Abstract
Aging is associated with widespread changes in genome-wide patterns of DNA methylation. Thousands of CpG sites whose tissue-specific methylation levels are strongly correlated with chronological age have been previously identified. However, the majority of these studies have focused primarily on cosmopolitan populations living in the developed world; it is not known if age-related patterns of DNA methylation at these loci are similar across a broad range of human genetic and ecological diversity. We investigated genome-wide methylation patterns using saliva- and whole blood-derived DNA from two traditionally hunting and gathering African populations: the Baka of the western Central African rain forest and the ≠Khomani San of the South African Kalahari Desert. We identified hundreds of CpG sites whose methylation levels are significantly associated with age, thousands that are significant in a meta-analysis, and replicate trends previously reported in populations of non-African descent. We confirmed that an age-associated site in the promoter of the gene ELOVL2 shows a remarkably congruent relationship with aging in humans, despite extensive genetic and environmental variation across populations. We also demonstrate that genotype state at methylation quantitative trait loci (meQTLs) can affect methylation trends at some age-associated CpG sites. Our study explores the relationship between CpG methylation and chronological age in populations of African hunter-gatherers, who rely on different diets across diverse ecologies. While many age-related CpG sites replicate across populations, we show that considering common genetic variation at meQTLs further improves our ability to detect previously identified age associations.
Keywords: DNA methylation, aging, epigenetics, diverse human populations
AGING is a degenerative process that is associated with changes in many molecular, cellular, and physiological functions. Research identifying biomarkers associated with these changes has the potential to generate accurate predictions of both chronological and biological age in humans for health care and forensic applications. Recent epigenomic studies have shown that patterns of DNA methylation change substantially with chronological age: genome-wide methylation levels decrease with increasing age, while certain genomic regions, such as CpG islands, become more methylated with increasing age (Heyn et al. 2012; Hannum et al. 2013; Johansson et al. 2013; Teschendorff et al. 2013; Jones et al. 2015). An epigenome-wide study examining >475,000 CpG sites found significant age-associated changes in DNA methylation at almost one-third of sites (Johansson et al. 2013), demonstrating the extensive and stereotypic effect of aging on the human epigenome. Many previously proposed molecular biomarkers for aging, including leukocyte telomere length (Blasco 2007), aspartic acid racemization (Helfman and Bada 1975), and expression levels of certain genes (Simm et al. 2008; Li et al. 2011; Holly et al. 2013), can be challenging for age estimation due to a lack of precision, instability over time, or difficulty in measuring the quantity of interest (Meissner and Ritz-Timme 2010). In contrast, DNA methylation values measured from relatively few (from three to up to a few hundred) age-associated CpG sites (a-CpGs) have been shown to yield highly precise and accurate estimates of chronological age (Bocklandt et al. 2011; Horvath 2013; Weidner et al. 2014). Continual technological improvements, in particular the introduction of the Illumina Infinium HumanMethylation450 BeadChip array, have greatly expanded the scope of epigenetics research. This platform increases the density of assayed CpG sites across the human genome compared to the older Infinium HumanMethylation27 array, leading to the discovery of several novel potential aging biomarkers (Garagnani et al. 2012).
Changes in DNA methylation at putative a-CpGs may be affected both by genetic and environmental factors, in addition to aging itself. Extrinsic environmental factors such as smoking, sun exposure, and obesity, for example, are associated with specific changes in DNA methylation patterns (Grönniger et al. 2010; Breitling et al. 2011; Almén et al. 2014; Vandiver et al. 2015). Intrinsic factors, such as genetic background, can also influence patterns of epigenetic aging, including “baseline” DNA methylation levels at a-CpGs and the rate of change with age (Bell et al. 2011; Gentilini et al. 2013; Hannum et al. 2013). Importantly, specific genetic variants occurring at different frequencies or involving population-specific gene-environment interactions, can lead to patterns of DNA methylation that differ between human ethnic groups (Fraser et al. 2012; Heyn et al. 2013; Fagny et al. 2015; Galanter et al. 2017) and drive divergent patterns of epigenetic aging. Few studies have explored epigenetic aging while also explicitly considering ancestry (but see Zaghlool et al. 2015 and Horvath et al. 2016), and most previous work has focused on cosmopolitan populations of European origin (Hannum et al. 2013; Johansson et al. 2013; Florath et al. 2014). However, it cannot be assumed that age-related DNA methylation trends identified in one human population will be the same in other populations. Further validation of potential DNA methylation-based aging biomarkers in cohorts of diverse ethnic backgrounds is therefore essential before they can be widely applied in the fields of health care, anthropology, and forensics (Meissner and Ritz-Timme 2010). It is also important to note that different human cell types exhibit significantly different genome-wide methylation patterns (Illingworth et al. 2008; Rakyan et al. 2008; Byun et al. 2009; Christensen et al. 2009), a factor that affects a-CpGs as well (Farré et al. 2015).
To explore the impact of genetic ancestry and cell specificity on epigenetic aging, we measured DNA methylation at >480,000 CpG sites in saliva and peripheral whole blood samples from 189 African hunter-gatherer individuals from two populations: the ≠Khomani San of the South African Kalahari Desert and the Baka rain forest hunter-gatherers (also known as “pygmies”; Verdu and Destro-Bisol 2012) of the western Central African rain forest. These two populations diverged early on from the ancestors of all other modern humans, and exhibit much greater genomic variation than other populations whose global DNA methylation patterns have been assayed so far (Verdu et al. 2009; Veeramah et al. 2012). The ≠Khomani San, in particular, are among the most genetically diverse populations in the world (Verdu et al. 2009; Henn et al. 2011; Veeramah et al. 2012). Furthermore, the ≠Khomani San and the Baka differ in terms of their nutritional subsistence, ecological environs (semidesert and equatorial rain forest, respectively), and physical activity levels from the widely studied cohorts of cosmopolitan populations. By using DNA methylation data from these populations, we are able to explore patterns of epigenetic aging across a greater range of human genetic diversity and test previously published epigenetic age-prediction models to determine their accuracy across ancestries and cell types.
Materials and Methods
DNA and ethnographic collection
Saliva was collected from 56 ≠Khomani San individuals (aged 27–91, median age 62) and 36 Baka individuals (aged 5–59, median age 30) using Oragene DNA self-collection kits (Supplemental Material, Figure S1). Blood was collected from 97 additional Baka individuals (aged 16–90 years, median age 44 years) for a previous study (Fagny et al. 2015) (Figure S1). DNA samples from the ≠Khomani San were collected with written informed consent and approval of the Human Research Ethics Committee of Stellenbosch University (N11/07/210), South Africa, and Stanford University (protocol 13829). ≠Khomani San participant ages were verified ethnographically on a case-by-case basis. Various documents, such as birth certificates, wedding certificates, school records, and other forms of identification (e.g., apartheid government identification documents), were cross-referenced to identify any inconsistencies. Local major events, such as the creation of the Kalahari National Park in 1931, were also used to verify participants’ life-history stage. DNA samples from the Baka were collected with informed consent from all participants and from both parents of any participants under the age of 18. Ethical approval for this study was obtained from the institutional review boards of Institut Pasteur, Paris, France (RBM 2008-06 and 2011-54/IRB/3). Baka participant ages were determined ethnographically by Alain Froment by comparing individuals from a single cohort to one another, and with reference to major historical events. Baka individual ages are estimated to be accurate to within 5 years. The Baka saliva sample was known to contain nine trios and nine unrelated individuals.
DNA methylation data generation and data processing
The 97 Baka whole blood samples were previously processed and published in Fagny et al. (2015), while the 92 saliva samples were newly generated for this study. DNA extracted from all samples was bisulfite converted, whole-genome amplified, fragmented, and hybridized to the Illumina Infinium HumanMethylation450 BeadChip. This array assays methylation levels at >485,000 CpG sites throughout the genome through allele-specific, single-base extension of the target probe with a fluorescent label. The saliva samples from both populations were assayed together in two batches, and the blood samples in six batches. Methylation data from Illumina methylation arrays often exhibit substantial batch effects; that is, samples from one run may vary systematically from the same samples on a different run due to technical artifacts. To partially account for this, we included one ≠Khomani San individual from the saliva data set on both runs, and observed that the overall correlation between β values from the first and second runs was >0.99. Technical replicates were also included in the whole blood assay, and the overall correlations of β values between repeat individuals were all >0.98. However, these high correlation values do not preclude the presence of significant batch effects that affect only a subset of the genome, or offset β values by a constant factor across the entire genome. We further corrected for these using principal components analyses (PCA), described below.
The intensity of fluorescence was used to calculate DNA methylation levels. We removed probes with a detection P-value >0.01 in at least one sample, probes that were found to map to multiple genomic regions or to the sex chromosomes, or to contain known SNPs; leaving 334,079 sites in the saliva data set and 364,753 sites in the blood data set for subsequent analysis. Probe SNPs were identified using the 450K array annotation file published by Price et al. (2013) and by cross-referencing the genomic coordinates of our samples’ genotype data and the DNA methylation microarray probes using bedtools (Quinlan and Hall 2010). These values were background and color corrected, and technical differences between type I and type II probes were corrected by performing quantile and subset-quantile within-array normalization (SWAN) using the lumi and minfi R packages (Du et al. 2008; Maksimovic et al. 2012; Aryee et al. 2014). For a discussion of the various technical issues inherent in the 450K array design, see Dedeurwaerder et al. (2013) and Makismovic et al. (2012). One Baka individual was flagged for having abnormally low bisulfite controls. We conducted PCA separately on the saliva and blood methylation data sets using the prcomp function in R (Figure S2). All analyses were performed using continuous β values for each CpG site, which range from 0 (indicating that the site is completely unmethylated) to 1 (completely methylated).
SNP genotype data
The DNA samples were genotyped on either the Illumina OmniExpress, OmniExpressExome, OmniOne, or HumanHap550 SNP array (Henn et al. 2011; Patin et al. 2014; Fagny et al. 2015; Uren et al. 2016). All Baka individuals and 48 of the 56 ≠Khomani San individuals were successfully genotyped. OmniExpress data from the Baka blood samples was imputed using the results of the OmniOne genotyping. The data sets were filtered using a genotyping threshold of 0.95 and a minor allele frequency threshold of 0.01.
Ancestry inference
We intersected the genotype data generated for the Baka and ≠Khomani San with genotype data from African and European populations (specifically the Biaka pygmies, Mbuti pygmies, Namibian San, southern Bantu speakers, Kenyan Bantu, Yoruba, French, and Italian) generated by the Human Genome Diversity Project on the Illumina HumanHap array (Li et al. 2008). We performed an unsupervised clustering analysis using ADMIXTURE (Alexander et al. 2009) on the resulting data set of 254,080 SNPs from 319 individuals to determine their global ancestry proportions.
Saliva epigenome-wide association study
We used the R package CpGassoc to conduct an epigenome-wide association study (EWAS) test for the Baka saliva data (Barfield et al. 2012). Related individuals were assigned a shared family identity variable, while the nine unrelated individuals in the cohort were each assigned a unique family identity. Family identity as a random effect, the second and fourth DNA methylation principal components (PCs), and percentage of Bantu ancestry were used as covariates in testing for association with age:
We used the program EMMAX with the dosage option to conduct the EWAS on the ≠Khomani San methylation data. After removing the outlier individuals, there were 44 genotyped ≠Khomani San individuals in our data set. We generated a Balding–Nichols kinship matrix using genotype data from these individuals, which was included in the model to correct for relatedness within the population. For the ≠Khomani San analysis, proportions of European and Bantu ancestry, methyltyping batch, and the first DNA methylation PC were used as covariates:
The combination of PC and historically appropriate ancestry covariates used in the EWAS was selected to minimize the genomic inflation factor (Figure S3). We note that these low genomic inflation factors were sometimes obtained by including PCs that were moderately correlated with age as covariates (Figure S4), which may decrease our power to detect a-CpGs. By minimizing the genomic inflation factor in this way, our EWAS analyses are likely to be overly conservative, especially given that a substantial fraction of the 450K array sites actually do become differentially methylated with age (Hannum et al. 2013; Johansson et al. 2013; Steegenga et al. 2014). CpGassoc was used on the Baka saliva data set because it allowed family identity to be included as a random covariate, which produced the lowest overall λ. The genotype kinship matrix did not account as effectively for the presence of many first-degree relatives in this data set. We applied a Benjamini–Hochberg correction to the P-values of both EWAS to identify CpG sites whose methylation levels vary significantly with age at a false discovery rate (FDR) of 5%.
Blood EWAS
We performed a correction for cell-type composition using the method described by Houseman et al. (2012), implemented in the minfi package. This compares the observed DNA methylation data from the Baka with reference profiles of each cell type. Proportions of these cell types can vary significantly with age, and, because each cell type has a distinct methylation profile, it is important to correct for this heterogeneity to avoid spurious correlations between DNA methylation and age in whole blood (Jaffe and Irizarry 2014). We used EMMAX with the dosage option to conduct the analysis on the Baka whole blood DNA methylation data (Kang et al. 2010). Genotype data were used to generate a Nichols–Balding kinship matrix of all the individuals, which was included in the model to correct for unknown relatedness within the population. The proportion of Bantu ancestry, methyltyping batch, first three PCs, as well as the estimated proportions of five blood-cell types (CD8+ T lymphocytes, CD4+ T lymphocytes, B lymphocytes, natural killer lymphocytes, and monocytes) were used as covariates:
The combination of PC and historically appropriate ancestry covariates used in the EWAS was selected to minimize the genomic inflation factor (Figure S3). We applied a Benjamini–Hochberg correction to the Baka blood EWAS P-values to identify CpG sites whose methylation levels vary significantly with age at a FDR of 5%.
Meta-analysis
We conducted a meta-analysis by combining P-values from both ≠Khomani San and Baka saliva EWAS using Fisher’s method (Evangelou and Ioannidis 2013). We applied a Benjamini–Hochberg correction to the Fisher’s P-values to identify CpG sites whose methylation levels vary significantly with age at a FDR of 5%.
Hyper- and hypo-methylation with age
Significant a-CpGs that were identified in the saliva meta-analysis, and which exhibited a concordant direction of effect in the two cohorts, were divided into hyper- and hypo-methylated sites. We used the Illumina 450K annotation file to determine the position of each significant a-CpG site relative to a CpG island or a gene region. To characterize the background distribution of island and gene region locations of the 334,079 CpG sites that were used in the saliva analyses, we drew a set of 10,000 CpG sites and calculated the percentage of them that fell into each category. We repeated this sampling 1000 times.
For each of these sites, we fit a linear model of DNA methylation level with age across the two saliva cohorts together using the R function lm. We then calculated the residual SE, multiple r2, and Akaike information criterion (AIC) value (using the R function AIC) of the linear model. We then fit a new model after first log transforming chronological age, and recalculated the residual SE, multiple r2, and AIC value. For every site, we then calculated the difference in the residual SE, multiple r2, and AIC value between the linear model and the log-linear model. For all three of these measures, we performed a t-test to compare sites that become hyper-methylated with age to those that become hypo-methylated with age. Note that models are only considered a significantly better fit if the difference in AIC values is greater than two (Akaike 1974). However, as our goal was to examine general trends in the characteristics of hyper-methylated and hypo-methylated sites, we included the entire distribution of AIC value differences.
Replication of previous studies
We compiled a comprehensive list of 163,170 significant a-CpGs published from 17 studies of DNA methylation and aging conducted in any tissue type (Rakyan et al. 2010; Teschendorff et al. 2010; Bocklandt et al. 2011; Alisch et al. 2012; Bell et al. 2012; Garagnani et al. 2012; Heyn et al. 2012; Cruickshank et al. 2013; Hannum et al. 2013; Johansson et al. 2013; Florath et al. 2014; Weidner et al. 2014; Xu and Taylor 2014; Fernández et al. 2015; Marttila et al. 2015; Zaghlool et al. 2015; Kananen et al. 2016). We compared the significant a-CpGs from our three EWAS and the meta-analysis to identify a set of a-CpGs that were unique to our study.
Age prediction
We applied a previously published multi-tissue epigenetic age calculator to estimate the chronological ages of our sampled individuals (Horvath 2013). The Horvath calculator accepts DNA methylation array data as input and outputs a DNA methylation-based age estimate. We used data sets that were not filtered for any probes because the normalization step of the algorithm would not run with a large quantity of missing data. We also tested 450K methylation data from a total of 60 European individuals that were freely available from the Gene Expression Omnibus (GEO) data repository [GSE30870 (Heyn et al. 2012) and GSE49065 (Steegenga et al. 2014)].
Methylation quantitative trait loci scan
We identified cis-methylation QTLs (cis-meQTLs) in the Baka blood samples by conducting linear regressions in R of the methylation value at each of the 346,753 CpG sites against the genotype of all SNPs that lay within 200 kb of that site, and had a minor allele frequency of at least 10% in the sample. Significant cis-meQTL associations were identified by applying a Benjamini–Hochberg correction to the P-values at a FDR of 1%, as determined by 100 permutations.
Conditional analysis
We performed a conditional association analysis for each a-CpG with a significant meQTL by including the genotype state of the associated SNP as an additional covariate in the model. Because EMMAX cannot handle missing values, and because some genotype information was missing, we repeated the baseline EWAS for the Baka whole blood data and performed the conditional analysis using CpGassoc, correcting for all the same covariates, but excluding the Balding–Nichols kinship matrix. We also performed a permutation analysis by pairing each CpG site with a randomly chosen meQTL SNP and repeating the conditional EWAS, where the genotype state of the “false meQTL” was included as a covariate instead of the “true” one. We permuted CpG-SNP associations a total of 100 times to build a distribution of effects of a random meQTL on general age-association trends.
Data availability
The data used in this article have been submitted to the European Genome-Phenome Archive (EGA) (www.ebi.ac.uk/ega/home) and GEO (https://www.ncbi.nlm.nih.gov/geo). The SNP and methylation array data for the Baka can be found under the EGA accession numbers EGAS00001001066 and EGAS00001002226. The SNP and methylation array data for the ≠Khomani San can be found under the GEO super series GSE99091.
Results
PCA and ADMIXTURE
We performed PCA to determine if there were factors other than age driving systematic differences in DNA methylation profiles. PCA were conducted on the saliva and blood data sets separately, since these tissues are expected to differ substantially in their DNA methylation profiles (Byun et al. 2009). Individuals clustered together by batch identity in biplots of the first and second PCs, demonstrating that batch effects were the strongest drivers of DNA methylation profile differences, as expected (Wilhelm-Benartzi et al. 2013); but neither population identity (for the saliva data set) nor sex appeared to drive clustering in the first two PCs (Figure S2). Six ≠Khomani San and one Baka individual were excluded from subsequent analyses because their DNA methylation profiles were extreme outliers. The latter was previously flagged as unreliable because it had abnormally low methyltyping bisulfite controls. We found a significant correlation between some PCs and age: in particular saliva PC 1 with ≠Khomani San age, and blood PCs 1 and 2 with Baka age (Figure S4).
Both the Baka and ≠Khomani San have experienced recent gene flow, to differing extents, from Bantu-speaking agriculturalists and additionally, for the ≠Khomani San, from Europeans (Quintana-Murci et al. 2008; Jarvis et al. 2012; Pickrell et al. 2012; Patin et al. 2014). Since DNA methylation patterns vary substantially across human populations, it is possible that ancestral makeup could also affect patterns of epigenetic aging in admixed individuals (Fraser et al. 2012; Heyn et al. 2013; Fagny et al. 2015; Galanter et al. 2017). To account for this, we inferred global ancestry proportions using ADMIXTURE for all the individuals in our data sets for whom SNP genotype array data were available (Alexander et al. 2009). Prior work has demonstrated an average ancestry of 6.5% from neighboring Bantu speakers in the Baka population (Patin et al. 2014), and an average of 11% for each of Bantu and European ancestry in the ≠Khomani San (Henn et al. 2011). We therefore expected distinct ancestral components corresponding to Pygmy, San, European, and Bantu-speaking populations in our data set, and assumed k = 4 ancestries when running the ADMIXTURE algorithm (Figure S5).
Both the ≠Khomani San and the Baka populations remain relatively endogamous, and, coupled with field-sampling bias, members of extended families are often collected together. Therefore, we also used the genotype data to generate Balding–Nichols (Balding and Nichols 1995) kinship matrices for the association analyses of the ≠Khomani San saliva and the Baka blood data sets to control for the degree of relatedness between individuals in subsequent analyses. Genetic relationship matrices have been shown to appropriately control for stratification in association studies (Kang et al. 2008).
EWAS
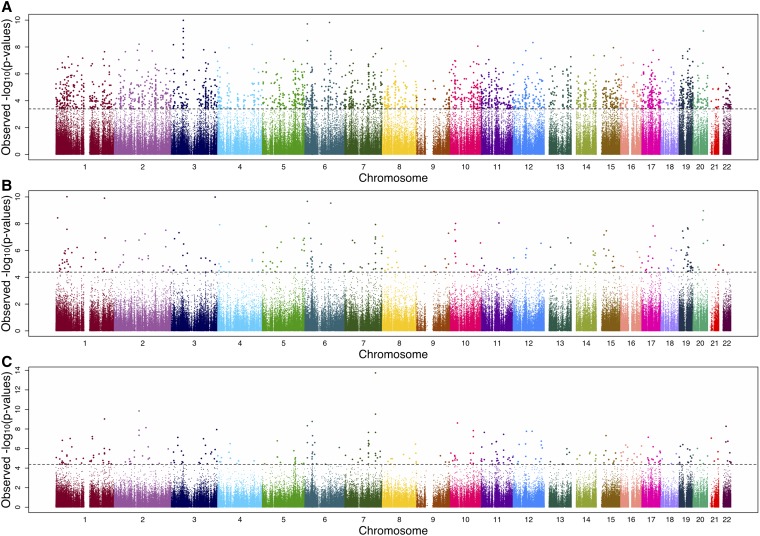
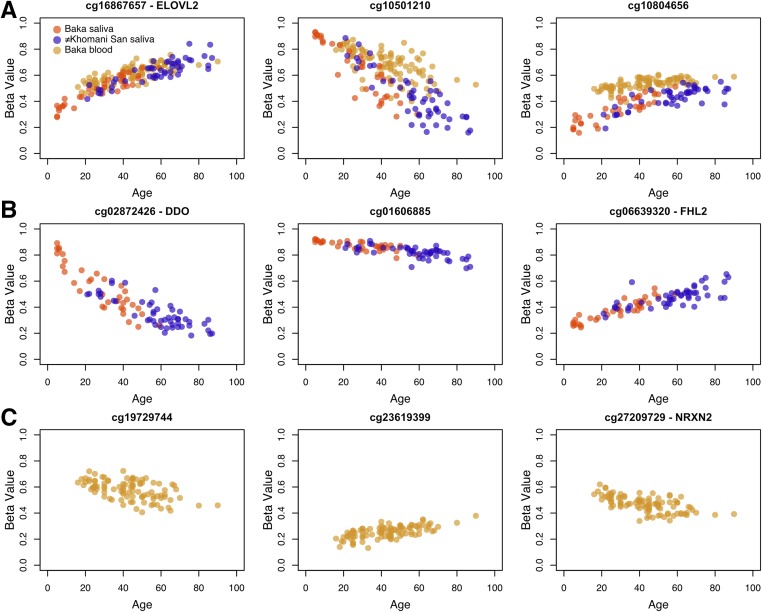
We conducted an EWAS of DNA methylation level and chronological age in each of the three data sets: the ≠Khomani San saliva, the Baka saliva, and the Baka blood. We identified 2714 CpG sites in the Baka saliva, 276 sites in the ≠Khomani San saliva, and 306 sites in the Baka blood that were significantly associated with age at an FDR of 5% (Figure 1, Table S2, Table S3, and Table S4). A total of 188 of these sites replicated independently in both saliva EWAS and 43 in all three EWAS (Figure 2).
Figure 1.
Manhattan plot of EWAS for age-associated CpGs. The −log10 P-values from the EWAS are plotted against the assayed autosomal genomic CpGs for (A) the Baka saliva data set, (B) the ≠Khomani San saliva data set, and (C) the Baka blood data set. All samples were assayed on the Illumina Infinium HumanMethylation450 BeadChips. The horizontal dashed line in each panel represents the Benjamini–Hochberg-corrected threshold for significance (FDR of 5%) for each EWAS.
Figure 2.
Scatterplots of β value vs. age for a-CpGs. Methylation levels as β values, which are continuous from 0 (indicating that the site is completely unmethylated) to 1 (indicating that the site is completely methylated), are plotted against age for three of the a-CpGs that were identified as significant in (A) all three EWAS, (B) only the two saliva EWAS, and (C) only the Baka blood EWAS. β values plotted here are not adjusted for the covariates included in each EWAS.
Meta-analysis of saliva EWAS
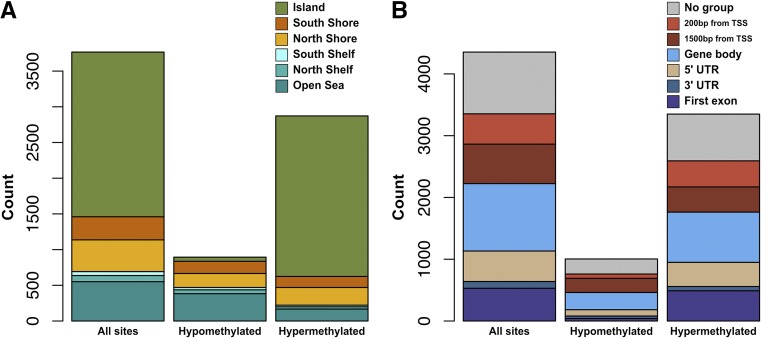
To improve our power to detect significant age associations in hunter-gatherer saliva, we performed a meta-analysis by calculating Fisher’s P-values from the P-values of both saliva EWAS. We identified 3842 CpG sites that were significantly associated with age at an FDR of 5% in the meta-analysis of our saliva studies. Of these, 2872 (74.7%) show a hyper-methylation trend (increasing β value) with age, 894 (23.3%) show a hypo-methylation trend (decreasing β value) with age, and 76 (2.0%) show opposite effects in each of the two cohorts (Table S5). Excluding the 76 discordant a-CpGs, we determined the location of each of the remaining sites relative to specific genes, genic features, and CpG islands using the 450K annotation file available from Illumina. Among these a-CpGs, 2306 (61.2%) fall in CpG islands, 141 in island shelves (3.7%; 86 “North” and 55 “South”), 768 in island shores (20.4%; 444 North and 324 South), and 551 (14.6%) in “open sea.” When considering all CpG sites assayed in the saliva EWAS, 33.2% of them fall in CpG islands, 9.0% in shelves, 23.6% in shores, and 34.2% in open sea. Most island a-CpGs showed a hyper-methylation trend with age (97.5%), while most open-sea sites showed a hypo-methylation trend with age (69.5%), which is broadly in line with previously reported trends (Figure 3A and Table 1) (Christensen et al. 2009; Heyn et al. 2012; Johansson et al. 2013). A total of 2767 a-CpGs were annotated to specific genes. Among these, we counted the number of sites in each of the following six genic regions: first exon, 3′ untranslated region (UTR), 5′ UTR, gene body, within 1500 bp of the transcriptional start site (TSS), and within 200 bp of the TSS (Figure 3B and Table 1). To determine if there was a significant enrichment of a-CpGs in any annotation category, we randomly sampled the background set of CpG sites 1000 times and compared it to our observation. In Table 1, the percentage point enrichment relative to the background is given in brackets, and one-sided empirical P-values are also shown.
Figure 3.
Locations of a-CpGs according to genic features and hyper- and hypo-methylation trends. Stacked bar plots indicate counts of a-CpGs and their physical positions relative to (A) CpG islands and (B) genes. In cases where a single CpG site was annotated to multiple gene regions, each region was counted independently. Annotations were based on the probe information file provided from Illumina. A total of 76% of a-CpGs become hyper-methylated (increase in β value) with age, and 78% of these lie in CpG islands. By contrast, only 6.5% of a-CpGs that become hypo-methylated (decrease in β value) with age lie in CpG islands
Table 1. Enrichment of CpG probe annotation categories relative to background.
| All sites | Hypo-methylated sites | Hyper-methylated sites | ||||
|---|---|---|---|---|---|---|
| Percentagea | P-valueb | Percentagea | P-valueb | Percentagea | P-valueb | |
| Relative to CpG islands | ||||||
| Open sea | 14.6 (−19.6) | <0.001 | 42.9 (+8.7) | <0.001 | 5.8 (−28.4) | <0.001 |
| North shelf | 2.3 (−30.9) | <0.001 | 6.6 (−26.5) | <0.001 | 1.1 (−32.1) | <0.001 |
| South shelf | 1.5 (−3.3) | <0.001 | 4 (−0.8) | <0.001 | 0.8 (−4) | <0.001 |
| North shore | 11.8 (−1.5) | <0.001 | 22 (+8.7) | <0.001 | 8.6 (−4.7) | <0.001 |
| South shore | 8.6 (+4.4) | <0.001 | 17.9 (+13.7) | <0.001 | 5.4 (+1.2) | <0.001 |
| Island | 61.2 (+50.8) | <0.001 | 6.6 (−3.8) | <0.001 | 78.3 (+67.9) | <0.001 |
| Relative to gene regions | ||||||
| First exon | 12.2 (+4.8) | <0.001 | 3.6 (−3.8) | <0.001 | 14.6 (+7.2) | <0.001 |
| 3′ UTR | 2.5 (−1) | <0.001 | 3.9 (+0.4) | <0.001 | 2.1 (−1.4) | <0.001 |
| 5′ UTR | 11.3 (−0.8) | 0.002 | 9.8 (−2.3) | <0.001 | 11.7 (−0.4) | 0.087 |
| Body | 25.1 (−6.2) | <0.001 | 29.9 (−1.3) | <0.001 | 24.3 (−7) | <0.001 |
| TSS 1500 | 14.7 (−0.5) | 5.0 | 23.7 (+8.5) | <0.001 | 12.2 (−3) | <0.001 |
| TSS 200 | 11.3 (−0.3) | 0.134 | 6.8 (−4.8) | <0.001 | 12.6 (+1) | 0.001 |
| No gene | 22.9 (+4) | <0.001 | 22.3 (+3.4) | <0.001 | 22.6 (+3.7) | <0.001 |
The percentage of CpG sites that were annotated to a specific category is shown, and in brackets the percentage point difference compared to the background distribution, determined by 1000 simulations, is shown.
One-tailed empirical P-values from 1000 simulations.
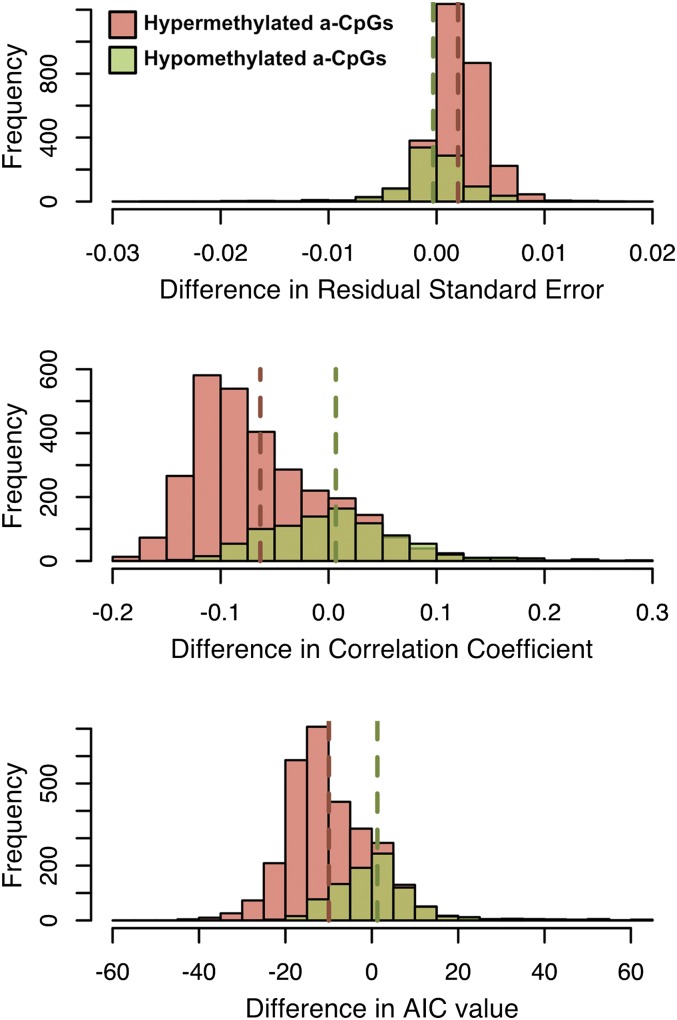
We observed that several a-CpGs exhibited a log-linear change in methylation level with age, and particularly in children, as previously reported by Alisch et al. (2012). Interestingly, we observed this pattern visually more frequently in a-CpGs that become hypo-methylated with age. Again excluding the 76 CpG sites with discordant effects in the Baka and ≠Khomani San, we systematically tested this observation by fitting a linear model to the β values at each of the remaining 3766 sites for both direct chronological age and a log transformation thereof. We found that these two classes of sites showed significantly different distributions of residual SE (P = 1.58 × 10−65) and multiple r2 (P = 2.06 × 10−146). We also calculated the difference in AIC values between the linear and log-linear models of methylation level and age (Akaike 1974). We then performed a t-test on the difference in AIC values for linear and log-linear models and found, again, that sites that become hyper-methylated with age are significantly different from sites that become hypo-methylated (P = 3.58 × 10−129). All three methods yielded the same general trend: hypo-methylated sites tended to be better fit by log-linear model (as demonstrated by their generally higher r2 values, lower residual SEs, and lower AIC values when fit by a log-linear rather than a strictly linear model), while hyper-methylated sites tended to be better fit by linear models (Figure 4).
Figure 4.
Evaluating the fit of log-linear vs. linear models of methylation level and age to hyper- and hypo-methylated a-CpGs. Each of the 3766 a-CpGs that were identified in the saliva meta-analysis was fit with both a linear and log-linear model of age with methylation level. The distribution of the differences in (A) residual SE, (B) correlation coefficient, and (C) AIC values between the linear model and the log-linear model are shown. The means of the distributions are indicated by dashed vertical lines of the same color. The linear model is a better fit for the relationship between methylation and age when differences in residual SE are large and positive, and when differences in correlation coefficient and AIC value are large and negative. By all three measures, a-CpGs that hyper-methylate with age (orange) are better fit by a linear model, and a-CpGs that hypo-methylate (green) by a log-linear model.
Replication of previous studies
We sought to determine the independent replication rate of significant a-CpGs that we identified by searching the literature for studies that quantitatively investigate the relationship between CpG methylation and chronological age. We included 17 studies conducted on either 27 or 450K array technologies in any human tissue (Rakyan et al. 2010; Teschendorff et al. 2010; Bocklandt et al. 2011; Alisch et al. 2012; Bell et al. 2012; Garagnani et al. 2012; Heyn et al. 2012; Cruickshank et al. 2013; Hannum et al. 2013; Johansson et al. 2013; Florath et al. 2014; Weidner et al. 2014; Xu and Taylor 2014; Fernández et al. 2015; Marttila et al. 2015; Zaghlool et al. 2015; Kananen et al. 2016). We found that >93% of the a-CpGs sites we identified in our analyses were reported in one of these previous studies. However, we also found 277 a-CpG sites that were uniquely identified in our study of African hunter-gatherer groups. For each of these 277 sites, we calculated the Pearson correlation coefficient between β value and chronological age and also fit a linear model to determine the slope and trend of the association (Table S1). To further narrow down this list to the most potentially useful aging markers, we focused on sites that either exhibited a high Pearson correlation value (absolute value >0.7) and a steep slope (absolute value >0.002 β value per year); 16 of these 277 a-CpGs met at least both of these criteria (Figure S6).
The site cg16867657, annotated to the promoter of ELOVL2, is significantly associated with chronological age in all three data sets, across populations and tissues. This site was first identified as a potential biomarker for age by Garagnani et al. (2012) and replicated in subsequent epigenetic aging studies of additional cohorts of European, Hispanic, and Arab descent (Hannum et al. 2013; Johansson et al. 2013; Florath et al. 2014; Zaghlool et al. 2015). By observing a signal of age association independently in three African cohorts following different lifestyles, and using DNA sourced from two different tissue types, we further validate the use of cg16867657 methylation as a true biological marker for age across the full spectrum of human diversity. We observe that the pattern of age-related methylation change is also remarkably congruent across blood and saliva (Garagnani et al. 2012; Zbieć-Piekarska et al. 2015).
In the saliva data sets, we observed a significant age-associated hypo-methylation signal in the TSS of the gene D-aspartate oxidase (DDO) at cg02872426 in both African populations (Figure 2B). This site was previously identified in a study of whole blood of Arab individuals (Zaghlool et al. 2015) and other CpG sites annotated to DDO have also been previously associated with age (Ali et al. 2015; Fernández et al. 2015; Zaghlool et al. 2015). We identified additional sites (cg00804078, cg06413398, and cg07164639) in the TSS of the gene DDO, which exhibit hypo-methylation with age at a relaxed significance threshold of P < 0.001 in all three data sets (Figure S7).
Testing an epigenetic aging predictor
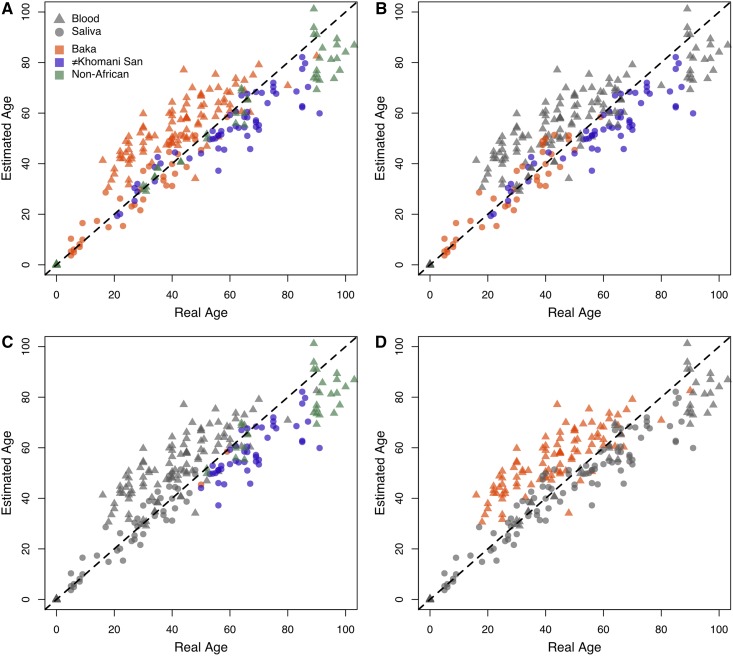
DNA methylation can be affected by genetic variation, as well as environmental and lifestyle variation during development. We therefore asked how accurately existing age-prediction models, developed primarily on DNA methylation data derived from individuals of European ancestry, would perform on our African data sets. We applied a multi-tissue age predictor developed by Horvath (2013) to all three data sets, hereafter referred to as the “Horvath model” (Figure 5A). This model uses a linear combination of methylation information from 353 sites, termed “clock-CpGs,” to produce an estimate of age. The DNA methylation-age estimates for the Baka saliva were very accurate, with a median absolute difference of 3.90 years between the true and estimated ages (r = 0.94), and the estimates for the ≠Khomani San saliva data set had an overall greater median absolute difference of 6.01 years (r = 0.90) (Figure 5B), typically underestimating the age in the older individuals. To investigate whether the reduction in accuracy was specific to the ≠Khomani San, we applied the age predictor to European DNA methylation data sets from blood [GEO data sets GSE30870 (Heyn et al. 2012) and GSE49064 (Steegenga et al. 2014)]. We observed a similar underestimation of age in older Europeans, suggesting that this observation in adults >50 years is not indicative of a ≠Khomani San-specific slowdown in the epigenetic aging rate (Figure 5C).
Figure 5.
Scatterplots of true age against estimated age as predicted by the Horvath model. Chronological age reported by individuals is plotted against estimated ages generated from epigenetic data using Horvath’s age-prediction model (Horvath 2013). All four panels show the age estimates for the Baka, ≠Khomani San, and European blood and saliva data sets, colored to emphasize different features of the data. Blood and saliva tissue sources are indicated by ▴ and ●, respectively. (A) All data points, colored based on population identity. (B) Only estimates for African saliva data are colored, all others are grayed out. (C) Only estimates for individuals whose true age is 50 or more, except for the Baka blood data, are colored. (D) Only estimates for Baka blood are colored. The dashed line represents perfectly accurate prediction of chronological age.
Finally, we observed a systematic overestimation of age from the DNA methylation profiles of Baka blood (median absolute difference of 13.06 years, r = 0.81) (Figure 5D). It is important to note that the correlation between chronological and estimated age remains high, and the discrepancy between the two may be indicative of technical artifacts or batch effects in the application of the arrays. However, it is not possible to rule out a biological driver that causes Baka blood to exhibit increased epigenetic age under the Horvath model (see Discussion).
meQTLs in age-related CpG sites
Given the observed differences in accuracy of age estimation in different human populations, we sought to further understand why age-related epigenetic patterns might not replicate across study cohorts. Even at lower significance thresholds, success in reconciling reported epigenetic signals of aging in different studies has been mixed (Hannum et al. 2013). Indeed, several age-related CpG sites that have been reported previously did not replicate in our populations. There are many potential reasons for this, including our smaller sample sizes, and the comparison of different tissue types which may exhibit tissue-specific patterns of methylation with age. However, it is also possible that intrinsic genetic factors may drive these differences.
To explore this, we tested whether meQTLs could drive variation in epigenetic aging patterns. meQTLs are genetic variants that are statistically associated with methylation levels at certain CpG sites (Smith et al. 2014). We scanned all CpG sites in the Baka blood data set for cis-meQTL associations, by fitting a linear model of methylation level by genotype state using chronological age, sex, and blood cell-type proportions as covariates. We identified 11,559 meQTLs at an FDR of 1% in the Baka blood data set. We also compiled a list of 18,229 a-CpGs identified in previous studies of blood methylation (Rakyan et al. 2010; Teschendorff et al. 2010; Alisch et al. 2012; Bell et al. 2012; Garagnani et al. 2012; Heyn et al. 2012; Cruickshank et al. 2013; Hannum et al. 2013; Florath et al. 2014; Weidner et al. 2014; Xu and Taylor 2014; Marttila et al. 2015; Zaghlool et al. 2015; Kananen et al. 2016). Interestingly, there is an overlap of 901 CpG sites that were identified as being associated with age in Europeans and are also associated with a specific cis genetic variant in the Baka. This is more overlap than would be expected by chance, as determined by randomly sampling and intersecting 18,229 CpG sites with the 11,559 significant meQTLs; after 10,000 simulations, the maximum overlap obtained under this null scenario was 662 (P < 0.001). Only 8 of these 901 sites were among the 306 significant a-CpGs identified in our EWAS of Baka blood methylation.
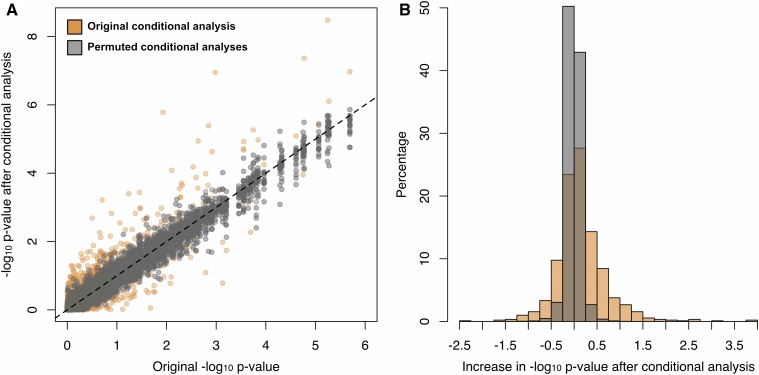
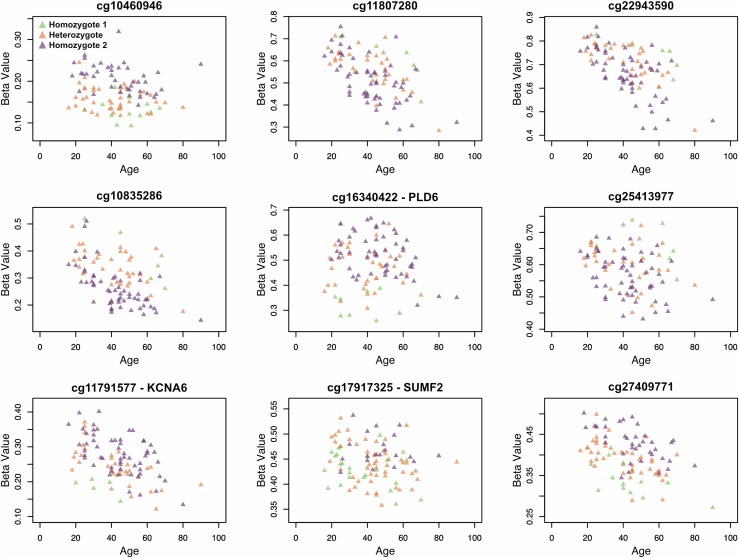
We performed a conditional analysis to determine if incorporating SNP genotype information recovers significant age association in the Baka at these CpG sites. For each of these 901 CpG sites, we included the genotype state at the associated meQTL as an additional covariate and repeated the association analysis. We also permuted all the CpG-SNP associations by assigning each of the 901 CpG sites a SNP selected at random from among the 11,559 identified meQTLs, and repeating this 100 times. In the true conditional analysis, we observed an overall upward shift in the distribution of −log10 P-values when meQTL-specific genotype data were included (Figure 6A), indicating that incorporation of the meQTL genotype generally improves the age-methylation association for these CpG sites (mean increase in −log10 P-value after conditional analysis of 0.15); this increase was not observed in our permutation analysis when a random SNP covariate was included (mean difference in −log10 P-value of −0.008) (Figure 6B). More specifically, 39 of the 901 CpGs (4.3%) that were not significantly associated with age at an FDR of 5% in the original EWAS recovered significance when true meQTL genotype was included, while this occurred only 0.17% of the time in the permutation analysis. We observe that a small number (15 out of 901, 1.7%) of CpGs decrease in significance by over one order of magnitude, which may be due to meQTL genotypes that are spuriously correlated with age in the discovery EWAS. We also observe that 6.1% of CpGs increase in significance by over one order of magnitude under the conditional analysis, while only 0.023% of CpGs increase by as much in the permutation analysis, and nine CpG sites (1%) become more significantly associated with age by over two orders of magnitude under the conditional analysis (Figure 7). These results suggest that, for some CpG sites, the genotype state of the true meQTL provides additional information for characterizing the relationship between methylation level and chronological age.
Figure 6.
Conditional analysis of meQTL-associated a-CpGs. (A) The −log10 P-values from an EWAS on Baka blood are plotted against the −log10 P-values from a conditional analysis in which an meQTL genotype state was included as an additional covariate for 2842 a-CpGs. (B) The distribution of effects of the conditional analysis are depicted as the difference in −log10 P-values before and after conditional analysis. The orange points and bars represent the results of the conditional analysis. The gray points and bars represent the results of 100 permutations of the conditional analysis where the CpG-meQTL associations were randomized.
Figure 7.
Scatterplots of a-CpGs with associated meQTL genotype states. Scatterplots of β value and age are shown for the nine CpG sites for which age association improves (i.e., P-value decreases) by over two orders of magnitude when SNP genotype information from a known meQTL is accounted for in the EWAS. Individuals are colored by their genotype (homozygous reference, alternative, or heterozygous) state, demonstrating genotype-specific trends between methylation and age exist at these CpG sites. β values plotted here are not adjusted for the covariates included in each EWAS.
Discussion
In this study, we investigated patterns of aging in the epigenome across an extended range of human genetic diversity by characterizing the DNA methylation profiles of saliva and whole blood tissues from two contemporary African hunter-gatherer populations using a large, comprehensive, genome-wide methylation array. We replicate several of the strongest signals of age-related DNA methylation change reported in previous studies of other populations, including cg16867657 in the gene ELOVL2, which supports the utility of this gene as a predictive marker of chronological aging in all humans as was previously suggested by Garagnani et al. (2012). We further demonstrate that this a-CpG replicates strongly in saliva, identifying it independently in both our hunter-gatherer data sets. ELOVL2 is part of a family of enzymes that are responsible for elongating polyunsaturated fatty acids, whose levels have been shown to decline with chronological age in human skin (Kim et al. 2010). It is possible that the continuous life-long increase in methylation of cg16867657 and the ELOVL2 promoter in general contributes to this trend. It is important to note that this aging biomarker has not been identified in skin tissue itself, but rather in whole blood and white blood cells (Johansson et al. 2013; Steegenga et al. 2014; Vandiver et al. 2015; Zaghlool et al. 2015).
We identified a strong hypo-methylation trend with age in the gene DDO, particularly in saliva tissue. The most significant DDO-annotated CpG site from our study, cg02872426, was found to be significantly age-related in a previous study of an Arab population (Zaghlool et al. 2015). The enzyme encoded by DDO deaminates D-aspartic acid, the enantiomer of L-aspartic acid, which is the optical form naturally synthesized by biological organisms (D’Aniello et al. 1993; Ritz-Timme and Collins 2002). Nonenzymatic accumulation of D-aspartic acid is age dependant in living tissues, and is so pronounced in tissues with low turnover that it has been proposed as a biomarker for aging (Helfman and Bada 1975). The role of DDO is to eliminate this abnormal version of aspartic acid in proteins and counteract the racemization process and, interestingly, its levels increase in the liver and kidneys with age (D’Aniello et al. 1993). The hypo-methylation trend we observe in the DDO promoter is compatible with these observations and previous age-related DNA methylation studies, and suggests a potential mechanism by which DDO expression levels are regulated throughout an organism’s lifetime. Our observations further suggest that the hypo-methylation of DDO, which may be related to its continued upregulation throughout life, is protective against the effects of age-accumulated protein damage and facilitates “healthy” aging.
A parallel can be drawn between DNA methylation at DDO and telomerase reverse transcriptase (TERT), their transcriptional regulation, and their function as biomarkers of aging. Shortened telomere length in lymphocytes, a commonly used indicator of biological age, is associated with decreased telomerase levels (Blasco 2007). Almén et al. (2014) observe hyper-methylation of TERT with age, and speculate that this epigenetic trend is what ultimately underlies the observed trend of telomere shortening with age. Age-related changes in DDO methylation may influence gene transcription, but unlike the relationship with TERT methylation and telomere shortening, increased levels of DDO in older individuals would counteract pathogenic accumulation of abnormal protein.
We also identify 277 significant a-CpGs across three EWAS and a meta-analysis that have not, to our knowledge, been reported in any previous study of DNA methylation and aging. Of these, 16 have high correlation coefficients or strong regression between methylation level and age. All but 12 of these are absent from the 27K array and therefore could not be identified in studies using only that technology. However, there exist other difficulties in replicating our results between populations and tissue types, even within our own study. For example, the site cg26559209 exhibits a clear hypo-methylation trend in saliva, but a slight hyper-methylation trend in whole blood (Figure S6). This may indicate a tissue-specific pattern of epigenetic aging that is further complicated by the cell-type heterogeneity of whole blood, which, despite bioinformatic correction algorithms, could introduce noise to the aging signal at certain a-CpGs. We also note that many of our novel a-CpGs change only slightly in methylation level over time. The aging signal at these sites may be too weak to be consistently detectable in other studies, or they may be false positives in our study.
We replicate general trends in the genomic features of a-CpGs, such as the differences in the CpG-island context of hyper-methylated and hypo-methylated classes of sites. Previous work on methylation and aging in pediatric cohorts found that dramatic changes in methylation patterns occur during childhood, and that most a-CpGs, both hyper- and hypo-methylated, are better modeled by a log-linear relationship between β value and age (Alisch et al. 2012). In our study, the analysis of Baka children allowed us to observe a similar pattern. However, we also found that hypo-methylated a-CpGs were significantly better fit by log-linear models than were hyper-methylated a-CpGs. Taken together, this suggests that hyper- and hypo-methylated a-CpGs in general are affected differently by aging, and that different biological mechanisms may underlie these epigenetic modifications. It has been generally accepted that the substantial changes in DNA methylation that occur over an organism’s lifetime are mainly a signal of dysregulation of the epigenetic machinery, which ultimately underlies an individual’s age-elevated risk for cellular damage and cancer (Jaenisch and Bird 2003; Teschendorff et al. 2013). In particular, the pervasive hypo-methylation with age of CpG sites that lie outside of CpG islands has been highlighted as an indication of this biological breakdown. Lifestyle and environmental factors can also affect the trajectory of changes in genomic methylation and can potentially compound or mitigate this risk (Li et al. 2011; Almén et al. 2014; Vandiver et al. 2015; Zannas et al. 2015). However, there are also certain regions of the genome where epigenetic changes appear to be tightly regulated throughout life, despite environmental and stochastic variation, and we speculate that these may be protective against the detrimental effects of aging or otherwise adaptive. We hypothesize that DDO is an example of a gene that is regulated in such a manner throughout an individual’s life. This is in agreement with the recently proposed conceptual distinction made by Jones et al. (2015) between random “epigenetic drift” that may occur due to loss of regulatory control with age, and the “epigenetic clock” that is much more precisely correlated with age in humans.
We tested the Horvath model of epigenetic-age prediction built on 353 clock-CpGs, which were selected only from sites present on both the 27 and 450K arrays, and was trained primarily on European tissue methylation data sets (Horvath 2013). This model was also tested on chimpanzees in an effort to demonstrate its wide applicability to all humans, and was found to produce accurate estimates in this closely related species, particularly from whole blood where the correlation between chronological and predicted age was 0.9 and the median error was 1.4 years (Horvath 2013). However, this type of validation does not account for the possibility of variation in DNA methylation profiles among diverse human populations, potentially resulting from divergent selection on meQTLs or unique environmental or nutritional factors.
We found that the Horvath model does not predict age accurately in our Baka whole blood methylation data set, and it yields an inflated estimate of epigenetic age. It has been previously observed that specific populations and cohorts often exhibit an offset between chronological and epigenetic age when using this algorithm (Marioni et al. 2015; Horvath et al. 2016). It is unclear what causes this discrepancy, but epigenetic age can be used as a proxy for biological age to predict longevity and health outcomes (Marioni et al. 2015; Horvath et al. 2016). Pygmies, such as the Baka, who reside in tropical jungles are known to have lower than average life expectancies. This has caused some to hypothesize that their small body size, earlier cessation of growth, and hastened fertility schedules are driven by developmental adaptations that maximize fitness under the high mortality rates that these populations experience (Migliano et al. 2007). Therefore, it is possible that the DNA methylation profiles of Baka whole blood are reflecting true increases in biological age compared to European whole blood.
We also considered alternative explanations for the inflated age estimates of this cohort. We rely on self-reported age in this study, and although it can often be challenging to determine true chronological age in the field, this does not appear to be a driving cause of the inflation observed in Baka blood, as saliva-derived DNA methylation profiles from the same population yield highly accurate estimates of age. As these DNA methylation arrays were run in several batches and separately from our saliva data sets, we considered that this result might be due to a technical artifact. We explored additional preprocessing pipelines and ComBat batch correction, but could not eliminate the overestimation effect. Ultimately, the overestimation of age of the Baka whole blood cohort could not be attributed to any particular factor. We also could not speculate on the reason that this pattern is not observed in saliva. These observations warrant further investigation to better understand their biological bases and consequences for longevity and health in this population.
We found that methylation levels at 901 previously reported a-CpGs are also significantly associated with the genotype state at a cis genetic variant. Only eight of these are also significant a-CpGs in our study, and we demonstrate that variation at the associated meQTL is a significant explanatory factor for this lack of replication. By performing a conditional analysis, which accounts for the genotype state of the meQTL, we were able to recover significant age association in >4% of these CpG sites. For nine CpG sites, including the genotype state at the meQTL increases the statistical age association by over two orders of magnitude (Figure 7). These sites may prove to be excellent candidates for aging biomarkers or components of an epigenetic age predictor when used in tandem with SNP data, as many of them exhibit large changes in β value and strong correlations with age. The results from our conditional analysis also offer an explanation for the difficulty in replicating a-CpGs from one study to another, namely that differences in the degree of genetic variation at meQTLs confounds the consistent identification of a-CpGs across cohorts, both between and within human populations. Identifying genetic variants that affect a-CpGs is a challenge because the noise introduced by this genetic variability makes it difficult to identify signals of age-related changes in methylation using standard statistical methods. The approach we use here, which identifies meQTLs at all assayed CpG sites in one cohort and finds overlap with a-CpGs identified in a separate cohort, makes it possible to identify these interactions.
In this study of African hunter-gatherer DNA methylation patterns, we demonstrate that CpG methylation changes with age are strongly conserved at specific a-CpGs across genetically diverse human populations and across tissues, and can be confirmed as reliable and universal biomarkers for human aging. We identify 277 novel a-CpGs, some of which could be useful aging biomarkers in these populations. We also observe that genetic variation in a population, particularly at meQTLs, can result in variation in patterns of age-related differential DNA methylation. This variation, if uncharacterized or unaccounted for in epigenetic age-prediction algorithms, can lead to poor estimates of age in different cohorts and populations. On the other hand, this variation can also be leveraged to improve the precision of age prediction. We conclude that DNA methylation patterns at a-CpGs constitute a promising suite of molecular biomarkers for age across diverse human groups, and that further characterizing these patterns in genetically and ecologically diverse cohorts will facilitate continued improvements in epigenetic age prediction.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.195594/-/DC1.
Acknowledgments
We thank Eileen Hoal and Marlo Möller for their assistance with fieldwork and ethical approval. We thank Christopher Gignoux for helpful discussions. We would also like to thank the Working Group of Indigenous Minorities in Southern Africa and the South African San Institute for their advice regarding community research. Finally, we thank the ≠Khomani San and Baka communities in which we have sampled; without their support, this study would not have been possible. S.G. is supported by the National Institute of Justice Graduate Research Fellowship in Science, Technology, Engineering and Mathematics award 2016-DN-BX-0011. Funding was provided to B.M.H. by a Stanford University Center on the Demography and Economics of Health and Aging seed grant (National Institutes of Health, National Institute on Aging P30 AG-017253-12); to L.Q.-M. by the Institut Pasteur, the Centre National de la Recherche Scientifique, a Centre National de la Recherche Scientifique Maladies Infectieuses et Environnement grant, and a Simone and Cino del Duca Foundation research grant; and to M.S.K. by the Canadian Institute for Advanced Research. M.S.K. is also the Canada Research Chair in Social Epigenetics. L.M.M. is supported by a Canadian Institute of Health Research Doctoral Research Award (F15-04283).
Footnotes
Communicating editor: P. Scheet
Literature Cited
- Akaike H., 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19: 716–723. [Google Scholar]
- Alexander D. H., Novembre J., Lange K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali O., Cerjak D., Kent J. W., James R., Blangero J., et al. , 2015. An epigenetic map of age-associated autosomal loci in northern European families at high risk for the metabolic syndrome. Clin. Epigenetics 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisch R. S., Barwick B. G., Chopra P., Myrick L. K., Satten G. A., et al. , 2012. Age-associated DNA methylation in pediatric populations. Genome Res. 22: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almén M. S., Nilsson E. K., Jacobsson J. A., Kalnina I., Klovins J., et al. , 2014. Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. Gene 548: 61–67. [DOI] [PubMed] [Google Scholar]
- Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., et al. , 2014. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 30: 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding D. J., Nichols R. A., 1995. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96: 3–12. [DOI] [PubMed] [Google Scholar]
- Barfield R. T., Kilaru V., Smith A. K., Conneely K. N., 2012. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics 28: 1280–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J. T., Pai A. A., Pickrell J. K., Gaffney D. J., Pique-Regi R., et al. , 2011. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 12: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J. T., Tsai P. C., Yang T. P., Pidsley R., Nisbet J., et al. , 2012. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 8: e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M. A., 2007. Telomere length, stem cells and aging. Nat. Chem. Biol. 3: 640–649. [DOI] [PubMed] [Google Scholar]
- Bocklandt S., Lin W., Sehl M. E., Sánchez F. J., Sinsheimer J. S., et al. , 2011. Epigenetic predictor of age. PLoS One 6: e14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling L. P., Yang R., Korn B., Burwinkel B., Brenner H., 2011. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet. 88: 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun H. M., Siegmund K. D., Pan F., Weisenberger D. J., Kanel G., et al. , 2009. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum. Mol. Genet. 18: 4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B. C., Houseman E. A., Marsit C. J., Zheng S., Wrensch M. R., et al. , 2009. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 5: e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank M. N., Oshlack A., Theda C., Davis P. G., Martino D., et al. , 2013. Analysis of epigenetic changes in survivors of preterm birth reveals the effect of gestational age and evidence for a long term legacy. Genome Med. 5: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aniello A., D’Onofrio G., Pischetola M., D’Aniello G., Vetere A., et al. , 1993. Biological role of D-amino acid oxidase and D-aspartate oxidase: effects of D-amino acids. J. Biol. Chem. 268: 26941–26949. [PubMed] [Google Scholar]
- Dedeurwaerder S., Defrance M., Bizet M., Calonne E., Bontempi G., et al. , 2013. A comprehensive overview of Infinium HumanMethylation450 data processing. Brief. Bioinform. 15: 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Kibbe W. A., Lin S. M., 2008. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24: 1547–1548. [DOI] [PubMed] [Google Scholar]
- Evangelou E., Ioannidis J. P. A., 2013. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 14: 379–389. [DOI] [PubMed] [Google Scholar]
- Fagny M., Patin E., MacIsaac J. L., Rotival M., Flutre T., et al. , 2015. The epigenomic landscape of African rainforest hunter-gatherers and farmers. Nat. Commun. 6: 10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré P., Jones M. J., Meaney M. J., Emberly E., Turecki G., et al. , 2015. Concordant and discordant DNA methylation signatures of aging in human blood and brain. Epigenetics Chromatin 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández A. F., Bayón G. F., Urdinguio R. G., Toraño E. G., Cubillo I., et al. , 2015. H3K4me1 marks DNA regions hypomethylated during aging in human stem and differentiated cells. Genome Res. 25: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florath I., Butterbach K., Müller H., Bewerunge-hudler M., Brenner H., 2014. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum. Mol. Genet. 23: 1186–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H. B., Lam L. L., Neumann S. M., Kobor M. S., 2012. Population-specificity of human DNA methylation. Genome Biol. 13: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanter J. M., Gignoux C. R., Oh S. S., Torgerson D., Pino-Yanes M., et al. , 2017. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. Elife 6: e20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagnani P., Bacalini M. G., Pirazzini C., Gori D., Giuliani C., et al. , 2012. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell 11: 1132–1134. [DOI] [PubMed] [Google Scholar]
- Gentilini D., Mari D., Castaldi D., Remondini D., Ogliari G., et al. , 2013. Role of epigenetics in human aging and longevity: genome-wide DNA methylation profile in centenarians and centenarians’ offspring. Age (Omaha) 35: 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönniger E., Weber B., Heil O., Peters N., Stäb F., et al. , 2010. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 6: e1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., et al. , 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49: 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman P. M., Bada J. L., 1975. Aspartic acid racemization in tooth enamel from living humans. Proc. Natl. Acad. Sci. USA 72: 2891–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B. M., Gignoux C. R., Jobin M., Granka J. M., Macpherson J. M., et al. , 2011. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl. Acad. Sci. USA 108: 5154–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H., Li N., Ferreira H. J., Moran S., Pisano D. G., et al. , 2012. Distinct DNA methylomes of newborns and centenarians. Proc. Natl. Acad. Sci. USA 109: 10522–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H., Moran S., Hernando-Herraez I., Sayols S., Gomez A., et al. , 2013. DNA methylation contributes to natural human variation. Genome Res. 23: 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly A. C., Melzer D., Pilling L. C., Henley W., Hernandez D. G., et al. , 2013. Towards a gene expression biomarker set for human biological age. Aging Cell 12: 324–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., 2013. DNA methylation age of human tissues and cell types. Genome Biol. 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., Gurven M., Levine M. E., Trumble B. C., Kaplan H., et al. , 2016. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 17: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E. A., Accomando W. P., Koestler D. C., Christensen B. C., Marsit C. J., et al. , 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R., Kerr A., Desousa D., Helle J., Ellis P., et al. , 2008. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 6: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R., Bird A., 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33: 245–254. [DOI] [PubMed] [Google Scholar]
- Jaffe A. E., Irizarry R. A., 2014. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis J. P., Scheinfeldt L. B., Soi S., Lambert C., Omberg L., et al. , 2012. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet. 8: e1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A., Enroth S., Gyllensten U., 2013. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS One 8: e67378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. J., Goodman S. J., Kobor M. S., 2015. DNA methylation and healthy human aging. Aging Cell 14: 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kananen L., Marttila S., Nevalainen T., Jylhävä J., Mononen N., et al. , 2016. Aging-associated DNA methylation changes in middle-aged individuals: the Young Finns study. BMC Genomics 17: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S.-Y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. J., Kim M., Jin X., Oh J., Kim J. E., et al. , 2010. Skin aging and photoaging alter fatty acids composition, including 11,14,17-eicosatrienoic acid, in the epidermis of human skin. J. Korean Med. Sci. 25: 980–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Absher D. M., Tang H., Southwick A. M., Casto A. M., et al. , 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Li Y., Daniel M., Tollefsbol T. O., 2011. Epigenetic regulation of caloric restriction in aging. BMC Med. 9: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic J., Gordon L., Oshlack A., 2012. SWAN: subset-quantile within array normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol. 13: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni R. E., Shah S., McRae A. F., Chen B. H., Colicino E., et al. , 2015. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 16: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttila S., Kananen L., Häyrynen S., Jylhävä J., Nevalainen T., et al. , 2015. Ageing-associated changes in the human DNA methylome: genomic locations and effects on gene expression. BMC Genomics 16: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner C., Ritz-Timme S., 2010. Molecular pathology and age estimation. Forensic Sci. Int. 203: 34–43. [DOI] [PubMed] [Google Scholar]
- Migliano A. B., Vinicius L., Lahr M. M., 2007. Life history trade-offs explain the evolution of human pygmies. Proc. Natl. Acad. Sci. USA 104: 20216–20219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patin E., Katherine J. S., Guillaume L., Hélène Q., Harmant C., et al. , 2014. The impact of agricultural emergence on the genetic history of African rainforest hunter-gatherers and agriculturalists. Nat. Commun. 5: 3163. [DOI] [PubMed] [Google Scholar]
- Pickrell J. K., Patterson N., Barbieri C., Berthold F., Gerlach L., et al. , 2012. The genetic prehistory of southern Africa. Nat. Commun. 3: 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. E., Cotton A. M., Lam L. L., Farré P., Emberly E., et al. , 2013. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L., Quach H., Harmant C., Luca F., Massonnet B., et al. , 2008. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc. Natl. Acad. Sci. USA 105: 1596–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan V. K., Down T. A., Thorne N. P., Flicek P., Kulesha E., et al. , 2008. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res. 18: 1518–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan V. K., Down T. A., Maslau S., Andrew T., Yang T. P., et al. , 2010. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 20: 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz-Timme S., Collins M. J., 2002. Racemization of aspartic acid in human proteins. Ageing Res. Rev. 1: 43–59. [DOI] [PubMed] [Google Scholar]
- Simm A., Nass N., Bartling B., Hofmann B., Silber R. E., et al. , 2008. Potential biomarkers of ageing. Biol. Chem. 389: 257–265. [DOI] [PubMed] [Google Scholar]
- Smith A. K., Kilaru V., Kocak M., Almli L. M., Mercer K. B., et al. , 2014. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics 15: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegenga W. T., Boekschoten M. V., Lute C., Hooiveld G. J., De Groot P. J., et al. , 2014. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age (Omaha) 36: 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E., Menon U., Gentry-Maharaj A., Ramus S. J., Weisenberger D. J., et al. , 2010. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 20: 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E., West J., Beck S., 2013. Age-associated epigenetic drift: Implications, and a case of epigenetic thrift? Hum. Mol. Genet. 22: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren C., Kim M., Martin A. R., Bobo D., Gignoux C. R., et al. , 2016. Fine-scale human population structure in southern Africa reflects ecogeographic boundaries. Genetics 204: 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandiver A. R., Irizarry R. A., Hansen K. D., Garza L. A., Runarsson A., et al. , 2015. Age and sun exposure-related widespread genomic blocks of hypomethylation in nonmalignant skin. Genome Biol. 16: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramah K. R., Wegmann D., Woerner A., Mendez F. L., Watkins J. C., et al. , 2012. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing data. Mol. Biol. Evol. 29: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu P., Destro-Bisol G., 2012. African Pygmies, what’s behind a name? Hum. Biol. 84: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu P., Austerlitz F., Estoup A., Vitalis R., Georges M., et al. , 2009. Origins and genetic diversity of pygmy hunter-gatherers from Western Central Africa. Curr. Biol. 19: 312–318. [DOI] [PubMed] [Google Scholar]
- Weidner C. I., Lin Q., Koch C. M., Eisele L., Beier F., et al. , 2014. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 15: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm-Benartzi C. S., Koestler D. C., Karagas M. R., Flanagan J. M., Christensen B. C., et al. , 2013. Review of processing and analysis methods for DNA methylation array data. Br. J. Cancer 109: 1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Taylor J. A., 2014. Genome-wide age-related DNA methylation changes in blood and other tissues relate to histone modification, expression and cancer. Carcinogenesis 35: 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghlool S. B., Al-Shafai M., Al Muftah W. A., Kumar P., Falchi M., et al. , 2015. Association of DNA methylation with age, gender, and smoking in an Arab population. Clin. Epigenetics 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannas A. S., Arloth J., Carrillo-Roa T., Iurato S., Röh S., et al. , 2015. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 16: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbieć-Piekarska R., Spólnicka M., Kupiec T., Makowska Ż., Spas A., et al. , 2015. Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci. Int. 14: 161–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this article have been submitted to the European Genome-Phenome Archive (EGA) (www.ebi.ac.uk/ega/home) and GEO (https://www.ncbi.nlm.nih.gov/geo). The SNP and methylation array data for the Baka can be found under the EGA accession numbers EGAS00001001066 and EGAS00001002226. The SNP and methylation array data for the ≠Khomani San can be found under the GEO super series GSE99091.