Abstract
Membrane proteins encapsulated by detergent micelles are widely used for structural study. Because of their amphipathic property, detergents have the ability to maintain protein solubility and stability in an aqueous medium. However, conventional detergents have serious limitations in their scope and utility, particularly for eukaryotic membrane proteins and membrane protein complexes. Thus, a number of new agents have been devised; some have made significant contributions to membrane protein structural studies. However, few detergent design principles are available. In this study, we prepared meta and ortho isomers of the previously reported para-substituted xylene-linked maltoside amphiphiles (XMAs), along with alkyl chain-length variation. The isomeric XMAs were assessed with three membrane proteins, and the meta isomer with a C12 alkyl chain was most effective at maintaining solubility/stability of the membrane proteins. We propose that interplay between the hydrophile–lipophile balance (HLB) and alkyl chain length is of central importance for high detergent efficacy. In addition, differences in inter-alkyl-chain distance between the isomers influence the ability of the detergents to stabilise membrane proteins.
Keywords: amphiphiles, detergents, membrane proteins, noncovalent interactions, protein solubilization, protein stabilization
Introduction
Membrane protein structural study has attracted tremendous attention since the emergence of the first crystal structure of the bacterial photosynthetic reaction centre in 1985.[1] As more than 50% of pharmaceuticals target membrane proteins, a detailed understanding of their three-dimensional structures is of major importance in drug development.[2] Such structural information provides fundamental insights into the molecular mechanisms of membrane proteins, including transporters and receptors. Despite their pharmaceutical and biological importance, however, the number of membrane proteins with known structure (~700) is far lower than that of soluble proteins (~100000).[3] There are several hurdles associated with membrane protein study, including protein expression, solubilisation, stabilisation and crystallisation in an aqueous environment. Detergents are essential for the manipulation of membrane proteins in non-native environments. n-Octyl-β-D-glucopyranoside (OG), n-dodecyl-β-D-maltopyranoside (DDM) and lauryldimethylamine-N-oxide (LDAO) are widely used conventional detergents for this purpose,[4] but membrane proteins encapsulated by these popular detergents have the propensity to aggregate and denature. This is particularly true for eukaryotic membrane proteins and membrane protein complexes, which are generally more unstable than prokaryotic/monomeric membrane proteins.[5] Thus, new detergents with enhanced protein stabilisation efficacy are necessary for successful structural study with these challenging membrane proteins/protein complexes.[6]
Since the emergence of peptitergents[7a] in 1993, a number of detergents with new architectures have been developed. Among these, peptide-based agents have been the most popular, mainly because of the variety in their physical properties and structures. Examples include lipopeptide detergents (LPDs)[7b] with an α-helical structure, short peptides comprising several amino acids[7c] and β-peptides (BPs),[7d] which have unusual octyl side chains. Nano-self-assemblies comprising a patch of lipid bilayer in the form of a nanodisc (ND)[8a] or nano-lipodisq[8b] are other recent innovations. Along with amphiphilic polymers (Apols),[8c,d] however, these peptide-based detergents and nano-self-assemblies have yet to contribute to high-resolution structure determination of membrane proteins. In contrast, small amphipathic molecules including tripod amphiphiles (TPAs),[9a–c] neopentyl glycols (NGs) comprising maltose neopentyl glycols (MNGs)[9d–f] and glucose neopentyl glycols (GNGs),[9,hg] and facial amphiphiles (FAs)[9i,j] derived from cholic acid have been shown to be useful for membrane protein structural study. More recently, glyco-diogenin (GDN),[10a] mannitol-based amphiphiles (MNAs),[10b] neopentyl glycol triglucosides (NDTs)[10c] and penta-saccharide amphiphiles (PSEs)[10d] have been shown to be effective at stabilising the structures of many membrane proteins. Despite the development of dozens of new agents, progress in membrane protein structural study is still slow. This is attributable, in part at least, to the limited number and narrow scope of utility of new/conventional detergents, compared to the wide diversity of membrane proteins in terms of their propensity to denature and aggregate. Thus, we need to continue to develop new detergents with distinct architectures as a means of coping with the large variation in membrane protein behaviour. It is notable that there are few clear detergent design principles, in spite of the large number of detergent studies. Therefore, efforts to establish these principles are necessary together with the development of new agents for membrane protein study.
In a previous study, we developed and characterised maltoside amphiphiles with a para-xylene linker (XMAs; Figure S1 in the Supporting Information).[11] Of the agents tested, XMA-4 (C11 alkyl chain) was the most effective at stabilising certain membrane proteins, but it was inferior to DDM for some membrane proteins, such as the leucine transporter (LeuT) and the β2 adrenergic receptor (β2AR). We hypothesise that the distance between the two alkyl chains of this XMA is too long to give effective molecular packing around certain membrane proteins. One strategy to address this is to use a linker that results in a shorter inter-chain distance (e.g., meta- or ortho-xylene). Thus, we prepared meta or ortho isomeric compounds of the para-XMAs and assessed these for protein stability/solubility with three membrane proteins. On the basis of the results obtained in this study, new design principles governing detergent efficacy for membrane protein stability are proposed.
Results and Discussion
The new isomeric agents can be categorised into two classes according to the geometry of the linker. The amphiphiles with the meta-xylene linker were designated M-XMAs, and those with the ortho-xylene linker were designated O-XMAs. Alkyl chain length within each class varied from C11 to C18, as reflected in the names of the new agents (Scheme 1). These agents commonly have two alkyl chains and two branched dimalto-sides (hydrophobic and hydrophilic groups, respectively). This structural feature is the same as in our previous XMAs (P-XMAs).[11] Because of the isomeric linker (para-, meta- or ortho-xylene), the new agents differ from the previous XMAs in terms of both inter-alkyl chain distance and inter-head group distance. O-XMAs have the shortest distance, and P-XMAs have the longest (M-XMAs are in between). The XMAs are architecturally different from MNGs: MNGs have neopentyl glycol as a linker; the xylene linker in the XMAs is more rigid.[9d]
Scheme 1.
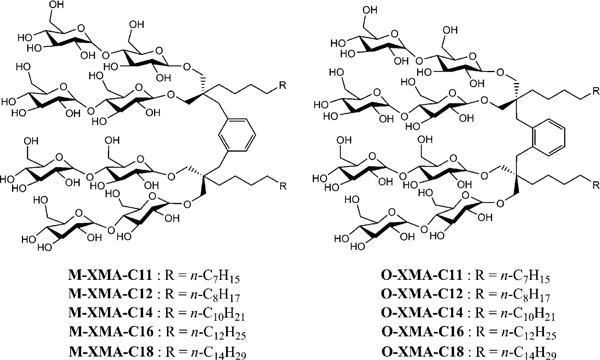
Chemical structures of M-XMAs and O-XMAs with alkyl chain lengths from C11 to C18. The number of carbons in the alkyl chain was used for detergent designation.
The M-XMAs and O-XMAs were prepared according to the same synthetic protocol as that used for P-XMAs. Briefly, mono-alkylated diethylmalonates with different alkyl chain lengths were introduced into a meta- or ortho-xylene core by an alkylation reaction. The resulting tetra-ethyl ester compounds were reduced to tetra-alcohols; glycosylation with a protected maltosyl bromide and a subsequent global deprotection afforded the individual M-/O-XMAs. Despite potential steric hindrance, the efficiency of O-XMA preparation was comparable to that for M-XMAs and P-XMAs. The overall yields of M-XMAs and O-XMAs were about 55 and 50%, respectively, thus making preparation of multigram amounts of material feasible.
In our previous study, the XMA with a C12 alkyl chain (P-XMA-C12, the longest P-XMA alkyl chain) showed limited water-solubility (~10%), and required sonication for complete dissolution.[11] It tended to precipitate over time in an aqueous medium. Surprisingly, the C12 M-XMAs and O-XMAs were completely soluble at concentrations of more than 20% (w/v) in water. M-XMAs and O-XMAs with C12, C14, C16 or C18 alkyl chains were water-soluble (at >20%, w/v), with no observed precipitation over time. This result indicates that micelles formed from these isomeric amphiphiles should have significantly different properties from those of P-XMAs, despite the structural similarity.
The new agents were characterised in terms of critical micelle concentration (CMC) and micelle size, by fluorophore encapsulation[12] and dynamic light scattering (DLS), respectively (Table 1). The CMC values decreased with increasing alkyl chain length. For instance, O-XMA-C11 (shortest alkyl chain) gave the largest CMC value (~10 μm) whereas O-XMA-C18 (longest chain) gave the smallest (~1.5 μm). In the isomeric comparison, the O-XMA CMC values were a little higher than those of the M-XMAs (e.g., ~10 μm for O-XMA-C11, ~6 μm for M-XMA-C11). Similar trends were observed for other comparisons between the O-XMA and M-XMA isomers.
Table 1.
Molecular weights (MWs) and CMCs of P-XMA-C11, M-XMAs, O-XMAs and DDM, and hydrodynamic radii of their micelles.
| Detergent | MW | CMC [μm] | CMC (wt%) | Rh[a] |
|---|---|---|---|---|
| P-XMA-C11 | 1860.0 | ~6.0 | ~0.0011 | 3.3 ±0.03 |
| M-XMA-C11 | 1860.0 | ~6.0 | ~0.0011 | 3.2±0.01 |
| O-XMA-C11 | 1860.0 | ~10 | ~0.0019 | 3.0±0.02 |
| M-XMA-C12 | 1888.1 | ~4.0 | ~0.0008 | 3.4±0.02 |
| O-XMA-C12 | 1888.1 | ~6.0 | ~0.0011 | 3.2±0.03 |
| M-XMA-C14 | 1994.2 | ~2.5 | ~0.0005 | 3.6±0.04 |
| O-XMA-C14 | 1994.2 | ~3.0 | ~0.0006 | 3.5±0.04 |
| M-XMA-C16 | 2000.3 | ~2.0 | ~0.0004 | 3.9 ±0.04 |
| O-XMA-C16 | 2000.3 | ~2.0 | ~0.0004 | 3.9±0.06 |
| M-XMA-C18 | 2056.4 | ~1.5 | ~0.0003 | 4.1 ±0.04 |
| O-XMA-C18 | 2056.4 | ~1.5 | ~0.0003 | 3.9±0.04 |
| DDM | 510.1 | ~170 | ~0.0087 | 3.4±0.02 |
Hydrodynamic radius (Rh) at 1.0% (w/v) detergent concentration measured by dynamic light scattering (n=5).
Detergent micelle size also depended on alkyl chain length: longer alkyl chains produced larger micelles, consistent with the observation for the P-XMAs with varying alkyl chain length.[11] For instance, O-XMA-C11 micelles had an estimated hydrodynamic radius (Rh) of 3.0 nm, whereas O-XMA-C18 micelles were estimated at 3.9 nm. The M-XMAs tended to form larger micelles than the O-XMA isomers (e.g., 3.2 vs 3.0 nm, respectively, for C11), thus indicating that the M isomers have a more cylindrical shape than the O isomers.
Thus, M-XMAs had lower CMC values and produced larger micelles than their ortho counterparts. The differences in CMC values (i.e., aggregation tendency) and micelle size are likely associated with differences in detergent stabilisation efficacy toward target membrane proteins. All the XMAs (1.5–10 μm) gave much lower CMC values than DDM (170 μm); their micelle sizes (3.0–4.1 nm) were smaller or larger than that of DDM (3.4 nm) depending on alkyl chain length. All the XMAs showed lower CMC values (1.5–10 μm) than for MNGs (10–150 μm),[9d,f] and their micelle sizes (3.0~4.1 nm) varied considerably less than those of MNGs (2.5~7.2 nm) with different alkyl chain length. All the new XMAs showed a single set of micelle population (in terms of size), indicative of high micelle homogeneity (Figure S2).
The M-XMAs and O-XMAs were evaluated first with the leucine transporter (LeuT) from Aquifex aeolicus.[13] The transporter was extracted from the membrane by using DDM and purified in the same detergent. LeuT (~1.2 mgmL−1) was diluted into buffer including individual XMAs (final detergent concentration: CMC+0.04%, w/v). P-XMA-C11 and DDM were used as a representative P-XMA and conventional detergent. (P-XMA-C11 was the most effective of five P-XMAs with different alkyl chains for LeuT stability.[11]) Substrate binding of the transporter was measured by a scintillation proximity assay (SPA) with radiolabelled substrate ([3H]Leu).[14] The measurements were carried out at regular intervals over 12 days with incubation at room temperature (Figure 1). As observed in the previous study, P-XMA-C11 was slightly inferior to DDM in maintaining substrate binding ability of the transporter. Similar results were obtained for M-XMA-C11 and O-XMA-C11. However, the C12 versions (M-XMA-C12 and O-XMA-C12) showed a substantial enhancement. M-XMA-C12 appeared to be slightly better than O-XMA-C12. (Note: P-XMA-12, like P-XMA-C11, was inferior to DDM for LeuT stability.)[11] With increasing XMA alkyl chain length from C12 to C18, substrate binding ability progressively decreased (worst results for M-XMA-C18 and O-XMA-C18). Thus, the C12 alkyl chain length in the XMA architecture appears to be optimal for this protein (Figure 1A). When detergent concentration was increased to CMC+0.2% (w/v), similar trends were observed (Figure 1B). M-XMA-C12 and O-XMA-C12 were better than M-XMA-C11, O-XMA-C11 and DDM at maintaining substrate binding (M-XMA-C12 better than O-XMA-12). However, transporter activity rapidly decreased for M-/O-XMAs with higher alkyl chain length (C14, C16 and C18). P-XMA-C11 was inferior to M-XMA-C11 and O-XMA-C11 at the higher detergent concentration. Overall, the new XMAs with C12 alkyl chains showed improved efficacy in preserving the substrate binding ability of LeuT compared to the previously reported XMA (P-XMA-C11) and a conventional detergent (DDM).
Figure 1.
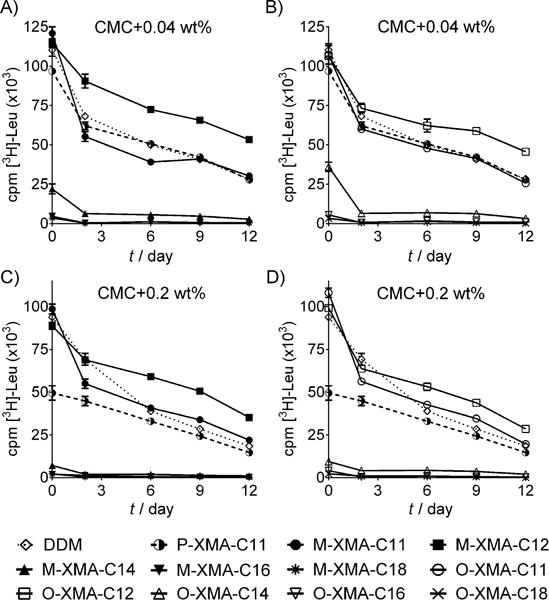
Long-term stability of LeuT solubilised in M-XMAs, O-XMAs, P-XMA-C11 and DDM. Two detergent concentrations were used: CMC+0.02 and +0.4% (w/v). Protein stability was assessed based on the substrate binding activity of LeuT by a scintillation proximity assay after incubation for up to 12 days at room temperature (mean ± SEM, n=2 or 3).
The new agents were next evaluated with Salmonella typhimurium melibiose permease (MelBs).[15] For this experiment, Escherichia coli membranes expressing MelBSt (10 mgmL−1) were treated with 1.5% (w/v) XMAs (M-XMAs, O-XMAs or P-XMA-C11) or DDM. In order to investigate how efficiently these XMAs extract MelBSt, membranes containing the protein were incubated with detergent for 90 min at 0 °C. After ultracentrifugation, the amounts of soluble MelBSt were estimated by SDS-PAGE and western blotting (Figures 2 and S3). In a previous study, DDM gave quantitative extraction at this temperature.[11] Although M-XMAs with longer alkyl chains (e.g., M-XMA-C16 and M-XMA-C18) were comparable to DDM, most XMAs were inferior (Figure 2). However, it is notable that all the new XMAs were better than P-XMA-C11 in this regard. Interestingly, the M-XMAs and O-XMAs showed a different trend with regard to alkyl chain length. MelB solubilisation efficiency tended to increase with increasing M-XMA chain length but to decrease with increasing O-XMA chain length (Figure S3). The overall trends were similar at 45°C. M-XMAs and O-XMAs were better than (or at least similar to) P-XMA-C11 at solubilising the protein but less efficient than DDM.[11]
Figure 2.
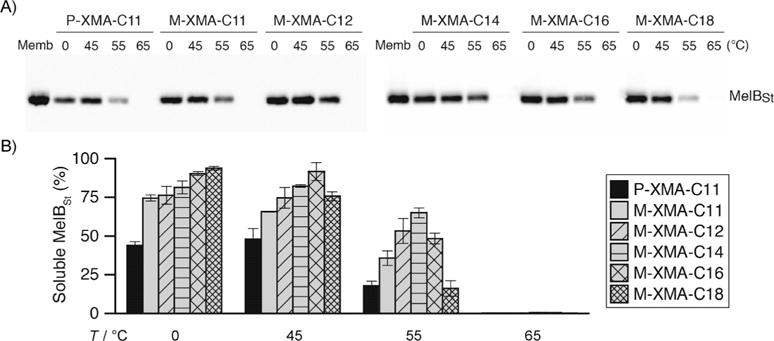
Thermostability of MelBSt solubilised in M-XMAs or P-XMA-C11. E. coli membranes containing MelBSt were treated with detergent (1.5%, w/v) for 90 min at 0, 45, 55, or 65 °C. The amounts of soluble MelBst after ultracentrifugation were estimated by A) SDS-PAGE and western blotting, then B) expressed as histograms (percentage of total MelBSt in untreated membrane sample). Band densities were measured by ImageQuant software (mean ± SEM, n=2–4).
However, at 55°C differences between the new agents as well as between the XMAs and DDM became more marked. Previously we showed that DDM failed to maintain the solubility of MelBSt in aqueous solution at this temperature, presumably because the protein aggregated or denatured.[11] In contrast, all the new XMAs (except the longest, M-XMA-C18 and O-XMA-C18) maintained substantial MelBSt in solution (Figures 2B and S3). M-XMA-C14 was the most effective (~70% retention); O-XMA-C11, O-XMA-C12 and O-XMA-C14 gave about 60% (significantly better than P-XMA-C11). At 65°C, none of the XMAs maintained MelBSt in a soluble state.
This thermostability assay allowed us to assess which of the detergents efficiently extracts MelBSt from the membranes and which effectively maintains MelBSt solubility. M-XMA-C18 was very good at maintaining MelBSt solubility at 0°C, but the efficacy dropped dramatically at higher incubation temperature, thus indicating that it efficiently solubilises MelBSt but is comparatively poor at stabilising the protein. In contrast, M-XMA-C14 was able to retained MelBSt in a soluble state over the range of temperatures tested, thus indicating that it is effective at both stabilisation and solubilisation. M-/O-XMAs with long alkyl chains (C16 and C18) were less effective at stabilising the protein, consistent with the results for LeuT. On the basis of these results, the optimum chain length for MelBSt appeared to be C14 for M-XMAs and C11 to C14 for O-XMAs.
The encouraging results obtained with LeuT and MelBSt prompted us to evaluate these agents with the β2 adrenergic receptor (β2AR), a member of the G protein-coupled receptor (GPCR) family.[16] In order to investigate detergent ability to stabilise β2AR, DDM-purified receptor was diluted into buffer solutions containing P-XMA-C11, M-XMAs, O-XMAs or DDM. Ligand binding was assessed with the antagonist [3H]dihydroalprenolol ([3H]DHA). M-XMA-C11 was slightly better than P-XMA-C11, but O-XMA-C11 was somewhat inferior (Figure 3). With C12 chain lengths, stabilisation noticeably increased relative to the respective C11 XMAs. For the C12 versions, M-XMA-C12 was superior to O-XMA-C12 for ligand binding (similar to DDM), but at C18, efficacy decreased dramatically (Figure 3A). Thus, for M-XMAs, C12 was optimal for receptor stability. Interestingly, the ability to bind ligand varied only slightly in the O-XMAs with different alkyl chain lengths (Figure 3B), so the optimal chain length for β2AR stability could not be determined. A similar trend was observed for MelBSt (C11, C12 and C14 O-XMAs were similar for permease stability).
Figure 3.
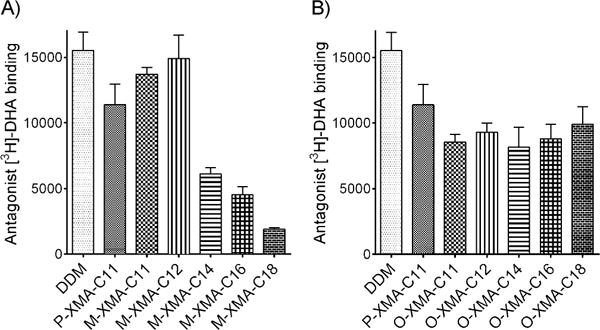
Ligand binding by β2AR solubilised in A) M-XMAs or B) O-XMAs, compared against P-XMA-C11 and DDM. The ligand-binding assay used the antagonist [3H]DHA with detergents at CMC +0.2% (w/v) (mean ±SEM, n = 3).
The different trends in detergent efficacy between M-XMAs and O-XMAs likely reflect differences in architecture, which can influence the binding mode to membrane proteins. Detergent behaviour strongly depends on the chemical nature of the head and tail groups. M-XMAs and O-XMAs are isomers both of each other and of P-XMAs, so this isomeric set enabled us to compare protein stabilisation efficacy of detergents with quite different overall architectures but with the same head and tail groups. For example, P-XMA-C11, M-XMA-C11 and O-XMA-C11 share two C11 alkyl chains and two branched dimaltoside groups, but differ in the distance between the chains and head groups that project from the xylene core. Thus, we compared detergent efficacy against inter-alkyl-chain distance without a change in the hydrophile–lipophile balance (HLB). Note that there are few comparative studies of geometrical isomers of new detergents with membrane proteins.[17]
We found a large variation in detergent efficacy toward membrane protein stabilisation, depending on the target membrane protein. Of the C11 alkyl-chain XMAs (P-XMA-C11, M-XMA-C11 and O-XMA-C11), the para isomer was the worst at stabilising LeuT and MelBSt whereas the ortho isomer was worst for β2AR activity. It is noticeable that the meta isomer (M-XMA-C11) was substantially better than (or at least comparable to) the other two isomers for all three membrane proteins. A similar trend was observed for the C12 versions. This indicates the favourable architecture of the meta isomer relative to the para and ortho isomers.
Para isomers (e.g., P-XMA-C11) have a long inter-alkyl-chain distance relative to the meta or ortho isomers. As a result, micelles formed by these isomers would have comparatively sparse interiors, thus making them less efficient at packing tightly around a target membrane protein. Because of the reduced inter-alkyl-chain distance, the meta isomer is likely to be more effective than the para isomer. The markedly higher water solubility of meta isomer relative to the para isomer could be a strong indication of favourable packing of its alkyl chains within the detergent micelles. We anticipate that this tight packing in the meta isomers decreases their mobility in the micelle, and thus produces an environment more closely resembling that of a lipid bilayer. It is interesting that a further decrease in the inter-alkyl-chain distance, corresponding to the chemical structures of the O-XMAs, did not further increase membrane protein stability. Rather, the O-XMAs were generally comparable to (or somewhat inferior to) the M-XMAs, depending on the target membrane protein, and both were better overall than the para isomers. It is hard to explain the reason for the suboptimal behaviour of the O-XMAs. Micelles formed by these isomers might have interiors that are too densely packed for protein stability. Alternatively, steric hindrance imposed by the exposed benzene ring in the ortho isomers, because of the biased substitution around the benzene ring, might induce a change in binding mode to membrane proteins. In-depth experimental study or molecular simulation is required for a full understanding of the differences between the efficacies of the isomers.
Detergent alkyl chain length strongly affects detergent efficacy in membrane protein stabilisation. In conventional detergents, optimal alkyl chain length appears to be different from one class to another. For instance, n-dodecyl alkyl chain (C12) is best for maltoside detergents (e.g., DDM), whereas n-octyl alkyl (C8) is generally best for glucoside detergents (e.g., OG).[4a] Thus, the dependency of optimal alkyl chain length on the head group can be understood in terms of detergent HLB. The maltoside head group is more hydrophilic than the glucoside head group, thus requiring a larger hydrophobic group (i.e., longer alkyl chain) to reach optimal HLB. The XMAs described here contain a branched dimaltoside head group, which is much more hydrophilic than the maltoside head group. Consequently, even the C18 XMAs (M-XMA-C18 and O-XMA-C18) are highly water-soluble. Thus, the XMA alkyl chain length for optimum HLB value is C16 to C20 (Table S1).[18] Paradoxically, the best behaviour in this study was generally observed for XMAs with C12 alkyl chains rather than longer chains. Furthermore, XMA stabilisation efficacy dramatically decreased with increasing alkyl chain length (C14 to C18) for two of the proteins assessed. These results imply additional factors governing detergent stabilisation efficacy that have not been explored in detail by other studies. We propose that this seemingly contradictory outcome from our study resulted primarily from a mismatch between detergent alkyl chain length and the dimensions of the membrane proteins. Specifically, the poor behaviours of the C16 and C18 M-XMAs and O-XMAs could be attributed to their long alkyl chains relative to the hydrophobic dimensions of a target membrane protein. This suggests there is an important interplay between detergent HLB and alkyl chain length in determining detergent behaviour toward membrane proteins, a connection that has not been fully characterised to date.
Conclusion
Evaluation of the prepared isomeric XMAs with three target membrane proteins revealed that M-XMA-C12 was the most effective among previously reported XMAs (P-XMAs) and the ten new XMAs (five M-XMAs and five O-XMAs). It was superior to (or at least comparable to) DDM in maintaining the integrity or solubility of all target membrane proteins; the previously reported P-XMA-C11 was inferior to DDM for two membrane proteins (LeuT and β2AR). Thus, the development of different detergent isomers could be effective for finding an optimal detergent for a particular membrane protein. More importantly, the study revealed a few detergent properties/features that together dictate detergent efficacy for membrane protein stabilisation (e.g., HLB, inter-alkyl chain distance and alkyl chain length). The importance of HLB was already known, but the importance of inter-alkyl chain distance and alkyl chain length as well as the interplay between these features (e.g., HLB and chain length) was recognised for the first time in this study and discussed in detail. Although we cannot completely explain how these features contribute to the effects of individual detergents, there is no doubt that these are important subjects for future study. The outcome of such studies would be essential to find useful new detergents, and to provide helpful guidelines for new amphiphile design.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grant number 2008–0061891 and 2016R1A2B2011257 to P.S.C. and K.H.C.).
Footnotes
Supporting information and the ORCID identification numbers for the authors of this article can be found under http://dx.doi.org/10.1002/cbic.201600429, including synthesis and characterisation of the new amphiphiles, and the membrane protein stability assay.
References
- 1.Woodbury NW, Becker M, Middendorf D, Parson WW. Biochemistry. 1985;24:7516–7521. doi: 10.1021/bi00347a002. [DOI] [PubMed] [Google Scholar]
- 2.a) Overington JP, Al-Lazikani B, Hopkins AL. Nat Rev Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]; b) Sanders CR, Myers JK. Annu Rev Biophys Biomol Struct. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- 3.http://blanco.biomol.uci.edu/mpstruc
- 4.a) Privé GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]; b) Chae PS, Laible PD, Gellman SH. Mol BioSyst. 2010;6:89–94. doi: 10.1039/b915162c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Proc Natl Acad Sci USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Newstead S, Ferrandon S, Iwata S. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) He Y, Wang K, Yan N. Protein Cell. 2014;5:658–672. doi: 10.1007/s13238-014-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Kang HJ, Lee C, Drew D. Int J Biochem Cell Biol. 2013;45:636–644. doi: 10.1016/j.biocel.2012.12.018. [DOI] [PubMed] [Google Scholar]; b) Zhang Q, Tao H, Hong WX. Methods. 2011;55:318–323. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Moraes I, Evans G, Sanchez-Weatherby J, Newstead S, Stewart PDS. Biochem Biophys Acta. 2014;1838:78–87. doi: 10.1016/j.bbamem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Schafmeister CE, Miercke LJW, Stroud RM. Science. 1993;262:734–738. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]; b) McGregor C-L, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Privé GG. Nat Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; c) Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Proc Natl Acad Sci USA. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Tao H, Lee SC, Moeller A, Roy RS, Siu FY, Zimmermann J, Stevens RC, Potter CS, Carragher B, Zhang Q. Nat Methods. 2013;10:759–761. doi: 10.1038/nmeth.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]; b) Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A. Nano Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]; c) Tribet C, Audebert R, Popot JL. Proc Natl Acad Sci USA. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Popot J-L, Althoff T, Bagnard D, Banères J-L, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, Crémel G, Dahmane T, Maza LM de La, Ebel C, Gabel F, Giusti F, Gohon Y, Goormaghtigh E, Guittet E, Kleinschmidt JH, et al. Annu Rev Biophys Bioeng. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- 9.a) McQuade DT, Quinn MA, Yu SM, Polans AS, Krebs MP, Gellman SH. Angew Chem Int Ed. 2000;39:758–761. [PubMed] [Google Scholar]; Angew Chem. 2000:112, 774–777. [Google Scholar]; b) Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chae PS, Cho KH, Wander MJ, Bae HE, Gellman SH, Laible PD. Biochem Biophys Acta. 2014;1838:278–286. doi: 10.1016/j.bbamem.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot JL, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Nat Methods. 2010;7:1003–1009. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Cho KH, Byrne B, Chae PS. ChemBioChem. 2013;14:452–455. doi: 10.1002/cbic.201200759. [DOI] [PubMed] [Google Scholar]; f) Cho KH, Husri M, Amin A, Gotfryd K, Lee HJ, Go J, Kim JW, Loland CJ, Guan L, Byrne B, Chae PS. Analyst. 2015;140:3157–3163. doi: 10.1039/c5an00240k. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Chae PS, Rana RR, Gotfryd K, Rasmussen SGF, Kruse AC, Cho KH, Capaldi S, Carlsson E, Kobilka B, Loland CJ, Gether U, Banerjee S, Byrne B, Lee JK, Gellman SH. Chem Commun. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Cho KH, Bae HE, Das M, Gellman SH, Chae PS. Chem Asian J. 2014;9:632–638. doi: 10.1002/asia.201301303. [DOI] [PubMed] [Google Scholar]; i) Lee SC, Bennett BC, Hong WX, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, Stout CD, Yeager MJ, Zhang Q. Proc Natl Acad Sci USA. 2013;110:E1203–E1211. doi: 10.1073/pnas.1221442110. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Chae PS, Gotfryd K, Pacyna J, Miercke LJW, Rasmussen SGF, Robbins RA, Rana RR, Loland CJ, Kobilka B, Stroud R, Byrne B, Gether U, Gellman SH. J Am Chem Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Kruse AC, Manglik A, Cho KH, Nurva S, Gether U, Guan L, Loland CJ, Byrne B, Kobilka BK, Gellman SH. Chem Eur J. 2012;18:9485–9490. doi: 10.1002/chem.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hussain H, Du Y, Scull NJ, Mortensen JS, Tarrasch J, Bae HE, Loland CJ, Byrne B, Kobilka BK, Chae PS. Chem Eur J. 2016;22:7068–7073. doi: 10.1002/chem.201600533. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sadaf A, Mortensen JS, Capaldi S, Tikhonova E, Hariharan P, de Castro Ribeiro O, Loland CJ, Guan L, Byrne B, Chae PS. Chem Sci. 2016;7:1933–1939. doi: 10.1039/c5sc02900g. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ehsan M, Du Y, Scull NJ, Tikhonova E, Tarrasch J, Mortensen JS, Loland CJ, Skiniotis G, Guan L, Byrne B, Kobilka BK, Chae PS. J Am Chem Soc. 2016;138:3789–3796. doi: 10.1021/jacs.5b13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho KH, Du Y, Scull NJ, Hariharan P, Gotfryd K, Loland CJ, Guan L, Byrne B, Kobilka BK, Chae PS. Chem Eur J. 2015;21:10008–10013. doi: 10.1002/chem.201501083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay A, London E. Anal Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 13.Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 14.a) Hart HE, Greenwald EB. Mol Immunol. 1979;16:265–267. doi: 10.1016/0161-5890(79)90065-8. [DOI] [PubMed] [Google Scholar]; b) Quick M, Javitch JA. Proc Natl Acad Sci USA. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Guan L, Nurva S, Ankeshwarapu SP. J Biol Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ethayathulla AS, Yousef MS, Amin A, Leblanc G, Kaback HR, Guan L. Nat Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Amin A, Ethayathulla AS, Guan L. J Bacteriol. 2014;196:3134–3139. doi: 10.1128/JB.01868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 17.Tao H, Fu Y, Thompson A, Lee SC, Mahoney N, Stevens RC, Zhang Q. Langmuir. 2012;28:11173–11181. doi: 10.1021/la3020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Griffin WC. J Soc Cosmet Chem. 1949;1:311–326. [Google Scholar]; b) Griffin WC. J Soc Cosmet Chem. 1954;5:249–256. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


