Abstract
Histone post-translational modifications play vital roles in a variety of nuclear processes, including DNA repair. It has been previously shown that histone H3K79 methylation is important for the cellular response to DNA damage caused by ultraviolet (UV) radiation, with evidence that specific methylation states play distinct roles in UV repair. Here, we report that H3K79 methylation is reduced in response to UV exposure in Saccharomyces cerevisiae. This reduction is specific to the dimethylated state, as trimethylation levels are minimally altered by UV exposure. Inhibition of this reduction has a deleterious effect on UV-induced sister chromatid exchange, suggesting that H3K79 dimethylation levels play a regulatory role in UV repair. Further evidence implicates an additional role for H3K79 dimethylation levels in error-free translesion synthesis, but not in UV-induced G1/S checkpoint activation or double-stranded break repair. Additionally, we find that H3K79 dimethylation levels are influenced by acetylatable lysines on the histone H4 N-terminal tail, which are hyperacetylated in response to UV exposure. Preclusion of H4 acetylation prevents UV-induced reduction of H3K79 dimethylation, and similarly has a negative effect on UV-induced sister chromatid exchange. These results point to the existence of a novel histone crosstalk pathway that is important for the regulation of UV-induced DNA damage repair.
Keywords: histone modification, DNA repair, methylation, sister chromatid exchange, histone crosstalk
CELL survival depends on the preservation of genomic integrity. Cells are perpetually exposed to intrinsic and extrinsic factors that chemically alter DNA, potentially causing genomic instability. One of the most prevalent environmental factors that challenge genome integrity is solar radiation, specifically wavelengths that fall within the ultraviolet (UV) spectrum (Rastogi et al. 2010). DNA absorbs UV radiation, leading to the formation of structurally deforming cyclo-butane pyrimidine dimers and 6–4 photoproducts. Such lesions can inhibit essential cellular operations, such as DNA replication and transcription, and can cause mutations. As a result, UV exposure is one of the greatest risk factors for environmentally associated cancer in humans (Friedberg et al. 2006).
UV-induced DNA damage is processed by a variety of molecular pathways. Initially, DNA-binding factors detect UV-induced DNA irregularities and activate cell cycle checkpoints at G1/S, mid-S, and G2/M (Sugasawa 2016). Nucleotide excision repair is the primary mechanism for repair of UV damage, in which the lesion is removed and replaced by nascent DNA (Prakash and Prakash 2000). Damage tolerance pathways also contribute to survival following UV exposure (Boiteux and Jinks-Robertson 2013). For example, “postreplication repair” describes a variety of processes that complete gaps in DNA that arise during replication of the damaged template, such as translesion synthesis (TLS), by low-fidelity DNA polymerases (Broomfield et al. 2001). Likewise, recombination mechanisms can be employed to fill replication-associated gaps via sister chromatid exchange, as well as to repair double-stranded breaks generated by UV damage (Kadyk and Hartwell 1993; Kupiec 2000; Gangavarapu et al. 2007).
In eukaryotic organisms, the recognition and repair of DNA damage occurs in the context of chromatin. Minimally, chromatin must be remodeled to accommodate the repair machinery, with an “access-repair-restore” model describing the changes to chromatin that are required for efficient repair (Polo and Almouzni 2015). Consequently, chromatin-associated proteins, particularly histones, are integral players in DNA repair mechanisms. Histones are subject to a wide array of post-translational modifications, many of which have been implicated in DNA repair (Cao et al. 2016). The roles of these modifications in repair include influence on DNA accessibility, recruitment of repair factors, establishment of interactions between homologous chromosomes and sister chromatids, regulation of repair-related gene expression, and modulation of cell cycle progression. Disruption of histone modifications causes various repair deficiencies, often leading to genomic instability, and, as a result, having important implications for cancer progression (Wang et al. 2016).
Methylation of histone H3 at lysine 79 (H3K79me) is important for UV repair, as loss of this modification causes a reduction in survival following UV exposure (Bostelman et al. 2007; Evans et al. 2008; Chaudhuri et al. 2009). Prior studies have implicated functions for H3K79me in DNA damage checkpoint activation and global NER (Giannattasio et al. 2005; Wysocki et al. 2005; Chaudhuri et al. 2009; Tatum and Li 2011; Rossodivita et al. 2014), as well as UV-induced sister chromatid exchange (Rossodivita et al. 2014). Furthermore, we have previously reported evidence indicating that specific H3K79 methylation states play distinct roles in UV repair in yeast. H3K79 can possess up to three methyl groups per residue (denoted H3K79me1, me2, and me3), catalyzed by histone methyltransferase Dot1 (Ng et al. 2002a; van Leeuwen et al. 2002), and further influenced via crosstalk with histone H2B K123 ubiquitylation (Ng et al. 2002b; Sun and Allis 2002; Shahbazian et al. 2005; Frederiks et al. 2008). Our prior studies revealed that while both the me2 and me3 states contribute to UV-induced checkpoint activation, the me3 state is uniquely required for sister chromatid exchange in response to UV exposure (Rossodivita et al. 2014).
H3K79 methylation has been suggested to act in a steady-state manner in the context of DNA repair by virtue of its ubiquitous presence in the genome (Huyen et al. 2004). However, it has been shown that H3K79me2 levels oscillate during the cell cycle (Schulze et al. 2009), raising the possibility that methylation state levels might be modulated in response to DNA damage. As we will describe below, we find that H3K79me2 levels are uniquely reduced in response to UV exposure during the G1/S checkpoint response in bakers yeast, dropping to roughly half of their pre-exposure levels. We will present additional evidence indicating that H3K79me2 acts within several DNA damage pathways, and is part of a novel histone crosstalk interaction involving histone H4 acetylation.
Materials and Methods
Yeast strain construction
Yeast strains used in this study are listed in Table 1. DOT1 was deleted by PCR-mediated gene disruption (Brachmann et al. 1998), using the KanMX marker. Replacement of the wild-type gene encoding histone H3 (HHT2) with various mutant alleles of H3 was done by a plasmid swapping method (Evans et al. 2008), and strains containing the ade2-n::TRP1::ade2-I reporter construct for sister chromatid exchange assays were constructed as previously described (Rossodivita et al. 2014).
Table 1. Saccharomyces cerevisiae strains used in this study.
| Strain name | Genotype | Source |
|---|---|---|
| JTY34 | MAT a ade2-101 his3Δ200 lys2-801 trp1Δ901 ura3-52 hht1,hhf1::LEU2 hht2,hhf2::HIS3 plus pJT34 (HHT2-HHF2 LYS2 CEN4 ARS1) | Thompson et al. (1994) |
| JTY308 | Isogenic to JTY34 except with pJTH3-8 (hht2-K79E LYS2 CEN4 ARS1) in place of pJT34 | Thompson et al. (2003) |
| JTY309 | Isogenic to JTY34 except with pJT309 (hht2-L70S LYS2 CEN4 ARS1) in place of pJT34 | Thompson et al. (2003) |
| JTY34D | Isogenic to JTY34 except dot1::kanMX4 | Bostelman et al. (2007) |
| JTY34b1 | Isogenic to JTY34 except bar1::URA3 | Rossodivita et al. (2014) |
| JTY309b1 | Isogenic to JTY34b1 except with pJT309 (hht2-L70S LYS2 CEN4 ARS1) in place of pJT34 | Rossodivita et al. (2014) |
| AKY34c1 | Isogenic to JTY34, except cac1::hisG-URA3-hisG | Bostelman et al. (2007) |
| JTY34r30 | Isogenic to JTY34 except rad30::URA3 | Bostelman et al. (2007) |
| JTY309r30 | Isogenic to JTY34r30 except with pJT309 (hht2-L70S LYS2 CEN4 ARS1) in place of pJT34 | This study |
| JTY34r52 | Isogenic to JTY34 except rad52::URA3 | Bostelman et al. (2007) |
| JTY34v1 | Isogenic to JTY34 except rev1::URA3 | Bostelman et al. (2007) |
| JTY309v1 | Isogenic to JTY34v1 except with pJT309 (hht2-L70S LYS2 CEN4 ARS1) in place of pJT34 | This study |
| JTY34ATA | Isogenic to JTY34 except ade2-n::TRP1::ade2-I | Rossodivita et al. (2014) |
| JTY309ATA | Isogenic to JTY309 except ade2-n::TRP1::ade2-I | Rossodivita et al. (2014) |
| JTY34DATA | Isogenic to JTY34ATA except dot1::kanMX4 | Rossodivita et al. (2014) |
| JTY309DATA | Isogenic to JTY309ATA except dot1::kanMX4 | This study |
| MEY34M | Isogenic to JTY34, except pHHT2HHF2 (HHT2, HHF2; Dion et al. (2005)) in place of pJT34 | Evans et al. (2008) |
| MEYK5812R | Isogenic to JTY34 except pK5,8,12R (hhf2-K5,8,12R; Dion et al. (2005)) in place of pHHT2HHF2 | Evans et al. (2008) |
| MEYK5816R | Isogenic to JTY34 except pK5,8,16R (hhf2-K5,8,16R; Dion et al. (2005)) in place of pHHT2HHF2 | Evans et al. (2008) |
| MEYK51216R | Isogenic to JTY34 except pK5,12,16R (hhf2-K5,12,16R; Dion et al. (2005)) in place of pHHT2HHF2 | Evans et al. (2008) |
| MEYK81216R | Isogenic to JTY34 except pK8,12,16R (hhf2-K8,12,16R; Dion et al. (2005)) in place of pHHT2HHF2 | Evans et al. (2008) |
| ABY34MD | Isogenic to MEY34M, except dot1::kanMX4 | This study |
| ABYK5812RD | Isogenic to MEY34K5812R, except dot1::kanMX4 | This study |
| ABYK5816RD | Isogenic to MEY34K5816R, except dot1::kanMX4 | This study |
| ABYK51216RD | Isogenic to MEY34K51216R, except dot1::kanMX4 | This study |
| ABYK81216RD | Isogenic to MEY34K81216R, except dot1::kanMX4 | This study |
| JTYTFATA | Isogenic to MEY34M except ade2-n::TRP1::ade2-I | This study |
| JTYTFDATA | Isogenic to JTYTFATA except dot1::kanMX4 | This study |
| JTYK5816RATA | Isogenic to JTYTFATA except with pK5,8,16R (hhf2-K5,8,16R; Dion et al. (2005)) in place of pHHT2HHF2 | This study |
| JTYK5816RDATA | Isogenic to JTYK5816RATA except dot1::kanMX4 | This study |
Western blot analysis of histone modifications following UV exposure
Yeast cultures were grown to log phase, early stationary phase, or arrested with α-factor, as indicated in the figure legends. Cells were subsequently exposed to UV radiation (254 nm), followed by nuclear protein isolation and western blot analysis, as previously described (Rossodivita et al. 2014). Various antibodies were used to monitor histone modifications (anti-H3, Abcam #1791, 1:7500; anti-H3 K79me2, Cell Signaling Technology #5427, 1:15,000; anti-H3 K79me3, Cell Signaling Technology #4260, 1:15,000; anti-H4, Millipore #05–858, 1:2000; anti-acetylated H4, Millipore #06–866, 1:4000). Antibody binding was detected with goat anti-rabbit IgG HRP antibody (Millipore #12–348, 1:3000), in conjunction with ECL2 Western Blotting Analysis System (GE Healthcare) and a FluroChem HD2 Chemilluminescent Workstation (Alpha Innotech). Densitometry analysis was done using ImageJ software (Schneider et al. 2012). Band intensity values were normalized relative to the general histone H3 or H4 antibody to adjust for gel loading variation, and were subsequently normalized relative to the unexposed control samples for each modification state examined. Each experiment was repeated at least five times for the various strains and/or conditions examined. Nuclei preps were run on two sets of gels for each antibody probing, and normalized densitometry values of the two runs were averaged for each trial. Data from the separate trials were averaged to generate the reported data, and results were statistically evaluated by Kruskal–Wallis one-way nonparametric analysis using JMP Pro software, version 12 (SAS Institute). Letters are displayed on graphs to connect data points whose differences are not statistically significant; differences between any pair of values within a given data set that do not share a letter are statistically significant (P value thresholds indicated in figure legends). Data sets presented without letters possess no significant differences.
DNA damage assays
UV cell survival, sister chromatid exchange, hydroxyurea, and methyl methanesulfonate sensitivity, checkpoint delay, budding assays were executed as previously described (Siede et al. 1993; Conde and San-Segundo 2008; Rossodivita et al. 2014). Each assay was completed a minimum of five times for each strain (three times for UV survival assays). For the sister chromatid exchange and checkpoint delay budding assays, averages for each condition were compared by a one-way ANOVA with Tukey HSD post hoc test using JMP Pro software. Statistical differences are indicated on sister chromatid exchange assay graphs using the lettering system as described above in the western blot methodology.
Sensitivity to double-stranded breaks was tested by transforming relevant strains with plasmid pJR1152 (Barnes and Rine 1985), possessing a galactose-inducible version of the gene encoding the EcoRI endonuclease. Transformants were inoculated into selectable broth containing glucose and allowed to grow for 24–48 hr at 30°. Each culture was diluted to an OD600 of 1.0 and serially diluted in 10-fold increments in a 96-well microtiter plate to a dilution of 10−4. Five microliter of each dilution was spot-plated onto glucose- and galactose-containing selectable media in duplicate. Plates were incubated over a period of ∼7 days and photographed at various times during the incubation. Each strain was tested a minimum of three times, and photographs shown represent typical observations.
Data availability
All strains constructed for this study are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
H3K79 dimethylation is reduced in response to UV exposure
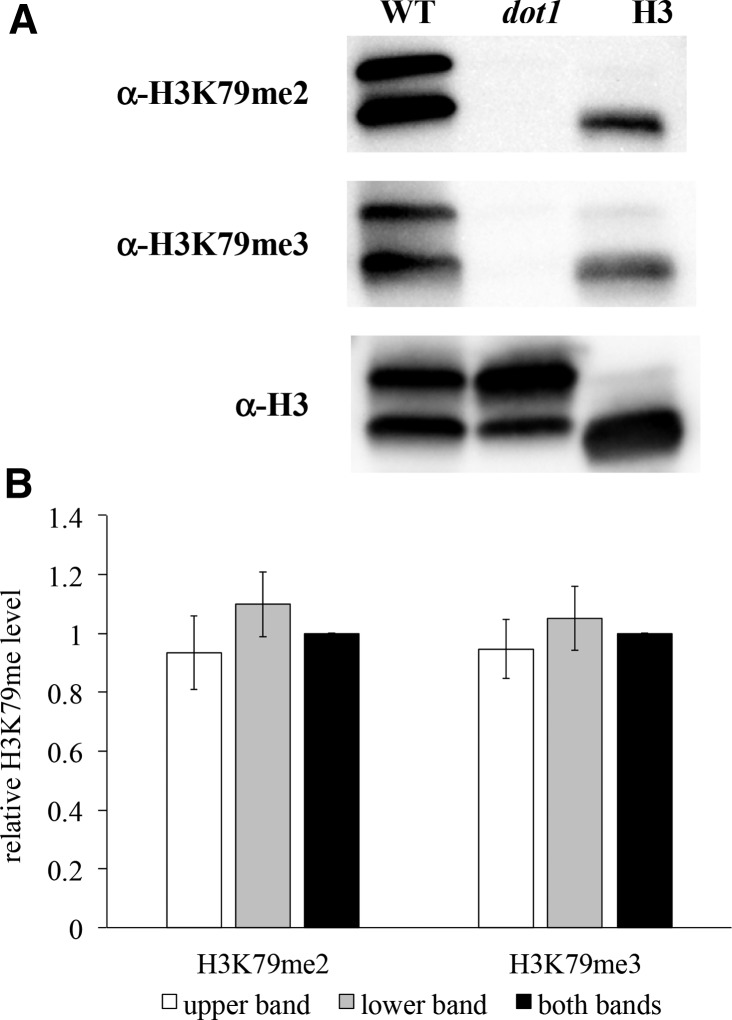
Histone H3K79 methylation state levels were examined by Western blot analysis, using H3K79me state-specific antibodies. Antibody specificity for this modification was confirmed using nuclear extracts from wildtype and H3K79me-deficient dot1 yeast strains (Figure 1A). We found that all three antibodies (α-H3, α-H3K79me2, and α-H3K79me3) detected a doublet in the wildtype strain, with a band of ∼15 kDa that comigrated with the primary band detected in purified calf thymus histones, and a second slower-migrating band, which is faintly observed in the purified histone sample. Both bands were detected by the general H3 antibody in the dot1 strain, but neither was detected by the H3K79me-specific antibodies. Since H3K79 is the only target for the Dot1 methyltransferase (van Leeuwen et al. 2002), this indicates that both bands represent H3, potentially arising from yeast-specific variations in charge-altering modifications or an artifact caused by the purification method employed (Georgieva and Sendra 1999; Glowczewski et al. 2004). Furthermore, we find that the relative H3K79me2 and H3K79me3 densitometry levels are comparable between the two bands (Figure 1B), thus both bands were used concurrently for the analyses described throughout this manuscript.
Figure 1.
Site-specificity confirmation of H3K79me2 and H3K79me3 antibodies. (A) Western blot analysis of wildtype (JTY34) and dot1 (JTY34D) strains, using α-H3, α-H3K79me2, and α-H3K79me3 antibodies, as described in Materials and Methods. Calf thymus histones (H3; Sigma #H6005) were also included for comparison (0.01 μg on the α-H3 blot, 0.1 μg on the α-H3K79me2, and 1 μg α-H3K79me3 blots). (B) Relative H3K79me2 and H3K79me3 levels, based on western blot densitometry. Values represent relative H3K79me2 or H3K79me3 band intensities in the wild-type strain, calculated by divided the intensity of the H3K79me2 and H3K79me3 bands (upper, lower, or both concurrently) by the intensity of the corresponding band(s) detected by the general H3 antibody. These values were then normalized relative to the corresponding value for the “both bands” measurement for each methylation state. Error bars represent 1 SE.
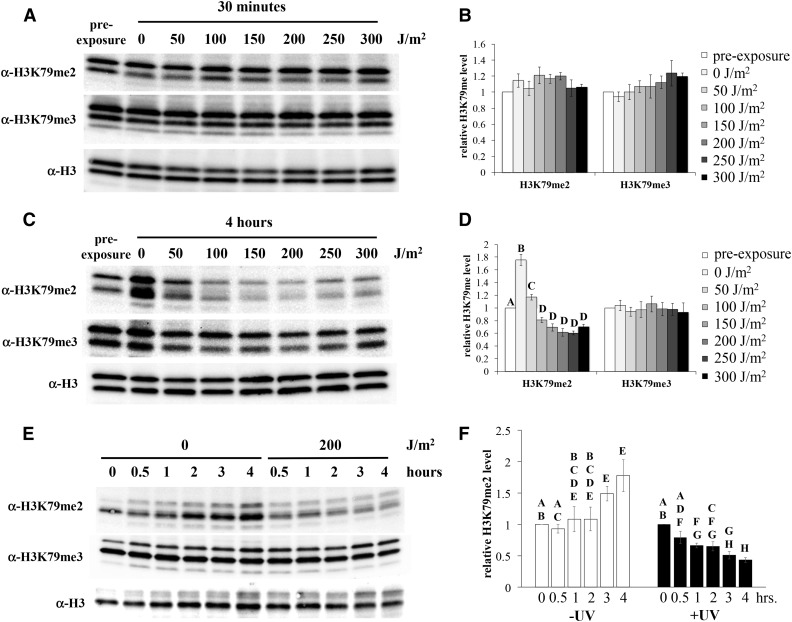
To examine the effect of UV on H3K79me state levels, cells were synchronized in stationary phase, and subsequently exposed to varying dosages of UV radiation. Samples were collected at select times after exposure, and nuclear extracts from the cells were evaluated for H3K79me state levels by western blot, as described above. No changes in H3K79me were observed in samples collected after 30 min (Figure 2, A and B), but significant decreases in H3K79me2 levels were detected 4 hr after UV exposure relative to the pre-exposed control (Figure 2, C and D). In contrast, H3K79me2 levels increased by ∼50% in unexposed cells during the incubation period, consistent with progression into S phase (Schulze et al. 2009). H3K79me2 levels were distinctly reduced in response to UV dosages of 100 J/m2 and higher, reaching a low of ∼60% of pre-exposure levels at 200–250 J/m2. However, no changes in H3K79me3 were observed in exposed or unexposed cells. At 200 J/m2, the decrease in H3K79me2 was apparent within 1 hr of UV exposure (Figure 2, E and F), with additional reductions observed over the subsequent 3 hr.
Figure 2.
Reduction of histone H3K79me levels in response to UV exposure. Wildtype yeast (JTY34) were synchronized by growth into early stationary phase, exposed to UV radiation (or mock exposed), and then incubated in fresh medium. Nuclear extracts from cells collected after various incubation times were analyzed by western blot, using H3K79me state-specific antibodies, as described previously (Rossodivita et al. 2014) and in Materials and Methods. Representative blots are shown. Histone H3 is observed as a doublet, as described in Figure 1. The faint, faster migrating band observed in some panels is a proteolytic product of H3 associated with this purification method (Shahbazian et al. 2005). Densitometry values represent means of at least five assays, normalized relative to the general H3 levels, and subsequently normalized to the pre-exposure levels; error bars represent 1 SE. Letters are displayed on graphs to connect data points whose differences are not statistically significant (P > 0.05); differences between any pair of values within a given data set that do not share a letter are statistically significant. Data sets without letters indicates that no significant differences were observed. (A, B) exposure to varying UV dosages, as indicated, followed by a 30-min incubation period. (C, D) same as (A) and (B), except incubated for 4 hr. P < 0.01 for significant differences denoted. (E, F) 200 J/m2 UV exposure, varying incubation times as indicated. P < 0.03 for significant differences denoted.
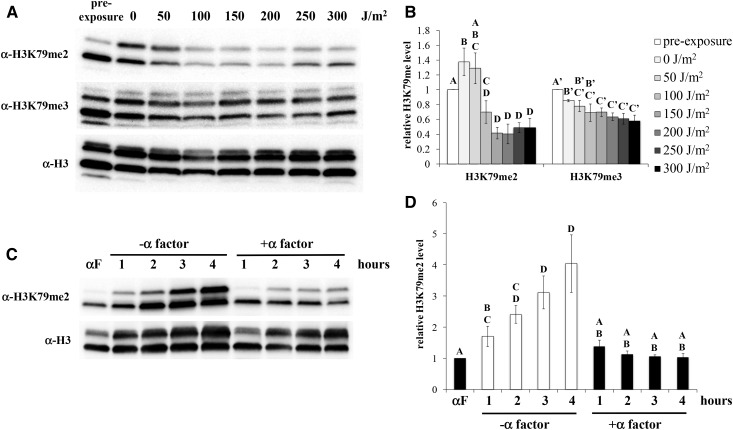
To determine if the observed changes in H3K79me2 were a result of the synchronization method, dosage response experiments were repeated using cultures synchronized in late G1 with yeast α-factor. Comparable to the stationary phase-synchronized cells, H3K79me2 levels fell to ∼40–50% of pre-exposure levels in cells exposed to UV at dosages of 100 J/m2 and higher (Figure 3, A and B), indicating that the reduction of this modification is a result of UV exposure, not the synchronization method. In contrast to the stationary phase observations, H3K79me3 levels were reduced to 60–80% of pre-exposure levels in all cultures following release from α-factor arrest. Modest differences (∼20–25%) were observed between the high dosage exposure cultures and the unexposed/incubated control, at the upper threshold of statistical significance (P = 0.04). Thus, UV exposure causes a small reduction in H3K79me3 under these synchronization conditions, but a notable proportion of the observed effect is simply due to the release from α-factor.
Figure 3.
UV-induced reduction of H3K79me2 occurs in α-factor arrested cells, but not in the absence of UV. (A, B) A bar1- strain (JTY34b1) was synchronized with α-factor, followed by release from α-factor, exposure to UV, postexposure incubation for 3 hr, and western blot analysis, as displayed in Figure 2. Error bars represent 1 SE. P < 0.01 for significant differences denoted, except H3K79me3 0 J/m2 vs. 150–300 J/m2, respectively (P = 0.04). (C, D) A bar1- culture was arrested with α-factor as in (A), and then transferred into fresh medium containing either α-factor or protease XIV (to degrade residual α-factor). Samples were collected at the indicated times and analyzed by western blot, as described in Figure 2. P < 0.03 for significant differences denoted.
We examined the possibility that the reduction of H3K79me2 was a product of extended time spent in G1 as a result of UV-induced G1/S checkpoint activation. Cells were arrested with α-factor as done in the experiments above, and then inoculated into fresh medium, with or without α-factor, for an additional 4 hr. As expected, removal of α-factor resulted in an increase in H3K79me2 levels (Figure 3, C and D), consistent with the previously reported increase in this modification state during the cell cycle (Schulze et al. 2009). In contrast, cells maintained in α-factor arrest displayed consistent levels of H3K79me2. Therefore, the observed reduction of H3K79me2 levels in the experiments above is a result of UV exposure, not simply due a prolonged G1 phase.
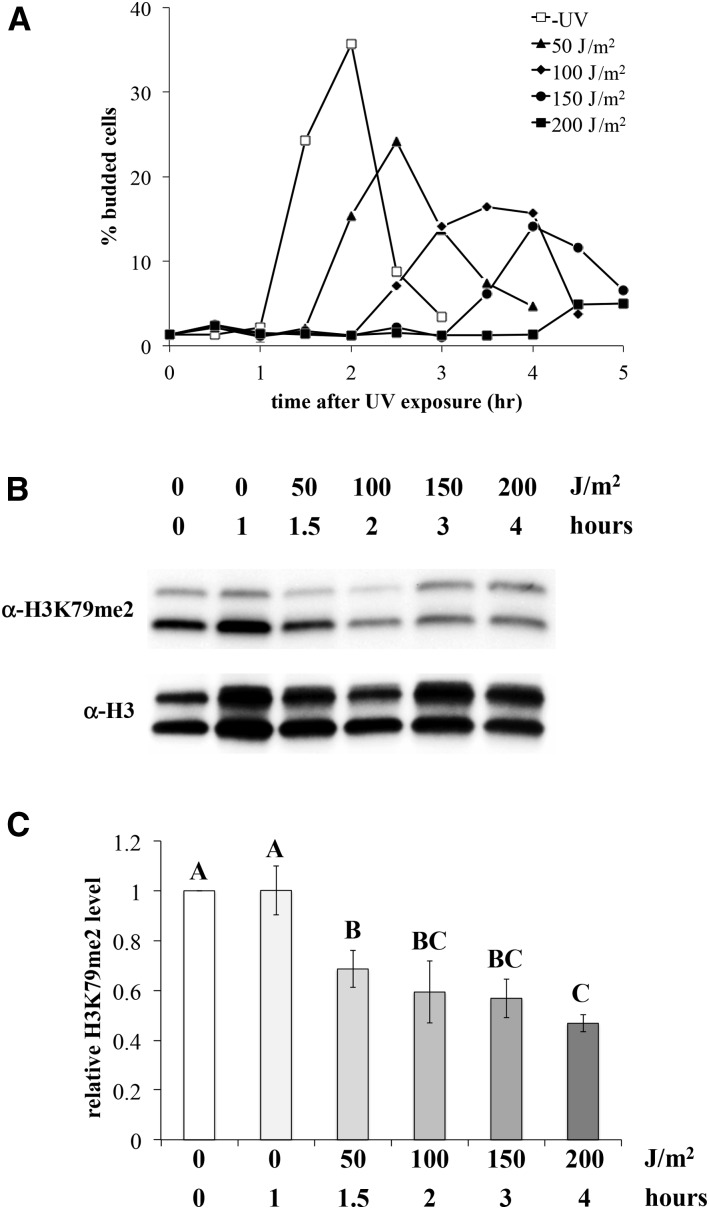
Our observations above indicate that the reduction in H3K79me2 does not occur in response to UV dosages <100 J/m2. However, H3K79me2 levels oscillate during the cell cycle, starting at a minimum during G1, and rising during the remainder of the cycle (Schulze et al. 2009). Since the length of the G1/S checkpoint delay is UV dosage-dependent, the elevated levels of H3K79me2 in the 50 J/m2-exposed cells reported above might be a product of cell cycle progression during the postexposure incubation period. To address this possibility, H3K79me2 levels were examined in UV-exposed cells near the end of the G1/S checkpoint. Stationary phase-arrested cells were exposed to varying dosages of UV, and cell cycle progression was monitored during the incubation period via the emergence of buds, which is tightly linked to entry into S phase (Pringle and Hartwell 1981). Samples collected at times immediately prior to bud emergence at each UV dosage were then analyzed by western blot. As expected, delayed emergence of buds was observed in UV-exposed cells, in a dosage-dependent manner (Figure 4A). Western blot analysis of pre-S phase samples indicated that reduction of H3K79me2 was observed at all dosages (Figure 4, B and C), ranging from ∼50 to 70% of pre-exposure levels. Modest differences were observed between the varying UV dosages, with significantly lower H3K79me2 levels in cells exposed to 200 J/m2 vs. 50 J/m2 of UV. Thus, we conclude that UV-induced reduction in H3K79me2 occurs across the range of UV dosages examined, with an inverse relationship between the UV dosage/checkpoint delay time and H3K79me2 levels.
Figure 4.
UV exposure induces H3K79me2 reduction during checkpoint arrest at all dosages. Yeast strain JTY34 was synchronized by growth into stationary phase, followed by UV exposure at varying dosages, and incubation in fresh medium over time. (A) Aliquots of UV-exposed cultures (and a mock-exposed control) were examined microscopically for the emergence of buds, as described in Materials and Methods, reported as the percentage of cells possessing small buds. A representative experiment is shown. (B) Samples were collected from the same UV-exposed culture as in (A), at the latest time point at which a given subculture had not yet experienced bud emergence (as noted on the figure labels). Western blot analysis on nuclear extracts was done as described and displayed in Figure 2. Error bars represent 1 SE. (C) Densitometry of western blot results, as displayed in Figure 2. P < 0.01 for significant differences denoted, except between 0 J/m2 (0 hr) and 50 J/m2 (P = 0.02), 0 J/m2 (1 hr) and 50 J/m2 (P = 0.02), 0 J/m2 (1 hr), and 100 J/m2 (P = 0.04) and 50 J/m2 and 200 J/m2 (P = 0.02).
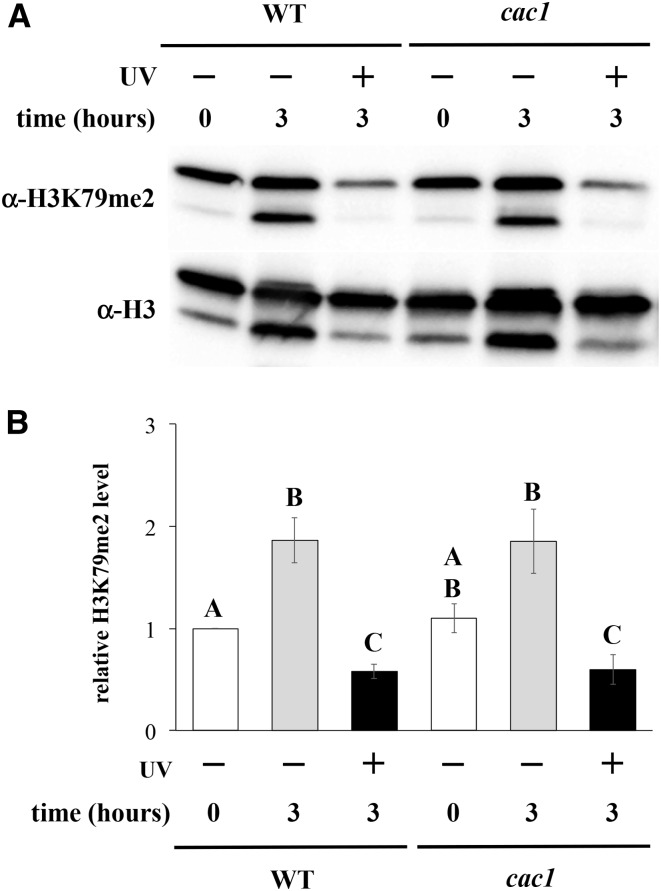
Histone replacement occurs in response to DNA damage, which can lead to altered histone modification levels. Histone chaperone CAF-1 has been shown to be important for damage-associated histone replacement (Polo et al. 2006), and is important for UV repair (Game and Kaufman 1999). Since histone H3 is unmethylated at K79 prior to incorporation into chromatin (van Leeuwen et al. 2002), we considered the possibility that UV-induced reduction in H3K79me2 might be the result of CAF-1-dependent histone replacement. To address this, we measured H3K79me2 levels in strains lacking Cac1, an essential subunit of CAF-1, following UV exposure. A comparable decrease in H3K79me2 levels was observed following UV exposure in the cac1 strain relative to that seen in the wildtype strain (Figure 5). Thus, these results indicate that CAF-1 is not required for the UV-induced decrease in H2K79me2.
Figure 5.
UV-induced reduction of H3K79me2 is independent of CAF-1. Yeast cells were exposed to 200 J/m2 UV (stationary phase synchronization, 3-hr post-exposure incubation), and H3K79me2 levels were analyzed by western blot, as described and displayed in Figure 2. Error bars represent 1 SE. Strains used: wildtype (JTY34) and cac1 mutant (AKY34c1). (A) Representative western blot. (B) Densitometry to relative H3K79me2 levels. P < 0.01 for all significant differences, except cac1 −UV 0 hr vs. cac1 +UV 3 hr (0.04).
H3K79me2 levels are important for UV-induced sister chromatid exchange and translesion synthesis, but not double-stranded break repair or G1/S checkpoint activation
To gain insight into the biological function of UV-induced reduction of H3K79me2, we examined repair processes in which H3K79me has been shown to participate. One such process is sister chromatid exchange (SCE), which serves as a DNA damage bypass mechanism. It has been proposed that such damage tolerance is achieved by the use of an undamaged sister chromatid to enable the completion of DNA replication of a damaged chromatid, through a template-switching mechanism (Gangavarapu et al. 2007). We have previously demonstrated that H3K79me is important for UV-induced SCE, predominantly mediated by the H3K79me3 state (Rossodivita et al. 2014).
To evaluate the influence of the UV-induced decrease of H3K79me2 on SCE, we measured UV-induced SCE frequencies in a yeast strain possessing the histone H3 mutation L70S, which causes elevated H3K79me levels (Evans et al. 2008). SCE was monitored through a previously described reporter construct containing two distinct nonfunctional alleles of the ADE2 gene, flanked around the TRP1 gene (Mozlin et al. 2008). An unequal sister chromatid exchange event between the ade2 alleles can produce a functional ADE2 gene. This can occur via a gene conversion event, with concomitant retention of the TRP1 gene, or by a crossover/“popout” event, leading to loss of TRP1 (Symington 2002).
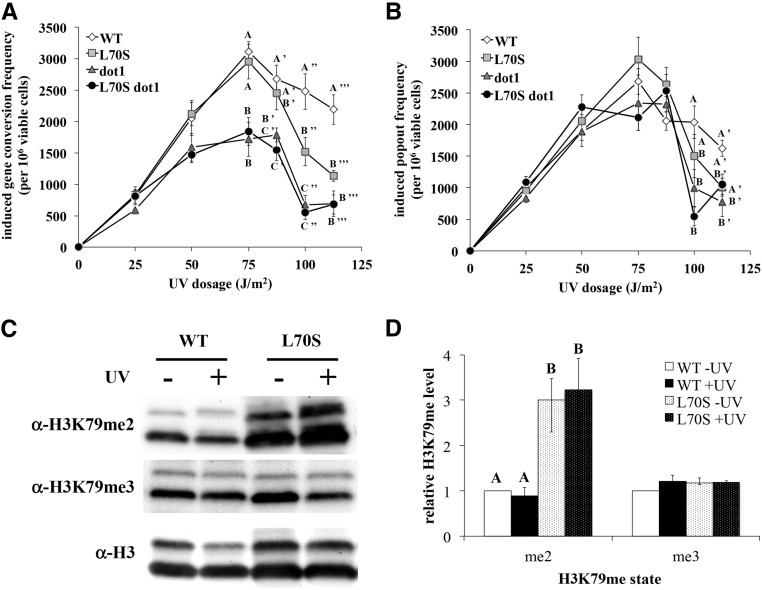
As previously observed (Rossodivita et al. 2014), dot1 strains lacking H3K79me exhibited reduced frequency of UV-induced gene conversion events, ∼20–50% of wildtype levels, depending on the UV dosage (Figure 6A). Popout events were modestly decreased in the dot1 strain as well, but only at the highest UV dosages (Figure 6B). In contrast, SCE levels were unaffected by the L70S mutation up to 75 J/m2, regardless of the mechanism, but gene conversion events were distinctly reduced to ∼50% of wildtype levels at the two highest UV dosages. The L70S dot1 double mutant strain exhibited reduced levels of gene conversion identical to that observed in the dot1 strain. To correlate these phenotypic observations with the effects of this mutation on H3K79me, we evaluated H3K79me state levels in the H3 L70S strain by western blot, using the experimental conditions employed in the SCE assay (log phase, UV at 125 J/m2). We found that H3K79me2 levels were increased around threefold in the L70S strain relative to wildtype levels, while no differences in H3K79me3 levels were observed (Figure 6, C and D). Furthermore, H3K79me2 levels remained elevated in the mutant strain following UV exposure. Thus, the combined results indicate that the H3 L70S mutation has a negative impact on gene conversion, potentially due to its effect on H3K79me2 levels.
Figure 6.
H3K79me2 levels affect UV-induced sister chromatid exchange. Sister chromatid exchange assays were done as described previously (Rossodivita et al. 2014). Data represent the means of UV-induced gene conversion (A) and popout (B) frequencies per 106 surviving cells, from at least five separate assays. Error bars represent 1 SE. Letters are displayed on graphs to identify data points at a given UV dosage whose differences are not statistically significant; values within a given dosage not sharing letters are statistically significant [P < 0.01, except WT vs. L70S gene conversion at 100 J/m2 (P = 0.03), and WT vs. dot1 popout at 100 J/m2 (P = 0.04)]; data lacking letters indicates that no significant differences were observed at that dosage. Strains used: JTY34ATA (WT), JTY309ATA (L70S), JTY34DATA (dot1), and JTY309DATA (H3 L70S dot1. (C, D) Western blot analysis of H3K79me levels in wild-type (JTY34) and H3 L70S (JTY309) strains in response to UV exposure (log phase cultures, 125 J/m2 exposure, 1-hr post-exposure incubation). Data are displayed as described in Figure 2. P < 0.03 for significant differences denoted.
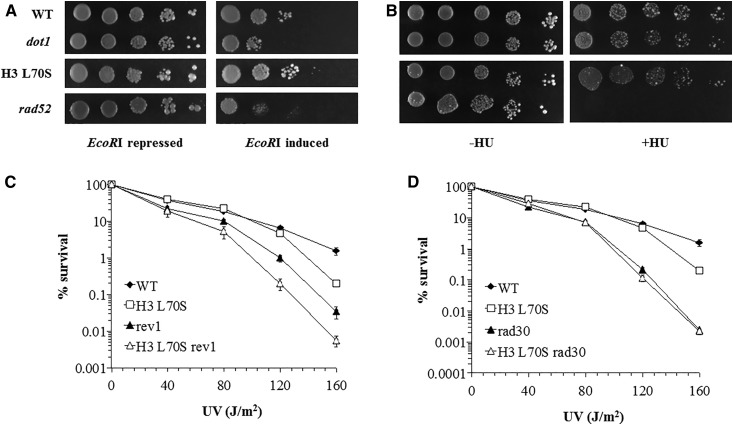
H3K79me is important for the repair of double-stranded breaks in DNA, as demonstrated by sensitivity to ionizing radiation in strains lacking this modification (Game et al. 2006). UV damage can induce double-stranded breaks in DNA as a result of replication fork collapse (Elvers et al. 2011); thus, we considered the possibility that the reduction of H3K79me2 might be important for the repair of UV-induced double-stranded breaks. We examined the H3 L70S mutation in a strain expressing an inducible version of the gene encoding for the restriction endonuclease EcoRI. We found that the dot1 strain exhibited modest sensitivity to EcoRI-induced breaks (Figure 7A), comparable to the degree of sensitivity to HO endonuclease previously observed in the dot1 strain (Rossodivita et al. 2014), indicating that H3K79me plays a minor role in double-stranded break repair. In contrast, no change in sensitivity to EcoRI was observed in the H3 L70S strain relative to the wild-type strain. We also examined sensitivity to hydroxyurea (HU), which can stall replication due to depletion of nucleotide pools. Neither loss of H3K79me, nor the H3 L70S mutation, affected HU sensitivity (Figure 7B), in contrast to the high sensitivity observed in the recombination repair-deficient rad52 strain. Therefore, these results indicate that increased levels H3K79me2 levels do not influence double-stranded break repair.
Figure 7.
The histone H3 L70S mutation does not affect sensitivity to EcoRI or hydroxyurea, but genetically acts through an error-free TLS pathway. (A) Strains were transformed with a plasmid possessing the gene encoding the EcoRI endonuclease, as described in Materials and Methods. Expression is regulated by the presence of glucose (repressed) or galactose (induced) in the growth medium. Cultures were serially diluted 10-fold, and spot plated on selectable medium containing either glucose or galactose. (B) Cultures were serially diluted 10-fold and spot plated on YEPD plates lacking (−HU) or containing (+HU) hydroxyurea at 150 mM. For experiments shown in (A) and (B), plates were photographed after ∼4–6 days of growth. Images shown are representative of the replicate trials executed. Labels on the left apply to both experiments. Strains used: JTY34 (WT), JTY34D (dot1), JTY309 (H3 L70S), and JTY34r52 (rad52). (C, D) UV survival assays were done as previously described (Bostelman et al. 2007). Values represent the mean of at least three assays; error bars represent 1 SE (not visible in many cases, due to the small size of the error). Strains used: JTY34 (WT), JTY309 (H3 L70S), JTY34v1 (rev1), JTY309v1 (H3 L70S rev1), JTY34r30 (rad30), and JTY309r30 (H3 L70S rad30).
TLS embodies DNA damage response pathways that employ lesion-tolerant DNA polymerases to complete replication gaps at sites of damage (Broomfield et al. 2001). H3K79me has been proposed to act as negative regulator of TLS, with loss of H3K79me resulting in hyper-resistance to the alkylating agent methyl methanesulfonate (Conde and San-Segundo 2008). In light of these observations, we considered the possibility that the H3K79me2 state might be important in UV-induced TLS. To address this, we evaluated cell survival in response to UV in strains lacking key genes involved in TLS. REV1 and RAD30 encode for error-prone and error-free DNA polymerases, respectively, both of which are important for UV damage tolerance (Nelson et al. 1996; Johnson et al. 1999; Acharya et al. 2006). We found that the H3 L70S/rev1 double mutant exhibited reduced survival after UV exposure compared to the rev1 mutant (Figure 7C), indicating that these two mutations act through separate pathways. In contrast, an epistatic relationship was observed between H3 L70S and rad30 (Figure 7D), as the double mutant strain exhibited the same survival frequencies as the rad30 strain. Since an epistatic relationship generally indicates that the two genes in question possess some form of interaction or connection (Haynes and Kunz 1981), these results indicate that the modest UV sensitivity conferred by the H3 L70S mutation acts through an error-free branch of TLS, thus suggesting an additional role for H3K79me2 in response to UV damage.
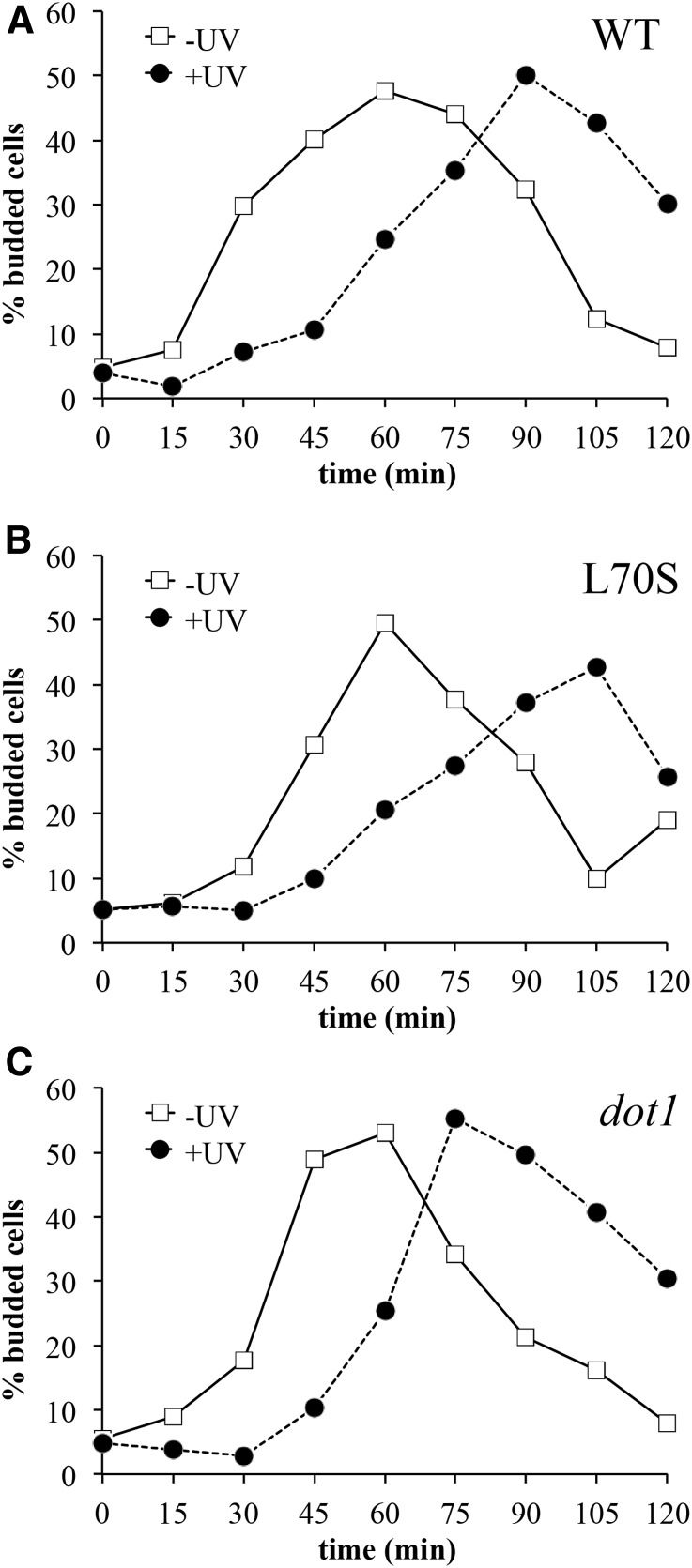
We also wished to determine if UV-induced reduction of H3K79me2 is important for G1/S checkpoint activation in response to DNA damage. It has been previously shown that H3K79me is required for this checkpoint (Giannattasio et al. 2005; Wysocki et al. 2005; Tatum and Li 2011), with loss of methylation eliminating the checkpoint in response to DNA damage. We previously found that the H3 L70S mutation did not affect checkpoint activation in response to UV at 50 J/m2 (Rossodivita et al. 2014), suggesting that elevated levels of H3K79me2 do not have an adverse effect on checkpoint activation. However, we also demonstrated that the dot1 strain displays a normal checkpoint response at 100 J/m2 of UV, indicating that H3K79 methylation is not required for checkpoint activation in response to high UV dosages. Thus we speculated that UV-induced reduction of H3K79me2 presented above might be required for checkpoint activation in response to high levels of UV damage. To address this possibility, we measured checkpoint delay times in the H3 L70S strain following exposure to UV at 100 J/m2 via a bud emergence assay, as described above. While the H3 L70S and dot1 strains had slightly shorter checkpoint delay times relative to the wildtype strain (Figure 8 and Table 2), none of the differences were statistically significant. Thus, elevated H3K79me2 levels do not affect UV-induced G1/S checkpoint activation at any of the UV dosages tested.
Figure 8.
H3K79me2 levels do not affect UV-induced G1/S checkpoint arrest. Strains were arrested with α-factor, exposed to UV at 100 J/m2, and the frequency of small budded cells was measured over time, as described in Figure 4 and Materials and Methods. Representative experiments are shown; quantitative compilation of data are displayed in Table 2. Strains utilized: (A) JTY34 (WT), (B) JTY309 (L70S), and (C) JTY34D (dot1).
Table 2. G1/S checkpoint delay times.
| Genotype | Time (min)a |
|---|---|
| WT | 34.0 ± 3.9 |
| H3 L70S | 27.0 ± 6.1 |
| dot1 | 27.0 ± 5.5 |
Difference in time between UV exposed (100 J/m2) and unexposed controls respectively reaching 20% budding frequency (corresponding to the linear range of the respective budding curves); values are mean ± 1 SE.
Histone H4 acetylation influences UV-induced changes in H3K79 methylation
Histone crosstalk is a phenomenon in which one histone modification influences another modification (Zhang et al. 2015), and prior work has suggested such an interaction between H3K79me and the H4 N-terminal tail (Altaf et al. 2007; Fingerman et al. 2007). In particular, we previously found that mutation of the acetylated lysines at positions 5, 8, 12, and 16 in H4 (H4ac) results in elevated H3K79me levels (Evans et al. 2008); however, it has not been determined if this effect is significant in the context of DNA repair. To determine if histone H4ac and H3K79me genetically interact in the context of UV exposure, survival assays were undertaken with strains deficient for either or both of these modifications. Inhibition of H4ac was achieved by lysine-to-arginine triple-substitution mutations at the aforementioned sites, to mimic the unacetylated state [a quadruple mutant was not tested, due to its lethal phenotype (Dion et al. 2005)]. Cells lacking H3K79me and/or H4ac were exposed to varying dosages of UV, and survival frequencies were determined and compared.
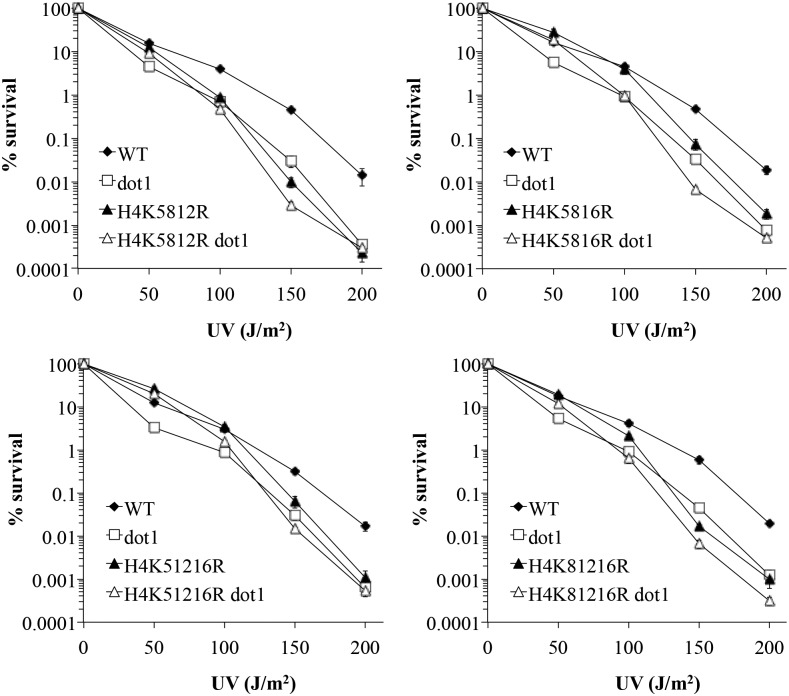
Consistent with prior studies (Bostelman et al. 2007; Evans et al. 2008), we found that loss of H3K79me or H4ac resulted in reduced survival in response to UV relative to the wildtype strain (Figure 9). The double mutant strains displayed a complex pattern of survival relative to the single mutant strains, with a suppressive effect observed at the lowest UV dosage, and a largely epistatic relationship at higher dosages. The same general pattern was observed regardless of the particular combination of H4 lysines that were mutated. Both suppressive and epistatic effects are indicative of interactions between genes (Haynes and Kunz 1981). Therefore, these observations implicate a functional relationship between H4ac and H3K79me in the context of UV repair.
Figure 9.
Epistasis analysis between H4ac and H3K79me. UV survival assays were done as previously described (Bostelman et al. 2007). Values represent the mean of at least three assays; error bars represent 1 SE (not visible in many cases, due to the small size of the error). Strains used: MEY34M (WT), ABY34MD (dot1), MEYK5812R (H4 K5812R), ABYK5812RD (H4 K5812R dot1), MEYK5816R (H4 K5816R), ABYK5816RD (H4 K5816R dot1), MEYK51216R (H4 K51216R), ABYK51216RD (H4 K51216R dot1), MEYK81216R (H4 K81216R), and ABYK81216RD (H4 K81216R dot1).
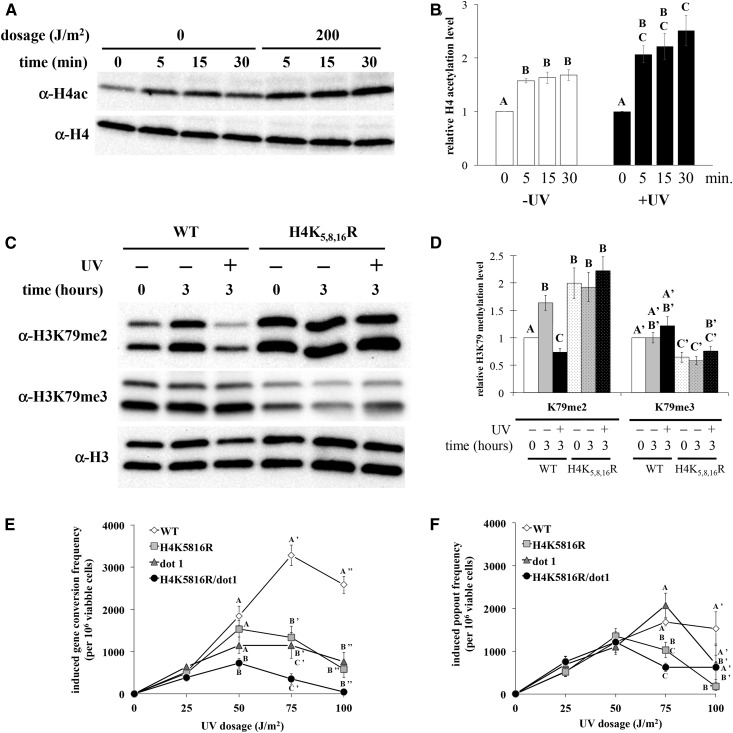
In light of the genetic relationship between H3K79me and H4ac, we postulated that H4ac drives the observed UV-induced change in H3K79me2. It has been previously reported that H4 tail lysines are hyperacetylated in response to UV (Yu et al. 2005), and, as an initial step in evaluating the potential for crosstalk, we measured H4ac levels following UV exposure, using the experimental conditions employed above. Stationary phase-arrested cells were exposed to UV at 200 J/m2, and nuclear extracts were evaluated by western blot, using an antibody specific for histone H4 acetylated at sites 5, 8, 12, and 16. Consistent with prior observations (Yu et al. 2005), H4ac levels increased ∼2.5-fold in response to UV. Elevated H4ac was detectable within 5 min, and achieved significantly higher levels compared to the unexposed controls within ∼30 min postexposure (Figure 10, A and B).
Figure 10.
H4ac is required for UV-induced reduction of H3K79me2 and UV-induced SCE. Yeast were synchronized in stationary phase, exposed to 200 J/m2 UV, and analyzed at varying times after exposure by western blot. Data displayed as described in Figure 2. Error bars represent 1 SE. (A, B) H4 acetylation levels at sites 5, 8, 12, and 16 were analyzed in strain JTY34. Postexposure incubation times are indicated. P < 0.02 for significant differences denoted. (C, D) H3K79me2 and H3K79me3 levels were analyzed in wildtype (MEY34M) and H4K5,8,16R (MEYK5816R) yeast strains, following a 3-hr post-exposure incubation. P < 0.01 for significant differences denoted, except H3K79me2 WT −UV 0 hr vs. K5,8,16R −UV 3 hr (P = 0.04), and H3K79me3 WT −UV 3 hr vs. K5,8,16R −UV 0 hr (P = 0.03). (E, F) Sister chromatid exchange assays were done as described in Materials and Methods, and reported as in Figure 6. (E) Gene conversion events. (F) Popout events. P < 0.01 for significant differences denoted, except gene conversion H4K5816R vs. H4K5816R/dot1 and popout WT vs. H4K5816R (P = 0.03 for both). Strains used: JTYTFATA (WT), JTYK5816RATA (H4K5816R), JTYTFDATA (dot1), and JTYK5816RDATA (H4K5816R/dot1).
In order to determine if H4ac is necessary for the decrease in H3K79me following UV exposure, H3K79me levels were assessed in the yeast strain possessing the H4 K-to-R substitution mutations at positions 5, 8, and 16 (stationary phase-arrested, UV at 200 J/m2, 3-hr incubation). Wildtype H3K79me2 levels increased in unexposed cells following the postexposure incubation, while UV-exposed H3K79me2 levels decreased (Figure 10, C and D), consistent with the observations reported above. In contrast, H3K79me2 levels were approximately twofold higher in the H4 mutant strain compared to wildtype levels in unexposed cells, while H3K79me3 levels were reduced to ∼60% of wild-type levels. Furthermore, H3K79me2 levels were unchanged following UV exposure in the mutant strain relative to the unexposed mutant control. Thus, mutation of H4 acetylatable lysines alters the steady-state distribution of H3K79me2 and me3 states in the absence of DNA damage, and further prevents the UV-induced reduction of H3K79me2.
Since the H4 K-R mutation causes changes in H3K79me2 and me3 levels, both of which are important for UV-induced SCE (Figure 6 and Rossodivita et al. 2014), we predicted that inhibition of H4ac would have a negative impact on SCE as well. As anticipated, UV-induced gene conversion frequency was reduced in the H4 mutant strain, comparable to that observed in the dot1 strain (Figure 10E). Modest differences in gene conversion frequencies were observed between the double mutant strain and the two single mutant strains at 50 and 75 J/m2, but not at 100 J/m2. Furthermore, the difference between the double mutant and the dot1 strain is technically not statistically significant (P = 0.45 at 50 J/m2, 0.14 at 75 J/m2). These results indicate a genetic relationship between these two modifications that lands somewhere between epistatic and additive with respect to gene conversion, suggesting that H4ac and H3K79me exert some effects through a common gene conversion pathway, while also possessing additional independent functions in SCE. With respect to popout events, the H4 mutant strain also displayed a reduced frequency at 100 J/m2 relative to the wildtype strain (Figure 10F), indicating an additional role in SCE beyond that in gene conversion. There was no statistically significant difference between the H4 mutant strain and either the dot1 or double mutant strains, suggesting that H4ac and H3K79me act through a common pathway with regards to popout events. However, popout events in the dot1 and double mutant strains were not statistically different from the wildtype strain at this dosage; thus, we cannot fully elucidate the genetic relationship between H4ac and H3K79me in the context of SCE popout events.
Discussion
The results presented here identify a novel histone modification change in response to DNA damage, joining a select number of other damage-induced modifications (Cao et al. 2016). The change in H3K79me is distinct, however, in that it entails the reduction of a specific modification, in contrast to increases observed in most other damage-induced modifications. Furthermore, the observed change is state-specific, in which dimethylation is uniquely reduced in response to UV, without any substantial changes to trimethylation levels. This provides further evidence for H3K79me state-specific roles in the context of DNA repair (Evans et al. 2008; Rossodivita et al. 2014). It is possible that UV-induced changes to H3K79me might also apply to the monomethylated state; however, the lack of a consistently reliable antibody against this modification state prevented us from addressing this point. However, it is worth noting that our prior investigations have suggested that H3K79me1 does not play a functional role in UV repair (Evans et al. 2008; Rossodivita et al. 2014).
In terms of functionality, the evidence is consistent with a model in which H3K79me2 levels regulate UV-induced SCE. Preclusion of the UV-induced decrease in H3K79me2 via the H3 L70S mutation caused a reduction in UV-induced gene conversion, suggesting that H3K79me2 has a negative effect on sister chromatid exchange. However, since these conclusions are based on the phenotypic effects of the L70S mutation, some caution must be exercised with regards to these conclusions. While the collective results support the role of H3K79me2 as a negative regulator of SCE, it is nonetheless plausible that the effect of the L70S mutation on SCE is a product of the altered stoichiometry of the H3K79me states, rather than due to elevated dimethylation levels. We also cannot definitively rule out the possibility that the L70S mutation itself exerts effects on UV-induced SCE independent of its influence on H3K79me2 levels (albeit through a pathway that includes dot1). Assuming that elevated H3K79me2 is responsible for the reduction in UV-induced SCE in the L70S mutant strain, it is important to note that impaired SCE in this strain is only observed at dosages of ∼100 J/m2 and higher, while UV-induced reduction of H3K79me2 is observed at all dosages tested. It is plausible that the effect of H3K79me2 on SCE is only triggered by reducing dimethylation levels below a specific threshold, as observed at higher UV dosages. Alternatively, some other factor may act in concert with the reduction in H3K79me2 to regulate high-dosage SCE.
These observations raise the question as to how the role of H3K79me2 in SCE relates to our prior conclusion that the H3K79me3 state is uniquely involved in this damage response pathway (Rossodivita et al. 2014). In light of these new results, we propose that both modification states participate in SCE, but in distinct manners. Our collective data suggest that the trimethylated state is required to promote UV-induced gene conversion in general, while the dimethylated state suppresses forms of gene conversion that are specifically employed in response to high levels of damage. It has been demonstrated that different forms of DNA damage are inflicted depending on the dosage of UV exposure (Yin and Petes 2013). Thus, reduction of H3K79me2 may enable cells to engage alternative damage tolerance or repair mechanisms to respond to distinct forms of damage created at high UV levels. The nature of such alternative processes remains to be determined, but our results indicate that it does not likely involve double-strand break repair.
In addition to the role of H3K79me2 in UV-induced SCE, our results point to an additional function in TLS in the context of UV repair. It has been previously shown that H3K79me negatively regulates TLS in response to methyl methanesulfonate, acting through an error-prone pathway (Conde and San-Segundo 2008). In contrast, we find that the H3 L70S mutation causes a modest reduction in UV survival through an error-free pathway involving Rad30. This is an unexpected observation, since mutations that act through error-free pathways are predicted to increase mutation frequencies; however, we have previously found that the L70S mutation does not affect UV-induced mutation frequencies (Evans et al. 2008). Given that H3K79me2 levels appear to exert effects on multiple DNA repair processes, it is possible that the L70S mutation influences error-free and error-prone pathways resulting in no net change in mutation frequencies. It is also important to note that the epistatic relationship between rad30 and H3 L70S may reflect the impact of the histone mutation itself on UV survival that is independent of its effect on H3K79me2 levels. Thus, further investigations into the influence of H3K79me2 on TLS will be needed to resolve these questions.
The H3K79me2-specific change in response to UV implies a targeting of particular genomic loci during repair. It has been previously demonstrated that H3K79me states are nonrandomly distributed across the yeast genome, with H3K79me2 and H3K79me3 states enriched in mutually exclusive coding regions (Schulze et al. 2009). H3K79me2-enriched genes tend to be G1-expressed, when H3K79me2 levels are at their lowest during the cell cycle. Our observations suggest that such loci are more highly affected by demethylation in response to high levels of UV damage, potentially altering expression of these genes during the damage response. While such an effect suggests that H3K79me2 demethylation regulates cell cycle progression in the presence of DNA damage, our results indicate that UV-induced H3K79me2 changes are not important for the G1/S checkpoint. Alternatively, recombination is induced by UV to higher levels in G1 compared to other cell cycle stages (Yin and Petes 2013); thus, given our proposed role for H3K79me2 in regulating UV-induced gene conversion, we might anticipate that certain H3K79me2-enriched genes influence recombination repair. Intriguingly, a variety of recombination-related genes have been shown to act as negative regulators of H3K79me (Vlaming et al. 2016), suggesting a potential feedback loop between recombination repair and H3K79me levels. Identification of the underlying targets of UV-induced reduction of H3K79me2 should prove insightful for understanding the role of this modification in UV repair.
We have also presented evidence implicating a novel crosstalk interaction between H3K79me2 and H4 N-terminal tail acetylation. Mutation of the acetylation sites prevents the reduction in H3K79me2, supporting a causal relationship between these two modification changes. It is possible that the effects caused by the H4 K-R mutations are due to influences of the specific amino acid substitutions beyond the inhibition of H4ac. However, such charge-conserved substitutions in histones generally serve as a reliable proxy for mimicking the unacetylated state (Dion et al. 2005); thus, we argue that the crosstalk relationship suggested by these findings likely pertains to acetylation at these sites.
Our observations point to two distinct dimensions of this crosstalk interaction. First, we find that, in the absence of DNA damage, H4ac promotes the conversion of H3K79me2 to H3K79me3. This is supported by our observation that inhibition of H4ac results in a concurrent increase in H3K79me2 and a decrease in H3K79me3, consistent with prior observations (Jung et al. 2015; Simoneau et al. 2015). These acetylatable lysines may be part of a neighboring domain of basic residues in H4 that also influence H3K79me (Altaf et al. 2007; Fingerman et al. 2007). However, mutations in this basic domain have different effects on H3K79me compared to our observations, suggesting that the H4 tail exerts multiple influences on H3K79me. Second, we find that H4ac is required for the UV-induced decrease of H3K79me2, as demonstrated by the persistent levels of H3K79me2 following UV exposure when H4ac is impaired. Our results do not enable us to determine if UV-induced H4 hyperacetylation is specifically required for reducing H3K79me2, although the sequential nature of these UV-induced changes leads us to speculate that the increase in H4ac is important for this effect.
In closing, our findings support a model in which UV-induced H4ac triggers a reduction in H3K79me2, in turn activating specific SCE pathways, as a means of increasing damage tolerance. The underlying mechanism for reduction of the H3K79me2 state remains to be elucidated, but the lack of a demethylase for this site (Sweet et al. 2010; Zee et al. 2010) suggests that the decrease is more likely explained by histone turnover and/or degradation (Vlaming et al. 2016; Hauer et al. 2017), albeit via an undefined CAF-1-independent pathway. The proposed crosstalk pathway is potentially part of a larger network of damage-related histone modification interactions. We have observed an epistatic relationship with respect to UV survival between mutations that inhibit histone H4ac and H2B K123 ubiquitylation (A.L.B and J.S.T, unpublished observations), a modification that is required for H3K79me3 (Ng et al. 2002b; Sun and Allis 2002; Shahbazian et al. 2005; Frederiks et al. 2008), and is important for the response to UV and regulating TLS (Rossodivita et al. 2014; Hung et al. 2017). Additionally, a functional relationship has been demonstrated between H4ac, H3K79me, and H3K56 acetylation with respect to exposure to several genotoxic drugs (Simoneau et al. 2015). Thus, further investigations are warranted to characterize the nature of these interactions, and the manner by which they regulate DNA repair.
Acknowledgments
We thank A. Albrecht, A. Cole, A. Karl, A. Rossodivita, J. Snee, and E. Vines for technical assistance, O. Rando and L. Symington for plasmids provided, and S. Cole for invaluable discussions. Funding was provided by the National Institutes of Health (1R15GM093849-01), the Reid and Polly Anderson Endowment, the Lisska Center for Scholarly Engagement, the Office of the Provost, and the Department of Biology at Denison University.
Footnotes
Communicating editor: N. Hunter
Literature Cited
- Acharya N., Johnson R. E., Prakash S., Prakash L., 2006. Complex formation with Rev1 enhances the proficiency of yeast DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol. Cell. Biol. 26: 9555–9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf M., Utley R. T., Lacoste N., Tan S., Briggs S. D., et al. , 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28: 1002–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes G., Rine J., 1985. Regulated expression of endonuclease EcoRI in Saccharomyces cerevisiae: nuclear entry and biological consequences. Proc. Natl. Acad. Sci. USA 82: 1354–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Jinks-Robertson S., 2013. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 193: 1025–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostelman L. J., Keller A. M., Albrecht A. M., Arat A., Thompson J. S., 2007. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst.) 6: 383–395. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Broomfield S., Hryciw T., Xiao W., 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486: 167–184. [DOI] [PubMed] [Google Scholar]
- Cao L. L., Shen C., Zhu W. G., 2016. Histone modifications in DNA damage response. Sci. China Life Sci. 59: 257–270. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S., Wyrick J. J., Smerdon M. J., 2009. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res. 37: 1690–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde F., San-Segundo P. A., 2008. Role of Dot1 in the response to alkylating DNA damage in Saccharomyces cerevisiae: regulation of DNA damage tolerance by the error-prone polymerases Polζ/Rev1. Genetics 179: 1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion M. F., Altschuler S. J., Wu L. F., Rando O. J., 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 102: 5501–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvers I., Johansson F., Groth P., Erixon K., Helleday T., 2011. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 39: 7049–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. L., Bostelman L. J., Albrecht A. M., Keller A. M., Strande N. T., et al. , 2008. UV sensitive mutations in histone H3 in Saccharomyces cerevisiae that alter specific K79 methylation states genetically act through distinct DNA repair pathways. Curr. Genet. 53: 259–274. [DOI] [PubMed] [Google Scholar]
- Fingerman I. M., Li H. C., Briggs S. D., 2007. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 21: 2018–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiks F., Tzouros M., Oudgenoeg G., van Welsem T., Fornerod M., et al. , 2008. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat. Struct. Mol. Biol. 15: 550–557. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., et al. , 2006. DNA Repair and Mutagenesis, Ed. 2 ASM Press, Washington, DC. [Google Scholar]
- Game J. C., Kaufman P. D., 1999. Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics 151: 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C., Williamson M. S., Spicakova T., Brown J. M., 2006. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics 173: 1951–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu V., Prakash S., Prakash L., 2007. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 7758–7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva E. I., Sendra R., 1999. Mobility of acetylated histones in sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Anal. Biochem. 269: 399–402. [DOI] [PubMed] [Google Scholar]
- Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M., 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280: 9879–9886. [DOI] [PubMed] [Google Scholar]
- Glowczewski L., Waterborg J. H., Berman J. G., 2004. Yeast chromatin assembly complex 1 protein excludes nonacetylatable forms of histone H4 from chromatin and the nucleus. Mol. Cell. Biol. 24: 10180–10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer M. H., Seeber A., Singh V., Thierry R., Sack R., et al. , 2017. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol. 24: 99–107. [DOI] [PubMed] [Google Scholar]
- Haynes R. H., Kunz B. A., 1981. DNA repair and mutagenesis, pp. 371–414 in Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hung S.-H., Wong R. P., Ulrich H. D., Kao C.-F., 2017. Monoubiquitylation of histone H2B contributes to the bypass of DNA damage during and after DNA replication. Proc. Natl. Acad. Sci. USA 114: E2205–E2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y., Zgheib O., Ditullio R. A., Jr., Gorgoulis V. G., Zacharatos P., et al. , 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Prakash S., Prakash L., 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science 283: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Jung I., Park J., Choi C., Kim D., 2015. Identification of novel trans-crosstalk between histone modifications via genome-wide analysis of maximal deletion effect. Genes Genomics 37: 693–701. [Google Scholar]
- Kadyk L. C., Hartwell L. H., 1993. Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics 133: 469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M., 2000. Damage-induced recombination in the yeast Saccharomyces cerevisiae. Mutat. Res. 451: 91–105. [DOI] [PubMed] [Google Scholar]
- Mozlin A. M., Fung C. W., Symington L. S., 2008. Role of the Saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics 178: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. R., Lawrence C. W., Hinkle D. C., 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731. [DOI] [PubMed] [Google Scholar]
- Ng H. H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., et al. , 2002a Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16: 1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Xu R. M., Zhang Y., Struhl K., 2002b Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277: 34655–34657. [DOI] [PubMed] [Google Scholar]
- Polo S. E., Almouzni G., 2015. Chromatin dynamics after DNA damage: the legacy of the access-repair-restore model. DNA Repair (Amst.) 36: 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S. E., Roche D., Almouzni G., 2006. New histone incorporation marks sites of UV repair in human cells. Cell 127: 481–493. [DOI] [PubMed] [Google Scholar]
- Prakash S., Prakash L., 2000. Nucleotide excision repair in yeast. Mutat. Res. 451: 13–24. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Hartwell L. H., 1981. The Saccharomyces cerevisiae cell cycle, pp. 97–142 in The Molecular Biology of the Yeast Saccharomyces, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Rastogi R. P., Richa A., Kumar M. B., Tyagi, Sinha R. P., 2010. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010: 592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossodivita A. A., Boudoures A. L., Mecoli J. P., Steenkiste E. M., Karl A. L., et al. , 2014. Histone H3 K79 methylation states play distinct roles in UV-induced sister chromatid exchange and cell cycle checkpoint arrest in Saccharomyces cerevisiae. Nucleic Acids Res. 42: 6286–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J. M., Jackson J., Nakanishi S., Gardner J. M., Hentrich T., et al. , 2009. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35: 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M. D., Zhang K., Grunstein M., 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19: 271–277. [DOI] [PubMed] [Google Scholar]
- Siede W., Friedberg A. S., Friedberg E. C., 1993. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90: 7985–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau A., Delgoshaie N., Celic I., Dai J., Abshiru N., et al. , 2015. Interplay between histone H3 lysine 56 deacetylation and chromatin modifiers in response to DNA damage. Genetics 200: 185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., 2016. Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair (Amst.) 44: 110–117. [DOI] [PubMed] [Google Scholar]
- Sun Z. W., Allis C. D., 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108. [DOI] [PubMed] [Google Scholar]
- Sweet S. M. M., Li M., Thomas P. M., Durbin K. R., Kelleher N. L., 2010. Kinetics of re-establishing H3K79 methylation marks in global human chromatin. J. Biol. Chem. 285: 32778–32786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatum D., Li S., 2011. Evidence that the histone methyltransferase Dot1 mediates global genomic repair by methylating histone H3 on lysine 79. J. Biol. Chem. 286: 17530–17535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Ling X., Grunstein M., 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369: 245–247. [DOI] [PubMed] [Google Scholar]
- Thompson J. S., Snow M. L., Giles S., McPherson L. E., Grunstein M., 2003. Identification of a functional domain within the essential core of histone H3 that is required for telomeric and HM silencing in Saccharomyces cerevisiae. Genetics 163: 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F., Gafken P. R., Gottschling D. E., 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756. [DOI] [PubMed] [Google Scholar]
- Vlaming H., Molenaar T. M., van Welsem T., Poramba-Liyanage D. W., Smith D. E., et al. , 2016. Direct screening for chromatin status on DNA barcodes in yeast delineates the regulome of H3K79 methylation by Dot1. eLife 5: e18919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xin M., Li Y., Zhang P., Zhang M., 2016. The functions of histone modification enzymes in cancer. Curr. Protein Pept. Sci. 17: 438–445. [DOI] [PubMed] [Google Scholar]
- Wysocki R., Javaheri A., Allard S., Sha F., Cote J., et al. , 2005. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 25: 8430–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Petes T. D., 2013. Genome-wide high-resolution mapping of UV-induced mitotic recombination events in Saccharomyces cerevisiae. PLoS Genet. 9: e1003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Teng Y., Liu H., Reed S. H., Waters R., 2005. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc. Natl. Acad. Sci. USA 102: 8650–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee B. M., Levin R. S., Xu B., LeRoy G., Wingreen N. S., et al. , 2010. In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 285: 3341–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Cooper S., Brockdorff N., 2015. The interplay of histone modifications – writers that read. EMBO Rep. 16: 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains constructed for this study are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.