Chromosomes are organized in the nucleus to accommodate multiple functions. Chu et al. show a functional link between a nuclear pore complex protein...
Keywords: meiosis, homologous recombination, chromosome organization, nucleoporin, synaptonemal complex
Abstract
Meiosis is a specialized cellular program required to create haploid gametes from diploid parent cells. Homologous chromosomes pair, synapse, and recombine in a dynamic environment that accommodates gross chromosome reorganization and significant chromosome motion, which are critical for normal chromosome segregation. In Saccharomyces cerevisiae, Ndj1 is a meiotic telomere-associated protein required for physically attaching telomeres to proteins embedded in the nuclear envelope. In this study, we identified additional proteins that act at the nuclear periphery from meiotic cell extracts, including Nup2, a nonessential nucleoporin with a known role in tethering interstitial chromosomal loci to the nuclear pore complex. We found that deleting NUP2 affects meiotic progression and spore viability, and gives increased levels of recombination intermediates and products. We identified a previously uncharacterized 125 aa region of Nup2 that is necessary and sufficient for its meiotic function, thus behaving as a meiotic autonomous region (MAR). Nup2-MAR forms distinct foci on spread meiotic chromosomes, with a subset overlapping with Ndj1 foci. Localization of Nup2-MAR to meiotic chromosomes does not require Ndj1, nor does Ndj1 localization require Nup2, suggesting these proteins function in different pathways, and their interaction is weak or indirect. Instead, several severe synthetic phenotypes are associated with the nup2Δ ndj1Δ double mutant, including delayed turnover of recombination joint molecules, and a failure to undergo nuclear divisions without also arresting the meiotic program. These data suggest Nup2 and Ndj1 support partially overlapping functions that promote two different levels of meiotic chromosome organization necessary to withstand a dynamic stage of the eukaryotic life cycle.
MEIOSIS is a cellular program that creates haploid gametes from diploid parent cells. Ploidy is reduced through two chromosome segregation events that follow a single round of DNA replication. Mutations affecting chromosome structure, organization, and recombination during meiosis I (MI) prophase often lead to segregation errors or the failure to execute the MI division (Zickler and Kleckner 2015). These errors are the leading cause of birth defects and developmental delays in humans (Hassold and Hunt 2001).
The transition from the mitotic to the meiotic cellular program involves gross chromosome rearrangements, from a configuration where centromeres cluster to one side of the nucleus to one where telomeres cluster instead (Trelles-Sticken et al. 1999; Jin et al. 2000). The events of meiotic prophase follow a specialized round of DNA replication when the meiotic chromosome axis is formed. The chromosome axis is composed of a linear array of loops of sister chromatids attached serially to an axial protein substrate (Zickler and Kleckner 1999; Kleckner 2006). This configuration directs nearly every chromosome-based event of meiotic prophase, including the regulation of double-strand break (DSB) formation, recombination partner choice, and acts as part of the meiotic checkpoint signaling apparatus (Kleckner et al. 2004). Over the course of meiotic prophase, the chromosome axes are aligned via Spo11-induced recombination interactions, and are ultimately joined along their lengths by the transverse element of the synaptonemal complex (SC), Zip1 (Sym et al. 1993; Keeney et al. 1997). Zip1 initially loads where DNA recombination intermediates have been stabilized during zygotene, and proceeds to polymerize along the full length of chromosomes marking pachytene (Borner et al. 2004; Tsubouchi et al. 2008).
In addition, chromosomes are organized in the MI prophase nucleus, with telomeres attached to a protein bridge composed of a trimer of SUN (Sad1/UNC-84) domain proteins that span the inner nuclear membrane, and a trimer of KASH (Klarsicht, ANC-1, Syne Homology) proteins that span the outer nuclear membrane (Sosa et al. 2012; Horn et al. 2013; Luxton and Starr 2014; Stewart and Burke 2014). This bridge directs telomere-led chromosome motion that is directed by cytoplasmic motor proteins (Conrad et al. 2008; Kosaka et al. 2008; Koszul et al. 2008; Wanat et al. 2008; Sonntag Brown et al. 2011; Lee et al. 2012). In budding yeast, Ndj1 accumulates at telomeres, and is required for bouquet formation via its interaction with Mps3 (Figure 1A; Chua and Roeder 1997; Conrad et al. 1997, 2007; Scherthan 2007). Loss of Ndj1 decreases the efficiency of homolog pairing and recombination, presumably due to the loss of rapid prophase movements (Chua and Roeder 1997; Conrad et al. 1997; Rockmill and Roeder 1998; Peoples-Holst and Burgess 2005; Wu and Burgess 2006a; Lee et al. 2012).
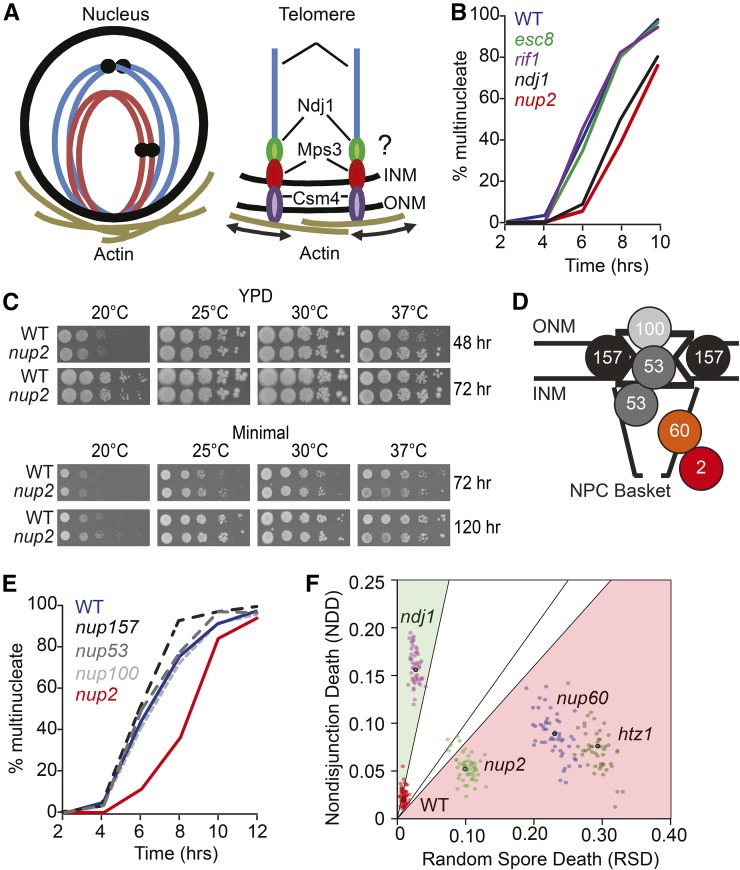
Figure 1.
Identification of Nup2 and its role in meiotic progression. (A) Schematic of the bouquet configuration of chromosomes during meiotic prophase I. The spatial arrangement of chromosomes with telomeres clustered and attached at the inner nuclear membrane is shown. These attachment sites are linked to actin-bundles that surround the nucleus via an Ndj1-Mps3-Csm4 protein bridge that spans the inner and outer nuclear membranes. The arrows depict the actin-directed motion that occurs through this linkage. (B) Kinetics of nuclear division in a time-course experiment from synchronized cells cultured in liquid SPM. Analyzed cells were removed from the culture at the indicated hours after their initial transfer from an overnight YPA culture. At least 200 cells were analyzed for the presence of one, or more than one, DAPI-staining body. The percent of spore viability for each strain is indicated. All strains are diploid, and are isogenic to the WT strain WT (Blue; SBY1903) except for esc8Δ (Green; SBY3942), rif1Δ (Purple; SBY3948), nup2Δ (Red; SBY3945), and ndj1Δ (Black; SBY1904). The kinetics for nuclear division of rif2Δ and the rif1Δ rif2Δ double mutant are shown in Figure S1. (C) Lack of growth phenotype for the nup2Δ mutant under different incubation temperatures on rich (YPD) and minimal media. WT is SBY4102. The growth phenotypes of all mutants are shown in Figure S2. (D) Schematic of the NPC with the relative positions of proteins analyzed in this study, including the outer-nuclear Nup membrane (ONM), the inner-nuclear membrane (INM), the central channel, and the nucleoplasmic NPC basket. (E) Kinetics of nuclear division from a time course experiment as described in (A). WT (Blue; SBY1903), nup157Δ (Black dashed; SBY5256), nup53Δ (Dark gray dashed; SBY5437), nup100Δ (Light gray dashed; SBY5549), and nup2Δ (Red; SBY4102). (F) Cause of spore death in WT (SBY1903) ndj1Δ (SBY2030), nup2Δ (SBY4102 and SBY3945), nup60Δ (SBY5216), and htz1Δ (SBY5268). TetFit was used to calculate the estimated contributions of spore death due to NDD and by RSD, which gives the best fit to the experimental data set (Chu and Burgess 2016). In this assay, precocious sister-chromatid separation, or defects in MII division would appear as RSD. A detailed description of the TetSim and TetFit R-Scripts with annotated instruction is in File 2. To analyze the variation of the output values from TetFit-A, we ran 50 independent simulations using TetFit.Test using the calculated NDD and RSD values for each genotype, and the corresponding number of experimentally dissected tetrads (Table S2 in File S2). The graphical output of the simulations in Figure 4B shows that each genotype gives well defined clusters representing NDD and RSD values for 50 simulated tetrad data sets.
In mitotically dividing cells, the region near the inner-nuclear membrane is enriched for specific DNA sequences, macromolecular complexes including the NPC, and chromatin silencing factors (Taddei and Gasser 2012). In this study, we identified Nup2 enriched in a pool of proteins that copurified with a TAP-tagged Ndj1 protein from meiotic extracts. Nup2 is a mobile nucleoporin found at the inner-nuclear envelope via binding to Nup60 (Hood et al. 2000; Solsbacher et al. 2000), yet is also present in the nucleoplasm (Loeb et al. 1993; Denning et al. 2001). Genome-wide analysis has shown that Nup2 binds near promoter regions of genes, and is required to transport activated genes to the nuclear pore complex (Casolari et al. 2004; Dilworth et al. 2005; Schmid et al. 2006; Brickner et al. 2012). Artificial targeting of Nup2 to a reporter gene promotes association of the locus with the pore, resulting in boundary activity that blocks heterochromatin spreading along the chromosome (Ishii et al. 2002). Whether or not Nup2 plays a role in meiotic chromosome dynamics is not known. Here, we show that a previously uncharacterized 125 aa region of Nup2 is necessary and sufficient to carry out a meiotic role for Nup2 that we call the MAR (meiotic autonomous region). The Nup2-MAR binds meiotic chromosomes, and is sufficient for localization to the nuclear periphery. The nup2Δ ndj1Δ double mutant gives a more severe meiosis defect compared to the single mutants, including a delay in the repair of recombination intermediates, and a block to nuclear divisions. Our results uncover an additional layer of chromosome organization in the meiotic nucleus that acts in parallel with Ndj1.
Materials and Methods
Strains
All strains in this study are derivatives of SK1, and are listed in Table 2. All media were generated as previously described (Ho and Burgess 2011; Lui et al. 2013). Gene knockouts and fluorescent tagging were constructed using standard tailed PCR based gene replacement and tagging techniques (Longtine et al. 1998; Goldstein and McCusker 1999; Sheff and Thorn 2004; Lee et al. 2013). All gene knockouts were confirmed by PCR and new alleles were confirmed by DNA sequencing. Double and triple mutants were created by tetrad dissection. All GFP and mRuby2 protein tags were added to the C-terminal end of the tagged protein with the exception of Zip1. Zip1 was tagged with GFP internally by integrating a ZIP1-GFP plasmid marked with URA3 at the endogenous ZIP1 locus and looping out the endogenous ZIP1 (Scherthan et al. 2007). Truncations were generated using two-step allele replacement (Rothstein 1991). Genomic preparations for PCR were generated as previously described (Danilevich and Grishin 2002).
Table 2. Yeast strains used in this study.
| Strain | Genotypea |
|---|---|
| SBY1903 | MATa/Matα ho::hisG/” leu2::hisG/” ura3/” his4-x::LEU2-(NBam)-URA3/HIS4::LEU2-(NBam) |
| SBY4081 | NDJ1-TAP::klTRP/” SBY1903 |
| SBY3945 | nup2Δ/” SBY1903 |
| SBY1904 | ndj1Δ/” SBY1903 |
| SBY3942 | esc8Δ/” SBY1903 |
| SBY3948 | rif1Δ/” SBY1903 |
| SBY4928 | rif2Δ/” SBY1903 |
| SBY5997 | rif1Δ/” rif2Δ/” SBY1903 |
| SBY4102 | nup2Δ/” SBY1903 |
| SBY5437 | nup53Δ/” SBY1903 |
| SBY5549 | nup100Δ/” SBY1903 |
| SBY5256 | nup157Δ/” SBY1903 |
| SBY5096 | nup2Δ556-720/” SBY1903 |
| SBY5108 | nup2Δ175-720/” SBY1903 |
| SBY5120 | nup2Δ1-50/” SBY1903 |
| SBY5120 | nup2Δ1-50/” SBY1903 |
| SBY5078 | nup2Δ1-175/” SBY1903 |
| SBY5242 | nup2-MAR/” SBY1903 |
| SBY5138 | NUP2-eGFP/” SBY1903 |
| SBY5385 | nup2-MAR-eGFP SBY1903 |
| SBY5629 | NUP2-eGFP/” nup60Δ/” SBY1903 |
| SBY5635 | nup2-MAR-eGFP nup60Δ/” SBY1903 |
| SBY4621 | NDJ1-eGFP/” nup2/” SBY1903 |
| SBY5612 × SBY5614 | nup2-MAR-eGFP/” ndj1/” SBY1903 |
| SBY5470 | SRP1-eGFP/” SBY1903 |
| SBY5476 | SRP1-eGFP/” nup2-MAR-mCherry/” SBY1903 |
| SBY5482 | SRP1-eGFP/” nup2Δ/” SBY1903 |
| SBY5488 | CSE1-eGFP/” SBY1903 |
| SBY5494 | CSE1-eGFP/” nup2Δ50Cnup2-MAR-mCherry/” SBY1903 |
| SBY5500 | CSE1-eGFP/” nup2Δ/” SBY1903 |
| SYB5419 | ndt80Δ/” ZIP1-GFP-700/” SBY1903 |
| SBY5425 | ndt80Δ/” nup2Δ/” ZIP1-GFP-700/” SBY1903 |
| SBY2611 | sae2Δ/” SBY1903 |
| SBY4185 | sae2Δ/” nup2Δ/” SBY1903 |
| SBY3945 | nup2Δ/” SBY1903 |
| SBY2030 | ndj1Δ/” SBY1903 |
| SBY4116 | nup2Δ/” ndj1Δ/” SBY1903 |
| SBY4040 | csm4Δ/” SBY1903 |
| SBY4045 | nup2Δ/” csm4Δ/” SBY1903 |
| SBY5102 | nup2Δ556-720/” ndj1Δ/” SBY1903 |
| SBY5114 | nup2Δ175-720/” ndj1Δ/” SBY1903 |
| SBY5126 | nup2Δ1-50/”ndj1Δ/” SBY1903 |
| SBY5084 | nup2Δ1-175/” ndj1Δ/” SBY1903 |
| SBY5249 | nup2-MAR/” ndj1Δ/” SBY1903 |
| SBY5216 | nup60Δ/” SBY1903 |
| SBY5268 | htz1Δ/” SBY1903 |
| SBY2249 | spo11Δ/” SBY1903 |
| SBY4029 | nup2Δ/” spo11Δ/” SBY1903 |
| SBY4026 | nup2Δ/” ndj1Δ/” spo11Δ/” SBY1903 |
| SBY4890 | spo11-HA-His6/” SBY1903 |
| SBY4980 | nup2Δ/” spo11-HA3-His6/” SBY1903 |
| SBY4914 | spo11-Y135F-HA3-His6/” SBY1903 |
| SBY4986 | nup2Δ/” spo11-Y135F-HA3-His6/” SBY1903 |
| SBY5044 | nup2Δ/” spo11Δ/” SBY1903 |
| SBY5963 | SPC42-eGFP/” HTB2-mRuby2/” SBY1903 |
| SBY5969 | nup2Δ/” SPC42-eGFP/” HTB2-mRuby2/” SBY1903 |
| SBY5987 | ndj1Δ/” SPC42-eGFP/” HTB2-mRuby2/” SBY1903 |
| SBY5993 | nup2Δ/” ndj1Δ/” SPC42-eGFP/” HTB2-mRuby2/” SBY1903 |
| SBY5975 | spo11Δ/” SPC42-eGFP/” HTB2-mRuby2/” SBY1903 |
| SBY5981 | nup2Δ/” spo11Δ/” SPC42-eGFP/” HTB2-mRuby2/” SBY1903 |
| SBY5020 | MATa/Matα ho::hisG/” leu2::hisG/” ura3(DSma-Pst)/” his4-x::LEU2-(NgoMIV; +ori)-URA3/HIS4::LEU2-(BamHI; +ori) |
| SBY5184 | nup2Δ/” SBY5020 |
| SBY5158 | ndj1Δ/” SBY5020 |
| SBY5204 | nup2Δ/” ndj1Δ/” SBY5020 |
| SBY5826 | MATa/Matα ho::hisG/” LEU2::tetR-GFP/” URA3::tetOx224/” his3::hisG/” ndt80Δ/” GAL3/” |
| SBY5832 | nup2Δ/” SBY5826 |
| SBY5838 | ndj1Δ/” SBY5826 |
| SBY5844 | nup2Δ/” ndj1Δ/” SBY5826 |
All disruptions are marked with natMx, hphMx or kanMx (Longtine et al. 1998; Goldstein and McCusker 1999). All fluorescent tags are marked with kanMx (Sheff and Thorn 2004; Lee et al. 2013).
Meiotic time course protocol
The time course protocol was followed as previously reported (Ho and Burgess 2011; Lui et al. 2013). Cells were synchronized for progression through meiosis by first patching and mating haploid cells taken from glycerol stocks stored at −80° on YP media plates (2% Bacto peptone, 1% Bacto yeast extract, 0.01% adenine sulfate, 0.004% tryptophan, and 0.002% uracil) supplemented with 3% glycerol and 2% bacto agar, for 15 hr at 30°. Cells were then streaked onto YPD plates (YP plus 2% dextrose and 2% bacto agar) for 2 days at 30°. Well-isolated diploid single colonies were used to inoculate 5 ml liquid YPD cultures (YPD w/o uracil supplementation), and incubated for 30 hr at 30° on a roller drum. The YPD liquid culture was then added to YPA (YP + 1% potassium acetate w/o uracil supplementation) to a final OD600 of 0.23 and grown for 14.5 hr at 30° on a roller drum. Cells were pelleted by brief centrifugation, washed with SPM (1% potassium acetate, 0.02% raffinose, and 0.009% SC dropout powder), and resuspended in SPM to a final OD600 of 3.0. Cells were removed from the culture at various time points thereafter (starting at t = 0 hr), fixed in 40% ethanol, and stained with DAPI to follow the staged kinetics of meiotic prophase (e.g., synapsis, DSB formation and repair, and nuclear divisions). Nuclear division was marked by the formation of cells with two well-differentiated DAPI-stained foci. At least 200 cells were analyzed for each time point. Lifespan analysis was carried out as previously described (Padmore et al. 1991; Wanat et al. 2008).
Purification of Ndj1-TAP tagged proteins
Ndj1 was C-terminally TAP tagged as previously described (Rigaut et al. 1999). Ndj1 was purified according to the method of Puig et al. (2001), with modifications: 4 liters of meiotic time course culture were harvested after 4 hr when bouquet formation peaks (Wanat et al. 2008). The cell pellet was resuspended in lysis buffer [50 mM HEPES pH 7.5, 150 mM NaCl, 20% glycerol, 0.1% NP-40, 1 mM DTT, 1 mM PMSF, 10 mM sodium pyrophosphate, 50 mM NaF, 60 mM β-glycerophosphate, 1× Halt Protease Inhibitor Cocktail (ThermoFisher Scientific; Conrad et al. 2008)]. The resuspended cells were bead beat using a Beadbeater Disrupter (Biospec) with 0.5 mm diameter zirconia/silica beads (Biospec) with eight cycles of 1 min of bead beating followed by 2 min of on ice. The cell homogenate was spun at 45,000 × g for 30 min at 4°, and the supernatant was transferred to a new tube. MgCl2 and DNaseI (Clontech) were then added to the samples with a final concentration of 5 mM and 1 U/50 μl, respectively, and the samples were incubated at 37° for 30 min, and then mixed with 0.5 ml of IgG sepharose 6 fast flow beads (GE) pre-equilibrated with lysis buffer for 3 hr at 4°. The bead slurry was run through a 0.8 × 4-cm Poly-Prep column (Bio-Rad). The beads were washed three times with 10 ml of lysis buffer, and once with 10 ml of TEV cleavage buffer (10 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 0.5 mM EDTA, 0.1% NP-40, 1 mM DTT). The beads were then incubated in 1 ml of TEV cleavage buffer with 100 units of AcTEV protease (ThermoFisher Scientific) rotating overnight at 4°. The digested products were eluted, and mixed with 3 ml of calmodulin binding buffer (10 mM Tris pH 8.0, 150 mM NaCl, 1 mM imidazole, 1 mM MgOAc, 2 mM CaCl2, 10% glycerol, 10 mM β-mercaptoethanol, 0.1% NP-40) and 3 μl of 1 M CaCl2. This solution was added to 0.5 ml of Calmodulin Affinity Resin (Agilent) preequilibrated with calmodulin binding buffer in a 0.8 × 4-cm Poly-Prep column (Bio-Rad) and mixed for 2 hr at 4°. The beads were washed three times with 10 ml of calmodulin binding buffer. Following the wash, the beads were directly digested, and analyzed by the UC Davis Proteomics core for mass spectroscopy. Two experimental samples with Ndj1 TAP tagged and one negative wild-type control without Ndj1 tagged were analyzed and reported in Supplemental Material, File S1.
Spore viability and nondisjunction analysis
Spore viability was determined as the percent viable spores from dissected tetrads. Single colonies from YPD plates were patched onto solid SPM media and incubated for 3 days at 30°, followed by spore dissection on YPD plates. For each strain, spore viabilities are from pooled tetrad data dissected from multiple colonies on multiple independent occasions. Sporulation efficiency was scored as a cell having two or more spores. A computational estimation of nondisjunction frequencies was determined using TetFit (Chu and Burgess 2016). Expected live:dead tetrad frequencies were determined using the R-suite (TetSim, TetFit, and TetFit.Test) with the following conditions: for TetFit, the number of nondisjunction intervals (ndint) was 3000, the number of random spore death intervals (rsdint) was 3000, ANID was 0.035, and the MI-ND multiplier (ndm) was 10. Detailed background and instructions, and examples of the outputs for using the R-scripts are provided in File S2. For multiple comparisons, the P values were corrected using the Benjamini and Hochberg method (Benjamini and Hochberg 1995).
Spotting assay for vegetative growth
Diploid cells were prepared for synchronization as described above, except that cells from the 5 ml YPD culture were diluted to an OD600 = 1.0 after 24 hr growth. Suspended cells were serially diluted 1:10 in a 96-well plate, and 2.5 μl of the diluted cultures were spotted onto YPD and minimal medium plates using a multichannel pipettor, followed by incubation at 20, 25, 30, and 37°. YPD plates were imaged after incubation for 48 and 72 hr and minimal medium plates (0.67% Difco yeast nitrogen base, 2% dextrose, and 2% agar) were imaged after incubation for 72 and 120 hr using an Epson Perfection 1200 U scanner.
Cell fixation and imaging
A 0.5 ml aliquot of cultured cells was pelleted by centrifugation and resuspended in 1 ml of ice-cold fixative (1% formaldehyde, 100 mM KPO4 pH 7.5, 4% sucrose) followed by incubation at 4° for 30 min while gently rotating. Cells were washed twice with 1 ml of ice-cold potassium acetate/sorbitol solution (1.2 M sorbitol, 100 mM KPO4 pH 7.5, 0.01% Na-Azide) and resuspended in 100 μl sodium acetate/sorbitol solution. Slide preparation for imaging cells was performed as previously described (Dresser 2009). Fixed intact cells were imaged between an SPM agar pad and cover slip. Imaging of TetR-GFP bound at the URA3::tetOx224 locus for the pairing assay was performed as previously described (Lui et al. 2013). A Hybrid Marianas confocal spinning disk 3D fluorescence wide-field microscope (Intelligent Imaging Innovations, 3i) was used for imaging using a 100 × 1.46 NA oil objective lens (Olympus), and with a Yokogawa spinning disk head at room temperature. Fluorescence microscopy data were acquired using an electron multiplying charge coupled device camera. Chromosome spreads were prepared according to Rockmill (2009) and imaged using a Nikon Structured Illumination super-resolution microscope. The fluorophores imaged in this paper were DAPI, eGFP, mRuby2, and mCherry (Sheff and Thorn 2004; Lee et al. 2013). Primary antibodies used in this study include rabbit polyclonal antibody to GFP (Noldus Information Technology NB600-308; 1:2000 dilution), mouse monoclonal antibody to mCherry (World Lab ATB-T5604; 1:2000 dilution). Secondary antibodies used include goat anti-rabbit 488 (Thermo Fisher Alexa Fluor A-11008; 1:2000 dilution), goat anti-rabbit 594 (Thermo Fisher Alexa Fluor A-11012; 1:2000 dilution). The number of Spc42-GFP foci was determined by visual inspection using a Zeiss Axioscope epifluorescence microscope equipped with TRITC, FITC, and Cascade blue filter sets (Chroma).
Colocalization analysis
To test for nonrandom colocalization, copies of the original Nup2-MAR-mCherry fluorescence channel were transformed and superimposed onto the unaltered Ndj1-GFP channel using a custom R-script (Coloc.mis, File S3) based on an overlay misorientation approach (Gasior et al. 1998). These transformations included rotation by 90, 180, and 270°, horizontally mirroring the image, and mirroring the image combined with a 90° rotation. Colocalization frequency was estimated by measuring the proportion of pixels containing a fluorescent signal in each image and detecting the overlapping pixels in the merged image. The overlap was then normalized by the total possible overlap, calculated from a merge of the same channel.
DNA analysis
DNA physical assays and meiotic progression analysis were performed as described previously (Oh et al. 2009). Analysis of crossover (CO) and noncrossover (NCO) products of recombination was carried out by digesting purified genomic DNA with XhoI, or XhoI and NgoMIV, respectively (Oh et al. 2009; Wu et al. 2010). The reported CO and NCO are based on measurements from three independent colonies. The mean and SD are reported, and P values were corrected for multiple comparisons using the “Holm” method in R (Holm 1979).
Data availability
Strains are available upon request. File S1 is an Excel spreadsheet showing the enrichment of proteins analyzed by mass spectrometry. File S2 contains the R-scripts with detailed instructions for using TetSim, TetFit, and TetFit.Test from Chu and Burgess (2016). File S3 contains the R-script with instructions for using Coloc.mis to calculate percent colocalization of pixels in two channels.
Results
Identification of proteins that copurify with Ndj1
We sought to identify telomere-associated proteins in yeast that act at the nuclear periphery during meiotic prophase. We reasoned that our proteins of interest would copurify with TAP-tagged Ndj1 by virtue of its association with telomeres in meiotic prophase extracts. Five proteins were identified: Rif1, Esc8, Yrf1, Nop56, and Nup2 (File S1). All five proteins were shown previously to localize in the nucleus in vegetatively growing cells (Huh et al. 2003). Rif1 is a nonessential protein that binds to the C-terminal region of Rap1, and is involved in telomere silencing and regulation of telomere length (Wotton and Shore 1997; Teixeira et al. 2004). Esc8 is a nonessential protein involved in telomeric and mating-type locus silencing, and interacts with Sir2 (Cuperus and Shore 2002). YRF1 is a repeated gene encoded by the Y′ element of subtelomeric regions, is highly expressed in mutants lacking the telomerase component TLC1, and the protein product is potentially phosphorylated by Cdc28 (Yamada et al. 1998; Ubersax et al. 2003). Nop56 is an evolutionarily conserved component of the box C/D snoRNP complex (Lafontaine and Tollervey 2000). Nup2 is a component of the nuclear pore complex (NPC), exhibits boundary activity, and localizes a subset of actively transcribed genes to the nuclear periphery (Loeb et al. 1993; Dilworth et al. 2005; Schmid et al. 2006; Light et al. 2010). Protein components of the ribosome (11), spliceosome (Prp31), and cytoplasmic proteins (Pet9, and Prr1) were not considered for further analysis (File S1). Notably, we did not isolate Mps3 or Csm4, which have previously been shown to form a complex with Ndj1 using a different purification method (Conrad et al. 2008; Kosaka et al. 2008; Li et al. 2015). Thus, identification of a nonoverlapping set of proteins in this study may reflect differences in the purification protocols.
We tested if deleting RIF1, ESC8, or NUP2 decreased sporulation efficiency, spore viability, or delayed meiotic progression, which are observed in ndj1Δ (Table 1). In all cases, the mutants sporulated with near wild-type (WT) efficiency (>91%, n = 200), yet only the nup2Δ mutation conferred decreased levels of spore viability compared to WT (86.1%, n = 1536 and 97.1%, n = 1152, respectively; Table 1). Since Nop56 is essential, we did not explore a possible role in meiosis. We also did not characterize a possible role of Yrf1 since it has multiple paralogs.
Table 1. Sporulation efficiency and spore viability for strains used in this study.
| Strain | % Sporulation Efficiency (n) | Tetrads Dissected | 4-SV | 3-SV | 2-SV | 1-SV | 0-SV | % Spore Viability |
|---|---|---|---|---|---|---|---|---|
| WT | 95.0 (200) | 288 | 269 | 10 | 6 | 1 | 2 | 97.1 |
| WTa | 89 (231) | 1199 | 1087 | 77 | 32 | 1 | 2 | 96.8 |
| nup2Δb | 92.0 (200) | 384 | 239 | 95 | 36 | 10 | 4 | 86.1 |
| esc8Δ | 91.5 (200) | 48 | 42 | 3 | 2 | 1 | 0 | 94.8 |
| rif1Δ | 94.0 (200) | 48 | 41 | 4 | 2 | 0 | 1 | 96.7 |
| nup53Δ | 94.0 (200) | 72 | 66 | 3 | 1 | 1 | 1 | 95.8 |
| nup100Δ | 93.0 (200) | 72 | 69 | 2 | 1 | 0 | 0 | 95.9 |
| nup157Δ | 94.5 (200) | 72 | 69 | 1 | 2 | 0 | 0 | 98.2 |
| nup2Δ557-720 | 92.5 (200) | 48 | 42 | 4 | 2 | 0 | 0 | 95.8 |
| nup2Δ176-720 | 91.5 (200) | 48 | 45 | 2 | 1 | 0 | 0 | 97.9 |
| nup2Δ2-50 | 93.0 (200) | 72 | 67 | 2 | 3 | 0 | 0 | 97.2 |
| nup2Δ2-175 | 94.0 (200) | 48 | 28 | 12 | 8 | 0 | 0 | 84.5 |
| nup2-MAR | 92.5 (200) | 96 | 83 | 11 | 2 | 0 | 0 | 96.1 |
| nup2-MAR-GFP | 94.0 (200) | 48 | 45 | 2 | 1 | 0 | 0 | 97.9 |
| nup60Δ | 71 (200) | 264 | 85 | 96 | 46 | 25 | 12 | 70.6 |
| htz1Δ | 67.0 (200) | 288 | 77 | 91 | 69 | 40 | 11 | 66.0 |
| ndj1Δ | 75.0 (200) | 192 | 136 | 14 | 19 | 6 | 17 | 82.0 |
| ndj1Δa | 59 (221) | 851 | 482 | 112 | 123 | 38 | 96 | 74.9 |
| nup2Δ ndj1Δ | 1.5 (200) | 144 | 83 | 40 | 15 | 6 | 0 | 84.7 |
| nup53Δ ndj1Δ | 75.0 (200) | 144 | 103 | 10 | 12 | 6 | 13 | 82.0 |
| nup100Δ ndj1Δ | 74.5 (200) | 72 | 48 | 3 | 14 | 1 | 6 | 79.9 |
| nup157Δ ndj1Δ | 72.5 (200) | 72 | 39 | 5 | 14 | 4 | 10 | 70.5 |
| csm4Δ | 77.5 (200) | |||||||
| nup2Δ csm4 | 1.0 (200) | |||||||
| spo11Δ | 93.5 (200) | |||||||
| nup2Δ spo11Δ | 0.5 (200) | |||||||
| spo11Δ | 83.5 (200) | |||||||
| nup2Δ spo11Δ | 3.0 (200) | |||||||
| SPO11-HA3-His6 | 79.0 (200) | |||||||
| Spo11-Y135F-HA3-His6 | 79.0 (200) | |||||||
| nup2Δ SPO11-HA3-His6 | 73.5 (200) | |||||||
| nup2Δ Spo11-Y135F-HA3-His6 | 8.0 (200) |
Data from Wanat et al. (2008) for comparison.
Tetrad data from SBY3945 (n = 98) and SBY4102 (n = 288) were pooled.
To measure the kinetics of MI division in rif1Δ, esc8Δ, and nup2Δ, we calculated the fraction of multinucleate cells taken from a synchronized cell culture for various hours following transfer to sporulation medium starting at t = 0 hr. Interestingly, the formation of binucleate cells in the nup2Δ mutant was delayed by ∼2 hr compared to WT, which is similar to the ndj1Δ phenotype (Figure 1B; Conrad et al. 1997). The rif1Δ and esc8Δ mutants, however, progressed through MI at the same rate as WT (Figure 1B). Since RIF1 and RIF2 are partially redundant (Wotton and Shore 1997), we analyzed rif2Δ and the rif1Δ rif2Δ double mutant, and also found no delay in nuclear divisions (Figure S1).
We performed a colony growth assay to test if nup2Δ gave decreased growth efficiency that could cause asynchrony in the cultures, and appear to delay MI progression. A spotting assay of serially diluted cells showed that nup2Δ growth was no less efficient than WT on either rich or minimal medium at low or high temperatures (Figure 1C). Growth efficiency of nup2Δ on YPD at 37° was slightly more efficient than WT. These data point to a role for Nup2 in promoting normal meiotic progression that is dispensable for normal mitotic growth.
We were unable to reconstitute an interaction between Ndj1 and Nup2 proteins purified from Escherichia coli or by yeast two-hybrid analysis (data not shown). One possibility is that a physical association between Nup2 and Ndj1 may be weak or only indirect. Alternatively, post-translational modifications of the proteins may be required for their interaction, or the proteins copurified by virtue of their localization at the nuclear periphery, and not by direct contact. The outcomes of experiments described below suggest that the latter may be the case.
Deletion of NUP60 and NUP84 give meiotic phenotypes that are distinct from nup2Δ
While Nup2 was the only Nup protein that copurified with Ndj1, we explored if deleting genes encoding other nonessential nups would give a similar meiotic phenotype to nup2Δ. We targeted Nup60 (hNup153) and Nup53, found primarily on the nuclear basket (Loeb et al. 1993; Fan et al. 1997; Hood et al. 2000), and Nup100, Nup157, and Nup84, which interact with other parts of the NPC (Aitchison and Rout 2012; Figure 1D). Neither nup53Δ, nup100Δ, nor nup157Δ showed a decrease in spore viability, delayed meiotic progression, or other obvious growth defects (Figure 1E and Figure S2). In the nup84Δ mutant, 70% of cells could undergo at least one nuclear division, yet <1% of cells gave two or more spores (n = 200 for both). In a previous study by Marston et al. (2004), nup84Δ was competent for sporulation. One difference between the two analyses could be the presence of a second-site suppressor mutation from the yeast deletion collection, which have been described for other mutations in other genes (Winzeler et al. 1999; Hughes et al. 2000). The nup60Δ mutant exhibited reduced sporulation efficiency (71%) and viability (70.6%) compared to WT (95% efficiency and 97.1% viability).
Since the nup60Δ and nup84Δ mutants showed less growth fitness compared to WT 30° using a spotting assay (Figure S2), we excluded them from time-course analysis due to the low likelihood that these cells could be synchronized in culture. Together, these data suggest that the nonessential nucleoporins Nup2, Nup60, and Nup84 are required for normal meiosis. Given its relationship to Ndj1 and Nup60 (below), we focused our attention on Nup2.
Unlike ndj1Δ, spore death in nup2Δ, nup60Δ, and htz1Δ is not due to MI nondisjunction (NDD)
We recently developed a suite of R-scripts that gives a computational estimate of the rate of MI nondisjunction for a given strain based on the fractional incidence of 4, 3, 2, 1, and 0 viable spore tetrads (Chu and Burgess 2016). We applied this approach to infer the causes of spore death in WT, ndj1Δ, nup2Δ, nup60Δ, and htz1Δ (Table 1). HTZ1 encodes the histone variant H2AZ, and has been shown previously to function with Nup60 and Nup2 in boundary activity (Dilworth et al. 2005). We found that the htz1Δ mutant exhibited reduced sporulation efficiency (66%) and viability (67%) compared to WT (95% efficiency and 97.1% viability; Table 1), yet it is not known if inviability is due to increased death due to nondisjunction (NDD). TetFit finds the best-fit values for calculated spore death due to NDD and random spore death (RSD). The NDD/RSD ratios for our WT and ndj1Δ data sets were 2.0%/0.9% and 15.8%/2.7%, respectively. In both cases, the cause of spore death was skewed to NDD. This is consistent with NDD/RSD outcomes of TetFit analysis of previously published data sets for these genotypes (Wanat et al. 2008), where the calculated NDD/RSD ratios for WT and ndj1Δ were 1.8%/1.8% and 20.8%/5.8%, respectively (Figure S3).
We used TetFit to analyze the observed tetrad distributions of nup2Δ, nup60Δ, and htz1Δ. In all cases, these strains gave greater NDD compared to WT, suggesting that spore death in these mutants can be attributed, in part, to MI-ND. However, the best-fit NDD/RSD ratios in these mutants were skewed to greater RSD values: nup2Δ was 5.2%/9.9%; nup60Δ was 8.9%/23.1%; and htz1Δ was 7.5%/29.4%. Thus, unlike ndj1Δ, the cause(s) of spore death in nup2Δ, nup60Δ, and htz1Δ appear to be predominantly due to RSD rather than MI-ND (Figure S3). RSD could arise either by improper partitioning of organelles or other essential cytoplasmic components or when defects in essential cellular processes lead to germination defects. However, precocious sister chromatid separation and MII segregation errors will also appear as RSD (Chu and Burgess 2016). Marston et al. (2004) showed that the segregation of homozygous GFP-tagged chromosome in intact tetrads gave increased levels of 3:1 GFP+:GFP- spores tetrads in the nup2Δ mutant compared to WT. Therefore, increased RSD could reflect PSS or MII segregation errors.
To analyze the confidence of the output values from TetFit, we ran 50 independent simulations of 4, 3, 2, 1, and 0 viable spore tetrads based on the calculated best fit NDD and RSD values found for each mutant and the number of experimentally dissected tetrads. Simulated dissection data were then analyzed by TetFit.Test to estimate the best fitting NDD and RSD. The output of these 50 simulations of all five strains is shown in Figure 1F. The distributions of simulated data support the notion that the cause of spore death is different in ndj1Δ compared to nup2Δ, nup60Δ, and htz1Δ mutants. All three annotated R-scripts for TetSim, TetFit, and TetFit.Test can be found in the Supplemental Methods.
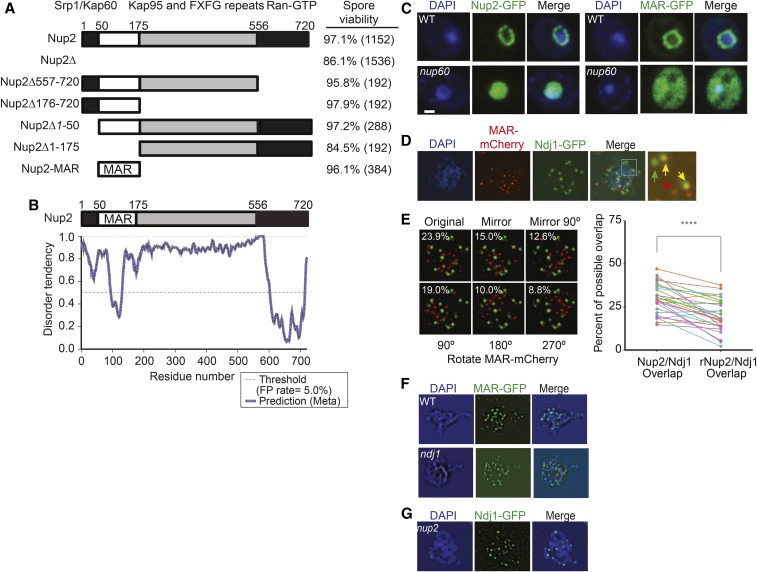
A 125 aa domain of Nup2 is both necessary and sufficient for its role in promoting meiotic progression and spore viability
To test if the nup2Δ meiotic phenotypes were due to a nuclear transport defect(s), we created a series of truncation mutations in one or more of Nup2’s three characterized transport domains (Figure 2A): (1) the first 50 aa N-terminal domain, which binds Srp1/Kap60 (importin-α; Hood et al. 2000; Matsuura et al. 2003), (2) the 154 aa C-terminus (aa 557–720) that binds the Ran-GTP homolog, Gsp2 (Booth et al. 1999; Hood et al. 2000; Matsuura et al. 2003), and (3) the 381 aa unstructured domain containing FXFG repeats, which binds to Kap95 (importin-β; aa 176–556; Solsbacher et al. 2000). We found that all three functional domains of Nup2 could be deleted either on their own, or together without negatively affecting spore viability (Figure 2A). Deleting the region encoding the first 175 aa of Nup2 (nup2Δ2-175) was the only other deletion mutation that conferred a nup2Δ-like meiotic phenotype. Together, these data point to an uncharacterized 125-aa region of the protein likely to have a role in meiosis. We expressed this region alone, and found it was both necessary and sufficient for supporting Nup2’s role in promoting spore viability and meiotic progression (Figure 2A, below). We refer to this region as the Nup2-MAR (meiotic autonomous region).
Figure 2.
Mapping the functional meiotic-autonomous region of Nup2, and its localization in fixed cells. (A) Schematic of the 720 aa Nup2 protein architecture and deletion of known functional domains involved in nucleocytoplasmic transport. The known protein-binding partners that have been associated with specific regions of Nup2 are indicated. All strains are in the SBY1903 background; nup2Δ557-720 (SBY5096), nup2Δ176-720 (SBY5108), nup2Δ1-50 (SBY5120), nup2Δ1-175 (SBY5078), and nup2-MAR (SBY5242).The spore viability and the number of spores analyzed from four-spore tetrads are given on the right. (B) Disorder profile plots using metaPrDOS of the full length Nup2 protein. A higher score indicates a region more likely to be disordered. The dashed line represents the 5% cutoff for false positives. (C) Localization of Nup2-GFP or Nup2-MAR-GFP in fixed cells taken from a mitotically dividing culture. Fixed cells were stained with DAPI and imaged using confocal spinning disk 3D fluorescence wide-field microscope. Images are from a single Z-slice taken from about the middle of the nucleus. Left: Representative cells with Nup2-GFP (SBY5138) and Nup2-GFP nup60Δ (SBY5629) strains are shown on the left. Right: Cells are Nup2-MAR (MAR-GFP; SBY5385) and the MAR-GFP nup60Δ (SBY5635). (D) Localization of Nup2-MAR-mCherry and Ndj1-GFP in a representative meiotic chromosome spread. Cells were taken from a meiotic time course experiment 6 hr after transfer to SPM. mCherry and GFP were detected using polyclonal antibodies to mCherry and GFP, respectively. Secondary antibodies were conjugated to fluorophores that matched the emission spectra of the two protein tags. (E) Estimation of the Nup2-Ndj1 colocalization frequency in meiotic chromosome spreads through overlay misorientation (File S3). To test for random colocalization, copies of the original Nup2-MAR-mCherry fluorescence channel were transformed and superimposed onto the unaltered Ndj1-GFP channel. The figure uses the nucleus from (D) to show the five different transformations that were used, including horizontally mirroring the image, and mirroring the image combined with a 90° rotation (top) and rotation by 90, 180, and 270° (bottom). Colocalization frequency was estimated by measuring the proportion of pixels containing a fluorescent signal in each image and detecting the overlapping pixels in the merged image. The graph shows the reduction in Nup2/Ndj1 overlap following overlay misorientation (rNup2/Ndj1). The statistically significant difference is noted as ****P ≤ 0.0001 (paired t-test). (F) Localization of GFP tagged Nup2-MAR in WT and ndj1Δ mutants (SBY5612xSBY5614) in a representative meiotic chromosome spread as in (D). In this case, the secondary antibody to GFP was conjugated to a red fluorophore for better signal. The signal was then pseudocolored green to maintain consistency with (D) and (F). (G) Same as in (D), except that NUP2 is deleted (SBY4621). These images show that Ndj1-GFP localizes to telomeres in the absence of Nup2.
Nup2-MAR likely folds into a discrete protein domain
A protein BLAST search of the Nup2-MAR sequence returned Nup2 orthologs from related fungal species, and weak matches to the Nup2 mammalian ortholog Nup50. No domains or motifs were found using the PROSITE or Pfam databases of protein families and domains (Finn et al. 2010; Sigrist et al. 2010). The MAR sequence is predicted to form a stable structure based on the Meta Protein DisOrder prediction System (metaPrDOS; Figure 2B; Ishida and Kinoshita 2008).
Nup2-MAR enrichment at the nuclear periphery depends on Nup60
To test if the Nup2-MAR exhibits a similar nuclear location pattern as Nup2, we expressed Nup2-GFP and Nup2-MAR-GFP and analyzed their intracellular localization in fixed cells from an exponentially dividing culture. We found that both Nup2-GFP and Nup2-MAR-GFP signal was enriched at the nuclear periphery (Figure 2C). The Nup2-MAR-GFP fusion protein also showed some diffuse signal in the cytoplasm not seen for Nup2-GFP, which would be expected due to its small size allowing it to diffuse across the NPC (41 kDa; Figure 2C; Shulga et al. 2000). Previous studies have demonstrated that Nup2’s localization to the nuclear basket requires Nup60; however, there are also low levels of Nup2 free of the nuclear basket in the nucleoplasm (Denning et al. 2001). We tested the cellular localization of Nup2-GFP and Nup2-MAR-GFP in a nup60Δ mutant background, and found that the nuclear signal was diffuse, with no obvious enrichment at the nuclear periphery in both cases (Figure 2C). Together, these data suggest that the Nup2-MAR sequence is sufficient to target Nup2 to the nuclear periphery, and that this localization requires Nup60.
The MAR forms foci on meiotic chromosome spreads, independent of Ndj1
Since we identified Nup2 in a pool of proteins copurifying with Ndj1-TAP, we tested if fluorescently tagged Nup2-MAR and Ndj1 proteins would colocalize on spread meiotic chromosome preparations. For this analysis, we synchronized cells coexpressing Nup2-MAR-mCherry and Ndj1-GFP, and removed samples 5 hr after transfer to SPM to enrich for cells in midmeiotic prophase. Both Nup2-MAR-mCherry and Ndj1-GFP foci were seen in spread chromosome preparations, showing they are present in the nucleus at the same time (Figure 2D).
Further analysis of the chromosome spreads (Coloc.mis, File S3), indicated that the level of Nup2-MAR-mCherry and Ndj1-GFP colocalization was, on average, 29.7% (n = 28 cells) of the possible spatial overlap (Figure 2E). Using an overlay misorientation approach (Gasior et al. 1998), we found that transforming one of the channels (by mirroring and/or rotation) reduced this overlap to 9.7% (95% CI: 6.5–12.9), allowing us to estimate that only 20% of this spatial overlap can be explained by random chance. A nonrandom colocalization component of Nup2-MAR-mcherry and Ndj1-GFP is consistent with direct or indirect contacts between Nup2 and Ndj1, and may explain how Nup2 copurified with Ndj1-TAP.
There was some cell-to-cell variation in the number of Nup2-MAR foci, suggesting that the number of foci may be temporally regulated. This is not surprising since the culture at t = 5 hr likely contains a mixed population of cells at various stages of meiotic prophase. Future studies will be directed at addressing this question. Notably, binding of Nup2-GFP did not depend on Ndj1, however, nor did binding of Ndj1-GFP depend on Nup2 (Figure 2, F and G). This is in contrast to another nuclear pore complex protein, Nup49, that has been shown to bind at telomeres in an Ndj1-dependent fashion (Lee et al. 2012).
The MAR sequence is required for normal meiotic progression independent of transport function
Since the Nup2-MAR is missing every known sequence element associated with transport function, we expected that Nup2’s contribution to transport function would be disrupted in the nup2-MAR mutant. In a previous study, both nup2Δ50 (an N-terminal deletion missing the Srp1/Kap60 binding domain and the MAR) and nup2Δ mutants were found to be defective for the enriched localization of Srp1 and Cse1 near the nuclear envelope (Booth et al. 1999; Solsbacher et al. 2000; Matsuura et al. 2003). We expected this would also be the case for the nup2-MAR allele. By tagging Srp1/Kap60 and Cse1 with GFP, we found that these proteins localized to the nuclear periphery, with some cytoplasmic and nuclear signal in WT cells. Localization of these fusion proteins at the nuclear periphery was disrupted in cells expressing only the Nup2-MAR, where their staining was more diffuse (Figure S4). These results support the notion that the Nup2-MAR functions during meiosis, independent of Nup2’s role in nucleocytoplasmic transport.
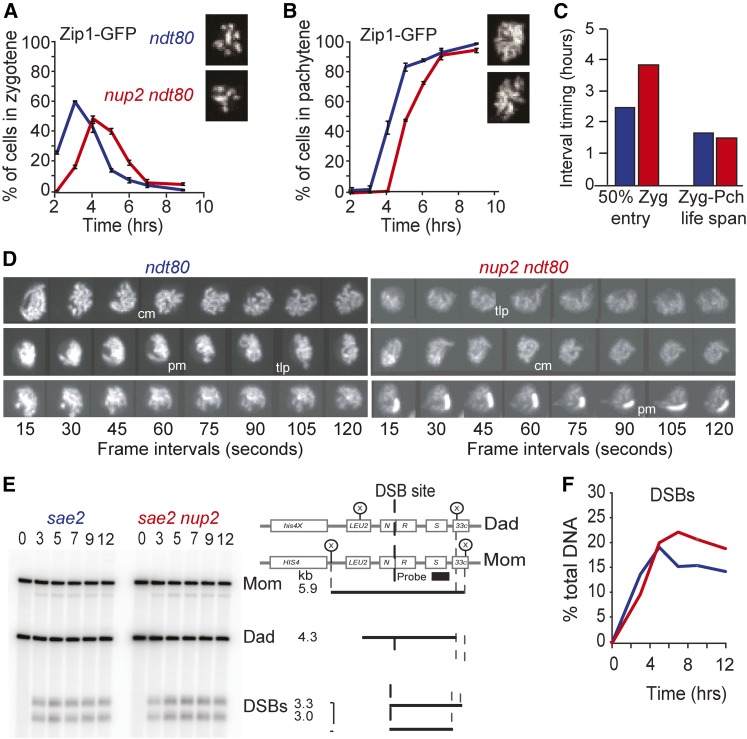
The onset of zygotene, but not its duration, is delayed in nup2Δ
We next examined if meiotic prophase events preceding nuclear division are delayed in the absence of Nup2. First, we examined the initiation of SC formation, which marks the time of the zygotene stage of meiosis. We did this by scoring the presence of patchy appearance of Zip1-GFP fusion protein (Scherthan et al. 2007) using a meiotic time-course assay as described above. As cells transition to full synapsis at the pachytene stage, the Zip1-GFP signal appears as lines (Figure 3, A and B). Synapsis is a terminal phenotype of these cells since NDT80, a regulatory gene required for cells to progress beyond the pachytene stage, was deleted (Xu et al. 1995). We found that the peak occurrence of zygotene in ndt80Δ and nup2Δ ndt80Δ cells was marked at 3 and 4 hr after transfer to sporulation medium, respectively (Figure 3A). By 12 hr, 99% of ndt80Δ and 94% of nup2Δ ndt80Δ cells reached the pachytene stage (Figure 3B). Thus, while Nup2 is required for timely entry into zygotene, it is not required for synapsis.
Figure 3.
Kinetics of zygotene and pachytene stages and DSB formation in the absence of Nup2. (A, B) Meiotic time course of ntd80Δ (Blue; SYB5419) and nup2Δndt80 (Red; SBY5425) strains expressing Zip1-GFP. Shown is the fraction of cells (n = 200) scored as Zygotene (A) and pachytene (B). Representative maximum intensity Z-projections of image stacks are shown on the right side of each figure. Error bars represent the average ± SD for three cultures run in parallel. Similar results were seen for time courses run on different days. (C) The calculated zygotene entry and lifespans from (A) and (B) of ntd80Δ (blue), nup2Δndt80 (red). The life spans of the zygotene stage (the time between which 50% of cells enter and 50% of cells exit) were calculated using data shown in (A) and (B). Shown on the left is the time at which 50% of cells have entered the zygotene stage after transfer to SPM medium. The “Zyg-Pch life span” is defined by the time interval marking 50% entry to zygotene stage and 50% entry into pachytene (hr). (D) Telomere-led movements in ntd80Δ (Blue; SYB5419) and nup2Δndt80 (Red; SBY5425). Shown are 2D projections of 3D image stacks taken at 15 sec intervals in live cells. Synapsed chromosomes are tagged with Zip1-GFP. The various types of movements seen in both WT and nup2Δ include coordinated movement (cm), telomere-led projections (tlp), and perinuclear movement (pm). (E) Southern blot of DNA isolated from sae2Δ (SBY2611) and nup2Δ sae2Δ (SBY4185) mutant cells in a meiotic time courses. Left: Migration of DNA products of a XhoI digest of genomic DNA after gel electrophoresis and probed with sequences that reside on the right side of the DSB hot spot after transfer to a nylon membrane. (F) Quantifications of DSB products (DSBs/Total DNA) from the Southern blot depicted in (D) of sae2Δ (blue) and nup2Δ sae2Δ (red). Both DSB bands were summed to calculate DSB as a percent of total DNA in the lane (Right). Data from one experiment is shown. Identical results were found in an independent replica experiment done on a different day.
Based on these data, we next calculated the time window, or lifespan, in which WT and mutant cells transit through zygotene to pachytene (Padmore et al. 1991). The lifespan of the zygotene stage can be derived from the area under the curve of the primary data (Hunter and Kleckner 2001; Wanat et al. 2008). An increase in lifespan compared to WT indicates a specific delay in passing through the corresponding stage (Hunter and Kleckner 2001). The calculated lifespans for zygotene in ndt80Δ and nup2Δ ndt80Δ show that deleting NUP2 does not cause a delay in this stage (1.7 and 1.5 hr, respectively; Figure 3C). Therefore, while zygotene entry was delayed by ≥1 hr in the absence of Nup2, the time cells took to transit from zygotene to pachytene was not delayed. These results suggest that Nup2 function is required for a meiotic process that acts prior to synapsis initiation.
Nup2 exhibits similar types of chromosome motion as WT
Since the nup2Δ mutant was proficient for synapsis we took advantage of the Zip1-GFP marker to measure chromosome motion as described by Scherthan et al. (2007). We found that nup2Δ cells exhibited the distinct types of movement previously described for WT cells (Koszul et al. 2008). These include coordinated movement of chromosomes in groups (cm), frequent telomere-led projections (tlp) that moved outward from the chromosome mass of the nucleus, and perinuclear movement (pm) of entire chromosomes (Figure 3D). We also observed rare instances where “orphan” or “maverick” chromosomes appeared to completely disjoin from the mass as seen previously (data not shown; Koszul et al. 2008). Thus on a gross level, nup2Δ cells displayed WT-like patterns of motion rather than the absence of telomere-led movement seen in the ndj1Δ mutant (Conrad et al. 2008; Wanat et al. 2008; Sonntag Brown et al. 2011).
DSB levels are elevated in the absence of Nup2
Since synapsis initiates at the sites of DSBs, we tested if the delay in entering the zygotene stage corresponded to a reduction and/or a delay in the formation of DSBs. For this, we measured the physical precursors and products of DSB formation at the well-characterized HIS4LEU2 hot-spot locus (Cao et al. 1990; Storlazzi et al. 1995; Hunter and Kleckner 2001). Following restriction digestion of DNA isolated from cells in a meiotic time course, the DNA fragments were resolved and probed by Southern blotting. The predicted migration of bands reflects the precursor, intermediates and products of the formation and repair of Spo11-induced DSBs. For this analysis we used NUP2 sae2Δ and nup2Δ sae2Δ strain in which the resection of DSB ends is prevented. This gives a readout of total DSBs formed without being turned over (McKee and Kleckner 1997). We found that peak DSB levels occurred at 5 and 7 hr for sae2Δ and nup2Δ sae2Δ strains, respectively. In the sae2Δ mutant, DSB levels reached ∼20% of total DNA levels, while, in the nup2Δ sae2Δ mutant, peak levels were 23% of total DNA (Figure 3, E and F). At later time points, the nup2Δ sae2Δ mutant also gave overall greater levels of DSBs compared to sae2Δ. A second independent time course experiment gave a similar result. The cause of the early delay phenotype was not explored further, yet it is consistent with the possibility that the nup2Δ mutation may disrupt meiotic DNA replication or early chromosome axis structure (Blitzblau and Hochwagen 2013). These results suggest that Nup2 is required for the timing and frequency of DSB formation at the HIS4LEU2 hotspot.
The nup2Δ ndj1Δ mutant exhibits a synthetic sporulation defect and a failure to form binucleate cells
Since we identified Nup2 as a possible interactor of Ndj1, we tested if Nup2 and Ndj1 act in the same or different pathways to promote timely meiotic progression. Since we were unable to detect a direct interaction between the two proteins, or their colocalization, we expected the latter to be the case. To our surprise, only 1.5% of nup2Δ ndj1Δ double mutant cells sporulated, while 92% of nup2Δ and 75% of ndj1Δ cells gave two or more spores (Table 1). These results suggest that Nup2 and Ndj1 function in separate pathways, yet with possible overlapping functions.
Both the nup2Δ and ndj1Δ mutants gave a modest reduction in spore viability among four-spore tetrads (86.1%, n = 1536, and 82.0%, n = 768, respectively) compared to WT (97.1%, n = 1152; Table 1). From the results above we also expected a synthetic reduction in spore viability among the very few tetrads that formed in the double mutant. To our surprise, spores from these tetrads were largely viable (84.7%, n = 576; Table 1). While these values could be highly skewed from the very low sporulation rate, they also lend some insight into the cause of the overall failure to form binucleate cells. For example, if segregation was blocked due to a defect in resolving double-Holliday junctions (Kaur et al. 2015; Tang et al. 2015), there may be a small fraction of cells that escape this block by successfully resolving all joint molecules (JMs). Therefore, the relatively high level of viability suggests that the inability of the double mutant to form binucleate cells is not due to a catastrophic defect in repairing Spo11-induced DSBs, but instead may affect a later step such as CO resolution (see below).
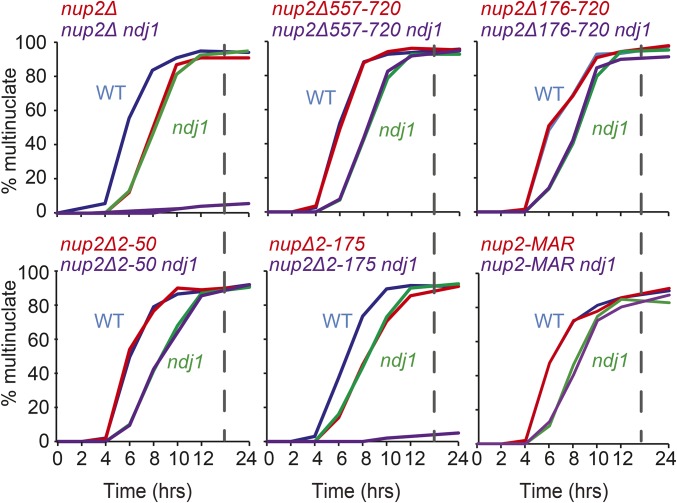
We next measured the kinetics of nuclear division in a meiotic time course of WT, nup2Δ, ndj1Δ, and double mutant combinations involving the truncations mutations described in Figure 2. Among the deletions, only the nup2Δ1-175 mutant gave delayed MI kinetics, similar to nup2Δ, and also failed to progress beyond the mononucleate stage when Ndj1 was absent (Figure 1 and Figure 4). On the other hand, expressing Nup2-MAR in the ndj1Δ background had no synthetic effect. This is another indication that the Nup2-MAR region alone is important for meiotic progression.
Figure 4.
Synthetic block to nuclear division in mutants carrying nup2 truncation mutations in the absence of Ndj1. Meiotic time courses were done as described in Figure 1B. Allele names are the same as shown in Figure 2A.
Since Csm4 is the likely KASH protein linking telomeres to cytoskeletal actin in budding yeast (Kosaka et al. 2008; Wanat et al. 2008; Fridolfsson and Starr 2010), we tested if the csm4Δ mutation in combination with nup2Δ would give a synthetic phenotype similar to nup2Δ ndj1Δ. First we looked at the sporulation efficiency of the single and double mutants, and found that while 92% of nup2Δ and 76% of csm4Δ cells produced two or more spores (n = 200), only 1% of nup2Δ csm4Δ double mutant cells (n = 200) produced spores, similar to what we observed for nup2Δ ndj1Δ (1.5%, n = 200; Table 1). Likewise, in a meiotic time-course experiment, nuclear divisions were nearly absent in the nup2Δ csm4Δ double mutant, which is similar to what we observed for nup2Δ ndj1Δ (Figure S5). Therefore, the synthetic phenotype caused by deletion of both Nup2 and Ndj1 extends to other mutants affecting the bouquet or chromosome motion. These results are consistent with the notion that Nup2 functions in a chromosome event of meiosis that is partially redundant with Ndj1 and Csm4.
Spindle-pole body duplication without nuclear divisions in the nup2Δ ndj1Δ double mutant
To better understand the cause of the failure of the nup2Δ ndj1Δ double mutant to form binucleate cells, we asked if this mutant could progress beyond prophase arrest, which would not be expected if the lesion activated the recombination checkpoint (Tsuchiya et al. 2014). As an output of cell cycle progression, we measured spindle-pole body (SPB) separation and reduplication. The SPB is the microtubule-organizing center in budding yeast, and is required for chromosome separation in meiosis and mitosis (Lim et al. 2009). During meiosis, SPBs duplicate in coordination with DNA replication (Moens and Rapport 1971), but do not separate at MI until the end of pachytene, when the Ndt80 transcription factor is activated (Xu et al. 1995; Chu and Herskowitz 1998). The segregation of homologous chromosomes at MI is followed by a second round of SPB duplication and separation to segregate sister chromatids.
For this analysis, we expressed the fluorescently tagged SPB component Spc42-GFP in WT and mutant cells (Shirk et al. 2011). The most striking feature that came from this analysis was that the nup2Δ ndj1Δ cells remained mononucleate, even though the Spc42-GFP foci separated and reduplicated to form four SPBs (Figure 5A). In the nup2Δ ndj1Δ mutant, 78% of cells gave ≥2 Spc42-GFP (n = 200) at 12 hr after transfer to SPM (Figure 5B). This is in stark contrast to the <5% of cells that were also binucleate (Figure 5C). These results suggest that the nup2Δ ndj1Δ double mutant progresses through the meiotic program without separating the DNA masses. SPB separation also rules out the possibility that the cell cycle is blocked due to a checkpoint-induced arrest at prophase.
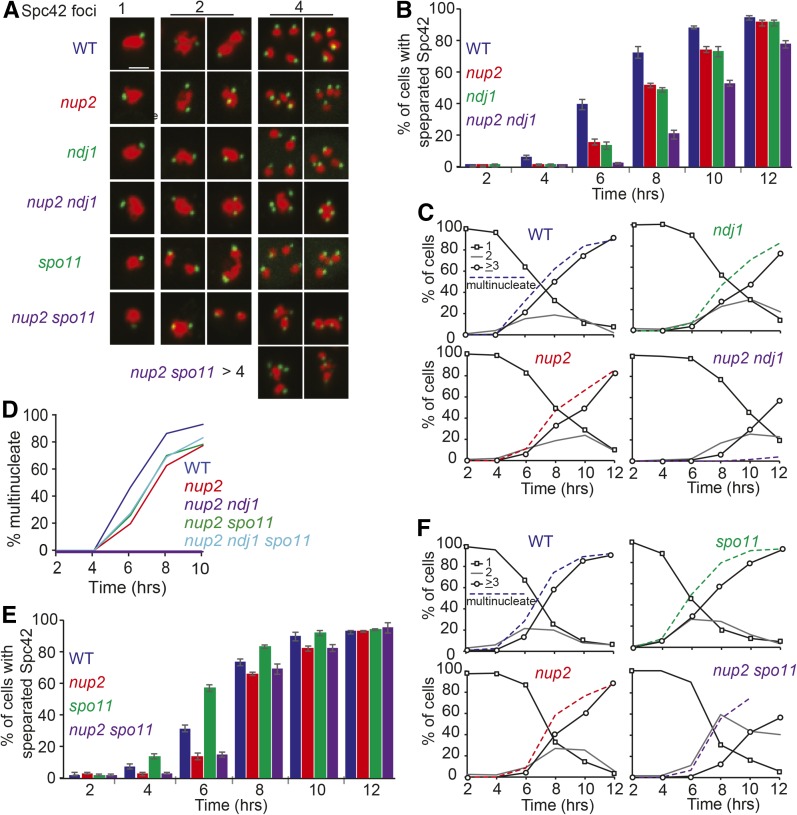
Figure 5.
Meiotic time course to assay SPB duplication and separation in WT, nup2Δ ndj1Δ, nup2Δ ndj1Δ, spo11Δ, and nup2Δ spo11Δ cells. (A) Representative images of with 1, 2, or 4 Spc42-GFP foci in fixed cells taken from a culture 12 hr after transfer to SPM. Spc42-GFP is shown in green and Htb2-mRuby2 is shown in red. WT (SBY5963), nup2Δ (SBY5969), ndj1Δ (SBY5987), nup2Δ ndj1Δ (SBY5993), spo11Δ (SBY5975), and nup2Δ spo11Δ (SBY5981). (B) Kinetics of the percent of cells in the strains above with two or more separated Spc42-GFP foci in a meiotic time course at the indicated hours after transfer to SPM. (C) Kinetics of the percent of cells in the strains above with separated Spc42-GFP foci in a meiotic time course at the indicated hours after transfer to SPM. Percentage of cells with one Spc42-GFP focus is shown in solid black line with open squares; cells with two Spc42-GFP foci are shown in gray; cells with ≥3 Spc42-GFP foci are shown black with open circles. Multinucleate cells were scored by DAPI staining of nuclei (dashed line). (D) Kinetics of nuclear division in a time course experiment with WT, nup2Δ (SBY3945), nup2Δ spo11Δ (SBY4029), nup2Δ ndj1Δ (SBY3983), and nup2Δ ndj1Δ spo11Δ (SBY4026). (E) Same as in (B) except that the strains are WT (SBY5963), nup2Δ (SBY5969), spo11Δ (SBY5975), and nup2Δ spo11Δ (SBY5981). (F) Same as in (C) except that the strains are the same as those used in (E).
Separation of Spc42-GFP foci in nup2Δ and ndj1Δ single mutants gave delayed kinetics compared to WT (Figure 5, B and C), which is not surprising given that they both show delays of meiotic prophase stages. This unusual phenotype allowed us to determine if the nup2Δ and ndj1Δ alleles show epistasis with respect to the timing of Spc42-GFP separation. We found that the time at which 50% of cells had separated Spc42-GFP foci was delayed ∼1 hr in the nup2Δ mutant, and ∼1.3 hr in the ndj1Δ mutant compared to WT, while the double mutant exhibited a delay of ∼2.9 hr (Figure 5C). This general lack of epistasis, compared to the other strong synthetic block to nuclear divisions, indicates that the primary defects in nup2Δ and ndj1Δ single mutants that lead to delayed SPB separation affect different cellular processes.
Deleting SPO11 bypasses the block to forming nuclear divisions
The severe synthetic phenotype of the double mutant appears to be neomorphic, and may arise by the formation of a poisonous intermediate, or product, that creates a physical impediment to chromosome separation, such as chromosome entanglements, or the inability to separate sister chromatids. To test if chromosomes are unable to separate in the nup2Δ ndj1Δ double mutant due to topological constraints caused by homologous recombination, we deleted SPO11 and measured the kinetics of nuclear divisions. We found that spo11Δ could indeed suppress the block to nuclear divisions in the nup2Δ ndj1Δ background (Figure 5D), indicating that the failure of nup2Δ ndj1Δ cells to undergo at least one nuclear division was likely due to a defect in processing one or more recombination intermediates (see below) that would bypass the prophase checkpoint.
The nup2Δ spo11Δ mutant exhibits a synthetic sporulation defect and a failure to form tetranucleate cells
Even though the nup2Δ ndj1Δ spo11Δ triple mutants could form binucleate cells, there was still a block to forming tetranucleate cells and normal spores, indicating a possible independent synthetic phenotype of nup2Δ and spo11Δ. We evaluated sporulation and Spc42-GFP separation and reduplication in the nup2Δ spo11Δ double mutant, and found that this strain also failed to sporulate, even though SPBs duplicated and reduplicated (Figure 5, A, E, and F and Table 2). More than four Spc42-GFP foci were seen in a small population of nup2Δ spo11Δ cells, indicating possible misregulation of SPB duplication or fragmentation (Figure 5A).
These results uncover an unexpected second synthetic phenotype associated with the nup2Δ spo11Δ double mutant. Since Spo11 also has apparent roles in axis structure that are independent of its ability to catalyze DSBs (Cha et al. 2000), we tested if the nup2Δ spo11Δ synthetic phenotype was due to the failure to make DSBs or a second DSB-independent role of Spo11. To distinguish between these possibilities, we introduced a catalytically inactivating mutation spo11-Y135F in the nup2Δ mutant background that would prevent DSB formation without disrupting these other roles for Spo11. We found that the nup2Δ spo11-Y135F double mutant also gave a synthetic sporulation defect and failure to form tetranucleate cells in the absence of Nup2 (Table 1). Thus, it appears that DSBs themselves may influence some feature of meiotic chromosome morphology that is disrupted in the absence of Nup2; however, it is not clear what this mechanism of action is.
DSB formation and repair in nup2Δ, ndj1Δ, and nup2Δ ndj1Δ
Since the inability of nup2Δ ndj1Δ cells to separate DNA depends on the formation of Spo11 induced DSBs, we tested if the molecular basis for this synthetic phenotype would be evident in one or more stages governing meiotic DSB repair. We thus measured the physical recombination intermediates and products in WT, nup2Δ, ndj1Δ, and nup2Δ ndj1Δ using the HIS4LEU2 DSB hotspot (Figure 6, A–C; Storlazzi et al. 1995; Hunter and Kleckner 2001).
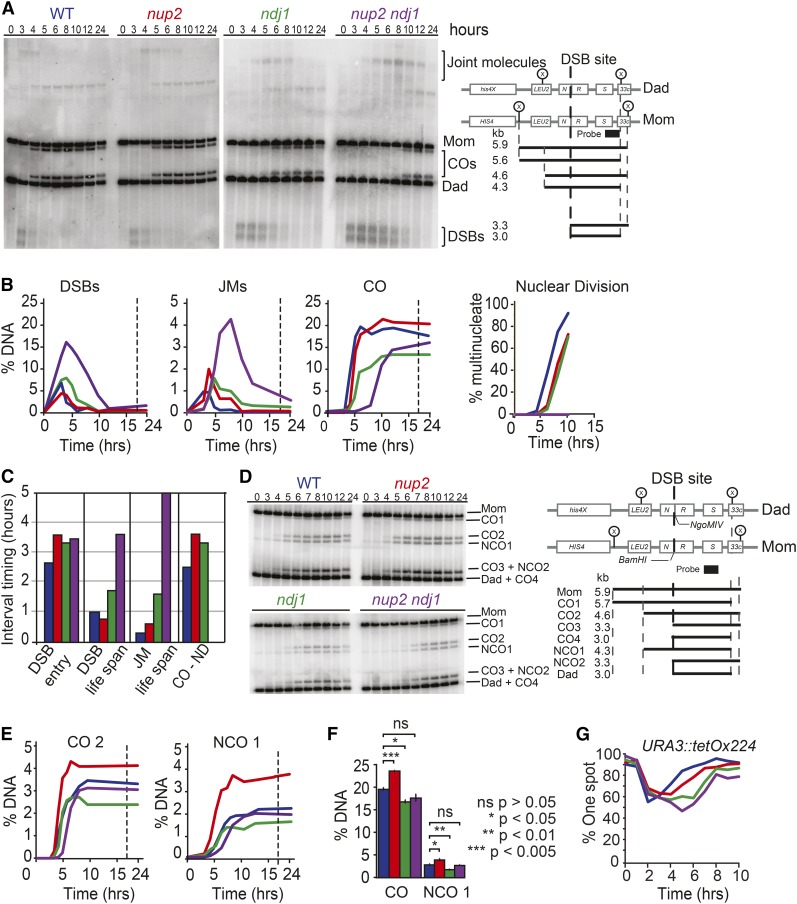
Figure 6.
Time-course analysis of precursors, intermediates and products of homologous recombination at the HIS4LEU2 hot spot in WT, nup2Δ ndj1Δ, and nup2Δ ndj1Δ cells. (A) Representative Southern blots and schematic of the HIS4LEU2 hotspot region showing the position of the Xho1 cut sites (x). The predicted sizes for the starting chromosomes from the HIS4LEU2 strain (mom) and his4XLEU2 strain (Dad), DSBs (3.3 and 3.0 kb), JMs, and CO products are shown. (B) Amounts of DSB, JM, and recombinant products as a fraction of total DNA for each lane in a time course assay for WT (Blue; SBY1903), nup2Δ (Red; SBY3945), ndj1Δ (Green; 1904), and nup2Δ ndj1Δ (Purple; SBY3983). Right: Kinetics of forming multinucleate cells in the same time course. (C) Lifespan analysis for DSB and JMs was performed as described in Figure 3. The interval between the time at which 50% of CO have formed, and the time at which 50% of cells have undergone nuclear division, is shown for all strains, except nup2Δ ndj1Δ, which does not undergo nuclear divisions. Lifespan analysis is based on maximum levels of DSBs measured for sae2Δ, nup2Δ sae2Δ, and the nup2Δ ndj1Δ sae2Δ double mutant (Padmore et al. 1991). For comparison, lifespans for ndj1Δ were normalized to sae2Δ levels (Wu and Burgess 2006a; Wanat et al. 2008). (D) Southern blot analysis of CO and NCO formation in WT, nup2Δ, ndj1Δ, and nup2Δ ndj1Δ. The left side shows a Southern blot of a meiotic time course of WT (Blue; SBY5826), nup2Δ (Red; SBY5832), ndj1Δ (Green; SBY5838), and nup2Δ ndj1Δ (Purple; SBY5844). The right side shows the schematic and recombination products of the HIS4LEU2 hotspot used. DNA was digested with XhoI and NgoMIV. (E) CO2 and NCO1 quantifications as percent of total DNA. (F) CO and NCO quantifications of WT (blue), nup2Δ (red), ndj1Δ (green), and nup2Δ ndj1Δ (purple) from Figure S6, A and B. Data are represented as mean of three independent replicates ±SD. P values are based on a t-test with multiple comparisons adjustment using the Holm method. Representative blots are shown in Figure S6. (G) Pairing analysis of strains containing TetO arrays integrated at URA3 and expressing TetR-GFP fusion protein. Homologs were scored as paired if only a single GFP focus could be observed, and unpaired if two GFP foci were observed. All strains are ndt80Δ. The pairing levels of WT (Blue; SBY5826), nup2Δ (Red; SBY5832), ndj1Δ (Green; SBY5838), and nup2Δ ndj1Δ (Purple; SBY5844) during a meiotic time course; 200 cells were scored for each time point.
Using the total DSB levels found for sae2Δ and nup2Δ sae2Δ strains above, we calculated the interval timing between the formation and repair of DSBs. We found that the “lifespan” of DSBs was similar in WT and nup2Δ, if not slightly shorter in nup2Δ (0.9 and 0.8 hr, respectively). This was consistent with the slightly shorter zygotene window we observed (above). As seen previously (Wanat et al. 2008), we found the DSB lifespan in the ndj1Δ mutant was about twice as long as WT (1.7 hr), and this transition in nup2Δ ndj1Δ was ∼3.6 hr. By simply adding the two effects we would expect a delay of <1 hr in DSB turnover in the nup2Δ ndj1Δ mutant, yet the delay we found was nearly 3 hr longer (Figure 6C). Considered together, these results point to a synthetic defect in the repair of DSBs in the double mutant as a possible contributing factor to the failure of this strain to form binucleate cells.
DNA JM turnover in nup2Δ, ndj1Δ, and nup2Δ ndj1Δ
DSBs are processed into JMs that include single-end invasions and double-Holliday junction intermediates (Hunter 2015; Lam and Keeney 2015). Disruption of the recombination program can also result in more complex forms of JMs (Oh et al. 2007; Kaur et al. 2015; Tang et al. 2015). We found that the JM lifespan in nup2Δ (0.6 hr) was longer than that in WT (0.3 hr), but not to the extent of ndj1Δ (1.6 hr; Figure 6C). The most dramatic effect on JM lifespan, however, was seen in the nup2Δ ndj1Δ double mutant (5.0 hr; Figure 6C). These results point to the accumulation of JMs as a possible contributing factor to the nuclear-division failure of the nup2Δ ndj1Δ mutant. Even though it appears that most of the JMs are turned over at the HIS4LEU2 hotspot, less efficient repair at other loci could impede chromosome separation. This could explain the relatively high spore viability of the nup2Δ ndj1Δ double mutant; if segregation was blocked due to a defect in resolving double-Holliday junctions (Kaur et al. 2015; Tang et al. 2015), there may be a small fraction of cells that escape this block by successfully resolving all JMs. Since SPB separation in the nup2Δ ndj1Δ mutant was delayed by only 2.9 hr compared to WT, but JM resolution was delayed by 5 hr, this suggests that SPB separation occurs even though JMs have not been resolved, thus pointing to JMs as potential physical barrier to nuclear division.
CO formation to MI division timing is delayed in nup2Δ and ndj1Δ compared to WT
The timing of CO formation to MI provides an additional landmark to account for the post-DSB contribution to the nup2Δ delay. The nup2Δ and ndj1Δ mutants exhibited a 0.9 and 0.6 hr delay from 50% CO levels to the time at which 50% of cells underwent nuclear division, respectively, compared to WT (Figure 6C). The nup2Δ ndj1Δ mutant could not be analyzed in this way due to the failure to segregate chromosomes. These data, and those reported above, suggest that there are at least three stages of meiotic prophase that are extended in the nup2Δ mutant compared to WT (approximate values in hours): a period prior to, or during, DSB formation (+0.9), the lifespan of JMs (+0.3), and the interval between CO formation and chromosome separation at MI (+0.9; Figure 5C). The ndj1Δ mutant also exhibits a delay in DSB and JM turnover as reported previously (Wu and Burgess 2006a; Wanat et al. 2008), and these intervals are greatly extended in nup2Δ ndj1Δ compared to WT (+2.7 and +4.7 hr, respectively; Figure 6C). Thus, by acting alone or with Ndj1, Nup2 appears to influence multiple steps of the meiotic program, from DSB formation to the separation of chromosomes at MI.
CO and NCO levels are elevated in nup2Δ compared to WT
In the time course study described above, the nup2Δ mutant gave slightly elevated CO levels compared to WT (Figure 6B). We followed up this observation by analyzing a modified HIS4LEU2 allele, whereby cutting by both XhoI and NgoMIV can recover NCO products that have undergone gene conversion but not exchange of flanking markers (Figure 6D). By analyzing the physical products of recombination at this locus in a time course assay, we found that CO and NCO levels of well separated products in nup2Δ were overall higher than in WT (Figure 6E). We next carried out a separate experiment done in triplicate using just XhoI to analyze total CO levels at 12 hr after transfer to SPM. Accordingly, we found the total CO levels in the nup2Δ mutant to be significantly higher than in WT (23.7% vs. 19.6%, respectively, t-test, P = 0.004; Figure 6F and Figure S6A). Using XhoI and NgoMIV, we found NCO levels in the nup2Δ mutant to be slightly higher than in WT: 3.3% vs. 2.4% (t-test, P = 0.04; Figure 6F and Figure S6B). Taken together, the increased total levels of DSBs and recombination products we see in the absence of Nup2 suggest that Nup2 may have a negative influence on regulating DSB formation (see Discussion).
Nup2 and Ndj1 contribute independently to the efficiency of homolog pairing
It is well established that homologous chromosome pairing is delayed in ndj1Δ and csm4Δ mutants (Chua and Roeder 1997; Peoples-Holst and Burgess 2005; Conrad et al. 2008; Lee et al. 2012; Lui et al. 2013). We used a previously described “one-spot, two-spot” assay (Brar et al. 2009; Sonntag Brown et al. 2011; Lui et al. 2013), in which chromosome V homologs, each with an array of tetO-repeats inserted at URA3, were visualized in individual cells expressing a TetR-GFP fusion protein. Binding of this protein to the tetO-repeats gives a one-focus “spot” if the loci are paired, and two foci if they are unpaired (Michaelis et al. 1997). This experiment was done in an ndt80Δ mutant strain background, where pairing persists since cells arrest in the pachytene stage. As seen previously, over 80% of cells gave a one-spot signal at the time of transfer to SPM (at the G0 stage; Figure 6G; Brar et al. 2009; Lui et al. 2013). At 2 hr, NUP2 NJD1, nup2Δ, ndj1Δ, and nup2Δ ndj1Δ strains exhibited timely disruption of G0 pairing, which coincides with DNA replication timing (Brar et al. 2009). Pairing was re-established in both ndj1Δ and nup2Δ mutants, albeit with some delay compared to WT (Figure 6G). Pairing in nup2Δ ndj1Δ cells was further delayed compared to either single mutant, suggesting that the contributions of Nup2 and Ndj1 to the timing of homolog pairing occur through independent pathways (Figure 6G).
Discussion
We show that a 125-aa region of the nonessential nucleoporin, Nup2, is necessary and sufficient for normal meiotic progression in budding yeast. The Nup2-MAR sequence alone was sufficient for localization at the nuclear periphery, and showed the same genetic dependency on Nup60 as full length Nup2. The Nup2-MAR also localized as foci on chromosome spreads from cells in meiotic prophase. Deletion of NUP2 resulted in a delay in MI nuclear division, including a delay in forming DSBs, and a modest decrease in spore viability. Synthetic genetic interactions between nup2Δ, and mutations in NDJ1 and SPO11 point to functionally redundant roles in meiotic chromosome organization. Together, these findings uncover a meiotic role for Nup2 in the normal progression of the chromosome events of meiosis at the level of nuclear organization.
While Nup2 was found in an enriched pool of proteins with purified Ndj1, several lines of evidence argue that this interaction may be weak or indirect. First, while Nup2-MAR-mCherry and Ndj1-GFP foci were associated with meiotic chromosome spreads during midprophase, most were spatially separated, although a subset of signals were overlapping. Second, Nup2-MAR binding to meiotic chromosomes did not require Ndj1, nor did Ndj1 binding require Nup2. Finally, we were unable to reconstitute a physical interaction biochemically, or by the yeast two-hybrid system. It is important to note that both proteins have been shown previously to organize specific chromosome sequences at the nuclear periphery. Ndj1 is required for attaching telomeres to the nuclear envelope, and formation of the bouquet in meiosis (Trelles-Sticken et al. 1999), while Nup2 anchors specific transcriptionally activated genes to the nuclear pore complex in nonmeiotic cells (e.g., the GAL locus in the presence of galactose; Schmid et al. 2006). Together, these results suggest that Nup2 and Ndj1 both reside at the nuclear periphery, yet their local environments may be distinct.
The phenotypes of the nup2Δ and ndj1Δ single mutants are also largely distinguished from one another, suggesting the Nup2 and Ndj1 proteins act independent of one another. First, our analysis using TetFit showed that spore inviability in the ndj1Δ mutant can be attributed to MI chromosome nondisjunction, while spore inviability in the nup2Δ mutant is likely due other causes, such as the misappropriation of cellular components, meiosis II segregation errors, or precocious sister chromatid separation. Second, the nup2Δ mutant exhibited previously described patterns of motion seen in wild-type cells, including telomere-led protrusions (Koszul et al. 2008) that are not seen in the ndj1Δ mutant (Conrad et al. 2008; Wanat et al. 2008; Sonntag Brown et al. 2011). Third, while the lifespan of DSBs was longer in the ndj1Δ mutant compared to WT, it was the same, or even faster, in the nup2Δ mutant. Fourth, deleting SPO11 suppressed delayed nuclear division in the absence of Ndj1, but not in the absence of Nup2. Thus, while the delay seen in ndj1Δ is due to the presence of unrepaired recombination intermediates that activate the recombination checkpoint (Wu and Burgess 2006a,b; Wanat et al. 2008), the delay in nup2Δ appears to be due to other defect(s) that precede DSB formation or synapsis. Finally, the timing of SPB separation, and the timing of homolog pairing in the nup2Δ ndj1Δ double mutant, reflected an additive effect of the two mutations.
Synthetic phenotypes of the nup2Δ ndj1Δ double mutant phenotype compared to each single mutant uncovered potentially redundant roles for Nup2 and Ndj1 in meiosis. The double mutant gave a severe reduction in the ability of cells to undergo the MI nuclear division and a dramatic accumulation of DNA joint molecules, suggesting a defect in the resolution of late-stage recombination intermediates. These two phenotypes are similar to the meiotic depletion alleles of top3/rmi1 that disrupt decatenase activity and lead to the accumulation of unresolved JMs. These mutations block nuclear divisions without disrupting the meiotic program, also known as “meiotic catastrophe” (Gangloff et al. 1999; Jessop and Lichten 2008; Oh et al. 2008; Kaur et al. 2015; Tang et al. 2015). Similar to the top3/rmi1 mutants, deletion of SPO11 or mutating the catalytic residue of SPO11 (SPO11-Y135F) suppressed the nup2Δ ndj1Δ block to nuclear divisions, presumably by eliminating a physical constraint caused by intermediates, or aberrant structures, associated with meiotic recombination. While there are other details that limit further comparison to the top3/rmi1 phenotype, the nup2Δ ndj1Δ phenotype is consistent with the presence of recombination-induced topological entanglements that prevent nuclear division.
A second way binucleate formation can be blocked is through the failure to lose connections between sister chromatids, as is seen in the absence Tid1, which causes abnormal persistence of the Mcd1 and Rec8 cohesin proteins (Kateneva et al. 2005). Like nup2Δ ndj1Δ, the tid1Δ single mutant also fails to undergo nuclear divisions even though the spindle program progresses. Kateneva et al. (2005) also showed that deletion of SPO11 could bypass the MI block in the absence of Tid1, but that these cells were unable to exit anaphase II. Similarly, we found that deleting SPO11 enabled nup2Δ ndj1Δ mutants to undergo the first nuclear division, but not the second. This was also true, however, for the nup2Δ spo11Δ and nup2Δ spo11-Y135F double mutants, pointing to a more complex relationship between the nup2Δ and spo11Δ(Y135F) mutations. In this respect, nup2Δ alone does not phenocopy tid1Δ. This interpretation suggests that DSBs would play a role in developing axis structure, or defining the relationship of sister chromatid cohesion, that is not supported in the absence of Nup2.
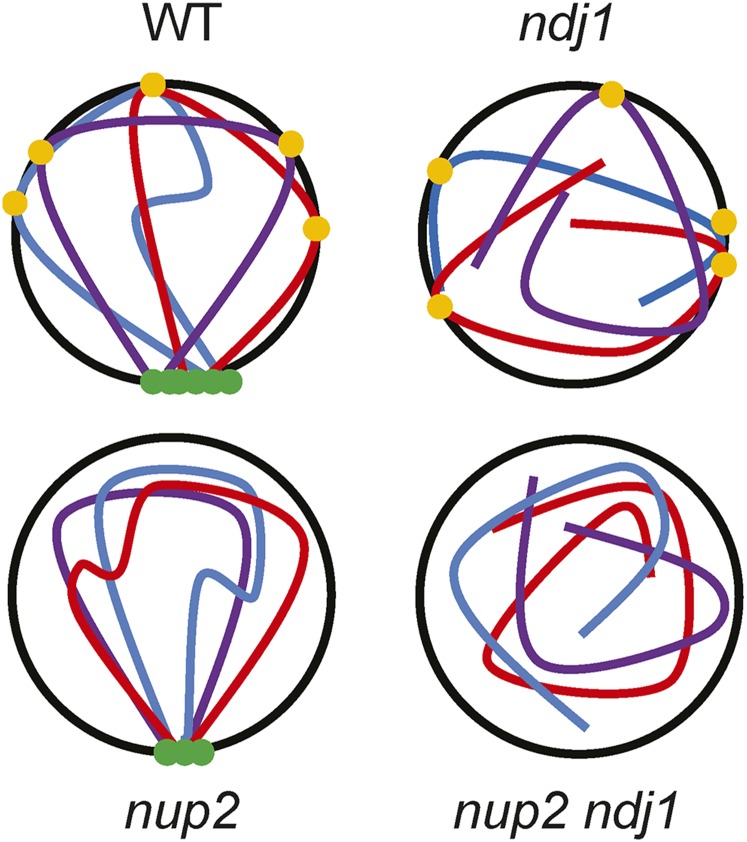
We propose that chromosome organization in the meiotic nucleus requires Nup2, which acts in parallel with Ndj1-mediated functions to support the dynamic chromosome interactions associated with homolog pairing, synapsis, and homologous recombination. Disruption of either configuration alone can be accommodated to complete meiosis, with relatively little impact on spore formation and spore viability; however, when both are disrupted, aberrant intersister and/or interhomolog interactions prevents their separation at MI or meiosis II (Figure 7). The constellation of phenotypes of nup2Δ ndj1Δ make it unique, thus pointing to a heretofore uncharacterized meiotic cellular process. For example, the interdependency of Nup2 and Ndj1 indicate that they may function to bridge the transition from the mitotic to the meiotic programs, when chromosomes transition from the Rabl orientation, with centromeres clustered, to the meiotic bouquet, with telomeres clustered. This dramatic reorientation of chromosomes in the nucleus is accompanied by myriad changes at other levels of organization that are critical for meiotic chromosome progression (Zickler and Kleckner 1998, 2015; Koszul and Kleckner 2009). Interestingly, chromosomal regions tethered at the nuclear periphery by Nup2 in nonmeiotic cells can vary with changes in the transcriptional program (Schmid et al. 2006; Ahmed et al. 2010; Light et al. 2010; Brickner et al. 2012). Entry to the meiotic program in yeast is also coupled to changes in the transcriptional profile across the genome (Chu and Herskowitz 1998). It is intriguing to speculate that Nup2 brings DNA sequences to the nuclear periphery that are functionally or transcriptionally relevant to meiosis.
Figure 7.
Model for two levels of chromosome organization in the meiotic nucleus supported by Ndj1 and Nup2. Loss of either pathway allows for meiotic progression, albeit with delay in nuclear division and progression through meiotic prophase. In the absence of both Ndj1 and Nup2, chromosome organization is impacted so that chromosomes are physically unable to separate. Another possibility is that the nup2Δ mutation impacts the ability of sister chromatids to separate, which is not mutually exclusive of the given model (not shown).
Other than the meiotic bouquet, little is known about how chromosomes are organized at the nuclear periphery in the meiotic nucleus, or how additional layers of organization may be modulated during the dynamic chromosome events of meiosis. This study uncovers a role for Nup2, which is involved in the organization of chromosomes during vegetative growth, and its functional relationship with Ndj1, which is involved in attaching telomeres to cytoskeletal motor proteins, and Spo11, which initiates meiotic recombination at the chromosome axis. These findings will lead to new lines of inquiry to better understand how multiple layers of nuclear organization are integrated to carry out the meiotic program.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.194555/-/DC1.
Acknowledgments
We would like to thank Dan Starr, JoAnne Engebrecht, Anne Britt, James McGehee, and An Nguyen for their insightful review of this manuscript, and Soni Lacefield for discussion. We thank Yana Blokhina for assistance with imaging and Kelsey Walters for assistance in strain construction. We thank the Kaback laboratory for the ZIP1-GFP plasmid, Neil Hunter for strains, and Akira Shinohara for sharing detailed protocols. The authors have declared that no conflicting interests exist. This work was funded by National Institutes of Health (NIH) R01 GM075119 award to S.M.B. and pilot funds from the University of California Davis Proteomics Core.
Footnotes
Communicating editor: J. Surtees
Literature Cited
- Ahmed S., Brickner D. G., Light W. H., Cajigas I., McDonough M., et al. , 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J. D., Rout M. P., 2012. The yeast nuclear pore complex and transport through it. Genetics 190: 855–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Blitzblau H. G., Hochwagen A., 2013. ATR/Mec1 prevents lethal meiotic recombination initiation on partially replicated chromosomes in budding yeast. eLife 2: e00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth J. W., Belanger K. D., Sannella M. I., Davis L. I., 1999. The yeast nucleoporin Nup2p is involved in nuclear export of importin alpha/Srp1p. J. Biol. Chem. 274: 32360–32367. [DOI] [PubMed] [Google Scholar]
- Borner G. V., Kleckner N., Hunter N., 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Brar G. A., Hochwagen A., Ee L. S., Amon A., 2009. The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol. Biol. Cell 20: 1030–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Ahmed S., Meldi L., Thompson A., Light W., et al. , 2012. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev. Cell 22: 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N., 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Casolari J. M., Brown C. R., Komili S., West J., Hieronymus H., et al. , 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Cha R. S., Weiner B. M., Keeney S., Dekker J., Kleckner N., 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14: 493–503. [PMC free article] [PubMed] [Google Scholar]
- Chu D. B., Burgess S. M., 2016. A computational approach to estimating nondisjunction frequency in Saccharomyces cerevisiae. G3 Bethesda 6: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., Herskowitz I., 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1: 685–696. [DOI] [PubMed] [Google Scholar]
- Chua P. R., Roeder G. S., 1997. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11: 1786–1800. [DOI] [PubMed] [Google Scholar]
- Conrad M. N., Dominguez A. M., Dresser M. E., 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276: 1252–1255. [DOI] [PubMed] [Google Scholar]
- Conrad M. N., Lee C. Y., Wilkerson J. L., Dresser M. E., 2007. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104: 8863–8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M. N., Lee C. Y., Chao G., Shinohara M., Kosaka H., et al. , 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133: 1175–1187. [DOI] [PubMed] [Google Scholar]
- Cuperus G., Shore D., 2002. Restoration of silencing in Saccharomyces cerevisiae by tethering of a novel Sir2-interacting protein, Esc8. Genetics 162: 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevich V. N., Grishin E. V., 2002. A new approach to the isolation of genomic DNA from yeast and fungi: preparation of DNA-containing cell envelopes and their use in PCR. Russ. J. Bioorganic Chem. 28: 136–146. [DOI] [PubMed] [Google Scholar]
- Denning D., Mykytka B., Allen N. P., Huang L., Al B., et al. , 2001. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154: 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth D. J., Tackett A. J., Rogers R. S., Yi E. C., Christmas R. H., et al. , 2005. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell Biol. 171: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M. E., 2009. Time-lapse fluorescence microscopy of Saccharomyces cerevisiae in meiosis. Methods Mol. Biol. 558: 65–79. [DOI] [PubMed] [Google Scholar]
- Fan F., Liu C. P., Korobova O., Heyting C., Offenberg H. H., et al. , 1997. cDNA cloning and characterization of Npap60: a novel rat nuclear pore-associated protein with an unusual subcellular localization during male germ cell differentiation. Genomics 40: 444–453. [DOI] [PubMed] [Google Scholar]
- Finn R. D., Mistry J., Tate J., Coggill P., Heger A., et al. , 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson H. N., Starr D. A., 2010. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol. 191: 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., de Massy B., Arthur L., Rothstein R., Fabre F., 1999. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 18: 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S. L., Wong A. K., Kora Y., Shinohara A., Bishop D. K., 1998. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 12: 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P., 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2: 280–291. [DOI] [PubMed] [Google Scholar]
- Ho H. C., Burgess S. M., 2011. Pch2 acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. PLoS Genet. 7: e1002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S., 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6: 65–70. [Google Scholar]
- Hood J. K., Casolari J. M., Silver P. A., 2000. Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-alpha export. J. Cell Sci. 113(Pt. 8): 1471–1480. [DOI] [PubMed] [Google Scholar]
- Horn H. F., Brownstein Z., Lenz D. R., Shivatzki S., Dror A. A., et al. , 2013. The LINC complex is essential for hearing. J. Clin. Invest. 123: 740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. R., Roberts C. J., Dai H., Jones A. R., Meyer M. R., et al. , 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25: 333–337. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., et al. , 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Hunter N., 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7: a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N., 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Ishida T., Kinoshita K., 2008. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics 24: 1344–1348. [DOI] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., Laemmli U. K., 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109: 551–562. [DOI] [PubMed] [Google Scholar]
- Jessop L., Lichten M., 2008. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell 31: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q. W., Fuchs J., Loidl J., 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113(Pt. 11): 1903–1912. [DOI] [PubMed] [Google Scholar]
- Kateneva A. V., Konovchenko A. A., Guacci V., Dresser M. E., 2005. Recombination protein Tid1p controls resolution of cohesin-dependent linkages in meiosis in Saccharomyces cerevisiae. J. Cell Biol. 171: 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H., De Muyt A., Lichten M., 2015. Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol. Cell 57: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kleckner N., 2006. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–94. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Zickler D., Jones G. H., Dekker J., Padmore R., et al. , 2004. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA 101: 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka H., Shinohara M., Shinohara A., 2008. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 4: e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kleckner N., 2009. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 19: 716–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K. P., Prentiss M., Kleckner N., Kameoka S., 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133: 1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. L., Tollervey D., 2000. Synthesis and assembly of the box C+ D small nucleolar RNPs. Mol. Cell. Biol. 20: 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I., Keeney S., 2015. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 7: a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Conrad M. N., Dresser M. E., 2012. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 8: e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lim W. A., Thorn K. S., 2013. Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One 8: e67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Shao Y., Jin H., Yu H. G., 2015. Ndj1, a telomere-associated protein, regulates centrosome separation in budding yeast meiosis. J. Cell Biol. 209: 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W. H., Brickner D. G., Brand V. R., Brickner J. H., 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A. Z incorporation and INO1 transcriptional memory. Mol. Cell 40: 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. H., Zhang T., Surana U., 2009. Regulation of centrosome separation in yeast and vertebrates: common threads. Trends Cell Biol. 19: 325–333. [DOI] [PubMed] [Google Scholar]
- Loeb J., Davis L., Fink G., 1993. NUP2, a novel yeast nucleoporin, has functional overlap with other proteins of the nuclear pore complex. Mol. Biol. Cell 4: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lui D. Y., Cahoon C. K., Burgess S. M., 2013. Multiple opposing constraints govern chromosome interactions during meiosis. PLoS Genet. 9: e1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G. W., Starr D. A., 2014. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr. Opin. Cell Biol. 28: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston A. L., Tham W. H., Shah H., Amon A., 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303: 1367–1370. [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Lange A., Harreman M. T., Corbett A. H., Stewart M., 2003. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 22: 5358–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A. H., Kleckner N., 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146: 797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K., 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45. [DOI] [PubMed] [Google Scholar]
- Moens P. B., Rapport E., 1971. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol. 50: 344–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Lao J. P., Hwang P. Y.-H., Taylor A. F., Smith G. R., et al. , 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Lao J. P., Taylor A. F., Smith G. R., Hunter N., 2008. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell 31: 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. D., Jessop L., Lao J. P., Allers T., Lichten M., et al. , 2009. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol. Biol. 557: 209–234. [DOI] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N., 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256. [DOI] [PubMed] [Google Scholar]
- Peoples-Holst T. L., Burgess S. M., 2005. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 19: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., et al. , 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., et al. , 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rockmill B., 2009. Chromosome spreading and immunofluorescence methods in Saccharomyces cerevisiae. Methods Mol. Biol. 558: 3–13. [DOI] [PubMed] [Google Scholar]
- Rockmill B., Roeder G. S., 1998. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev. 12: 2574–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194: 281–301. [DOI] [PubMed] [Google Scholar]
- Scherthan H., 2007. Telomere attachment and clustering during meiosis. Cell. Mol. Life Sci. 64: 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Wang H., Adelfalk C., White E. J., Cowan C., et al. , 2007. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104: 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., et al. , 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21: 379–391. [DOI] [PubMed] [Google Scholar]
- Sheff M. A., Thorn K. S., 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21: 661–670. [DOI] [PubMed] [Google Scholar]
- Shirk K., Jin H., Giddings T. H., Winey M., Yu H.-G., 2011. The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J. Cell Sci. 124: 2891–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., Mosammaparast N., Wozniak R., Goldfarb D. S., 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149: 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J., Cerutti L., de Castro E., Langendijk-Genevaux P. S., Bulliard V., et al. , 2010. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38: D161–D166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J., Maurer P., Vogel F., Schlenstedt G., 2000. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol. Cell. Biol. 20: 8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag Brown M., Zanders S., Alani E., 2011. Sustained and rapid chromosome movements are critical for chromosome pairing and meiotic progression in budding yeast. Genetics 188: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B. A., Rothballer A., Kutay U., Schwartz T. U., 2012. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149: 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. L., Burke B., 2014. The missing LINC: a mammalian KASH-domain protein coupling meiotic chromosomes to the cytoskeleton. Nucleus 5: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A., Xu L., Cao L., Kleckner N., 1995. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc. Natl. Acad. Sci. USA 92: 8512–8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Engebrecht J. A., Roeder G. S., 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- Taddei A., Gasser S. M., 2012. Structure and function in the budding yeast nucleus. Genetics 192: 107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Wu M. K. Y., Zhang R., Hunter N., 2015. Pervasive and essential roles of the Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol. Cell 57: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. T., Arneric M., Sperisen P., Lingner J., 2004. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 117: 323–335. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E., Loidl J., Scherthan H., 1999. Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112(Pt. 5): 651–658. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Macqueen A. J., Roeder G. S., 2008. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 22: 3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya D., Yang Y., Lacefield S., 2014. Positive feedback of NDT80 expression ensures irreversible meiotic commitment in budding yeast. PLoS Genet. 10: e1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., et al. , 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864. [DOI] [PubMed] [Google Scholar]
- Wanat J. J., Kim K. P., Koszul R., Zanders S., Weiner B., et al. , 2008. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 4: e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wotton D., Shore D., 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11: 748–760. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Burgess S. M., 2006a Ndj1, a telomere-associated protein, promotes meiotic recombination in budding yeast. Mol. Cell. Biol. 26: 3683–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Burgess S. M., 2006b Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr. Biol. 16: 2473–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Ho H. C., Burgess S. M., 2010. Mek1 kinase governs outcomes of meiotic recombination and the checkpoint response. Curr. Biol. 20: 1707–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N., 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Hayatsu N., Matsuura A., Ishikawa F., 1998. Y′-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J. Biol. Chem. 273: 33360–33366. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N., 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N., 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N., 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7: a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. File S1 is an Excel spreadsheet showing the enrichment of proteins analyzed by mass spectrometry. File S2 contains the R-scripts with detailed instructions for using TetSim, TetFit, and TetFit.Test from Chu and Burgess (2016). File S3 contains the R-script with instructions for using Coloc.mis to calculate percent colocalization of pixels in two channels.