Goiffon et al. introduce the optimal population value (OPV) as a new metric for genomic selection. OPV is based on the maximum possible haploid value...
Keywords: GenPred, shared data resource, genetic gain, genomic selection, optimal haploid value, optimal population value, population-based selection
Abstract
Genomic selection (GS) identifies individuals for inclusion in breeding programs based on the sum of their estimated marker effects or genomic estimated breeding values (GEBVs). Due to significant correlation between GEBVs and true breeding values, this has resulted in enhanced rates of genetic gain as compared to traditional methods of selection. Three extensions to GS, weighted genomic selection (WGS), optimal haploid value (OHV) selection, and genotype building (GB) selection have been proposed to improve long-term response, and to facilitate the efficient development of doubled haploids. In separate simulation studies, these methods were shown to outperform GS under various assumptions. However, further potential for improvement exists. In this paper, optimal population value (OPV) selection is introduced as selection based on the maximum possible haploid value in a subset of the population. Instead of evaluating the breeding merit of individuals as in GS, WGS, and OHV selection, the proposed method evaluates the breeding merit of a set of individuals as in GB. After testing these selection methods extensively, OPV and GB selection were found to achieve greater responses than GS, WGS, and OHV, with OPV outperforming GB across most percentiles. These results suggest a new paradigm for selection methods in which an individual’s value is dependent upon its complementarity with others.
IN genomic selection (GS), genome-wide genetic markers and phenotypic observations are used to estimate marker effects that can subsequently be used to accurately predict breeding values of individuals that have only been genotyped (Meuwissen et al. 2001). GS improves upon marker-assisted selection by more effectively capturing the effects of all quantitative trait loci (QTL). Because GS typically more accurately identifies superior parents than traditional selection methods, and decreases the amount of necessary phenotyping, it has been recognized as a way to increase the rate of genetic gain in cultivar improvement programs (Bernardo and Yu 2007), and to allow for more breeding cycles per unit time (Heffner et al. 2010).
Three extensions have been proposed to improve GS. The first extension, weighted genomic selection (WGS), was proposed to increase the frequency of rare favorable alleles in the population to maximize long-term response (Goddard 2009). In a simulation study, WGS was shown to increase response relative to GS (Jannink 2010). The second extension, optimal haploid value (OHV) selection, calculates the best possible future breeding value of an individual when doubled haploids (DHs) are produced from it. This method was shown to improve response in a simulated wheat program using DHs (Daetwyler et al. 2015). By taking the maximum haplotype GEBV at each segment, Daetwyler et al. (2015) demonstrated the utility of maintaining seemingly unfavorable genome segments, because subsequent recombination can release favorable alleles. Selecting only those individuals with the highest overall genomic estimated breeding values (GEBVs) can lead to the loss of rare favorable alleles in the population, but an individual whose GEBV falls below the truncation point may be more favorable in the long-term because it harbors rare favorable alleles.
GS, WGS, and OHV selection perform truncation selection on individual breeding values (of varying definitions). However, the genetic merit of a single individual also depends on the genetic merit of the individuals with which it may be mated. After several generations of crossing and recombination, individuals will contain genetic material from multiple founder lines. Thus, the third extension, genotype building (GB) selection, uses the best two haplotype blocks of a subset of the founder population to derive a combined fitness value for that subset (Kemper et al. 2012). As such, it represents a shift from selecting superior individuals to selecting the set of individuals that are more likely to produce superior progeny when crossed with each other. Since plant breeders often seek to develop high performing inbred lines, a genotype building strategy can be applied without the constraints on coancestry used by Kemper et al. (2012).
In this article, we propose optimal population value (OPV) selection as a combination of GB and OHV selection. Like OHV selection, OPV considers the haploid values of individual selection candidates but evaluates the merit of potential progeny of a subset of selection candidates like GB selection. First, we mathematically defined the OPV, GS, WGS, OHV, and GB selection approaches. Then, a simulation study based on empirical data obtained from a set of maize inbreds was used to analyze OPV’s relative ability to improve response. The objectives of this paper were to (i) improve genetic gain, and (ii) investigate the efficacy of population-based selection methods.
Materials and Methods
In this section, we first present OPV selection. Then, four existing selection methods (GS, WGS, OHV, and GB selection) are mathematically defined. The following definitions will be used in subsequent sections to describe the five selection methods compared in this paper:
L: the number of marker loci.
M: the ploidy of the plants.
N: the number of individuals in the population.
a binary matrix indicating whether the allele at locus l on the mth copy of a chromosome of individual n is the major () or minor () allele.
the effect of having the major allele at locus l.
q: the number of individuals to be selected for reproduction in each generation.
Additionally, for OHV, GB, and OPV selection, adjacent markers are likely to segregate together, and may be grouped into representative haplotype blocks. For these three methods, the following definitions will also be used:
B: the number of haplotype blocks per chromosome.
the set of marker loci that belong to haplotype block b.
Optimal population value selection
OPV is defined as the GEBV of the best possible progeny produced by a breeding population after an unlimited number of generations, assuming markers segregate in the haplotype blocks considered. This can be interpreted as a generalization of the upper selection limit (Cole and VanRaden 2011) to varying haplotype lengths, rather than just haplotypes with a length of one marker. For a given breeding population selected from the entire population, the OPV is defined as
| . | (1) |
The challenge is to select the optimal breeding population S so that is maximized.
Genomic selection
This method selects the q individuals with the largest GEBVs (Meuwissen et al. 2001). The GEBV of individual n is defined as:
| . | (2) |
Weighted genomic selection
WGS has been proposed as a variation of GS (Goddard 2009; Jannink 2010):
| . | (3) |
Here, the estimated marker effect was weighted using the frequency of the most beneficial allele in the population at that locus, denoted by This procedure is outlined in detail in Jannink (2010). Since is undefined when a rule was set to assume for these cases. It is important to note that, when every member in the population has the same allele. Thus, assuming has an equal effect on all lines.
Optimal haploid value selection
The OHV of individual n is the GEBV of the best possible DH individual derived from it (Daetwyler et al. 2015):
| . | (4) |
Qualitatively, OHV is the sum of the maximum GEBVs of predefined blocks along a chromosome. This method selects the q individuals with the highest OHV.
Genotype building selection
This strategy was initially designed to select founder individuals of the next generation such that the founders possessed the best possible combinations of blocks for each block across the genome. In the general context of genomic selection, and using the definitions made in this paper, the genotype building selection approach is trying to select a subset to maximize the sum of estimated allelic effects across M chromosomes at each block, which can be formulated as
| , | (5) |
such that
| , | (6) |
and
| . | (7) |
Optimization of OPV and GB selection metrics
Here, an integer programming model is formulated to select a breeding population of q individuals with maximal OPV or GB values. The decision variables are and for all blocks and individuals which are all binary. Variable indicates whether individual n is selected () or not (), and variable indicates whether () or not () individual n makes the largest contributions to block b among all selected individuals. The integer programming model can be formulated as follows.
| , | (8) |
such that
| , | (9) |
| , | (10) |
| , | (11) |
and
| (12) |
Here, the parameter K determines the type of metric to be optimized. If Equation (8) represents the OPV metric; if it represents the GB metric. Constraint (11) defines the number of individuals to be selected. Constraint (11) limits the number of best chromosomes for each block to be used in the metric. Constraint (11) means that only selected individuals can be used for the calculation of the metrics. This model can be solved using standard integer programming solvers. However, the computation time could increase exponentially as the dimensions of the model grow (e.g., the numbers of blocks, B, and individuals, N).
Alternatively, we propose a two-step heuristic algorithm to solve this model:
Step 1: Randomly select a breeding population with q individuals, and calculate the desired metric.
Step 2: Propose pairwise swaps between a selected individual and an unselected one, and calculate the resulting metric. Accept the proposed swap if the metric is improved. Repeat this step until no more swaps improve the metric.
The essence of this heuristic algorithm is to treat the breeding population selection problem as a complex multidimensional nonlinear optimization problem. The solution from this heuristic algorithm is guaranteed to be at least locally optimal.
Performance enhancement strategies for OHV, OPV, and GB
The performances of OHV, OPV, and GB are sensitive to two parameters, which can be optimized to enhance their effectiveness. The first parameter, B, represents a tradeoff between short-term and long-term gains by changing the number of haplotype blocks considered on each chromosome. If then OHV, OPV, and GB consider one haplotype block per chromosome, and, therefore, focus on short-term gain similar to GS. If B is a large number, then the algorithms focus on long-term gain by evaluating progeny with up to B recombination events. The second parameter, F, specifies the removal of the of individuals with the lowest GEBVs before optimizing the selected population. If then OHV, OPV, and GB selection are equivalent to GS, whereas a smaller value of F would put more emphasis on long-term gain through the potential selection of individuals with lower overall GEBVs that harbor rare favorable alleles.
Simulation
Data:
SNPs from the 369 maize inbred lines used by Leiboff et al. (2015) were merged with additional SNPs genotyped using tGBS (Schnable et al. 2013), and phased using Beagle (Browning and Browning 2008). This produced ∼1.4 million SNPs distributed across the 10 maize chromosomes. The 369 SAM volume phenotypes from Leiboff et al. (2015) were used to estimate marker effects, using the BayesB model (Meuwissen et al. 2001) implemented in GenSel (Fernando and Garrick 2009). We assumed that marker effects were known without error, and that inaccuracies in marker effect estimation affected all selection methods equally.
Recombination rates in this population were estimated using the genetic map developed from the maize nested association mapping (NAM) (Yu et al. 2008) population. Of the 1144 genetic markers in the NAM population, 133 were removed because the orderings between physical and genetic positions were inconsistent. The remaining 1011 markers were used to estimate the genetic positions of the SNPs by linear interpolation between the NAM markers flanking each SNP. Once the genetic positions of the SNPs were estimated, recombination rates were calculated between adjacent SNPs using Haldane’s mapping function. This produced a genetic map of 1544 cM.
In silico breeding process model:
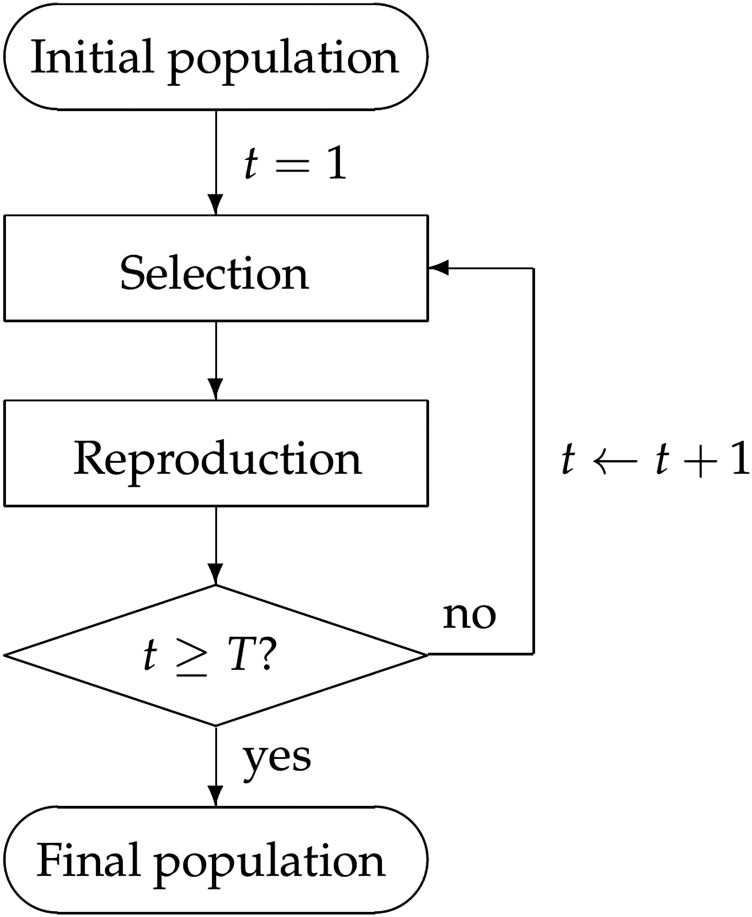
The generic plant breeding program considered in this paper is illustrated in Figure 1. Starting with an initial population, the program iterates over the selection, and reproduction steps ten times () to simulate ten generations of breeding. In each simulation, 200 individuals were randomly selected from the 369 lines to form the initial population, and a breeding population of individuals was selected in each generation. Each initial population was used once for all five selection approaches.
Figure 1.
Diagram of the simulation process.
Five selection approaches were used in the selection step: GS, WGS, OHV, OPV, and GB. To determine appropriate parameter setting for the latter three approaches, 100 replicates of the generic plant breeding process were performed using 60 different combinations of the parameters: and For these three approaches, the best combinations ( for OHV; for GB; and for OPV) were used in 2000 additional simulation replicates of all five approaches. Due to the large dimension of the data, the heuristic algorithm was used for OPV and GB in all simulations.
In the reproduction step, we use the following two substeps to simulate the next generation.
Substep 1: Pair the 20 selected individuals to make 10 crosses in descending order of their GEBVs, i.e., the individual with the highest GEBV is crossed with the second, the third highest with the fourth, etc.
- Substep 2: Produce 20 progeny from each of the 10 crosses to maintain a population size of 200. Let r denote the vector of recombination frequencies and the genotype matrix of a random offspring from crossing individuals and Then P is determined as
Here, are two identical and independent random vectors following the inheritance distribution with recombination frequency vector r. The inheritance distribution was defined in Han et al. (2017) as follows.
- Definition 0.1: Han et al. (2017) We say that a random binary vector follows an inheritance distribution with parameter vector if
(13) (14) Here, “w.p.” stands for “with probability.”
This process was implemented in silico in Octave (Eaton et al. 2015).
Results
Each selection approach was used in 2000 independent simulation runs. Since each random initial population was used on all five selection methods, the rankings of their population maximums in the 10th generations reveal their comparative effectiveness. Table 1 summarizes the frequencies of these rankings across the 2000 replications. These results suggest that OPV outperformed the other four approaches in achieving genetic gains in the first 10 generations.
Table 1. Frequencies of relative rankings of the five selection approaches on the population maximum in the 10th generation across 2000 replications.
| GS (%) | WGS (%) | OHV (%) | GB (%) | OPV (%) | |
|---|---|---|---|---|---|
| First | 1.5 | 6.3 | 4.6 | 22.1 | 65.7 |
| Second | 11.6 | 6.4 | 27.6 | 46.8 | 7.7 |
| Third | 31.5 | 13.5 | 39.9 | 7.7 | 7.5 |
| Fourth | 33.9 | 19.5 | 25.5 | 14.1 | 7.2 |
| Fifth | 21.6 | 54.4 | 2.5 | 9.5 | 12.1 |
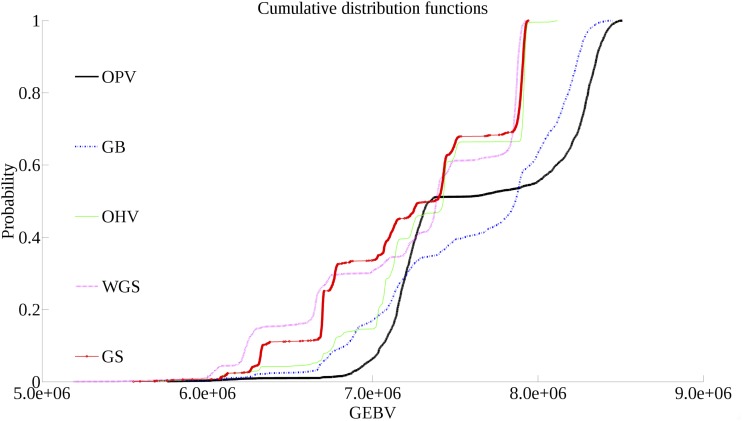
To provide a more insightful assessment of different selection approaches, we also compared the cumulative distribution functions (CDFs) of the population maximums in the 10th generation from each replicate (Figure 2). Since the vertical value of a point on a CDF curve indicates the percentage of random outcomes that have a lower GEBV than its horizontal value, the best performing selection approach at a given vertical point will be the farthest right. GS and WGS perform similarly, although GS outperforms WGS below the 30th percentile. Above the 40th percentile, OHV performs similarly to GS and WGS. Below this percentile, however, OHV outperforms both of these methods, matching the performance of GB. OPV outperforms GB below the 30th percentile and above the 50th percentile.
Figure 2.
Cumulative distribution functions of population maximums after 10 generations of selection over 2000 replications for each selection approach.
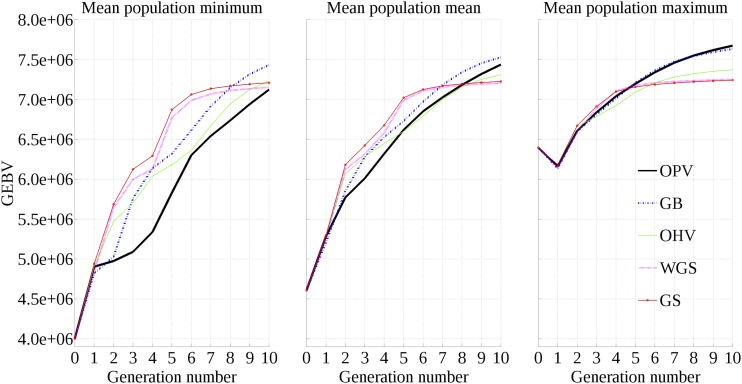
Figure 3 displays the means of population minimum, mean, and maximum of GEBV over 10 generations for all selection approaches. As expected, GS and WGS show quick and substantial improvement in GEBV before reaching a plateau at generation 4. OHV sacrifices short-term gain for long-term gain, exceeding the performance of GS and WGS by generation 9, 8, and 6 with respect to mean population minimum, mean, and maximum, respectively. GB demonstrates a similar growth pattern as OHV, with enhanced performance across all three criteria. OPV has the slowest increases in mean population minimum; its mean population mean grows slowly in the first five generations but finishes second to GB in generation 10; its mean population maximum initially improves more slowly than GS and WGS but eventually surpasses all other four approaches in generation 10.
Figure 3.
Mean population minimum, mean, and maximum over 10 generations.
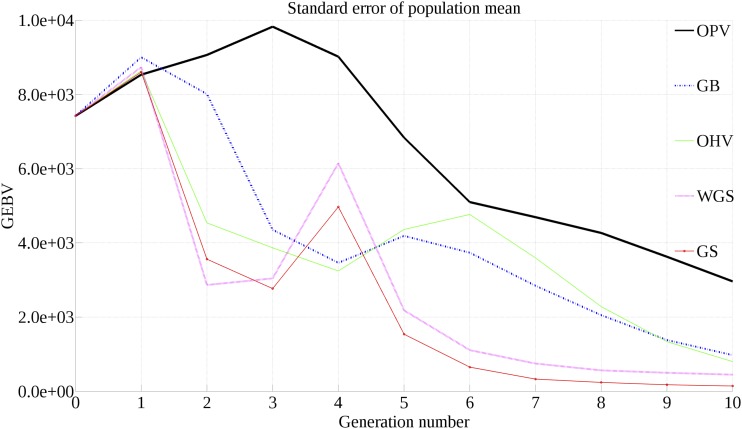
Figure 4 displays the SE of population mean over 10 generations for all selection approaches. As expected, OHV, GB, and OPV maintain genetic variance in the breeding population longer than either GS or WGS. However, OPV maintains substantially more genetic variance than either GB or OHV, indicating that there is greater room for population improvement after 10 generations.
Figure 4.
SE of population mean over 10 generations.
Discussion
In this study, we have introduced a new selection approach, optimal population value selection (OPV), which evaluates the genetic merit of a set of selection candidates instead of performing truncation selection on an individual metric. OPV is similar to optimal haploid value (OHV) and genotype building (GB) in that all three methods consider the effects of recombination on the genetic merit of future individuals. By focusing on the GEBV of segments of the genome instead of total GEBV, these three methods can identify and select for segments that contain rare favorable alleles but whose favorable effects would be discarded by truncation methods such as GS and WGS.
In our simulations, the primary advantage of OHV over GS and WGS is the decreased incidence of individuals with low GEBVs. The mean best individual over 10 generations is significantly better than the same individual from GS and WGS on average, although Figure 2 shows that there are rare cases where GS and WGS can match the performance of OHV. Daetwyler et al. (2015) reported that OHV maintained genetic variance longer than GS and hypothesized that this was due to the ability of OHV to maintain rare alleles in the population. OPV is similar to OHV with one important difference. After calculation of optimal haploid values, OHV selection selects a fraction of individuals with the highest OHVs—a form of truncation selection. OPV selects a subset of individuals based on the maximum haploid value at individual segments across the selected individuals. Thus, there is still the possibility that OHV will discard individuals with rare favorable alleles that are masked by a large number of unfavorable alleles. OPV allows for the inclusion of individuals that have low genetic merit but contain favorable alleles that are rare in the rest of the selected individuals.
There are three principal differences between our implementation of OHV and that of Daetwyler et al. (2015). First, the population sizes used in our study (200) are much smaller than those of Daetwyler et al. (2015) (55,000). One of the benefits of both OPV and OHV is their ability to maintain rare favorable alleles in breeding populations. Their ability to identify alleles is independent of population size, which increases the probability of obtaining favorable recombinants. Changes in population size are expected to affect both methods equally. Second, we have not implemented the use of DHs in our simulated breeding program as in Daetwyler et al. (2015). Despite this omission, we observe that OHV outperforms GS. While a comparison of OPV and OHV when using DHs would be valuable, our results suggest that the advantage of OHV over GS is maintained without the use of DHs. Third, we remove of individuals with the lowest GEBVs prior to selection, which is a strategy that has been shown to enhance the efficiency of OHV, OPV, and GB.
OPV is similar to GB, but it makes two improvements. First, the OPV metric exclusively measures the long-term potential of the most outstanding progeny in the population, whereas the GB metric also values the GEBV of the breeding parents like GS does. Second, we presented an integer programming model and a heuristic algorithm for selecting an optimal subset of individuals to maximize the OPV or GB metric, which is an improvement over the algorithm presented in Kemper et al. (2012). As Figure 3 shows, OPV and GB produce similar response curves for the average of the best individuals. However, when we consider the distributions of these best individuals, we see that the worst of the best individuals produced by OPV are better than those of GB, and the best of the best individuals produced by OPV are better than those of GB. GB only outperforms OPV between the 30th and 50th percentiles. This indicates that while there is a chance that GB produces better individuals than OPV, in the best and worst cases, OPV is expected to be the superior method. One possible explanation for this phenomenon is that OPV maintains genetic variance longer than all other methods (Figure 4).
Conclusions
In this paper, a new selection method, optimal population value (OPV) selection, was presented. Instead of using evaluations of individual lines to select the breeding population, a candidate breeding population was selected as a unit. While this presents some computational challenges, such as solving a combinatorial optimization problem, it was shown to outperform existing methods in a series of simulation experiments that spanned 10 generations, and used empirical data from an inbred maize population. Future research into genomic selection approaches should focus on selecting sets of individuals as a unit, rather than how to better evaluate individual breeding values prior to performing truncation selection.
Acknowledgments
This work was supported in part by the National Science Foundation grant IOS-1238142, the United States Department of Agriculture National Institute of Food and Agriculture (NIFA) Award 2017-67007-26175, and the Plant Sciences Institute at Iowa State University.
Appendix
Mean of maximal GEBV in the 10th generation for 100 random simulations using OHV (Table A1), GB (Table A2), and OPV (Table A3) selection. Optimal parameters are highlighted.
Table A1. Mean of maximal GEBV in the 10th generation for 100 random simulations using OHV selection.
| F | ||||||
|---|---|---|---|---|---|---|
| B | ||||||
| 1 | 7.30 | 7.20 | 7.21 | 7.24 | 7.19 | 7.28 |
| 2 | 7.23 | 7.15 | 7.14 | 7.22 | 7.22 | 7.28 |
| 3 | 7.24 | 7.34 | 7.43 | 7.43 | 7.45 | 7.26 |
| 6 | 7.23 | 7.26 | 7.46 | 7.46 | 7.42 | 7.37 |
| 12 | 7.34 | 7.36 | 7.38 | 7.50 | 7.40 | 7.35 |
| 20 | 7.33 | 7.33 | 7.41 | 7.39 | 7.47 | 7.31 |
| 30 | 7.27 | 7.32 | 7.36 | 7.44 | 7.45 | 7.25 |
| 40 | 7.27 | 7.39 | 7.39 | 7.40 | 7.43 | 7.28 |
| 50 | 7.28 | 7.29 | 7.39 | 7.41 | 7.41 | 7.31 |
| 60 | 7.31 | 7.30 | 7.39 | 7.41 | 7.49 | 7.26 |
Table A2. Mean of maximal GEBV in the 10th generation for 100 random simulations using GB selection.
| F | ||||||
|---|---|---|---|---|---|---|
| B | ||||||
| 1 | 7.71 | 7.69 | 7.69 | 7.70 | 7.57 | 7.27 |
| 2 | 7.26 | 7.50 | 7.53 | 7.46 | 7.47 | 7.30 |
| 3 | 7.33 | 7.45 | 7.50 | 7.67 | 7.64 | 7.28 |
| 6 | 7.25 | 7.37 | 7.52 | 7.65 | 7.76 | 7.42 |
| 12 | 7.18 | 7.39 | 7.56 | 7.71 | 7.71 | 7.34 |
| 20 | 7.15 | 7.35 | 7.54 | 7.70 | 7.73 | 7.30 |
| 30 | 7.19 | 7.29 | 7.61 | 7.73 | 7.72 | 7.20 |
| 40 | 7.12 | 7.34 | 7.59 | 7.68 | 7.70 | 7.31 |
| 50 | 7.10 | 7.29 | 7.55 | 7.67 | 7.64 | 7.27 |
| 60 | 7.17 | 7.22 | 7.64 | 7.74 | 7.77 | 7.26 |
Table A3. Mean of maximal GEBV in the 10th generation for 100 random simulations using OPV selection.
| F | ||||||
|---|---|---|---|---|---|---|
| B | ||||||
| 1 | 7.86 | 7.73 | 7.83 | 7.67 | 7.52 | 7.28 |
| 2 | 7.65 | 7.68 | 7.71 | 7.73 | 7.55 | 7.27 |
| 3 | 7.36 | 7.48 | 7.62 | 7.70 | 7.67 | 7.26 |
| 6 | 7.14 | 7.28 | 7.53 | 7.70 | 7.67 | 7.40 |
| 12 | 7.14 | 7.34 | 7.46 | 7.69 | 7.61 | 7.36 |
| 20 | 7.17 | 7.33 | 7.46 | 7.73 | 7.76 | 7.28 |
| 30 | 7.12 | 7.29 | 7.53 | 7.68 | 7.72 | 7.22 |
| 40 | 7.08 | 7.38 | 7.60 | 7.72 | 7.62 | 7.27 |
| 50 | 7.08 | 7.25 | 7.64 | 7.74 | 7.66 | 7.30 |
| 60 | 7.08 | 7.27 | 7.50 | 7.68 | 7.71 | 7.27 |
Footnotes
Communicating editor: D. J. de Koning
Literature Cited
- Bernardo R., Yu J., 2007. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 47: 1082–1090. [Google Scholar]
- Browning B., Browning S., 2008. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J., VanRaden P., 2011. Use of haplotypes to estimate Mendelian sampling effects and selection limits. J. Anim. Breed. Genet. 128: 446–455. [DOI] [PubMed] [Google Scholar]
- Daetwyler H., Hayden M., Spangenberg G., Hayes B., 2015. Selection on optimal haploid value increases genetic gain and preserves more genetic diversity relative to genomic selection. Genetics 200: 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton, J. W., D. Bateman, S. Hauberg, and R. Wehbring, 2015 GNU Octave version 4.0.0 manual: a high-level interactive language for numerical computations. Available at: http://www. gnu.org/software/octave/doc/interpreter. Accessed: June 13, 2017.
- Fernando, R., and D. Garrick, 2009 Gensel—user manual for a portfolio of genomic selection related analyses. Technical report. Available at: http://bigs.ansci.iastate.edu/bigsgui. Accessed: June 13, 2017. [Google Scholar]
- Goddard M., 2009. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136: 245–257. [DOI] [PubMed] [Google Scholar]
- Han Y., Cameron J., Wang L., Beavis W., 2017. The predicted cross value for genetic introgression of multiple alleles. Genetics 205: 1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner E. L., Lorenz A. J., Jannink J.-L., Sorrells M. E., 2010. Plant breeding with genomic selection: gain per unit time and cost. Crop Sci. 50: 1681–1690. [Google Scholar]
- Jannink J.-L., 2010. Dynamics of long-term genomic selection. Genet. Sel. Evol. 42: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper K. E., Bowman P. J., Pryce J. E., Hayes B. J., Goddard M., 2012. Long-term selection strategies for complex traits using high-density genetic markers. J. Dairy Sci. 95: 4646–4656. [DOI] [PubMed] [Google Scholar]
- Leiboff S., Li X., Hu H.-C., Todt N., Yang J., et al. , 2015. Genetic control of morphometric diversity in the maize shoot apical meristem. Nat. Commun. 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T., Hayes B., Goddard M., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P., Liu S., Wu W., 2013. Genotyping by next-generation sequencing. U.S. Patent Application No. 13/739,874.
- Yu J., Holland J., McMullen M., Buckler E., 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]