCR rates in the classical GCR assay. Column 4: the rad1 and rad10 single mutants. Column 5: double mutants whose GCR rates are not affected by rad1 or rad10 mutations. Column 6: single mutations that are not suppressed by rad1 or rad10 mutations. Suppression of the formation of GCRs likely indicates a role for the cleavage of ssDNA overhangs during formation of many types of GCRs involving steps where regions of microhomology anneal to each other (Figure 2). The lack of suppression of the pif1-m2 GCR rate by rad1 or rad10 mutations could be consistent with reports that an endonucleolytic activity of telomerase can cleave non-TG-containing portions of ssDNA tails (Collins and Greider 1993; Cohn and Blackburn 1995; Melek et al. 1996). The lack of suppression of the exo1 GCR rate by rad1 or rad10 mutations could be consistent with defects in resection of double-strand breaks and other substrates in exo1 mutant strains (Zhu et al. 2008).
In the “suppression/repair model,” one gene plays a role in preventing metabolic errors during DNA replication and another gene plays a role in the repair of the damage after it has occurred (Figure 9). An obvious gene that plays a role in a damage suppression pathway is TSA1, which encodes the major thioredoxin peroxidase that protects S. cerevisiae from oxidative stress (Chae et al. 1994; Park et al. 2000). Defects in TSA1 cause increased GCRs and cause synergistic interactions with defects in multiple DNA repair pathways (Huang et al. 2003; Huang and Kolodner 2005).
Analyses of GCR data must also take into account several additional complications. (1) Genes that might be expected to fit the biochemical model for interactions may only be partially redundant, and consequently mutations in such genes may show both synergistic interactions and also result in distinct rates of accumulating GCRs, have distinct sets of interactions with mutations in other genes, and/or give rise to distinct classes of GCRs. An example of this model is the partial redundancy between TEL1 and MEC1 (Myung et al. 2001c) discussed above, which is consistent with the view that Mec1 and Tel1 likely act on distinct but overlapping sets of targets (Morrow et al. 1995; Sanchez et al. 1996; Vialard et al. 1998). (2) Genes may have dual roles in repairing DNA damage and preventing the formation of GCRs as a result of DNA damage, which complicates any analysis by the damage/response model. An example of this type of gene is RAD52, which suppresses GCRs by mediating allelic HR, but also promotes the formation of GCRs in duplication-mediated assays by mediating nonallelic HR (Figure 8B). Hence, rad52 mutations cause increased GCR rates in single-copy sequence-mediated assays but decreased GCR rates in duplication-mediated assays (Chen and Kolodner 1999; Putnam et al. 2009a). (3) The formation of a selectable GCR requires both DNA damage and formation of an aberrant chromosome that is stable enough so that cells containing the GCR can survive under selective conditions. The generation of GCRs is thus a form of “nonconservative” DNA repair that does not restore the structure and sequence of the original undamaged chromosomes. As such, the formation of GCRs is dependent on DNA repair mechanisms, and inactivation of pathways required to generate GCRs, as suggested for the Rad1-Rad10 endonuclease (Figure 10B) (Hwang et al. 2005), can play crucial roles on the effects of individual mutations or combinations of mutations have on the recovery of GCRs.
A global view of genome instability suppressing genes
Considerable effort has been focused on the identification of Genome Instability Suppressing (GIS) genes. To date, these studies have identified 171 nonessential S. cerevisiae genes in which mutations cause increased GCR rates in normally growing cells [discussed in Putnam et al. (2016)], and 29 essential genes in which mutations potentially cause increased GCR rates (Table 1). Validation studies suggest that this list is close to a complete list of the nonessential genes that act to suppress GCRs (Putnam et al. 2016). In contrast, there have been very limited studies on genes that act to suppress GCRs that are induced by exogenous DNA-damaging agents (Myung and Kolodner 2003) or by defects in essential genes (Chen and Kolodner 1999; Huang and Koshland 2003; Shah et al. 2012; Albuquerque et al. 2013; Y. Zhang et al. 2013). Advances in these areas may come from studies examining the remodeling of growth-based genetic interaction networks by DNA-damaging agents (Bandyopadhyay et al. 2010; Srivas et al. 2016) and systematic screening of hypomorphic alleles of essential genes. Known GIS genes play significant roles in DNA replication, DNA repair, DNA damage checkpoints, telomere maintenance, response to oxidative stress, cell cycle control, protein sumoylation, subsets of the nuclear pore, and chromatin assembly (see below).
Table 1. Genes implicated in suppressing genome instability in S. cerevisiae.
| ORF | Gene | ORF | Gene | ORF | Gene |
|---|---|---|---|---|---|
| YML086C | ALO1 | YGL087C | MMS2 | YER070W | RNR1 |
| YOR141C | ARP8 | YEL019C | MMS21 | YIL066C | RNR3 |
| YJL115W | ASF1 | YBR098W | MMS4 | YHR200W | RPN10 |
| YPL115C | BEM3 | YIR002C | MPH1 | YHR031C | RRM3 |
| YBR290W | BSD2 | YCL061C | MRC1 | YLR357W | RSC2 |
| YML102W | CAC2 | YMR224C | MRE11 | YHR056C | RSC30 |
| YMR038C | CCS1 | YOL090W | MSH2 | YOR014W | RTS1 |
| YJL194W | CDC6 | YDR097C | MSH6 | YJL047C | RTT101 |
| YDL017W | CDC7 | YBR195C | MSI1 | YER104W | RTT105 |
| YFR036W | CDC26 | YGR257C | MTM1 | YHR154W | RTT107 |
| YCR094W | CDC50 | YDR386W | MUS81 | YLL002W | RTT109 |
| YLR418C | CDC73 | YHL023C | NPR3 | YDR159W | SAC3 |
| YDL164C | CDC9 | YDR288W | NSE3 | YGL175C | SAE2 |
| YGL003C | CDH1 | YKR082W | NUP133 | YBR171W | SEC66 |
| YBR274W | CHK1 | YAR002W | NUP60 | YDR363W-A | SEM1 |
| YPL008W | CHL1 | YDL116W | NUP84 | YOR140W | SFL1 |
| YOR039W | CKB2 | YML060W | OGG1 | YMR190C | SGS1 |
| YPR119W | CLB2 | YBR060C | ORC2 | YHL006C | SHU1 |
| YPR120C | CLB5 | YLL004W | ORC3 | YDR078C | SHU2 |
| YPL256C | CLN2 | YNL261W | ORC5 | YLR079W | SIC1 |
| YIL132C | CSM2 | YDR113C | PDS1 | YDR227W | SIR4 |
| YMR048W | CSM3 | YOR386W | PHR1 | YDR409W | SIZ1 |
| YMR078C | CTF18 | YML061C | PIF1 | YHR206W | SKN7 |
| YPR135W | CTF4 | YBL051C | PIN4 | YKL108W | SLD2 |
| YHR191C | CTF8 | YNL102W | POL1 | YIL105C | SLM1 |
| YJL006C | CTK2 | YDL102W | POL3 | YLR135W | SLX4 |
| YDR052C | DBF4 | YBL035C | POL12 | YDL013W | SLX5 |
| YCL016C | DCC1 | YBR088C | POL30 | YER116C | SLX8 |
| YPL194W | DDC1 | YJR043C | POL32 | YLR383W | SMC6 |
| YOR080W | DIA2 | YIR008C | PRI1 | YDR011W | SNQ2 |
| YHR164C | DNA2 | YKL045W | PRI2 | YCR033W | SNT1 |
| YOR005C | DNL4 | YKL116C | PRR1 | YAL009W | SPO7 |
| YDR440W | DOT1 | YOL146W | PSF3 | YMR179W | SPT21 |
| YJL090C | DPB11 | YML095C | RAD10 | YLR055C | SPT8 |
| YGL043W | DST1 | YOR368W | RAD17 | YML034W | SRC1 |
| YDL101C | DUN1 | YCR066W | RAD18 | YJL092W | SRS2 |
| YDR359C | EAF1 | YER173W | RAD24 | YHR064C | SSZ1 |
| YOR144C | ELG1 | YKL113C | RAD27 | YCL032W | STE50 |
| YKL048C | ELM1 | YDR419W | RAD30 | YDR082W | STN1 |
| YMR219W | ESC1 | YER162C | RAD4 | YJR046W | TAH11 |
| YDR363W | ESC2 | YNL250W | RAD50 | YNL273W | TOF1 |
| YLR233C | EST1 | YLR032W | RAD5 | YKR010C | TOF2 |
| YLR318W | EST2 | YER095W | RAD51 | YLR234W | TOP3 |
| YIL009C-A | EST3 | YML032C | RAD52 | YML028W | TSA1 |
| YOR033C | EXO1 | YPL153C | RAD53 | YDR092W | UBC13 |
| YNL153C | GIM3 | YGL163C | RAD54 | YGR184C | UBR1 |
| YPL137C | GIP3 | YDR076W | RAD55 | YML088W | UFO1 |
| YKL017C | HCS1 | YDR004W | RAD57 | YBR173C | UMP1 |
| YGL194C | HOS2 | YDL059C | RAD59 | YLR373C | VID22 |
| YKL101W | HSL1 | YDR014W | RAD61 | YMR077C | VPS20 |
| YOR025W | HST3 | YGL058W | RAD6 | YJL029C | VPS53 |
| YDR191W | HST4 | YDR217C | RAD9 | YHR134W | WSS1 |
| YDR225W | HTA1 | YBR073W | RDH54 | YDR369C | XRS2 |
| YPL017C | IRC15 | YIL139C | REV7 | YML007W | YAP1 |
| YDR332W | IRC3 | YAR007C | RFA1 | YCL026C | YCL026C |
| YBR245C | ISW1 | YNL312W | RFA2 | YNL064C | YDJ1 |
| YJR054W | KCH1 | YJL173C | RFA3 | YDL162C | YDL162C |
| YDR499W | LCD1 | YJR068W | RFC2 | YHL026C | YHL026C |
| YDR439W | LRS4 | YOL094C | RFC4 | YJL218W | YJL218W |
| YBL023C | MCM2 | YBR087W | RFC5 | YKR023W | YKR023W |
| YLR274W | MCM5 | YLR453C | RIF2 | YMR284W | YKU70 |
| YBR136W | MEC1 | YPR018W | RLF2 | YMR106C | YKU80 |
| YLR288C | MEC3 | YDR255C | RMD5 | YML002W | YML002W |
| YIL128W | MET18 | YPL024W | RMI1 | YML020W | YML020W |
| YMR167W | MLH1 | YEL050C | RML2 | YGR270W | YTA7 |
| YLL061W | MMP1 | YDR279W | RNH202 | YMR273C | ZDS1 |
| YPR164W | MMS1 | YLR154C | RNH203 |
Note that quantitative and semiquantitative Gross Chromosomal Rearrangement rate data are not available for all of the essential genes listed in this table.
Additionally, some genes like TLC1 and DNL4 are not GIS genes, but defects in these genes do cause genome instability in combination with other mutations, and we have termed them cooperating GIS (cGIS) genes. cGIS genes likely play redundant or accessory roles with other genes in suppressing GCRs. Systematic genetic interaction analysis has led to the identification of 438 cGIS genes (Putnam et al. 2016). However, additional analyses are required to both validate these interactions and to test predicted interactions involving genes encoding these complexes and other pathways. It will also be of interest to extend these genetic interactions to mutations in essential genes, although such studies are likely to be complicated by growth defects caused by mutations in essential genes. Known cGIS genes define a much broader set of biological functions than those implicated by the GIS genes. These functions include multiple complexes involved in transcription, mRNA processing, and protein degradation, as well as additional pathways in DNA repair and cell cycle control (see below). Moreover, defects in cGIS genes can also have important impacts on the spectrum of observed GCR structures (Myung et al. 2001c; Putnam et al. 2014).
Finally, ∼9200 double mutants with reduced GCR rates have been identified (Putnam et al. 2016). However, most of these double mutants will require extensive validation because genetic interactions that result in reduced growth rates can appear as if they result in reduced GCR rates in semiquantitative patch test assays. By carefully validating and extending the systematic genetic interaction studies performed to date and possibly incorporating genetic interaction data from other types of studies, it should be possible to ultimately define in detail the genetic network that suppresses the formation of GCRs and the pathways that promote the formation of GCRs in mutants with high GCR rates or after treatment with DNA-damaging agents.
Pathways implicated in the suppression of spontaneous genome rearrangements
DNA repair pathways
DNA repair pathways were implicated in the suppression of GCRs by the identification of the first mutants with increased GCR rates (Tishkoff et al. 1997; Chen et al. 1998; Chen and Kolodner 1999; Myung et al. 2001a,b; Myung and Kolodner 2002; Putnam et al. 2009a), and subsequent studies identifying GIS and cGIS genes (Huang et al. 2003; Smith et al. 2004; Kanellis et al. 2007; Stirling et al. 2011; Putnam et al. 2016) have supported the view that DNA repair pathways are central to the suppression of GCRs. The central role of DNA damage and how that damage is processed in the suppression of genome instability is also emphasized by the increase in GCR rates observed when cells are treated with DNA-damaging agents (Myung and Kolodner 2003).
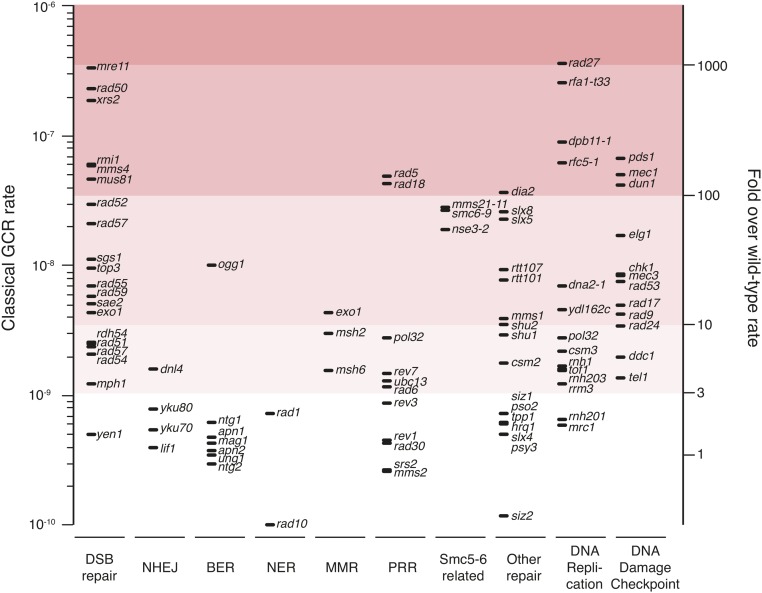
Despite the general importance of DNA repair pathways in suppressing GCRs, not all DNA repair pathways contribute equally (Figure 11). For example, base-excision repair (BER) genes generally play little or no role in suppressing spontaneous GCRs. A possible exception is OGG1 (Huang and Kolodner 2005), which encodes 8-oxoguanine glycosylase/lyase; however, OGG1 deletions might increase the rates of forming de novo telomere addition GCRs due to effects of deletion of OGG1 on the adjacent PIF1 gene. Similarly, defects in nucleotide-excision repair (NER) do not generally affect GCR rates; however, those that do also affect NER genes that play roles in other processes (Hwang et al. 2005; Putnam et al. 2016). Remarkably, DNA repair defective mutations that have little effect in GCR assays tend to have fewer genetic interactions causing synthetic growth interactions under normal growth conditions than mutations having larger effects in GCR assays (Tong et al. 2004; Collins et al. 2007; Costanzo et al. 2010). The relationship between genome instability and genetic interactions might reflect the relative importance of different repair pathways in the repair of the spontaneous DNA damage that underlies the formation of GCRs (see section DNA replication).
Figure 11.
Gross Chromosomal Rearrangements (GCR) rates of single mutants in DNA repair, DNA replication, and DNA damage checkpoint pathways. GCR rates in the classical GCR assay caused by individual mutations are indicated by the positions of the horizontal lines grouped according to pathway. GCR rates that are less than threefold above the wild-type rate are typically not statistically significant. Data are from a summary of the previous literature (Putnam et al. 2012).
HR acts to both suppress and generate GCRs. Both HR and BIR depend on Rad52 and have two major subpathways defined by dependence on the Rad51 pairing and strand exchange protein or the Rad59 strand annealing protein [for a review, see Krogh and Symington (2004)]. During normal growth, HR acts in error-free repair of DSBs (and potentially other damage) in S- or G2-phase using the intact sister chromatid as a donor for repair of the DSB; this type of allelic sister chromatid HR suppresses the formation of GCRs (Figure 8). Thus, loss of HR, either by a rad52 mutation or the combination of the rad51 and rad59 mutations, will substantially increase the GCR rate in assays that only have single-copy sequences in the assay breakpoint region (Chen and Kolodner 1999; Myung et al. 2001a; Putnam et al. 2009a). The GCRs selected in the classical GCR assay are a mixture of de novo telomere additions, translocations, and interstitial deletions, which appear to be formed by NHEJ, based on the short or lack of homologous sequences at the breakpoint junctions (Chen and Kolodner 1999; Myung et al. 2001a; Putnam et al. 2005). Mutations in HR pathway genes that affect only one HR subpathway often have intermediate effects on GCR rates (Figure 11) (Chen and Kolodner 1999; Myung et al. 2001a). In contrast, GCR assays with homologies to ectopic sites in the breakpoint region can select for translocation, duplication, or deletion GCRs that are generated by nonallelic HR (Figure 8B). In these homology-containing GCR assays, a rad52 mutation decreases the rate of accumulating GCRs due to loss of nonallelic HR; however, the rate is not reduced to wild-type levels because error-free allelic HR that suppresses some GCRs is also eliminated (Putnam et al. 2009a; Chan and Kolodner 2011). In a Ty-containing GCR assay, deletion of RAD51 or RAD59 alone caused a small increase or no increase in GCR rate, respectively, but substantially altered the spectrum of translocations targeting different Ty elements in the genome (Chan and Kolodner 2011, 2012). In contrast, deletion of RAD52 caused a greater decrease in the rate of Ty-mediated GCRs because deletion of RAD52 causes greater defects in HR. These results indicate that not only are the two HR subpathways partially redundant but that HR with individual Ty targets shows a unique dependence on one or the other of the HR subpathways.
The Mre11-Rad50-Xrs2 (MRX) complex plays an important role in initiating resection at DSBs during HR in S. cerevisiae [for a review, see Krogh and Symington (2004)] and likely promotes error-free sister chromatid HR (Hartsuiker et al. 2001), in addition to having roles in NHEJ, the intra-S checkpoints, and telomere length maintenance (Moore and Haber 1996; Boulton and Jackson 1998; D’Amours and Jackson 2001). It also plays an important role in sensing DNA damage in different DNA damage checkpoints (see below). Deletions of genes encoding the MRX complex cause dramatic increases in GCR rates in multiple GCR assays (Figure 11), including assays with and without homologies in the breakpoint regions (Chen and Kolodner 1999; Putnam et al. 2009a; Chan and Kolodner 2011). Defects in the MRX complex also alter the GCR spectrum by reducing the formation of de novo telomere addition GCRs (Chen and Kolodner 1999; Putnam et al. 2009a; Chan and Kolodner 2011). Mutations in MRE11 that affect the MRX nuclease activity, which acts in end resection, and deletion of SAE2, which acts in conjunction with MRX to cleave DNA hairpins at DSBs, also alter the types of GCRs formed (Smith et al. 2005; Putnam et al. 2014; Deng et al. 2015). Thus, in contrast to the deletion of MRE11, Mre11 nuclease defects and deletion of SAE2 result in large increases in isoduplications, which are thought to be mediated by the formation of hairpin structures at DSBs (Lobachev et al. 2002; Putnam et al. 2014; Deng et al. 2015). It seems likely that mutations affecting the MRX complex increase the formation of GCRs due to a variety of reasons, including defects in promoting sister chromatid HR, in DNA damage checkpoints, in NHEJ, in telomere maintenance, and in hairpin cleavage.
Multiple proteins process HR intermediates, including the Mph1-Mte1 complex that mediates dissolution of D-loops, the Sgs1-Top3-Rmi1 complex that unwinds double Holliday junctions, the Yen1 Holliday junction resolvase, and the Slx1-Slx4 and Mms4-Mus81 structure-selective endonucleases whose human homologs act cooperatively in the cleavage of double Holliday junctions (Fabre et al. 2002; Krogh and Symington 2004; Munoz-Galvan et al. 2012; Castor et al. 2013; Garner et al. 2013; Mazon and Symington 2013; Wyatt et al. 2013; Silva et al. 2016; Xue et al. 2016; Yimit et al. 2016). These proteins can also play roles in DNA replication, including termination of replication (Boddy et al. 2001; Mundbjerg et al. 2015), establishing BIR from DSBs (Pardo and Aguilera 2012) and promoting resection of DSBs (Gravel et al. 2008; Zhu et al. 2008). Defects in YEN1 do not cause increased GCR rates in the assays studied to date (Figure 11) (Smith et al. 2004; Doerfler et al. 2011; Putnam et al. 2016), consistent with the observation that Yen1 plays little if any role in HR when Mus81-Mms4 is present (Blanco et al. 2010). Defects in the genes encoding the other protein complexes cause increased GCR rates (Figure 11) (Myung et al. 2001b; Hwang et al. 2005; Putnam et al. 2009a, 2012; Chan and Kolodner 2011; Doerfler et al. 2011; Allen-Soltero et al. 2014). Interestingly, defects in the genes encoding these complexes, particularly the Sgs1-Top3-Rmi1 complex, cause larger increases in GCR rates in duplication-mediated GCR assays compared to single-copy sequence-mediated GCR assays such as the classical GCR assay; defects in SLX1, SLX4, and MPH1 cause little or no increase in GCR rates in single-copy sequence-mediated GCR assays (Putnam et al. 2009a). These results suggest that nonallelic HR is minimized by unwinding D-loops (Mph1-Mte1) or by reversing double Holliday junctions (Sgs1-Top3-Rmi1). Remarkably, the absence of Sgs1 promotes GCRs formed by HR between CAN1 and its divergent homologs ALP1 and LYP1, which likely reflects template switching during nonallelic HR and a relaxation of heteroduplex rejection (Myung et al. 2001b; Spell and Jinks-Robertson 2004; Schmidt et al. 2006b; Smith et al. 2007). Defects in Sgs1-Top3-Rmi1 and Mms4-Mus81 could also cause defects in processing HR intermediates during error-free repair, which could allow damaged DNAs to be acted on by GCR-generating processes such as de novo telomere addition or NHEJ.
NHEJ by itself appears to play only a small role in suppressing the spontaneous formation of GCRs (Figure 11) (Myung et al. 2001a; Putnam et al. 2012, 2014, 2016). Deletion of DNL4 or LIF1, which encode the DNA ligase involved in NHEJ, caused little if any increase in GCR rates in GCR assays that only contain single-copy DNA sequences in the assay chromosome breakpoint region (Myung et al. 2001a; Putnam et al. 2014) and tended to reduce the GCR rates of mutants that have increased GCR rates in the classical GCR assay, consistent with a role for NHEJ in generating GCRs such as translocations selected in these assays (Myung et al. 2001a). However, deletion of DNL4 did cause a modest increase in GCR rates in a duplication-mediated GCR assay, consistent with the possibility that NHEJ channels some DNA damage away from HR, which normally promotes the formation of GCRs by nonallelic HR (Putnam et al. 2014). Deletion of YKU70 or YKU80 also caused only small increases in GCR rates, most notably in duplication-mediated GCR assays, similar to the effect of a DNL4 deletion (Chen and Kolodner 1999; Myung et al. 2001a; Putnam et al. 2014). However, because Ku70 and Ku80 play a role in the synthesis of telomeres by telomerase, they are required for the formation of de novo telomere addition GCRs and, as a consequence, yku70 and yku80 mutations suppress the increased GCR rate caused by a pif1 mutation (Figure 10A) (Myung et al. 2001a).
DNA mismatch repair (MMR) corrects mispaired bases that arise due to errors during DNA replication, and defects in MMR underlie both inherited and sporadic cancers [reviewed in Lagerstedt Robinson et al. (2007), Fishel (2015), Reyes et al. (2015), Heinen (2016)]. MMR also repairs mispaired bases that are formed in heteroduplex HR intermediates (White et al. 1985; Bishop et al. 1987; Borts et al. 1990; Reenan and Kolodner 1992; Haber et al. 1993; Alani et al. 1994; Tham et al. 2016). In addition, MMR has also been implicated in suppressing the formation of GCRs (Figure 11) (Myung et al. 2001b; Putnam et al. 2009a), primarily due to the role of MMR in suppressing HR between divergent homologous sequences, sometimes called homeologous recombination (Bailis and Rothstein 1990; Datta et al. 1996; Myung et al. 2001b; Spell and Jinks-Robertson 2004; Sugawara et al. 2004; Tham et al. 2016). msh2 and msh6 mutations, which inactivate the Msh2-Msh6 mispair recognition complex, resulted in increased GCR rates in duplication-mediated GCR assays with homologies to ectopic sites in the breakpoint region. In contrast, an msh3 mutation, which inactivates the Msh2-Msh3 mispair recognition complex, caused little increase in GCR rates in duplication-mediated GCR assays (Putnam et al. 2009a). The importance of Msh2-Msh6 relative to Msh2-Msh3 in suppressing the formation of these duplication-mediated GCRs is consistent with a heteroduplex rejection mechanism; heteroduplexes formed in this assay are predicted to primarily contain base:base mispairs, which are more readily recognized by Msh2-Msh6 than by Msh2-Msh3 [reviewed in Reyes et al. (2015) and Groothuizen and Sixma (2016)]. Similarly, an msh2 mutation also increased the rate of GCRs selected in GCR assays that only contain single-copy sequences in the breakpoint region, and a fraction of the GCRs recovered were translocations with extended regions of imperfect homology at their breakpoints (Myung et al. 2001b). Another key MMR protein complex, Mlh1-Pms1, at best had a minor role in the suppression of duplication-mediated GCRs (Putnam et al. 2009a), consistent with the fact that Mlh1-Pms1 plays a major role in mispair correction [reviewed in Reyes et al. (2015), Groothuizen and Sixma (2016)] but only plays a minor role in suppression of homeologous recombination (Datta et al. 1996; Sugawara et al. 2004). Sgs1 also plays an important role in both the suppression of homeologous recombination and the suppression of duplication-mediated GCRs, although it may act at a different mechanistic step to Msh2-Msh6, as combining sgs1 and msh2 mutations resulted in a synergistic increase in the rate of both homeologous recombination and duplication-mediated GCRs (Myung et al. 2001b; Spell and Jinks-Robertson 2004; Sugawara et al. 2004; Putnam et al. 2009a).
Postreplication repair (PRR), which is a DNA damage tolerance pathway rather than a DNA repair pathway [reviewed in Branzei (2011), Branzei and Szakal (2016)], plays different roles in the suppression and formation of GCRs that can be identified using different types of GCR assays. Mutations in the upstream genes, RAD6 and RAD18, which encode a ubiquitin E2 conjugase and a ubiquitin E3 ligase, respectively, result in dramatically increased GCR rates in a duplication-containing GCR assay (Putnam et al. 2010). These increased GCR rates caused by rad6 and rad18 mutations are largely similar to the increased GCR rates caused by a mutation that eliminates the Rad6-Rad18-dependent PCNA monoubiquitination site (Hoege et al. 2002; Putnam et al. 2010); these mutations causing increased GCR rates are also epistatic to deletion of the SRS2 antirecombinase, which is upstream of PRR (Lawrence and Christensen 1979). Several subpathways exist downstream of Rad6 and Rad18, including RAD5-dependent subpathways that may act to regress replication forks or to mediate cross-fork template switching (Goldfless et al. 2006; Blastyak et al. 2007; Branzei et al. 2008), and are more important for suppressing GCRs than the downstream translesion polymerases that act in bypassing lesions during DNA replication (Motegi et al. 2006; Putnam et al. 2010). Rad5 is a DNA helicase and a ubiquitin E3 ligase, and both activities are required for repair of UV damage (Gangavarapu et al. 2006); however, only the helicase activity of Rad5 is important in suppressing the formation of duplication-mediated GCRs (Putnam et al. 2010). In contrast, defects in PRR genes by themselves cause little or no increase in GCR rates in GCR assays that only contain single-copy sequences in the assay chromosome breakpoint region (though differing effects have been observed for rad5 and rad18 deletions in different studies), and suppress the increased GCR rates caused by other mutations in these assays (Figure 11) (Motegi et al. 2006; Kats et al. 2009; Putnam et al. 2010). A simple explanation for the assay-specific effects of PRR defects is that PRR downregulates HR in response to DNA replication-induced DNA damage; consequently, PRR defects would result in increased nonallelic HR and increased GCRs selected in duplication-mediated GCR assays, as well as increased allelic HR and suppression of GCRs selected in single-copy sequence-mediated GCR assays.
Mutations in most NER genes, except for genes encoding the Rad1-Rad10 endonuclease, have little or no effect on GCR rates in any of the GCR assays tested (Figure 11) (Hwang et al. 2005; Putnam et al. 2016). However, deletions of RAD1 and RAD10 cause increased rates in duplication-mediated GCR assays. In contrast, these deletions strongly suppress GCR rates in single-copy sequence-mediated GCR assays caused by many other mutations (Figure 10B) (Hwang et al. 2005; Putnam et al. 2009a). Rad1-Rad10 plays roles in other processes besides NER, including processing of HR intermediates (Schiestl and Prakash 1988, 1990; Sugawara et al. 1997), resolving interstrand cross-links (Niedernhofer et al. 2004), and microhomology-mediated end-joining (Ma et al. 2003). Because Rad1-Rad10 is thought to trim unmatched ssDNA overhangs in different types of HR and NHEJ intermediates as well as cleave ssDNA branches, Rad1-Rad10 may facilitate the formation of GCRs by processing DSBs so that they can participate in NHEJ and de novo telomere addition reactions (Hwang et al. 2005). In contrast, similar cleavage of branched intermediates formed during nonallelic HR might reduce the formation of these HR-dependent GCRs.
DNA replication
Spontaneous errors during DNA replication are an important source of the DNA damage that underlies increased genome instability (Flores-Rozas and Kolodner 2000; Michel 2000). For example, DNA replication defects can result in increased accumulation of HR intermediates (Zou and Rothstein 1997), and dysregulation of replication origins results in increased rates of accumulating GCRs (Lengronne and Schwob 2002; Watanabe et al. 2002). Several lines of indirect evidence are also consistent with the notion that DNA replication errors play a major role in the formation of GCRs. (1) Multiple DNA repair pathways, such as HR and PRR (see section DNA repair pathways), which play important roles in suppressing the formation of GCRs, also act in the repair of damaged DNA replication forks and in the formation of new replication fork-like structures during BIR, which is a type of DSB repair [reviewed in Anand et al. (2013), Mehta and Haber (2014)]. (2) S-phase DNA damage and replication checkpoints both suppress spontaneous GCRs and stabilize damaged replication forks (Lopes et al. 2001; Myung et al. 2001c; Myung and Kolodner 2002) (see section S-phase checkpoints). Finally, (3) DNA repair-defective mutations cause increased GCR rates and tend to have large numbers of synthetic genetic interactions with other mutations resulting in growth defects (Tong et al. 2004; Collins et al. 2007; Costanzo et al. 2010), suggesting that repair of DNA damage, possibly occurring during DNA replication, is required during normal growth. Moreover, defects in some essential DNA replication genes have been shown to cause increased GCR rates (Figure 11) (Chen and Kolodner 1999; Putnam et al. 2009a; Chan and Kolodner 2011; Stirling et al. 2011; Shah et al. 2012; Y. Zhang et al. 2013), although definitive analysis of all DNA replication genes has not yet been performed. It should be noted that, in many cases, the defects in essential genes that have been reported to cause increased GCR rates have not been validated using quantitative GCR rate assays that definitively detect the formation of GCRs.
Hypomorphic alleles and alleles that reduce the expression of proteins involved in origin recognition and firing—ORC2, ORC3, ORC5, SLD2, PSF3, CDC7, DBF4, CDC6, and TAH11—have been implicated as causing increased genomic instability (Huang and Koshland 2003; Stirling et al. 2011; Y. Zhang et al. 2012, 2013). Additionally, hypomorphic alleles of genes encoding DNA polymerases and primases, e.g., POL1, POL12, POL2, POL3, PRI1, and PRI2, and the replicative helicases MCM2 and MCM5, have been implicated in causing increased genome instability (Putnam et al. 2009a; Stirling et al. 2011; Shah et al. 2012; Zhang et al. 2012, 2013b). At least one mutation in POL30, which encodes the S. cerevisiae homolog of PCNA, caused a modest increase in GCR rates whereas at least two other pol30 mutations did not cause increased GCR rates (Chen et al. 1998; Chen and Kolodner 1999); because POL30 is an essential gene and because PCNA functions in many processes besides DNA replication, additional studies will be required to determine if replication defects caused by pol30 mutations cause increased GCR rates. Similarly, hypomorphic alleles of RFC2, RFC4, and RFC5, which affect replication factor C, which loads PCNA onto DNA, also cause increased GCR rates (Chen and Kolodner 1999; Myung et al. 2001c; Stirling et al. 2011; Y. Zhang et al. 2012, 2013). Temperature-sensitive and other hypomorphic alleles of RFA1 and truncations of RFA2 and RFA3, which encode subunits of the single-stranded DNA-binding Replication Protein A (RPA), also cause large increases in GCR rates (Chen et al. 1998; Chen and Kolodner 1999; Wang et al. 2005; Y. Zhang et al. 2012, 2013); however, RPA plays multiple roles during DNA metabolism, so these effects may not be specific to DNA replication defects. Deletion of RAD27, which encodes the S. cerevisiae homolog of human Flap Endonuclease 1 (FEN1) and is required for processing the 5′-ends of Okazaki fragments, causes a large increase in genome instability in multiple GCR assays (Chen and Kolodner 1999; Putnam et al. 2009a; Chan and Kolodner 2011). The synthetic lethality observed between rad27 mutations and HR defects implies that the failure to correctly process Okazaki fragments results in DSBs that then result in increased levels of GCRs (Tishkoff et al. 1997; Symington 1998; Loeillet et al. 2005). Some hypomorphic or reduced-expression alleles of DNA2, which encodes a nuclease–helicase involved in Okazaki fragment maturation and has additional roles in regulating telomere length, cause a modest increase in GCR rates (Budd et al. 2006; Stirling et al. 2011; Y. Zhang et al. 2012, 2013). A temperature-sensitive allele of CDC9, which encodes the replicative DNA ligase that primarily functions during lagging strand synthesis, also causes a large increase in GCR rate (Chan and Kolodner 2011; Stirling et al. 2011). Deletions of MRC1 and TOF1, which encode proteins implicated in stabilizing stalled replication forks (Katou et al. 2003) and mediating sister chromatid cohesion (Xu et al. 2007), cause modest increases in spontaneous GCR rates in single-copy sequence-mediated GCR assays and larger increases in GCR rates in duplication-containing GCR assays (Pennaneach and Kolodner 2009; Putnam et al. 2009a, 2012). Moreover, consistent with the genetic evidence for MRC1 and TOF1 acting in parallel pathways in sister chromatid cohesion (Xu et al. 2007), mrc1tof1 double-mutant strains have increased GCR rates relative to the mrc1 and tof1 single-mutant strains (Putnam et al. 2009a, 2012). Based on the observed effects of mutations in the limited number of replication genes analyzed to date, it will be of interest to more exhaustively analyze different defects in essential replication genes for their effects in different quantitative GCR assays.
S-phase checkpoints
S-phase checkpoints were originally identified as pathways that promote cell cycle delay or arrest in S-phase in response to treatment with exogenous DNA-damaging agents (Weinert and Hartwell 1988; Lowndes and Murguia 2000; Michelson and Weinert 2000; Zhou and Elledge 2000; Putnam et al. 2009b). Normally, S-phase checkpoints prevent cells from entering mitosis with unrepaired DNA damage; however, in the presence of some types of long-lived and unrepairable damage, S. cerevisiae cells can undergo a process called adaptation in which cell division can occur even in the presence of S-phase checkpoint signaling (Sandell and Zakian 1993; Toczyski et al. 1997; Lee et al. 1998). Triggering the S-phase checkpoints has multiple cellular consequences: (1) stalled replication forks are maintained in a state that allows them to resume DNA synthesis (Lopes et al. 2001; Tercero and Diffley 2001); (2) late origins are prevented from firing (Santocanale and Diffley 1998; Shirahige et al. 1998); (3) DNA replication is slowed (Paulovich et al. 1997a); (4) cell morphological events and polarized cell growth are delayed (Enserink et al. 2006; Smolka et al. 2006); (5) a transcriptional response driving the production of dNTPs is induced (Allen et al. 1994); (6) progression into anaphase is prevented (Sanchez et al. 1999); and (7) genome-wide postreplicative cohesion is triggered (Strom et al. 2007; Unal et al. 2007). Strikingly, mutations affecting S-phase checkpoints, and in particular the replication checkpoint, cause increases in genome instability in multiple GCR assays (Figure 11) (Myung et al. 2001c; Myung and Kolodner 2002; Huang and Koshland 2003; Putnam et al. 2009a; Chan and Kolodner 2011), consistent with a role for DNA replication errors in the formation of GCRs. In contrast, defects in the classical G1 and G2 DNA damage checkpoints and the mitotic spindle checkpoints had little if any effect on the rate of accumulating GCRs (Myung et al. 2001c).
S-phase checkpoints appear to comprise two separate checkpoints: the DNA replication checkpoint and the intra-S checkpoint. The DNA replication checkpoint is triggered by defects in the replication fork and appears to help maintain damaged replication forks in a state that can resume replication; this stabilization may either be due to suppression of HR acting on stalled replication forks or suppression of mechanisms that generate HR substrates as a result of stalled replication forks (Lisby et al. 2004; Lambert et al. 2007). In contrast, the intra-S checkpoint causes reduced rates of DNA replication and slower cell cycle progression in response to treatment with DNA-damaging agents (Paulovich et al. 1997b; Lowndes and Murguia 2000). Some components of the S-phase checkpoints appear to be specific to the replication checkpoint (RFC5, MRC1, and DPB11) or the intra-S checkpoint (RAD9, RAD17, RAD24, MEC3, and SGS1) (Lowndes and Murguia 2000; Michelson and Weinert 2000; Zhou and Elledge 2000). After phosphorylation by Mec1 or Tel1, Mrc1 and Rad9 separately bind and activate Rad53 and act in the replication checkpoint and intra-S checkpoint, respectively. In contrast, many S-phase checkpoint components act in both pathways (Paulovich et al. 1997a,b; Santocanale and Diffley 1998; Shirahige et al. 1998; Frei and Gasser 2000; Lowndes and Murguia 2000; Zhou and Elledge 2000; Myung and Kolodner 2002), particularly the signal transduction kinases and the effector functions of the checkpoint pathways. Moreover, the two S-phase checkpoints likely have some amount of overlap, as damaged replication forks that are not properly stabilized by the replication checkpoint could undergo modification into substrates that trigger the intra-S checkpoint. Strains with defects in both pathways, such as combining an rfc5-1 or dpb11-1 mutation with a mutation in the rad9, rad17, rad24, or mec3 group, have increased rates of accumulating GCRs (Myung and Kolodner 2002).
The types of GCRs formed in strains with different S-phase checkpoint defects are distinct. In single-copy sequence-mediated GCR assays, the replication checkpoint-specific defect rfc5-1 and the downstream kinase defect mec1 cause the accumulation of only de novo telomere addition GCRs (Myung et al. 2001c; Putnam et al. 2014). This distribution may be due to the inability of strains with defects in Mec1 activation to phosphorylate Cdc13, a protein that plays a role in telomere maintenance (see below), and prevent the recruitment of telomerase to DSBs thereby preventing de novo telomere addition at DSBs (Zhang and Durocher 2010). In duplication-containing GCR assays, mec1 mutations cause increased GCR rates that are higher than in single-copy sequence-mediated GCR assays (Putnam et al. 2009a), suggesting that mec1 defects suppress GCRs through multiple mechanisms whereas the increase in de novo telomere additions in single-copy sequence-mediated GCR assays is likely due only to an increase in the efficiency of de novo telomere addition. Interestingly, strains with a dun1 mutation, which affects a step downstream of mec1, also only accumulate de novo telomere addition-mediated GCRs (Myung et al. 2001c), which could suggest additional levels of control of de novo telomere addition or the accumulation of damage that is readily recognized by telomere maintenance proteins.
In contrast, tel1 mutants do not have increased GCR rates but have an altered GCR spectrum as tel1 mutants do not accumulate de novo telomere addition GCRs (Myung et al. 2001c; Putnam et al. 2014), but rather show an increase in the accumulation of translocations and isoduplications relative to wild-type strains (Myung et al. 2001c; Putnam et al. 2014). As for mec1 mutants, these changes in distribution likely represent changes in the efficiency of different repair pathways, with tel1 mutations causing decreases in the efficiency of de novo telomere additions and a decrease in the efficiency of the pathways that cleave DNA hairpins that can form at DSBs (Putnam et al. 2014). Supporting the view that tel1 mutations do not eliminate de novo telomere additions, tel1 mutations do not suppress the increased GCR rates of pif1 mutants (Myung et al. 2001a; Putnam et al. 2014). Unlike the mec1, tel1, dun1, and rfc5-1 defects, checkpoint defects caused by rad9, mec3, rad53, and chk1 mutations do not dramatically alter the spectrum of GCRs recovered (Myung et al. 2001c; Myung and Kolodner 2002), consistent with the possibility that these defects do not alter telomerase activity.
Chromatin assembly, remodeling, and modification
DNA replication and chromatin assembly are coordinated in eukaryotic cells (Nelson et al. 2002), and failure to complete chromatin assembly during DNA replication causes S-phase arrest (Ye et al. 2003). The chromatin-assembly factor I (CAF-I) and replication-coupling assembly factor (RCAF) complexes assemble chromatin after DNA synthesis (Ransom et al. 2010). CAF-I, which is comprised of Rlf2/Cac1, Cac2, and Msi1/Cac3 in S. cerevisiae, acts as a histone H3-H4 chaperone and also binds PCNA, which targets CAF-I to the replication fork. RCAF consists of Asf1 and a dimer of histones H3 and H4, and RCAF also forms a complex with Rtt109 that acts as an acetyltransferase that promotes the acetylation of histone H3 on K56 (Recht et al. 2006; Driscoll et al. 2007; Han et al. 2007; Tsubota et al. 2007; Ransom et al. 2010). CAF-I and RCAF also function to terminate the DNA damage checkpoint (Kim and Haber 2009). Deletions of RLF2, CAC2, MSI1, and ASF1 all caused increased GCR rates, and asf1 mutations (and to a lesser extent rlf2 mutations) synergized with defects in HR, suggesting that HR suppresses the formation of GCRs in rlf2 and asf1 mutants (Myung et al. 2003). asf1 mutant strains, and to a lesser extent rlf2 mutant strains, have increased levels of Ddc2-GFP foci (Kats et al. 2006), which are a cytological marker of checkpoint activation and DSBs (Melo et al. 2001). This result suggests that defects in replication-associated chromatin assembly result in damaged and possibly broken chromosomes that could underlie the formation of GCRs. This conclusion is consistent with the synergistic increase in GCR rate seen when rlf2 or asf1 mutations are combined with a pif1 mutation (Myung et al. 2003), which increases the efficiency of healing broken chromosomes by de novo telomere addition, resulting in GCRs (see section Telomere maintenance). Interestingly, deletion of RLF2 resulted in synergistic increases in GCR rates when combined with defects in the DNA damage checkpoint but not the DNA replication checkpoint, whereas the deletion of ASF1 resulted in a modest synergistic increase in GCR rates when combined with DNA damage checkpoints defects and a much stronger synergistic increase in GCR rates when combined with DNA replication checkpoint defects (Myung et al. 2003). Similarly, S-phase progression of asf1 mutants showed a dependence on the DNA replication checkpoint, whereas S-phase progression of rlf2 mutants did not show clear checkpoint dependence (Kats et al. 2006). These results suggest that in the absence of RCAF, replication fork instability due to reduced nucleosome disassembly in front of the fork or due to defects of nucleosome assembly after the fork has passed results in increased GCR rates, whereas CAF-I defects may result in some type of DNA damage that persists or occurs after DNA replication is completed and results in high GCR rates.
Control of replication-associated H3-K56 acetylation appears to play an important role in proper chromatin assembly. This histone mark is added to Asf1-presented H3-H4 dimers by Rtt109 in S-phase (Recht et al. 2006; Driscoll et al. 2007; Han et al. 2007; Tsubota et al. 2007), is removed in G2-phase by the Hst3 and Hst4 histone deacetylases in S. cerevisiae (Celic et al. 2006; Maas et al. 2006), and plays a role in promoting expression of some S-phase genes including those encoding histones (Xu et al. 2005). Deletion of ASF1 or RTT109 results in similar levels of increased GCR rates, increased sensitivity to DNA-damaging agents, slowed growth, increased checkpoint activation, increased Rad52 foci [a cytological marker of HR intermediates (Lisby et al. 2001, 2003)], and increased sister chromatid HR (Myung et al. 2003; Kats et al. 2006; Driscoll et al. 2007; Putnam et al. 2009a, 2012, 2016; Chan and Kolodner 2011, 2012; Munoz-Galvan et al. 2013). Similarly, mutations in the genes encoding histone H3, which prevent acetylation of H3-K56, also resulted in increased GCR rates albeit not as high as caused by asf1 or rtt109 mutations (Chan and Kolodner 2011, 2012). Thus, in aggregate, these results suggest that acetylation of histone H3-K56 accounts for some of the role of RCAF in suppressing GCRs. In addition, strains with an asf1 mutation, an rtt109 mutation, or mutations in the genes encoding histone H3 preventing acetylation of H3-K56, had increased levels of aneuploidy, predominantly involving duplication of chromosomes XII and VII (Chan and Kolodner 2011, 2012). In contrast, the Asf1-Vps75 histone acetyltransferase did not appear to play a role in suppressing either GCRs or aneuploidy (Chan and Kolodner 2012).
Cells lacking H3-K56 acetylation have very similar phenotypes to those with hyperacetylation of H3-K56 resulting from defects in HST3 and HST4, including increased GCR rates, sensitivity to DNA-damaging agents, increased checkpoint activation, increased Rad52 foci, and increased sister chromatid HR (Celic et al. 2008; Kadyrova et al. 2013; Munoz-Galvan et al. 2013; Che et al. 2015; Putnam et al. 2016). In addition, hst3hst4 double-mutant strains appear to have defects in sister chromatid cohesion and in BIR due to inhibition of repair DNA synthesis (Thaminy et al. 2007; Che et al. 2015). Remarkably, at least some defects in the hst3hst4 double-mutant strain can be suppressed by overexpression of Rfc1 (Celic et al. 2008), which is a subunit of the PCNA clamp loader, and by inactivation of the alternative clamp loaders Ctf18, Rad24, and Elg1, which are involved in activation of the DNA damage checkpoint and removal of PCNA [reviewed in Kupiec (2016)]. These results could be consistent with the ability of increased recruitment of CAF-I by PCNA to suppress the defect caused by hyperacetylation of H3-K56. Taken together, the similarities between the hyperacetylation and hypoacetylation of H3-K56 argue that they may affect the same process, and possibly that DNA replication and/or repair of DNA replication errors depends upon having unmarked histones preceding the replication fork and marked histones following it.
In addition to CAF-I and RCAF, there are many other proteins and protein complexes that remodel and/or modify chromatin [reviewed in Cairns (2009), Gerhold et al. (2015)]. Most of these have been discovered through studies of transcription and gene regulation. Some, like the Swr1 complex and the Ino80 complex, have been implicated in DNA repair, as have histone modifications that occur in response to treating cells with DNA-damaging agents (Morrison et al. 2004; van Attikum et al. 2004, 2007). However, defects affecting some of these proteins and protein modifications have been reported to only modestly increase sensitivity to DNA-damaging agents, to only modestly decrease excision from DSBs (Morrison et al. 2004; van Attikum et al. 2004, 2007; Chen et al. 2012; Costelloe et al. 2012), and in some cases to cause increased GCR rates (Myung et al. 2003; Putnam et al. 2009a, 2012, 2016). However, the observed increases in GCR rates reported were relatively small, and only a small number of the genes encoding any individual chromatin modification/remodeling complexes were identified in genetic screens for GCR-suppressing genes, consistent with only minor roles in suppressing GCRs. In contrast, many more genes encoding such complexes were identified in the genetic screen for cGIS genes (Putnam et al. 2016). Therefore, with the exception of CAF-I and RCAF, most chromatin remodeling and modifying complexes likely play minor roles in suppressing GCRs by themselves but may cooperate with other complexes and pathways to suppress GCRs.
Telomere maintenance
In S. cerevisiae, telomeres are maintained by the reverse transcriptase telomerase, consisting of the proteins Est1, Est2, and Est3, and the RNA subunit TLC1 [reviewed in Kupiec (2014)]. In the absence of telomerase, telomeres undergo continuous shortening, which eventually leads to senescence (Lundblad and Szostak 1989; Singer and Gottschling 1994; Shore 1998). The onset of senescence occurs when erosion of at least one chromosome leads to activation of a DNA damage checkpoint response (d’Adda di Fagagna et al. 2003; Abdallah et al. 2009; Xu et al. 2013). Within the senescing cell population, survivors can arise that maintain their chromosome ends by one of two different HR-mediated processes that can be distinguished due to differences in the resulting telomeric structures (amplification of either Y’ subtelomeric regions or telomeric repeats) and the genetic requirements for the formation of each type of survivor [reviewed in McEachern and Haber (2006)].
McClintock first demonstrated that telomeres protect chromosome ends, preventing breakage–fusion–bridge cycles (McClintock 1939; de Lange 2002). In S. cerevisiae strains that have recovered from senescence caused by loss of telomere maintenance, the absence of telomerase activity does not cause increased GCR rates because, under these conditions, HR maintains telomeres (Myung et al. 2001a; McEachern and Haber 2006). Similarly, the deletion of many genes encoding factors required for optimal telomere maintenance result in shortened telomeres (e.g., tel1, rnh201, sin3, soh1, ctk1, nam7, and xrn1) (Askree et al. 2004; Gatbonton et al. 2006; Ungar et al. 2009) and do not result in increased GCR rates (Putnam et al. 2016), although some deletions (e.g., mre11, xrs2, and rad50) that result in shorter telomeres as well as other defects do cause increased GCR rates (Chen and Kolodner 1999). Synergistic increases in GCR rates are seen when telomerase defects (e.g., tlc1 and est2) or defects resulting in shorter telomeres reflecting reduced efficiency of telomere maintenance (e.g., tel1) are combined with defects in other pathways including HR (e.g., rad51 and rad59) and the DNA damage checkpoints (e.g., mec1), but not the replication checkpoint (Myung et al. 2001a). Analysis of the structure of the GCRs recovered from these types of double-mutant strains has revealed the formation of monocentric translocations and circular chromosomes as well as dicentric translocations including translocations mediated by telomere-to-telomere fusions, telomere to broken chromosome end fusions, broken chromosome end-to-end fusions, and dicentric isoduplications, all of which are subsequently resolved to monocentric translocations by additional rounds of rearrangement (Myung et al. 2001a; Craven et al. 2002; Pennaneach and Kolodner 2004, 2009). The observation of GCRs that did not appear to involve telomere-to-telomere or telomere-to-DSB fusions suggests that, in addition to GCRs mediated by aberrant telomeres, there may also be increased general chromosome fragmentation in strains with telomerase dysfunction-driven genome instability. Many of the same types of GCRs, as well as truncated chromosomes potentially healed by de novo telomere addition, can be seen in senescing est1Δ cells that have been stabilized by the reintroduction of EST1 (Hackett et al. 2001).
Together, these results provide some insight into how genome instability is driven by telomere dysfunction. Erosion of telomeres past a critical length eliminates the protective features that keep telomeres from being recognized as DNA damage and allows them to be acted upon by other DNA repair pathways [reviewed in Eckert-Boulet and Lisby (2010)]. HR is the most efficient pathway that acts on the resulting chromosome ends, channeling them into alternative telomere maintenance pathways. However, when HR or the DNA damage checkpoints are compromised, the deprotected telomeres and telomeres formed by HR can be acted on by other repair pathways leading to GCRs (Pennaneach and Kolodner 2004, 2009). In addition, Exo1 and potentially other enzymes can degrade the deprotected ends to produce substrates for BIR (Dewar and Lydall 2010); these substrates can lead to both nonreciprocal monocentric and dicentric translocations with other chromosomes and can undergo intramolecular hairpin formation leading to dicentric isoduplications (Pennaneach and Kolodner 2004, 2009). Mutations that result in inefficient telomere maintenance leading to shortened telomeres (e.g., tel1) also show similar genetic interactions with HR and checkpoint defects, resulting in increased rates of accumulating GCRs, particularly those mediated by telomere-to-telomere fusion (Craven et al. 2002; Pennaneach and Kolodner 2004, 2009). Consistent with these results, expression of a Cdc13-Est2 fusion, which allows telomere extension in the absence of Tel1 and Mec1 (Tsukamoto et al. 2001), reduced the frequency of chromosomal rearrangements in a mec1tel1 double mutant (McCulley and Petes 2010). It should be noted that 205 mutations have been identified as causing shortened telomeres (Askree et al. 2004; Gatbonton et al. 2006; Ungar et al. 2009); however, many of these mutations have not yet been studied in GCR assays to determine if they cause increased GCR rates by themselves or in combination with other mutations.
Suppression of inappropriate telomere addition
A key problem for cells with functional telomerase is to ensure that telomere addition is properly targeted to the chromosome ends and does not occur at DSBs. The earliest studies of GCRs observed GCRs that appeared to be formed by chromosome breakage followed by healing of the broken chromosomes by de novo telomere addition; these telomere additions did not appear to target any type of significant telomere seed sequence (Chen et al. 1998; Chen and Kolodner 1999). At least three mechanisms suppress de novo telomere additions at DSBs, thereby facilitating the repair of DSBs by HR.
The first mechanism is the regulation of telomerase by the Pif1 DNA helicase (Schulz and Zakian 1994; Zhou et al. 2000; Mangahas et al. 2001). The Pif1 DNA helicase was identified in a genetic screen to detect telomere maintenance functions (Schulz and Zakian 1994; Zhou et al. 2000), and mutations in PIF1 result in a 240- to 1000-fold increase in the rate of accumulating spontaneous GCRs in which terminal chromosome deletions are healed by de novo telomere addition (Myung et al. 2001a). Thus, Pif1 defines an enzymatic pathway that suppresses de novo telomere additions and de novo telomere addition-driven genome instability (Schulz and Zakian 1994; Zhou et al. 2000; Myung et al. 2001a); however, other components of this pathway, if any exist, have not yet been identified. Consistent with this result, the increase in GCR rate caused by pif1 mutations can be suppressed by mutations in genes encoding proteins and RNA required for normal telomerase activity (est1, est2, est3, and tlc1), Cdc13 (cdc13), and Ku (yku70 and yku80) (Figure 10A) (Myung et al. 2001a). The GCR spectrum in pif1 mutant strains is most consistent with a role of Pif1 in suppressing de novo telomere addition by removal of telomerase from DSBs (Eugster et al. 2006; Boule and Zakian 2007; Li et al. 2014) and not the recently discovered role of Pif1 in recombination-coupled DNA synthesis (Saini et al. 2013; Wilson et al. 2013); however, both roles might act to promote de novo telomere addition, as the failure of recombination-coupled DNA synthesis in pif1 mutants might generate substrates for telomerase or block their processing by other pathways like BIR.
The second mechanism is the inhibition of the action of telomerase at DSBs by the DNA damage checkpoint kinase Mec1, which phosphorylates Cdc13, preventing the accumulation of Cdc13 at DSBs (Zhang and Durocher 2010) (see section S-phase checkpoints). Because Cdc13 facilitates the recruitment of telomerase at DSBs where de novo telomere additions occur (Bianchi et al. 2004), Mec1 activity downregulates de novo telomere additions and de novo telomere addition-driven GCRs.
The third mechanism is that de novo telomere addition may be limited by the normal cell cycle regulation of telomerase activity. Telomere maintenance functions act on normal telomeres starting in late S-phase (Marcand et al. 2000), and the activity of telomere maintenance functions on telomere “seed” sequences located near an HO-induced DSB appears to be upregulated in G2 (Diede and Gottschling 1999). Spontaneous GCRs appear to result from errors or damage that occur during S-phase (Myung et al. 2001c; Myung and Kolodner 2002); therefore, this normal regulation of telomerase activity, combined with the activation of S-phase checkpoints by DNA damage potentially reduces de novo telomere addition-driven GCRs.
The Hrq1 helicase has also been suggested to play a role in suppressing de novo telomere additions (Bochman et al. 2014). This conclusion was based on the observation that the GCR spectrum in the classical assay in a wild-type strain had no de novo telomere additions (0%), and the GCR spectrum of the hrq1Δ mutant was dominated by de novo telomere additions (77%) (Paeschke et al. 2013; Bochman et al. 2014). However, in other studies, the GCR spectrum of the wild-type strain in the classical GCR assay is typically dominated by de novo telomere additions (Chen and Kolodner 1999; Putnam et al. 2004). Moreover, while the hrq1Δ mutation causes increased GCR rates, it did not cause the same GCR rate in both duplication-mediated and single-copy sequence-mediated GCR assays like a pif1 mutation that results in increased de novo telomere additions (Putnam et al. 2010). Thus, it seems unlikely that Hrq1 plays a Pif1-like role in suppressing de novo telomere addition.
Smc5-6 and protein sumoylation
S. cerevisiae contains three complexes containing members of the structural maintenance of chromosome (SMC) family [reviewed in Jeppsson et al. (2014), Kschonsak and Haering (2015)]. Cohesin (Smc1-Smc3) and condensin (Smc2-Smc4) play important roles during mitosis in sister chromatid cohesion and chromosome condensation. The third complex, made up of Smc5-Smc6 and a number of non-Smc proteins (Nse1-Nse6), is important for some types of DNA repair including promoting error-free sister-chromatid HR (De Piccoli et al. 2009). Consistent with these results, mutations in several genes encoding the Smc5-Smc6 cohesion complex cause increased GCR rates (Figure 11) (De Piccoli et al. 2006; Hwang et al. 2008; Stirling et al. 2011; Albuquerque et al. 2013). The most studied allele, a hypomorphic allele of SMC6, smc6-9, causes increased rates of accumulating GCRs that were primarily nonreciprocal translocations with microhomology breakpoints; the formation of these translocations was dependent on HR and Pol32, which is a subunit of DNA polymerase δ, suggesting the involvement of BIR in their formation (De Piccoli et al. 2006; Hwang et al. 2008; Stirling et al. 2011). Mutations in MMS21/NSE2 and NSE3 cause increased GCR rates in quantitative assays (Hwang et al. 2008; Albuquerque et al. 2013), whereas other SMC family complexes have not been as extensively investigated (Huang and Koshland 2003; Stirling et al. 2011).
Mms21/Nse2 is a small ubiquitin-like modifier (SUMO) E3 ligase that is a component of the Smc5-Smc6 complex and mediates the addition of the ubiquitin-like SUMO protein onto different target proteins (Zhao and Blobel 2005). The MMS21 gene is essential in S. cerevisiae; however, its sumoylation activity is dispensable for viability when the mitotic SUMO E3 ligases, Siz1 and Siz2, are functional (Reindle et al. 2006). Mms21 has a different subset of sumoylation targets and plays a more important role in suppressing GCRs than Siz1 or Siz2 (Albuquerque et al. 2013). Moreover, ESC2, which encodes a protein with multiple SUMO-like domains (Novatchkova et al. 2005), functions in conjunction with Smc5-Smc6 in the repair of DNA damage during replication (Mankouri et al. 2009; Choi et al. 2010) and plays an important role in suppressing GCRs (Putnam et al. 2009a; Allen-Soltero et al. 2014). Esc2 is also a positive regulator of protein sumoylation by Mms21 (Albuquerque et al. 2013), suggesting that esc2 and mms21 mutants share a common defect. Mms21 targets include nucleolar proteins, such as RNA polymerase I, Fob1, and Tof2, as well as cohesin and condensin subunits (Albuquerque et al. 2013); however, the specific sumoylation events that are responsible for suppressing GCRs have not yet been determined.
Slx5-Slx8 is a SUMO-targeted E3 ubiquitin ligase complex (Prudden et al. 2007; Xie et al. 2007) that localizes preferentially to the nuclear pores where long-lived DSBs and eroded telomeres are relocalized and repair by HR is suggested to occur (Nagai et al. 2008). This relocalization appears to be dependent on sumoylation of proteins bound to the damaged DNA (Chung et al. 2015; Churikov et al. 2016; Horigome et al. 2016). Consistent with a role in this process, mutations affecting some of the nuclear pore subcomplexes, such as nup84, nup120, and nup133, cause increased GCR rates (Putnam et al. 2012, 2016), and cause lethality when combined with mutations such as rad27, which are thought to cause increased levels of the DNA damage that can underlie GCRs (Loeillet et al. 2005). Deletion of SLX5 or SLX8 causes a large increase in GCR rates in duplication-mediated but not single-copy sequence-mediated GCR assays (Putnam et al. 2009a) and an increase in the general level of sumoylated proteins, with the strongest influence being on the level of Mms21 targets (Albuquerque et al. 2013). Both the positive regulators of sumoylation of Mms21 targets (MMS21 and ESC2) and the negative regulators of sumoylation of Mms21 targets (SLX5 and SLX8) play roles in suppressing GCRs. Thus, regulating the levels of sumoylation by Mms21 and potentially the dynamics of these events is likely important in maintaining genome stability.
Oxidative stress response
Increased levels of oxidative stress also underlie increased genome instability. Deletions of TSA1, encoding the major thioredoxin peroxidase that scavenges hydrogen peroxide in S. cerevisiae, and SKN7 and YAP1, encoding transcription factors that control an oxidative stress response (Lee et al. 1999), were identified in a systematic screen for mutator mutants and caused increased rates of accumulating GCRs (Huang et al. 2003). In contrast, both targeted genetic analysis and systematic screens for GCR-suppressing genes did not identify other potential oxidative stress response genes such as those encoding superoxide dismutases, catalases, and thioredoxins as GCR-suppressing genes (Huang et al. 2003; Smith et al. 2004; Kanellis et al. 2007; Stirling et al. 2011; Y. Zhang et al. 2013; Putnam et al. 2016). The idea that TSA1 likely suppresses some type of DNA damage is underscored by the observations that tsa1 mutations result in: (1) synthetic lethal or synthetic slow growth interactions with rad52, mre11, rad50, xrs2, sgs1, rad6, rad18, rad5, and mec1 mutations (Huang and Kolodner 2005); (2) increased levels of Rad52-YFP foci (Ragu et al. 2007); (3) increased levels of intracellular reactive oxygen species (Wong et al. 2002); (4) modestly increased sensitivity to DNA-damaging agents (Tang et al. 2009); (5) elevation of intracellular dNTP pools (Tang et al. 2009); and (6) activation of the DNA damage checkpoint (Tang et al. 2009). In addition, a tsa1 deletion mutation resulted in a synergistic increase in GCR rate when combined with a pif1 mutation or an ogg1 mutation, which resulted in increased healing of broken DNAs by de novo telomere addition and decreased BER of oxidative DNA damage, respectively (Huang et al. 2003; Huang and Kolodner 2005). Moreover, anaerobic growth of S. cerevisiae suppressed the increased GCR rate caused by deletion of TSA1 and some but not all DNA repair genes, and alleviated the synthetic growth interactions between a tsa1 deletion and deletions of different DNA repair genes (Ragu et al. 2007). Additionally, a mutation in the Skn7- and Yap1-activated gene TRR1, which encodes thioredoxin reductase, was found to suppress the synthetic lethality between deletions of TSA1 and RAD51, potentially by both decreasing intracellular reactive oxygen species through Yap1 activation and by reducing intracellular dNTP pools by reducing the activity of ribonucleotide reductase (Ragu et al. 2014). It is unclear why TSA1 is more important than other enzymes involved in detoxifying reactive oxygen species; however, the basal level of expression of TSA1 is much higher than its paralog TSA2, which could account for its relative importance (Wong et al. 2002).
R-loop formation
R-loops are three-stranded RNA–DNA hybrids in which a stretch of RNA displaces one strand of a complementary dsDNA molecule and are formed during transcription [reviewed in Costantino and Koshland (2015), Santos-Pereira and Aguilera (2015), Sollier and Cimprich (2015)]. In transcription, R-loops are thought to be mostly transient; however, features of the displaced ssDNA strand, such as its propensity to form secondary structure, have been suggested to facilitate the formation of long-lived R-loops [reviewed in Costantino and Koshland (2015)]. Similarly, defects in RNA processing and transcriptional elongation, including indirect topological defects induced by loss of the Top1 or Top2 topoisomerases, have been linked to the formation of R-loops (El Hage et al. 2014). In the “thread-back model,” transient underwinding of dsDNA behind the transcription machinery was proposed to promote pairing with the nascent RNA molecule (Liu and Wang 1987). In addition, recent experiments have indicated that the HR machinery can utilize RNA molecules and that Rad51-dependent HR can promote the formation of R-loops in strains with RNA metabolism defects (Wahba et al. 2013; Keskin et al. 2014), suggesting that RNA–DNA hybrids do not exclusively result from long-lived transcription intermediates.
R-loops can be removed in several ways. First, helicases, such as Sen1 (homolog of human senataxin), can unwind RNA–DNA hybrids (Mischo et al. 2011). Second, the RNA strand of RNA–DNA hybrids can be degraded by RNase H enzymes [reviewed in Cerritelli and Crouch (2009)]. RNase H1, which is encoded by RNH1, can only degrade stretches of consecutive ribonucleotides, and RNase H2, which is encoded by RNH201, RNH202, and RNH203, can cleave single misincorporated ribonucleotides in addition to degrading longer stretches of RNA [reviewed in Cerritelli and Crouch (2009)]. Third, RNA export, such as that mediated by the THO complex, and RNA degradation, such as that mediated by the RNA exosome, also reduce the level of R-loops, potentially through interactions with unpaired portions of the RNA molecules that are not in the RNA–DNA hybrid (Wahba et al. 2011, 2013; Luna et al. 2012).
Several lines of evidence indicate that R-loops can be a source of DNA damage leading to GCRs. First, many mutations that cause increased R-loop formation also cause increased formation of Rad52-GFP foci, which in many cases can be suppressed by overexpression of RNase H1 (Gomez-Gonzalez et al. 2009; Mischo et al. 2011; Wahba et al. 2011; Stirling et al. 2012; Castellano-Pozo et al. 2013). While these data suggest that R-loops may be processed to DSBs, leading to the formation of HR intermediates, it is possible that in some cases Rad52 foci could reflect R-loop formation by HR. Second, a number of mutations that cause accumulation of R-loops also cause synthetic growth defects when combined with mutations in S-phase checkpoint genes and some HR genes (Gomez-Gonzalez et al. 2009; Mischo et al. 2011). Third, some mutations that cause accumulation of R-loops also cause increased rates of HR of direct-repeat recombination substrates (Huertas and Aguilera 2003; Mischo et al. 2011; Castellano-Pozo et al. 2013), plasmid loss (Castellano-Pozo et al. 2013), small increases in GCR rates in the classical GCR assay (deletion of HPR1) (Gomez-Gonzalez et al. 2009), increased GCRs in a S. cerevisiae artificial chromosome (YAC)-based GCR assay (Wahba et al. 2011), and an increase in LOH on chromosomes III and XII in diploid strains, mediated by nonreciprocal translocations between homologous chromosomes (Wahba et al. 2011). Fourth, loss of both RNase H activities causes increased accumulation of damage in the G2–M-phase of the cell cycle (Amon and Koshland 2016); however, increases in mitotic recombination in strains with RNase H deficiencies has been alternately attributed to primarily R-loops (O’Connell et al. 2015), only ribonucleotide misincorporation by DNA polymerases (Conover et al. 2015), or both (Cornelio et al. 2017). Fifth, overexpression of SPT2, which appears to result in increased R-loop formation, causes increased accumulation of GCRs (Sikdar et al. 2008). In many cases, these increased levels of genome instability can be suppressed by overexpression of RNH1. How R-loops cause DNA damage is unclear. Current models include the possibility of collisions between replication forks and R-loops as well as through cleavage of the R-loop by nucleases, such as those involved in NER [reviewed in Sollier and Cimprich (2015)], resulting in a DSB. In contrast to the accumulated data suggesting that R-loops mediate genome instability, one recent report suggests that transient RNA–DNA hybrids are formed at resected DSBs and help promote repair, potentially by promoting further resection via disruption of the adjacent chromatin structure (Ohle et al. 2016). These results suggest that RNA–DNA hybrids may both promote genome instability and promote DSB repair, depending on the precise context of the hybrid.
The following pathways have been implicated in suppressing R-loop-mediated genome instability: (1) transcription initiation [BUR2; Wahba et al. (2011); (2) transcription elongation by the PAF1 complex [CDC73 and LEO1; Wahba et al. (2011, 2013)] and Spt2 (Sikdar et al. 2008; Wahba et al. 2011); (3) transcriptional repression by the RPD3 histone deacetylase complex [SIN3, RPD3, and SDS3; Wahba et al. (2011); Chan et al. (2014)], Not5, and Stb3 (Wahba et al. 2011); (4) mediator functions [MED1, MED5, MED12, MED13, and CDK8; Wahba et al. (2011, 2013)]; (5) transcriptional termination by CF1A [CLP1, PCF11, RNA15, CFT2, and FIP1; Stirling et al. (2012)], Pbp1 (Salvi et al. 2014), Sen1 (Mischo et al. 2011), and Rtt103 (Stirling et al. 2012); (6) RNA transport by the THO complex (THO1, HPR1, MFT1, and THP2) and Npl3 (Huertas and Aguilera 2003; Gomez-Gonzalez and Aguilera 2007; Gomez-Gonzalez et al. 2009; Wahba et al. 2011; Stirling et al. 2012; Castellano-Pozo et al. 2013; Pfeiffer et al. 2013); (7) RNA degradation by Kem1/Xrn1, Rrp6, and Trf4 (Wahba et al. 2011, 2013); and (8) the Srm1 Ran guanyl-nucleotide exchange factor (Stirling et al. 2012). Consistent with the observations that most mutations causing the accumulation of R-loops that have been tested in GCR assays cause only small increases in GCR rates (Gomez-Gonzalez et al. 2009), most of these genes and pathways were not identified in a genome-wide screen for genes that suppress the accumulation of GCRs (Putnam et al. 2016). However, several of these genes and pathways were identified in a screen for cGIS genes (see section A global view of genome instability suppressing genes) (Putnam et al. 2016). Thus, it seems likely that mutations causing the accumulation of R-loops result in DNA damage that is acted on by different pathways including checkpoints and HR, which suppress the formation of GCRs that might otherwise result from R-loop formation.
Ribonucleotide misincorporation
Misincorporation of single ribonucleotide bases by replicative DNA polymerases has recently been identified as a source of DNA damage. An estimated 10,000 ribonucleotides are removed during each cell division via a process called ribonucleotide excision repair (Nick McElhinny et al. 2010b; Sparks et al. 2012; Chon et al. 2013). These repair events are dependent upon the ability of RNase H2 to cleave single ribonucleotides in DNA (Jeong et al. 2004) in conjunction with nick-directed DNA synthesis by DNA polymerase δ, flap cleavage by Rad27/FEN1, and ligation of the flap-excised product by DNA ligase (Stith et al. 2008; Burgers 2009; Sparks et al. 2012). RNase H2-defective mutants have a weak mutator phenotype, primarily due to the accumulation of two-base deletion mutations in repeat sequences (Nick McElhinny et al. 2010a; Kim et al. 2011; Allen-Soltero et al. 2014). The mutations appear to result from the cleavage of the DNA strand containing the misincorporated ribonucleotide by the topoisomerase Top1, leading to formation of a ssDNA gap, followed by realignment of the DNA strands and repair of the gap (Kim et al. 2011). Mutations in the genes encoding RNase H2 also cause little or no increase in the rate of accumulating GCRs (Huang et al. 2003; Sikdar et al. 2008; Putnam et al. 2016; Allen-Soltero et al. 2014); however, there is evidence for increased mitotic recombination in diploid strains due to misincorporated ribonucleotides (Conover et al. 2015; O’Connell et al. 2015; Cornelio et al. 2017). In addition, RNase H2 mutations cause decreased growth rates, altered cell cycle distribution, and aberrant cell morphology when combined with mutations affecting DNA damage signaling (e.g., rad53), PRR (e.g., rad5 and rad6), or HR (e.g., rad52, rad51, sgs1, mre11, and mus81) (Allen-Soltero et al. 2014). In some cases, the decreased growth rates of the double mutants were suppressed by a rad51 mutation, implicating recombination intermediates as a cause of the slow growth. Moreover, many of the rnh double mutants with slow growth phenotypes also had synergistic increases in GCR rates, and in some, but not all, of the double mutants the increased GCR rates as well as the aberrant cell morphology could be suppressed by top1 and rad51 mutations (Allen-Soltero et al. 2014). Thus, RNase H2-defective mutations appear to act as damage-generating defects in the damage/response GCR model and primarily lead to increased accumulation of GCRs when combined with defects in pathways that process this damage. In some cases, these increased levels of GCRs appear to result from the cleavage of misincorporated ribonucleotides by Top1, potentially leading to the formation of inappropriate HR intermediates.
Perspectives
In the 20 years since the identification of the first S. cerevisiae mutants with increased rates of accumulating GCRs and the development of the first quantitative GCR assays, considerable insights have been obtained into how spontaneous GCRs arise and are prevented. As discussed in this review, these include: (1) the identification of genes and pathways that suppress the formation of GCRs, (2) the identification of pathways that form GCRs, and (3) the initial identification of an extensive genetic network that functions in the suppression of GCRs. However, there are several aspects of the genome instability problem that are not as well-understood.
First, the spectrum of GCRs has only been determined for a small proportion of strains containing either individual or combinations of GCR-causing mutations, despite the fact that the structures of these GCRs provide important clues into the underlying mechanisms by which GCRs can be formed. Moreover, even for better-studied mutant strains with altered GCR rates and altered GCR structures, the number of GCRs analyzed has been relatively small (< 20); it is unclear whether increasing the number of GCRs analyzed per strain would provide greater insight. Although the analysis of the structures of GCRs is still time-consuming and expensive, modern techniques like next-generation whole-genome sequencing have improved our ability to analyze more GCRs to thoroughly characterize more mutant strains and more samples per mutant.
Second, only a limited analysis of the role of essential genes in suppressing GCRs has been performed. These studies are complicated by several technical factors: (1) different hypomorphic alleles of an individual gene often cause different phenotypes and the available collection of alleles for any gene might not encompass all possible defects that might result from mutations in that gene; and (2) strains containing defects in essential genes often grow poorly, which complicates systematic screening efforts. Despite these problems, some essential genes are known that act in processes that are known or suspected to play roles in suppressing GCRs or in which defects might be expected to increase GCR rates. Analysis of existing mutations in these genes and screening for new mutations that cause altered GCR rates and altered GCR spectra should provide important insights and useful tools for understanding DNA metabolic errors that underlie the formation of GCRs.
Third and finally, identification of genetic interactions that cause increased GCR rates in both hypothesis-driven and systematic studies is in its infancy given the large numbers of GIS and cGIS genes identified to date [for example, see Myung et al. (2001a), Hwang et al. (2005), Putnam et al. (2016)]. Despite the technical challenges, fully characterizing even small portions of the total network of these interactions, particularly in conjunction with fully characterizing the structures of the GCRs that result from genetic interactions that cause altered GCR rates, has the potential to greatly improve our understanding of how these pathways function to preserve the structure of the genome.
The ultimate goal of studying the pathways that suppress and promote the formation of GCRs is to understand the underlying mechanisms that generate the DNA damage that initiates the formation of GCRs. One of the challenges of such studies is that the rates of accumulating GCRs are low even in mutants with high GCR rates. Thus, it is currently impossible to follow a single GCR-generating event from initiating damage to final GCR. However, a clearer understanding is likely to emerge from the analysis of the structure of GCRs, a full characterization of the genetic interactions between GCR-causing/-altering mutations, and integration of these data with data from other mechanistic studies.
Studies of the pathways that prevent or promote GCRs in S. cerevisiae are particularly relevant to our understanding of genome instability in cancer. The accumulation of genome rearrangements or GCRs is characteristic of many cancers (Lengauer et al. 1998; Thompson and Compton 2011; Vogelstein et al. 2013; Kass et al. 2016). Whether there is a genetic basis for the accumulation of GCRs in cancer, either inherited or somatic, has not been well-established for all cancers that appear to show ongoing genome instability. There are some clear examples of genetic defects that underlie human cancers with genome instability where the comparable defect in S. cerevisiae or other model organisms, including human cell lines, causes genome instability. For example, defects in the mediator protein BRCA2, which is essential for HR because it loads RAD51 onto DNA [reviewed in West (2003)] likely result in genome instability similar to loss of the S. cerevisiae mediator protein Rad52 (Tutt et al. 1999; Yu et al. 2000). Defects in other genes that act in the BRCA2-dependent HR and DNA damage response pathways, such as BRCA1, the genes encoding BRCA1- and BRCA2-interacting proteins, and the Fanconi Anemia genes, also appear to result in increased genome instability [reviewed in Moldovan and D’Andrea (2009), Konstantinopoulos et al. (2015)]. In the case of the S. cerevisiae genes encoding homologs or functional analogs of these proteins, defects in these genes are known to cause increased GCR rates (Chen and Kolodner 1999; Myung et al. 2001b,c; Yan et al. 2010; Chan and Kolodner 2011; Putnam et al. 2016). Other examples include the BLM gene, DNA damage response genes such as ATM and ATR, and the MMR genes (Gobbini et al. 2013; Sarbajna and West 2014; Lee et al. 2016; Schmidt and Pearson 2016); MMR suppresses the accumulation of mutations as well as GCRs that are mediated by HR between divergent sequences (Putnam et al. 2009a; Chan and Kolodner 2011). It is difficult to directly screen for GCR-suppressing genes in mammalian cells due to the lack of convenient genetic tests. However, mining cancer genomics data using a list of the human homologs of S. cerevisiae GIS genes has identified many GIS genes that are potentially defective in cancers with genome instability, further establishing S. cerevisiae as a useful tool for obtaining insights into genome instability in cancer (Putnam et al. 2016).
Acknowledgments
The authors would like to thank Anjana Srivatsan for helpful comments on the manuscript and Binzhong Li for the Pulsed-Field Gel Electrophoresis image. Work in the authors’ laboratory was supported by the Ludwig Institute for Cancer Research and National Institutes of Health grant R01 GM26017.
Footnotes
Communicating editor: R. Rothstein
Literature Cited
- Abdallah P., Luciano P., Runge K. W., Lisby M., Geli V., et al. , 2009. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 11: 988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Admire A., Shanks L., Danzl N., Wang M., Weier U., et al. , 2006. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 20: 159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenova A. Y., Greenwell P. W., Dominska M., Shishkin A. A., Kim J. C., et al. , 2013. Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc. Natl. Acad. Sci. USA 110: 19866–19871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Reenan R. A., Kolodner R. D., 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque C. P., Wang G., Lee N. S., Kolodner R. D., Putnam C. D., et al. , 2013. Distinct SUMO ligases cooperate with Esc2 and Slx5 to suppress duplication-mediated genome rearrangements. PLoS Genet. 9: e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. B., Zhou Z., Siede W., Friedberg E. C., Elledge S. J., 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8: 2401–2415. [DOI] [PubMed] [Google Scholar]
- Allen-Soltero S., Martinez S. L., Putnam C. D., Kolodner R. D., 2014. A Saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol. Cell. Biol. 34: 1521–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon J. D., Koshland D., 2016. RNase H enables efficient repair of R-loop induced DNA damage. Elife 5: e20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R. P., Lovett S. T., Haber J. E., 2013. Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 5: a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R. P., Tsaponina O., Greenwell P. W., Lee C. S., Du W., et al. , 2014. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 28: 2394–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Westmoreland J., Mieczkowski P. A., Gawel M., Petes T. D., et al. , 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105: 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneric M., Lingner J., 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 8: 1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree S. H., Yehuda T., Smolikov S., Gurevich R., Hawk J., et al. , 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis A. M., Rothstein R., 1990. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair dependent process. Genetics 126: 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S., Mehta M., Kuo D., Sung M. K., Chuang R., et al. , 2010. Rewiring of genetic networks in response to DNA damage. Science 330: 1385–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera M. A., Petes T. D., 2006. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103: 12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Negrini S., Shore D., 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell 16: 139–146. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Williamson M. S., Fogel S., Kolodner R. D., 1987. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature 328: 362–364. [DOI] [PubMed] [Google Scholar]
- Blanco M. G., Matos J., Rass U., Ip S. C., West S. C., 2010. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst.) 9: 394–402. [DOI] [PubMed] [Google Scholar]
- Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., et al. , 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M. L., Paeschke K., Chan A., Zakian V. A., 2014. Hrq1, a homolog of the human RecQ4 helicase, acts catalytically and structurally to promote genome integrity. Cell Rep. 6: 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Gaillard P. H., McDonald W. H., Shanahan P., Yates J. R., III, et al. , 2001. Mus81-Eme1 are essential components of a holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Leung W. Y., Kramer W., Kramer B., Williamson M., et al. , 1990. Mismatch repair-induced meiotic recombination requires the pms1 gene product. Genetics 124: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G., Haber J. E., 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150: 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule J. B., Zakian V. A., 2007. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 35: 5809–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P., 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17: 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., 2011. Ubiquitin family modifications and template switching. FEBS Lett. 585: 2810–2817. [DOI] [PubMed] [Google Scholar]
- Branzei D., Szakal B., 2016. DNA damage tolerance by recombination: molecular pathways and DNA structures. DNA Repair (Amst.) 44: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Vanoli F., Foiani M., 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920. [DOI] [PubMed] [Google Scholar]
- Budd M. E., Reis C. C., Smith S., Myung K., Campbell J. L., 2006. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol. Cell. Biol. 26: 2490–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M., 2009. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 284: 4041–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R., 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198. [DOI] [PubMed] [Google Scholar]
- Campbell P. J., Yachida S., Mudie L. J., Stephens P. J., Pleasance E. D., et al. , 2010. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467: 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Ley T. J., Miller C., Ding L., Raphael B. J., Mungall A.J., et al. , 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368: 2059–2074 (erratum: N. Engl. J. Med. 369: 98). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper, A. M., P. W. Greenwell, W. Tang, and T. D. Petes, 2009 Chromosome aberrations resulting from double-strand DNA breaks at a naturally occurring yeast fragile site composed of inverted ty elements are independent of Mre11p and Sae2p. Genetics 183: 423–439, 421SI–426SI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M., Santos-Pereira J. M., Rondon A. G., Barroso S., Andujar E., et al. , 2013. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell 52: 583–590. [DOI] [PubMed] [Google Scholar]
- Castor D., Nair N., Declais A. C., Lachaud C., Toth R., et al. , 2013. Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81–EME1 nucleases. Mol. Cell 52: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celic I., Masumoto H., Griffith W. P., Meluh P., Cotter R. J., et al. , 2006. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 16: 1280–1289. [DOI] [PubMed] [Google Scholar]
- Celic I., Verreault A., Boeke J. D., 2008. Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics 179: 1769–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli S. M., Crouch R. J., 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276: 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H. Z., Chung S. J., Rhee S. G., 1994. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269: 27670–27678. [PubMed] [Google Scholar]
- Chan J. E., Kolodner R. D., 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet. 7: e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. E., Kolodner R. D., 2012. Rapid analysis of Saccharomyces cerevisiae genome rearrangements by multiplex ligation-dependent probe amplification. PLoS Genet. 8: e1002539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. A., Aristizabal M. J., Lu P. Y., Luo Z., Hamza A., et al. , 2014. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 10: e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Arneric M., Lingner J., 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21: 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che J., Smith S., Kim Y. J., Shim E. Y., Myung K., et al. , 2015. Hyper-acetylation of histone H3K56 limits break-induced replication by inhibiting extensive repair synthesis. PLoS Genet. 11: e1004990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Kolodner R. D., 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23: 81–85. [DOI] [PubMed] [Google Scholar]
- Chen C., Umezu K., Kolodner R. D., 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2: 9–22. [DOI] [PubMed] [Google Scholar]
- Chen X., Cui D., Papusha A., Zhang X., Chu C. D., et al. , 2012. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489: 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. M., 2015. The Saccharomyces genome database: a tool for discovery. Cold Spring Harb. Protoc. 2015: pdb.top083840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Szakal B., Chen Y. H., Branzei D., Zhao X., 2010. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell 21: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H., Sparks J. L., Rychlik M., Nowotny M., Burgers P. M., et al. , 2013. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 41: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Dietrich F. S., Fink G. R., 1988. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55: 413–425. [DOI] [PubMed] [Google Scholar]
- Chung D. K., Chan J. N., Strecker J., Zhang W., Ebrahimi-Ardebili S., et al. , 2015. Perinuclear tethers license telomeric DSBs for a broad kinesin- and NPC-dependent DNA repair process. Nat. Commun. 6: 7742. [DOI] [PubMed] [Google Scholar]
- Churikov D., Charifi F., Eckert-Boulet N., Silva S., Simon M. N., et al. , 2016. SUMO-dependent relocalization of eroded telomeres to nuclear pore complexes controls telomere recombination. Cell Rep. 15: 1242–1253. [DOI] [PubMed] [Google Scholar]
- Ciccia A., Elledge S. J., 2010. The DNA damage response: making it safe to play with knives. Mol. Cell 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M., Blackburn E. H., 1995. Telomerase in yeast. Science 269: 396–400. [DOI] [PubMed] [Google Scholar]
- Collins K., Greider C. W., 1993. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 7: 1364–1376. [DOI] [PubMed] [Google Scholar]
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., et al. , 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810. [DOI] [PubMed] [Google Scholar]
- Connolly B., White C. I., Haber J. E., 1988. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2342–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover H. N., Lujan S. A., Chapman M. J., Cornelio D. A., Sharif R., et al. , 2015. Stimulation of chromosomal rearrangements by ribonucleotides. Genetics 201: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelio D. A., Sedam H. N., Ferrarezi J. A., Sampaio N. M., Argueso J. L., 2017. Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability. DNA Repair (Amst.) 52: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L., Koshland D., 2015. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 34: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T., Louge R., Tomimatsu N., Mukherjee B., Martini E., et al. , 2012. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489: 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R. J., Greenwell P. W., Dominska M., Petes T. D., 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics 161: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P. M., Clay-Farrace L., Fiegler H., Carr P., et al. , 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198. [DOI] [PubMed] [Google Scholar]
- D’Amours D., Jackson S. P., 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15: 2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Adjiri A., New L., Crouse G. F., Jinks Robertson S., 1996. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T., 2002. Protection of mammalian telomeres. Oncogene 21: 532–540. [DOI] [PubMed] [Google Scholar]
- De Piccoli G., Cortes-Ledesma F., Ira G., Torres-Rosell J., Uhle S., et al. , 2006. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat. Cell Biol. 8: 1032–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G., Torres-Rosell J., Aragon L., 2009. The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res. 17: 251–263. [DOI] [PubMed] [Google Scholar]
- Deem A., Barker K., Vanhulle K., Downing B., Vayl A., et al. , 2008. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179: 1845–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L., Batzer M. A., 1999. Alu repeats and human disease. Mol. Genet. Metab. 67: 183–193. [DOI] [PubMed] [Google Scholar]
- Deng S. K., Yin Y., Petes T. D., Symington L. S., 2015. Mre11-Sae2 and RPA collaborate to prevent palindromic gene amplification. Mol. Cell 60: 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar J. M., Lydall D., 2010. Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping. EMBO J. 29: 4020–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede S. J., Gottschling D. E., 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99: 723–733. [DOI] [PubMed] [Google Scholar]
- Doerfler L., Harris L., Viebranz E., Schmidt K. H., 2011. Differential genetic interactions between Sgs1, DNA-damage checkpoint components and DNA repair factors in the maintenance of chromosome stability. Genome Integr. 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R., Hudson A., Jackson S. P., 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert-Boulet N., Lisby M., 2010. Regulation of homologous recombination at telomeres in budding yeast. FEBS Lett. 584: 3696–3702. [DOI] [PubMed] [Google Scholar]
- El Hage A., Webb S., Kerr A., Tollervey D., 2014. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 10: e1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J. M., Smolka M. B., Zhou H., Kolodner R. D., 2006. Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J. Cell Biol. 175: 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster A., Lanzuolo C., Bonneton M., Luciano P., Pollice A., et al. , 2006. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat. Struct. Mol. Biol. 13: 734–739. [DOI] [PubMed] [Google Scholar]
- Fabre F., Chan A., Heyer W. D., Gangloff S., 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99: 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R., 2015. Mismatch repair. J. Biol. Chem. 290: 26395–26403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H., Kolodner R. D., 2000. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem. Sci. 25: 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C., Gasser S. M., 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14: 81–96. [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., et al. , 2006. DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- Gangavarapu V., Haracska L., Unk I., Johnson R. E., Prakash S., et al. , 2006. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 7783–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R., 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E., Kim Y., Lach F. P., Kottemann M. C., Smogorzewska A., 2013. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced holliday junctions. Cell Rep. 5: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatbonton T., Imbesi M., Nelson M., Akey J. M., Ruderfer D. M., et al. , 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold C. B., Hauer M. H., Gasser S. M., 2015. INO80-C and SWR-C: guardians of the genome. J. Mol. Biol. 427: 637–651. [DOI] [PubMed] [Google Scholar]
- Gibson W. J., Hoivik E. A., Halle M. K., Taylor-Weiner A., Cherniack A. D., et al. , 2016. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat. Genet. 48: 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbini E., Cesena D., Galbiati A., Lockhart A., Longhese M. P., 2013. Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair (Amst.) 12: 791–799. [DOI] [PubMed] [Google Scholar]
- Goldfless S. J., Morag A. S., Belisle K. A., Sutera V. A., Jr, Lovett S. T., 2006. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol. Cell 21: 595–604. [DOI] [PubMed] [Google Scholar]
- Gomez-Gonzalez B., Aguilera A., 2007. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc. Natl. Acad. Sci. USA 104: 8409–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B., Felipe-Abrio I., Aguilera A., 2009. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol. Cell. Biol. 29: 5203–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin D. A., Resnick M. A., 1998. Yeast ARMs (DNA at-risk motifs) can reveal sources of genome instability. Mutat. Res. 400: 45–58. [DOI] [PubMed] [Google Scholar]
- Gordenin D. A., Lobachev K. S., Degtyareva N. P., Malkova A. L., Perkins E., et al. , 1993. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol. 13: 5315–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. J., Resio B., Pellman D., 2012. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13: 189–203. [DOI] [PubMed] [Google Scholar]
- Gravel S., Chapman J. R., Magill C., Jackson S. P., 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22: 2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B. M., Finn K. J., Li J. J., 2010. Loss of DNA replication control is a potent inducer of gene amplification. Science 329: 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groothuizen F. S., Sixma T. K., 2016. The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst.) 38: 14–23. [DOI] [PubMed] [Google Scholar]
- Gundem G., Van Loo P., Kremeyer B., Alexandrov L. B., Tubio J. M., et al. , 2015. The evolutionary history of lethal metastatic prostate cancer. Nature 520: 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., Thorburn P. C., 1984. Healing of broken linear dicentric chromosomes in yeast. Genetics 106: 207–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., Ray B. L., Kolb J. M., White C. I., 1993. Rapid kinetics of mismatch repair of heteroduplex DNA that is formed during recombination in yeast. Proc. Natl. Acad. Sci. USA 90: 3363–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J. A., Feldser D. M., Greider C. W., 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106: 275–286. [DOI] [PubMed] [Google Scholar]
- Han J., Zhou H., Horazdovsky B., Zhang K., Xu R. M., et al. , 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655. [DOI] [PubMed] [Google Scholar]
- Hartman J. L. t., Garvik B., Hartwell L., 2001. Principles for the buffering of genetic variation. Science 291: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Hartsuiker E., Vaessen E., Carr A. M., Kohli J., 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20: 6660–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D. C., 1963. A deletion in yeast and its bearing on the structure of the mating type locus. Genetics 48: 1727–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen C. D., 2016. Mismatch repair defects and lynch syndrome: the role of the basic scientist in the battle against cancer. DNA Repair (Amst.) 38: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. T., Petes T. D., 1993. Instability of a plasmid-borne inverted repeat in Saccharomyces cerevisiae. Genetics 134: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka M., Watanabe K., Umezu K., Maki H., 2000. Spontaneous loss of heterozygosity in diploid Saccharomyces cerevisiae cells. Genetics 156: 1531–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S., 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Horigome C., Bustard D. E., Marcomini I., Delgoshaie N., Tsai-Pflugfelder M., et al. , 2016. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev. 30: 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Koshland D., 2003. Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev. 17: 1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. E., Kolodner R. D., 2005. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol. Cell 17: 709–720. [DOI] [PubMed] [Google Scholar]
- Huang M. E., Rio A. G., Nicolas A., Kolodner R. D., 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100: 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Aguilera A., 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12: 711–721. [DOI] [PubMed] [Google Scholar]
- Hwang J. Y., Smith S., Myung K., 2005. The Rad1-Rad10 complex promotes the production of gross chromosomal rearrangements from spontaneous DNA damage in Saccharomyces cerevisiae. Genetics 169: 1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. Y., Smith S., Ceschia A., Torres-Rosell J., Aragon L., et al. , 2008. Smc5-Smc6 complex suppresses gross chromosomal rearrangements mediated by break-induced replications. DNA Repair (Amst.) 7: 1426–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaki K., Liu E. T., 2012. Structural mutations in cancer: mechanistic and functional insights. Trends Genet. 28: 550–559. [DOI] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J. E., 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., Medema R. H., 2013. Genetic instability: tipping the balance. Oncogene 32: 4459–4470. [DOI] [PubMed] [Google Scholar]
- Jeong H. S., Backlund P. S., Chen H. C., Karavanov A. A., Crouch R. J., 2004. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 32: 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson K., Kanno T., Shirahige K., Sjogren C., 2014. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 15: 601–614. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S., Petes T. D., 1986. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics 114: 731–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. B., Hall D. W., 2004. Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected. Genetics 168: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova L. Y., Mertz T. M., Zhang Y., Northam M. R., Sheng Z., et al. , 2013. A reversible histone H3 acetylation cooperates with mismatch repair and replicative polymerases in maintaining genome stability. PLoS Genet. 9: e1003899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W. G., Jr, 2005. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 5: 689–698. [DOI] [PubMed] [Google Scholar]
- Kanellis P., Gagliardi M., Banath J. P., Szilard R. K., Nakada S., et al. , 2007. A screen for suppressors of gross chromosomal rearrangements identifies a conserved role for PLP in preventing DNA lesions. PLoS Genet. 3: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass E. M., Moynahan M. E., Jasin M., 2016. When genome maintenance goes badly Awry. Mol. Cell 62: 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., et al. , 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Kats E. S., Albuquerque C. P., Zhou H., Kolodner R. D., 2006. Checkpoint functions are required for normal S-phase progression in Saccharomyces cerevisiae RCAF- and CAF-I-defective mutants. Proc. Natl. Acad. Sci. USA 103: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kats E. S., Enserink J. M., Martinez S., Kolodner R. D., 2009. The Saccharomyces cerevisiae Rad6 postreplication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants. Mol. Cell. Biol. 29: 5226–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil R. L., McWilliams A. D., 1993. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 135: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrest A., Anand R. P., Sundararajan R., Bermejo R., Liberi G., et al. , 2009. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat. Struct. Mol. Biol. 16: 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin H., Shen Y., Huang F., Patel M., Yang T., et al. , 2014. Transcript-RNA-templated DNA recombination and repair. Nature 515: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. A., Haber J. E., 2009. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc. Natl. Acad. Sci. USA 106: 1151–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Huang S. N., Williams J. S., Li Y. C., Clark A. B., et al. , 2011. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332: 1561–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. T., Lim D. S., Canman C. E., Kastan M. B., 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274: 37538–37543. [DOI] [PubMed] [Google Scholar]
- Klein H. L., 2001. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics 159: 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinopoulos P. A., Ceccaldi R., Shapiro G. I., D’Andrea A. D., 2015. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 5: 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Caburet S., Dujon B., Fischer G., 2004. Eukaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23: 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer K. M., Brock J. A., Bloom K., Moore J. K., Haber J. E., 1994. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell. Biol. 14: 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B. O., Symington L. S., 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38: 233–271. [DOI] [PubMed] [Google Scholar]
- Kschonsak M., Haering C. H., 2015. Shaping mitotic chromosomes: from classical concepts to molecular mechanisms. Bioessays 37: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M., 2014. Biology of telomeres: lessons from budding yeast. FEMS Microbiol. Rev. 38: 144–171. [DOI] [PubMed] [Google Scholar]
- Kupiec M., 2016. Alternative clamp loaders/unloaders. FEMS Yeast Res. 16: fow084. [DOI] [PubMed] [Google Scholar]
- Kupiec M., Petes T. D., 1988. Allelic and ectopic recombination between Ty elements in yeast. Genetics 119: 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerstedt Robinson K., Liu T., Vandrovcova J., Halvarsson B., Clendenning M., et al. , 2007. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J. Natl. Cancer Inst. 99: 291–299. [DOI] [PubMed] [Google Scholar]
- Lambert S., Watson A., Sheedy D. M., Martin B., Carr A. M., 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702. [DOI] [PubMed] [Google Scholar]
- Lambert S., Froget B., Carr A. M., 2007. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst.) 6: 1042–1061. [DOI] [PubMed] [Google Scholar]
- Laureau R., Loeillet S., Salinas F., Bergstrom A., Legoix-Ne P., et al. , 2016. Extensive recombination of a yeast diploid hybrid through meiotic reversion. PLoS Genet. 12: e1005781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., Christensen R. B., 1979. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 139: 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Godon C., Lagniel G., Spector D., Garin J., et al. , 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274: 16040–16046. [DOI] [PubMed] [Google Scholar]
- Lee K., Tosti E., Edelmann W., 2016. Mouse models of DNA mismatch repair in cancer research. DNA Repair (Amst.) 38: 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. E., Moore J. K., Holmes A., Umezu K., Kolodner R. D., et al. , 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409. [DOI] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Lobachev K., Petes T. D., 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598. [DOI] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Kokoska R. J., Petes T. D., 2008. Reduced levels of DNA polymerase delta induce chromosome fragile site instability in yeast. Mol. Cell. Biol. 28: 5359–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K. W., Vogelstein B., 1998. Genetic instabilities in human cancers. Nature 396: 643–649. [DOI] [PubMed] [Google Scholar]
- Lengronne A., Schwob E., 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9: 1067–1078. [DOI] [PubMed] [Google Scholar]
- Li J. R., Yu T. Y., Chien I. C., Lu C. Y., Lin J. J., et al. , 2014. Pif1 regulates telomere length by preferentially removing telomerase from long telomere ends. Nucleic Acids Res. 42: 8527–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda D. E., Winston F., 2006. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature 443: 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S., Shalit P., Picologlou S., 1981. Ty elements are involved in the formation of deletions in DEL1 strains of Saccharomyces cerevisiae. Cell 26: 401–409. [DOI] [PubMed] [Google Scholar]
- Lisby M., Rothstein R., Mortensen U. H., 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98: 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Mortensen U. H., Rothstein R., 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5: 572–577. [DOI] [PubMed] [Google Scholar]
- Lisby M., Barlow J. H., Burgess R. C., Rothstein R., 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C., 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84: 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K. S., Stenger J. E., Kozyreva O. G., Jurka J., Gordenin D. A., et al. , 2000. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 19: 3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K. S., Gordenin D. A., Resnick M. A., 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Loeillet S., Palancade B., Cartron M., Thierry A., Richard G. F., et al. , 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amst.) 4: 459–468. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., et al. , 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561. [DOI] [PubMed] [Google Scholar]
- Lowndes N. F., Murguia J. R., 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10: 17–25. [DOI] [PubMed] [Google Scholar]
- Luna R., Rondon A. G., Aguilera A., 2012. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim. Biophys. Acta 1819: 514–520. [DOI] [PubMed] [Google Scholar]
- Lundblad V., Szostak J. W., 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643. [DOI] [PubMed] [Google Scholar]
- Lydall D., Weinert T., 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270: 1488–1491. [DOI] [PubMed] [Google Scholar]
- Ma J. L., Kim E. M., Haber J. E., Lee S. E., 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23: 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas N. L., Miller K. M., DeFazio L. G., Toczyski D. P., 2006. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23: 109–119. [DOI] [PubMed] [Google Scholar]
- Macintyre G., Ylstra B., Brenton J. D., 2016. Sequencing structural variants in cancer for precision therapeutics. Trends Genet. 32: 530–542. [DOI] [PubMed] [Google Scholar]
- Mangahas J. L., Alexander M. K., Sandell L. L., Zakian V. A., 2001. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell 12: 4078–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri H. W., Ngo H. P., Hickson I. D., 2009. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 20: 1683–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Brevet V., Mann C., Gilson E., 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10: 487–490. [DOI] [PubMed] [Google Scholar]
- Maringele L., Lydall D., 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16: 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L., Lydall D., 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18: 2663–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G., Symington L. S., 2013. Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol. Cell 52: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA 25: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley J. L., Petes T. D., 2010. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc. Natl. Acad. Sci. USA 107: 11465–11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M. J., Haber J. E., 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75: 111–135. [DOI] [PubMed] [Google Scholar]
- Mehta A., Haber J. E., 2014. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6: a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melek M., Greene E. C., Shippen D. E., 1996. Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol. Cell. Biol. 16: 3437–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J. A., Cohen J., Toczyski D. P., 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15: 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25: 173–178. [DOI] [PubMed] [Google Scholar]
- Michelson R. J., Weinert T., 2000. Closing the gaps among a web of DNA repair disorders. Bioessays 22: 966–969. [DOI] [PubMed] [Google Scholar]
- Mieczkowski P. A., Mieczkowska J. O., Dominska M., Petes T. D., 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100: 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikus M. D., Petes T. D., 1982. Recombination between genes located on nonhomologous chromosomes in Saccharomyces cerevisiae. Genetics 101: 369–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo H. E., Gomez-Gonzalez B., Grzechnik P., Rondon A. G., Wei W., et al. , 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell 41: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F., Johansson B., Mertens F., 2006. Database of chromosome aberrations and gene fusions in cancer, http://cgap.nci.nih.gov/Chromosomes/Mitelman
- Mitelman F., Johansson B., Mertens F., 2007. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7: 233–245. [DOI] [PubMed] [Google Scholar]
- Mizuno K., Lambert S., Baldacci G., Murray J. M., Carr A. M., 2009. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 23: 2876–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G. L., D’Andrea A. D., 2009. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 43: 223–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. K., Haber J. E., 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A. J., Highland J., Krogan N. J., Arbel-Eden A., Greenblatt J. F., et al. , 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775. [DOI] [PubMed] [Google Scholar]
- Morrow D. M., Tagle D. A., Shiloh Y., Collins F. S., Hieter P., 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82: 831–840. [DOI] [PubMed] [Google Scholar]
- Motegi A., Kuntz K., Majeed A., Smith S., Myung K., 2006. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundbjerg K., Jorgensen S. W., Fredsoe J., Nielsen I., Pedersen J. M., et al. , 2015. Top2 and Sgs1-Top3 act redundantly to ensure rDNA replication termination. PLoS Genet. 11: e1005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Galvan S., Tous C., Blanco M. G., Schwartz E. K., Ehmsen K. T., et al. , 2012. Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol. Cell. Biol. 32: 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Galvan S., Jimeno S., Rothstein R., Aguilera A., 2013. Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet. 9: e1003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Kolodner R. D., 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 4500–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Kolodner R. D., 2003. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair (Amst.) 2: 243–258. [DOI] [PubMed] [Google Scholar]
- Myung K., Chen C., Kolodner R. D., 2001a Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076. [DOI] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., Kolodner R. D., 2001b SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27: 113–116. [DOI] [PubMed] [Google Scholar]
- Myung K., Datta A., Kolodner R. D., 2001c Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408. [DOI] [PubMed] [Google Scholar]
- Myung K., Pennaneach V., Kats E. S., Kolodner R. D., 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 100: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M. B., Roberts T. M., et al. , 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan V., Mieczkowski P. A., Kim H. M., Petes T. D., Lobachev K. S., 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125: 1283–1296. [DOI] [PubMed] [Google Scholar]
- Nelson D. M., Ye X., Hall C., Santos H., Ma T., et al. , 2002. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22: 7459–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. W., Liu Y., Hasselblatt K. T., Mok S. C., Berkowitz R. S., 1999. A new human topoisomerase III that interacts with SGS1 protein. Nucleic Acids Res. 27: 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S. A., Kumar D., Clark A. B., Watt D. L., Watts B. E., et al. , 2010a Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 6: 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundstrom E. B., et al. , 2010b Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA 107: 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L. J., Odijk H., Budzowska M., van Drunen E., Maas A., et al. , 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24: 5776–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M., Bachmair A., Eisenhaber B., Eisenhaber F., 2005. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2–NIP45) family. BMC Bioinformatics 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell P. C., 1976. The clonal evolution of tumor cell populations. Science 194: 23–28. [DOI] [PubMed] [Google Scholar]
- O’Connell K., Jinks-Robertson S., Petes T. D., 2015. Elevated genome-wide instability in yeast mutants lacking RNase H activity. Genetics 201: 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohle C., Tesorero R., Schermann G., Dobrev N., Sinning I., et al. , 2016. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167: 1001–1013 e1007. [DOI] [PubMed] [Google Scholar]
- Ooi S. L., Pan X., Peyser B. D., Ye P., Meluh P. B., et al. , 2006. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 22: 56–63. [DOI] [PubMed] [Google Scholar]
- Oughtred R., Chatr-aryamontri A., Breitkreutz B. J., Chang C. S., Rust J. M., et al. , 2016. BioGRID: a resource for studying biological interactions in yeast. Cold Spring Harb. Protoc. 2016: pdb.top080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek A. L., Kaochar S., Jones H., Elezaby A., Shanks L., et al. , 2009. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 23: 2861–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K., Bochman M. L., Garcia P. D., Cejka P., Friedman K. L., et al. , 2013. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 497: 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B., Aguilera A., 2012. Complex chromosomal rearrangements mediated by break-induced replication involve structure-selective endonucleases. PLoS Genet. 8: e1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. G., Cha M. K., Jeong W., Kim I. H., 2000. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 275: 5723–5732. [DOI] [PubMed] [Google Scholar]
- Paulovich A. G., Margulies R. U., Garvik B. M., Hartwell L. H., 1997a RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 145: 45–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich A. G., Toczyski D. P., Hartwell L. H., 1997b When checkpoints fail. Cell 88: 315–321. [DOI] [PubMed] [Google Scholar]
- Payen C., Koszul R., Dujon B., Fischer G., 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V., Kolodner R. D., 2004. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast. Nat. Genet. 36: 612–617. [DOI] [PubMed] [Google Scholar]
- Pennaneach V., Kolodner R. D., 2009. Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS One 4: e6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V., Crittin J., Grolimund L., Lingner J., 2013. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 32: 2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A., Serero A., Boule J. B., Legoix-Ne P., Lopes J., et al. , 2012. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS Genet. 8: e1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., et al. , 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26: 4089–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Pennaneach V., Kolodner R. D., 2004. Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 13262–13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Pennaneach V., Kolodner R. D., 2005. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol. Cell. Biol. 25: 7226–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Hayes T. K., Kolodner R. D., 2009a Specific pathways prevent duplication-mediated genome rearrangements. Nature 460: 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Jaehnig E. J., Kolodner R. D., 2009b Perspectives on the DNA damage and replication checkpoint responses in Saccharomyces cerevisiae. DNA Repair (Amst.) 8: 974–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Hayes T. K., Kolodner R. D., 2010. Post-replication repair suppresses duplication-mediated genome instability. PLoS Genet. 6: e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Allen-Soltero S. R., Martinez S. L., Chan J. E., Hayes T. K., et al. , 2012. Bioinformatic identification of genes suppressing genome instability. Proc. Natl. Acad. Sci. USA 109: E3251–E3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Pallis K., Hayes T. K., Kolodner R. D., 2014. DNA repair pathway selection caused by defects in TEL1, SAE2, and de novo telomere addition generates specific chromosomal rearrangement signatures. PLoS Genet. 10: e1004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Srivatsan A., Nene R. V., Martinez S. L., Clotfelter S. P., et al. , 2016. A genetic network that suppresses genome rearrangements in Saccharomyces cerevisiae and contains defects in cancers. Nat. Commun. 7: 11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragu S., Faye G., Iraqui I., Masurel-Heneman A., Kolodner R. D., et al. , 2007. Oxygen metabolism and reactive oxygen species cause chromosomal rearrangements and cell death. Proc. Natl. Acad. Sci. USA 104: 9747–9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragu S., Dardalhon M., Sharma S., Iraqui I., Buhagiar-Labarchede G., et al. , 2014. Loss of the thioredoxin reductase Trr1 suppresses the genomic instability of peroxiredoxin tsa1 mutants. PLoS One 9: e108123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M., Dennehey B. K., Tyler J. K., 2010. Chaperoning histones during DNA replication and repair. Cell 140: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J., Tsubota T., Tanny J. C., Diaz R. L., Berger J. M., et al. , 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 103: 6988–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan R. A., Kolodner R. D., 1992. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics 132: 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindle A., Belichenko I., Bylebyl G. R., Chen X. L., Gandhi N., et al. , 2006. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 119: 4749–4757. [DOI] [PubMed] [Google Scholar]
- Reyes G. X., Schmidt T. T., Kolodner R. D., Hombauer H., 2015. New insights into the mechanism of DNA mismatch repair. Chromosoma 124: 443–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K. B., Mallory J. C., Petes T. D., 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R., 1980. DNA rearrangements associated with a transposable element in yeast. Cell 21: 239–249. [DOI] [PubMed] [Google Scholar]
- Rosen D. M., Younkin E. M., Miller S. D., Casper A. M., 2013. Fragile site instability in Saccharomyces cerevisiae causes loss of heterozygosity by mitotic crossovers and break-induced replication. PLoS Genet. 9: e1003817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., 1979. Deletions of a tyrosine tRNA gene in S. cerevisiae. Cell 17: 185–190. [DOI] [PubMed] [Google Scholar]
- Rothstein R., Helms C., Rosenberg N., 1987. Concerted deletions and inversions are caused by mitotic recombination between delta sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M., Tuzon C. T., Zakian V. A., 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell 27: 550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N., Ramakrishnan S., Elango R., Ayyar S., Zhang Y., et al. , 2013. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502: 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi J. S., Chan J. N., Szafranski K., Liu T. T., Wu J. D., et al. , 2014. Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA hybrids. Dev. Cell 30: 177–191. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Desany B. A., Jones W. J., Liu Q., Wang B., et al. , 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Bachant J., Wang H., Hu F., Liu D., et al. , 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286: 1166–1171. [DOI] [PubMed] [Google Scholar]
- Sandell L. L., Zakian V. A., 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75: 729–739. [DOI] [PubMed] [Google Scholar]
- Santocanale C., Diffley J. F., 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618. [DOI] [PubMed] [Google Scholar]
- Santos-Pereira J. M., Aguilera A., 2015. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16: 583–597. [DOI] [PubMed] [Google Scholar]
- Sarbajna S., West S. C., 2014. Holliday junction processing enzymes as guardians of genome stability. Trends Biochem. Sci. 39: 409–419. [DOI] [PubMed] [Google Scholar]
- Schacherer J., de Montigny J., Welcker A., Souciet J. L., Potier S., 2005. Duplication processes in Saccharomyces cerevisiae haploid strains. Nucleic Acids Res. 33: 6319–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J., Tourrette Y., Potier S., Souciet J. L., de Montigny J., 2007. Spontaneous duplications in diploid Saccharomyces cerevisiae cells. DNA Repair (Amst.) 6: 1441–1452. [DOI] [PubMed] [Google Scholar]
- Scherer S., Mann C., Davis R. W., 1982. Reversion of a promoter deletion in yeast. Nature 298: 815–819. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Prakash S., 1988. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol. Cell. Biol. 8: 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Prakash S., 1990. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol. Cell. Biol. 10: 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K. H., Pennaneach V., Putnam C. D., Kolodner R. D., 2006a Analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 409: 462–476. [DOI] [PubMed] [Google Scholar]
- Schmidt K. H., Wu J., Kolodner R. D., 2006b Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom’s syndrome protein. Mol. Cell. Biol. 26: 5406–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. H., Pearson C. E., 2016. Disease-associated repeat instability and mismatch repair. DNA Repair (Amst.) 38: 117–126. [DOI] [PubMed] [Google Scholar]
- Schouten J. P., McElgunn C. J., Waaijer R., Zwijnenburg D., Diepvens F., et al. , 2002. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz V. P., Zakian V. A., 1994. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76: 145–155. [DOI] [PubMed] [Google Scholar]
- Segre D., Deluna A., Church G. M., Kishony R., 2005. Modular epistasis in yeast metabolism. Nat. Genet. 37: 77–83. [DOI] [PubMed] [Google Scholar]
- Serero A., Jubin C., Loeillet S., Legoix-Ne P., Nicolas A. G., 2014. Mutational landscape of yeast mutator strains. Proc. Natl. Acad. Sci. USA 111: 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K. A., Shishkin A. A., Voineagu I., Pavlov Y. I., Shcherbakova P. V., et al. , 2012. Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2: 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., et al. , 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621. [DOI] [PubMed] [Google Scholar]
- Shore D., 1998. Telomeres–unsticky ends. Science 281: 1818–1819. [DOI] [PubMed] [Google Scholar]
- Sikdar N., Banerjee S., Zhang H., Smith S., Myung K., 2008. Spt2p defines a new transcription-dependent gross chromosomal rearrangement pathway. PLoS Genet. 4: e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S., Altmannova V., Luke-Glaser S., Henriksen P., Gallina I., et al. , 2016. Mte1 interacts with Mph1 and promotes crossover recombination and telomere maintenance. Genes Dev. 30: 700–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. S., Gottschling D. E., 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409. [DOI] [PubMed] [Google Scholar]
- Smith C. E., Llorente B., Symington L. S., 2007. Template switching during break-induced replication. Nature 447: 102–105. [DOI] [PubMed] [Google Scholar]
- Smith S., Hwang J. Y., Banerjee S., Majeed A., Gupta A., et al. , 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 9039–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Gupta A., Kolodner R. D., Myung K., 2005. Suppression of gross chromosomal rearrangements by the multiple functions of the Mre11-Rad50-Xrs2 complex in Saccharomyces cerevisiae. DNA Repair (Amst.) 4: 606–617. [DOI] [PubMed] [Google Scholar]
- Smolka M. B., Chen S. H., Maddox P. S., Enserink J. M., Albuquerque C. P., et al. , 2006. An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J. Cell Biol. 175: 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J., Cimprich K. A., 2015. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 25: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Dominska M., Greenwell P. W., Petes T. D., 2014. Genome-wide high-resolution mapping of chromosome fragile sites in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 111: E2210–E2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks J. L., Chon H., Cerritelli S. M., Kunkel T. A., Johansson E., et al. , 2012. RNase H2-initiated ribonucleotide excision repair. Mol. Cell 47: 980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell R. M., Jinks-Robertson S., 2004. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics 168: 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivas R., Shen J. P., Yang C. C., Sun S. M., Li J., et al. , 2016. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol. Cell 63: 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Charles J., Hazkani-Covo E., Yin Y., Andersen S. L., Dietrich F. S., et al. , 2012. High-resolution genome-wide analysis of irradiated (UV and gamma-rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics 190: 1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P. C., Bloom M. S., Solanki-Patil T., Smith S., Sipahimalani P., et al. , 2011. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 7: e1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P. C., Chan Y. A., Minaker S. W., Aristizabal M. J., Barrett I., et al. , 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stith C. M., Sterling J., Resnick M. A., Gordenin D. A., Burgers P. M., 2008. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 283: 34129–34140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L., Karlsson C., Lindroos H. B., Wedahl S., Katou Y., et al. , 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317: 242–245. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Szostak J. W., 1983. Recombination between sequences in nonhomologous positions. Proc. Natl. Acad. Sci. USA 80: 5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Paques F., Colaiacovo M., Haber J. E., 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94: 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Goldfarb T., Studamire B., Alani E., Haber J. E., 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101: 9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surosky R. T., Tye B. K., 1985. Resolution of dicentric chromosomes by Ty-mediated recombination in yeast. Genetics 110: 397–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26: 5589–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. M., Siu K. L., Wong C. M., Jin D. Y., 2009. Loss of yeast peroxiredoxin Tsa1p induces genome instability through activation of the DNA damage checkpoint and elevation of dNTP levels. PLoS Genet. 5: e1000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson R. B., Ebran N., Herrera A. E., Lindsley J. E., 2002. A novel selection system for chromosome translocations in Saccharomyces cerevisiae. Genetics 160: 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J. A., Diffley J. F., 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557. [DOI] [PubMed] [Google Scholar]
- Tham K. C., Kanaar R., Lebbink J. H., 2016. Mismatch repair and homeologous recombination. DNA Repair (Amst.) 38: 75–83. [DOI] [PubMed] [Google Scholar]
- Thaminy S., Newcomb B., Kim J., Gatbonton T., Foss E., et al. , 2007. Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J. Biol. Chem. 282: 37805–37814. [DOI] [PubMed] [Google Scholar]
- Thompson S. L., Compton D. A., 2011. Chromosomes and cancer cells. Chromosome Res. 19: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower D. A., Stemple J., Yeh E., Bloom K., 2003. Nuclear oscillations and nuclear filament formation accompany single-strand annealing repair of a dicentric chromosome in Saccharomyces cerevisiae. J. Cell Sci. 116: 561–569. [DOI] [PubMed] [Google Scholar]
- Tishkoff D. X., Filosi N., Gaida G. M., Kolodner R. D., 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88: 253–263. [DOI] [PubMed] [Google Scholar]
- Toczyski D. P., Galgoczy D. J., Hartwell L. H., 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90: 1097–1106. [DOI] [PubMed] [Google Scholar]
- Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., et al. , 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., et al. , 2007. Histone H3–K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25: 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Taggart A. K., Zakian V. A., 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11: 1328–1335. [DOI] [PubMed] [Google Scholar]
- Tutt A., Gabriel A., Bertwistle D., Connor F., Paterson H., et al. , 1999. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 9: 1107–1110. [DOI] [PubMed] [Google Scholar]
- Uchi R., Takahashi Y., Niida A., Shimamura T., Hirata H., et al. , 2016. Integrated multiregional analysis proposing a new model of colorectal cancer evolution. PLoS Genet. 12: e1005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K., Hiraoka M., Mori M., Maki H., 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E., Heidinger-Pauli J. M., Koshland D., 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248. [DOI] [PubMed] [Google Scholar]
- Ungar L., Yosef N., Sela Y., Sharan R., Ruppin E., et al. , 2009. A genome-wide screen for essential yeast genes that affect telomere length maintenance. Nucleic Acids Res. 37: 3840–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Hohn B., Gasser S. M., 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788. [DOI] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Gasser S. M., 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26: 4113–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialard J. E., Gilbert C. S., Green C. M., Lowndes N. F., 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17: 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V. E., Zhou S., Diaz L. A., Jr, et al. , 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Amon J. D., Koshland D., Vuica-Ross M., 2011. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell 44: 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Gore S. K., Koshland D., 2013. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife 2: e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Putnam C. D., Kane M. F., Zhang W., Edelmann L., et al. , 2005. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat. Genet. 37: 750–755. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Morishita J., Umezu K., Shirahige K., Maki H., 2002. Involvement of RAD9-dependent damage checkpoint control in arrest of cell cycle, induction of cell death, and chromosome instability caused by defects in origin recognition complex in Saccharomyces cerevisiae. Eukaryot. Cell 1: 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H., 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322. [DOI] [PubMed] [Google Scholar]
- West S. C., 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4: 435–445. [DOI] [PubMed] [Google Scholar]
- White J. H., Lusnak K., Fogel S., 1985. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature 315: 350–352. [DOI] [PubMed] [Google Scholar]
- Williams J. S., Smith D. J., Marjavaara L., Lujan S. A., Chabes A., et al. , 2013. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell 49: 1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A., Kwon Y., Xu Y., Chung W. H., Chi P., et al. , 2013. Pif1 helicase and polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 502: 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. M., Zhou Y., Ng R. W., Kung Hf H. F., Jin D. Y., 2002. Cooperation of yeast peroxiredoxins Tsa1p and Tsa2p in the cellular defense against oxidative and nitrosative stress. J. Biol. Chem. 277: 5385–5394. [DOI] [PubMed] [Google Scholar]
- Wyatt H. D., Sarbajna S., Matos J., West S. C., 2013. Coordinated actions of SLX1–SLX4 and MUS81–EME1 for holliday junction resolution in human cells. Mol. Cell 52: 234–247. [DOI] [PubMed] [Google Scholar]
- Xie Y., Kerscher O., Kroetz M. B., McConchie H. F., Sung P., et al. , 2007. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282: 34176–34184. [DOI] [PubMed] [Google Scholar]
- Xu F., Zhang K., Grunstein M., 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121: 375–385. [DOI] [PubMed] [Google Scholar]
- Xu H., Boone C., Brown G. W., 2007. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics 176: 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Duc K. D., Holcman D., Teixeira M. T., 2013. The length of the shortest telomere as the major determinant of the onset of replicative senescence. Genetics 194: 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Papusha A., Choi K., Bonner J. N., Kumar S., et al. , 2016. Differential regulation of the anti-crossover and replication fork regression activities of Mph1 by Mte1. Genes Dev. 30: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Delannoy M., Ling C., Daee D., Osman F., et al. , 2010. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell 37: 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Franco A. A., Santos H., Nelson D. M., Kaufman P. D., et al. , 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell 11: 341–351. [DOI] [PubMed] [Google Scholar]
- Yimit A., Kim T., Anand R. P., Meister S., Ou J., et al. , 2016. MTE1 Functions with MPH1 in double-strand break repair. Genetics 203: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu V. P., Koehler M., Steinlein C., Schmid M., Hanakahi L. A., et al. , 2000. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 14: 1400–1406. [PMC free article] [PubMed] [Google Scholar]
- Yuen K. W., Warren C. D., Chen O., Kwok T., Hieter P., et al. , 2007. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc. Natl. Acad. Sci. USA 104: 3925–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zeidler A. F., Song W., Puccia C. M., Malc E., et al. , 2013. Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae. Genetics 193: 785–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Durocher D., 2010. De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev. 24: 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Shishkin A. A., Nishida Y., Marcinkowski-Desmond D., Saini N., et al. , 2012. Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol. Cell 48: 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Saini N., Sheng Z., Lobachev K. S., 2013. Genome-wide screen reveals replication pathway for quasi-palindrome fragility dependent on homologous recombination. PLoS Genet. 9: e1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Blobel G., 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D. Q., Zhang K., Wu X. C., Mieczkowski P. A., Petes T. D., 2016. Global analysis of genomic instability caused by DNA replication stress in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 113: E8114–E8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. B., Elledge S. J., 2000. The DNA damage response: putting checkpoints in perspective. Nature 408: 433–439. [DOI] [PubMed] [Google Scholar]
- Zhou J., Monson E. K., Teng S. C., Schulz V. P., Zakian V. A., 2000. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289: 771–774. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyev A., Kuperstein I., Barillot E., Heyer W. D., 2013. Synthetic lethality between gene defects affecting a single non-essential molecular pathway with reversible steps. PLoS Comput. Biol. 9: e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Rothstein R., 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90: 87–96. [DOI] [PubMed] [Google Scholar]