dsRNA-mediated gene silencing is heritable in Caenorhabditis elegans, termed RNAi inheritance. Spracklin et al. describe the results of a forward genetic...
Keywords: epigenetic inheritance, small RNAs, Caenorhabditiselegans
Abstract
Gene silencing mediated by dsRNA (RNAi) can persist for multiple generations in Caenorhabditis elegans (termed RNAi inheritance). Here we describe the results of a forward genetic screen in C. elegans that has identified six factors required for RNAi inheritance: GLH-1/VASA, PUP-1/CDE-1, MORC-1, SET-32, and two novel nematode-specific factors that we term here (heritable RNAi defective) HRDE-2 and HRDE-4. The new RNAi inheritance factors exhibit mortal germline (Mrt) phenotypes, which we show is likely caused by epigenetic deregulation in germ cells. We also show that HRDE-2 contributes to RNAi inheritance by facilitating the binding of small RNAs to the inheritance Argonaute (Ago) HRDE-1. Together, our results identify additional components of the RNAi inheritance machinery whose conservation provides insights into the molecular mechanism of RNAi inheritance, further our understanding of how the RNAi inheritance machinery promotes germline immortality, and show that HRDE-2 couples the inheritance Ago HRDE-1 with the small RNAs it needs to direct RNAi inheritance and germline immortality.
FOR most of the last century, it was widely believed that all epigenetic information was erased at, or near, the start of each new generation. It is now clear that this is not the case and that epigenetic information can be passed from parent to progeny. In some cases, epigenetic information can be transmitted from parent to progeny for multiple generations (termed transgenerational epigenetic inheritance, TEI). Examples of TEI include paramutation in plants, the inheritance of acquired traits in mammals, and RNA interference in nematodes [reviewed in Miska and Ferguson-Smith (2016)]. In many eukaryotes, double-stranded RNA (dsRNA) triggers silencing of cellular messenger RNAs (mRNAs), which contain sequence homology to the dsRNA, via a process known as RNA interference (RNAi). Interestingly, RNAi is heritable in Caenorhabditis elegans: the progeny of animals exposed to dsRNA continue to silence genes that were targeted by dsRNA in previous generations (termed RNAi inheritance). In some cases, RNAi inheritance can perdure for many generations (>5 generations) (Vastenhouw et al. 2006). RNAi inheritance is not accompanied by changes in DNA sequence, indicating that RNAi inheritance is an epigenetic phenomenon and a robust and unambiguous example of TEI.
dsRNAs are processed into small interfering RNAs (siRNAs) by the RNase III-like enzyme Dicer (Bernstein et al. 2001). siRNAs are bound by a conserved family of proteins termed Argonautes (Agos). Together, Ago proteins and their associated siRNAs regulate gene expression by binding and inhibiting complementary cellular RNAs. In some organisms, like C. elegans, RNA-dependent RNA polymerases (RdRPs) use mRNA templates to amplify siRNA populations (termed 2° siRNAs) (Motamedi et al. 2004; Pak and Fire 2007; Sijen et al. 2007). In many animals, siRNAs enter the nucleus and regulate gene expression cotranscriptionally (termed nuclear RNAi) (Martienssen and Moazed 2015). In C. elegans, the nuclear-localized Agos HRDE-1 (germline) and NRDE-3 (soma) bind 2° siRNAs in the cytoplasm, move to the nucleus, and interact with complementary nascent transcripts via base pairing of their associated 2° siRNAs. The nuclear Agos then recruit the nuclear RNAi factors (NRDE-1/2/4) to genomic sites of RNAi. Together, the nuclear RNAi machinery (by unknown mechanisms) directs the deposition of repressive chromatin marks such as H3K9me3 at RNAi-targeted loci and induce cotranscriptional inhibition of RNA Polymerase II (Guang et al. 2008, 2010; Burkhart et al. 2011). In the fission yeast Schizosaccharomyces pombe, nuclear RNAi triggers H3K9 methylation via the histone methyltransferase (HMT) Clr4 (Hall et al. 2002; Volpe et al. 2002). H3K9me contributes to nuclear RNAi by recruiting RNAi silencing factors to sites of nuclear RNAi (Motamedi et al. 2004). Thus, in S. pombe, noncoding RNAs and H3K9 methylation are part of a feed-forward loop that maintains stable states of nuclear gene silencing (Martienssen and Moazed 2015). It is not clear if or how RNAi-directed H3K9 methylation contributes to gene silencing in C. elegans.
Nuclear RNAi and RNAi inheritance require the same factors in C. elegans and nuclear gene-silencing events are needed for RNAi silencing signals to be inherited (Burton et al. 2011; Ashe et al. 2012; Buckley et al. 2012; Shirayama et al. 2012). Thus, nuclear RNAi and RNAi inheritance are related processes. HRDE-1 is an Ago that drives both nuclear RNAi and RNAi inheritance in germ cells (Ashe et al. 2012; Buckley et al. 2012; Luteijn et al. 2012; Shirayama et al. 2012). HRDE-1 is a member of an expanded clade of 12 C. elegans Agos (termed worm-specific Agos or WAGOs). The WAGOs are thought to bind endogenous small RNAs termed the 22G endo-siRNAs to mediate gene silencing of endogenous genes and RNAs (termed endogenous RNAi) (Billi et al. 2014). The degree to which the WAGOs act independently or redundantly during endogenous RNAi is unclear. It is also unclear to what extent WAGOs bind the same siRNAs or if systems exist to direct subsets of 22G siRNAs to subsets of WAGOs.
HRDE-1 binds 22G endo-siRNAs that direct nuclear RNAi at hundreds of germline-expressed genes during the normal course of development (termed HRDE-1 target genes) (Buckley et al. 2012). HRDE-1 target genes include conserved genes, cryptic loci, and retrotransposons. Of these, LTR retrotransposons appear to be the most dramatically regulated loci (Ni et al. 2014). 21U-RNAs, which are the C. elegans piwi-interacting RNAs (piRNAs), likely direct RdRPs to interact with mRNAs to generate a subset of the 22G endo-siRNAs that bind HRDE-1 (Bagijn et al. 2012). However, many endo-siRNAs that associate with HRDE-1 are not piRNA dependent and the signals that direct RdRPs to produce these HRDE-1 siRNAs remain unknown (Ni et al. 2014). HRDE-1(−) animals (and animals lacking other known components of nuclear RNAi machinery) exhibit a mortal germline (Mrt) phenotype: Disabling the nuclear RNAi/RNAi inheritance machinery by mutation does not cause fertility defects immediately; however, after 3–5 generations, mutant animals become sterile (Buckley et al. 2012). The following events occur coincidently with the loss of germ cell viability; H3K9me3 marks are progressively lost at HRDE-1 target genes while expression of many HRDE-1 genes progressively increases (Buckley et al. 2012). Therefore, the RNAi inheritance machinery may promote germline immortality by maintaining proper epigenetic states at HRDE-1 target genes to fine-tune germline gene expression programs that are necessary for germ cell totipotency. The RNAi machinery is also known to silence parasitic genetic elements such as DNA transposons (Ketting et al. 1999). Therefore, it is also possible that Mrt could be caused by the accrual of genetic damage over generations as transposons or retrotransposons are mobilized in the germline of RNAi-inheritance-defective animals.
Here we report on the results of a genetic screen to identify factors required for RNAi inheritance in C. elegans. This screen identified six RNAi inheritance factors: GLH-1/VASA, a conserved RNA helicase, PUP-1/CDE-1, a nucleotidyltransferase, SET-32, a putative histone methyltransferase, MORC-1, a conserved GHKL ATPase, and two novel nematode-specific factors that we term HRDE-2 and HRDE-4. We explore the role of these new RNAi inheritance factors in germline immortality and investigate in detail the mechanism by which one of these factors, HRDE-2, contributes to RNAi inheritance.
Materials and Methods
Strains
N2, (YY513) pkIs32[pie-1p::gfp::h2b], (YY528) hrde-1(tm1200);pkIs32, (YY1063) hrde-2(gg517);pkIs32, (YY673) hrde-3(gg266);pkIs32, (YY676) hrde-4(gg300);pkIs32, (YY494) glh-1(ok439);pkIs32, (YY1217) pup-1(gg574);pkIs32, (YY714) morc-1(gg430);pkIs32;dpy-17(e164), (YY538) hrde-1(tm1200), (YY1013) hrde-2(gg517), (YY902) set-32(gg266), (YY1197) hrde-4(gg300), (YY976) pup-1(gg519), (YY1195) glh-1(gg408), (YY890) morc-1(tm6048), (YY626) hrde-2(gg268), (YY657) hrde-1(tm1200);hrde-2(gg268), (YY909) ggSi2[hrde-1p::3xflag::gfp::hrde-2];unc-119(ed3), (YY913) nrde-2(gg518), (YY934) hrde-2(gg517);nrde-2(gg518), (YY946) rde-1(ne219);nrde-2(gg518), (YY947) hrde-1(tm1200);nrde-2(gg518), (YY584) ggSi1[hrde-1p::3xflag::gfp::hrde-1], (YY921) rde-1(ne219);ggSi1, (YY922) hrde-2(gg517);ggSi1, (YY636) hrde-2(gg268);ggSi1, (SX1316) mjIs144[mex-5p::gfp::his-58::21UR-1::tbb-2 3′UTR];unc-119(ed3), (YY1041) hrde-2(gg517);mjIs144, (YY742) hrde-2(gg252), (YY737) hrde-2(gg399), (GR1747) mut-15(tm1358), (YY626) hrde-2(gg268);pkIs32, (YY1143) hrde-2(gg517);ggSi3[hrde-1p::gfp::hrde-2(AAAA)], (YY910) hrde-2(gg252);ggSi2, (YY634) hrde-1(gg265);pkIs32, (YY631) hrde-1(gg257);pkIs32, (YY562) hrde-1(tm1200);oma-1(zu405), (TX20) oma-1(zu405), (YY635) hrde-2(gg268);oma-1(zu405), (SX461) mjIs31[pie-1::gfp::h2b];unc-119(ed3), (YY720) hrde-1(tm1200);mjIs31, (YY722) hrde-2(gg268);mjIs31, (YY721) hrde-1(tm1200);hrde-2(gg268);mjIs31, (YY1246) ppw-3(gg589), (YY1340) hrde-2(gg517);ppw-3(gg589), (YY1312) hrde-1(gg594), (YY1192) hrde-1(gg594);hrde-2(gg517).
Construction of plasmids and transgenic strains
For 3XFLAG::GFP::HRDE-1 (ggSi1) (referred to as FLAG::HRDE-1 when assaying HRDE-1 immunoprecipitation) the hrde-1 promoter, coding region, and predicted 3′ UTR were amplified by PCR from genomic N2 DNA and cloned into pCFJ151 using Gibson cloning. For GFP::3XFLAG::HRDE-2 (ggSi2), the hrde-2 coding region and predicted 3′UTR, along with the hrde-1 promoter were amplified by PCR and assembled using Gibson cloning into pCFJ151. DEVD sequence (A.A. 99–102) in HRDE-2 was mutated to AAAA in ggSi3. The aforementioned plasmids were injected into EG6699 and integrated into the genome using the MosSCI technique (Frøkjaer-Jensen et al. 2008). Insertions were verified using PCR and sequencing.
NRDE-2::3XFLAG::HA (nrde-2(gg518)) (referred to as FLAG::NRDE-2 when assaying NRDE-2 immunoprecipitations) CRISPR strain was generated with 50 ng/μl pDD162 (Cas9), 25 ng/μl pRB1017-derived guide RNA (gRNA), and 500 nM single-stranded oligo (IDT 4-nmol Ultramer) using protocol as previously described (Farboud and Meyer 2015). A gRNA targeting unc-58 and repair oligo (AF-JA-76) were used as a coconversion marker. Insertions were verified using PCR and sequencing.
CRISPR deletions (hrde-2(gg517), set-32(gg545), set-32(gg546), hrde-4(gg558), pup-1(gg519)) were generated using the protocol above with the exception of an extra gRNA added to cause two dsDNA breaks.
RNAi
RNAi experiments were conducted as previously described (Timmons and Fire 1998; Burton et al. 2011). All clones were taken from the Ahringer library (Kamath et al. 2003).
RNA Immunoprecipitation
Adult worms were flash frozen then sonicated (2 min, 30 sec ON 30 sec OFF, 30% amplitude) three times in sonication buffer [20 mM Tris pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 10% glycerol, RNaseOUT, Proteinase Inhibitor (Roche)] in a QSonica 800R sonicator. Samples were then rotated for 15 min at 4° then spun at 14K rpm for 15 min at 4°. Lysates were precleared with 30 μl protein-A/salmon sperm DNA agarose beads (16–157, Millipore, Bedford, MA) for 30 min at 4° then spun at 14K for 5 min at 4°. Supernatant was then transferred to a fresh tube with 2 μl of anti-Flag M2 conjugated to agarose (A2220, Sigma, St. Louis, MO) and rotated at 4° for 4 hr – /N. Six washes were carried out with 1 ml RNA Immunoprecipitation (RIP) buffer (20 mM Tris pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 10% glycerol) and spun at 2K for 1 min. After the washes, 100 μl of elution buffer [1 ml sonication buffer, 3 μl 3xFLAG peptide (5 mg/ml, F4799, Sigma)] was then added and rotated for 30 min at 4°. The supernatant was transferred to a new tube, 5 μl saved for protein assays, and the sample was then DNaseI treated MgCl2 added) at 37° for 20 min. Samples were then added to 400 μl TRIzol and standard isopropanol precipitation was carried out followed by resuspension in 20 μl dH2O.
Chromatin Immunopecipitation
Gravid adult animals were cross-linked in 2% formaldehyde/M9 solution for 30 min at room temperature, and fixation was stopped by adding glycine (0.125 M) and incubating for 5 min. Gravid adult animals were washed twice with M9 and stored at −80°. Fixed gravid adult animals were resuspended in 1 ml FA buffer [50 mM Tris/HCl pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 150 mM NaCl, with proteinase inhibitor (Roche)]. Samples were sonicated for 30 sec ON 30 sec OFF, 70% amplitude for 20 min in a QSonica 800R sonicator. Lysates were clarified by centrifugation for 10 min at 4°. Supernatant was transferred to a new tube and precleared with salmon sperm DNA-coated protein-A agarose beads (16–157, Millipore). Precleared lysates were incubated with 2 μl antitrimethylated Histone 3 lysine 9 (Upstate 07–523) for 2 hr – O/N. BSA and single-stranded DNA (ssDNA) pretreated protein-A agarose beads were added. Beads were pelleted and washed twice in FA buffer, twice in FA-500 buffer (50 mM Tris/HCl pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 500 mM NaCl), once in LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0), and once in TE buffer. Chromatin was eluted with elution buffer (1% SDS, 0.1 M NaHCO3) for 30 min each at room temperature. Thirty microliters of 5 M NaCl was added, and cross-link reversal carried out by heating at 65° O/N. DNAs were further purified by the QIAquick PCR purification kit (QIAGEN, Valencia, CA), and analyzed by quantitative real-time PCR (qPCR). ChIP data were normalized to co-immunoprecipitated eft-3.
Complementary DNA (cDNA) preparation
RNAs were converted to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, 170-8890, Hercules, CA) following the manufacturer’s protocol.
TaqMan
Total RNA was isolated from adult worms by vortexing flash frozen extracts for 10 min in TRIzol, followed by isopropanol precipitation. oma-1 small RNAs were reverse-transcribed by microRNA reverse-transcription kits (4366596; Applied Biosystems, Foster City, CA). cDNA was quantified by qPCR using the custom TaqMan small RNA assay from Applied Biosystems (4398987; assay IDs CSKAJ9W, CSLJIF4, CSMSGMC, CSN1ESK). Taqman sequences were the same as those used in Buckley et al. (2012).
Small RNA cloning and computational analysis
Small RNAs were enriched with a mirVana miRNA Isolation Kit (Ambion, AM1560). Two micrograms of small RNAs were cloned using a 5′-mono-phosphate independent small RNA protocol as previously described by Gent et al. (2009). Libraries were multiplexed with a 4nt 5′ barcode and a 6 nt 3′ barcode and pooled for HTS on a NextSeq500 (Biopolymers Facility, HMS).
Fast × 0.0.13 was used to separate reads that contained the 3′ adapter and filter low quality reads for further analysis. Custom scripts were used to separate samples based on the in-line 5′ barcode. Reads >17 nt were mapped to the C. elegans genome (WS220) using Bowtie 1.1.1 “-f -v 3 -p 4 -a–best–strata -m 400” [described in Shirayama et al. (2014)]. The read counts were normalized to the total number of reads that matched the genome (in millions). To account for differences in sequencing volume between samples, reads were normalized to nonstructural RNAs. Custom scripts were used to draw the single nucleotide plots. All scripts are available upon request.
Mrt assay
At each generation, six larval stage (L3) animals were picked to a single plate. Average brood sizes were calculated by counting the total number of progeny per plate.
oma-1 RNAi assay
Six larval stage (L1) were singled onto bacteria expressing oma-1 dsRNA and propagated at 20° (restrictive temperature). These animals were designated the P0/RNAi generation. Six L1 progeny (F1) animals were then transferred to bacteria without oma-1 dsRNA. Brood size was obtained from counting F1 gravid adults remaining on the P0 plates. This process was repeated at every generation.
Primers
oma-1 1F/R: (5′-TTGTTAAGCATTCCCTGCAC-3′) and (5′-TCGATCTTCTCGTTGTTTTCA-3′)
oma-1 2F/R: (5′-GTGAGTTTAACAAAATTCAAATGAAAA-3′) and (5′-AGTGGTGAAACGGGCAAAGT-3′)
spr-2 RT 1F/R: (5′-AAACTTTCGCAATGTAACTCTTCC-3′) and (5′-CTGGTAGTGCTGCGTACGTG-3′)
C09G9.5 RT 1F/R: (5′-CCAACGAAGCTTTGTGGATA-3′) and (5′-GCGTGTGGGAACCTAGAAAA-3′)
C27B7.2 1F/R: (5′-TGGGTTAACTGAAACAACTTGAA-3′) and (5′-TCACAAACAACGTCTTTTCGAT-3′)
cars-2/crs-2: (5′-GCCGGAAGTTGAAAACTACATC-3′) and (5′-TTCGTCCAAATCCTCGAATC-3′)
Y61A9LA.12: (5′-TGGAGAGCACTCTGAAAAATTG-3′) and (5′-TCTATTCCAGCAATTTCTTATCTCAA-3′)
bath-45: (5′-CTGATTCAAAGCTATTCGGAGAA-3′) and (5′-CATCGATCAGGAAAGCATCA-3′)
R11A8.1: (5′-TCTTCGGCGCTAATCTTTTC-3′) and (5′-TTGGTAGAAGCTGCATCACTTT-3′)
tlk-1: (5′-TCATTTTGGCGACTTCATCA-3′) and (5′-TTTGTTATTGAATGTGACGTG-3′)
lit-1: (5′-TCAATTTTTGCCATGGAATC-3′) and (5′-ATGTCCTTCTCCCACTTTGG-3′)
eef-1A.1/eft-3: (5′-ACTTGATCTACAAGTGCGGAGGA-3′) and (5′-CGGGTGAGAAAATCTTTCAAACTA-3′).
Data availability
Illumina data are available from GEO under the accession number GSE92840.
Results
A genetic screen identifies RNAi inheritance factors
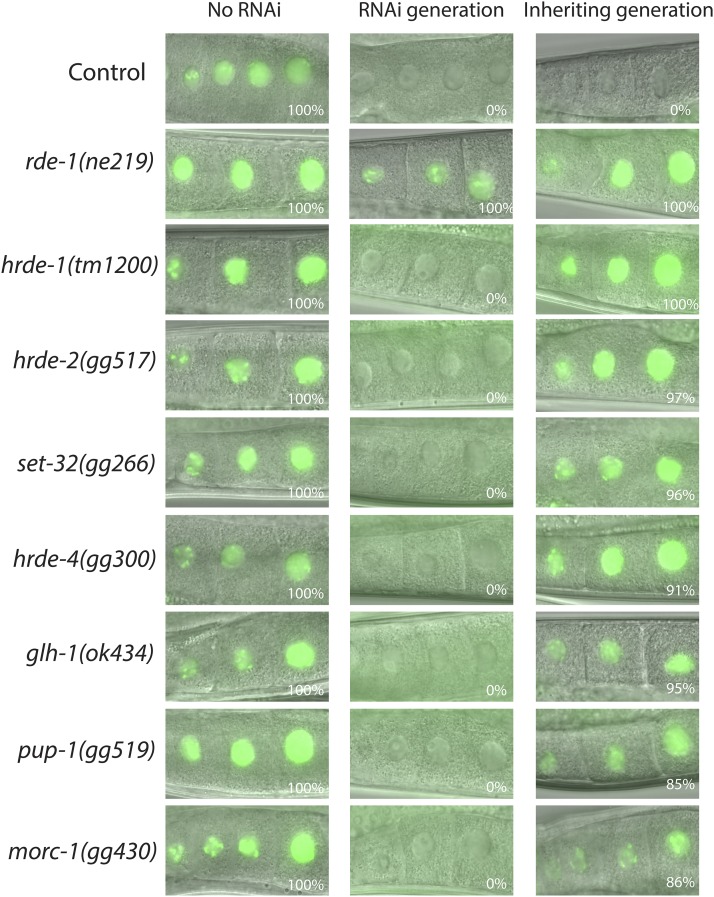
Exposure of wild-type C. elegans to gfp dsRNA causes silencing of a germline-expressed gfp transgene and this silencing is heritable (Figure 1) (Vastenhouw et al. 2006; Buckley et al. 2012). To further our understanding of the mechanism of RNAi inheritance, we conducted a forward genetic screen looking for mutations that specifically disrupted the inheritance phase of RNAi. Briefly, animals expressing GFP in germ cells were EMS mutagenized. F2 progeny of mutagenized animals were exposed to gfp dsRNA. We isolated mutant animals that were able to silence GFP in response to gfp RNAi but failed to inherit GFP silencing in subsequent generations. We screened ∼6,000,000 haploid genomes and identified 57 heritable RNAi defective (hrde) alleles (Supplemental Material, Figure S1 in File S1). Previously, this screen identified the nuclear Ago HRDE-1/WAGO-9 (henceforth referred to as HRDE-1) and the nuclear RNAi factors NRDE-1/2/4 (Buckley et al. 2012). The following approaches were used to focus our efforts on novel RNAi inheritance factors. First, animals lacking NRDE-1/2/4 respond idiosyncratically to dsRNAs targeting the pos-1 and lin-26 genes (Burkhart et al. 2011; Buckley et al. 2012). Mutant strains that behaved like nrde-1/2/4 mutant animals, when treated with pos-1 and lin-26 RNAi, were discarded as many of these animals likely harbored mutations in nrde-1/2/4. Second, hrde alleles were complemented with hrde-1(tm1200) animals, and those alleles that failed to complement hrde-1 were not analyzed further (Figure S2 in File S1). The remaining hrde alleles (23/57) were subjected to whole genome sequencing. Combinations of the following approaches, which varied for each hrde gene, were used to assign molecular identities: positional mapping, complementation analysis, genetic rescue, and in silico complementation analysis (Minevich et al. 2012) (see below). Finally, additional mutant alleles (generated by CRISPR/CAS9, or obtained from the Caenorhabditis Genome Center) of candidate hrde genes were tested for RNAi inheritance defects in order to confirm identities. Evidence establishing the identity of each hrde gene is summarized in Table 1 and is described in detail in Figures S2–S8 in File S1. In addition, a detailed description of the identification of hrde-2 is presented in the main text below. Together, these experiments show that GLH-1/VASA, PUP-1/CDE-1, SET-32, MORC-1, and two nematode-specific factors, HRDE-2 and HRDE-4, are required for RNAi inheritance in C. elegans (Figure 1). The causative mutations in three hrde strains remain to be identified.
Figure 1.
Genetic screen identifies hrde genes. Animals of the indicated genotype expressing a pie-1p::gfp::h2b transgene were exposed to bacteria producing gfp dsRNA. F1 embryos were isolated by hypochlorite treatment and grown on bacteria not expressing gfp dsRNA. GFP expression in oocytes of animals exposed to gfp dsRNA (RNAi generation) and the progeny of these animals (inheriting generation) were visualized by fluorescence microscopy. Percentage of animals expressing GFP is indicated (n > 50). Note: morc-1 is marked with dpy-17(e164).
Table 1. Genetic screen identifies seven genes as required for RNAi inheritance.
| Gene | Predicted function | Alleles identifieda | Additional allelesb | Complementation groupc | |
|---|---|---|---|---|---|
| hrde-1 | wago-9 | Argonaute | 25 | tm1200 | tm1200, gg265, gg257 |
| hrde-2 | t01c3.9 | Unknown | 4 | gg517 | gg268, gg252, gg399 |
| hrde-3 | set-32 | H3K9 methyltransferase | 9 | gg545, gg546, ok1457 | gg374, gg375, gg376 |
| hrde-4 | t10b11.7 | Unknown | 4 | gg558 | in silico |
| hrde-5 | glh-l | VASA/Helicase | 1 | ok439 | N/A |
| hrde-6 | pup-1 | poly(U)polymerase | 1 | tm936, tm1021, gg519 | N/A |
| hrde-7 | morc-1 | ATPase | 1 | tm6048, gk1754232 | N/A |
Number of alleles identified from the forward genetic screen outlined in Figure S1 in File S1.
Independently isolated alleles in the corresponding gene (obtained via Caenorhabditis Genetics Center or CRISPR/Cas9).
Defining members of each complementation group were determined by exposing F1 trans-heterozygotes to gfp dsRNA and assaying percentage of F2 animals that failed to inherit GFP silencing.
RNAi inheritance factors promote germline immortality
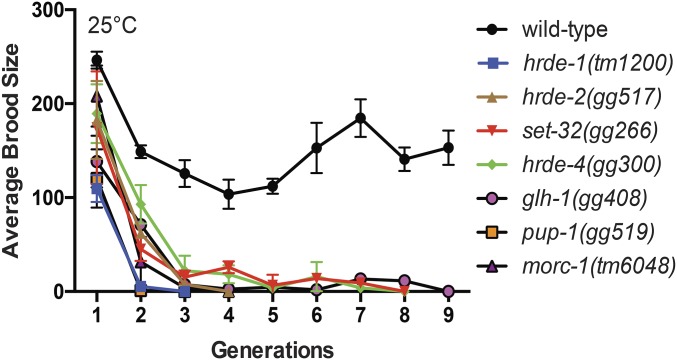
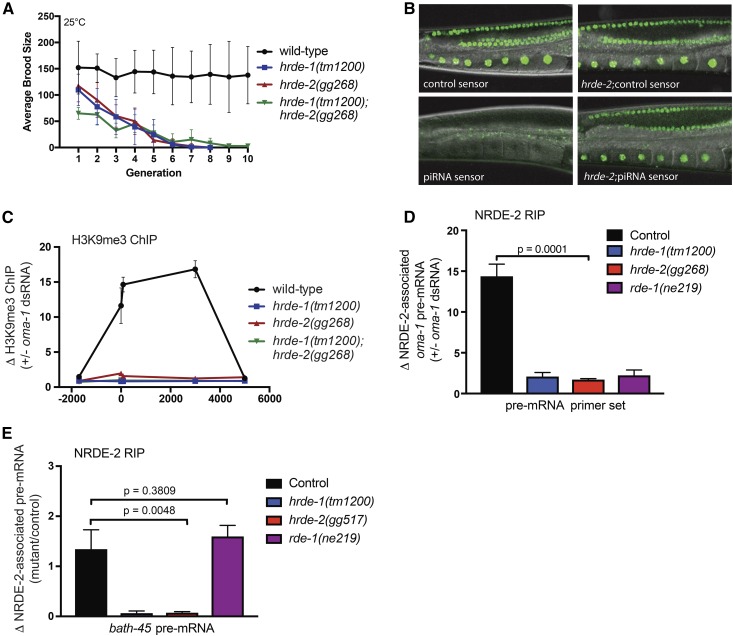
The germline is immortal in the sense that, unlike somatic cells, germ cells and their descendants are able to divide in perpetuity. Germline immortality is under genetic control (Ahmed and Hodgkin 2000; Buckley et al. 2012; Sakaguchi et al. 2014; Simon et al. 2014). Previously known components of the RNAi inheritance machinery are required for germline immortality: animals that lack NRDE-1/2/4 or HRDE-1 show near wild-type fertility; however, the descendants of these animals become sterile after 3–5 generations at 25° (Buckley et al. 2012). This transgenerational loss of fertility is referred to as a mortal germline (Mrt) phenotype (Ahmed and Hodgkin 2000). We asked if animals lacking the newly identified components of the RNAi inheritance machinery were Mrt. Note, the Mrt phenotype of RNAi inheritance defective animals (such as nrde-2 or hrde-1 mutants) is temperature sensitive: growth at 25° induces Mrt; however, growth at 20° does not induce Mrt (Buckley et al. 2012). Therefore, we shifted glh-1, pup-1, set-32, morc-1, hrde-2, and hrde-4 mutant animals to growth at 25° and monitored the fertility of these animals over generations. As expected, wild-type animals were not Mrt, and hrde-1 mutant animals were Mrt (Figure 2). glh-1, pup-1, set-32, morc-1, hrde-2, and hrde-4 mutant animals were Mrt (Figure 2). We tested a second allele of each of these genes (except for morc-1) and found that these strains also exhibited Mrt phenotypes. These data show that a general property of RNAi inheritance factors is to promote germline immortality.
Figure 2.
hrde genes are required for germline immortality. Brood sizes for animals of indicated genotype were counted as detailed in Materials and Methods. Data are mean ± SD (n = 3).
The RNAi inheritance machinery likely promotes germline immortality by maintaining the C. elegans epigenome
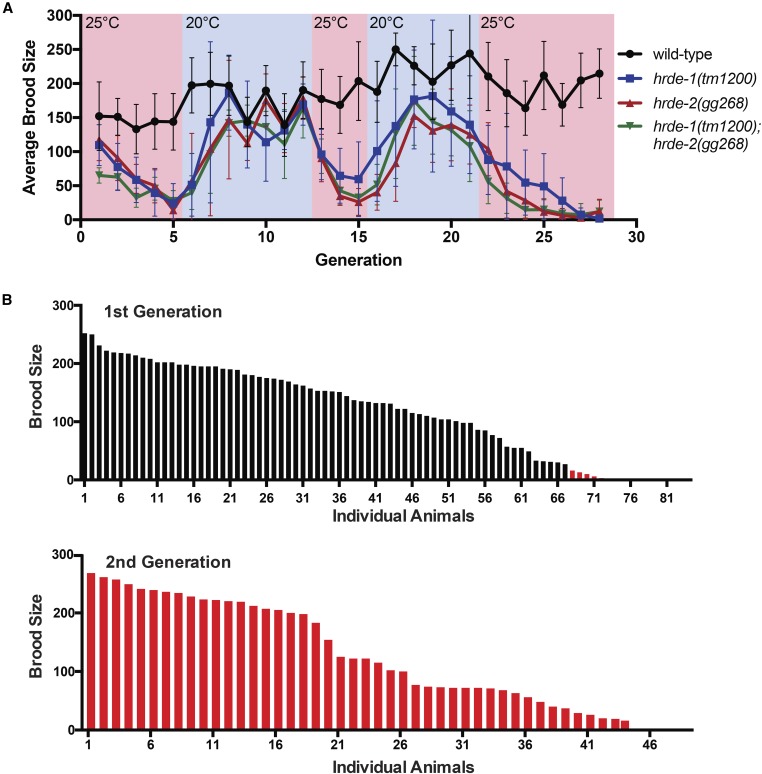
The RNAi machinery regulates the expression of both protein-coding genes and parasitic nucleic acids such as transposons (Meister 2013). Therefore, the Mrt phenotype of RNAi inheritance defective animals might be caused by progressive changes in gene expression or by genetic damage resulting from transposon mobilization. The following experiments show that the cause of germline mortality in RNAi inheritance defective animals is unlikely to be genetic damage. After 2–3 generations of growth at the nonpermissive (25°) temperature, the brood size of hrde-1 or hrde-2 mutant animals was <25% that of wild-type animals (Figure 3A). We transferred these low-fertility animals to the permissive temperature (20°) and found that after three generations, fertility in these populations was restored to near wild-type levels (Figure 3A and Figure S9 in File S1). Moving these animals back to the nonpermissive temperature again resulted in a progressive decline in fertility, which, again, could be reversed by a shift to permissive temperatures (Figure 3A and Figure S9 in File S1). These data show that the Mrt phenotype of hrde-1 or hrde-2 mutant animals is reversible and these data are consistent with recently published reports (Sakaguchi et al. 2014; Simon et al. 2014; Ni et al. 2016). We considered the possibility that rare individuals, which had not incurred DNA damage, might be present in hrde populations undergoing Mrt. These animals might, over generations, overtake sicker DNA-damaged siblings, thus, reversing Mrt. To address this issue, we quantified the brood of 84 hrde-1/-2 double mutant animals from populations that were almost Mrt at permissive temperatures. We found that animals from a Mrt population exhibited a spectrum of fertility rates ranging from zero to near wild type (Figure 3B, top). Importantly, individuals in the population that had the lowest (nonzero) brood sizes gave rise to progeny that had wild-type fertility (Figure 3B, bottom). We conclude that Mrt is reversible and that the accumulation of DNA damage does not cause Mrt in RNAi inheritance defective animals.
Figure 3.
Genetic damage is not likely to be the cause of Mrt in RNAi inheritance defective animals. (A) Brood sizes for animals of indicated genotype were counted as detailed in Materials and Methods. Shift in temperature is indicated by changing color scheme (n = 6, ± SD). (B) hrde-1;hrde-2 animals from the fifth generation at 25° were singled and grown at 20° and their brood sizes were scored (B, top). Animals whose parents had a low brood size (indicated by red bars) were singled, grown at 20°, and their brood sizes were scored (B, bottom).
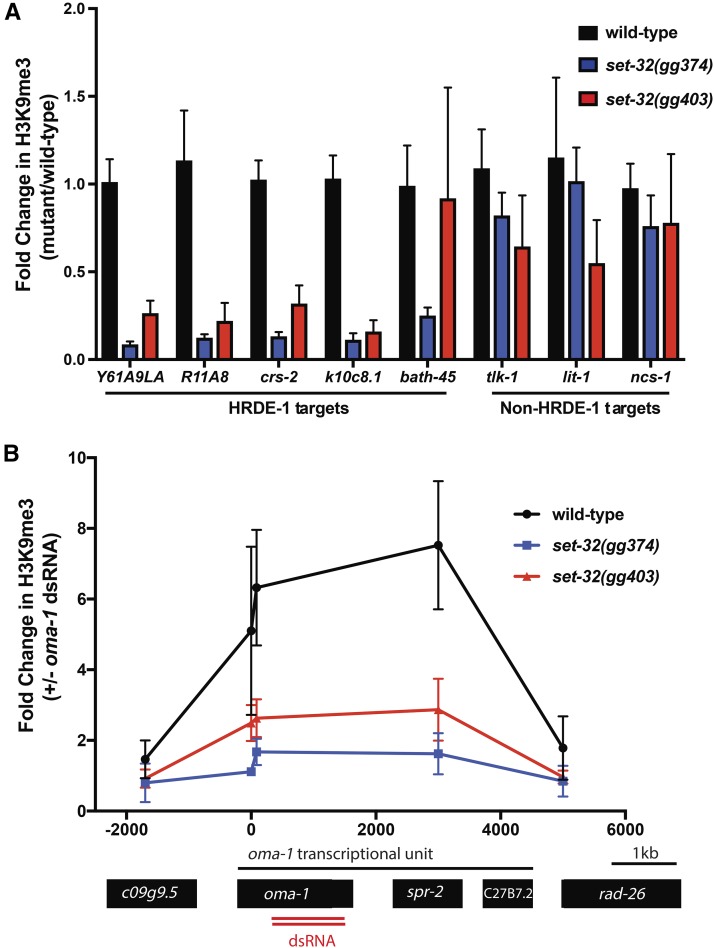
How then does the RNAi inheritance machinery promote germline immortality? It has been proposed that endogenously expressed siRNAs may promote germline immortality by maintaining a germline H3K9me3 pattern that is necessary for germ cell health (Buckley et al. 2012). Our inheritance screen identified nine alleles of the gene set-32, which encodes a SET domain protein with homology to S. pombe Clr4 and fly Su(var)3–9. Clr4 and Su(var)3–9 are founding members of a family of enzymes that methylate H3K9 (Rea et al. 2000). Thus, SET-32 is a candidate H3K9 methyltransferase. Additionally, set-32 mutant animals are Mrt, consistent with the idea that maintenance of germline H3K9me3 patterns by SET-32 is required for germline immortality (Figure 2). HRDE-1 is the Ago that engages nuclear RNAi to maintain germline immortality and the gene targets of HRDE-1 are known (Buckley et al. 2012). To test the idea that SET-32-directed H3K9me3 may promote germline immortality, we asked if SET-32 contributed to H3K9me3 at HRDE-1 target genes. Animals lacking SET-32 had less H3K9me3 at HRDE-1 target genes than wild-type animals (Figure 4A). H3K9me3 at control (non-HRDE-1 target) genes was unaffected by loss of SET-32 (Figure 4A). Exposing C. elegans to experimental dsRNAs mimics the effects of endogenous siRNAs and induces H3K9me3 at genomic sites that exhibit sequence homology to dsRNAs (Guang et al. 2010). SET-32 was required for experimentally supplied oma-1 dsRNA to trigger H3K9me3 at the oma-1 locus (Figure 4B). Thus, SET-32 is required for H3K9me3 to accumulate at genes targeted by exogenous and endogenous sources of siRNAs in C. elegans. These data suggest that SET-32 is an siRNA-directed H3K9 methyltransferase and that siRNA-directed H3K9 methylation is an obligate step during RNAi inheritance. Additionally, these data support the model that the RNAi inheritance machinery promotes germline immortality by establishing an epigenome conducive to germ cell function.
Figure 4.
SET-32 is required for nuclear RNAi-directed H3K9me3. (A) H3K9me3 ChIP was conducted on animals of indicated genotype as described in Materials and Methods. qPCR of H3K9me3 coprecipitating DNA using primers detecting five genes thought to be targeted by nuclear RNAi and three genes not thought to be targeted by nuclear RNAi. Data are relative to coprecipitating eft-3 DNA and expressed as a ratio of signal from indicated mutant to signal from wild-type (n = 6, ± SD). (B) Animals exposed to oma-1 dsRNA (+ oma-1 dsRNA) or control bacteria with L4440 vector (− oma-1 dsRNA) were subjected to H3K9me3 ChIP. Coprecipitating oma-1 DNA was quantified by qPCR. Data were normalized to coprecipitating eft-3 DNA and expressed as a ratio of signals in animals subjected to oma-1 RNAi over signals from no RNAi (−) animals (“1” denotes no change) (n = 3, ± SD). On the x-axis, 0 denotes the predicted start codon of oma-1.
Molecular identification of hrde-2
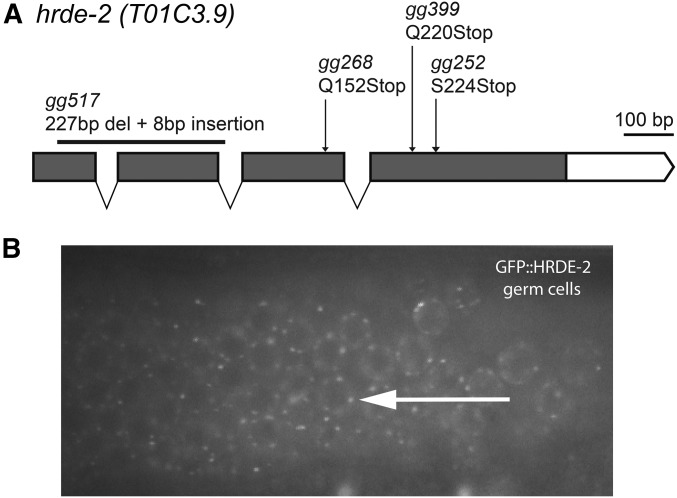
The remainder of this paper describes the identification and characterization of hrde-2. hrde-2 was identified in our genetic screen for heritable silencing of a gfp transgene. We wondered if HRDE-2 was required for RNAi inheritance at endogenous genes. oma-1 and pos-1 are two germline-expressed genes and RNAi silencing of these genes is heritable. hrde-2 animals were defective for the inheritance of oma-1 and pos-1 RNAi gene silencing, indicating that HRDE-2 is generally required for RNAi inheritance in C. elegans (Figure S10, A and B in File S1). The following lines of evidence show that hrde-2 is t01c3.9, a nematode-specific factor required for RNAi inheritance. We mapped hrde-2 to a 2-cM interval on chromosome V (Figure S3B in File S1). Within this interval, we identified loss-of-function mutations in the open reading frame of t01c3.9 in three out of our three hrde-2 strains, which previous work had established were members of the hrde-2 complementation group (Figure 5A and Figure S3A in File S1 and Table 1). We generated a deletion in t01c3.9 with CRISPR/Cas9 and found that animals harboring this deletion allele were defective for RNAi inheritance (Figure 1). Finally, a wild-type copy of t01c3.9 was sufficient to rescue the Mrt phenotype of hrde-2 mutant animals (Figure S11 in File S1). We conclude that hrde-2 is t01c3.9, a nematode-specific protein that is required for RNAi inheritance. Note, t01c3.9 is cotranscribed in an operon with the gene mut-15/rde-5, which is known to be involved in RNAi silencing in C. elegans (Sijen et al. 2007; Phillips et al. 2012). hrde-2(gg268) complemented mut-15(tm1358) for RNAi defects, indicating that mutation of t01c3.9 does not cause RNAi inheritance defects by affecting expression of mut-15 (Figure S3C in File S1).
Figure 5.
hrde-2 encodes a germline factor required for RNAi inheritance. (A) Predicted hrde-2 gene structure. Arrows indicate mutant alleles identified in genetic screen and black bar indicates deletion allele created by CRISPR/CAS9. (B) HRDE-2 localizes to cytoplasmic foci in germ cells. Fluorescence microscopy of the hermaphrodite germline in adult animals expressing gfp::hrde-2. Arrow shows perinuclear puncta.
HRDE-2 localizes to cytoplasmic foci in germ cells
The following observations argue that HRDE-2 is a germline-expressed RNAi inheritance factor. First, we created a gfp::hrde-2 fusion gene, which we integrated into the genome using MosSCI technology (Frøkjaer-Jensen et al. 2008). gfp::hrde-2 rescued the Mrt phenotype of hrde-2 mutant animals, suggesting that gfp::hrde-2 encodes a functional protein whose expression likely reflects that of endogenous HRDE-2 (Figure S11 in File S1). We observed GFP::HRDE-2 fluorescence throughout the male and female germline in gfp::hrde-2 animals (Figure 5B and Figure S12 in File S1). GFP::HRDE-2 appeared concentrated in perinuclear foci that surrounded nuclei (see Discussion). In addition, microarray and RNA-seq based studies show that the hrde-2 mRNA expression is enriched/restricted to the germline (Reinke et al. 2004; Ortiz et al. 2014). Together, these data argue that HRDE-2 is a germline-expressed RNAi inheritance factor.
HRDE-2 is a component of the C. elegans nuclear RNAi pathway in the germline
The C. elegans nuclear RNAi machinery is required for RNAi inheritance (Burton et al. 2011; Ashe et al. 2012; Buckley et al. 2012; Shirayama et al. 2012). We wondered if HRDE-2 might function as part of the nuclear RNAi pathway. To address this question, we first conducted a genetic analysis with hrde-2 and the nuclear RNAi Ago hrde-1 to ask if these genes acted in the same genetic pathway. Animals lacking both HRDE-1 and HRDE-2 became Mrt at a similar rate to animals lacking either HRDE-1 or HRDE-2, individually (Figure 6A). These data suggest that hrde-2 acts in the same genetic pathway as hrde-1 to promote germline immortality.
Figure 6.
HRDE-2 is part of the nuclear RNAi pathway. (A) Brood sizes for animals of indicated genotype were counted as detailed in Materials and Methods. hrde-1;hrde-2 double mutant animals do not show an additive fertility defect at 25° when compared to the single mutant animals, suggesting HRDE-1 and HRDE-2 act in the same pathway (n = 6, ± SD). (B) Fluorescence images of oocytes from wild-type animals expressing a piRNA sensor transgene (mjIs144), which contains a binding site for the piRNA 21U-1, and a control piRNA sensor transgene (mjIs145) which lacks the 21U-1 site [as described in Bagijn et al. (2012)]. Right panel, hrde-2(gg517) animals harboring mjIs145 (top) and mjIs144 (bottom). (C) Animals of the indicated genotypes were exposed to oma-1 dsRNA (+ oma-1 dsRNA) or empty L4440 vector (− oma-1 dsRNA) and subjected to H3K9me3 ChIP. Coprecipitating oma-1 DNA was quantified by qPCR. Data were normalized to coprecipitating eft-3 DNA and expressed as a ratio ±oma-1 RNAi (“1” denotes no change) (n = 3, ± SD). On the x-axis, 0 denotes the predicted start codon of oma-1. (D and E) FLAG-NRDE-2 was precipitated from animals of indicated genotypes with an anti-Flag antibody and NRDE-2 coprecipitating RNA was isolated by TRIzol extraction. RNA was converted to cDNA and quantified by qRT-PCR with primers detecting oma-1 pre-mRNA or bath-45 pre-mRNA as described in Materials and Methods. Comparisons were performed using t-tests (Prism) (E). Data are expressed as fold change ±oma-1 RNAi, or (E) as fold change mutant/control. Mean ± SD are shown (n = 3).
The following data provide further evidence that HRDE-2 is a component of nuclear RNAi pathway. First, piRNA-mediated gene silencing of a gfp reporter transgene (mjIs144) requires nuclear RNAi (Bagijn et al. 2012). We found that, like the known components of the nuclear RNAi pathway, HRDE-2 was required for piRNA-mediated gene silencing of this reporter gene (Figure 6B). Second, as described above, exposing C. elegans to dsRNAs is sufficient to induce H3K9me3 at genomic sites that exhibit sequence homology to dsRNAs (Guang et al. 2010). We found that, like the other known components of the nuclear RNAi pathway, HRDE-2 was required for oma-1 dsRNA to trigger H3K9 methylation at the oma-1 locus (Figure 6C and Figure S13 in File S1). The nuclear RNAi factor NRDE-2 binds nascent RNAs (pre-mRNA) when genes are targeted by dsRNA, and this step of nuclear RNAi requires the Ago HRDE-1 (Guang et al. 2010; Buckley et al. 2012). We found that, like HRDE-1, HRDE-2 was required for dsRNA to induce NRDE-2/pre-mRNA association (Figure 6D). Similarly, we found that HRDE-2 was required for NRDE-2 to bind the pre-mRNA of an endogenous target of nuclear RNAi, bath-45 (Figure 6E). Taken together, these data show that HRDE-2 is a component of the nuclear RNAi pathway and that HRDE-2 acts upstream of NRDE-2/pre-mRNA binding and NRDE-dependent H3K9 methylation during nuclear RNAi.
HRDE-2 is required for the Ago HRDE-1 to bind siRNAs following RNAi
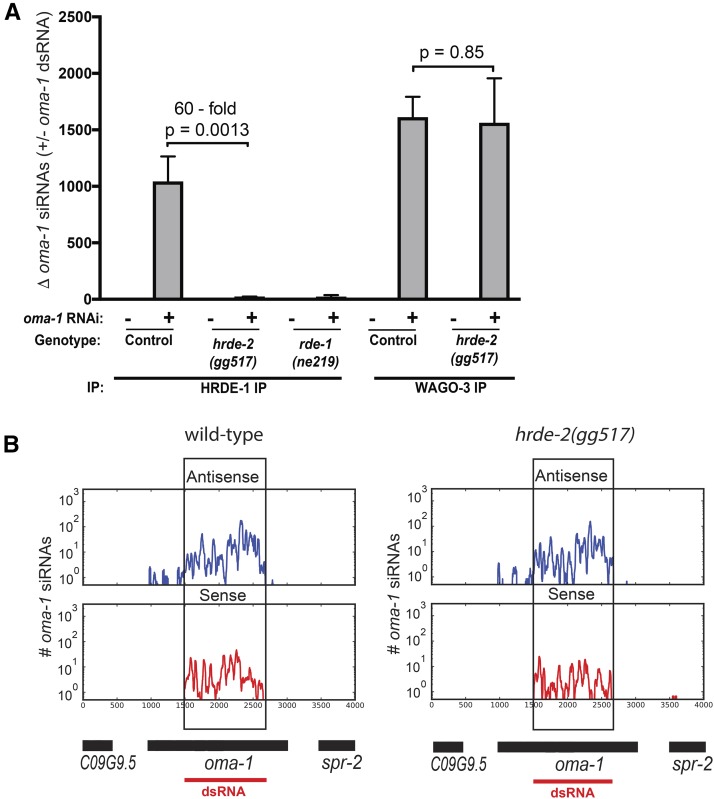
HRDE-2 acts in the nuclear RNAi pathway upstream of NRDE-2/pre-mRNA binding and NRDE-dependent H3K9 methylation. HRDE-1 is the major Ago that drives germline nuclear RNAi in C. elegans (Buckley et al. 2012). To further pinpoint how HRDE-2 contributes to nuclear RNAi, we asked if HRDE-2 was required for HRDE-1 to bind siRNAs. HRDE-1 binds endogenous small RNAs (termed 22G siRNAs), which are produced by RdRPs acting upon cytoplasmic mRNA templates (Pak and Fire 2007; Sijen et al. 2007; Buckley et al. 2012). For unknown reasons, 22G siRNAs are predominantly antisense to the RNAs used by RdRPs for their production. We treated wild-type and hrde-2 mutant animals with oma-1 dsRNA, immunoprecipitated HRDE-1, and isolated coprecipitating RNAs. We then used four sets of TaqMan probes, which were designed to detect oma-1 22G siRNAs (antisense to the oma-1 mRNA), to ask if HRDE-2 was required for HRDE-1 to associate with siRNAs. Two of these TaqMan primer sets detect antisense oma-1 siRNAs derived from sequences covered by the oma-1 dsRNA and two primer sets detect antisense oma-1 siRNAs derived from sequences not covered by the oma-1 dsRNA. Using all four probe sets, we found that HRDE-1 failed to associate with oma-1 siRNAs in hrde-2 mutant animals (Figure 7A and Figure S14 in File S1). Note: Expression of HRDE-1 was reduced (1.4-fold) in a hrde-2 mutant background (Figure S15 in File S1); however, this reduction is unlikely to explain the 60-fold reduction in HRDE-1 coprecipitating small RNAs that we observe in hrde-2 mutant animals (Figure 7A). Together, these data suggest that the reason HRDE-2 is required for RNAi inheritance is that HRDE-2 is required for the Ago HRDE-1 to bind siRNAs.
Figure 7.
HRDE-2 is required for HRDE-1 to associate with siRNAs. (A) FLAG::HRDE-1- or HA::ppw-3-expressing animals were treated with oma-1 dsRNA (+oma-1 dsRNA) or with empty L4440 vector (-oma-1 dsRNA). HRDE-1 and ppw-3 were immunoprecipitated with anti-Flag and anti-HA antibodies respectively, and coprecipitating RNA was isolated by TRIzol extraction. oma-1 coprecipitating siRNAs were quantified with an oma-1 TaqMan probe set that detected a 22G RNA antisense to the oma-1 mRNA. TaqMan signals shown relative to Taqman signals from non-RNAi-treated control animals, which was defined as one. Relative enrichment following RNAi treatment is shown (n = 3, ± SD). Comparisons were performed using t-tests (Prism). (B) Wild-type and hrde-2(gg517) animals were treated with oma-1 dsRNA. Small RNAs were sequenced as described in Materials and Methods. Small RNAs that mapped to the oma-1 locus are shown. Sense siRNA (red) and antisense siRNA (blue). Data were normalized by the total number of sequenced reads for each genotype, reads per million (rpm). oma-1 siRNAs sequenced from control (non-RNAi) samples are shown in Figure S16 in File S1. A cartoon representing the oma-1 genomic regions is shown below the graphs. The region of the oma-1 locus targeted by dsRNA is demarcated with a red line and a black box.
HRDE-1 is one of 12 members of a clade of C. elegans Agos collectively referred to as the worm-specific Agos or WAGOs (Yigit et al. 2006). Like HRDE-1, the other 11 WAGOs are thought to bind RdRP-produced 22G siRNAs (Yigit et al. 2006; Gu et al. 2009). We wondered if HRDE-2 was generally required for all WAGOs to bind siRNAs or if HRDE-2 function might be restricted to a subset of the WAGOs. To address this question, we used CRISPR/Cas9 to tag the endogenous ppw-3/ppw-2 locus (hereafter referred to as ppw-3) with an HA-epitope and then used this reagent to ask if HRDE-2 was required for ppw-3 to associate with siRNAs. We chose ppw-3 for these experiments because, like HRDE-1, ppw-3 is expressed in the germline (data not shown). After exposure to oma-1 dsRNA, HA::ppw-3 associated with similar amounts of oma-1 siRNAs in wild-type and hrde-2 mutant animals (Figure 7A). These data show that the role of HRDE-2 in WAGO/siRNA binding is limited to a subset of the WAGOs.
HRDE-2 might be required for producing HRDE-1 siRNAs or for facilitating the binding of HRDE-1 to siRNAs. To begin to address this question, we sequenced total small RNAs from wild-type and hrde-2 mutant animals that were exposed to oma-1 dsRNA or empty vector. Wild-type and hrde-2 mutant animals produced similar levels of oma-1 siRNAs (Figure 7B). The similarity in oma-1 siRNA patterns extended to regions of the oma-1 mRNA not directly targeted by oma-1 dsRNA, establishing that RdRP activity is present and apparently normal in hrde-2 mutant animals (Figure 7B). In empty vector-treated animals, very few oma-1 siRNAs were identified (<0.5% of oma-1 RNAi-treated animals) (Figure S16 in File S1). These results indicate that hrde-2 mutant animals do not exhibit obvious defects in generating the type of siRNA to which HRDE-1 binds. Taken together, our results suggest that the function of HRDE-2 is to facilitate the binding of HRDE-1 to the siRNA cofactors it needs to direct RNAi inheritance and germline immortality.
Discussion
Here we report the identification of six RNAi inheritance factors: GLH-1/VASA, PUP-1/CDE-1, SET-32, MORC-1, HRDE-2, and HRDE-4. Five of these factors have not previously been implicated in the inheritance of RNAi. HRDE-2 is required for the Ago HRDE-1 to bind siRNAs after RNAi treatment. Interestingly, HRDE-2 does not appear to be needed for the production of the siRNAs to which HRDE-1 binds, suggesting that the function of HRDE-2 is to assist HRDE-1 in binding siRNAs. It is unclear if HRDE-2 has a direct or indirect role in promoting this HRDE-1/siRNA interaction. In S. pombe, the Argonaute small interfering RNA chaperone (ARC) complex facilitates Ago1 siRNA binding (Holoch and Moazed 2015). C. elegans HRDE-2 may function analogously to ARC complex to directly mediate the loading of siRNAs into HRDE-1. Alternatively, HRDE-2 may promote HRDE-1/siRNA binding by an indirect mechanism. For instance, HRDE-2 could be needed to activate a factor that itself is required for HRDE-1/siRNA binding. Favoring (but not proving) this model, is the fact that at steady state, HRDE-2 localizes to the cytoplasm (Figure 5B and Figure S12 in File S1) while HRDE-1 localizes to the nucleus (Buckley et al. 2012; Shirayama et al. 2012). Future in vivo interaction and in vitro biochemical studies may help differentiate these models. Interestingly, we find that HRDE-2 is not required for other Agos (ppw-3) to bind siRNAs in response to dsRNA treatment, indicating some degree of specificity for HRDE-2 function during Ago/siRNA binding. C. elegans possesses ∼27 Agos and >12 of these Agos are thought to bind chemically indistinguishable 22G siRNAs. If or how these AGOs associate with subsets of the total cellular pool of 22G siRNAs is unknown. Our results suggest that HRDE-2 could be one mechanism by which different WAGOs are coupled to different 22G siRNAs. Interestingly, the C. elegans genome encodes two paralogs of hrde-2 (t24c2.2 and f43c11.9), which may act as specificity factors for other C. elegans Argonaute proteins.
H3K9me3, RNAi inheritance, and germline immortality
There are 38 potential histone-tail methyltransferases (HMTs) encoded in the C. elegans genome (Andersen and Horvitz 2007). Our genetic screen identified nine alleles of one of these HMTs, SET-32. These results suggest that SET-32 is the major HMT contributing to germline RNAi inheritance in C. elegans. SET-32 is required for H3K9me3 to accumulate at genomic sites targeted by endogenous and exogenous siRNAs (Figure 4). Together, these data argue that SET-32 is likely an H3K9 methyltransferase and that siRNA-directed H3K9 methylation is likely an important step for transgenerational gene silencing. Consistent with this idea, previous work has implicated SET-32 in piRNA mediated epigenetic inheritance (Ashe et al. 2012). In addition, we show that SET-32 is required for germline immortality, suggesting that siRNA-directed H3K9 methylation is likely an important facet of small RNA-mediated germline immortality.
How might SET-32 promote RNAi inheritance? In S. pombe, Ago1 directs the HMT Clr4 to genomic sites targeted by siRNAs so that Clr4 can methylate H3K9 (Volpe et al. 2002). Additionally, the chromodomain protein Chp1 binds H3K9me3 and recruits Ago1 to genomic sites of RNAi (Partridge et al. 2002; Verdel et al. 2004; Hayashi et al. 2012; Rougemaille et al. 2012). Thus, in S. pombe, small RNAs and H3K9 methylation act together in a feed-forward loop to maintain stable states of nuclear gene silencing (Martienssen and Moazed 2015). It is possible that a similar feed-forward mechanism may be at play in C. elegans. The nuclear RNAi machinery could direct SET-32 to chromatin to deposit H3K9me3, which could then help stabilize the nuclear RNAi machinery at repressive chromatin. It will, therefore, be of interest to ask if SET-32 physically associates with chromatin in response to RNAi, and if SET-32 and the nuclear RNAi factors require each other for stable association with chromatin. Finally, it may be worth noting that our screen identified a mutation in SET-32 that mutates a serine residue to phenylalanine (Ser170Phe), hinting that SET-32 might be regulated by phosphorylation. If this turns out to be the case, such a system might provide an elegant mechanism by which C. elegans could fine tune RNAi inheritance programs in response to changing environmental conditions.
VASA, germ granules, and RNAi inheritance
VASA is a conserved DEAD-box RNA helicase that contributes to germ cell function in most metazoans (Voronina et al. 2011). Here we show that C. elegans lacking GLH-1/VASA are defective for RNAi inheritance (Figure 1). In insects, VASA helps to silence transposable elements (TEs) by binding TE RNAs and processing TE RNAs into intermediates that are fed to Ago proteins (Guzzardo et al. 2013; Xiol et al. 2014). The association between VASA-like helicases and Ago proteins may be conserved: RDE-12, Belle, and MVH are all VASA homologs that associate with Ago proteins in worms, flies, and mice, respectively (Kirino et al. 2010; Pek and Kai 2011; Shirayama et al. 2014; Yang et al. 2014). Together, these observations hint that GLH-1/VASA helicases may be conserved mediators of small RNA-based gene-silencing pathways in metazoans and that VASA-like enzymes might fulfill this role by linking RNAi factors to RNAs in need of silencing.
A previous study failed to detect RNAi defects in glh-1/vasa mutant animals (Spike et al. 2008b). We speculate that this is likely due to the fact that while glh-1/vasa mutant animals are defective for RNAi inheritance, they respond normally when exposed directly to dsRNA (Figure 1). Why would the loss of GLH-1/VASA specifically affect the inheritance phase of RNAi? Germ granules are self-assembling nonmembrane-bound organelles found in the germ cells of most metazoans (Voronina et al. 2011). In C. elegans, germ granules are referred to as P granules. P granules segregate with the germ lineage during embryonic cell divisions (Strome and Wood 1983). GLH-1 (like VASA homologs in most animals) localizes to germ granules (Voronina et al. 2011). The concentration of GLH-1/VASA into P granules, and the segregation of these granules with the germline, position GLH-1/VASA perfectly to mediate heritable gene-silencing events. It is important to note, however, that GLH-1/VASA activity is required for normal P granule formation (Spike et al. 2008b). Several core RNAi factors (such as Dicer, Agos, and RdRPs) localize to P granules (Claycomb et al. 2009; Gu et al. 2009; Conine et al. 2010; Vasale et al. 2010; Beshore et al. 2011). It is possible, therefore, that failure to properly concentrate core RNAi factors in P granules could be responsible for the RNAi inheritance defects observed in GLH-1(−) animals. Consistent with this latter idea, DEPS-1 and PGL-1 are two germline factors that are required for wild-type P granule assembly and animals lacking either of these factors exhibit RNAi defects (Robert et al. 2005; Spike et al. 2008a).
RNA uridylation and RNAi inheritance
Poly(u) polymerase-1 (pup-1 also known as cde-1 or cid-1) encodes a poly(U) polymerase that adds short tracts of uracil to the 3′ termini of RNAs in C. elegans (Kwak and Wickens 2007; van Wolfswinkel et al. 2009). PUP-1 is thought to uridylate the endo-siRNAs that bind a germline Ago called CSR-1 (van Wolfswinkel et al. 2009). PUP-1 uridylation is thought to destabilize CSR-1 siRNAs (van Wolfswinkel et al. 2009). Thus, in animals lacking pup-1 more CSR-1 siRNAs are available for binding to CSR-1. Unlike most Agos (which negatively regulate target RNAs), CSR-1, and its associated siRNAs, is thought to activate gene expression (Seth et al. 2013; Wedeles et al. 2013; Cecere et al. 2014). Therefore, one model to explain why PUP-1 is required for RNAi inheritance is the following: in pup-1 mutant animals, CSR-1 is hyperactive and the enhanced antisilencing activity of CSR-1 prevents RNAi inheritance by overriding the prosilencing activity of the RNAi inheritance machinery. Alternatively, PUP-1 may also play a direct role in RNAi inheritance. In S. pombe, a homolog of PUP-1 (Cid12) is part of a complex of proteins termed the RNA-directed RNA polymerase complex (RDRC). RDRC consists of Cid12, as well as an RNA helicase (Hrr1), and an RdRP (Motamedi et al. 2004). The RDRC is thought to produce silencing RNAs needed to perpetuate stable long-term heterochromatic states in S. pombe (Martienssen and Moazed 2015). Thus, PUP-1-like enzymes have now been associated with long-term gene-silencing processes in both yeast and worms. Perhaps RNA uridylation in C. elegans is a signal that shunts RNAs into long-term gene-silencing pathways. For instance, in somatic cells of C. elegans, the putative poly(U) polymerase mut-2 is thought to add a uracil to the 3′ end of mRNA fragments produced in response to RNAi (Tsai et al. 2015). It has been suggested that these 3′ uracils may serve as beacons to recruit RdRPs, which amplify small RNA populations and reinforce gene silencing (Tsai et al. 2015). PUP-1 could function analogously by marking germline RNAs as templates for RdRP’s activity each generation during RNAi inheritance. By so doing, PUP-1 could maintain small RNA populations at levels sufficient to trigger gene silencing over multiple generations.
MORC-1
MORC-1 is a member of a conserved family of GHKL ATPases named after the mouse Microorchidia (Morc) gene. In plants, MORC family members help silence repetitive genomic elements such as TEs. Plant MORCs are thought to be recruited to TE genes by RNA-directed DNA methylation (Liu et al. 2014). Once localized to TEs, MORC enzymes may regulate gene expression by compacting chromatin into higher order structures that are not conducive to transcription (Moissiard et al. 2012). Interestingly, in mice, MORC also represses TEs, hinting that the silencing of genome parasites may be a conserved feature of the MORC family of ATPases (Pastor et al. 2014). The C. elegans MORC homolog (MORC-1) has been previously implicated in RNAi silencing processes (Kim et al. 2005; Robert et al. 2005; Moissiard et al. 2012). Our results suggest that MORC-1 activity may be particularly important during the inheritance phase of RNAi. If MORC-1 were a component of the nuclear RNAi machinery, one would expect such an inheritance-specific defect. Perhaps nuclear RNAi directs MORC-1 to genes undergoing heritable gene silencing in C. elegans in order to compact chromatin into states not compatible with gene expression. Asking if nuclear RNAi compacts chromatin in C. elegans, and whether MORC-1 is needed for this compaction, will be an important test of this model.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.198812/-/DC1.
Acknowledgments
We thank P. Anderson and F. O'brien for helpful discussions. This work was supported by grants from the National Institutes of Health (NIH) GM088289 and GM104232 (S.K), a NIH-sponsored predoctoral training fellowship to the University of Wisconsin Genetics Training Program GM007133 (G.S. and B.F), and NSF Pre-doctoral fellowships (B.F. and A.S). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Ahmed S., Hodgkin J., 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403: 159–164. [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Horvitz H. R., 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134: 2991–2999. [DOI] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E.-M., Mitchell J., Bagijn M. P., et al. , 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn M. P., Goldstein L. D., Sapetschnig A., Weick E.-M., Bouasker S., et al. , 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J., 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Beshore E. L., McEwen T. J., Jud M. C., Marshall J. K., Schisa J. A., et al. , 2011. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Dev. Biol. 350: 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi A. C., Fischer S. E. J., Kim J. K., 2014. Endogenous RNAi pathways in C. elegans. WormBook 7: 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A., et al. , 2012. A nuclear argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart K. B., Guang S., Buckley B. A., Wong L., Bochner A. F., et al. , 2011. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 7: e1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton N. O., Burkhart K. B., Kennedy S., 2011. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108: 19683–19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G., Hoersch S., O’Keeffe S., Sachidanandam R., Grishok A., 2014. Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat. Struct. Mol. Biol. 21: 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C. C., Batista P. J., Gu W., Claycomb J. M., Chaves D. A., et al. , 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107: 3588–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B., Meyer B. J., 2015. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J. I., Schvarzstein M., Villeneuve A. M., Gu S. G., Jantsch V., et al. , 2009. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics 183: 1297–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Jr, Vasale J., Batista P. J., et al. , 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Bochner A. F., Pavelec D. M., Burkhart K. B., Harding S., et al. , 2008. An argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Bochner A. F., Burkhart K. B., Burton N., Pavelec D. M., et al. , 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzardo P. M., Muerdter F., Hannon G. J., 2013. The piRNA pathway in flies: highlights and future directions. Curr. Opin. Genet. Dev. 23: 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I. M., Shankaranarayana G. D., Noma K.-I., Ayoub N., Cohen A., et al. , 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hayashi A., Ishida M., Kawaguchi R., Urano T., Murakami Y., et al. , 2012. Heterochromatin protein 1 homologue Swi6 acts in concert with Ers1 to regulate RNAi-directed heterochromatin assembly. Proc. Natl. Acad. Sci. USA 109: 6159–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., Moazed D., 2015. Small-RNA loading licenses argonaute for assembly into a transcriptional silencing complex. Nat. Struct. Mol. Biol. 22: 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Ketting R. F., Haverkamp T. H., van Luenen H. G., Plasterk R. H., 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99: 133–141. [DOI] [PubMed] [Google Scholar]
- Kim J. K., Gabel H. W., Kamath R. S., Tewari M., Pasquinelli A., et al. , 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. [DOI] [PubMed] [Google Scholar]
- Kirino Y., Vourekas A., Kim N., de Lima Alves F., Rappsilber J., et al. , 2010. Arginine methylation of vasa protein is conserved across phyla. J. Biol. Chem. 285: 8148–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J. E., Wickens M., 2007. A family of poly(U) polymerases. RNA 13: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.-W., Shao C.-R., Zhang C.-J., Zhou J.-X., Zhang S.-W., et al. , 2014. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 10: e1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn M. J., van Bergeijk P., Kaaij L. J. T., Almeida M. V., Roovers E. F., et al. , 2012. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31: 3422–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R., Moazed D., 2015. RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 7: a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., 2013. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 14: 447–459. [DOI] [PubMed] [Google Scholar]
- Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., 2012. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192: 1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E. A., Ferguson-Smith A. C., 2016. Transgenerational inheritance: models and mechanisms of non-DNA sequence-based inheritance. Science 354: 59–63. [DOI] [PubMed] [Google Scholar]
- Moissiard G., Cokus S. J., Cary J., Feng S., Billi A. C., et al. , 2012. MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336: 1448–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi M. R., Verdel A., Colmenares S. U., Gerber S. A., Gygi S. P., et al. , 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802. [DOI] [PubMed] [Google Scholar]
- Ni J. Z., Chen E., Gu S. G., 2014. Complex coding of endogenous siRNA, transcriptional silencing and H3K9 methylation on native targets of germline nuclear RNAi in C. elegans. BMC Genomics 15: 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Z., Kalinava N., Chen E., Huang A., Trinh T., et al. , 2016. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. A., Noble D., Sorokin E. P., Kimble J., 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 4: 1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Fire A., 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244. [DOI] [PubMed] [Google Scholar]
- Partridge J. F., Scott K. S. C., Bannister A. J., Kouzarides T., Allshire R. C., 2002. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 12: 1652–1660. [DOI] [PubMed] [Google Scholar]
- Pastor W. A., Stroud H., Nee K., Liu W., Pezic D., et al. , 2014. MORC1 represses transposable elements in the mouse male germline. Nat. Commun. 5: 5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek J. W., Kai T., 2011. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc. Natl. Acad. Sci. USA 108: 12007–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Montgomery T. A., Breen P. C., Ruvkun G., 2012. MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O’Carroll D., Strahl B. D., Sun Z. W., et al. , 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599. [DOI] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S., Kazmer K., 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- Robert V. J. P., Sijen T., van Wolfswinkel J., Plasterk R. H. A., 2005. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19: 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M., Braun S., Coyle S., Dumesic P. A., Garcia J. F., et al. , 2012. Ers1 links HP1 to RNAi. Proc. Natl. Acad. Sci. USA 109: 11258–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A., Sarkies P., Simon M., Doebley A.-L., Goldstein L. D., et al. , 2014. Caenorhabditis elegans RSD-2 and RSD-6 promote germ cell immortality by maintaining small interfering RNA populations. Proc. Natl. Acad. Sci. USA 111: E4323–E4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M., Shirayama M., Gu W., Ishidate T., Conte D., Jr, et al. , 2013. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell 27: 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.-C., Gu W., Ishidate T., et al. , 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Stanney W., III, Gu W., Seth M., Mello C. C., 2014. The Vasa Homolog RDE-12 engages target mRNA and multiple argonaute proteins to promote RNAi in C. elegans. Curr. Biol. 24: 845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Steiner F. A., Thijssen K. L., Plasterk R. H. A., 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247. [DOI] [PubMed] [Google Scholar]
- Simon M., Sarkies P., Ikegami K., Doebley A.-L., Goldstein L. D., et al. , 2014. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Bader J., Reinke V., Strome S., 2008a DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development 135: 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C., Meyer N., Racen E., Orsborn A., Kirchner J., et al. , 2008b Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics 178: 1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Wood W. B., 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Tsai H.-Y., Chen C.-C. G., Conte D., Jr, Moresco J. J., Chaves D. A., et al. , 2015. A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Claycomb J. M., Batista P. J., Mello C. C., Berezikov E., et al. , 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148. [DOI] [PubMed] [Google Scholar]
- Vasale J. J., Gu W., Thivierge C., Batista P. J., Claycomb J. M., et al. , 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/argonaute pathway. Proc. Natl. Acad. Sci. USA 107: 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw N. L., Brunschwig K., Okihara K. L., Müller F., Tijsterman M., et al. , 2006. Gene expression: long-term gene silencing by RNAi. Nature 442: 882. [DOI] [PubMed] [Google Scholar]
- Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., et al. , 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I. S., et al. , 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Voronina E., Seydoux G., Sassone-Corsi P., Nagamori I., 2011. RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 3: a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeles C. J., Wu M. Z., Claycomb J. M., 2013. Protection of germline gene expression by the C. elegans argonaute CSR-1. Dev. Cell 27: 664–671. [DOI] [PubMed] [Google Scholar]
- Xiol J., Spinelli P., Laussmann M. A., Homolka D., Yang Z., et al. , 2014. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157: 1698–1711. [DOI] [PubMed] [Google Scholar]
- Yang H., Vallandingham J., Shiu P., Li H., Hunter C. P., et al. , 2014. The DEAD box helicase RDE-12 promotes amplification of RNAi in cytoplasmic foci in C. elegans. Curr. Biol. 24: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E., Batista P. J., Bei Y., Pang K. M., Chen C.-C. G., et al. , 2006. Analysis of the C. elegans argonaute family reveals that distinct argonautes act sequentially during RNAi. Cell 127: 747–757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina data are available from GEO under the accession number GSE92840.