Abstract
Gene duplication provides spare genetic material that evolution can craft into new functions. Sox2 and Sox3 are evolutionarily related genes with overlapping and unique sites of expression during embryogenesis. It is currently unclear whether SOX2 and SOX3 have identical or different functions. Here, we use CRISPR/Cas9-assisted mutagenesis to perform a gene-swap, replacing the Sox3 ORF with the Sox2 ORF to investigate their functional equivalence in the brain and testes. We show that increased expression of SOX2 can functionally replace SOX3 in the development of the infundibular recess/ventral diencephalon, and largely rescues pituitary gland defects that occur in Sox3 null mice. We also show that ectopic expression of SOX2 in the testes functionally rescues the spermatogenic defect of Sox3 null mice, and restores gene expression to near normal levels. Together, these in vivo data provide strong evidence that SOX2 and SOX3 proteins are functionally equivalent.
Keywords: SOXB1 genes, CRISPR/CAS9 mutagenesis, gene swap
ONE of the driving forces for the evolution of complex life is the duplication of genes, chromosomes, or entire genomes, providing the genetic material upon which natural selection can operate. Evolutionary theory predicts that having duplicated, a gene pair will be relieved from selective constraints, thereby enabling the accumulation of genetic alterations that can alter protein function (Force et al. 1999; Lynch and Conery 2000). The consequences of this are thought to favor loss of one copy (nonfunctionalization). Alternatively, gene functions can be divided between the paralogues (subfunctionalization), or one copy can acquire a novel advantageous function (neofunctionalization) while the other copy retains its original function (Force et al. 1999; Lynch and Conery 2000). Under this paradigm, it is expected that shared function within a given tissue will not be preserved by natural selection, and should therefore be lost over time. This stands in contrast to many observations of genetic redundancy that have emerged in the age of molecular genetics as gene deletions in seemingly important genes routinely yield no, or mild, phenotypes, and appear to be compensated for by paralogous partner genes (Wagner 2005). Estimates suggest that as many as 10–15% of mouse gene knockouts may have no or mild phenotypes (Barbaric et al. 2007). What forces allow the persistence of genetic redundancy are unclear, but genetic robustness that acts to maintain and bolster important processes may play a role (Force et al. 1999; Wagner 2005; Barbaric et al. 2007).
Persistent genetic redundancy is particularly striking in the SoxB1 subfamily, which consists of Sox1, Sox2, and Sox3. These genes share highly similar sequences, both within and, to a lesser extent, outside of the DNA-binding HMG box. Several studies suggest that SOXB1 proteins have similar, if not identical, functional capabilities. For example, overexpression of chick or mouse SoxB1 genes in chick neural tube results in inhibition of neural differentiation with cells retaining a progenitor identity (Bylund et al. 2003; Graham et al. 2003). Similarly, mouse Sox1 and Sox3 are able to replace Sox2 for reprogramming of iPS cells (Nakagawa et al. 2008). Loss of function studies also generally support functional equivalence, particularly in the developing CNS where the SoxB1 genes exhibit extensive overlapping expression. For example, Sox3 deletion in mice results in relatively mild neural defects, indicating that SOX2 and/or SOX1 can compensate for the absence of SOX3 in most neuroprogenitor contexts. However, one notable exception is the infundibulum, a ventral evagination of the ventral diencephalon that is responsible for induction of the anterior pituitary primordium (Rathke’s Pouch). Despite coexpression of Sox2 and Sox3, pituitary induction and development is severely compromised in Sox2 and Sox3 single mutants (Rizzoti et al. 2004; Kelberman et al. 2006). It is thought that this is due to reduced dosage of SOX2 or SOX3, as opposed to unique roles of these proteins (Zhao et al. 2012). However, to date, experimental approaches that distinguish between these possibilities have not been published.
Restricted zones of SoxB1 expression outside of the nervous system have also been described, many of which are in stem/progenitor cells of developing organs. For example, Sox3 is uniquely expressed in the spermatogonial stem/progenitor cells of the postnatal testes (Rizzoti et al. 2004; Raverot et al. 2005). Consistent with a model of limited subfunctionalization, more severe phenotypes occur in knockout mice at sites of unique expression. For example, Sox3 null mice have spermatogenic defects likely due to the absence of Sox1 and Sox2 (Raverot et al. 2005). However, it is not known whether SoxB1 genes are functionally interchangeable at these unique zones of expression.
Herein, we describe an in vivo gene swap experiment in which Sox3 open reading frame (ORF) was deleted and replaced with Sox2 ORF to investigate their functional similarities. We show that SOX2 can functionally replace SOX3 in both the developing pituitary and testes, thereby rescuing phenotypes associated with SOX3-null mice.
Materials and Methods
Generation of CRISPR/Cas9 modified mice
CRISPR gRNAs were designed either side of the Sox3 ORF (5′-CCTGATGCGTTCTCTCGAGC-3′ and 5′-GACAGTTACGGCCAAACTTT-3′) using CRISPR Design tool (http://crispr.mit.edu/) and generated according to the protocol described in Wang et al. (2013). gRNA IVT was performed using HiScribe T7 Quick High Yield RNA Synthesis Kit. Cas9 mRNA was generated by IVT using the mMESSAGE mMACHINE T7 ULTRA Transcription Kit (Ambion) from pCMV/T7-hCas9 (Toolgen) digested with Xho1. gRNAs and Cas9 mRNA were purified using a MEGAclear Transcription Clean-Up Kit (Ambion). Our previously published Sox3 targeting vector (Hughes et al. 2013) was modified to replace Sox3 ORF with Sox2 ORF. Cas9 mRNA (100 ng/μl), gRNAs (50 ng/μl each) and donor plasmid (200 ng/μl) were injected into the cytoplasm of C57BL/6N zygotes using a FemtoJet microinjector, transferred to pseudo pregnant recipients, and allowed to develop to term. Homology directed repair (HDR) from the vector resulted in the Sox3Sox2KI mice carrying a neomycin resistance cassette 1 kb downstream from the Sox2-KI stop codon. The Sox3Sox2KI mice also contain a 2 bp deletion in the 5′UTR at the upstream gRNA site, and a 1 bp in the 3′UTR at the downstream gRNA site, presumably as a result of CRISPR/Cas9 recutting after HDR.
Microarray analysis
Microarray expression profiling was performed using Affymetrix GeneChip Mouse Gene 1.0 ST Arrays on three Sox3 null and three Sox3Sox2KI 2-week testes. A total of six wild type age matched samples was included, comprising two groups of three matched to the same genetic background as the Sox3 null and Sox3Sox2KI samples. Two way-ANOVA, using batch as a factor, was used to identify the significantly regulated genes. ANOVA was performed comparing to matched WT samples and comparing to pooled WT samples with similar results. We have presented data comparing two pooled WT samples.
Sperm counting
Cauda epididymis were isolated and minced in 1 ml of 37° DMEM media. Sperm were allowed to disperse for 10–15 min at 37°; 10 μl of the resuspension was diluted with 10 μl of 1 M Tris pH 9.5 solution to immobilize sperm before counting with a hemocytometer.
Data availability
Microarray data has been submitted to Gene Expression Omnibus (GEO) with accession number GSE96805. All other reagents can be made available upon request. Supporting data can be found in Supplemental Material, File S1.
Results
Generation of Sox3Sox2KI mice using CRISPR/Cas9 mutagenesis
To investigate the functional redundancy within the SoxB1 subgroup, we replaced the Sox3 ORF with that of Sox2 while leaving the remaining native Sox3 flanking sequences including the promoter and untranslated regions (UTR) intact. This mouse model, which we refer to as Sox3Sox2KI, therefore lacks SOX3, and expresses SOX2 from the Sox3 locus on the X-chromosome. To generate Sox3Sox2KI mice, we initially modified an existing Sox3 KO targeting construct (Rizzoti et al. 2004; Hughes et al. 2013) by replacing the Sox3 ORF with Sox2. Attempts to generate Sox3Sox2KI mice by conventional gene targeting in mouse ES cells failed to produce any chimeras despite multiple rounds of injections using germline competent cells (data not shown). To circumvent this issue, we employed CRISPR/Cas9 technology to generate Sox3Sox2KI mice by zygotic injection of Cas9 mRNA, as well as gRNA pairs targeting either side of the Sox3 ORF, and the donor (targeting) plasmid (Figure 1A). PCR screening of 17 founders identified a Sox3Sox2KI female harboring the intended replacement event (Figure 1B). The other allele of this Sox3Sox2KI female, as well as seven other founders, lacked the entire Sox3 ORF due to deletion of the interval between the two gRNA cuts. These were used to generate Sox3-null mice (Sox3KO). No gross abnormalities were observed in Sox3Sox2KI or Sox3KO adult mice (data not shown), and their body weights were comparable with wild type (WT) littermates (Figure S1 in File S1).
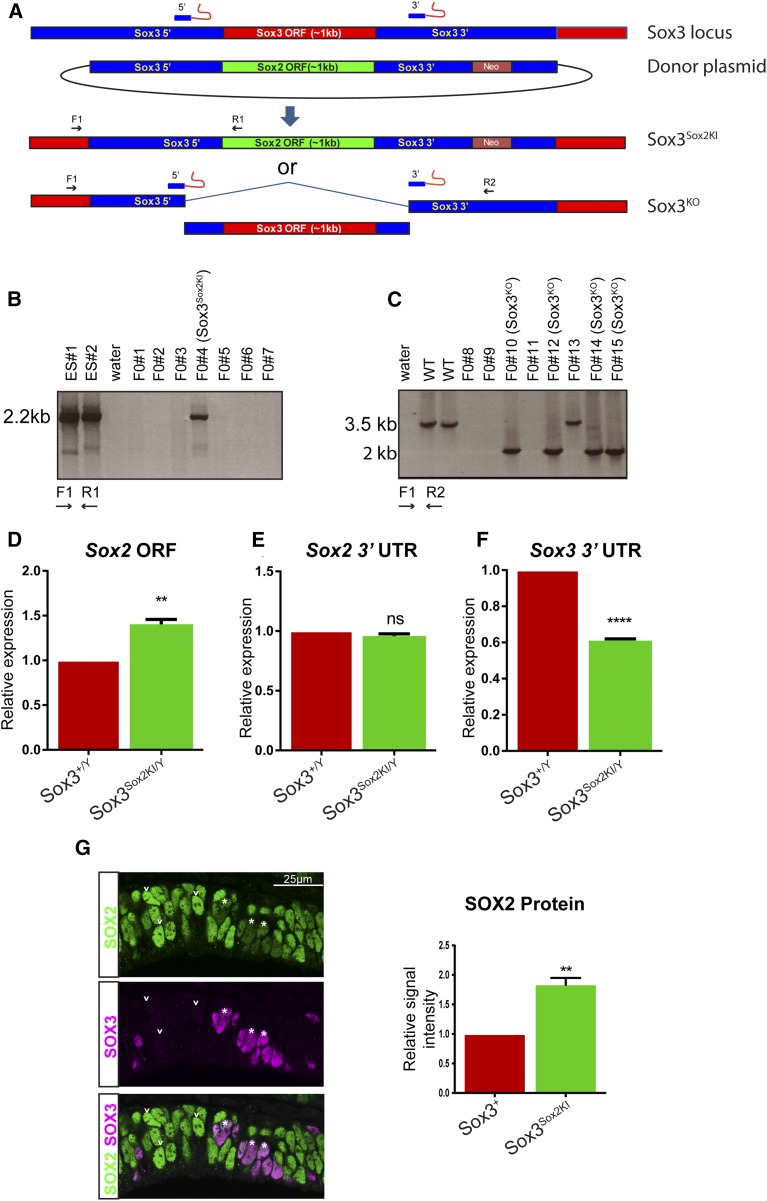
Figure 1.
Generation of Sox3Sox2KI and Sox3KO mice with CRISPR mutagenesis. (A) Schematic showing the strategy for generation of the Sox3Sox2KI mice. (B, C) PCR screening of founder mice. (D) Sox2 transcript levels were measured in 11.5 dpc brains by qPCR with primers located in the Sox2 ORF. (E) qPCR showing expression from the endogenous Sox2 locus using primers located in the 3′UTR. (F) Transcription from the Sox3 locus was reduced in Sox3Sox2KI/Y as determined by qPCR with primers located in the Sox3 3′UTR. (G) SOX3 and SOX2 coimmunostaining in the ventral diencephalon of 12.5 dpc Sox3Sox2KI/+ embryos. Asterisks indicate SOX3 positive cells, and arrowheads indicate SOX3 negative cells. Quantification of SOX2 staining intensity in SOX3 positive and SOX3 negative cells (n = 3 embryos).
To determine whether Sox2 was expressed from the Sox3 locus in the developing brain, we performed qRT-PCR using primers that amplify the Sox2 ORF, the Sox2 3′UTR, and the Sox3 3′UTR. The level of Sox2-ORF-containing transcripts was significantly elevated in Sox3Sox2KI/Y embryos vs. Sox3+/Y (Figure 1D). Sox2 3′UTR-containing transcript levels were equivalent (Figure 1E), indicating that the lack of Sox3 in Sox3Sox2KI embryos does not result in a compensatory increase in Sox2 transcript levels. Notably, the level of transcripts from the Sox3 locus is lower in Sox3Sox2KI embryonic heads compared to Sox3+/Y heads (Figure 1F). This reduction may reflect small differences in transcription rate caused by transgene elements, or reduction in the stability of the Sox3Sox2KI/Y chimeric transcript.
To confirm that the observed increase in Sox2 transcript levels in Sox3Sox2KI/Y embryos resulted in increased protein levels, we performed immunostaining on the ventral diencephalon, where SOX3 and SOX2 are coexpressed in the infundibulum (Wood and Episkopou 1999; Rogers et al. 2013). We examined Sox3Sox2KI/+ embryos in which random X-inactivation results in a mixture of cells expressing either the Sox2KI or Sox3 allele. SOX2 immunostaining intensity was significantly elevated in Sox2KI cells (SOX3 negative) in comparison to neighboring SOX3 positive cells, consistent with additional expression of SOX2 from Sox3Sox2KI allele (Figure 1G). Collectively, these results demonstrate successful gene replacement in our Sox3Sox2KI mice, and show that Sox3 has been removed and replaced with a copy of Sox2 that is regulated in a Sox3-specific fashion.
Rescue of pituitary induction defect in Sox3Sox2KI mice
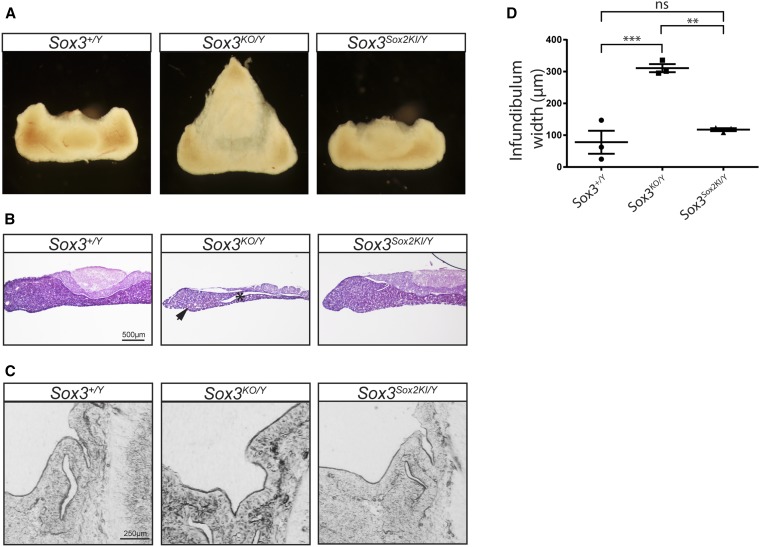
We next sought to assess whether SOX2 protein could functionally rescue Sox3-null phenotypes. Given that pituitary development is extremely sensitive to SoxB1 gene dosage (Zhao et al. 2012), we examined adult pituitaries to determine whether the replacement of Sox3 with Sox2 was able to rescue Sox3 null pituitary defects (Rizzoti et al. 2004). To assess rescue by Sox2KI, we collected pituitaries from 8-week-old mice. As expected, we found malformations in Sox3KO/Y pituitaries; indeed, these were even more severe than previously reported on a mixed genetic background (Figure 2A) (Rizzoti et al. 2004; Hughes et al. 2013). The dorsal aspect of Sox3 null pituitaries was triangular, such that the neural lobe was rostrally displaced forming a tenuous connection with the anterior lobe. Sectioning revealed hypoplasia of the anterior lobe, and deep clefting of the residual lumen of Rathke’s Pouch in Sox3KO/Y pituitaries, similar to previous reports (Figure 2B) (Rizzoti et al. 2004; Hughes et al. 2013). In contrast, the gross morphology of Sox3Sox2KI/Y pituitaries was indistinguishable from Sox3+/Y controls (Figure 2A), apart from very subtle clefting between the intermediate and anterior lobe (Figure 2, A and B). We next compared anterior pituitary induction in Sox3KO/Y and Sox3KI/Y embryos. Sox3KO/Y embryos had an infundibular recess that was significantly wider than Sox3+/Y embryos, and a dysmorphic Rathke’s Pouch with extensive branching, some of which (three of four) had failed to detach from the oral cavity (Figure 2, C and D and Figure S2 in File S1). In contrast, the width of the infundibulum in Sox3Sox2KI embryos was completely rescued. Furthermore, Rathke’s Pouch was always separated from the oral cavity, although a mild branching phenotype was observed (Figure S2 in File S1). Together, this data indicates that SOX2 is able to functionally replace SOX3 in the developing brain.
Figure 2.
Increased expression of Sox2 can rescue Sox3 null pituitary defects. (A) Adult pituitaries showing dorsal displacement of the neural lobe in Sox3KO/Y mice. (B) Haemotoxylin and eosin staining of 8-week-old pituitary coronal sections. Asterisk highlights abnormal clefting. Arrowhead indicates hypoplastic anterior lobe. (C) Phase contrast images of 12.5 dpc developing pituitaries (saggital sections). (D) Quantification of infundibulum width of 12.5 dpc embryos (n = 3). One-way ANOVA using Tukey’s Multiple comparison test. Mean ± SEM.
Ectopic SOX2 in the testes can regulate SOX3 target genes
We next assessed whether SOX2 is able to replace SOX3 in a tissue where it is not normally expressed. For these experiments, we turned to the testes, where Sox3 is the only SoxB1 member expressed in spermatogonial stem/progenitor cells, and is functionally required for normal spermatogenesis (Raverot et al. 2005). After first confirming the absence of Sox1 and Sox2 in the testes by qRT-PCR (Figure S3 in File S1), we then examined testes from Sox3Sox2KI/Y mice to determine whether Sox2 was expressed in a Sox3-specific manner. Robust expression of Sox2 was detected by qRT-PCR in Sox3Sox2KI/Y testes, but not in Sox3+/Y testes (Figure 3A). To determine whether the level of Sox2KI expression in Sox3Sox2KI/Y mice was similar to endogenous Sox3 expression in Sox3+/Y mice, we compared Sox3 3′ UTR transcript levels in testes (as Sox3Sox2KI/Y retains an intact Sox3 UTR; Figure 1F). Sox3 locus expression in Sox3Sox2KI/Y was slightly lower than levels seen in Sox3+/Y, but was a closer match than that observed in the developing brain (Figure 1F and Figure 3B, respectively). In addition, SOX2 immunostaining could not be detected in Sox3+/Y and Sox3KO/Y testes, but was present in Sox3Sox2KI/Y testes in undifferentiated spermatogonia (Figure 3C). SOX3 immunostaining was present in Sox3+/Y testes in undifferentiated spermatogonia, but, as expected, not in Sox3KO/Y and Sox3Sox2KI/Y testes (Figure 3C).
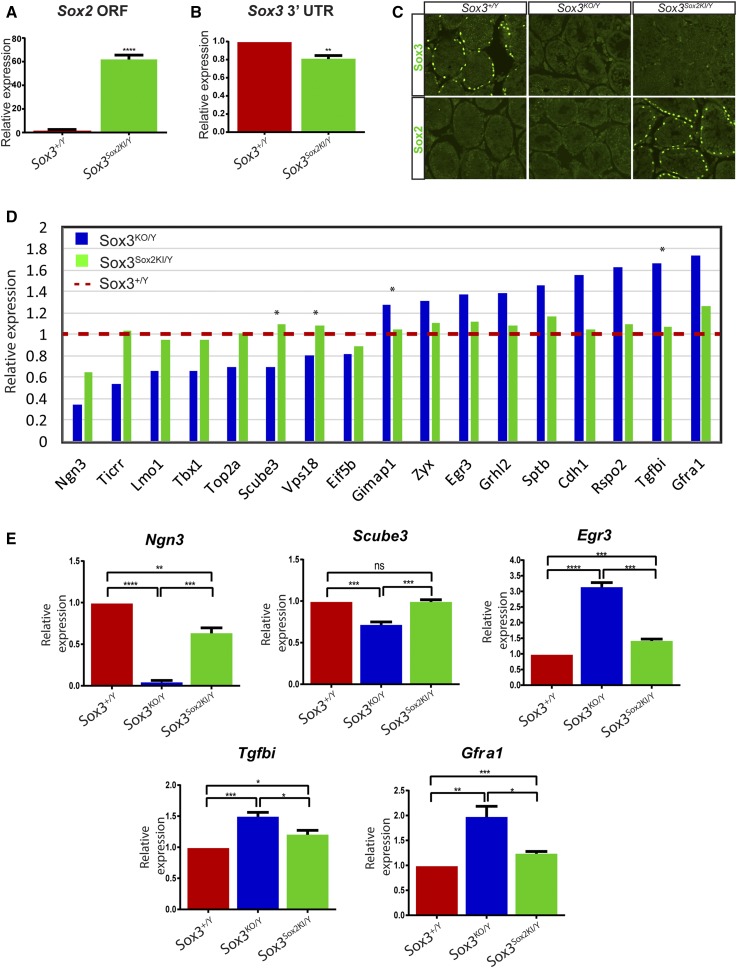
Figure 3.
SOX2 regulates SOX3 target genes in the mouse testes. (A) Sox2 qPCR analysis in 2 week Sox3+/Y and Sox3Sox2KI/Y testes [Student’s two tailed unpaired t-tests (****<0.0001)]. (B) qPCR analysis of Sox3 3′ UTR in 2 week Sox3+/Y and Sox3Sox2KI/Y testes [Student’s two-tailed unpaired t-tests (**<0.01)]. (C) SOX2 and SOX3 immunostaining on Sox3+/Y, Sox3KO/Y and Sox3Sox2KI/Y 4-week-old testes. (D) Microarray analysis was performed on 2 week testes from Sox3+/Y, Sox3KO/Y and Sox3Sox2KI/Y, and two-way comparisons were performed between Sox3+/Y and either Sox3Sox2KI/Y or Sox3KO/Y. Genes presented were significantly different between Sox3+/Y and Sox3KO/Y testes (Step-up P value <0.05). * <0.05 step-up P value between Sox3KO/Y and Sox3Sox2KI/Y. (E) qPCR validation of SOX2 regulation of SOX3 target genes in 2 week testes. ANOVA multiple comparisons with Bonferroni’s correction were performed using Graphpad Prism (****<0.0001, ***<0.001, **<0.01, *<0.05, ns, not significant).
Since SOX2 and SOX3 bind and regulate target genes via highly similar HMG-box DNA binding domain (Bergsland et al. 2011), we reasoned that SOX2 could be capable of regulating SOX3 target genes in the testes of Sox3Sox2KI/Y mice. To test this possibility, we performed microarray analysis comparing Sox3+/Y, Sox3KO/Y and Sox3Sox2KI/Y 2-week testes. A total of 17 genes (excluding Sox3) were significantly altered when comparing Sox3KO/Y to Sox3+/Y (step-up P value ≤0.05), of which nine were upregulated and eight were downregulated. These genes included Ngn3, which has previously been shown to be downregulated in Sox3 null testes (Raverot et al. 2005). Expression levels of all 17 genes were statistically returned to Sox3+/Y levels in Sox3Sox2KI/Y testes (Figure 3D), although only four were significantly different in Sox3KO/Y vs. Sox3Sox2KI/Y testes. As step-up P values are known to be a very strict measure of microarray changes, and often underestimate changes in order to avoid high false discovery rates (Benjamini and Hochberg 1995), we sought to validate the degree of expression rescue using qPCR on independent biological samples. Five of the 17 genes were selected for validation by qPCR, and fold changes were all in the same direction as the microarray data (Figure 3E). Notably, all five genes assessed by qPCR were statistically closer to Sox3+/Y levels in Sox3Sox2KI/Y in comparison to Sox3KO/Y, but, in four of five cases fell short of complete restoration, with only Scube3 showing no significant difference between Sox3+/Y and Sox3Sox2KI/Y. Collectively, this analysis indicates that ectopically expressed SOX2 is able to regulate SOX3 genes in the testes, and can largely restore normal gene expression patterns. However, a small subset of those genes most heavily deregulated by the loss of Sox3 fail to achieve complete rescue in Sox3Sox2K/Y mice, likely due to the slightly decreased levels of SOX2 expression in the testes.
Sox2 can functionally replace Sox3 in the testes
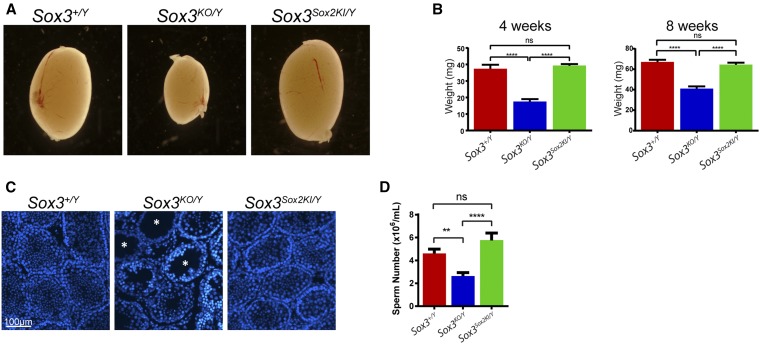
Given the high degree of expression restoration in Sox3Sox2KI/Y testes, we next sought to assess whether this correlated with functional rescue of testes size and sperm production, which has previously been reported for Sox3KO/Y animals (Rizzoti et al. 2004; Raverot et al. 2005). Sox3KO/Y mice exhibited significantly reduced testes size and weight at 4 and 8 weeks (Figure 4, A and B). Empty seminiferous tubules were observed in sections of 4-week-old Sox3KO/Y testes (Figure 4C), and Sox3KO/Y mice had a significantly reduced sperm count at 8 weeks of age (Figure 4D). In contrast, Sox3Sox2KI/Y males had normal testes size and sperm count with no evidence of empty tubules (Figure 4, A–D). Thus, testes morphology and function were completely rescued in Sox3Sox2KI/Y mice. These data provide unequivocal evidence of SoxB1 functional redundancy in vivo.
Figure 4.
SOX2 can functionally replace SOX3 in the testes. (A) Gross morphology of Sox3+/Y, Sox3KO/Y and Sox3Sox2KI/Y 4-week testes. (B) Quantification of testes weight at 4 and 8 weeks old [at least 18 testes were weighed for each genotype, and results were compared using one way ANOVA multiple comparisons with Bonferroni’s correction (****<0.0001, ns, not significant)]. (C) DAPI staining of Sox3+/Y, Sox3KO/Y and Sox3Sox2KI 4 week old testes. Asterisks indicate empty tubules. (D) Sperm counts from 8-week Sox3+/Y, Sox3KO/Y and Sox3Sox2KI/Y epididymis [at least 14 samples were counted for each genotype, and results were compared using one way ANOVA multiple comparisons with Bonferroni’s correction (****<0.0001, **<0.01 ns, not significant)].
Discussion
Gene swap experiments provide an ideal approach to investigate functional overlap of related genes under physiological conditions. To our knowledge, this is the first report describing the use of CRISPR/Cas9-assisted mutagenesis to perform an in vivo gene-swap of two closely related genes. By cutting either side of Sox3 in the presence of a Sox2-containing repair template, we were able to simultaneously remove Sox3 and insert the Sox2 ORF in its place. From a practical standpoint, this provides a feasible and rapid pathway to generate gene-swap mice. It should be noted that the efficiency of generation was lower than anticipated based on previous reports of CRISPR/Cas9-assisted insertional mutagenesis (Yang et al. 2013), with only a single Sox3Sox2KI founder generated from a total of 17 live born mice. Generation of null alleles was more efficient, with almost half of the founders having a large deletion extending between the two CRISPR cut sites. The reason for the low efficiency of Sox3Sox2KI alleles most likely reflects low efficiency of HDR in comparison to nonhomologous end joining, as noted by others (Yang et al. 2013). In the future, it will be interesting to test whether strategies to promote HDR, such as knockdown of the Ku protein, can be used to increase knock-in efficiency (Basu et al. 2015). Any future optimization should be sure to allow the coincidental production of KO alleles, as this enables simultaneous assessment of KI and KO phenotypes, and, as such, circumvents the significant confounding influence of genetic background. Indeed, we have illustrated the importance of this consideration with our description of a more severe pituitary phenotype in Sox3 null mice on a C57Bl/6 background in comparison with the previously published phenotype on a mixed genetic background (Rizzoti et al. 2004).
The testes provide a particularly tractable setting for this investigation, as SOX3 is the only SOXB1 protein to be expressed in this tissue, and its absence results in a relatively severe phenotype. Importantly, we have shown that the morphogenetic and spermatogenic defects that result from Sox3 deletion were completely rescued when Sox2 was ectopically expressed in its place. At the molecular level, these data indicate that ectopic SOX2 is able to bind and regulate SOX3 target genes in spermatogonial stem/progenitor cells. This finding is consistent with published ChIP-seq studies showing that the binding sites of these proteins overlap extensively in cultured neural progenitor cells (Bergsland et al. 2011). While a modest difference in spermatogonial marker expression remained in Sox3Sox2KI/Y testes, it seems likely this results from the slightly lower level of Sox3Sox2KI mRNA compared with wild type Sox3. However, potential differences in the affinity of SOX2 and SOX3 antibodies make it near impossible to directly compare levels of SOX3 and SOX2 protein in Sox3+/Y and Sox3Sox2KI/Y testes, respectively. Therefore, we cannot completely exclude the possibility that the residual differences in Sox3Sox2KI/Y testes are due to slightly different functions of SOX3 and SOX2 proteins. It is also conceivable that functional differences may be exposed under nonlaboratory conditions as observed in HoxA1/B1 gene swap mice (Ruff et al. 2015). Nevertheless, these gene swap data provide the most compelling evidence to date that SOX2 and SOX3 are functionally interchangeable proteins. Given the complete rescue of SOX3 morphological defects by SOX2, we predict that this functional redundancy should operate in both directions, such that SOX3 should be equally well suited to replacing SOX2 if expressed at the correct time and place. Thus, it would be interesting to perform the complementary gene swap to determine whether, for example, Sox3 expression from the Sox2 locus is capable of rescuing the early lethality phenotype of Sox2 null mice.
Unlike the testis, where a single SOXB1 protein (SOX3) is expressed in a small population of cells, all three SOXB1 proteins are expressed in neural progenitors across the developing CNS. Direct comparison of SOX3 and SOX2 expression in the embryonic brain has shown that virtually all SOX3+ cells also express SOX2 (Rogers et al. 2013; Cheah and Thomas 2015). Given our gene swap data showing SOX2/3 functional redundancy in the testes, as well as overexpression data indicating their functional equivalence (Bylund et al. 2003; Graham et al. 2003), it is not surprising that most of the CNS develops normally in Sox3 null mice. However, a notable exception is the developing ventral diencephalon where Sox3 loss-of-function results in CNS/pituitary defects in both mice and humans despite expression of Sox2. Here, we show that these Sox3 null defects are almost completely rescued by expression of Sox2 from the Sox3 locus, indicating that SOX2 and SOX3 proteins are functionally interchangeable in pituitary induction. It is likely that the residual minor clefting in the Sox3Sox2KI adult pituitary is due to a decreased level of Sox3Sox2KI mRNA, reinforcing that pituitary induction is extremely sensitive to Sox2/3 levels, which, in Sox3Sox2KI/Y brains, are reduced by only ∼16% compared to wild type (see File S1). Alternatively, these minor abnormalities may be due to a slight difference in the functionality of SOX3 and SOX2 in pituitary development.
In the mouse, only a handful of examples exist in which paralogous gene function has been examined using a KI gene swap approach. In these examples, the degree of functional equivalence has been complete (Otx1/2, En1/2, Osr1/2, and HoxA3/D3) (Hanks et al. 1995; Greer et al. 2000; Acampora et al. 2003; Gao et al. 2009), partial (Sox8/10 and HoxA1/B1) (Kellerer et al. 2006; Ruff et al. 2015), or limited (Phox2a/2b)(Coppola et al. 2005). Given the profound rescue of testes defects in Sox3Sox2KI mice, we suggest that SOX3 and SOX2 proteins are biochemically equivalent. The observed differences in their null mutant phenotypes therefore likely reflects unique zones of expression, presumably generated through evolutionary subfunctionalization. However, why two biochemically equivalent proteins have retained widespread overlapping expression within the developing brain across an extensive evolutionary period remains an intriguing question. This does not appear to be an accident of evolution, as ancestral SOX B proteins in Drosophila (SoxNeuro and Diachete) exhibit similar partial redundancy, and active conservation of binding sites that allow both paralogues to bind (Carl and Russell 2015). One explanation is that it may be difficult to fully disentangle regulatory elements that direct Sox3 expression in the ventral diencephalon from other neuroprogenitor zones. Further analysis of SoxB1 gene regulation, for example, though CRISPR/Cas9-mediated enhancer deletion, may reveal insights into this interesting biological phenomenon.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.202549/-/DC1.
Acknowledgments
We thank Robin Lovell-Badge for the Sox3 gene targeting plasmid, Todd Norton for technical assistance, Sandra Piltz for technical expertise in the generation of KI and KO mice, and Eileen McLaughlin for provision of the spermatogonia RNA. F.A. was supported by a scholarship from Beasiswa Unggulan DIKTI (Directorate General of Higher Education, Indonesian Government). Funding from the Australian Research Council supported this study. The authors declare no competing financial interests. All of the experiments involving animal use have been approved by the University of Adelaide Animal Ethics Committee. All studies were conducted in accordance with the principles of animal replacement and reduction and experimental refinement.
Author contribution: F.A., J.H. and P.T. conceived the project. F.A. performed all the experiments apart from the quantitative SOX2 immunofluorescence (performed by D.P.) and the SoxB1 qPCR comparison (performed by D.M.). J.H., P.T., and F.A. drafted the manuscript, which was reviewed and edited by all authors.
Footnotes
Communicating editor: T. R. Magnuson
Literature Cited
- Acampora D., Annino A., Puelles E., Alfano I., Tuorto F., et al. , 2003. OTX1 compensates for OTX2 requirement in regionalisation of anterior neuroectoderm. Gene Expr. Patterns GEP 3: 497–501. [DOI] [PubMed] [Google Scholar]
- Barbaric I., Miller G., Dear T. N., 2007. Appearances can be deceiving: phenotypes of knockout mice. Brief. Funct. Genomics Proteomics 6: 91–103. [DOI] [PubMed] [Google Scholar]
- Basu S., Aryan A., Overcash J. M., Samuel G. H., Anderson M. A. E., et al. , 2015. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc. Natl. Acad. Sci. USA 112: 4038–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57: 289–300. [Google Scholar]
- Bergsland M., Ramsköld D., Zaouter C., Klum S., Sandberg R., et al. , 2011. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 25: 2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B. G., Muhr J., 2003. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat. Neurosci. 6: 1162–1168. [DOI] [PubMed] [Google Scholar]
- Carl S. H., Russell S., 2015. Common binding by redundant group B Sox proteins is evolutionarily conserved in Drosophila. BMC Genomics 16: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah P.-S., Thomas P. Q., 2015. SOX3 expression in the glial system of the developing and adult mouse cerebellum. Springerplus 4: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola E., Pattyn A., Guthrie S. C., Goridis C., Studer M., 2005. Reciprocal gene replacements reveal unique functions for Phox2 genes during neural differentiation. EMBO J. 24: 4392–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., et al. , 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Lan Y., Ovitt C. E., Jiang R., 2009. Functional equivalence of the zinc finger transcription factors Osr1 and Osr2 in mouse development. Dev. Biol. 328: 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P., Pevny L., 2003. SOX2 functions to maintain neural progenitor identity. Neuron 39: 749–765. [DOI] [PubMed] [Google Scholar]
- Greer J. M., Puetz J., Thomas K. R., Capecchi M. R., 2000. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature 403: 661–665. [DOI] [PubMed] [Google Scholar]
- Hanks M., Wurst W., Anson-Cartwright L., Auerbach A. B., Joyner A. L., 1995. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science 269: 679–682. [DOI] [PubMed] [Google Scholar]
- Hughes J., Piltz S., Rogers N., McAninch D., Rowley L., et al. , 2013. Mechanistic insight into the pathology of polyalanine expansion disorders revealed by a mouse model for X linked hypopituitarism. PLoS Genet. 9: e1003290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelberman D., Rizzoti K., Avilion A., Bitner-Glindzicz M., Cianfarani S., et al. , 2006. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J. Clin. Invest. 116: 2442–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellerer S., Schreiner S., Stolt C. C., Scholz S., Bösl M. R., et al. , 2006. Replacement of the Sox10 transcription factor by Sox8 reveals incomplete functional equivalence. Development 133: 2875–2886. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., et al. , 2008. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26(1): 101–106. [DOI] [PubMed] [Google Scholar]
- Raverot G., Weiss J., Park S. Y., Hurley L., Jameson J. L., 2005. Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev. Biol. 283: 215–225. [DOI] [PubMed] [Google Scholar]
- Rizzoti K., Brunelli S., Carmignac D., Thomas P. Q., Robinson I. C., et al. , 2004. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat. Genet. 36: 247–255. [DOI] [PubMed] [Google Scholar]
- Rogers N., Cheah P.-S., Szarek E., Banerjee K., Schwartz J., et al. , 2013. Expression of the murine transcription factor SOX3 during embryonic and adult neurogenesis. Gene Expr. Patterns GEP 13: 240–248. [DOI] [PubMed] [Google Scholar]
- Ruff J. S., Saffarini R. B., Ramoz L. L., Morrison L. C., Baker S., et al. , 2015. Fitness assays reveal incomplete functional redundancy of the HoxA1 and HoxB1 paralogs of mice. Genetics 201: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A., 2005. Distributed robustness vs. redundancy as causes of mutational robustness. Bioessays 27: 176–188. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., et al. , 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H. B., Episkopou V., 1999. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 86: 197–201. [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., et al. , 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zevallos S. E., Rizzoti K., Jeong Y., Lovell-Badge R., et al. , 2012. Disruption of SoxB1-dependent Sonic hedgehog expression in the hypothalamus causes septo-optic dysplasia. Dev. Cell 22: 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data has been submitted to Gene Expression Omnibus (GEO) with accession number GSE96805. All other reagents can be made available upon request. Supporting data can be found in Supplemental Material, File S1.