Abstract
In all organisms, the majority of traits vary continuously between individuals. Explaining the genetic basis of quantitative trait variation requires comprehensively accounting for genetic and nongenetic factors as well as their interactions. The growth of microbial cells can be characterized by a lag duration, an exponential growth phase, and a stationary phase. Parameters that characterize these growth phases can vary among genotypes (phenotypic variation), environmental conditions (phenotypic plasticity), and among isogenic cells in a given environment (phenotypic variability). We used a high-throughput microscopy assay to map genetic loci determining variation in lag duration and exponential growth rate in growth rate-limiting and nonlimiting glucose concentrations, using segregants from a cross of two natural isolates of the budding yeast, Saccharomyces cerevisiae. We find that some quantitative trait loci (QTL) are common between traits and environments whereas some are unique, exhibiting gene-by-environment interactions. Furthermore, whereas variation in the central tendency of growth rate or lag duration is explained by many additive loci, differences in phenotypic variability are primarily the result of genetic interactions. We used bulk segregant mapping to increase QTL resolution by performing whole-genome sequencing of complex mixtures of an advanced intercross mapping population grown in selective conditions using glucose-limited chemostats. We find that sequence variation in the high-affinity glucose transporter HXT7 contributes to variation in growth rate and lag duration. Allele replacements of the entire locus, as well as of a single polymorphic amino acid, reveal that the effect of variation in HXT7 depends on genetic, and allelic, background. Amplifications of HXT7 are frequently selected in experimental evolution in glucose-limited environments, but we find that HXT7 amplifications result in antagonistic pleiotropy that is absent in naturally occurring variants of HXT7. Our study highlights the complex nature of the genotype-to-phenotype map within and between environments.
Keywords: HXT7, bulk segregant mapping, microcolony, quantitative trait locus, yeast
HERITABLE traits that vary quantitatively are the most pervasive class of phenotypic variation. Resolving the genotype–phenotype relationship for quantitative traits remains the central challenge for genetics in the genomic era. The trait value of any given individual can be considered as the result of three distinct sets of factors: (1) genetic factors that confer heritable differences within an environment, (2) environmental factors that modulate the effects of genotypes, and (3) the inherent variability of the trait given that genetic and environmental factors are identical. Our ability to predict phenotypes from genotypes requires a detailed understanding of how each of these factors contributes to quantitative trait variation.
A variety of genetic mapping techniques have been developed for identifying loci underlying quantitative trait variation in diverse organisms. Most studies focus solely on the additive effects of loci, but accurately mapping genotype-to-phenotype requires incorporation of both additive and epistatic effects and an understanding of how these vary with environment. Indeed, as the number of identified causative loci grows, it is becoming increasingly apparent that many genetic effects are conditionally dependent on variation at other loci or in the environment (Mackay et al. 2009; Liti and Louis 2012). More precise definition of phenotypes and incorporation of additional information about environmental conditions might reveal additional genetic determinants and enable more accurate estimation of effect size (Robinson et al. 2014). Unbiased genome-wide scans for epistatic effects on trait variation must overcome the limits on statistical power imposed by testing the many possible combinations of loci (Carlborg and Haley 2004). However, technological and analytical advances are making studies of epistasis increasingly feasible.
Individuals of identical genotypes can exhibit differences in trait values within the same environment. Although these differences are not genetically encoded, the extent to which genetically identical individuals differ in a given environment is influenced by genetic factors (Geiler-Samerotte et al. 2013). Hence, predicting phenotypic values on the basis of genotype (and quantifying the confidence in that prediction) depends not only on the genetic architecture that underlies average differences between genotypes, but also on the genetic factors that influence the phenotypic variability of a trait. Despite an increasing appreciation of the importance of phenotypic variability (Pelkmans 2012; Yvert 2014), many questions remain regarding its genetic and molecular basis (Geiler-Samerotte et al. 2013). Specifically, the extent to which the loci that determine phenotypic variability and those that determine phenotypic variation (i.e., differences in average trait values) overlap remains poorly understood (Hall et al. 2007; Yang et al. 2012; Ayroles et al. 2015).
Complex networks of interacting genetic and environmental factors regulate cell growth in microbes and multicellular organisms, making cell growth an ideal system to dissect the genetic basis of complex traits. Moreover, variation in cell growth is important from an evolutionary perspective as it is a major component of fitness in microbes (Blomberg 2011). Therefore, dissecting the genetic basis of cell growth variation in ecologically relevant environments may also illuminate adaptive variation in natural populations. The rate at which a cell grows is the result of myriad cellular processes including nutrient sensing and transport, signal transduction, macromolecular synthesis, and metabolism. In microbes, culture growth can be separated into three distinct phases: (1) a lag phase, which is a period of physiological adaptation during which cells do not grow; (2) an exponential growth phase, in which cells grow at a constant rate; and (3) a stationary phase, in which cell growth stops due to exhaustion of an essential nutrient from the environment (Monod 1949). These growth phases vary independently in natural populations in different conditions (Cubillos et al. 2011; Ziv et al. 2013b). Recently, we have developed a high-throughput microcolony growth rate assay (Levy et al. 2012; Ziv et al. 2013b) that accurately quantifies each of these phases in thousands of individuals and different environmental conditions, facilitating high-resolution dissection of cell growth phenotypes.
Budding yeast is an ideal model for the analysis of complex traits through genetic analysis (Liti and Louis 2012). Quantitative trait loci (QTL) underlying variation in complex traits can be mapped by analyzing genotype–phenotype relationships in segregants of crosses. The detection of QTL depends on the effect sizes and frequencies of alleles whereas resolving causative variants to individual genes depends on the frequency of recombination (Mackay et al. 2009). Individual segregant analysis involves genotyping and phenotyping individual segregants and testing for associations (Steinmetz et al. 2002; Gerke et al. 2009; Cubillos et al. 2011; Bloom et al. 2013). Alternatively, bulk segregant analysis involves selecting a subset of the population of segregants based on extreme trait values and looking for a deviation in allele frequency from that of the entire population (Michelmore et al. 1991; Ehrenreich et al. 2010; Swinnen et al. 2012). The advantage of bulk segregant mapping is that the increased sample size increases the power to detect loci with small effects (Ehrenreich et al. 2010). However, analysis of individual genotypes is required to identify genetic interactions (Wilkening et al. 2014). Resolving QTL to causative variants using bulk segregant mapping can be improved by using an advanced intercross population, as increased recombination decreases linkage between adjacent sites (Darvasi and Soller 1995; Parts et al. 2011; Cubillos et al. 2013; Illingworth et al. 2013).
In this study, we dissected the genetic architecture of cell growth using a combination of classical interval mapping (a form of individual segregant analysis) and sequencing under selection (a form of bulk segregant analysis) to identify QTL. We defined the genetic architecture of cell growth in two related environments: minimal chemically defined carbon-limiting media containing growth rate-limiting (0.22 mM) and growth rate-nonlimiting (4.44 mM) glucose concentrations. The latter concentration of glucose supports maximal growth rates, despite being more than an order of magnitude lower than that contained in standard lab media (111 mM) (Ziv et al. 2013b). These environments are ecologically relevant, as growth differences in low-glucose conditions distinguish sympatric Saccharomyces cerevisiae strains isolated from different niches within the same local area (Clowers et al. 2015). We decomposed cell growth by quantifying exponential growth rate and lag duration distributions, and mapped loci determining both the phenotypic variation and phenotypic variability of the traits using interval mapping in individual F2 segregants. We then used an advanced intercross population and bulk segregant analysis to increase the mapping resolution for QTL.
We find numerous QTL for cell growth, some of which depend on the environment. We also find a prevalence of genetic interactions underlying differences in the phenotypic variability of traits that do not affect average trait values. We validated in detail linkage to a QTL on chromosome IV. Allele replacements confirm the effect of natural variation in the glucose transporter HXT7, a known target of selection in experimental evolution (Brown et al. 1998; Gresham et al. 2008; Kao and Sherlock 2008; Koschwanez et al. 2013; Selmecki et al. 2015) on cell growth rates. We demonstrate the existence of both intergenic and intragenic interactions that impact the effect of variation in HXT7. These results highlight the intricate nature of genotype-to-phenotype mapping and illustrate the necessity of identifying the relevant genetic loci underlying both variation and variability, and how the environment modulates their effects, for accurate phenotypic prediction.
Materials and Methods
Yeast strains and growth analysis
Parental oak (BC248) and vineyard (BC241) strains and the panel of segregants (Gerke et al. 2006) were obtained from the lab of Barak Cohen (Washington University). There are a total of 480 segregants arrayed in six 96-well plates, of which 374 were previously genotyped (Gerke et al. 2009). All segregants were phenotyped using the microcolony assay; only genotyped strains were used for interval mapping. The F2 pool used in chemostat experiments consisted of all segregants. NCYC3606 and NCYC3591 (Cubillos et al. 2009) were used during allele replacements. Strains shown in Supplemental Material, Figure S9 in File S2 are from Gresham et al. (2008); the evolved clone with HXT6/HXT7 amplification is from population G2. All media were minimal chemically defined carbon-limiting media (Saldanha et al. 2004; Brauer et al. 2005) without amino acid or nucleotide supplements. Growth conditions, microscopy, and analysis of growth profiles were performed as described (Ziv et al. 2013b).
Data normalization
For interval mapping, growth rate assays were performed in 96-well plates with each parental strain present in four wells per plate. We found that, although the difference between the parents was clear on each plate, the absolute growth values and the magnitude of the difference changed slightly between plates (between 0.11 and 0.13 for growth rate in limiting glucose, 0.04–0.05 for growth rate in nonlimiting glucose, and 1.25–3.7 for lag duration in limiting glucose). Estimates for mean growth rate and median lag duration within each well on a plate were corrected for plate effects by subtracting the mean phenotype of the two parents and dividing by the difference between parent phenotypes for each plate. This has the effect of scaling segregants across plates, where 0.5 and −0.5 are the values of the vineyard and oak parents (Figure S1 in File S2). Parental phenotypes were calculated as the mean of all well estimates for a given parent on a given plate. Plate corrected values gave similar LOD profiles to unnormalized data (Pearson correlation > 0.98), only with sharper peaks. For variability traits, residuals of a loess regression (SD regressed against mean growth rate or median absolute deviation (MAD) regressed against median lag duration) were used. For a discussion on the use of loess residuals for variability estimation, see Geiler-Samerotte et al. (2013). Residuals were calculated using the “loess” function in R (R Development Core Team 2012), with default parameters. For regression analyses, we used unscaled values for each segregant. All segregants, including those that were not genotyped, were included in regression analyses for each environment. Use of different measures of central tendency and variability had little effect on the results. For example, we used within-plate ranks of dispersion to confirm that plate effects do not bias the results for variability traits. We calculated genetic variance proportions by subtracting from one, the ratio of the average parental replicate variance (calculated for each parental strain across 24 wells) to the F2 phenotypic variance (calculated for all segregants, including those that were not genotyped).
QTL mapping using R/qtl
We used the R package R/qtl for interval mapping (Broman et al. 2003). To identify individual additive QTL, we performed genome scans with a single-QTL model (“scanone” function), using a normal phenotype model, a 1-cM step size, and the Haley-Knott (HK) algorithm for all traits. To determine experimentwise significance thresholds for each QTL scan, we employed the method of Churchill and Doerge (1994) as implemented in R/qtl (Broman et al. 2003). Briefly, for each QTL scan we performed 10,000 permutations of the trait-specific data. From each permutation, the genome-wide maximum LOD score was retained and the resulting 10,000 values used to define the experimentwise null distribution. This approach has the effect of creating an empirical null distribution for a particular trait while controlling the overall type I error to be α or less across the multiple hypotheses tested in a genome-wide screen. Only one QTL per chromosome can be identified using the single-QTL scan, corresponding to the position with the maximum LOD score. Empirical null distributions generated from extreme values were used to compute the P-value of the maximum LOD score for each chromosome (Figure S10 in File S2). QTL were identified using significance thresholds, based on an α of 0.05 or 0.1, in an attempt to balance type I and type II errors. Because LOD profiles showed evidence for more than one QTL on some chromosomes, we also performed genome scans with a two-QTL model (“scantwo” function) using a 5-cM step size. The presence of a second additive QTL on the same chromosome was identified from the two-dimensional scan by considering both the additive and the difference between additive and single-QTL LOD scores (considering only self–self pairs of chromosomes), and comparison with an empirical null distribution generated using 1000 two-dimensional permutations at an α of 0.05 or 0.1 (Figure S11 in File S2). To identify significant interactions between pairs of loci, we used the scantwo function to compute the LOD scores for a pairwise interaction model (the difference between an additive and full model per pair of loci) (Broman et al. 2003), and determined threshold LOD values at an α of 0.05 or 0.1 based on an empirical null distribution generated using 1000 two-dimensional permutations (Figure S12 in File S2).
Final models included all identified additive loci and genetic interactions. QTL models were fit using the “fitqtl” function to determine the estimated effect sizes and percent of variance explained. Both within and between traits, QTL were considered the same locus if they were within 30 cM from one another. Average chromosome length is 250 cM (range: 67–537 cM).
Creation of an advanced intercross population
We created an advanced intercross population by 11 rounds of sporulation and mating starting with a hybrid (F1) of the oak and vineyard strains. As both parental strains are homothallic, we sought to maximize intertetrad mating as follows. Typically, 2.5 × 108 cells were sporulated for an average of 9 days. Cells were sporulated at room temperature in 1% potassium acetate at a density of 5 × 107 cell/ml. For mating, the sporulated culture was resuspended in equal amounts of water and ether, and vortexed for 10 min to kill unsporulated cells. Spores were separated using centrifugation, washed with water, and incubated in 1 mg/ml Zymolyase for 10 min at 30°. Spores were resuspended in a large volume of 0.01% Triton and vortexed to increase separation of spores. Spores were subsequently concentrated and plated at high density on multiple YPD plates (∼1.5 × 108 spores per plate). After 19 hr of growth, cells were scraped off the plates and resuspended in 1% potassium acetate to begin a new round of sporulation. Following the final round of sporulation, spores were resuspended in liquid YPD and incubated overnight to facilitate selfing via mating type switching and subsequent mating between mother and daughter cells, resulting in a mapping population comprising homozygous diploids.
Whole-genome sequencing and analysis
Libraries for DNA sequencing were prepared and multiplexed using standard protocols and sequenced using an Illumina HiSeq to an average depth of 40–175 ×. Reads were aligned to the reference genome (genome version R64-1-1 released 2/3/2011, also known as sacCer3) using BWA (Li and Durbin 2009) and single-nucleotide polymorphisms (SNPs) were identified using SAMtools (Li et al. 2009). SNP alleles, position, quality, and the number of high-quality reads mapping to the reference or alternate alleles were extracted from VCF files and analyzed in R. Oak- and vineyard-specific alleles were identified in each sample by comparing to SNPs found by sequencing the oak and vineyard strains. The read depth at each locus was calculated as the sum of reads mapping to reference and alternate alleles. The number of crossover events was identified in the advanced intercross F12 clones by identifying transitions between oak and vineyard SNPs. We required at least two adjacent SNPs from the same parent to define a crossover. For the panel of F2 segregants, numbers of crossovers were based on single transitions in marker genotypes.
QTL mapping using MULTIPOOL
To analyze sequencing under selection data we used MULTIPOOL, which performs genetic mapping from pooled sequencing experiments using a discrete dynamic Bayesian network (Edwards and Gifford 2012). The software takes SNP positions per chromosome as input and the read counts of each allele. MULTIPOOL analysis was performed on filtered data. SNPs with minor allele frequency < 10% were excluded. Samples used for comparative analysis of SNP frequencies were separated by 12–14 generations (low dilution rate chemostat) or 20–26 generations (high dilution rate chemostat). Replicate chemostats were analyzed separately and by combining reads at each SNP. Analyses were run in ‘contrast’ mode, with the exception of the advanced intercross results shown in Figure S5 in File S2. “Contrast” mode identifies significant differences between two experiments based on the null hypothesis that the underlying allele frequencies across the genome are the same between the two experiments. Each comparison was run with parameters n = 1000 or n = 200 (number of individuals) and r = 1000 or r = 2500 (length of centimorgan in bases). We found that different parameter combinations did not change the overall shape of the LOD profile; however, the n parameter has a large effect on the magnitude of LOD scores. To assess statistical significance, we performed null comparisons between replicate chemostats assayed prior to selection in the chemostat. The null comparisons had LOD scores ranges of −0.3 to 0.74 for n = 200 and 0.4–3.03 for n = 1000.
Analysis of variation in HXT6 and HXT7
The HXT6 and HXT7 genes were amplified individually using locus-specific PCR primers from the oak and vineyard strains and cloned in plasmids. Plasmid inserts were Sanger sequenced to catalog genetic variation between the oak and vineyard genes and used as templates for generating allele replacements. For the structure homology model, the vineyard HXT7 was modeled from PDB entry 4GBZ (Sun et al. 2012) using ModPipe (in ModBase) (Pieper et al. 2014). Reciprocal hemizygote strains were created by first replacing the HXT6 or HXT7 locus with a construct containing the G418 resistance marker (kanMX) in the homozygous oak and vineyard parental strains (BC248 and BC241). These strains were then mated to the opposite parental strain to create heterozygous gene deletions in the hybrid background. Allele replacements were created in haploid strains of the oak and vineyard genetic backgrounds [NCYC3606 and NCYC3591 (Cubillos et al. 2009)]. Overlapping PCR was used to create alleles containing single-amino acid modifications. For each allele replacement, HXT6 or HXT7 was first replaced by the URA3 gene and subsequently replaced by the modified allele. These strains were then crossed to the original oak and vineyard strain (BC248 and BC241) of the same background. The mated strains were sporulated and tetrads were screened to identify diploid homothallic prototrophs (i.e., containing functional HO and URA3 genes) exhibiting sensitivity to G418 and resistance to hygromycin, indicating inheritance of the sporulation marker from the original parental strains. The genotype at the HXT7 locus was determined by Sanger sequencing. Final allele replacement strains were also crossed to the opposite parental background to create homozygous allele replacements in the hybrid background.
Data availability
Strains and mapping populations are available upon request. Data and scripts used for analysis are available through the Open Science Framework: https://osf.io/p5z3f/ (DOI 10.17605/OSF.IO/P5Z3F) and bulk segregant sequencing data are available through the Sequence Read Archive (SRP101668).
Results
Distinct genetic architectures determine growth rate and lag duration distributions between environments
We have previously shown that two wild yeast isolates, isolated from an oak tree (BC248, hereafter “oak”) and from a vineyard (BC241, hereafter “vineyard”) differ in exponential growth rate (hereafter “growth rate”) and lag duration (hereafter “lag”) in media containing different glucose concentrations (Ziv et al. 2013b). On average, oak cells grow faster and lag for shorter amounts of time than vineyard cells, consistent with the general pattern that oak-associated strains grow better than vineyard strains in low-glucose conditions (Clowers et al. 2015). The difference in the response to increasing nutrient concentration can be characterized by growth in two different glucose environments: 0.22-mM (“growth rate-limiting” or “limiting”) and 4.44-mM (“growth rate-nonlimiting” or “nonlimiting”) glucose. To identify QTL that underlie variation in growth rate and lag, we used a panel of 374 recombinant segregants (Gerke et al. 2006) genotyped at 225 loci throughout the genome (Gerke et al. 2009). Each segregant was phenotyped in both growth rate-limiting and growth rate-nonlimiting glucose environments using a high-throughput microscopy-based microcolony assay (Levy et al. 2012; Ziv et al. 2013b).
The microcolony assay enables estimation of both the central tendency and dispersion of growth rate and lag for each genotype by measuring the growth of hundreds to thousands of genetically identical microcolonies within a single well of a microtiter plate. Growth rate distributions for wild isolates in both glucose environments are approximately normal and the central tendency, and dispersion, are characterized by the mean and SD, respectively. By comparison, lag distributions tend to be asymmetric and therefore the central tendency and dispersion are characterized by their median and MAD. Cell growth commences in < 1 hr of inoculation in nonlimiting glucose, so microcolonies do not have measurable lag times in that medium using our assay (Ziv et al. 2013b). As microcolonies collide prior to running out of glucose, our assay does not enable quantification of the stationary phase. Therefore, our phenotypic analysis resulted in three traits defined by central tendency and three traits defined by dispersion that are amenable to genetic mapping (Figure S1 in File S2).
Phenotypic analysis of recombinant segregants recapitulated three known correlations between phenotypes (Ziv et al. 2013b): (1) a strong positive correlation between median lag and lag MAD; (2) weak correlations between mean growth rate and growth rate SD; and (3) a negative correlation between mean growth rate and median lag (Figure S2 in File S2). There is also a positive correlation for mean growth rate in the limiting and nonlimiting glucose concentrations. As there are frequently relationships between the central tendency and dispersion of trait values, quantifying phenotypic variability requires estimates that are independent of the central tendency (Geiler-Samerotte et al. 2013). Therefore, we performed loess regression of estimates of dispersion (i.e., SD or MAD) as a function of estimates of central tendency (i.e., mean or median) and defined the residuals of the regression as phenotypic variability values used for mapping purposes (Figure S1 in File S2, Materials and Methods).
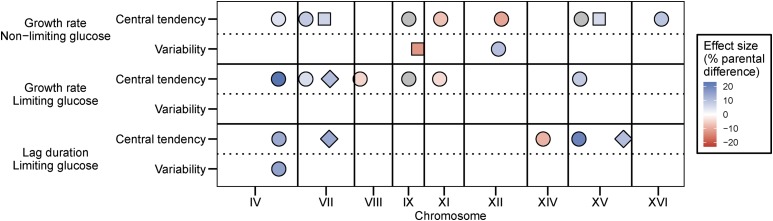
We identified QTL, defined as statistically significant associations between genotypes and traits, using the R package R/qtl (Broman et al. 2003) (Materials and Methods). We first searched for additive QTL and identified multiple such QTL for most traits (Figure 1 and Figure S3 in File S2). Genotype probabilities and LOD scores were calculated at each cM position (total map length is 4076 cM) and the average distance between genotyped markers is 19.4 cM (Materials and Methods). Both within and between traits, QTL were considered the same locus if they were within 30 cM of one another. We determined genome-wide significance levels for each QTL scan by determining the distribution of maximum LOD scores from 10,000 permutations of the data (Churchill and Doerge 1994; Broman et al. 2003). For mean growth rate in nonlimiting glucose, mean growth rate in limiting glucose, and median lag in limiting glucose, we found (using a genome-wide significance threshold of α = 0.05) seven, six, and five QTL, respectively (Figure 1). As expected based on trait correlations, some QTL are common between traits, but we also identified unique QTL for each trait (Figure 1 and Figure S3 and Table S1 in File S2). The amount of overlap between traits may be underestimated by eliminating loci that are close to significance. Increasing the significance threshold to 0.1 resulted in the identification of three additional loci. Although the new QTL were shared between phenotypes, each trait still retained unique loci (Figure 1). It should be noted that the overlap between correlated traits might also be overestimated due to correlated errors. The estimated effect of each QTL ranged from 4 to 23% of the difference in parental phenotypes (Figure S4 in File S2). Alleles at QTL that increase the trait value are found in both parents for each trait (Figure 1 and Figure S4 in File S2). We identified fewer QTL with additive effects for phenotypic variability traits: two for growth rate in nonlimiting glucose, none for growth rate in limiting glucose, and one for lag in limiting glucose (Figure 1). No additional QTL for variability traits were identified when the significance threshold was increased to 0.1 (Figure 1). Two of the three additive QTL contributing to phenotypic variability were also found to underlie phenotypic variation (Figure 1 and Table S1 in File S2). One of these two, a locus on chromosome IV, had an additive effect on four traits: mean growth rate in both glucose environments as well as median lag and lag variability in limiting glucose.
Figure 1.
Multiple QTL underlie phenotypic variation and variability of cell growth. Additive QTL identified for all analyzed traits are depicted. Chromosome size and QTL position correspond to genetic distance (in cM). Colors depict estimated effect sizes. Different shapes are used to distinguish distinct QTL on the same chromosome. Positive effects (blue) correspond to oak alleles that increase growth rate traits or decrease lag duration traits, whereas negative effects (red) correspond to vineyard alleles that increase growth rate traits or decrease lag duration traits. All loci were significant at α = 0.05 except those shown in gray, which were significant at α = 0.1.
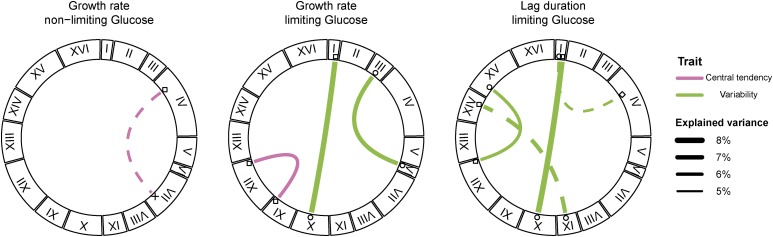
We tested for genetic interactions using two-dimensional genome scans (Materials and Methods). We identified (using a genome-wide significance threshold of α = 0.05) a total of five significant interactions across all traits (Figure 2 and Table S2 in File S2). The majority of interactions were found for variability traits (4/5). This pattern did not change when the significance threshold was increased to 0.1 (Figure 2), which results in the identification of three additional interactions (6/8 interactions for variability traits). We identified a case of sign epistasis, between a locus on chromosome I and a locus on chromosome X, that was common to two phenotypic variability traits: growth rate variability and lag variability in limiting glucose (Figure 3A). In this case, oak–oak and vineyard–vineyard combinations of the two QTL result in low variability whereas both combinations of oak–vineyard QTL result in high variability (Figure 3B). This symmetrical relationship means that the QTL exhibit no additive effects across the panel of segregants. Additionally, three loci that interact to contribute to lag variability, on chromosomes IX, XII, and XV, have interactive and additive effects on other traits. The chromosome XII locus interacts to affect mean growth rate in the limiting glucose concentration, whereas the loci on chromosomes IX and XV have additive effects on mean growth rate and median lag (Figure 1 and Figure 2). The effect sizes of interacting loci (given the genotype of the interaction partner) are comparable to the effect sizes of additive loci (Figure S4 in File S2).
Figure 2.
Genetic interactions contribute to phenotypic variation and variability of cell growth. Significant genetic interactions identified for phenotypic variation and variability traits are shown as lines connecting loci. Chromosome size and QTL position correspond to genetic distance (in cM). Line widths correspond to the percent of variance explained when only the two interacting loci are modeled. Interactions underlying heritable variation in central tendency (pink) and heritable variation in variability (green) were identified. Different shapes distinguish distinct loci on the same chromosome and correspond to shapes in Figure 1. Interactions were significant at α = 0.05 except those indicated by dashed lines, which were significant at α = 0.1.
Figure 3.
A sign epistatic interaction underlies heritable variation in growth rate variability. (A) LOD scores corresponding to additive (two-QTL) or full (two-QTL and interaction) models for each combination of chromosome I and X positions for linkage to phenotypic variability in growth rate measured in limiting glucose. (B) Average growth rate variability in the limiting glucose condition for the four genotype combinations corresponding to the two markers with the maximum LOD score difference shown in (A). Error bars represent SEs.
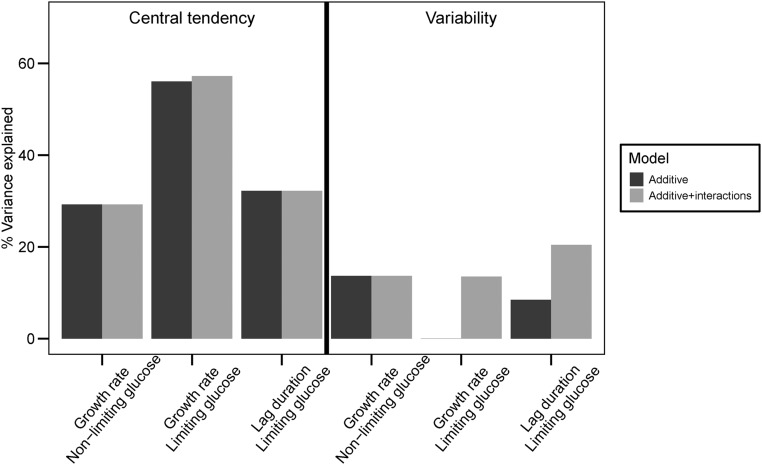
Central tendency traits differ from variability traits in the proportion of total trait variance explained by additive QTL compared with interactions. For central tendency traits, additive effects account for 29–56% of the total variance and genetic interactions only account for an additional 0–1.2% of the variance. By contrast, additive QTL effects only account for 0–14% of the variance in variability traits whereas interactions explain an additional 12–14% of the variance (Figure 4). It is not surprising that we explain less of the variance for variability traits overall as measuring variability is inherently noisier than measuring central tendency. Using estimates of variance between replicate wells of the parental strains, we estimated the proportion of phenotypic variance between the F2s that is genetic (Figure S1 in File S2, Materials and Methods). For central tendency traits, we find 83, 94, and 94% of the variance for growth rate in nonlimiting glucose, growth rate in limiting glucose, and lag duration in limiting glucose is genetic. As expected, the proportions for variability traits are lower; 14, 24, and 44% of the variance is genetic for the respective conditions.
Figure 4.
Variance in phenotypic variation is mainly explained by additive QTL whereas variance in phenotypic variability is mainly explained by genetic interactions. Percent of total trait variance explained per trait by a model comprising all identified additive QTL (black) or comprising all identified additive QTL and genetic interactions (gray) is shown.
Sequencing an advanced intercross population under selection increases mapping resolution
One of the challenges of QTL mapping is resolving loci to the causative gene and variant. We sought to improve the resolution of QTL mapping by using a method of bulk segregant mapping in which a pooled advanced intercross population is subjected to selection for the trait of interest and sequenced to identify changes in allele frequencies (Parts et al. 2011). We created an advanced intercross population starting from an oak/vineyard F1 using 11 rounds of meiosis and random mating (Materials and Methods). Allele frequencies and linkage in the advanced intercross population were determined by sequencing the final (F12) population and isolated clones. Allele frequencies in the F12 population deviated from the expectation of 0.5 at a number of loci (Figure S5 in File S2). A possible cause of this deviation is inadvertent selection during creation of the population. Indeed, two of the three major sporulation efficiency QTL known to segregate in this cross (Gerke et al. 2009) show strong nonrandom deviations in the F12 population (Figure S5 in File S2). Despite inadvertent selection during generation of the advanced intercross population, > 85% of SNPs still segregated with minor allele frequencies > 10% throughout the genome. Linkage in the intercrossed population was decreased compared with the F2 segregants as an average of 79.6 ± 19 crossover events were identified in three F12 clones (Figure S5 in File S2, Materials and Methods), compared to an average of 31.9 ± 5.6 in the 374 F2 segregants (Materials and Methods). The increase in recombination frequency is consistent with the genetic map expansion observed in previous studies (Parts et al. 2011).
Bulk segregant mapping requires a means of selecting, or enriching, genotypes with extreme phenotypes. We have previously shown that the effect of glucose concentration on the mean growth rates of the oak and vineyard strains can be recapitulated using glucose-limited chemostats (Ziv et al. 2013b). Therefore, to enrich for QTL conferring increased mean growth rate, we maintained the advanced intercross population in replicate glucose-limited chemostats at a low (D = 0.18 hr−1) or high (D = 0.35 hr−1) dilution rate. A lower dilution rate results in a lower steady-state glucose concentration in the chemostat (Ziv et al. 2013a). Therefore, the high dilution rate serves as a proxy for the nonlimiting glucose environment and the low dilution rate serves as a proxy for the limiting glucose environment. To directly assess the contribution of the advanced intercross population to QTL mapping resolution, we also pooled the panel of F2 segregants and grew them in the same chemostat conditions (Materials and Methods). We collected and sequenced multiple samples from each chemostat over 20–40 generations to minimize the chance of de novo mutations contributing to the selection. We determined allele frequencies using whole-genome population sequencing and identified QTL by comparing the allele frequencies between early and late time points using MULTIPOOL (Edwards and Gifford 2012) (Materials and Methods) (Figure S6 in File S2).
To directly compare results from the advanced intercross population with the genetic map used for interval mapping, we computed the maximal LOD score for each genomic interval flanked by markers used for interval mapping. We defined 209 intervals, with a median physical distance of 50 kb (range: 14–139 kb). A number of intervals with high LOD scores are common to all three QTL analyses (interval mapping of F2s, bulk segregant mapping of F2s, and bulk segregant mapping of the F12 intercross) (Figure S3 and Figure S6 and Table S3 in File S2). Among these intervals are those that contain the shared or environment-specific additive QTL for mean growth rate on chromosomes IV, VIII, and XVI. Additionally, the growth rate QTL at the left tip of chromosome VII, originally identified in both glucose concentrations, was also found using bulk segregant approaches, but only in the low-dilution condition. A failure to identify a QTL using the advanced intercross can be explained by inadvertent selection in the same region during creation of the advanced intercross, for example, the middle of chromosome VII (Figure S5 in File S2). However, additional regions were also missing from the analysis of the advanced intercross (but common for the bulk segregant and interval mapping of the F2s), specifically, regions of chromosomes VII, XI, XII, and XV (Table S3 in File S2). The small sample size for F2 segregants used for sequencing under selection could potentially confound the analysis as nonrandom associations between true QTL and unlinked loci may result in spurious linkage signals. This is supported by the observation of regions with high LOD scores for the F2 pool not shared by the advanced intercross or the interval mapping of the F2 segregants (Table S3 in File S2). As the differences between the interval mapping of F2s and the bulk segregant mapping of the advanced intercross may be due to technical or biological reasons, we focused on loci identified by both methods, which represent strong candidates for loci that are important for growth in low glucose environments.
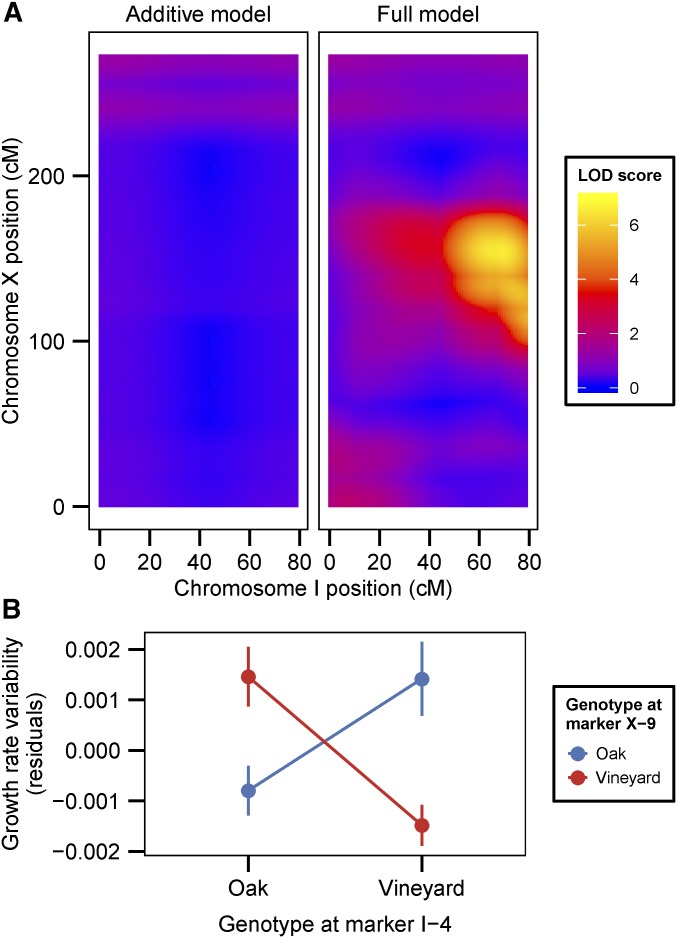
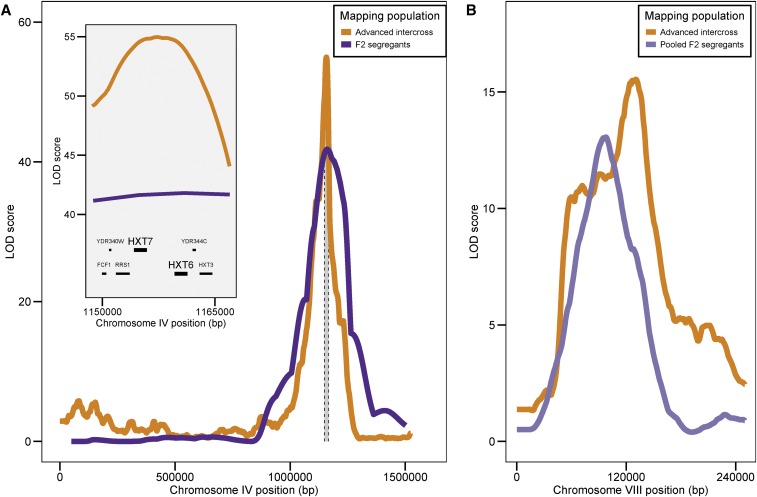
Decreased linkage in the advanced intercross population has two consequences for QTL resolution exemplified by two QTL. First, LOD scores decreased rapidly from their peaks at individual QTL (Figure 5A). The size of 2-LOD drop intervals for the chromosome IV QTL decreased from 67 kb (limiting) and 245 kb (nonlimiting) using interval mapping with F2s to 9.3 kb (low dilution) and 31.8 kb (high dilution) using the advanced intercross population. Most of the increased resolution was due to the use of the advanced intercross and not the bulk segregant approach, as the interval for the pooled F2s ranged between 22.9 and 78.8 kb in low glucose and between 66.2 and 203.4 kb in high glucose, depending on the choice of MULTIPOOL parameters (Materials and Methods). The increased resolution enabled us to identify HXT6 and HXT7, which encode high-affinity glucose transporters, as candidate causative genes at the chromosome IV QTL (Figure 5A). Second, linkage analysis in the advanced intercross population indicates that a QTL on chromosome VIII is composed of multiple linked QTL (Figure 5B). Analysis of the F2 pool suggests only a single QTL at this locus, which may be an example of a ghost QTL (Doerge 2002), in which two linked QTL result in a maximum LOD score at a location between the two loci due to insufficient mapping resolution (Figure 5B).
Figure 5.
Increased QTL resolution due to increased recombination in an advanced intercross population. (A) LOD score profiles for all of chromosome IV obtained by interval mapping using F2 segregants (dark purple) or sequencing under selection of an advanced intercross population (orange). For the interval mapping profile, genetic distances were converted to physical distances based on marker positions. The inset shows LOD profiles for a 15-kb region (gray region in main plot) centered on the peak LOD score and the corresponding physical position of genes within the QTL. (B) LOD score profiles for a region of chromosome VIII analyzed by sequencing under selection of a F2 pool (light purple) or using an advanced intercross population (orange). MULTIPOOL parameters for depicted data are n = 1000 and r = 1000.
Sequence variation in HXT7 contributes to variation in growth
HXT6 and HXT7 encode nearly identical high-affinity glucose transporters, making them plausible candidate genes underlying the growth rate QTL on chromosome IV. De novo amplifications of these genes are frequently selected during experimental evolution in glucose-limited chemostats (Brown et al. 1998; Gresham et al. 2008; Kao and Sherlock 2008); however, whole-genome sequencing of the oak and vineyard parental strains did not identify copy number variation at this locus. As high-throughput short-read sequencing is unable to accurately resolve nucleotide variation in duplicate genes, we cloned and sequenced HXT6 and HXT7 from each parent (File S1, Materials and Methods). To assess the contribution of natural variation in HXT6 and HXT7 to variation in growth rate and lag duration, we created all possible reciprocal hemizygotes, in which one parental allele of HXT6 or HXT7 is knocked out, in the F1 hybrid. Hemizygosity for either parental HXT6 allele results in no phenotypic difference in the F1, whereas the F1 containing only the oak HXT7 allele shows significant phenotypic differences from the F1 containing only the vineyard HXT7 allele, suggesting that variation in HXT7 alone contributes to variation in growth (Figure S7 in File S2).
Sequence analysis of the oak and vineyard HXT7 alleles revealed 79 SNPs and 26 aa differences in the 1713-bp (571 aa) open reading frame (ORF). To test the effect of sequence variation in HXT7, we performed allele swaps in parental strains and in the F1. We created HXT7 allele replacement strains that contained no additional genetic modifications except the replaced allele, which included the entire ORF and 530 bp of upstream sequence (which contained three SNPs and a 2-bp indel) (Materials and Methods). By considering sequence conservation and the nature of the amino acid changes, we identified a variant amino acid, T469Q, as potentially affecting protein function. This was further supported by using a protein structure model based on the crystal structure of the bacterial homolog XylE (Sun et al. 2012) (Materials and Methods). The structural homology model showed the amino acid pointing into the transporter channel, in close proximity to amino acids known to interact with the translocating sugar molecule (Sun et al. 2012; Madej et al. 2014). To test the effect of the T469Q variant, we also created single-amino acid replacement alleles in each genetic background.
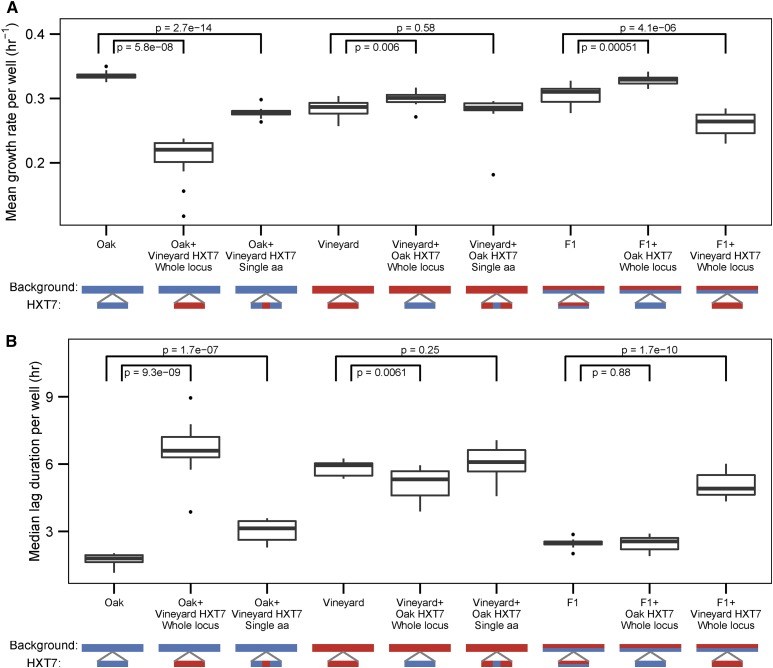
Phenotypic analyses revealed significant differences attributable to allele replacements in the growth-limiting glucose concentration consistent with effects determined by linkage mapping. Namely, the oak HXT7 allele confers enhanced mean growth rate (Figure 6A) and decreased median lag (Figure 6B). Interestingly, the effect of either the oak or vineyard HXT7 allele is dependent on the genetic background. A strain containing the vineyard allele in the otherwise oak background grows slower and lags for a longer time than the vineyard parent. In contrast, the oak allele in the vineyard background caused a small but significant increase in growth rate and decrease in lag duration. Similar effects were found in the F1 background (Figure 6). This is consistent with genome background effects modifying the effect of either parental HXT7 allele. We found that the single-amino acid modification T469Q in the oak allele significantly reduces the growth rate of the oak parent, but has a smaller effect than replacement with the entire HXT7 vineyard allele for both mean growth rate (Figure 6A) and median lag (Figure 6B), suggesting that additional variation within HXT7 affects growth. Conversely, engineering the reciprocal Q469T modification into the vineyard HXT7 allele does not alter any of the growth phenotypes in a vineyard genetic background consistent with intra-allelic variation in the oak HXT7 allele mediating the effect of the T469Q variant. Consistent with a smaller effect size of the QTL in the nonlimiting glucose concentration as determined using linkage mapping, replacing the entire oak HXT7 allele with the vineyard HXT7 allele in the oak background is required to detect a decrease in growth rate in nonlimiting conditions (Figure S8A in File S2). Although the chromosome IV QTL containing HXT7 is linked to variability in lag, we did not find a significant effect on lag variability in allele-swapped strains (Figure S8B in File S2). However, this is likely due to the small number of strains used in the regression to estimate median-independent lag variability, which reduces our power to detect a statistically significant difference (Levy and Siegal 2008).
Figure 6.
Background-dependent effects on HXT7 contribute to cell growth rate variation. Distributions of (A) mean growth rate and (B) median lag duration for allele replacement strains grown in limiting glucose. P-values are for two sample t-tests (n = 12 for each analyzed strain). The diploid strain genotype and the genotype at the HXT7 locus are of either oak (blue) or vineyard (red) parental origin. Single-amino acid allele replacements within HXT7 are depicted as mosaics of oak and vineyard genotypes at the HXT7 locus.
Although we have not found evidence for adaptive evolution at the HXT7 locus in natural populations, it is noteworthy that HXT7 is a recurrent target of selection in long-term experimental evolution in glucose-limited environments. Copy number variation containing HXT6/HXT7, but not nucleotide variation, has been reported in multiple experimental evolution studies (Brown et al. 1998; Gresham et al. 2008; Kao and Sherlock 2008; Koschwanez et al. 2013; Selmecki et al. 2015). To study the effect of an HXT6/HXT7 amplification on glucose-dependent growth, we measured growth rates of an evolved clone in which HXT6/HXT7 is amplified. (Gresham et al. 2008) at different glucose concentrations using the microcolony assay. We find that a HXT6/HXT7 amplification has a growth-rate advantage only at the limiting glucose concentration but results in decreased growth rate at higher glucose concentrations (Figure S9 in File S2), consistent with antagonistic pleiotropy. By contrast, the naturally occurring HXT7 allele in the oak strain is beneficial in both limiting and nonlimiting glucose environments.
Discussion
The field of quantitative genetics was established nearly 100 years ago, reconciling the inheritance of continuously distributed traits with Mendelian genetics (Nelson et al. 2013b). Large-scale genetic mapping of quantitative traits first became feasible following the utilization of molecular polymorphisms as genetic markers (Botstein et al. 1980) and development of analytical methods (Lander and Botstein 1989). Recently, with increased sample sizes and resources, genetic mapping studies are detecting more QTL but also uncovering surprising complexity, including gene-by-environment interactions, epistasis, and linkage between causative loci (Mackay et al. 2009). The results of our study serve to emphasize the prevalence of these characteristics even when considering closely related traits and environments. Furthermore, we show that genetic interactions and gene-by-environment interactions underlie variation in both the central tendency and variability of traits.
Gene-by-environment interactions are a result of genetic loci that have different effects on phenotypic variance in different conditions. Gene-by-environment interactions are frequently found in QTL studies, and in one study were shown to affect the expression of a third of yeast genes (Smith and Kruglyak 2008). We find both shared loci with different effect sizes as well as environment-specific loci and genetic interactions when quantifying cell growth in two environments that differ in glucose concentration 20-fold. Measuring changes in QTL effect sizes over a finer gradient of glucose concentration may be informative, analogous to dose-dependent effects observed for chemical resistance (Wang and Kruglyak 2014). For example, the contribution of HXT7 to growth rate variation might indicate the extent of control that the nutrient transport step has on growth rate, potentially relating the transporter Km to the Monod constants (Ks) of the oak and vineyard strains (Ziv et al. 2013b). We find that natural variation in HXT7 does not exhibit antagonistic pleiotropy, whereas amplification of HXT7, which is repeatedly observed in experimental evolution studies, confers a cost at higher glucose concentrations. Identification of a target of selection in laboratory evolution experiments that also shows causative natural variation suggests that experimental evolution serves as an important window into evolutionary adaptation in nature. Contrasting the outcomes between experimental evolution in the lab and natural variation may prove fruitful for understanding the causes and consequences of short-term adaptation on allelic variation, as compared with long evolutionary histories.
Recently, there has been increased interest in searching for so-called variance QTL (vQTL), by analyzing the difference in variance between genotypic classes, instead of the difference in means (Ronnegard and Valdar 2011; Shen et al. 2012; Yadav et al. 2016). vQTL are not the same as variability QTL, because for vQTL the relevant variance is computed across strains (specifically those strains with a particular allele at a particular locus). Therefore, vQTL analyses are unlikely to uncover loci that affect within-strain variance (variability), and instead they uncover loci with alleles that suppress or enhance the mean effects of genetic differences at other loci (Nelson et al. 2013a; Yadav et al. 2016). When clonal data and repeated measurements of different individuals with the same genotype are available, phenotypic variability can be directly estimated and mapped as a quantitative trait. Alleles determining variability segregate in natural populations and have been mapped in yeast (Ansel et al. 2008), flies (Mackay and Lyman 2005; Ayroles et al. 2015), plants (Hall et al. 2007; Jimenez-Gomez et al. 2011), and mice (Fraser and Schadt 2010). To our knowledge, our study is the first to find genetic interactions determining phenotypic variability. Moreover, we find that genetic interactions make up a larger proportion of explained trait variance for variability traits compared to traits of central tendency. This is consistent with the observation that phenotypic stabilizers (genes that increase phenotypic variability when absent or impaired) tend to be interaction hubs (Levy and Siegal 2008; Bauer et al. 2015). In particular, stabilizers are characterized by many synthetic lethal genetic interactions (Levy and Siegal 2008). However, these studies have also shown that single-gene mutations can affect phenotypic variability. It is tempting to think that, in our study, the parental strains have evolved to have low variability and the combination of disparate alleles leads to higher variability. This is supported by the effects of some but not all of the identified genetic interactions, including a sign epistatic interaction in which the interaction between two loci results in high phenotypic variability when the alleles are inherited from different parents and low variability when the alleles are from the same parent. It will be interesting to see if this observation will generalize to variability in different phenotypes and systems.
Our study highlights the advantage of using different mapping approaches to dissect the genetic basis of complex traits. Although individual segregant analysis has the advantage of detecting genetic interactions, the increased resolution of our bulk segregant approach was remarkable. The increased resolution is due to both use of an advanced intercross population and the increased sampling due to bulk segregant analysis. Importantly, our implementation of bulk segregant analysis did not require extensive parental-strain construction. Although previous studies used genetically engineered strains (Ehrenreich et al. 2010; Cubillos et al. 2013), we created our advanced intercross population using homothallic diploid strains without the need for auxotrophic or drug resistance markers. This approach should be readily adaptable to other strains and related species, enabling bulk segregant approaches without extensive strain manipulation.
We confirmed the effect of sequence variation in HXT7 on variation in growth rate and lag duration. However, we identified important distinctions between the segregant analysis and the allele replacements. Specifically, the differential effect of the allele depending on the genetic background indicates the presence of additional genetic interactions not identified by our two-dimensional scan. These effects might reflect higher order interactions involving more than two loci (Taylor and Ehrenreich 2014). Alternatively, they might be the result of an accumulation of undetected pairwise interactions. Although we did not find significant interactions involving the chromosome IV QTL position for mean growth rate and median lag duration, the estimated effect of the locus was consistently smaller in the vineyard background when considering the genotype at other additive loci, particularly in the higher glucose concentration.
Our study emphasizes the inherent challenges of accurately predicting phenotype from genotype. It is necessary to account for genetic interactions, environmental variation, and phenotypic variability, as the estimated marginal additive effect of an allele may not reflect the actual effect in any specific genetic background.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.195180/-/DC1.
Acknowledgments
We thank members of the Gresham and Siegal laboratories, as well as Matt Rockman, Fred Cross, and Gloria Corruzi, for feedback and helpful comments. We thank the Genomics Core Facility at NYU CGSB for Illumina sequencing, Barak Cohen for providing yeast strains, and Evan Baugh for assistance with homology structural modeling. This work was supported by National Institutes of Health grants R01GM086673, R01GM097415, R35GM118170 (to M.L.S.) and R01GM1007466 (to D.G.), and National Science Foundation grant MCB1244219 (to D.G.).
Footnotes
Communicating editor: J. B. Wolf
Literature Cited
- Ansel J., Bottin H., Rodriguez-Beltran C., Damon C., Nagarajan M., et al. , 2008. Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genet. 4: e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Buchanan S. M., O’Leary C., Skutt-Kakaria K., Grenier J. K., et al. , 2015. Behavioral idiosyncrasy reveals genetic control of phenotypic variability. Proc. Natl. Acad. Sci. USA 112: 6706–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. R., Li S., Siegal M. L., 2015. Essential gene disruptions reveal complex relationships between phenotypic robustness, pleiotropy, and fitness. Mol. Syst. Biol. 11: 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg A., 2011. Measuring growth rate in high-throughput growth phenotyping. Curr. Opin. Biotechnol. 22: 94–102. [DOI] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W. T., Lite T.-L. V., Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W., 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32: 314–331. [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Saldanha A. J., Dolinski K., Botstein D., 2005. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell 16: 2503–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Todd K. M., Rosenzweig R. F., 1998. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 15: 931–942. [DOI] [PubMed] [Google Scholar]
- Carlborg Ö., Haley C. S., 2004. Opinion: epistasis: too often neglected in complex trait studies? Nat. Rev. Genet. 5: 618–625. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowers K. J., Heilberger J., Piotrowski J. S., Will J. L., Gasch A. P., 2015. Ecological and genetic barriers differentiate natural populations of Saccharomyces cerevisiae. Mol. Biol. Evol. 32: 2317–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Louis E. J., Liti G., 2009. Generation of a large set of genetically tractable haploid and diploid Saccharomyces strains. FEMS Yeast Res. 9: 1217–1225. [DOI] [PubMed] [Google Scholar]
- Cubillos F. A., Billi E., Zörgö E., Parts L., Fargier P., et al. , 2011. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol. Ecol. 20: 1401–1413. [DOI] [PubMed] [Google Scholar]
- Cubillos F. A., Parts L., Salinas F., Bergstrom A., Scovacricchi E., et al. , 2013. High-resolution mapping of complex traits with a four-parent advanced intercross yeast population. Genetics 195: 1141–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A., Soller M., 1995. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge R. W., 2002. Multifactorial genetics: mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 3: 43–52. [DOI] [PubMed] [Google Scholar]
- Edwards M. D., Gifford D. K., 2012. High-resolution genetic mapping with pooled sequencing. BMC Bioinformatics 13(Suppl. 6): S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Torabi N., Jia Y., Kent J., Martis S., et al. , 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464: 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H. B., Schadt E. E., 2010. The quantitative genetics of phenotypic robustness. PLoS One 5: e8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte K., Bauer C., Li S., Ziv N., Gresham D., et al. , 2013. The details in the distributions: why and how to study phenotypic variability. Curr. Opin. Biotechnol. 24: 752–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J. P., Chen C. T. L., Cohen B. A., 2006. Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics 174: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J., Lorenz K., Cohen B., 2009. Genetic interactions between transcription factors cause natural variation in yeast. Science 323: 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D., Desai M. M., Tucker C. M., Jenq H. T., Pai D. A., et al. , 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. C., Dworkin I., Ungerer M. C., Purugganan M., 2007. Genetics of microenvironmental canalization in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 13717–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C. J. R., Parts L., Bergström A., Liti G., Mustonen V., 2013. Inferring genome-wide recombination landscapes from advanced intercross lines: application to yeast crosses. PLoS One 8: e62266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Gomez J. M., Corwin J. A., Joseph B., Maloof J. N., Kliebenstein D. J., 2011. Genomic analysis of QTLs and genes altering natural variation in stochastic noise. PLoS Genet. 7: e1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao K. C., Sherlock G., 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschwanez J. H., Foster K. R., Murray A. W., 2013. Improved use of a public good selects for the evolution of undifferentiated multicellularity. Elife 2: e00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. F., Siegal M. L., 2008. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 6: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. F., Ziv N., Siegal M. L., 2012. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 10: e1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Louis E. J., 2012. Advances in quantitative trait analysis in yeast. PLoS Genet. 8: e1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Lyman R. F., 2005. Drosophila bristles and the nature of quantitative genetic variation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1513–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Stone E. A., Ayroles J. F., 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10: 565–577. [DOI] [PubMed] [Google Scholar]
- Madej M. G., Sun L., Yan N., Kaback H. R., 2014. Functional architecture of MFS D-glucose transporters. Proc. Natl. Acad. Sci. USA 111: E719–E727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V., 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J., 1949. The growth of bacterial cultures. Annu. Rev. Microbiol. 3: 371–394. [Google Scholar]
- Nelson R. M., Pettersson M. E., Li X., Carlborg Ã., 2013a Variance heterogeneity in Saccharomyces cerevisiae expression data: trans-regulation and epistasis. PLoS One 8: e79507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R. M., Pettersson M. E., Carlborg Ö., 2013b A century after Fisher: time for a new paradigm in quantitative genetics. Trends Genet. 29: 669–676. [DOI] [PubMed] [Google Scholar]
- Parts L., Cubillos F. A., Warringer J., Jain K., Salinas F., et al. , 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 21: 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L., 2012. Using cell-to-cell variability–a new era in molecular biology. Science 336: 425–426. [DOI] [PubMed] [Google Scholar]
- Pieper U., Webb B. M., Dong G. Q., Schneidman-Duhovny D., Fan H., et al. , 2014. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 42: D336–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. R., Wray N. R., Visscher P. M., 2014. Explaining additional genetic variation in complex traits. Trends Genet. 30: 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnegard L., Valdar W., 2011. Detecting major genetic loci controlling phenotypic variability in experimental crosses. Genetics 188: 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A. J., Brauer M. J., Botstein D., 2004. Nutritional homeostasis in batch and steady-state culture of yeast. Mol. Biol. Cell 15: 4089–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A. M., Maruvka Y. E., Richmond P. A., Guillet M., Shoresh N., et al. , 2015. Polyploidy can drive rapid adaptation in yeast. Nature 519: 349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Pettersson M., Rönnegård L., Carlborg Ö., 2012. Inheritance beyond plain heritability: variance-controlling genes in Arabidopsis thaliana. PLoS Genet. 8: e1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. N., Kruglyak L., 2008. Gene–environment interaction in yeast gene expression. PLoS Biol. 6: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. [DOI] [PubMed] [Google Scholar]
- Sun L., Zeng X., Yan C., Sun X., Gong X., et al. , 2012. Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 490: 361–366. [DOI] [PubMed] [Google Scholar]
- Swinnen S., Schaerlaekens K., Pais T., Claesen J., Hubmann G., et al. , 2012. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res. 22: 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. B., Ehrenreich I. M., 2014. Genetic interactions involving five or more genes contribute to a complex trait in yeast. PLoS Genet. 10: e1004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2012. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Wang X., Kruglyak L., 2014. Genetic basis of haloperidol resistance in Saccharomyces cerevisiae is complex and dose dependent. PLoS Genet. 10: e1004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening S., Lin G., Fritsch E. S., Tekkedil M. M., Anders S., et al. , 2014. An evaluation of high-throughput approaches to QTL mapping in Saccharomyces cerevisiae. Genetics 196: 853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A., Dhole K., Sinha H., 2016. Differential regulation of cryptic genetic variation shapes the genetic interactome underlying complex traits. Genome Biol. Evol. 8: 3559–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Loos R. J. F., Powell J. E., Medland S. E., Speliotes E. K., et al. , 2012. FTO genotype is associated with phenotypic variability of body mass index. Nature 490: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G., 2014. “Particle genetics”: treating every cell as unique. Trends Genet. 30: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv N., Brandt N. J., Gresham D., 2013a The use of chemostats in microbial systems biology. J. Vis. Exp. 80: 50168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv N., Siegal M. L., Gresham D., 2013b Genetic and nongenetic determinants of cell growth variation assessed by high-throughput microscopy. Mol. Biol. Evol. 30: 2568–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and mapping populations are available upon request. Data and scripts used for analysis are available through the Open Science Framework: https://osf.io/p5z3f/ (DOI 10.17605/OSF.IO/P5Z3F) and bulk segregant sequencing data are available through the Sequence Read Archive (SRP101668).