Abstract
Perturbations in synaptic function could affect the normal behavior of an animal, making it important to understand the regulatory mechanisms of synaptic signaling. Previous work has shown that in Caenorhabditis elegans an immunoglobulin superfamily protein, RIG-3, functions in presynaptic neurons to maintain normal acetylcholine receptor levels at the neuromuscular junction (NMJ). In this study, we elucidate the molecular and functional mechanism of RIG-3. We demonstrate by genetic and BiFC (Bi-molecular Fluorescence Complementation) assays that presynaptic RIG-3 functions by directly interacting with the immunoglobulin domain of the nonconventional Wnt receptor, ROR receptor tyrosine kinase (RTK), CAM-1, which functions in postsynaptic body-wall muscles. This interaction in turn inhibits Wnt/LIN-44 signaling through the ROR/CAM-1 receptor, and allows for maintenance of normal acetylcholine receptor, AChR/ACR-16, levels at the neuromuscular synapse. Further, this work reveals that RIG-3 and ROR/CAM-1 function through the β-catenin/HMP-2 at the NMJ. Taken together, our results demonstrate that RIG-3 functions as an inhibitory molecule of the Wnt/LIN-44 signaling pathway through the RTK, CAM-1.
Keywords: C. elegans, neuromuscular junction, Wnt, RIG-3, immunoglobulin domain
WNTS are lipid-modified glycoproteins that play a role in a number of biological processes including embryonic development, cellular differentiation, and tissue homeostasis, along with neuronal and synapse development (reviewed in Chien et al. 2009; Mulligan and Cheyette 2012). Since Wnts play a key role in various physiological and developmental processes in most organisms, deregulation of the Wnt signaling pathway could lead to developmental defects, neurodegenerative diseases, and cancer (Clevers and Nusse 2012; Inestrosa et al. 2012; Hikasa and Sokol 2013; Oliva et al. 2013a; Chiurillo 2015). Although Wnt/s have been implicated in various developmental processes, it has only been shown recently that the Wnt signaling pathway is also required for normal circuit formation, synapse development, maintenance, and functioning, and is involved in aspects of learning and memory (reviewed in Salinas and Zou 2008; Korkut and Budnik 2009; Budnik and Salinas 2011; Maguschak and Ressler (2011); Henriquez and Salinas 2012; Jensen et al. 2012a; Park and Shen 2012; Dickins and Salinas 2013; Oliva et al. 2013a).
Given the importance of Wnt functioning in an organism, it is critical to fully understand the regulatory mechanisms involved in Wnt signaling. In this study, we show that an Immunoglobulin Superfamily molecule, RIG-3, functions to modulate the Wnt signaling pathway at the Caenorhabditis elegans neuromuscular junction (NMJ).
RIG-3 contains three Ig domains and a C-terminal GPI an-chor, and its expression is restricted to the nervous system. Previous work has shown that in C. elegans, RIG-3 is not required for normal nervous system development, but affects synaptic function in the presence of increased activity (Schwarz et al. 2009; Babu et al. 2011). It has been previously shown that mutants in rig-3 are hypersensitive to the acetylcholine esterase inhibitor, aldicarb. This hypersensitivity is brought about by the increased levels of functional acetylcholine receptor, ACR-16, at the postsynaptic site in mutants treated with aldicarb. In the absence of aldicarb, rig-3 mutants do not show an increase in synaptic AChR/ACR-16, indicating that the mutants show increased potentiation only when exposed to increased activity that is brought about by aldicarb treatment (Babu et al. 2011).
CAM-1 is a Ror receptor tyrosine kinase (RTK) that is expressed in the ventral nerve cord and muscles, and is particularly localized to the neuromuscular synapses (Francis et al. 2005). A number of previous studies have implicated CAM-1 as a nonconventional Wnt receptor that is required for normal WNT-dependent nervous system functions (Kim and Forrester 2003; Forrester et al. 2004; Green et al. 2007, 2008; Zinovyeva et al. 2008; Hayashi et al. 2009; Kennerdell et al. 2009; Song et al. 2010; Modzelewska et al. 2013; Chien et al. 2015). A more recent study by Jensen et al. (2012b) has shown that CAM-1 functions through the WNT signaling pathway, and, more specifically, through the Wnt, CWN-2, and its Frizzled receptor (LIN-17) to regulate postsynaptic receptor levels at the C. elegans NMJ. Previous work has also shown that RIG-3 and CAM-1 could work together to regulate signaling at the NMJ. Moreover, RIG-3 functions through CAM-1 to maintain normal levels of the Acetylcholine Receptor (AChR), ACR-16 at the C. elegans NMJ, in the presence of increased presynaptic activity, brought about by exposure to the acetylcholine esterase inhibitor, aldicarb (Babu et al. 2011).
In this study we show that presynaptic RIG-3 interacts directly with postsynaptic CAM-1 across the NMJ. CAM-1 has been previously shown to act as a nonconventional receptor for Wnt ligands (Green et al. 2008), indicating that the effects of RIG-3 on synaptic function could be a result of changes in Wnt signaling at the NMJ. Our work provides genetic evidence to show that CAM-1 functions as a receptor for the Wnt ligand, LIN-44. We further confirm and extend these results by showing that RIG-3 functions through both Wnt/LIN-44 and CAM-1. We have also characterized the downstream signaling components of the RIG-3 signaling pathway, and have identified the β-Catenin, HMP-2, as a possible effector of RIG-3 signaling that allows for regulation of normal AChR/ACR-16 levels at the NMJ.
Materials and Methods
Strains
All the strains used were maintained at 20° as previously described (Brenner 1974). Feeding of C. elegans was done using the Escherichia coli strain, OP50. The wild-type strain used was the Bristol N2 strain.
The mutant strains used in this study were: rig-3(ok2156), cam-1(ak37), lin-44(n1792), and hmp-2(qm39). A brief description of these alleles is given in Supplemental Material, Table S1 in File S1.
Constructs and transgenes
All the constructs were generated in pPD49.26 or pPD95.75 (Addgene). A description of the plasmids used in this study, along with the primers used, and the sequence of the constructs used for the BiFC study, are given in File S1. Briefly, The CAM-1 and LIN-44 rescue or expression plasmids used in this study are: Pmyo-3::CAM-1(FL)::GFP (BAB#0013), Pmyo-3::CAM-1(ΔIg)::GFP (BAB#014), Pmyo-3::CAM-1(FL) (BAB#038), Pmyo-3::CAM-1(ΔIg) (BAB#0039), Punc-17::LIN-44 (BAB#0021), and Punc-129::LIN-44::mCherry (BAB#0023). To obtain the CAM-1(FL) constructs, the cam-1a isoform (sequence from www.wormbase.org) was cloned under the myo-3 promoter. In order to generate the CAM-1(ΔIg) construct, amino acids (aa) 55–111 were deleted from CAM-1(FL). To achieve this, N-terminal (aa 1–55) and C-terminal (aa 111 until the stop codon) regions were separately amplified, and the fragment with the required deletion was obtained by overlap extension PCR, and then cloned. The Wnt/LIN-44 construct was engineered by cloning the Wnt/LIN-44 full-length cDNA into pPD49.26 having either the unc-17 or the unc-129 promoter in the construct. In order to design the Punc-129::LIN-44::mCherry construct, mCherry was cloned in the KpnI site downstream of Wnt/LIN-44. All constructs were confirmed for their sequence and orientation by sequencing, and injected into C. elegans as arrays. The rig-3::mCherry construct that was used for the rescue experiments and the localization of RIG-3, the Punc-17::RFP (nuIs321), and the Pmyo-3::AChR/ACR-16::GFP (nuIs299) lines were as previously described in Babu et al. (2011). The Pcam-1::CAM-1::GFP (cwIs6) line was previously used in Kim and Forrester (2003). All the constructs, transgenes, and lines used in this study are listed in Tables S2–S4 in File S1.
Behavioral assays
Aldicarb assay:
The aldicarb assay was performed as previously described with minor modifications (Harada et al. 1994; Miller et al. 1996; Nurrish et al. 1999). Briefly, 1 mM aldicarb (Sigma Aldrich; Cat. # 33386) plates were prepared and allowed to dry at room temperature (RT); 20–25 young adult C. elegans were picked and placed on an aldicarb plate, the animals were prodded every 10 min with a toothpick to test for paralysis. Animals showing <2 body bends immediately after prodding were considered as paralyzed. The assay was done in triplicate and repeated three to four times. A graph was plotted with the average percentage of C. elegans paralyzed at 80 min.
The rate of aldicarb-based paralysis was found to vary in a couple of the experiments, although the concentration of aldicarb used in all the experiments was the same. This variation is likely because of the use of different batches of aldicarb for these sets of experiments. However, this does not affect the overall outcome of the result drawn from these assays as the change in rate of paralysis seen in these assays was consistent for all the strains used in these assays.
Fluorescence microscopy and quantitative analysis
The cam-1::GFP and rig-3::mCherry localization experiments were done using a Leica TCS SP8 confocal microscope. All other imaging was done using a Zeiss fluorescence microscope Axio Imager Z2 with an Axiocam MRm camera. In order to visualize C. elegans, they were immobilized with 30 mg/ml BDM on 2% agarose pads as previously described (Sieburth et al. 2005). Imaging after aldicarb treatment was done with untreated animals, or after an 80-min exposure to 1 mM aldicarb. The ACR-16::GFP imaging was done along the VNC just in front of the vulva of young adult animals. Imaging was done with untreated animals or after an 80-min exposure to 1 mM aldicarb. Images were analyzed using Image J and Zen software. The fluorescence intensity obtained from 25–35 animals was averaged, and was used to plot the graph. Similarly for the Split GFP/BiFC experiments, fluorescence was quantitated along the dorsal cord of the animals, and the different strains generated were compared. Coelomocyte imaging was done in the mid and posterior coelomocytes that were imaged in adult animals. To quantitate the intensity of LIN-44::mCherry, the maximum intensity stack was utilized, and equal size regions of interest (ROI) were drawn along the coelomocytes; the average intensity obtained from each such ROI was used for analysis.
BiFC/ split GFP
RIG-3::spYFP (RIG-3::VN173), RIG-3(TMD)::spYFP (RIG-3(TMD)::VN173), CAM-1(FL)::spYFP (CAM-1(FL)::VC155), CAM-1(ΔIg)::spYFP (CAM-1(ΔIg)::VC155), and CAM-1(ΔKR, ΔCRD)+2Ig::spYFP (CAM-1(ΔKR, ΔCRD)+2Ig::VC155), were generated by custom gene synthesis (Thermofisher GENEART). The RIG-3::VN173 and RIG-3(TMD)::VN173 transgenes were cloned into NheI and KpnI sites in pPD49.26 under unc-17 or unc-129 promoters. The CAM-1(FL)::VC155, CAM-1(ΔIg)::VC155 and CAM-1(ΔKR, ΔCRD)+2Ig::VC155 inserts were cloned under myo-3 promoter in pPD49.26 by utilizing XbaI and KpnI sites. The VN173 and VC155 sequences were as described previously (Shyu et al. 2008). Linkers were inserted between the split fluorescent tag and the gene as previously described (Hiatt et al. 2008). The sequences used for this experiment are listed in File S1. For control experiments, CAM-1(FL)::VC155 and CAM-1(ΔIg)::VC155 constructs were injected at 20 μg into N2 animals, and the RIG-3::VN173 construct was injected into the rig-3 mutant C. elegans. The injection marker used was pCFJ90 (Pmyo-2::mCherry). The experimental protocol involved coinjection of 20 μg of RIG-3::VN173 with 20 μg of either CAM-1(FL)::VC155 or CAM-1(ΔIg)::VC155 in order to identify the interactions between RIG-3 and CAM-1. The presence of transgenes in the C. elegans was confirmed by genotyping. The interaction between RIG-3 and CAM-1 was observed by fluorescent imaging.
Immunoprecipitation
To perform immunoprecipitation experiments, CAM-1::GFP expressing C. elegans were grown on 20 plates 140 mm in diameter. The animals were collected using M9 buffer (∼5 ml) into 2 ml tubes. The C. elegans were allowed to settle, and the supernatant was removed; this washing process was repeated three more times. Finally, the C. elegans were suspended in a 2× volume of lysis buffer (B-PER with Enzymes Bacterial Protein Extraction Kit; Pierce# 90078) with protease inhibitors (Protease inhibitor cocktail setI -539131-1VLCN; Merck Millipore) and 1 mM PMSF. C. elegans were lysed by sonication; four bursts of (8–10 W) for 5 sec each, centrifuged at 14,000 rpm for 5 min, and the supernatant was taken to perform the pull-down assay. A 5 mg sample of protein was taken in a 500 μl volume of lysis buffer; 4 μl GFP antibody (EMD Millipore # MAB3580 (IF)) was added and incubated at 4° for 1 hr. Then, 50 μl of protein preequilibrated A/G beads (MERCK Millipore #16–125) were added to the lysate, and incubated further another 3 hr. Next, we gave five, 5-min washes with 1 ml ice-cold lysis buffer. The protein was then eluted in 100 μl of sample buffer by boiling for 5 min (Chan et al. 2015). Western blotting was performed by using 1:2000 dilution of affinity purified (Protein A beads) rabbit RIG-3 Polyclonal antibodies (raised through Merck, see File S1). We used anti-rabbit secondary Ab (1:2000; goat anti-rabbit, IgG-AP # sc-2007, Santa Cruz) to probe the western blot. Protein detection was done with Pierce Alkaline phosphatase substrate (1-Step NBT/BCIP Solution# 34042).
Statistical analysis
To perform statistical analysis, Excel 2010 with Analysis Toolpak (Microsoft, Seattle, WA) was used. Averages, percentages, and SEM were calculated from data generated after performing assays or quantitation. P values were identified by one-way or two-way ANOVA analysis across test populations, followed by post hoc analysis using two-tailed Student’s t-test with unequal variance.
Data availability
Strains and plasmids are available upon request. Tables S3 and S4 in File S1 contain all the plasmids and strains that have been used in this study.
Results
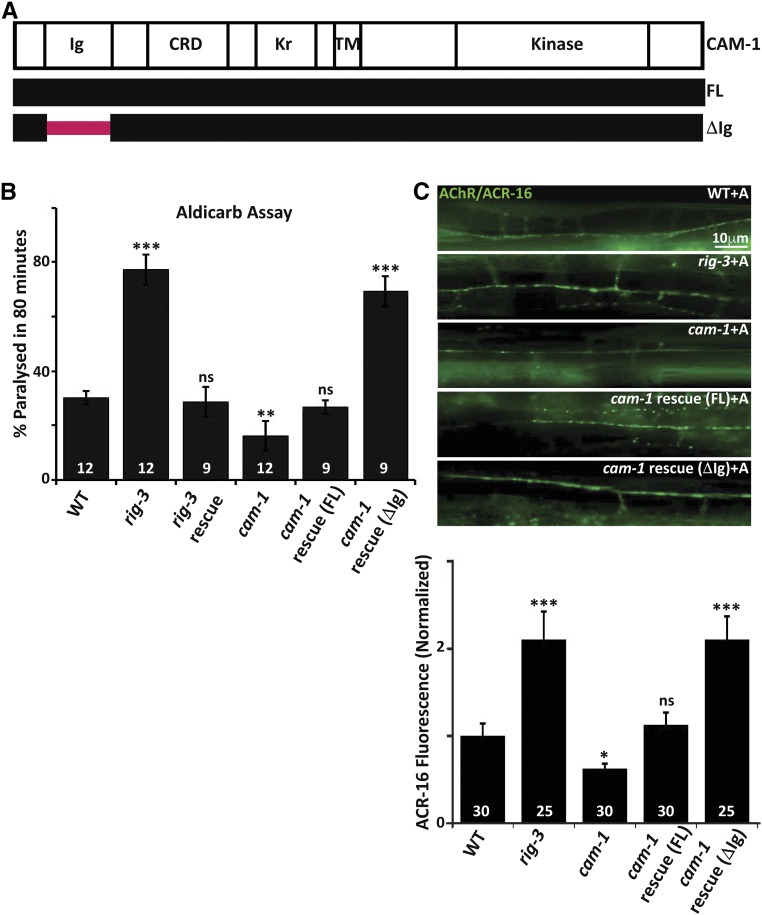
Overexpression of CAM-1 without the Ig domain (ΔIg) generates a phenotype similar to rig-3 mutants
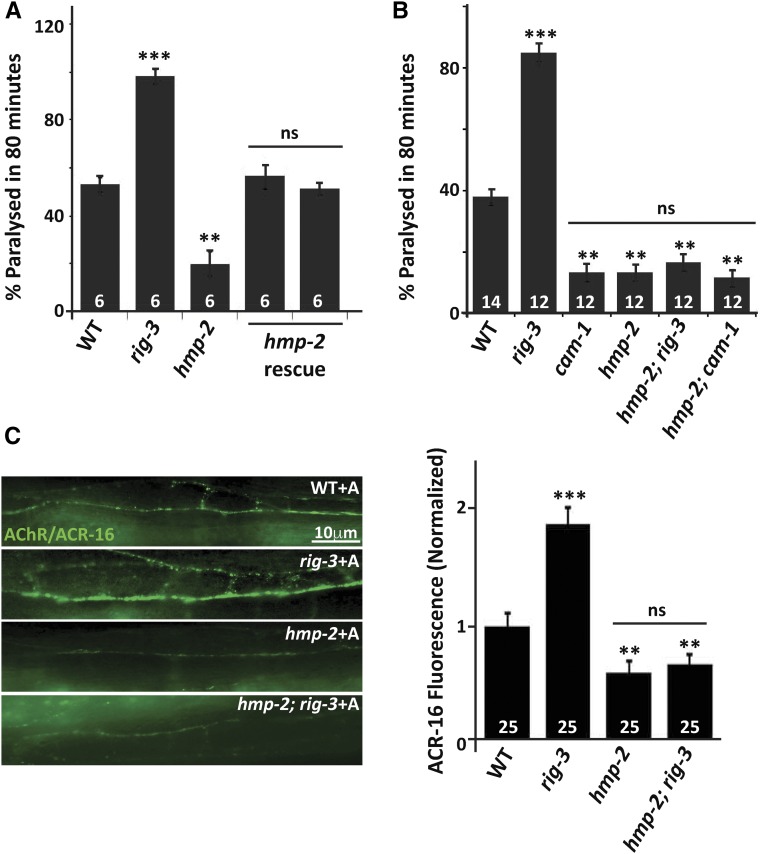
Previously published data have shown that RIG-3 genetically interacts with the Ror tyrosine kinase, CAM-1, the first evidence being the result that increased postsynaptic AChR/ACR-16 receptor levels, and the increased postsynaptic current phenotypes seen in rig-3 mutants treated with the cholinesterase inhibitor aldicarb, were completely suppressed by mutations in cam-1. The second is the result that rig-3 mutants show increased CAM-1::GFP punctate fluorescence at the NMJ (Babu et al. 2011). CAM-1 has an intracellular kinase domain and an extracellular region consisting of a Kringle domain, a Cysteine Rich Domain (CRD), and an Ig domain (Green et al. 2007; schematized in Figure 1A). A previous study in C. elegans has shown that the function of CAM-1 at the NMJ is largely dependent upon its extracellular domain (ECD) (Jensen et al. 2012b). Further, various studies have indicated the importance of Ig domains in protein–protein interactions, and have shown that these domains interact with each other in a homophilic or heterophilic manner (reviewed in Bork et al. 1994; Halaby and Mornon 1998; de Wit and Ghosh 2016).
Figure 1.
RIG-3 functions through the Ig domain of CAM-1. (A) Schematic of CAM-1: the panel indicates schematic representations of CAM-1 domains for Full Length (FL) CAM-1, and the CAM-1 coding sequence lacking the Immunoglobulin domain, i.e., CAM-1(ΔIg). CRD, Cysteine Rich Domain; Kr, Kringle domain; TM, Transmembrane domain. (B) Aldicarb assays of rig-3 and cam-1 mutants and their rescue lines: this panel indicates a bar- graph showing the percentage of C. elegans paralyzed on aldicarb at the end of 80 min. In all figures, mutant and rescue lines were tested against control WT animals for significant differences. * P < 0.05, ** P < 0.01, and *** P < 0.001. The numbers at the base of the bars for all figures showing aldicarb assays indicates number of times the assay was performed with 15–25 animals used for each trial. (C) Quantitation of AChR/ACR-16 in rig-3 and cam-1 mutants along with the CAM-1 rescue lines: the top portion of the panel shows images of AChR/ACR-16 tagged with GFP in control WT, rig-3, and cam-1 mutants, and CAM-1(FL) and CAM-1(ΔIg) rescue lines. The lower part of the panel shows quantitation of AChR/ACR-16 levels along the Ventral Nerve Cord (VNC). The numbers analyzed for each genotype are indicated. In all figures “+A” indicates “+Aldicarb,” where 1 mM aldicarb treatment was done for 80 min unless otherwise mentioned. In (B and C), the CAM-1 rescue experiments were done by expressing CAM-1 specifically in the body-wall muscle with the myo-3 promoter. In (B) and (C), RIG-3 rescue was done by expressing RIG-3 under it’s own promoter, which is expressed in cholinergic motor neurons. Error bars indicate SEM in all panels.
Since RIG-3 is localized extracellularly on presynaptic neurons (through a GPI anchor) (Babu et al. 2011), and CAM-1 is a transmembrane domain protein that functions postsynaptically in muscles (Francis et al. 2005; Jensen et al. 2012b), we reasoned that the extracellular Ig domain of CAM-1 might be showing an association with the Ig domains of RIG-3. If this was the case, deletion of the Ig domain (∆Ig) in CAM-1 (schematized in Figure 1A) could lead to loss of association between RIG-3 and CAM-1, and the cam-1 mutants expressing CAM-1(∆Ig) would behave like rig-3 mutants. We evaluated this hypothesis by rescuing the cam-1 mutant phenotypes using full length (FL) CAM-1 and the CAM-1(∆Ig) constructs, both expressed in the body-wall muscles of the animal. Previous work has shown that cam-1 mutants are resistant to aldicarb as they have less AChR/ACR-16 expressed at the neuromuscular synapse; conversely, rig-3 mutants are hypersensitive to aldicarb, as they show an increase in AChR/ACR-16 levels at the NMJ in the presence of aldicarb (Francis et al. 2005; Babu et al. 2011). Upon transforming the cam-1 mutants with CAM-1(FL), we were able to rescue the aldicarb-resistant phenotype of the mutant animals (Figure 1B), the rescue with CAM-1(∆Ig), however, showed hypersensitivity to aldicarb that was similar to that seen in rig-3 mutants (Figure 1B). The expression of the CAM-1 rescue lines was quantified by RT-PCR analysis, and found to be approximately the same in both cases (Figure S1A in File S1). In order to test if expressing CAM-1(ΔIg) would cause a change in the abundance of CAM-1 itself, we assayed for CAM-1::GFP levels upon expression of CAM-1(∆Ig), and found no change in CAM-1::GFP levels in these animals when compared to control C. elegans (Figure S1B in File S1).
The phenotype seen in rig-3 mutants was completely rescued by expressing a genomic fragment of rig-3 tagged with mCherry, which has previously been used to rescue the rig-3 hypersensitivity to aldicarb phenotype (Babu et al. 2011).
Since aldicarb affects the acetylcholine levels at the NMJ, and it was previously found that RIG-3 affects the levels of AChR/ACR-16 at the NMJ in an aldicarb-dependent manner (Babu et al. 2011), we tested the effect of these mutants on synaptic AChR/ACR-16 in the presence of aldicarb using a previously described ACR-16::GFP line (Babu et al. 2011). We found that the CAM-1(FL) rescued the decreased AChR/ACR-16 levels at the synapse that is seen in cam-1 mutants, while CAM-1(∆Ig) caused the cam-1 mutant animals to show increased synaptic AChR/ACR-16 upon aldicarb treatment, similar to what is seen in rig-3 mutants (Figure 1C).
RIG-3 and CAM-1 interact directly across the NMJ
To further analyze the association between RIG-3 and CAM-1 at the NMJ, we analyzed the localization pattern of RIG-3 and CAM-1 at the NMJ using previously described rig-3::mCherry and cam-1::GFP lines, both of which are genomically tagged genes with their own promoters (Forrester et al. 1999; Babu et al. 2011). Since CAM-1 and RIG-3 are likely to be functioning at the NMJ, we analyzed their localization pattern at the NMJ. We found that RIG-3 was localized to the presynaptic regions of the NMJ, whereas CAM-1 was expressed in the corresponding postsynaptic region (muscle) (Figure S2A in File S1). However, RIG-3 expression was diffuse; this could be due to the fact that RIG-3 is present both at the presynaptic terminal and is also secreted, as has been shown previously (Babu et al. 2011). CAM-1 expression was also found in the regions where muscle arms make contact with the ventral cord. These results further strengthened our hypothesis that there could be an association between RIG-3 and CAM-1 at the NMJ.
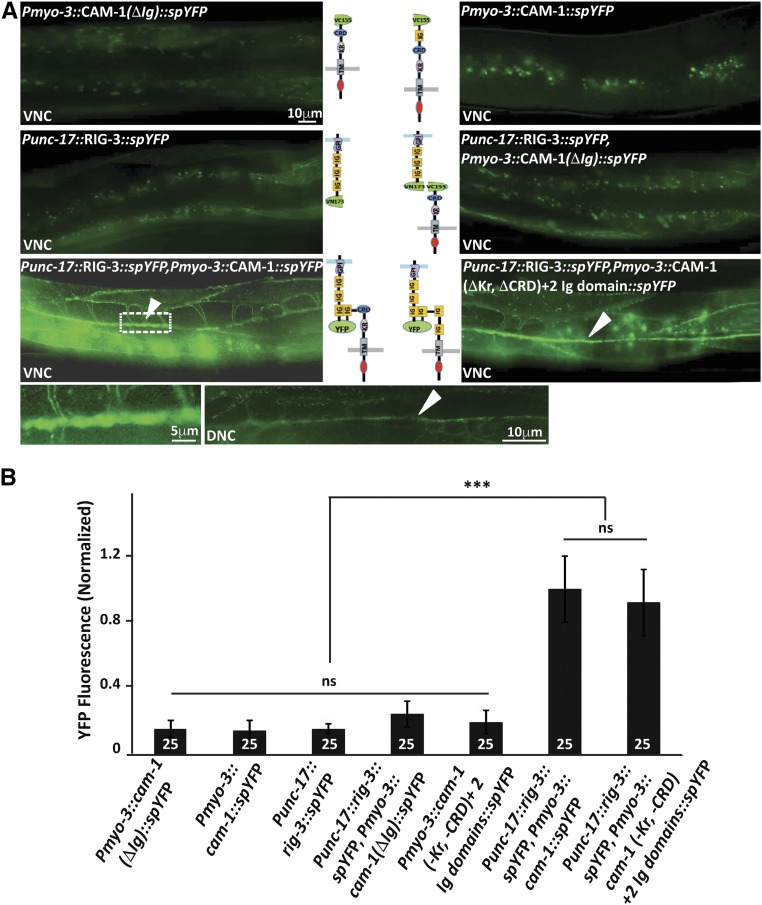
To further confirm that RIG-3 and CAM-1 interact directly across the NMJ, we performed the previously described method of split GFP/YFP or bimolecular fluorescence complementation (BiFC) (Hiatt et al. 2008; Shyu et al. 2008). The transgenic constructs with split variants of YFP were designed for RIG-3, CAM-1(FL), and CAM-1(ΔIg). The signal sequences were identified for the proteins, and the split variant fragments of venus yellow fluorescent protein (YFP) were fused along with linkers to obtain RIG-3::spYFP (RIG-3::VN173), CAM-1(FL)::spYFP (CAM-1(FL)::VC155), and CAM-1ΔIg::spYFP (CAM-1(ΔIg)::VC155). The RIG-3 transgene derived expression from the presynaptic unc-17 promoter while the CAM-1 transgenes were cloned under the body-wall muscle, myo-3, promoter. The split YFP tags were extracellular for all the transgenes. All the transgenes were tested for functionality by injecting into rig-3 and cam-1 mutants, and performing aldicarb rescue assays (Figure S2, B and C in File S1). The RIG-3::spYFP construct rescued the aldicarb hypersensitivity seen in rig-3 mutants, while the CAM-1(FL)::spYFP could also rescue the aldicarb resistant phenotype seen in the cam-1 mutants. Further, the CAM-1(ΔIg)::spYFP construct caused the cam-1 mutants to mimic the rig-3 hypersensitive phenotype that was previously seen (Figure 1B and Figure S2C in File S1). The above constructs were then injected individually, or CAM-1 constructs were coinjected with RIG-3. Upon analysis, we observed that the RIG-3 and CAM-1(FL and ΔIg) by themselves exhibited a basal level of fluorescence, whereas the coinjected RIG-3 and CAM-1(FL) fusion proteins showed a marked increase in the fluorescence at the NMJ (Figure 2, B and C, arrowhead in Figure 2B). Since RIG-3 is driven using the unc-17 promoter, which shows expression in both the dorsal DA, DB and ventral VA, VB motor neurons, we expected that YFP reconstitution would occur in both the ventral and dorsal cord, which is what we see, as depicted in Figure 2A. However, coinjection of RIG-3 and CAM-1(ΔIg) did not show the increased fluorescence seen with RIG-3 and CAM-1(FL) (Figure 2, A and B). However, we could not rule out the possibility that deleting the Ig domain from CAM-1 would cause the protein to be of a smaller size, which could in fact be the reason for loss of interaction between CAM-1 and RIG-3. In order to address this issue, we replaced the Ig domain of CAM-1 with a nonfunctional domain. This construct was also unable to reconstitute YFP with RIG-3, indicating that the Ig domain in CAM-1 could be necessary for the interaction between RIG-3 and CAM-1 (Figure S2D in File S1; panel 1). Our results so far indicate that the Ig domain of CAM-1 is necessary for interaction between RIG-3 and CAM-1. In order to see if any of the other extracellular domains of CAM-1 are also required for this interaction, we deleted the CRD and Kr domains individually in the CAM-1(FL) gene, and still observed interaction between RIG-3 and CAM-1 (Figure S2C in File S1; panels 2 and 3). Further, when we deleted both Kr and CRD domains from CAM-1 and replaced them with the Ig domain of CAM-1, we again found reconstitution of YFP at the NMJ of these animals (Figure 2, A and B). Taken together, these experiments show that the Ig domain of CAM-1 is necessary and sufficient for interaction with RIG-3. Previous work has shown that RIG-3 is both anchored to the cell membrane and is a secreted molecule (Babu et al. 2011), the secreted RIG-3 would also be expected to interact with CAM-1 in the muscle. We detected fluorescence along the muscle arms in the reconstituted BiFC line (Figure 2A), indicating that the secreted RIG-3 also interacted with CAM-1. As a control, the RIG-3 region required for attachment of the GPI anchor was replaced with the transmembrane domain of NLG-1 in the RIG-3(TMD) construct. This construct also showed reconstitution of YFP with CAM-1; however, the muscle fluorescence was reduced in this line as indicated in Figure S2D in File S1; panel 4. Our results show that RIG-3 tethered to the membrane can interact with CAM-1, and that this interaction is more specific to the NMJ, as less RIG-3 is secreted in this line, as has been shown previously (Babu et al. 2011).
Figure 2.
RIG-3 directly interacts with CAM-1. (A) Bimolecular fluorescence complementation (BiFC) analysis of CAM-1 and RIG-3 interaction at the NMJ: the figure shows representative fluorescent images (YFP) of the C. elegans Dorsal Nerve Cord (DNC) expressing the transgenic protein indicated. The top four panels display the controls showing basal YFP levels with Pmyo-3::CAM-1(ΔIg)::VC155, Pmyo-3::CAM-1(FL)::VC155, Punc-17::RIG-3::VN173, and Punc-17::RIG-3::VN173 coinjected with Pmyo-3::CAM-1(ΔIg)::VC155. The lower two panels that are adjacent to the illustrations of the constructs used in the BiFC experiments show the image where Punc-17::RIG-3::VN173 is coinjected with Pmyo-3::CAM-1(FL)::VC155 and Punc-17::RIG-3::VN173 is coinjected with Pmyo-3::CAM-1(ΔKr, ΔCRD)+ 2Ig::VC155 respectively. A clear increase over the basal levels of YFP is seen in these panels. The image to the bottom right of the figure is a small area along the VNC that has been zoomed into from the image on the left (dotted box). The image next to it shows expression of YFP along the DNC in the same animals that show YFP expression in the VNC, this was seen in all eight animals that were assayed to look at the DNC. VNC indicates ventral nerve cord, and DNC indicates dorsal nerve cord, respectively. CAM-1 was expressed in the body-wall muscle with the myo-3 promoter, while RIG-3 was expressed in cholinergic neurons using the unc-17 promoter. (B) Graph of the bimolecular fluorescence images: In order to quantify the YFP fluorescence levels, 25 animals for each genotype where imaged, and their fluorescence along the ventral nerve cord quantified and plotted in the graph. The fluorescence intensity was normalized against C. elegans coexpressing Punc-17::RIG-3::VN173 and Pmyo-3::CAM-1(FL)::VC155. Error bars indicate SEM.
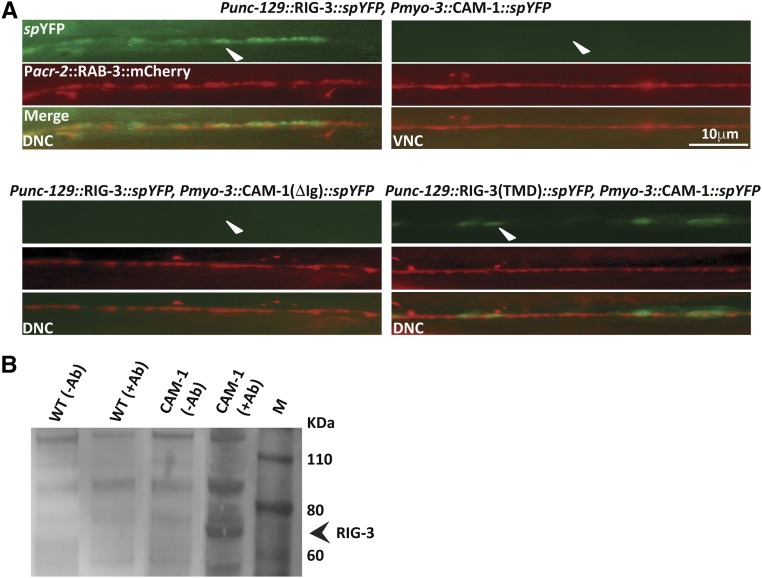
Since the unc-17 promoter allows for expression in both the DA and VA neurons among others, while the unc-129 promoter shows a more restricted expression in DA neurons (Rand 2007), we hypothesized that expressing the RIG-3::spYFP construct under the unc-129 promoter would show YFP reconstitution in the dorsal cord, and not the ventral nerve cord. We performed this experiment in the background of Pacr-2:: mCherry::RAB-3, which shows expression in both the ventral and dorsal cords. Our results showed that YFP expression was indeed seen only along the dorsal cord upon expressing Punc-129::RIG-3::spYFP and Pmyo-3::CAM-1::spYFP in the same animals (Figure 3A, top panels). Further, similar to the previous experiment, where we saw a more compact (less diffuse) expression of YFP in the dorsal cord, here, too, YFP expression was more restricted. The control Pmyo-3::CAM-1(ΔIg)::spYFP construct along with RIG-3::spYFP did not show any YFP reconstitution (Figure 3A). We also performed a similar experiment with the RIG-3 (TMD)::spYFP construct expressed under the unc-129 promoter, coinjected with Pmyo-3::CAM-1::spYFP, and again found YFP reconstitution only in the dorsal cord and not the ventral cord (Figure 3A, lower right panels).
Figure 3.
Interaction between RIG-3 and CAM-1. (A) YFP reconstitution across the dorsal cord in the background of Pacr-2::mCherry::RAB-3 that shows expression along both the cords: RIG-3 expressed under the unc-129 promoter interacts with CAM-1 expressed in body-wall muscle (myo-3 promoter) along the dorsal cord, but not along the ventral cord (top rows; arrowheads). The lower left set of images show the control, where RIG-3 does not interact with CAM-1(ΔIg). RIG-3(TMD) also shows interaction with CAM-1 along the dorsal cord (lower right panels, arrowhead). (B) Coimmunoprecipitation experiment: the panel shows the coimmunoprecipitation (Co-IP) blot. Protein extracts isolated from WT and CAM-1::GFP animals were allowed to interact with GFP antibody and pulled down using ProteinA/G beads. As a control, the same protein extracts were allowed to bind to beads but without GFP antibody. The coimmunoprecipitated complex was probed with the anti-RIG-3 antibody. The RIG-3 containing complex was pulled down only in the lane containing the CAM-1::GFP animals, and not in any of the other three control lanes (arrowhead). Co-IP analysis indicates interaction between CAM-1 and RIG-3.
Finally, we complemented this data with in vitro pull-down experiments, where we used the GFP antibody to pull down the CAM-1::GFP interacting complex. As a control, protein from WT C. elegans was allowed to interact with the GFP Ab. Upon blotting with an antibody against the RIG-3 protein (Figure S3 in File S1), we found that RIG-3 was pulled down with CAM-1::GFP complex, whereas, in the case of WT samples, no band was found corresponding to the RIG-3 protein. A similar result was seen with controls where the GFP antibody was not added (Figure 3B). Taken together, these results show that RIG-3 interacts with CAM-1 at the NMJ.
RIG-3 regulates Wnt/LIN-44 signaling through CAM-1
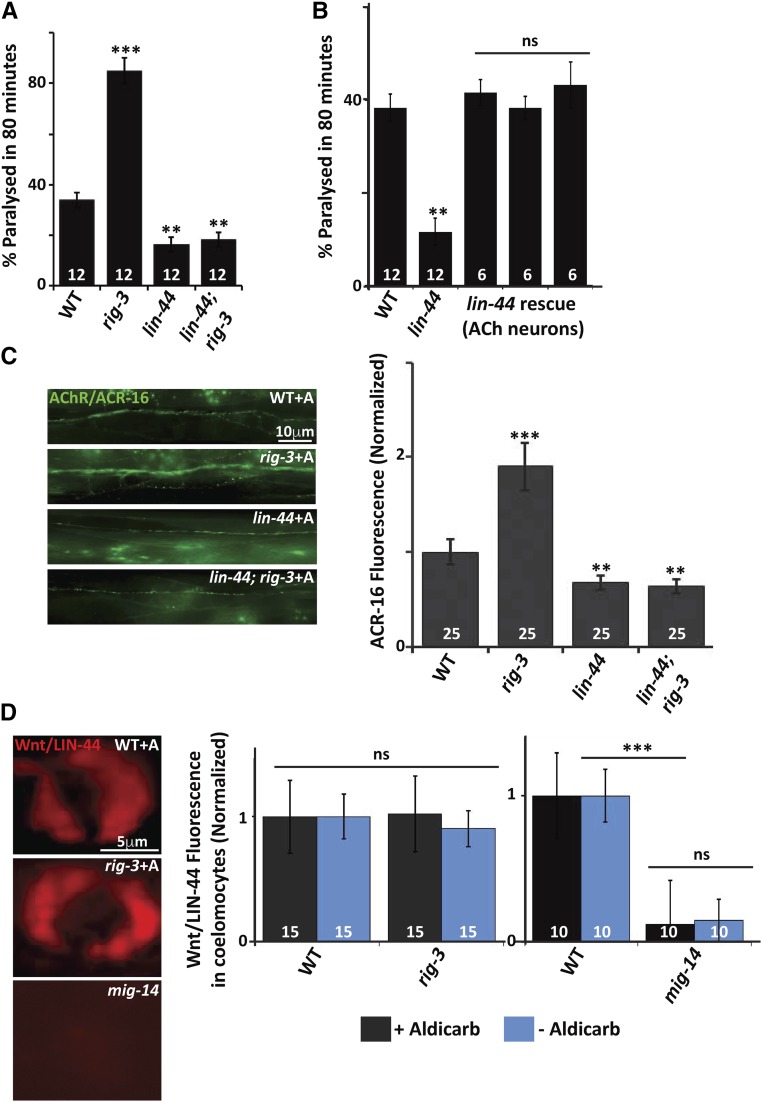
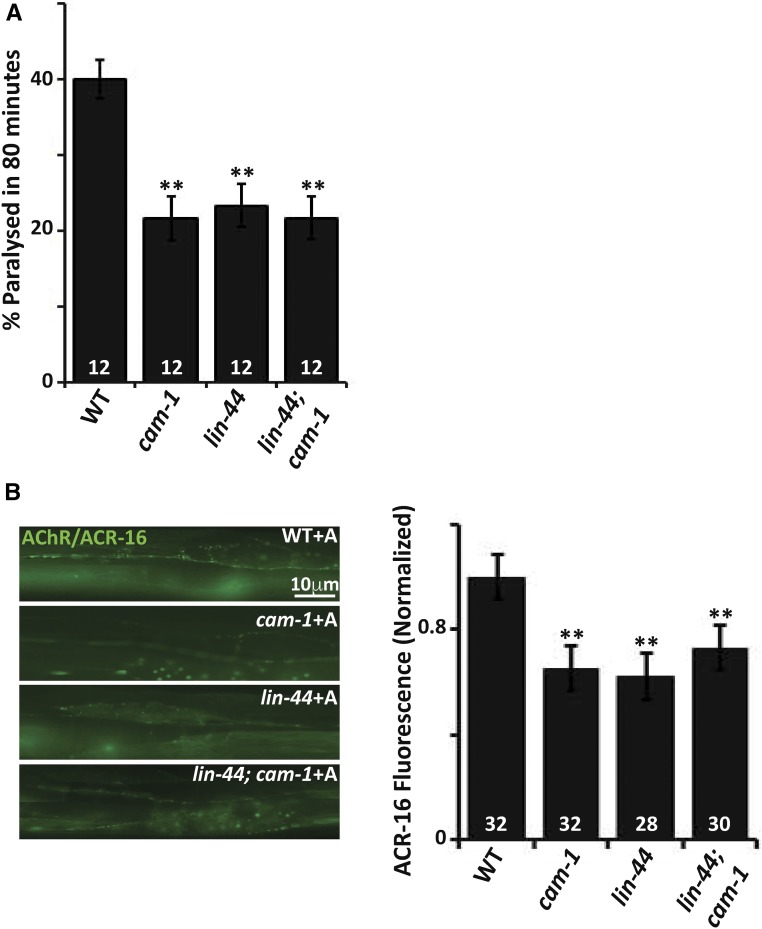
It has been previously reported that mutants in the Wntless homolog of C. elegans, mig-14, are resistant to aldicarb, and completely suppresses the hypersensitivity to aldicarb phenotype seen in rig-3 mutants (Babu et al. 2011). In order to identify the Wnt for the RIG-3 regulated signaling pathway at the NMJ, we tested the interaction between rig-3 and Wnts in aldicarb assays, we found that the Wnt/lin-44 mutant was resistant to aldicarb and completely suppressed the hypersensitivity to aldicarb seen in rig-3 mutants (Figure 4A). Since it has previously been shown that RIG-3 regulates signaling at the NMJ from the cholinergic neurons, we first investigated if the expression of Wnt/LIN-44 in cholinergic neurons would rescue the resistance to aldicarb phenotype seen in the mutants. Our results indicated that expressing Wnt/LIN-44 in the cholinergic neurons could indeed rescue the resistance to aldicarb phenotype (Figure 4B). Since Wnt/LIN-44 has only been shown to express at the posterior end of the C. elegans, we expressed Wnt/LIN-44 in cholinergic neurons and saw that it did rescue the aldicarb phenotype of Wnt/lin-44 mutants, indicating that the Wnt/LIN-44 may be expressed in other tissues like motor neurons. However, the fact that Wnt/LIN-44 could rescue the aldicarb phenotype by expression in any tissue type of the C. elegans body cannot be excluded. Next, we examined Wnt/lin-44 mutants for defects in cholinergic and GABAergic neuronal development, and found that, at a gross level, there were no defects in neuronal development and/or axon guidance in these neurons (Figure S4, A and B in File S1). A previous study has shown that LIN-44 and EGL-20 locally determined the presynaptic assembly and specification of the posterior DA9 synapses, and that Wnt/lin-44 mutants show synapse development defects only in the posterior DA9 neuromuscular synapses, and not the more anterior synapses (Klassen and Shen 2007). Similar to what the authors show, on analyzing both cholinergic and GABAergic synapse development using the SYD-2::GFP marker, we found that synapses anterior to the DA9 synapses were largely normal in the Wnt/lin-44 mutant animals (Figure S4, C and D in File S1).
Figure 4.
RIG-3 functions through the Wnt/LIN-44 at the NMJ. (A) Aldicarb assay for Wnt/lin-44; rig-3 mutants, and control strains: paralysis on aldicarb was assayed in WT control animals, rig-3, Wnt/lin-44, and Wnt/lin-44; rig-3 double mutants. (B) Rescue of the Wnt/lin-44 mutant phenotype: Graph indicates rescue of the aldicarb resistance phenotype seen in the Wnt/lin-44 mutants. The rescue constructs of all three rescuing lines expressed Wnt/LIN-44 in the cholinergic neurons using the unc-17 promoter. The three bars indicate three independent rescuing lines of Wnt/LIN-44. (C) The increased AChR/ACR-16 expression seen in rig-3 mutants is suppressed by Wnt/lin-44 mutation: the left panel displays images of WT, rig-3, Wnt/lin-44, and Wnt/lin-44; rig-3 double mutants showing expression of AChR/ACR-16 tagged with GFP along the VNC of C. elegans after treating the animals with aldicarb. The right side of the panel shows a graph of AChR/ACR-16 quantitation, which has been normalized against the average AChR/ACR-16 fluorescence values in WT animals. (D) RIG-3 does not affect Wnt/LIN-44 secretion: the left side of the panel shows the images of coelomocytes with Wnt/LIN-44 tagged to the mCherry marker in WT, rig-3 and mig-14 mutants. The coelomocytes show uptake of the Wnt/LIN-44 that has been secreted from cholinergic neurons where it was originally expressed. The right side of the panel shows the quantitation of the coelomocyte fluorescence normalized to WT. The black bars indicate normalized coelomocyte fluorescence in WT and rig-3 mutants before aldicarb treatment, while the blue bars indicate the normalized fluorescent values after aldicarb treatment. The number at the base of the graph indicates the number of animals that were tested. The second bar shows a similar quantitation where the coelomocyte fluorescence is compared between WT and mig-14 animals. Error bars indicate SEM in all panels.
Since rig-3 mutants showed higher AChR/ACR-16 levels at the NMJ upon increased activity, i.e., aldicarb treatment (Babu et al. 2011), and previous work has shown that Wnt/lin-44 is involved in the subcellular positioning of the DA9 presynaptic specializations at the NMJ (Klassen and Shen 2007), indicating a role for Wnt/LIN-44 at the NMJ, we were prompted to check if Wnt/lin-44 mutants showed defects in AChR/ACR-16 at the NMJ. We found that Wnt/lin-44 mutants displayed a decrease in AChR/ACR-16 levels at the NMJ upon treatment with aldicarb; furthermore, the mutants completely suppressed the increased AChR/ACR-16 levels seen in rig-3 mutants (Figure 4C). These results further indicated that Wnt/LIN-44 could function downstream of RIG-3 at the NMJ.
Since RIG-3 functions through Wnt/LIN-44 at the NMJ, we wanted to examine the affect of RIG-3 on the secretion of the Wnt/LIN-44 ligand. In order to test this, we made a line with Wnt/LIN-44 tagged with the mCherry marker, and expressed in a subset of cholinergic neurons. As previously described for other Wnts, the Wnt/LIN-44 was also secreted into phagocytic bodies, called coelomocytes, that are present along the body of C. elegans and could be detected using the mCherry signal (Jensen et al. 2012b). We quantitated the mCherry signal from control wild-type animals and rig-3 mutants, before and after treatment with aldicarb, and found that there was no significant difference in the fluorescence intensity of coelomocytes when quantitated in wild type (WT) and rig-3 mutant animals, with or without treatment with aldicarb (Figure 4D). Wnts are known to require Wntless proteins for their secretion (reviewed in Das et al. 2012). In order to ascertain that the LIN-44::mCherry construct also required Wntless/MIG-14 for its secretion, we assayed mCherry fluorescence in coelomocytes of mig-14 animals that were expressing Wnt/LIN-44 tagged to mCherry. We found a marked reduction of fluorescence in the coelomocytes of mig-14 mutant animals when compared to WT animals (Figure 4D), indicating that the LIN-44::mCherry requires Wntless/MIG-14 for its secretion into coelomocytes. These results suggest that RIG-3 is not required for normal secretion of Wnt/LIN-44 from cholinergic neurons, but is required for regulating the pathway that allows for maintaining normal AChR/ACR-16 levels through the Wnt/LIN-44 signaling pathway.
Wnt/LIN-44 functions through the RTK, CAM-1 at the NMJ
Previous studies have shown that CAM-1 acts as a nonconventional receptor or an inhibitor of the Wnt signaling pathway, and, in doing so, binds to Wnts through its CRD domain (Green et al. 2007). Moreover, a more recent study has shown that CAM-1, along with LIN-17, acts as a heteromeric receptor for the Wnt ligand CWN-2 and participates in synaptic transmission (Jensen et al. 2012b). Our data indicated that CAM-1 and Wnt/LIN-44 behaved similarly and antagonistically to RIG-3; we thus speculated that Wnt/LIN-44 could function through CAM-1. We investigated the aldicarb dependent paralysis phenotype of Wnt/lin-44; cam-1 double mutants, and found the phenotype of cam-1, Wnt/lin-44 and the double mutants to be similar and not additive (Figure 5A). In order to further analyze the effect of the Wnt/LIN-44 mediated signaling, we checked the levels of AChR/ACR-16 at the NMJ. We found that both Wnt/lin-44 and cam-1 mutants showed a similar reduction in AChR/ACR-16 levels at the NMJ, and that the double mutant was again similar to the single mutants and not additive (Figure 5B). These results indicate that Wnt/LIN-44 could function through CAM-1, and regulate AChR/ACR-16 levels at the NMJ in the presence of increased activity brought about by aldicarb treatment.
Figure 5.
Wnt/LIN-44 could function through CAM-1 at the NMJ. (A) Wnt/LIN-44 and CAM-1 genetically interact with each other: the graph indicates paralysis on aldicarb for Wnt/lin-44; cam-1 double mutants and control animals. The assay was performed on 1 mM aldicarb containing plates. The percentage of animals that paralyzed at 80 min were plotted, and statistical analysis was performed to ascertain significant difference of mutant values from the WT C. elegans. (B) Wnt/LIN-44 could function through CAM-1 to maintain AChR/ACR-16 levels at the NMJ: the left side of the panel shows images of AChR/ACR-16 tagged with GFP along the VNC. The right side of the panel shows the quantitation of the AChR/ACR-16 fluorescence levels that are normalized to control WT fluorescent values. Error bars indicate SEM in all panels.
If RIG-3 is indeed binding to CAM-1 and preventing the Wnt/LIN-44 from functioning through CAM-1, we speculated that expression of CAM-1(ΔIg) that cannot bind RIG-3 would show increased sensitivity to aldicarb. Similarly, increased expression of Wnt/LIN-44 would also show a similar increased sensitivity to aldicarb. To test our hypothesis, we went on to examine the C. elegans paralysis in response to aldicarb when overexpressing CAM-1(ΔIg) in the body-wall muscle of C. elegans, and Wnt/LIN-44 in the cholinergic neurons of the animals. Both sets of C. elegans showed similar hypersensitivity to aldicarb, again indicating that Wnt/LIN-44 could be functioning through CAM-1 at the NMJ (Figure S5 in File S1).
β-catenin/HMP-2 functions downstream of RIG-3
Wnts can function in a β-catenin dependent (canonical pathway) or independent (noncanonical) manner. In the canonical Wnt pathway, Wnts function through Frizzled receptors. However, in some cases, CAM-1 has been shown to act as a nonconventional receptor, or a coreceptor, for Wnts (Green et al. 2007; Jensen et al. 2012b).
We decided to test if RIG-3 functions through the known β-catenin genes, which are the final effector genes of Wnt signaling pathways that result in the activation of target genes, and are known to function downstream of frizzled genes or cam-1 (Jackson and Eisenmann 2012). In C. elegans, there are four genes that encode for β-Catenins (wrm-1, bar-1, sys-1, and hmp-2). Upon testing the mutants of these genes for aldicarb phenotypes, we found that only the β-catenin/hmp-2 mutant strain was resistant to aldicarb (Figure 6A). Further, we could rescue the resistance to aldicarb phenotype of the β-catenin/hmp-2 mutant by expressing β-Catenin/HMP-2 in the body-wall muscle of C. elegans (Figure 6A). In order to examine the possibility that β-catenin/hmp-2 may function downstream of rig-3 and cam-1, we made double mutants of β-catenin/hmp-2 with rig-3 and cam-1, respectively. It was evident from the aldicarb assay that β-catenin/HMP-2 could be functioning downstream of both RIG-3 and CAM-1 (Figure 6B). These data indicate that HMP-2 could be the β-catenin through which RIG-3 and CAM-1 function at the NMJ to maintain AChR levels. In order to test the possibility that β-catenin/hmp-2 mutants could affect AChR/ACR-16 levels, we examined AChR/ACR-16 levels in β-catenin/hmp-2 mutants in the presence of aldicarb, and found that there was a significant decrease in AChR/ACR-16 in β-catenin/hmp-2 mutants (Figure 6C, panel 3). We next checked the levels of ACR-16::GFP in hmp-2;rig-3 double mutant animals that were treated with aldicarb, and found that the hmp-2 mutation completely suppressed the increased AChR/ACR-16 levels seen in the rig-3 mutants (Figure 6C). Taken together, these results indicated that β-Catenin/HMP-2 functions downstream of RIG-3 to maintain AChR/ACR-16 levels at the C. elegans NMJ.
Figure 6.
RIG-3 functions through the β-catenin/HMP-2 at the C. elegans NMJ. (A) The resistance to aldicarb phenotype seen in β−catenin/hmp-2 mutants is rescued by expressing β−Catenin/HMP-2 in the body-wall muscle: This panel shows a graph of aldicarb induced paralysis shown for WT, rig-3 controls, β−catenin/hmp-2 mutants, and rescue of β−catenin/hmp-2 by expressing HMP-2 in the body-wall muscles of C. elegans using the myo-3 promoter. The HMP-2 rescue was done using two independent array lines, and data from both these lines are represented in this panel as separate bars. (B) Both RIG-3 and CAM-1 function through the β-catenin/HMP-2 at the NMJ: this panel shows a graph of aldicarb induced paralysis shown for β−catenin/hmp-2; rig-3 and β−catenin/hmp-2; cam-1 double mutants with controls. (C) The β−catenin/hmp-2 mutant shows decreased AChR/ACR-16 levels: this panel shows decreased AChR/ACR-16 in β−catenin/hmp-2 mutants along the VNC after treatment of the animals with aldicarb. WT and rig-3 mutants treated with aldicarb are shown as controls. Error bars indicate SEM in all panels.
Discussion
The Ig superfamily protein, RIG-3, upon deletion, has been shown previously to display increased AChR/ACR-16 levels at the NMJ when treated with an acetyl cholinesterase inhibitor (Babu et al. 2011). We have elucidated the molecular mechanism of the RIG-3 regulated signaling at the NMJ in the presence of the acetylcholine inhibitor, aldicarb. Previous work has shown that aldicarb does not effect AChR/ACR-16 levels or mEPSC amplitude at the NMJ, and that rig-3 mutants behave in a similar manner to wild-type C. elegans in the absence of aldicarb (Babu et al. 2011); hence, most of the experiments in this study have been performed in the presence of increased activity brought about by aldicarb, illustrated in Figure S6 in File S1.
Our results allow us to make two important conclusions. First, the presynaptic protein RIG-3 signals through the ROR RTK, CAM-1 by directly interacting with CAM-1 postsynaptically to maintain normal synaptic function at the NMJ (schematized in Figure S6D in File S1). Second, our results strongly support the idea that RIG-3 inhibits LIN-44 signaling. Hypomorphic mutants of Wnt/lin-44 and rig-3 mutations have opposite effects on aldicarb sensitivity and ACR-16 synaptic abundance. These data indicate that RIG-3 could prevent increased AChR/ACR-16 levels at the NMJ by inhibiting Wnt/LIN-44 signaling in the presence of increased activity. In the absence of rig-3, and presence of higher activity, increased Wnt/LIN-44 signaling causing enhanced AChR/ACR-16 at the NMJ (schematized in Figure S6 in File S1). Taken together, these results imply that RIG-3 acts as an inhibitor of Wnt/LIN-44 signaling through CAM-1. Below, we elaborate on the implications of these findings, and also discuss the role of Wnt signaling at the synapse.
Regulation of Wnt signaling through the ECD of the nonconventional Wnt receptor, CAM-1
Studies have shown that the nonconventional Wnt receptor, CAM-1, binds secreted Wnt ligands, and functions as a Wnt receptor, or as a Wnt antagonist, by inhibiting signaling through other Wnt receptors (Green et al. 2007, 2008; Chien et al. 2015). In a fairly recent study, Inestrosa and colleagues have shown that Wnt-5a, a Wnt that functions through the noncanonical Wnt signaling pathway is required to upregulate postsynaptic currents in rat hippocampal slices, and increase postsynaptic activity in cultured hippocampal neurons (Cerpa et al. 2008; Farias et al. 2009; Varela-Nallar et al. 2010). Furthermore, Wnt-5a is thought to function through the RTK Ror2 for its function in the noncanonical Wnt pathway (Oishi et al. 2003). Rors are thought to mediate aspects of nervous system development, and function through Wnts (Petrova et al. 2014). Here, we show that a similar mechanism occurs in C. elegans, where a Wnt, LIN-44, functions to maintain normal synaptic and neuronal function through CAM-1.
A previous study has shown that RIG-3 affects the postsynaptic AChR/ACR-16 receptors in the presence of increased activity at the NMJ. Furthermore, RIG-3 has been shown to genetically function through CAM-1 at the NMJ (Babu et al. 2011). In order to gain further insight into the interaction between RIG-3 and CAM-1, we utilized genetic and in vivo protein–protein interaction studies to show that the Ig domain of CAM-1 is important for its interaction with RIG-3, and for the regulation of postsynaptic AChR/ACR-16 levels. BiFC studies further implicated the role of Ig domain since the in vivo interaction between RIG-3 and CAM-1 was abolished upon deletion of the CAM-1 Ig domain. Previous studies have clearly shown that the ECD is important for AChR/ACR-16 localization and ACh/ACR-16 mediated behavior, and that the ECD is required for interaction with the Frizzled receptor, LIN-17, at the NMJ (Jensen et al. 2012b). Furthermore, it has been shown, both in C. elegans and in mammals, that the RTKs CAM-1 and Ror2 interact with Wnts through the extracellular CRD domains (Xu and Nusse 1998; Oishi et al. 2003; Forrester et al. 2004; Mikels and Nusse 2006). The Kringle domain is also rich in cysteine residues; however, it has not been reported previously to interact with Ig domains. The results from our studies indicate the importance of CAM-1 Ig domain in RIG-3 binding. These results also show that, in addition to the Extracellular domain, and specifically the CRD domain required for Wnt and Frizzled binding, the extracellular Ig domain is required for binding to the presynaptic protein, RIG-3. Since the CRD domain and the Ig domain of CAM-1 are adjacent to each other on CAM-1, it is possible that RIG-3 binds to CAM-1 and prevents binding of Wnt/Lin-44 to CAM-1 during aldicarb mediated stress (schematized in Figure S6 in File S1). Therefore, our studies have brought to light another important binding partner of the nonconventional Wnt receptor, CAM-1 through the extracellular Ig domain.
RIG-3 functions through CAM-1, Wnt/LIN-44 and β-catenin/HMP-2 at the NMJ
Components of the Wnt signaling pathway have multiple roles in the development of the nervous system, and are involved in neuronal migration, polarity, and axon outgrowth (Killeen and Sybingco 2008; Yang and Luo 2011; Salinas 2012; Bilimoria and Bonni 2013; Ackley 2014; Bielen and Houart 2014). Further, Wnts have fairly conserved roles in the synaptic development of Drosophila, C. elegans, and vertebrates (Koles and Budnik 2012). Although we have some knowledge about the role of Wnt signaling during nervous system development, it is only recently that the role for Wnt signaling in synaptic plasticity has begun to emerge (Speese and Budnik 2007; Korkut and Budnik 2009; Budnik and Salinas 2011; Dickins and Salinas 2013; Oliva et al. 2013a,b; Stamatakou and Salinas 2014). In this study, we have identified a Wnt signaling pathway, through which the Immunoglobulin Superfamily protein, RIG-3, functions.
Wnt/LIN-44 has been shown to participate in multiple developmental functions, like axon guidance of motor neurons, dendrite development, neuronal polarity of mechanosensory neurons, and synapse development (Herman et al. 1995; Hilliard and Bargmann 2006; Maro et al. 2009; Kirszenblat et al. 2011). Here, we show that mutants of Wnt/LIN-44 show defects in AChR/ACR-16 levels, and a corresponding decrease in muscle activity at the NMJ in the presence of the acetylcholine esterase inhibitor, aldicarb. To date, the Wnt/LIN-44 has been shown to express only in the C. elegans tail, and recent work has shown that it could form an anterior–posterior gradient through expression in the tail (Herman et al. 1995; Mizumoto and Shen 2013). It is conceivable that the motor neurons that are more posterior experience more Wnt/LIN-44 signaling and show a stronger phenotype. Another possibility is that Wnt/LIN-44 is expressed at very low, nondetectable, levels in motor neurons, and that secretion from these neurons affects AChR/ACR-16 levels through the muscle CAM-1.
This and other studies (Babu et al. 2011; Jensen et al. 2012b) implicate the Wnt signaling pathway in maintaining normal AChR/ACR-16 levels, and show the importance of studying Wnt signaling at the synapses. An interesting outcome of this study is the fact that Wnt/LIN-44 is being implicated in synaptic signaling function for the first time.
Jensen et al. (2012b) have shown that another Wnt ligand, CWN-2, functions through the heteromeric receptor complex of LIN-17/CAM-1 and the downstream Wnt effector protein Dishevelled/DSH-1 to maintain normal AChR/ACR-16-mediated currents at the NMJ. In contrast to their work, we have shown that Wnt/LIN-44 functions downstream of RIG-3 at the C. elegans NMJ in a CAM-1 dependent manner, and leads to an apparent decrease in the levels of AChR/ACR-16, which results in decreased activity levels at the NMJ. Further, RIG-3 does not appear to be affecting the secretion of Wnt/LIN-44. These data allow us to postulate that RIG-3 could be interrupting the interaction between LIN-44 and CAM-1, and affecting the synaptic signaling through CAM-1 (schematized in Figure S6 in File S1).
One question that our work raises is why RIG-3 does not function through Wnt/CWN-2, Frizzled/LIN-17, and Dishevelled/DSH-1. One possible reason could be the fact that RIG-3 affects AChR/ACR-16 levels only in the presence of increased activity brought about by the acetylcholine esterase inhibitor, aldicarb. To follow up on these initial findings, our experiments were done in the presence of aldicarb, while Jensen et al. (2012a) performed their studies in wild-type conditions. Thus, in the wild-type scenario Wnt/CWN-2 functions through Frizzled/LIN-17 and CAM-1 followed by Dishevelled/DSH-1 to maintain normal AChR/ACR-16 levels and normal activity at the NMJ. The loss of any one of these components gives rise to decreased AChR/ACR-16 at the NMJ (Francis et al. 2005; Jensen et al. 2012b); this process is likely to be independent of RIG-3. However, in the presence of increased activity brought about by aldicarb, RIG-3 functions through CAM-1, Wnt/LIN-44, and the β-catenin/HMP-2 to maintain normal AChR/ACR-16 levels at the NMJ. This signaling pathway gives rise to increased AChR/ACR-16 in the absence of rig-3, and the presence of aldicarb (schematized in S6). It is likely that the RIG-3-mediated pathway does not come into effect in the absence of increased activity (absence of aldicarb). Although our work shows the existence of a different Wnt signaling pathway that functions through RIG-3, it also shows that CAM-1 is an important component of the pathway required to maintain AChR/ACR-16 levels at the NMJ, as has been shown previously (Francis et al. 2005; Jensen et al. 2012b).
It is interesting to find that Wnt/LIN-44 may also utilize CAM-1 as a receptor in addition to the traditional Frizzled receptor, LIN-17. Wnt/LIN-44 is known to signal through Frizzled/LIN-17 during physiological processes such as asymmetric cell division (Sawa et al. 1996), PLM neuronal polarity (Hilliard and Bargmann 2006), dendrite development (Kirszenblat et al. 2011), and regulation of NMJ formation (Klassen and Shen 2007). Hence, understanding how Wnts selects one kind of receptor over another under various physiological conditions and growth stages could open interesting new avenues for Wnt signaling.
Other important molecules that contribute to Wnt mediated signaling are β-catenins. Unlike in vertebrates or Drosophila, the C. elegans genome encodes fewer Wnts and multiple β-catenin genes (Eisenmann 2005), which appear to have distinct functions in Wnt signaling and cell adhesion. As compared to other identified β-catenins, the β-catenin/HMP-2 shares maximum homology to human β-catenins (Jackson and Eisenmann 2012). In this study, we have identified β-catenin/HMP-2 as a component functioning downstream of the nonconventional Wnt receptor, CAM-1. Further, we could rescue the resistance to aldicarb phenotype of the β-catenin/hmp-2 mutant by expressing β-Catenin/HMP-2 in the body-wall muscle of C. elegans. Double mutants of β-catenin/hmp-2 with rig-3 and cam-1, respectively, demonstrated that the β-catenin/HMP-2 was functioning downstream of RIG-3 and CAM-1 to maintain normal AChR/ACR-16 levels at the C. elegans NMJ.
The next question that this study raises is what is the downstream target of the β-Catenin/HMP-2 that allows for changes in AChR/ACR-16 transcription. Previous yeast two-hybrid and immunoprecipitation studies have shown that β-Catenin/HMP-2 does not interact with the POP-1 (TCF/LEF) transcription factor that alters the transcription of target genes in the Wnt signaling pathway (Korswagen et al. 2000). However, recent studies indicate that β-Catenin/HMP-2 could interact with POP-1 and is involved in the localization and activity of the POP-1 transcription factor (Natarajan et al. 2004; Putzke and Rothman 2010; Sumiyoshi et al. 2011). Hence, one speculation could be that POP-1 could be the final target that results in the regulation of AChR/ACR-16 receptor levels at the NMJ. However, a recent study by Jensen et al. (2012b) found that POP-1 RNAi does not affect the AChR/ACR-16::GFP levels, making us rethink the fact that POP-1 could be the final effector of AChR/ACR-16. In future, studies could be oriented toward the understanding of transcription factors functioning downstream of β-Catenin/HMP-2. Such studies are required to better understand the regulatory mechanism existing at the NMJ normally, or when exposed to stress such as aldicarb.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.195297/-/DC1.
Acknowledgments
We thank Teresa Craft and Wayne Forrester for the cam-1::GFP lines, Mike Francis for the Pacr-2::mCherry::RAB-3 line, Yingsong Hao and Josh Kaplan for a number of marker lines, and Zhitao Hu and Josh Kaplan for discussions and suggestions. Several mutant strains were provided by Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work is supported by the Wellcome Trust/ Department of Biotechnology India Alliance (WT-DBT IA) grant (IA/I/12/500516), and part-funded by a DBT-Innovative Young Biotechnologist Award (DBT-IYBA) grant (BT/05/IYBA/2011) to K.B. K.B. also acknowledges support from Indian Institute of Science Education and Research (IISER) Mohali intramural funds. We also acknowledge use of the IISER Mohali confocal facility. P.P. acknowledges financial support from the DBT-Bio-CARe grant and Fellowship (DBT/Bio-CARe/01/10167), IISER Mohali, and postdoctoral support from the WT-DBT IA grant to K.B. A.B. is funded by Indian Council of Medical Research (ICMR)- Senior Research Fellowship (SRF). We thank the members of the Babu laboratory for comments on the manuscript, and Ankit Negi (supported on funds from a WT-DBT IA grant to K.B.) for technical help.
Footnotes
Communicating editor: B. Goldstein
Literature Cited
- Ackley B. D., 2014. Wnt-signaling and planar cell polarity genes regulate axon guidance along the anteroposterior axis in C. elegans. Dev. Neurobiol. 74: 781–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu K., Hu Z., Chien S. C., Garriga G., Kaplan J. M., 2011. The immunoglobulin super family protein RIG-3 prevents synaptic potentiation and regulates Wnt signaling. Neuron 71: 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei Y., Hogan J., Berkowitz L. A., Soto M., Rocheleau C. E., et al. , 2002. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev. Cell 3: 113–125. [DOI] [PubMed] [Google Scholar]
- Bielen H., Houart C., 2014. The Wnt cries many: Wnt regulation of neurogenesis through tissue patterning, proliferation, and asymmetric cell division. Dev. Neurobiol. 74: 772–780. [DOI] [PubMed] [Google Scholar]
- Bilimoria P. M., Bonni A., 2013. Molecular control of axon branching. Neuroscientist 19: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Holm L., Sander C., 1994. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242: 309–320. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V., Salinas P. C., 2011. Wnt signaling during synaptic development and plasticity. Curr. Opin. Neurobiol. 21: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerpa W., Godoy J. A., Alfaro I., Farias G. G., Metcalfe M. J., et al. , 2008. Wnt-7a modulates the synaptic vesicle cycle and synaptic transmission in hippocampal neurons. J. Biol. Chem. 283: 5918–5927. [DOI] [PubMed] [Google Scholar]
- Chan K. K., Seetharaman A., Selman G., Roy P., 2015. Immunoprecipitation of proteins in Caenorhabditis elegans. Bio Protoc. Vol 5. DOI: doi.org/10.21769/BioProtoc.1436. [Google Scholar]
- Chien A. J., Conrad W. H., Moon R. T., 2009. A Wnt survival guide: from flies to human disease. J. Invest. Dermatol. 129: 1614–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien S. C., Gurling M., Kim C., Craft T., Forrester W., et al. , 2015. Autonomous and nonautonomous regulation of Wnt-mediated neuronal polarity by the C. elegans Ror kinase CAM-1. Dev. Biol. 404: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurillo M. A., 2015. Role of the Wnt/beta-catenin pathway in gastric cancer: an in-depth literature review. World J. Exp. Med. 5: 84–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Nusse R., 2012. Wnt/beta-catenin signaling and disease. Cell 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Das S., Yu S., Sakamori R., Stypulkowski E., Gao N., 2012. Wntless in Wnt secretion: molecular, cellular and genetic aspects. Front. Biol. (Beijing) 7: 587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J., Ghosh A., 2016. Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 17: 22–35. [DOI] [PubMed] [Google Scholar]
- Dickins E. M., Salinas P. C., 2013. Wnts in action: from synapse formation to synaptic maintenance. Front. Cell. Neurosci. 7: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D. M., 2005. Wnt signaling. (June 25, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.7.1, http://www.wormbook.org. [Google Scholar]
- Farias G. G., Alfaro I. E., Cerpa W., Grabowski C. P., Godoy J. A., et al. , 2009. Wnt-5a/JNK signaling promotes the clustering of PSD-95 in hippocampal neurons. J. Biol. Chem. 284: 15857–15866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W. C., Dell M., Perens E., Garriga G., 1999. A C. elegans Ror receptor tyrosine kinase regulates cell motility and asymmetric cell division. Nature 400: 881–885. [DOI] [PubMed] [Google Scholar]
- Forrester W. C., Kim C., Garriga G., 2004. The Caenorhabditis elegans Ror RTK CAM-1 inhibits EGL-20/Wnt signaling in cell migration. Genetics 168: 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M. M., Evans S. P., Jensen M., Madsen D. M., Mancuso J., et al. , 2005. The Ror receptor tyrosine kinase CAM-1 is required for ACR-16-mediated synaptic transmission at the C. elegans neuromuscular junction. Neuron 46: 581–594. [DOI] [PubMed] [Google Scholar]
- Green J. L., Inoue T., Sternberg P. W., 2007. The C. elegans ROR receptor tyrosine kinase, CAM-1, non-autonomously inhibits the Wnt pathway. Development 134: 4053–4062. [DOI] [PubMed] [Google Scholar]
- Green J. L., Kuntz S. G., Sternberg P. W., 2008. Ror receptor tyrosine kinases: orphans no more. Trends Cell Biol. 18: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaby D. M., Mornon J. P., 1998. The immunoglobulin superfamily: an insight on its tissular, species, and functional diversity. J. Mol. Evol. 46: 389–400. [DOI] [PubMed] [Google Scholar]
- Harada S., Hori I., Yamamoto H., Hosono R., 1994. Mutations in the unc-41 gene cause elevation of acetylcholine levels. J. Neurochem. 63: 439–446. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Hirotsu T., Iwata R., Kage-Nakadai E., Kunitomo H., et al. , 2009. A trophic role for Wnt-Ror kinase signaling during developmental pruning in Caenorhabditis elegans. Nat. Neurosci. 12: 981–987. [DOI] [PubMed] [Google Scholar]
- Henriquez J. P., Salinas P. C., 2012. Dual roles for Wnt signalling during the formation of the vertebrate neuromuscular junction. Acta Physiol. (Oxf.) 204: 128–136. [DOI] [PubMed] [Google Scholar]
- Herman M. A., Vassilieva L. L., Horvitz H. R., Shaw J. E., Herman R. K., 1995. The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell 83: 101–110. [DOI] [PubMed] [Google Scholar]
- Hiatt S. M., Shyu Y. J., Duren H. M., Hu C. D., 2008. Bimolecular fluorescence complementation (BiFC) analysis of protein interactions in Caenorhabditis elegans. Methods 45: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikasa H., Sokol S. Y., 2013. Wnt signaling in vertebrate axis specification. Cold Spring Harb. Perspect. Biol. 5: a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard M. A., Bargmann C. I., 2006. Wnt signals and frizzled activity orient anterior-posterior axon outgrowth in C. elegans. Dev. Cell 10: 379–390. [DOI] [PubMed] [Google Scholar]
- Inestrosa N. C., Montecinos-Oliva C., Fuenzalida M., 2012. Wnt signaling: role in Alzheimer disease and schizophrenia. J. Neuroimmune Pharmacol. 7: 788–807. [DOI] [PubMed] [Google Scholar]
- Jackson B. M., Eisenmann D. M., 2012. Beta-catenin-dependent Wnt signaling in C. elegans: teaching an old dog a new trick. Cold Spring Harb. Perspect. Biol. 4: a007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M., Brockie P. J., Maricq A. V., 2012a Wnt signaling regulates experience-dependent synaptic plasticity in the adult nervous system. Cell Cycle 11: 2585–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M., Hoerndli F. J., Brockie P. J., Wang R., Johnson E., et al. , 2012b Wnt signaling regulates acetylcholine receptor translocation and synaptic plasticity in the adult nervous system. Cell 149: 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell J. R., Fetter R. D., Bargmann C. I., 2009. Wnt-Ror signaling to SIA and SIB neurons directs anterior axon guidance and nerve ring placement in C. elegans. Development 136: 3801–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen M. T., Sybingco S. S., 2008. Netrin, Slit and Wnt receptors allow axons to choose the axis of migration. Dev. Biol. 323: 143–151. [DOI] [PubMed] [Google Scholar]
- Kim C., Forrester W. C., 2003. Functional analysis of the domains of the C elegans Ror receptor tyrosine kinase CAM-1. Dev. Biol. 264: 376–390. [DOI] [PubMed] [Google Scholar]
- Kirszenblat L., Pattabiraman D., Hilliard M. A., 2011. LIN-44/Wnt directs dendrite outgrowth through LIN-17/Frizzled in C. elegans Neurons. PLoS Biol. 9: e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen M. P., Shen K., 2007. Wnt signaling positions neuromuscular connectivity by inhibiting synapse formation in C. elegans. Cell 130: 704–716. [DOI] [PubMed] [Google Scholar]
- Koles K., Budnik V., 2012. Wnt signaling in neuromuscular junction development. Cold Spring Harb. Perspect. Biol. 4: a008045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut C., Budnik V., 2009. WNTs tune up the neuromuscular junction. Nat. Rev. Neurosci. 10: 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen H. C., Herman M. A., Clevers H. C., 2000. Distinct beta-catenins mediate adhesion and signalling functions in C. elegans. Nature 406: 527–532. [DOI] [PubMed] [Google Scholar]
- Liu P., Ge Q., Chen B., Salkoff L., Kotlikoff M. I., et al. , 2011. Genetic dissection of ion currents underlying all-or-none action potentials in C. elegans body-wall muscle cells. J. Physiol. 589: 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguschak K. A., Ressler K. J., 2011. Wnt signaling in amygdala-dependent learning and memory. J. Neurosci. 31: 13057–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro G. S., Klassen M. P., Shen K., 2009. A beta-catenin-dependent Wnt pathway mediates anteroposterior axon guidance in C. elegans motor neurons. PLoS One 4: e4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels A. J., Nusse R., 2006. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 4: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Alfonso A., Nguyen M., Crowell J. A., Johnson C. D., et al. , 1996. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. USA 93: 12593–12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Shen K., 2013. Two Wnts instruct topographic synaptic innervation in C. elegans. Cell Rep. 5: 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewska K., Lauritzen A., Hasenoeder S., Brown L., Georgiou J., et al. , 2013. Neurons refine the Caenorhabditis elegans body plan by directing axial patterning by Wnts. PLoS Biol. 11: e1001465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan K. A., Cheyette B. N., 2012. Wnt signaling in vertebrate neural development and function. J. Neuroimmune Pharmacol. 7: 774–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan L., Jackson B. M., Szyleyko E., Eisenmann D. M., 2004. Identification of evolutionarily conserved promoter elements and amino acids required for function of the C. elegans beta-catenin homolog BAR-1. Dev. Biol. 272: 536–557. [DOI] [PubMed] [Google Scholar]
- Nurrish S., Segalat L., Kaplan J. M., 1999. Serotonin inhibition of synaptic transmission: Galpha(0) decreases the abundance of UNC-13 at release sites. Neuron 24: 231–242. [DOI] [PubMed] [Google Scholar]
- Oishi I., Suzuki H., Onishi N., Takada R., Kani S., et al. , 2003. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells 8: 645–654. [DOI] [PubMed] [Google Scholar]
- Oliva C. A., Vargas J. Y., Inestrosa N. C., 2013a Wnt signaling: role in LTP, neural networks and memory. Ageing Res. Rev. 12: 786–800. [DOI] [PubMed] [Google Scholar]
- Oliva C. A., Vargas J. Y., Inestrosa N. C., 2013b Wnts in adult brain: from synaptic plasticity to cognitive deficiencies. Front. Cell. Neurosci. 7: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Shen K., 2012. WNTs in synapse formation and neuronal circuitry. EMBO J. 31: 2697–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova I. M., Malessy M. J., Verhaagen J., Fradkin L. G., Noordermeer J. N., 2014. Wnt signaling through the Ror receptor in the nervous system. Mol. Neurobiol. 49: 303–315. [DOI] [PubMed] [Google Scholar]
- Putzke A. P., Rothman J. H., 2010. Repression of Wnt signaling by a Fer-type nonreceptor tyrosine kinase. Proc. Natl. Acad. Sci. USA 107: 16154–16159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, J. B., 2007 Acetylcholine. WormBook (January 30, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.131.1, http://www.wormbook.org.
- Salinas P. C., 2012. Wnt signaling in the vertebrate central nervous system: from axon guidance to synaptic function. Cold Spring Harb. Perspect. Biol. 4: pii: a008003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas P. C., Zou Y., 2008. Wnt signaling in neural circuit assembly. Annu. Rev. Neurosci. 31: 339–358. [DOI] [PubMed] [Google Scholar]
- Sawa H., Lobel L., Horvitz H. R., 1996. The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila frizzled protein. Genes Dev. 10: 2189–2197. [DOI] [PubMed] [Google Scholar]
- Schwarz V., Pan J., Voltmer-Irsch S., Hutter H., 2009. IgCAMs redundantly control axon navigation in Caenorhabditis elegans. Neural Dev. 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu Y. J., Suarez C. D., Hu C. D., 2008. Visualization of ternary complexes in living cells by using a BiFC-based FRET assay. Nat. Protoc. 3: 1693–1702. [DOI] [PubMed] [Google Scholar]
- Sieburth D., Ch’ng Q., Dybbs M., Tavazoie M., Kennedy S., et al. , 2005. Systematic analysis of genes required for synapse structure and function. Nature 436: 510–517. [DOI] [PubMed] [Google Scholar]
- Song S., Zhang B., Sun H., Li X., Xiang Y., et al. , 2010. A Wnt-Frz/Ror-Dsh pathway regulates neurite outgrowth in Caenorhabditis elegans. PLoS Genet. 6: pii: e1001056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese S. D., Budnik V., 2007. Wnts: up-and-coming at the synapse. Trends Neurosci. 30: 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakou E., Salinas P. C., 2014. Postsynaptic assembly: a role for Wnt signaling. Dev. Neurobiol. 74: 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi E., Takahashi S., Obata H., Sugimoto A., Kohara Y., 2011. The beta-catenin HMP-2 functions downstream of Src in parallel with the Wnt pathway in early embryogenesis of C. elegans. Dev. Biol. 355: 302–312. [DOI] [PubMed] [Google Scholar]
- Varela-Nallar L., Alfaro I. E., Serrano F. G., Parodi J., Inestrosa N. C., 2010. Wingless-type family member 5A (Wnt-5a) stimulates synaptic differentiation and function of glutamatergic synapses. Proc. Natl. Acad. Sci. USA 107: 21164–21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. K., Nusse R., 1998. The frizzled CRD domain is conserved in diverse proteins including several receptor tyrosine kinases. Curr. Biol. 8: R405–R406. [DOI] [PubMed] [Google Scholar]
- Yang G. Y., Luo Z. G., 2011. Implication of Wnt signaling in neuronal polarization. Dev. Neurobiol. 71: 495–507. [DOI] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Yamamoto Y., Sawa H., Forrester W. C., 2008. Complex network of Wnt signaling regulates neuronal migrations during Caenorhabditis elegans development. Genetics 179: 1357–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. Tables S3 and S4 in File S1 contain all the plasmids and strains that have been used in this study.