This study concerns building high-density genetic maps in situations with intrachromosomal recombination rate heterogeneity and differences in genotyping
Keywords: marker space, cosegregating markers, twin groups, skeletal markers, marker clustering, genotyping errors, missing data, marker filtration
Abstract
The study is focused on addressing the problem of building genetic maps in the presence of ∼103–104 of markers per chromosome. We consider a spectrum of situations with intrachromosomal heterogeneity of recombination rate, different level of genotyping errors, and missing data. In the ideal scenario of the absence of errors and missing data, the majority of markers should appear as groups of cosegregating markers (“twins”) representing no challenge for map construction. The central aspect of the proposed approach is to take into account the structure of the marker space, where each twin group (TG) and singleton markers are represented as points of this space. The confounding effect of genotyping errors and missing data leads to reduction of TG size, but upon a low level of these effects surviving TGs can still be used as a source of reliable skeletal markers. Increase in the level of confounding effects results in a considerable decrease in the number or even disappearance of usable TGs and, correspondingly, of skeletal markers. Here, we show that the paucity of informative markers can be compensated by detecting kernels of markers in the marker space using a clustering procedure, and demonstrate the utility of this approach for high-density genetic map construction on simulated and experimentally obtained genotyping datasets.
IN recent years, new genotyping technologies based on DNA-arrays (chips) and next generation sequencing (NGS) have become widely available for scoring thousands of single nucleotide polymorphic markers (SNPs) in a wide spectrum of model and nonmodel organisms. These datasets pose new challenges for building high-density genetic maps. With large-scale chip-based SNP genotyping data, genotyping-by-sequencing (GBS) or specific-locus-amplified-fragment-sequencing data (SLAF-Seq) (e.g., Qi et al. 2014), building genetic maps with 105–106 markers per genome (or 103–104 markers per chromosome) requires new algorithms. Indeed, the dramatic increase in the number of markers is only one of the challenges. Among other difficulties with such an amount of markers are genotyping errors, missing data, and small population size. If the mapping algorithms cannot efficiently cope with these problems, generating big SNP marker sets for building ultradense maps will not achieve the goal. Obviously, the population size sets an upper limit to the number of markers per chromosome that can be resolved by recombination; genotyping errors and missing data calls may complicate deducing the correct marker order in the chromosome.
Usually a two-phase approach is applied for genetic mapping: clustering of all markers into linkage groups (LGs) and ordering the markers within each LG. Earlier algorithms and software packages for genetic mapping were based on a few approaches suitable in a situation when the number of markers per population was relatively small, e.g., a few tens or hundreds per chromosome. In both phases, a full distance matrix for the chromosome markers is required. In case of a significantly increased dimension of the problem (n ∼ 104–106), the existing algorithms for genetic mapping cannot solve the problem in reasonable computer time, i.e., even using simple optimization algorithms of order O(n2). Moreover, a huge computer memory (RAM) for the distance matrix is required on the clustering and map construction phases (but see Strandova-Neeley et al. 2015). With big data, even more challenging are the difficulties caused by missing scores and genotyping errors. Usually markers with considerable data missing (e.g., 10–20%) are removed from the dataset at the pretreatment stage, while markers with genotyping errors are not removed. Instead, their positions are slightly corrected by corresponding maximum likelihood (ML) algorithms (Wu et al. 2008; Rastas et al. 2013, 2016). The inability of existing map construction algorithms to cope with these factors, rather unexpectedly for the wide genetics community, posed a serious problem. In some cases maps of 400–800 cM have been obtained and required additional “rescaling” correction in order to correspond to the expectations based on cytogenetic analysis of meiosis (Wang et al. 2014).
Several algorithms, proposed to deal with big mapping datasets, employ the ideas of k-means (Arthur and Vasilevskii 2007) or k-nearest neighbors (Liu et al. 2014) for marker clustering into LGs followed by the ordering of each LG. The genetic map is represented as a linear sequence of ordered marker subsets Si (bins), with the number of markers in each subset defined by a neighborhood of radius ri around a center ci (Liu et al. 2014). Ordering is conducted for bins rather than markers. In certain cases, genetic mapping can be reduced to the Minimum Spanning Tree (MST) problem (Wu et al. 2008) instead of a more traditional reduction to the Traveler Salesperson Problem (TSP) (Mester et al. 2003). The MST algorithm gives fast and good solutions for low-noise data and simple geometry of the spanning tree, i.e., when the majority of “leaves” of the tree are interconnected (via linkage) in a linear-like structure corresponding to the organization of the eukaryotic chromosome and only a small part of markers appears in the tree branches (Rastas et al. 2013). But if the number of markers in the branches is large, the maximal MST path may inadequately represent the chromosome. In such cases, MST can serve only as a source of an initial solution that should be complemented by markers from the branches. Thus, the Lep-Map algorithm (Rastas et al. 2013, 2016) imitates MST construction in finding a feasible initial order (path of maximum length) and then inserts markers from MST branches into the path via TSP heuristics. After this step, local changes in the order are applied to maximize the likelihood of the final order. However, the MST approach cannot manage situations with large numbers of markers in the presence of genotyping errors and missing data.
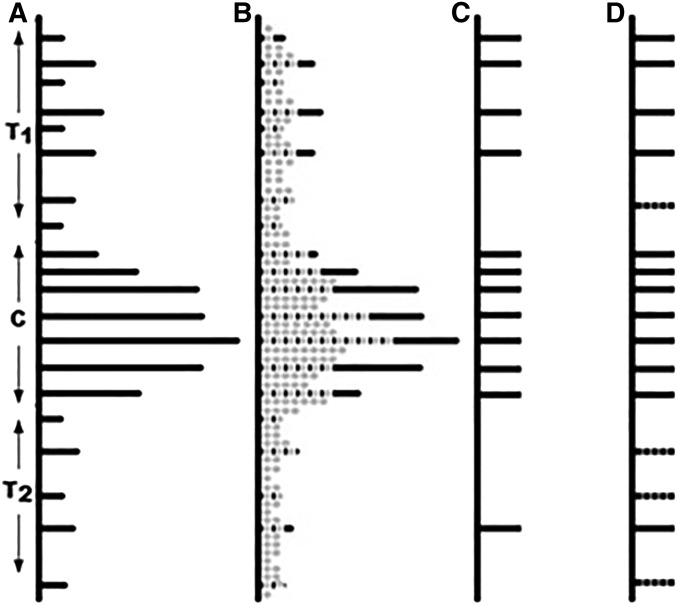
Another approach to solve the problem was first described in our short report (Ronin et al. 2015). Its central idea is to take into account the structure of the marker space of the mapping problem, where each point represents a marker with n coordinates corresponding to the marker alleles of n genotypes of the population. With this approach, in addition to routine filtering of markers based on segregation distortion and level of missing data, we suggested a heuristic procedure of selecting high-quality markers. It is based on the assumption that error-free markers are more abundant among groups of cosegregating (twin) markers, which should have priority during the selection of “skeletal markers” for inclusion into the genetic map. If the error rate is low (e.g., pe ∼ 0.01–0.02), a sufficient number of such markers can be selected to build a high-quality map. Here we propose a new approach for constructing genetic maps using big genotyping data (with up to 103–104 markers per chromosome), which extends the method by Ronin et al. (2015) and includes an additional filtering step to cope with a higher level of errors (say, pe ∼ 0.02–0.04 or more). Obviously, with the higher error rates, the quality of the maps is supposed to decrease. We show that the increase in the error rate can be compensated by the availability of a large number of markers allowing for building good-quality maps. In our algorithm, the procedure of choosing reliable marker candidates from noisier subsets of markers is applied after the best candidates, representing twin groups (TGs), have already been selected. The remaining markers are clustered and the representatives of such clusters, satisfying certain conditions, are appended to the set of the best candidates representing the TGs. The choice between the two approaches or usage of a hybrid strategy integrating both approaches for any dataset can be based on preliminary filtering/clustering cycles as described in the paper. The chromosomal distribution of markers suitable to be tried as candidates for the skeletal map at the consequent stages of analysis is shown in Figure 1.
Figure 1.
Selecting candidate markers for the skeletal map in the presence of strong regional heterogeneity of recombination, genotyping errors, and missing data. (a) Marker positions along the chromosome in an error-free situation. The continuous black horizontal bars represent groups of cosegregating markers (twins) unresolvable by recombination due to low local recombination rate and small population size. In the simulated example, much higher marker density is shown for the peri-centromeric (C) compared to subtelomeric regions (Т1 and Т2). (b) Disturbed distribution of markers due to genotyping errors that lead to disruption of some twin groups; markers with genotyping errors (shown in gray) may cause map length inflation if included in the map. (c) Skeletal map based on representatives of twin groups remained despite the “losses” of markers caused by genotyping errors. (d) Recovering a part of dissipated twin groups by cluster analysis.
For the ordering of the selected candidate markers, various optimization algorithms can be applied, for example, GES (Lin and Kernighan 1973; Helsgaun 2000; Applegate et al. 2003; Mester et al. 2010; Ronin et al. 2010) and Concord (Applegate et al. 2001). The efficiency of our approach for the selection of the most informative candidates was studied here on simulated and real datasets. Ordering the selected candidates, testing, and stepwise improving of the genetic map is then conducted using the effective scheme described in our previous publications (Mester et al. 2003, 2004, 2010; Korol et al. 2009; Ronin et al. 2010, 2012, 2015).
Materials and Methods
Simulation of mapping data
For testing the algorithms, we employed simulated and real mapping populations of doubled haploid (or backcross) type, with a population size n = 200, and the number of markers Nm = 10,000, 20,000, and 40,000 per chromosome. In many organisms, the distribution of recombination events along chromosomes is highly heterogeneous, with very high differences between peri-centromeric and subtelomeric regions due to the centromere and heterochromatin effects on recombination (Korol et al. 1994; Akhunov et al. 2003; Backström et al. 2010; Roesti et al. 2013; Sharma et al. 2013; Hill et al. 2015; Wang et al. 2015; Nambiar and Smith 2016; Tsai et al. 2016). Another contributing factor may be correlation between recombination rate and chromosomal variation in GC content along chromosomes (Duret and Arndt 2008). Obviously, upon an even distribution of polymorphic markers with respect to DNA physical length, the density of markers per unit of recombination in the regions with a low recombination rate will be much higher than in high-recombination regions. Therefore, in our simulations, three different regions were considered with respect to the proportion of simulated markers and genetic map length L (centimorgan). Namely, the peri-centromeric and the two subtelomeric regions included 80 and 20% of Nm, while the contribution of the peri-centromeric part to the genetic map length was much lower compared to the subtelomeric parts (Supplemental Material, Table S1 in File S1). Simulation of recombination distances between adjacent markers for each region was conducted by sampling the distance values from the preset region-specific ranges of very small, small, and moderate distances (dvs, ds, and dm, respectively) (Table S2 in File S1). The average characteristics of the resulting maps constructed for error-free data are presented in Table S3 in File S1. For all notations in the text and the tables and figures see File S2 in File S1 (Glossary).
Construction of a skeletal map in the case of a low level of genotyping errors using the “twin” method
The major part in the proposed approach for building dense genetic maps is selecting informative skeletal markers. Depending on the level of genotyping errors (unknown a priori), we developed and evaluated two procedures to address this problem. The first procedure is based on the fact that under low levels of genotyping errors (e.g., pe = 0.01–0.02 per marker locus), large numbers of markers per LG (e.g., Nm ∼ 104) and relatively small sizes of the mapping population (n ∼ 102), a considerable proportion of markers will appear as large groups of twins (TGs) (Ronin et al. 2015). For such situations, a simple and efficient principle for selecting reliable skeletal markers is to take representative (“delegate”) markers with the minimal rate of missing data for each TG (see File S3).
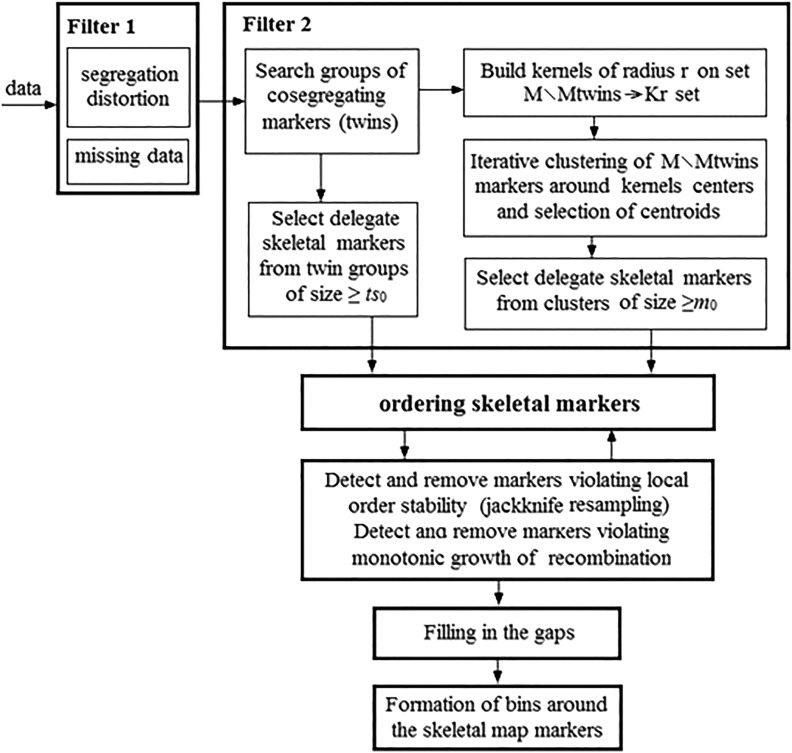
The process of constructing a skeletal map includes three stages. The first stage is to select a threshold value ts0 for the size ts of TGs, which will be represented by their delegate markers in the initial variant of the skeletal map (Ronin et al. 2015). In our approach, the selected markers are then ordered based on the reduction of the mapping problem to TSP using the Evolutionary Strategy heuristic optimization (Mester et al. 2003, 2004, 2010; Ronin et al. 2010). The second step is testing map quality using jackknife resampling followed by the deletion of markers violating local map stability and monotonicity (i.e., increase in recombination rate between a marker and its subsequent neighbors) (Ronin et al. 2010). After this step, we can insert in the resulting map additional markers representing TGs with smaller sizes compared to the chosen ts0 (as well as suitable singleton markers not causing map inflation), and then check the map quality again. This step may be helpful for filling in the gaps in the genetic map. Such cycles can be applied repeatedly (Figure 2).
Figure 2.
The scheme of the proposed strategy for building ultradense genetic maps. The major difference from other approaches and our previous algorithm is the proposed premapping filtering (Filter 2) based on twins and clusters of tightly linked markers.
Skeletal map construction in the presence of high genotype-calling error rates using clustering
The second approach is designed to manage situations with higher levels of errors (e.g., up to pe = 0.02–0.04). The decrease in the proportion of error-free markers leads to the erosion of most of the TGs. Thus, for a marker with pe = 0.03, the probability that in a population with n = 200 genotypes none of the marker scores could be erroneous is P = (1−0.03)200 ≈ e−6 ≈ 0.0024. Here, the average number of errors is six, implying that two markers inseparable-by-recombination (upon an error-free situation) will show a “distance” of ∼6 cM. In order to extend the twin-based filtration idea to such situations, when the number of TGs is not sufficient for covering the chromosome even at a low marker density, we employ marker clustering. An important geometrical fact is that in the n-dimensional space of markers, many groups of cosegregating markers that should be represented in this space as one point per group, as a result of errors will turn into clouds (clusters) of close points. The midpoint of such a cloud is geometrically close to the position of the corresponding (error-free) set of completely linked markers or twins. Thus, in the space of markers, noisy markers geometrically represent a “fuzzy” set of varying density, with higher density in the vicinity of the residence point of the original error-free markers. Bearing this in mind, we complement the procedure based on using TGs with zero intragroup distances, by a clustering procedure that dissects the entire set of markers into clusters in such a way that the sum of distances of markers to the means of their clusters, taken over all clusters, will be minimal (File S4). This enables covering the chromosomal regions with relatively high recombination rates, where the joint effect of an increased proportion of false (due to errors) “recombinants” and lower real density of markers per centimorgan may lead to a negligible proportion of the remaining TGs. With this approach, we select a representative marker from each cluster (the marker closest to the centroid) and use these representatives as additional candidate markers. This approach enables good-quality maps to be built even under the paucity or absence of TGs (due to genotyping errors) in datasets with high error rate. After selection of markers representing the obtained clusters, the next steps are identical to those described above for the twin approach, i.e., resampling-based detection and removal of markers violating map stability and monotonicity combined with saturating the skeletal map by filling in the gaps wherever possible (see Figure 2).
Although a priori we cannot know which of the two foregoing situations (low or high level of genotypic errors) is characteristic of the target mapping project, it is easy to address this question by a trial analysis and evaluate the number of TGs and the chromosomal distribution of twin sizes, and thereby to assign the project to the first or second class. Obviously, before starting the mapping analysis, trivial preliminary removal of low-quality data is needed based on the level of missing data and deviation of segregation ratios from the expected ones. In reality, the foregoing two situations, with high vs. negligible number of twins, may take place simultaneously within the same chromosome. As noted above, this may happen due to the effect of centromere and heterochromatin blocks on recombination. To deal with such situations we employ a hybrid procedure (see Figure 2).
Characterizing the quality of constructed maps
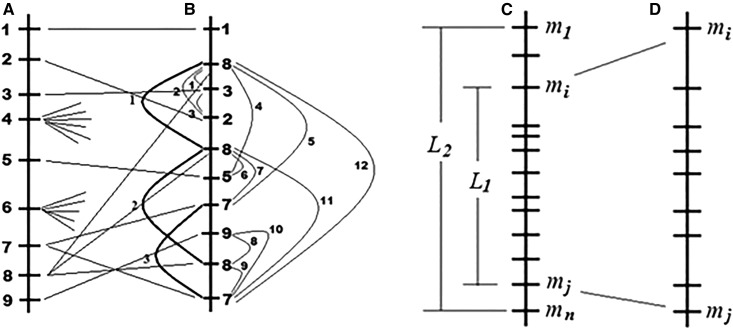
In the analysis of simulated data, to characterize the quality of maps constructed using different algorithms or different parameter settings of the same algorithm, we employ as a reference the map representing the “true” (simulated) order of markers, corresponding to the ideal error-free case with no missing data points. Then, the best algorithm is the one that generates a solution closest to the simulated order. Several parameters were used to assess the map quality. Bearing in mind that in a simulation study we do know the true order, the simplest score of map quality would be the coefficient of recovery (Cr), as described in Mester et al. (2003). Upon error-free genotyping, many markers are expected to cosegregate due to a high ratio of the total number of markers to population size, leading to the limited use of Cr because it does not take into account the fact that genotyping errors and missing marker scores can lead to fissions of TGs into groups of a smaller size (see Results). Thus, instead of Cr we employ two other characteristics: (a) ne, the number of errors in the order of markers compared to the simulated order (we consider as an error each situation when the marker’s original rank in the constructed skeletal map is higher than the rank of the next marker in the map); and (b) nr, the number of “repeats” resulting from the separation of the initial groups of cosegregating markers into subgroups due to genotyping errors and missing marker calls; such repeats will appear in the constructed skeletal map at separate (usually, but not necessarily, adjacent) positions. Figure 3, a and b illustrates the calculation of ne and nr. Numbers from 1 to 9 are the numbers of cosegregating groups in the simulated map; the figure shows the estimated order of noisy marker data. But the degree of deviation from the true order in the skeletal map is only a partial characteristic of the map quality; map coverage (mc) is another important score (Figure 3, c and d). Additional quality scores used in our analysis included: loss factor lf (%), which is the percentage of lost (noncharacterized) unique marker positions in the constructed map compared to the simulated map: lf = 100 [Nskef − (Nsk − nr)]/Nskef, where Nskef and Nsk are the number of intervals in the skeletal maps built for the simulated error-free and noisy markers.
Figure 3.
Illustration of complicating factors causing the need for introduced parameters (ne, nr, and mc) as characteristics of map quality for simulated data: errors and repeats as indicated by thin and thick lines, respectively (here, ne = 12 and nr = 3). (a) A fragment of a simulated map with nine ordered markers each representing a twin group. (b) The simulated noise (missing data points and genotyping errors) cause the following changes: (i) some twin groups undergo weak or zero disturbance implying stability of the corresponding skeletal markers (markers #1 and #5); (ii) for some markers (#2 and #3) the disturbance caused a slight change in the order; (iii) the noise caused disintegration of twin groups (represented by markers #7 and #8 in the simulated map) into subgroups with sizes fitting the condition ts ≥ ts0 for selection of candidate skeletal markers; the generated repeat markers contributed to the number of errors in the map; and (iv) disintegration of twin groups in such a way that none of the resulting subgroups obeys the condition ts ≥ ts0 (markers #4 and #6). (c) Simulated genetic map (with no disturbances); L1 and L2 represent the distances (in centimorgans) between markers mi and mj, and between m1 and mn, respectively. (d) Constructed genetic map where the first position is represented by a marker that in the simulated map was at the ith position (marker mi), while the last position is represented by a marker that in the simulated map was at the jth position (marker mj). Map coverage for a map constructed either for error-free or noisy simulated data is calculated as mc (%) = 100L1/L2.
In addition to simulated data, we demonstrated our approaches on several wheat chromosomes using data generated using the 90 K iSelect SNP genotyping assay for 150 doubled haploid (DH) wheat lines (Wang et al. 2014).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and in the Supplemental Material.
Results
Both procedures considered in this paper are based on selecting a subset of most informative markers, referred to as “skeletal” markers, as a basis for constructing a quality genetic map (Mester et al. 2003, 2004, 2010; Ronin et al. 2010). In the first procedure, the selected subset of markers for the skeletal map is comprised of “delegate” markers representing TGs. In the second procedure, when the proportion of TGs becomes small, we cluster markers into nonoverlapping subsets, e.g., using k-means or other algorithms (Bishop 1995; Strandova-Neeley et al. 2015); markers with minimal average distance to all other markers of such a subset serve as delegates. In reality, upon moderate level of genotyping errors the two situations may coexist within the same project, e.g., with considerable and very low proportions of cosegregating markers in near-centromeric regions (due to low centimorgan/megabase pair) and subtelomeric regions (due to high centimorgan/megabase pair), respectively. Therefore, in the present examples, we illustrate the proposed analytical schemes on the two types of practically important situations: (i) with the low level of genotyping errors (hence high proportion of TGs); and (ii) with a relatively high level of errors (hence much lower proportion of TGs) justifying the efforts to look for the representatives of “fuzzy TGs” by using cluster analysis. For both situations, we consider simulated data as well as real data on wheat.
Analysis of simulated data
Although one of the effects of missing data in error-free genotyping data is the reduction in the number of TGs, a nonzero but small level of errors leads to an increase in the number of TGs (Table S7; for comparison see File S5, File S6, Table S5, and Table S6 in File S1 with results for “no-missing” situations). Nevertheless, a further increase in the error rate reverses the direction of changes in the number and size of TGs, and thereby in the number of and confidence in the selected candidate skeletal markers. Thus, qualitatively the effect of missing genotype calls does not change the main conclusions reached for “no missing” situation. With the increased level of genotyping errors, a more liberal threshold ts0 should be chosen compared to the situations with low error rate. In Table 1, for a population of size n = 200 with two levels of errors (1 and 2%), we show how the results of the described map construction procedure depend on the choice of ts0 for different numbers of available markers per chromosome. As expected, for datasets with higher pe, a more liberal choice of ts0 should be recommended compared to the datasets with lower pe. The observed distribution of the numbers of groups of size 2, 3, 4 etc. can serve as a diagnostic characteristic of the underlying real situation.
Table 1 Effect of the selected threshold for minimal twin group size ts0 on the skeletal map length under low and moderate rates of genotyping errors and 20% of missing data points.
| Map | Initial | After cleaning | After saturation | δ | |||||
|---|---|---|---|---|---|---|---|---|---|
| pe | Nm | ts0 | Nsk | L | Nsk | L | Nsk | L | |
| 0.01 | 104 | 4 | 349 | 384.2 | 81 | 172.1 | 99 | 171.8 | 1.8 |
| 6 | 128 | 221.0 | 65 | 173.8 | 77 | 174.9 | 2.3 | ||
| 8 | 108 | 237.0 | 47 | 175.5 | 65 | 174.2 | 2.7 | ||
| 0.02 | 3 | 237 | 397.8 | 58 | 207.3 | 71 | 187.9 | 2.7 | |
| 4 | 102 | 213.8 | 38 | 128.8 | 47 | 126.6 | 2.8 | ||
| 0.01 | 2 × 104 | 6 | 398 | 472.1 | 97 | 216.1 | 111 | 217.1 | 2.0 |
| 8 | 256 | 357.9 | 91 | 219.9 | 104 | 217.4 | 2.1 | ||
| 10 | 178 | 307.3 | 71 | 205.8 | 83 | 201.7 | 2.5 | ||
| 0.02 | 4 | 305 | 406.4 | 85 | 208.0 | 103 | 200.9 | 2.0 | |
| 5 | 175 | 333.6 | 57 | 207.6 | 73 | 194.3 | 2.7 | ||
| 6 | 110 | 200.6 | 42 | 135.1 | 56 | 132.5 | 2.4 | ||
| 0.01 | 4 × 104 | 10 | 377 | 461.7 | 140 | 244.7 | 150 | 253.2 | 1.7 |
| 12 | 281 | 399.4 | 110 | 244.7 | 120 | 243.5 | 2.0 | ||
| 16 | 195 | 339.0 | 95 | 245.0 | 108 | 248.4 | 2.3 | ||
| 0.02 | 6 | 289 | 446.0 | 89 | 262.4 | 115 | 249.1 | 2.2 | |
| 8 | 157 | 364.9 | 67 | 272.9 | 83 | 254.4 | 3.1 | ||
| 10 | 94 | 323.6 | 50 | 283.0 | 67 | 252.2 | 3.8 | ||
Simulated map length was L = 191.3, 214.6, and 273 cM for populations with Nm = 104, 2 × 104, and 4 × 104 markers, respectively; pe, level of genotyping errors per marker locus; ts, twin group size, ts0, threshold ts value: skeletal markers should obey the condition ts ≥ ts0; Nsk, number of intervals in the map; Nm, number of markers per LG in the initial dataset; δ, map density (the ratio of the entire chromosome map length to the number of intervals).
A more detailed analysis of the joint effect of genotyping errors and missing data on the quality of the skeletal map and its regions with high and low recombination density (subtelomeric vs. peri-centromeric) is provided in Table S8. For that, in addition to the regional characteristics of map length (L, Lt1, Lt2, and Lc) and number of markers (Nsk, Nt1sk, Nt2sk, and Ncsk), we also employed the proportion of lost unique map positions (lf) compared to the simulated map; the coverage level of the skeletal map (mc); the number of errors and repeats (ne and nr); and map density represented by the ratio of the map length (for the entire chromosome and/or its segments) to the number of intervals (δ and/or δt1, δt2, δc) (for explanation see Figure 3 and File S2). The main findings can be summarized as follows:
Proportion of lost unique positions in the skeletal map (lf):
With increase in the level of genotyping errors (from 0.005 to 0.02) and missing data-points (from 0 to 20%), the lost factor lf increased monotonically with ts0 and varied from 10 to 70% independently on the number of markers within the analyzed range Nm = 104 − 4 × 104 (see also Table S6).
Map coverage (mc):
For the entire range of parameters, mc was higher than 97%, excluding the cases with simultaneously maximal values of pe and ts0. In such cases, the proportion of TGs may become very small (not sufficient to recruit candidates for a skeletal map), especially in regions with high centimorgan/megabase values. Such situations require the complementation of twin markers by markers representing the clusters of tightly linked but not cosegregating markers (see below).
Map density:
Map density decreased (δ increased) with the increased level of genotyping errors pe and the chosen threshold ts0, especially in the subtelomeric regions, due to a higher level of error-driven degradation of TGs. On average, a 2–2.5 increase in δ was observed for the maximal considered level of genotyping errors pe = 0.02.
Number of “repeats” (nr) in the map:
As explained in the Materials and Methods and illustrated in Figure 3, genotyping errors and missing data calls may lead to dissociation of a part of the initial TGs into subgroups; representatives of such subgroups appear in the constructed skeletal map at separate (usually, but not necessarily, adjacent) positions. Table S8 shows that an increase in the chosen ts0 results in lower nr, implying more efficient filtration in favor of better markers.
Errors in the order of markers (ne):
In the resulting skeletal maps, ne was rather small. Yet, attempts to compensate the effect of high pe by choosing too stringent ts0 may lead to a considerable increase of ne due to a drop in the number of skeletal markers and map coverage.
Two approaches can be used if a small number of TGs (i.e., candidate skeletal markers) and low coverage were obtained for the chosen ts0: (i) reduce ts0, thereby increasing the number of TGs; and (ii) recruit additional candidate markers based on the k-means or similar clustering procedures (see Materials and Methods). With a low level of genotyping errors, the first approach may be sufficient, at least for genomic regions with a relatively low centimorgan/megabase ratio; in combination with the second approach it may enable solving the problem for the whole genome. However, with a high level of errors, the number of TGs may be too small even at ts0 = 2. In this case, the second approach can be used as a major source of candidate skeletal markers. These considerations are reflected in the examples present in Table 2, where combined analysis enabled considerable improvement of the map coverage and reducing the proportion of lost map unique positions.
Table 2. Improvement of the skeletal map using the hybrid approach (tw_cl) for selecting candidate markers as compared to twin selection.
| Method | pe | ts0 | Lt1 | Nt1sk | δt1 | Lc | Ncsk | δc | Lt2 | Nt2sk | δt2 | L | Nsk | δ | ne | nr | mc% | lf % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nm =104 | ||||||||||||||||||
| tw | 0.01 | 8 | 57 | 11 | 5.2 | 38 | 34 | 1.1 | 79 | 19 | 4.2 | 174 | 64 | 2.7 | 2 | 3 | 0.96 | 57 |
| tw_cl | 0.01 | 8 | 61 | 20 | 3.1 | 41 | 39 | 1.0 | 82 | 25 | 3.3 | 184 | 84 | 2.2 | 2 | 8 | 0.98 | 47 |
| tw | 0.02 | 4 | 30 | 7 | 4.3 | 48 | 33 | 1.4 | 49 | 6 | 8.2 | 127 | 46 | 2.8 | 1 | 8 | 0.72 | 74 |
| tw_cl | 0.02 | 4 | 74 | 17 | 4.4 | 47 | 33 | 1.4 | 87 | 22 | 4.0 | 208 | 72 | 2.9 | 1 | 15 | 0.97 | 61 |
| Nm = 2 × 104 | ||||||||||||||||||
| tw | 0.02 | 6 | 58 | 12 | 4.8 | 49 | 36 | 1.4 | 26 | 7 | 3.7 | 133 | 55 | 2.4 | 6 | 4 | 0.72 | 72 |
| tw_cl | 0.02 | 6 | 62 | 14 | 4.4 | 48 | 34 | 1.4 | 89 | 20 | 4.5 | 199 | 68 | 2.9 | 5 | 6 | 0.98 | 65 |
| Nm = 4 × 104 | ||||||||||||||||||
| tw | 0.02 | 10 | 87 | 15 | 5.8 | 57 | 38 | 1.5 | 108 | 13 | 8.3 | 252 | 66 | 3.8 | 16 | 5 | 0.92 | 71 |
| tw_cl | 0.02 | 10 | 92 | 21 | 4.4 | 74 | 37 | 2.0 | 106 | 25 | 4.2 | 255 | 83 | 3.1 | 2 | 6 | 0.98 | 64 |
pe, level of genotyping errors per marker locus; Nm, number of markers per LG in the initial dataset; ts, twin group size; ts0, threshold ts value: skeletal markers should obey the condition ts ≥ ts0; in all tw_cl variants, the threshold kernel radius was r = 0.04 (for description see File S4); L, Lt1, Lt2, and Lc, the estimated genetic lengths (in centimorgans) of the entire chromosome map and its subtelomeric and near-centromeric regions, while Nsk, Nt1sk, Nt2sk, and Ncsk are the corresponding numbers of intervals of the entire map and its subtelomeric and peri-centromeric regions; δ, δt1, δt2, and δc, map density (centimorgan/interval) of the entire map and its subtelomeric and peri-centromeric regions; ne, the number of errors in the estimated order of markers compared to the simulated order; nr, the number of “repeats” caused by fission of the initial TGs into subgroups due to genotyping errors and missing marker scores; mc (%), map coverage, which represents the proportion of the constructed skeletal map length relative to the simulated map length; lf (%), loss factor, the percentage of lost (noncharacterized) map unique positions in the constructed skeletal map compared to the simulated map; it is calculated as lf = 100 [Nskef − (Nsk − nr)]/Nskef, = 100 (Nskef − Nsk + nr)/Nskef, where Nskef represents the number of intervals in the skeletal maps built for the simulated error-free data, while Nsk and (Nsk − nr) represent the number of noisy markers in the skeletal map, noncorrected and corrected for the number of repeats, respectively.
The decrease in the number of TGs may also be caused by an increase in the population size (n). Indeed, we have noted above that even for the small level of genotyping errors, an increase in n leads to a decrease in the size of the TGs. In Table 3, we show the effect of population size on map characteristics for error-free data and moderate level of genotyping errors (pe = 0.01) and missing data points (ms = 20%). For n = 200, the twin approach was sufficient to build a good-quality map, but for n = 500 it became impractical due to a catastrophic decrease in the number of TGs in the subtelomeric regions. Therefore, the map was constructed using the combination of twin approach and clustering. And finally, for n = 1000, only a clustering approach was suitable. It is noteworthy that despite the growing proportion of lost markers lf (%), the number of markers in and the genetic length of the constructed skeletal maps also grow with the population size. The last effect results from the known fact that genotyping errors lead to map length inflation.
Table 3. The effect of population size (n) on map characteristics.
| ms% | pe | ts0 | r | Lt1 | Nt1sk | δt1 | Lc | Ncsk | δc | Lt2 | Nt2sk | δt2 | L | Nsk | δ | ne | nr | mc% | lf% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nm = 104, n = 200 | |||||||||||||||||||
| 0 | 0 | 67 | 41 | 1.6 | 41 | 64 | 0.6 | 82 | 39 | 2.1 | 191 | 144 | 1.3 | 0 | 0 | 100 | 0 | ||
| 20 | 0.01 | 6 | 58 | 18 | 3.2 | 39 | 40 | 1.0 | 78 | 18 | 4.3 | 175 | 76 | 2.3 | 3 | 7 | 99.5 | 52 | |
| Nm = 104, n = 500 | |||||||||||||||||||
| 0 | 0 | 84 | 47 | 1.8 | 37 | 118 | 0.3 | 85 | 52 | 1.6 | 206 | 217 | 0.9 | 0 | 0 | 100 | 0 | ||
| 20 | 0.01 | 3 | 0.01 | 94 | 33 | 2.8 | 38 | 42 | 0.9 | 89 | 28 | 3.2 | 238 | 103 | 2.3 | 1 | 10 | 100 | 57 |
| Nm = 104, n = 103 | |||||||||||||||||||
| 0 | 0 | 73 | 55 | 1.3 | 40 | 194 | 0.2 | 85 | 65 | 1.3 | 198 | 314 | 0.6 | 0 | 0 | 100 | 0 | ||
| 20 | 0.01 | 2 | 0.01 | 86 | 27 | 3.2 | 85 | 66 | 1.3 | 107 | 38 | 2.8 | 278 | 131 | 2.1 | 2 | 12 | 100 | 62 |
pe, level of genotyping errors per marker; ms, simulated rate of missing data per marker; Nm, number of markers per LG in the initial dataset; ts, twin group size, ts0, threshold ts value: skeletal markers should obey the condition ts ≥ ts0; r, kernel radius; L, Lt1, Lt2, and Lc, the estimated genetic lengths (in centimorgans) of the entire chromosome map and its subtelomeric and near-centromeric regions, while Nsk, Nt1sk, Nt2sk, and Ncsk are the corresponding numbers of intervals of the entire map and its subtelomeric and peri-centromeric regions; δ, δt1,δt2, and δc, map density (centimorgan/interval) of the entire map and its subtelomeric and peri-centromeric regions; ne, the number of errors in the estimated order of markers compared to the simulated order; nr, the number of “repeats” caused by fission of the initial TGs into subgroups due to genotyping errors and missing marker scores; mc (%), map coverage, which represents the proportion of the constructed skeletal map length relative to the simulated map length; lf (%), loss factor, the percentage of lost (noncharacterized) map unique positions in the constructed skeletal map compared to the simulated map; it is calculated as lf = 100 [Nskef − (Nsk − nr)]/Nskef, = 100 (Nskef − Nsk + nr)/Nskef, where Nskef represents the number of intervals in the skeletal maps built for the simulated error-free data, while Nsk and (Nsk − nr) represent the number of noisy markers in the skeletal map, noncorrected and corrected for the number of repeats, respectively.
Analysis of empirical datasets
As was shown above, genotyping errors can result in a decrease in the number of TGs reducing the number of candidate markers for the skeletal map. Therefore, the usage of some of the standard mapping algorithms with the large number of markers can result in maps with inflated intermarker distances. The ability of our approaches to effectively deal with the high-density marker datasets was demonstrated by comparing the maps constructed for several wheat chromosomes using the MST algorithm (Wang et al. 2014) and the twin approach (Table 4). The lengths of MST-maps were 2–3 times longer than those constructed using the twin method and strongly disagreed with chiasma frequencies (∼1.7–2.5/bivalent) as cytogenetics characteristics of meiotic recombination in wheat (e.g., Feldman 1966; Koul et al. 2000). For chromosome 2A, in spite of the high estimated coefficient of MST-map coverage (0.994) by the markers from the UDM-map, we observed substantial differences in the estimates of genetic distances between the markers (Table S9). Moreover, the number of identified unique recombination map intervals (bins) was substantially higher in the MST-map (220 bins) than in the UDM-map (125 bins). More than half of the MST-map bins contained a single marker, whereas on the UDM-map only three bins had single markers. Considering the size of the DH population, it is unlikely that the inferred number of unique recombination bins on the MST-map is real; most likely it is caused by genotyping errors resulting in the overestimation of the recombination rate by MST. Therefore, hundreds of bins or more on the MST-map are represented by the replicated cosegregating markers that should be excluded from the map. While it is possible that some of these single markers do capture unique recombination events not accounted for in the final UDM-map, the usage of our approach would exclude hundreds of erroneously identified unique recombination events.
Table 4. Comparison of genetic maps constructed using the MST and twin approach implemented in the MultiPoint-UDM (MUDM) software.
| MST bins | L (cM) | |||||
|---|---|---|---|---|---|---|
| Chr | Nm | All | Singletons | MST | MUDM | Nsk |
| 2A | 862 | 217 | 125 | 813.9 | 181.7 | 60 |
| 2B | 1674 | 317 | 176 | 795.4 | 190.0 | 81 |
| 5A | 1556 | 296 | 167 | 787.7 | 156.7 | 67 |
| 7A | 930 | 269 | 151 | 633.8 | 225.7 | 78 |
L, map length using Kosambi metric; Nsk, number of intervals of the map; Nm, number of markers per LG in the initial dataset.
As a further complication of the considered example, the set of 26,000 markers generated based on the 90 K iSelect platform was merged with a set of ∼421,000 markers obtained using genotyping-by-sequencing (GBS) for the same population (Saintenac et al. 2013). Some of the GBS markers were used as usual two-allele SNPs, but the majority were of presence-absence type, representing either M6 or Opata alleles. The combined dataset was filtered to exclude markers with too high a level of missing scores (>40) and too high segregation distortion (χ2 > 35), leaving ∼130,400 for further analysis. This set was analyzed using a twin approach with ts0 = 4. After removal of markers violating map stability and monotonicity (Ronin et al. 2010), followed by map saturation with markers from smaller size TGs and singleton markers, the total number of skeletal markers was 1481. As an illustration, we provide here only the results for the 2B chromosome. The number of skeletal markers in the obtained map of 158.5 cM length was 81 (total number of markers was 526 when cosegregating markers were taken into account) (Figure S1). If we also attach markers for which intervals of 2B are their best location (but their inclusion in the skeletal markers would considerably reduce the map quality), then the total number of markers associated with 81 markers of the 2B skeletal map will be 2241 (Table S10).
Comparing MST and UDM algorithms on simulated data
For comparison, we employed a double haploid mapping population with M = 2000 markers positioned on 84 separable by recombination positions (for population size n = 200) of the simulated chromosome of L = 136.8 cM (Kosambi metric). As can be seen from the results in Table 5, for both levels of missing data (0 and 10%), the MST map undergoes an increase in the number of map bins and inflation of the map length growing with the rate of genotyping errors. This was the case even for error rates as small as 0.001: i.e., 30–35% for L and 70–110% for bin number increases as compared to simulated parameters. For a more realistic error level (1%), the corresponding numbers were: 400–450% for L and 750–900% for bin number. Unlike MST, the length of maps constructed with our approach practically does not vary with the error rate and remains remarkably close to the simulated map despite one order of magnitude in variation of genotyping error rate. Interestingly, the obtained results fit rather well the patterns observed in the above examples on real data from wheat, especially for chromosomes 2B and 5A (see Table 4), suggesting that the rate of genotyping errors in that data could be ∼1–2%. Additional important criteria that we used to assess the quality of maps constructed for simulated data were nr (the number of “repeats”) and lf (percentage of lost unique positions in the constructed skeletal map compared to the simulated map) (see Materials and Methods and File S2). As can be seen from Table 5, even in the worst cases, the number of repeats does not exceed the rate of one repeat per 3–4 skeletal markers. For MST, a lower bound estimation of nr varies with the number of bins, from 2 to 10 repeats per marker. With our approach, the proportion of lost unique positions in the considered examples is a growing function of the error rate and missing data calls (lf varied from 5 to 40%). Obviously, the increase of lf is an unavoidable cost for the noise caused by errors when our attitude is to employ error filtration for getting maps with a minimum number of errors in the reconstructed order of markers. An attempt to keep as many markers as possible leads to maps with unrealistic length and rather questionable marker order.
Table 5. Comparison of the proposed approach implemented in MultiPoint-UDM (MUDM) software with MST on a simulated double haploid population (of size 200 with 2000 markers per chromosome).
| ms% | 0 | 10 | |||||
|---|---|---|---|---|---|---|---|
| pe | 0.001 | 0.005 | 0.01 | 0.001 | 0.005 | 0.01 | |
| MST | LcM | 180 | 294 | 750 | 186 | 408 | 691 |
| Bins | 146 | 291 | 708 | 177 | 513 | 862 | |
| MUDM | LcM | 134 | 134 | 137 | 131 | 135 | 132 |
| Nsk | 81 | 75 | 58 | 75 | 86 | 64 | |
| nr | 1 | 3 | 1 | 6 | 20 | 14 | |
| lf % | 4.8 | 14.3 | 32.1 | 17.9 | 21.4 | 40.5 | |
pe, level of genotyping errors per marker; ms, simulated rate of missing data per marker; LcM, map length (in centimorgans) of a chromosome or LG; lf (%), loss factor, which represents the percentage of lost (noncharacterized) map unique positions in the constructed skeletal map compared to the simulated map; it is calculated as lf = 100 [Nskef − (Nsk − nr)]/Nskef, = 100 (Nskef − Nsk + nr)/Nskef, where Nskef is the number of intervals in the map built for the simulated error-free data, while Nsk and (Nsk − nr) are the number of noisy markers in the skeletal map, noncorrected and corrected for the number of repeats, respectively; nr, the number of “repeats” resulting from fission of the initial groups of cosegregating markers into subgroups due to genotyping errors and missing marker scores; such repeats will appear in the constructed map at separate (usually, but not necessarily, adjacent) positions.
Discussion
The increase in the number of markers by orders of magnitude achievable by the new technologies (GBS, RAD-seq, RNA-seq, etc.) was perceived as a breakthrough that enables building quality ultradense genetic maps. This expectation, being basically correct, may practically be far from reality, due to difficulties caused by genotyping errors, missing marker calls, strong intrachromosomal variation in recombination density, etc. These factors may lead to biased estimates of recombination rates and wrong marker orders, especially for markers with increased error load. Obviously, if the marker set is small, there are few possibilities for filtering and most of the markers will have to remain in the map even if the map lengths are inflated. However, high-throughput genotyping provides an ample amount of data that can be filtered to obtain high-quality datasets. The application of technology-specific filtering and use of appropriate quality controls is the first step for generating reliable data usable for map construction. At the level of genotyping datasets, data filtering not only allows the detection and removal of markers with high segregation distortion and massive losses of data but it is also possible to detect and remove markers violating local map stability by using jackknife resampling (Mester et al. 2003; Ronin et al. 2010). Another quality test is detection of nonmonotonicity of recombination rates along the map, although in some cases such deviation may be caused by negative interference (Denell and Keppy 1979; Peng et al. 2000; Korol et al. 2009; Aggarwal et al. 2015). The method of premapping filtering described in this paper is based on the idea that with very large numbers of scored markers, many markers remaining irresolvable by recombination and appearing as twin groups (TGs) exceeding certain minimal preset threshold size ts0 can be trusted more than singleton markers or markers from smaller size TGs. Simple analysis shows the importance of making proper decisions about ts0 for selecting trustable markers for the skeletal map. We are rather skeptical with respect to some alternative approaches that first build a trial genetic map and then apply different ways of marker correction followed by subsequent map correction. With a higher level of errors, the proportion of error-free markers may become negligible. To cover such situations as well, we employ a generalization of the twin approach based on analysis of the geometry of the n-dimensional space of markers of the mapping population with each marker being presented as a point. With such presentation, the genotyping errors lead to dissipation of TGs, so that the resulting marker agglomerations are “blurred” around the positions of the (unobservable because of errors) initial points corresponding to the error-free situation. Therefore, with a higher level of errors we employ an additional filtration: after the TGs exceeding the threshold size ts0 are selected as candidates for the skeletal map, we conduct clustering of the remaining markers by a procedure similar to the k-means algorithm. Then, representative markers of clusters are added to the set of selected candidates for building the skeletal map. The next steps include resampling-based detection and removing markers of violation of local map stability and monotonicity combined with saturation of the skeletal map by filling in the gaps wherever possible (Ronin et al. 2010). An important factor in getting a high-quality map from the available data given the known parameters (population size, total number of markers, and missing data) is the choice of threshold value (ts0) for TG size and initial radius r in the clustering procedure. Although both these parameters should depend on a priori unknown rate of genotyping errors pe, several trials should usually be enough to clarify the situation and allow a rational choice to be made (as illustrated in Results).
Like other approaches, our analysis starts from premapping filtering based on simple criteria (segregation distortion and missing data). Like other approaches, we also reduce the size of the target dataset by removing redundancy (by representing TGs by single markers). However, unlike others, we take advantage of the information on marker quality hidden in the structure of the multidimensional marker space, in particular in the sizes of TGs as well as in “derivatives” of such groups resulting from dissipation due to genotyping errors and missing data. Depending on genotyping quality, the number of available markers, and the population size, the number of confidently ordered skeletal markers may vary from several tens to several hundreds per chromosome. Yet, markers “represented” by the skeletal markers (i.e., their cosegregants) plus centroid markers from the blurred clusters and markers attached to the closest intervals of the skeletal map (but not included, to prevent map length inflation) may reach tens or even hundreds of thousands. The described system is implemented in the interactive user-friendly software MultiPoint-UDM (MUDM) for building ultradense genetic maps for controlled crosses (backcross, doubled haploids, F2, RIL populations); further development will also include F1 progeny of outbred species and multi-parental populations. Our approach for premapping filtering, together with previously developed algorithms (Mester et al. 2003; Ronin et al. 2010) implemented in the MultiPoint software, enable quality genetic maps to be built, with realistic map length and reliable marker orders (e.g., Avni et al. 2014; Reddy et al. 2014). A trial version for Windows with simulated examples can be downloaded using the link: http://evolution.haifa.ac.il/images/stories/Software/MultiPointUltradense_Demo.zip.
The proposed approach for building high-quality dense genetic maps with a possibility of dealing with big datasets of SNPs may be helpful in addressing a wide range of genetic and genomic problems. We list below just a few for illustration. Although several tens of markers per chromosome may be enough for usual applications of linkage mapping of trait loci (Mendelian or quantitative) based on biparental mapping populations, for association mapping and genomic selection the requirements are much more challenging, especially in situations with steep decay of linkage disequilibrium. Similarly, for map-based cloning, a high density of markers is needed to get as close as possible to the target candidate gene, implying the availability of an accurate dense map. Dense genetic maps were successfully used for anchoring physical contigs (Raats et al. 2013) and sequence scaffolds (Mascher and Stein 2014) to LGs and controlling the quality of sequence assembly (Hedgecock et al. 2015; Zeng et al. 2017). High-coverage shotgun sequencing in combination with new analytical tools of sequence scaffolding, ultradense mapping, and three-dimensional chromosome-conformation-capture-sequencing data was successfully used for high-quality sequencing of such a big and complex genome as wheat (https://www.wheatgenome.org/News2/RefSeq-v1.0-URGI). High-quality dense genetic maps have become a powerful tool for detailed analysis of recombination genomic distribution, sex dependence, genetic variation, and genetic control (Bauer et al. 2013; Rodgers-Melnicka et al. 2015; Ross et al. 2015; Li et al. 2016; Tsai et al. 2016), and genome comparisons of related species (Hill et al. 2015). Obviously, high-quality dense genetic maps are vital for successful use of genome mapping in these and numerous other applications in nonmodel organisms, where validated genome sequences are currently not available. We believe that such studies will benefit from using the approach described here.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.197491/-/DC1.
Acknowledgments
We thank X. Chen, S. Lonardi, and M. Moscou for providing us with original sources of the MergeMap and IPLMap programs. We thank G. Churchill and two anonymous referees for helpful comments and suggestions on the first version of the manuscript. This work was partially supported by the Israel Science Foundation (grant #800/10), US-Israel Binational Agricultural Research and Development Fund (grant #IS-4137-08), European FP7 Programme TriticeaeGenome (grant agreement number FP7-212019), and MultiQTL Ltd.
Footnotes
Communicating editor: G. A. Churchill
Literature Cited
- Aggarwal D., Rashkovetsky E., Michalak P., Cohen I., Ronin Y., et al. , 2015. Experimental evolution of recombination and crossover interference in Drosophila caused by directional selection for stress-related traits. BMC Biol. 13(1): 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov E. D., Goodyear A. W., Geng S., Qi L., Echalier B., et al. , 2003. The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 13: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate D., Bixby R., Chvatal V., Cook W., 2001. TSP Cuts Which Do Not Conform to the Template Paradigm, pp. 261–303 in: Computational Combinatorial Optimization. Lecture Notes in Computer Science, volume 2241, edited by M. Jünger, D. Naddef. Springer, Berlin, Heidelberg. [Google Scholar]
- Applegate D., Cook W., Rohe A., 2003. Chained Lin-Kernighan for large traveling salesman problems. INFORMS J. Comput. 15: 82–92. [Google Scholar]
- Arthur D., Vasilevskii S., 2007. OPOTICS: k-means++ the advantage of careful seeding, pp. 1027–1035 in Proceedings of ACM SIAM Symposium on Discrete Algorithms Society for Industrial and Applied Mathematics, Philadelphia, PA. [Google Scholar]
- Avni R., Nave M., Eilam T., Sela H., Alekperov C., et al. , 2014. Ultra-dense genetic map of durumwheat × wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Mol. Breed. 34: 1549–1562. [Google Scholar]
- Backström N., Forstmeier W., Schielzeth H., Mellenius H., Nam K., et al. , 2010. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 20: 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E., Falque M., Walter H., Bauland C., Camisan C., et al. , 2013. Intraspecific variation of recombination rate in maize. Genome Biol. 14(9): R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C., 1995. Neural Networks for Pattern Recognition. Oxford University Press, Oxford. [Google Scholar]
- Denell R. E., Keppy D. O., 1979. The nature of genetic recombination near the third chromosome centromere of Drosophila melanogaster. Genetics 93: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Arndt P., 2008. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4(5): e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., 1966. The effect of chromosomes 5A, 5B and 5D on chromosomal pairing in Triticum aestivum. Proc. Natl. Acad. Sci. USA 55: 1447–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock D., Shin G., Gracey A. Y., Van Den Berg D., Samanta M. P., 2015. Second-generation linkage maps for the pacific oyster Crassostrea gigas reveal errors in assembly of genome scaffolds. G3 5: 2007–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsgaun K., 2000. An effective implementation of the lin–kernighan traveling salesman heuristic. Eur. J. Oper. Res. 126(1): 106–130. [Google Scholar]
- Hill T., Ashrafi H., Chin-Wo S. R., Stoffel K., Truco M.-J., et al. , 2015. Ultra-high density, transcript-based genetic maps of pepper define recombination in the genome and synteny among related species. G3 5: 2341–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korol A., Preygel I., Preygel S., 1994. Recombination Variability and Evolution. Chapman & Hall, London. [Google Scholar]
- Korol A., Mester D., Frenkel Z., Ronin Y., 2009. Methods for genetic analysis in the Triticeae, pp. 163–200 in Genetics and Genomics of the Triticeae, edited by Feuillet C., Muehlbauer G. J. Springer Science, New York. [Google Scholar]
- Koul K. K., Nagpal R., Sharma A., 2000. Chromosome behaviour in the male and female sex mother cells of wheat (Triticum aestivum L.), oat (Avena sativa L.) and pearl millet (Pennisetum americanum L.). Caryologia 53: 175–183. [Google Scholar]
- Li C., Li Y., Shi Y., Song Y., Zhang D., et al. , 2016. Analysis of recombination QTLs, segregation distortion, and epistasis for fitness in maize multiple populations using ultra-high-density markers. Theor. Appl. Genet. 129: 1775–1784. [DOI] [PubMed] [Google Scholar]
- Lin S., Kernighan B., 1973. An effective heuristic algorithm for the traveling salesman problem. Oper. Res. 21: 498–516. [Google Scholar]
- Liu D., Ma C., Hong W., Huang L., Liu M., et al. , 2014. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS One 9(6): e98855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Stein N., 2014. Genetic anchoring of whole-genome shotgun assemblies (mini review). Front. Genet. 5: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester D., Ronin Y., Minkov D., Nevo E., Korol A., 2003. Constructing large scale genetic maps using an evolutionary strategy algorithm. Genetics 165: 2269–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester D., Korol F., Nevo E., 2004. Fast and high precision algorithms for optimization in large scale genomic problems. Comput. Biol. Chem. 28: 281–290. [DOI] [PubMed] [Google Scholar]
- Mester D., Ronin Y., Korostishevsky M., Frenkel Z., Bräysy O., et al. , 2010. Discrete optimization for some TSP-like genome mapping problems, pp. 1–40 in Handbook of Optimization Theory, edited by Varela J., Acuna S. Nova Science Publishers, New York. [Google Scholar]
- Nambiar M., Smith G. R., 2016. Repression of harmful meiotic recombination in centromeric regions. Semin. Cell Dev. Biol. 54: 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. H., Korol A. B., Fahima T., Röder M. S., Ronin Y. I., et al. , 2000. Molecular genetic maps in wild emmer wheat, Triticum dicoccoides: genome-wide coverage, massive negative interference, and putative quasi-linkage. Genome Res. 10: 1509–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Huang L., Zhu R., Xin D., Chunyan L., et al. , 2014. A high-density genetic map for soybean based on specific length amplified fragment sequencing. PLoS One 9(8): e104871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raats D., Frenkel Z., Krugman T., Dodek I., Hanan S., et al. , 2013. The physical map of wheat chromosome 1BS provides insights into its gene space organization and evolution. Genome Biol. 14: R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P., Paulin L., Hanski I., Lehtonen R., Auvinen P., 2013. Lep-Map: fast and accurate linkage map construction for large SNP datasets. Bioinformatics 29: 3128–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P., Calboli F. C. F., Baocheng G., Takahito S., Juha M., 2016. Construction of ultradense linkage maps with Lep-MAP2: stickleback F2 recombinant crosses as an example. Genome Biol. Evol. 8(1): 78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy U. K., Nimmakayala P., Levi A., Abbur V. L., Saminathan T., et al. , 2014. High-resolution genetic map for understanding the effect of genome-wide recombination rate on nucleotide diversity in watermelon. G3 4: 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnicka E., Bradburya P. J., Elshirea R. J., Glaubitz J. C., Acharya C. B., et al. , 2015. Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc. Natl. Acad. Sci. USA 112: 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesti M., Moser D., Berner D., 2013. Recombination in the threespine stickleback genome—patterns and consequences. Mol. Ecol. 22: 3014–3027. [DOI] [PubMed] [Google Scholar]
- Ronin Y., Mester D., Minkov D., Korol A. B., 2010. Building reliable genetic maps: different mapping strategies may result in different maps. Nat. Sci. 2: 576–589. [Google Scholar]
- Ronin Y., Mester D., Minkov D., Belotserkovski R., Jackson B. N., et al. , 2012. Two-phase analysis in consensus genetic mapping. G3 5: 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronin Y., Mester D., Minkov D., Akhunov E., Korol A., 2015. Building ultra-dense genetic maps in the presence of genotyping errors and missing data, pp. 127–133 in Advances in Wheat Genetics: From Genome to Field (Proceedings of the 12th International Wheat Genetics Symposium), edited by Ogihara Y., Takumi S., Handa H. Springer, Yokohama. [Google Scholar]
- Ross C. R., DeFelice D. S., Hunt G. J., Ihle K. E., 2015. Genomic correlates of recombination rate and its variability across eight recombination maps in the western honey bee (Apis mellifera L.). BMC Genomics 16: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C., Jiang D., Wang S., Akhunov E., 2013. Sequence-based mapping of the polyploid wheat genome. G3 3(7): 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. K., Bolser D., de Boer J., Sønderkær M., Amoros W., et al. , 2013. Construction of reference chromosome-scale pseudomolecules for potato: integrating the potato. G3 3: 2031–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandova-Neeley V., Buluc A., Chapmen J., Gilbert G. R., Gonzalez J., et al. , 2015. Efficient data reduction for large-scale genetic mapping, pp. 126–135 in Proceedings of the 6th ACM Conference on Bioinformatics and Computational Biology. Atlanta, GA. [Google Scholar]
- Tsai H. Y., Robledo D., Lowe N. R., Bekaert B. M., Taggart J. B., et al. , 2016. Construction and annotation of a high density SNP linkage map of the Atlantic salmon (Salmo salar) genome. G3 6: 2173–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wong D., Forrest K., Allen A., Chao S., et al. , 2014. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chen J., Zhang W., Hu Y., Chang L., et al. , 2015. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 16: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Bhat P., Close T., Lonardi S., 2008. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of the graph. PLoS Genet. 10: 10.1371/journal.pgen.1000212e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., Fu Q., Li Y., Waldbieser G., Bosworth B., et al. , 2017. Development of a 690 K SNP array in catfish and its application for genetic mapping and validation of the reference genome sequence. Sci. Rep. 7: 40347 10.1038/srep40347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and in the Supplemental Material.