Abstract
Flaxseed is a rich source of the plant lignan secoisolariciresinol diglucoside (SDG) which is metabolized into mammalian lignans enterodiol (ED) and enterolactone (EL) in the digestive tract. The anti-cancer properties of these lignans have been demonstrated for various cancer types, but have not been studied for lung cancer. In this study we investigated the anti-cancer effects of EL for several non-small cell lung cancer (NSCLC) cell lines of various genetic backgrounds. EL inhibited the growth of A549, H441, and H520 lung cancer cells in concentration- and time-dependent manners. The anti-proliferative effects of EL for lung cancer cells were not due to enhanced cell death, but rather due to G1-phase cell cycle arrest. Molecular studies revealed that EL- decreased mRNA or protein expression levels of the G1-phase promoters cyclin D1, cyclin E, cyclin-dependent kinases (CDK)-2, -4, and -6, and p-cdc25A; decreased phosphorylated retinoblastoma (p-pRb) protein levels; and simultaneously increased levels of p21WAF1/CIP1, a negative regulator of the G1-phase. The results suggest that EL inhibits the growth of NSCLC cell lines by down-regulating G1-phase cyclins and CDKs, and up-regulating p21WAF1/CIP1, which leads to G1-phase cell cycle arrest. Therefore, EL may hold promise as an adjuvant treatment for lung cancer therapy.
Keywords: flaxseed, cell proliferation, cyclin D1, retinoblastoma, NSCLC, A549, H441, H520
Introduction
Lung cancer accounts for about 38% of all cancer-related deaths in the US, which is more than the combined mortality due to breast, colon, and prostate cancers [1]. Conventional lung cancer treatment modalities such as surgery, chemotherapy, and radiotherapy have shown modest success; however, these treatment options often inflict toxicity to healthy tissues [2]. Recently, bioactive compounds isolated from natural food sources have gained importance in the field of cancer treatment due to their selective action against cancer cells [3].
Lignans are naturally occurring phytoestrogens that exhibit health-promoting effects in several diseases including cancer [4]. Flaxseed has an exceptionally high lignan content and has shown chemopreventive and chemotherapeutic benefits in an observational study of breast cancer [5], human clinical trials [6–8], and animal models of breast [9–11], colon [12, 13], ovarian [14], and prostate [15] cancers. A number of in vivo studies have linked the observed anticancer effects of flaxseed to the presence of its primary lignan, secoisolariciresinol diglucoside (SDG) [10–12, 16, 17]. Upon consumption, SDG is metabolized by a consortium of intestinal bacteria into mammalian lignans, ED and EL. The anti-carcinogenic effects of flaxseed and SDG have been attributed to these mammalian lignans in mammary carcinogenesis [18]. Furthermore, in vitro studies of breast, colon, and prostate cancers demonstrate that EL has growth-inhibitory properties. Several studies suggest that the anti-proliferative potential of EL may be due to its effects on the cell cycle and induction of cell death [19–21]. The cytotoxic effects of EL appear to be selective for cancer cells as EL (25–75 μM for 24 and 48 h) inhibited the growth of LNCaP human prostate cancer cells, but EL treatment at the same concentrations and time points did not affect the viability of normal prostate epithelial cells (CRL-2221) [19].
Progression through the four sequential phases of the cell cycle (G1, S, G2, and M) is mediated by interactions between cyclins and cyclin dependent kinases (CDKs) and their inhibitors [22]. Cyclin D1 interacts and forms complexes with CDK4 and CDK6 to regulate the G1-phase, while cyclin E forms a complex with CDK2 to regulate the G1-S transition of the cell cycle [22]. These complexes phosphorylate/inactivate retinoblastoma (pRb) protein, leading to the release of bound transcription factors, such as E2F, and promote expression of genes such as cell cycle division cycle 25 (cdc25), necessary to facilitate the G1-S phase transition [23, 24]. Additionally, CDK inhibitors (CDKIs), such as p21(WAF1/CIP1) and p27(KIP1), act as negative regulators of the cell cycle by binding to and inhibiting cyclin-CDK activity [24]. Disruption in CDKI/CDK/cyclin interactions may interfere with pRb/E2F complex formation, thereby, inhibiting progression of cells from G1 to S-phase of the cell cycle [23]. EL has been shown to down-regulate E2F mRNA expression in LNCaP prostate cancer cells [25]. In addition, EL-treatment has been found to alter CDKI/CDK/cyclin mRNA expressions in MDA-MB-231 breast and in LNCaP prostate cancer cells leading to S-phase cell arrest [21, 25].
There is also evidence that consumption of lignan-rich flaxseed significantly alters the expression of cell cycle regulatory genes in mouse lung tissue [26]. In addition, flaxseed is known to protect lungs against oxidative damage and inflammation in murine models, two major factors contributing towards lung cancer development and progression [27–29]. Therefore, we hypothesized that EL, a flaxseed-derived mammalian lignan, could suppress lung cancer cell growth by inducing cell cycle arrest and cell death. To test this hypothesis, we conducted growth, cytotoxicity, and cell cycle analysis assays for three NSCLC cell lines (A549, H441, and H520) representing unique genetic backgrounds.
MATERIALS AND METHOD
Materials
Purified EL (99.2% pure) was purchased from ChromaDex, Inc. (Irvine, CA). EL was dissolved in 100% dimethysulfoxide (DMSO; Corning Cellgro®; Manassas, VA) at a stock solution concentration of 200 mM. The stock solution was later diluted in PBS (pH 7.2) to result in working solutions of EL (0–100 μM) with a maximum DMSO concentration of 0.2%. alamarBlue® was obtained from AbD Serotech (Raleigh, NC), and trypan blue was purchased from Amresco (Solon, OH). Propidium iodide (PI) was purchased from Sigma Aldrich Chemical Co. (St. Louis, MO). Annexin V-FITC/PI apoptosis detection kit was purchased from Thermo Fisher Scientific (Eugene, OR). Antibodies for cyclin D1, p-pRb, CDK2, CDK4, CDK6, p21/Cip1, p27/Kip1, t-cdc25A, and GAPDH were purchased from Cell Signaling Technology (Danvers, MA). Antibodies for cyclin E, and p-cdc25A were obtained from Santa Cruz Biotechnology Inc. (Dallas, TX). Anti-rabbit and anti-mouse HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Labs Inc. (West Grove, PA).
Cell lines and cell culture
The human NSCLC cell lines (A549, H441, and H520) and normal lung fibroblast cell line (Hs888Lu) were obtained from the American Type Culture Collection (ATCC; Manassas, VA). The NSCLC cell lines were maintained in RPMI-1640 medium (Sigma Aldrich; St. Louis, MO), with 10% (v/v) fetal bovine serum (FBS; Atlanta Biologicals; Flowery Branch, GA). The normal lung cell line was maintained in DMEM medium (Sigma Aldrich) supplemented with 10% (v/v) FBS. All cell lines were incubated at 37°C in a humidified atmosphere of 95% air and 5% carbon dioxide. Cell lines were sub-cultured by enzymatic digestion with 0.25% trypsin/1mM EDTA solution when they reached approximately 70% confluency.
alamarBlue® cell viability assay
A549, H441, and H520 cells were seeded into individual wells of 96-well plates at a density of 5×103 cells per well. Twenty-four hours later, 10% alamarBlue® (10 μl/well) was added to each well, and the plate was incubated at 37°C for 4 h protected from direct light. After incubation, absorbance readings were taken at 570 and 600 nm using a plate reader (Microplate XMark™ spectrophotometer; Bio-Rad; Hercules, CA). The cells were then washed with PBS and fresh RPMI-1640 medium supplemented with 10% FBS was added. The cells were then treated with EL (0–100 μM). Every 24 h, 10% alamarBlue® was added, and absorbency readings were taken after 4 h. Cell viability was measured by calculating the percent reduction of alamarBlue® in response to each treatment for a period of 4 days using the formula given below:
In the formula, 117,216 and 80,586 are constants representing the molar extinction coefficients of alamarBlue® at 570 and 600 nm, respectively, in the oxidized form; whereas 155,677 and 14,652 are constants representing the molar extinction coefficients of alamarBlue® at 570 and 600 nm, respectively, in the reduced form. A1 and A2 represent absorbance of wells treated with EL at 570 and 600 nm, respectively. B1 and B2 represent absorbance of wells treated with vehicle control (0.2%, DMSO) at 570 and 600 nm, respectively. The % cell viability relative to control ± standard deviation in eight replicate wells per treatment for three independent experiments was determined.
Clonogenic survival assay
A549, H441, and H520 cells were seeded into individual wells of 6-well plates for 24 hours, rinsed with PBS, followed by addition of fresh RPMI-1640 media supplemented with variable concentrations of EL (0, 10, and 100 μM). The cells were then left undisturbed and allowed to grow and form colonies for 14 days. After 14 days, the colonies were fixed with a methanol and acetic acid (3:1) solution and stained with 0.5% crystal violet. The blue-stained colonies were then counted manually. Three independent experiments were performed for each cell line with two replicates per treatment. The % cell colonies relative to control ± standard deviation was determined.
Trypan blue exclusion assay
A549, H441, and H520 cells were seeded at a density of 20 × 103 cells/well in 24-well plates. The cells were treated with EL (0, 10, 50, and 100 μM) each day for 24, 48, and 72 h. After treatment, the cells were harvested by trypsinization, stained with trypan blue, and the number of live and dead (blue staining) cells were counted on a hemacytometer under a light microscope. The average live/dead cell count from three independent experiments was determined.
In addition, Hs888Lu normal lung fibroblast cells were seeded at a density of 15 × 103 cells/well in 6-well plates. The cells were treated with EL (0, 10, and 100 μM) each day for 24 and 48 h. After treatment, the cells were harvested by trypsinization, stained with trypan blue, and the number of live cells were counted on a hemacytometer under a light microscope. The % cell viability relative to control ± standard deviation for two independent experiments was determined.
Apoptosis detection by Annexin V-fluorescein isothiocyanate/Propidium iodide (annexin V-FITC/PI) staining
A549 cells were seeded at a density of 2 × 105 cells/well in 60 mm dishes. Twenty-four hours later, the cells were treated with EL (0, 10, 50, and 100 μM) each day for 24, 48, and 72 h. After treatment, the cells were harvested by trypsinization, washed with PBS, and re-suspended in 1X binding buffer. The cells were stained with 5 μl of FITC-Annexin V and 5 μl PI (100 μg/ml) for 10 min at room temperature. Finally, 400 μl of 1× binding buffer was added to the cells and data for live and apoptotic percentage of 10,000 cells were collected using a FACScan flow cytometer. The viable cells are negative for both annexin-V and PI staining; apoptotic cells are positive for annexin-V staining while negative for PI staining, and necrotic cells are positive for both annexin-V and PI staining. Three independent experiments were performed for each treatment group and time-point.
Cell cycle analysis
The cell cycle distribution was assessed by staining the cellular DNA content with propidium iodide (PI). A549, H441, and H520 cells were synchronized by incubating them in serum-deprived medium for 24 h followed by treatment with either the vehicle control (0.2%, DMSO) or EL (10 and 100 μM). The cells were then harvested by trypsinization, centrifuged at 200 rpm for 5 min, washed with PBS (pH 7.2), and centrifuged again at 200 rpm for 5 min. The resultant cell pellet was then re-suspended in cold 70% ethanol and fixed overnight at 4°C. Next day, the cells were collected by centrifugation at 200 rpm for 5 min, and the resultant cell pellet was re-suspended in 500 μl of PBS (pH 7.2) containing 100 μg/ml RNase and PI (40 μg/ml) for 30 min at 37°C protected from light. After staining with PI, 10,000 cells per sample were run through a FACScan flow cytometer equipped with a 488 nm argon laser, and the data were analyzed using FlowJo software. The average percentage of cells in each phase of the cell cycle per treatment for three independent experiments was determined.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from EL-treated (100 μM; 24 h) A549, H441, and H520 cells (1 X 106 cells) using the Fisher SurePrep Kit (Waltham, MA) as per the manufacturer’s instructions. The 260/280 and 260/230 OD ratios were measured using NanoDrop1000 spectrophotometer to assess the purity of RNA samples with respect to protein contamination and residual organic solvent, respectively. RNA samples with 260/280 and 260/230 OD ratios between 1.8 and 2 were used for RT-qPCR. Fifty nanograms of RNA were reverse transcribed into cDNA using the qScript cDNA synthesis kit (Quanta Biosciences; Gaithersburg, MD). Primers against cyclin D1 (5′-TGCGCTGCTACCGTTGACT-3′ and 5′-AGCGATGTGAATATTTCCAAACC-3′), CDK2 (5′-GGTCCTCCACCGAGACCTTAA-3′ and 5′-CAGGGACTCCAAAAGCTCTGG-3′), CDK4 (5′-CAGTGTACAAGGCCCGTGATC-3′ and 5′-ACGAACTGTGCTGATGGGAAG-3′), cdc25A (5′-CCCCAAAGGAACCATTGAGA-3′ and 5′-CTGATGTTTCCCAGCAACTG-3′), p21WAF1/CIP1 (5′-GGACAGCAGAGGAAGACCATGT-3′ and 5′-GCCGTTTTCGACCCTGAGA-3′), p27KIP1 (5′-CTATCTGCTGCGCGGTTACC-3′ and 5′-CCTGACAAGCCACGCAGTAG-3′), and 18S rRNA (5′-GGCCCTGTAATTGGAATGAGTC-3′ and 5′-CCAAGATCCAACTACGAGCTT-3′) and were designed using Primer Express software (version 2.0, Applied Biosystems), and synthesized by Integrated DNA Technologies (Coralville, IA). Steady-state mRNA levels for the cell cycle-related genes were evaluated by real-time PCR using PerfeCTa SYBR Green FastMix (Quanta Biosciences). The cycling parameters were: 95°C for 10 min followed by 40 cycles at 95°C for 30 s and 60°C for 1 min, and a dissociation program that included 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s ramping up at 0.2 °C/s. One distinct peak was observed for each primer set, suggesting target specificity. Duplicate wells were run for each experiment and the experiment was performed in triplicate. The relative change in gene expression was calculated using 2−ΔΔCt method using housekeeping gene 18S rRNA as internal control.
Western blotting analysis
A549, H441, and H520 cells (1×106) were treated with EL (100 μM) for variable times. Following treatment (0–24 h), the cells were collected by trypsinization, washed with PBS, and lysed using an SDS lysis buffer (Cell Signaling Technologies; Danvers, MA) containing protease and phosphatase inhibitors (Roche; Indianapolis, IN). The cell pellets were briefly sonicated to dissociate cell membranes. Thirty micrograms of total protein isolated from these cells were electrophoresed on 7.5% SDS-polyacrylamide gels at 100 V for 1 h. Proteins were then transferred to PVDF membranes at 100 V for 70 min at 4°C. The blots were then probed overnight at 4 °C with primary antibodies (1:1000) for cyclin D1, cyclin E, p-pRb, p-cdc25A, t-cdc25A, CDK2, CDK4, CDK6, p21WAF1/CIP1, p27KIP1, and GAPDH. The next day, the blots were rinsed with 1X TBS-tween (0.1%) and probed with anti-rabbit and anti-mouse HRP-conjugated secondary antibodies (1:5000) for 1 h at room temperature. The western blots were analyzed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific; Rockford, IL) and the images were captured using the MultiImage™ Light Cabinet (Alpha Innotech; San Leandro, CA). Target protein expression levels were normalized to GAPDH expression levels except for p-cdc25A protein expression levels, which were normalized to t-cdc25A protein levels. Western blotting was performed in triplicate. The densitometry results were obtained using ImageJ software.
Statistical analysis
Data are presented as means ± standard deviation for at least 3 independent experiments. The statistical significance of difference between the control and treatment groups was determined by paired t-test or two-way ANOVA. p ≤ 0.05 were considered statistically significant.
RESULTS
EL inhibits short-term proliferation of lung cancer cells in a concentration- and time-dependent manner but does not affect proliferation of normal lung cells
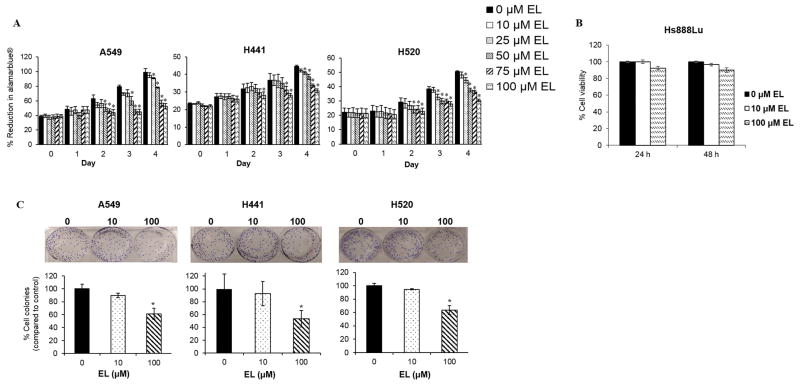
We investigated the anti-proliferative effects of EL for three human NSCLC cells: A549, H441, and H520. The short-term growth-inhibitory effects of EL on NSCLC cells were examined using an alamarBlue® cell viability assay. We found that EL-treatment led to a concentration- and time-dependent inhibition of proliferation of all lung cancer cell lines tested. We observed that EL ≥50 μM significantly (p≤0.05) inhibited the growth of A549 and H520 cells by day 2, while, similar growth inhibition was observed in H441 cells treated with 100 μM EL. By day 4, A549, H441, and H520 cells treated with EL (100 μM) showed a significant (p≤0.05) growth inhibition by 47%, 31%, and 30%, respectively (Fig. 1A).
Figure 1.
EL inhibits growth of lung cancer cell lines. A. The short-term growth of A549, H441, and H520 was measured by the percentage of alamarBlue® reduction in the presence of EL (0–100 μM) over a period of 4 days. The data represent the average ± standard deviation of eight replicate wells for three independent experiments for every cell line. In A549 and H520 cells, EL-treatment (≥50 μM) and in H441 cells, EL (100 μM) resulted in a significant inhibition of growth in a time-dependent manner. B. Cell viability of Hs888Lu cells was examined by a trypan blue exclusion assay following EL (0, 10, and 100 μM) treatment for 24 and 48 h. The bar graph represents the average cell count for two independent experiments. The error bars represent standard deviation from two independent experiments. C. The long-term growth of lung cancer cells in the presence of EL (10 and 100 μM) was determined by a clonogenic survival assay. The fraction of cells surviving EL treatment was examined after 14 days. The image shown is representative of one typical experiment. The data shown in the bar graph represent the average ± standard deviation for the fraction of surviving cells for three independent experiments for each cell line. (* designates p ≤ 0.05 as compared to control).
In addition, we investigated if EL affected proliferation of Hs888Lu, normal lung fibroblast cells. Hs888Lu cells were treated EL (0, 10, and 100 μM) for 24, and 48 h and cell viability was determined using trypan blue exclusion assay. EL-treatment (10 μM) showed no inhibition in cell proliferation after 24 h and a slight 3% inhibition of cell viability was observed after 48 h (Fig. 1B). Similarly, EL-treatment (100 μM) showed a minimal 8 and 7% inhibition of cell proliferation after 24 and 48 h, respectively (Fig. 1B). This indicates that EL has little or no effect on the proliferation of normal lung cells.
EL inhibits long-term proliferation of lung cancer cells in a concentration-dependent manner
Next we examined the long-term growth-inhibitory effects of EL on these cells using a clonogenic survival assay. Clonogenic survival assays are used to determine the ability of single cells to survive and form colonies after exposure to radiation, a chemotherapeutic agent or a combination of the two. After 14 days, EL treatment (100 μM) significantly reduced the size and the number of A549, H441, and H520 cell colonies by 61%, 53%, and 63% (p≤0.05), respectively, compared to untreated cells (Fig. 1B). As with the short-term proliferation studies, a low concentration of EL (10 μM) failed to influence proliferation.
EL-mediated inhibition of lung cancer cell proliferation is not caused by enhanced cell death
We carried out a trypan blue exclusion assay to determine whether inhibition of lung cancer cell proliferation was a result of enhanced cell death, as indicated by increased cell membrane permeability for trypan blue. A549, H441, and H520 cells were treated EL (0, 10, 50, and 100 μM) for 24, 48, and 72 h. These concentrations were chosen to show a range of effects (low-moderate-high) for EL-mediated lung cancer cell death. We did not observe significant cell death, as measured by the number of trypan blue stained cells, for EL-treated cells at any time point or concentration (Fig. 2A). A slight, but statistically insignificant increase in H520 cell death was noted with increasing concentrations of EL after 48 h.
Figure 2.
EL exposure results in G1-phase cell cycle arrest, but not cell death. A. Cell viability of A549, H441, and H520 cells was examined by a trypan blue exclusion assay following EL (0–100 μM) treatment for 24, 48, and 72 h. The bar graph represents the average total cell count, inclusive of live (black) and dead (white) cells, for three independent experiments for each cell line. The error bars represent standard deviation of the total number of dead cells from three independent experiments for each cell line. B. An Annexin V-FITC/PI apoptosis assay was performed to further assess the potential cytotoxic effects of EL. A549 cells were treated with EL (0, 10, 50, and 100 μM) for 24, 48, and 72 h. The treated cells were then stained with Annexin V/PI and flow cytometric analysis was performed. The representative data from three independent experiments are shown in dot plots. C. EL-induced G1-phase cell cycle arrest in lung cancer cells was examined by flow cytometry. The histograms show the cell cycle distribution for A549, H441, and H520 treated with the vehicle control or EL (10 and 100 μM) for 48 h. The data represent the average percentage of cells in each phase of the cell cycle for three independent experiments for each cell line.
To confirm the results from trypan blue exclusion assay we carried out an Annexin V-FITC/PI apoptosis assay. A549 cells were treated EL (0, 10, 50, and 100 μM) for 24, 48, and 72 h. Compared to the control, we did not observe any increase in cell death in A549 cells, as measured by the percentage of Annexin V positive/PI positive stained cells, in response to EL-treatment at any time point or concentration (Fig. 2B).
EL suppresses lung cancer cell proliferation through G1-phase cell cycle arrest
An alternative mechanism for reduced cell proliferation is through arrest of the cell cycle. Therefore, we investigated the effect of EL on cell cycle progression in NSCLC cells. A549, H441, and H520 cells that were synchronized by serum starvation for 24 h were treated with EL (10 and 100 μM) for 48 h. Cell cycle analysis was carried out using PI staining. EL-treatment led to an increase in the percentage of G1-phase cells from 64% to 76% for A549 (p≤0.05), from 35% to 46% for H441 (p≤0.05), and from 45% to 65% for H520 (p≤0.05) cells (Fig. 2C). Simultaneously, the percentage of the cells in the S phase decreased.
EL modulates G1-phase cell cycle regulatory genes in lung cancer cells
To explain how EL might arrest lung cancer cells in the G1-phase of the cell cycle, we used RT-qPCR to measure mRNA transcripts levels for cell cycle regulatory genes. A549, H441, and H520 lung cancer cells treated with 100 μM EL for 24 h, and the mRNA expression values for cyclin D1, CDK2 and CDK4, cdc25A, p21WAF1/CIP1 and p27KIP1 genes were determined. We observed significant changes in mRNA expression levels of CDK2 (p≤0.05) and p21WAF1/CIP1 (p≤0.05) in A549 cells; cyclin D1 (p≤0.05), CDK4 (p≤0.05), cdc25A (p≤0.05) in H441 cells; and cyclin D1 (p<0.05), CDK2 (p≤0.05), CDK4 (p≤0.05), cdc25A (p≤0.05) and p21WAF1/CIP1 (p≤0.05) in H520 cells. mRNA expression of p27KIP1 showed differential responses in the three cell lines with a slight, but not significant increase in A549 cells, and a significant decrease in H441(p<0.05), and H520 (p<0.05) cells (Fig. 3).
Figure 3.
EL regulates the expression of G1-phase related genes. A549, H441, and H520 cells were treated with EL (100 μM) for 24 h. Relative mRNA expression levels for cyclin D1, CDK2, CDK4, cdc25A, p21WAF1/CIP1, and p27KIP1 were determined by quantitative PCR. The relative mRNA levels for all genes were normalized to 18S rRNA. The data shown in the bar graphs represent the average ± standard deviation of relative mRNA expression for each gene for three independent experiments for each cell line. (* designates p ≤ 0.05 as compared to control).
EL alters the expression of G1-phase cell cycle regulatory proteins in lung cancer cells
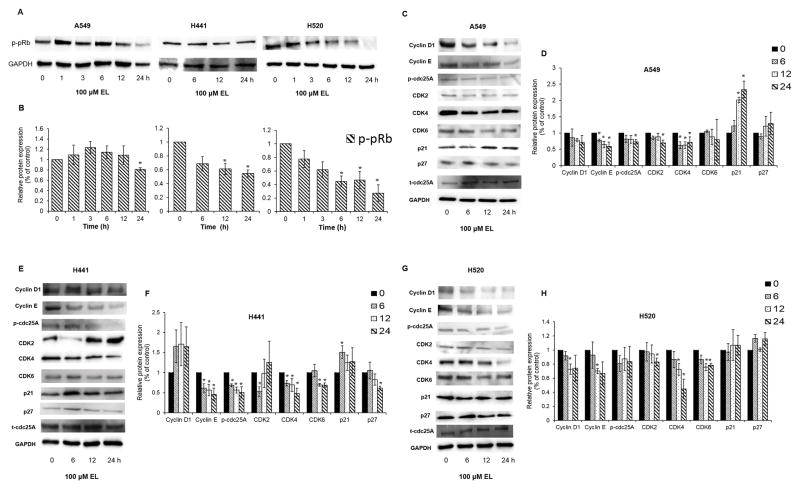
Next, we used western blotting to determine if EL-mediated changes in mRNA transcripts of G1-phase regulating cyclins, CDKs, and CDKI correlated with changes at their protein levels. EL treatment (100 μM, 0–24 h) led to reduced expression of p-pRb protein in the three lung cancer cell lines, with the most notable decreases in H441 and H520 cells (Fig. 4A and 4B). Further, EL-treatment affected the expression levels of cell cycle-promoting proteins, such as cyclins, CDKs, and p-cdc25A proteins and cell cycle inhibitory protein, p21WAF1/CIP1 in lung cancer cell lines. In A549, EL treatment led to a time-dependent decrease in cyclin D1, cyclin E1 (p≤0.05, 6, 12 and 24 h), p-cdc25A (p≤0.05, 24 h), CDK2 (p≤0.05, 24 h), CDK4 (p≤0.05, 6, 12, and 24 h), CDK6, and an increase in p21WAF1/CIP1 (p≤0.05, 12, and 24 h), and p27KIP1 protein levels (Fig. 4C and 4D). Similarly in H441, EL treatment led to a time-dependent decrease in cyclin E (p≤0.05, 6 12, and 24 h), p-cdc25A (p≤0.05, 6, 12, 24 h), CDK2 (p≤0.05, 6 h), CDK4 (p≤0.05, 6,12, and 24 h), CDK6 (p≤0.05, 12 and 24 h), and a significant increase in p21WAF1/CIP1 (p≤0.05, 6 h), and decrease in p27KIP1 (p≤0.05, 24 h) protein levels (Fig. 4E and 4F). While in H520, EL treatment led to a time-dependent decrease in cyclin D1 (p≤0.05, 12 h), cyclin E1 (p≤0.05, 12 h), p-cdc25A, CDK2 (p≤0.05, 24 h), CDK4 (p≤0.05, 12 and 24 h), and CDK6 (p≤0.05, 12 and 24 h) (Fig. 4G and 4H).
Figure 4.
EL induces cell cycle arrest by modulating the expression of G1-phase related proteins. A. A549 and H520 cells were treated with EL (100 μM) for 0, 1, 3, 6, 12, and 24 h. H441 cells were treated with EL (100 μM) for 0, 6, 12, and 24 h. The cells were lysed to collect proteins for western blotting. 30 μg protein were loaded on the gels, and blots were probed with p-pRb. B. Quantification of western blotting results was done using ImageJ. The data shown are the average of three independent experiments and are represented as mean ± S.E.M. A549 (C), H441 (E), and H520 (G) cells were treated with EL (100 μM) for 0, 6, 12, and 24 h. The cells were then lysed to collect proteins for western blotting. 30 μg protein were loaded on the gels, and blots were probed with cyclin D1, cyclin E, p-cdc25A, CDK2, CDK4, CDK6, p21WAF1/CIP1, and p27KIP1 G1-phase cell cycle regulatory proteins. Total cdc25A (t-cdc25A) protein levels were used to normalize phosphorylated cdc25A (p-cdc25A) protein levels. Quantification of western blotting results for A549 (D), H441 (F), and H520 (H) was done using ImageJ. The data shown are the average of three independent experiments and are represented as mean ± S.E.M. GAPDH was used as a loading control for the remaining western blots. (* designates p ≤ 0.05 as compared to control).
DISCUSSION
In this study, we evaluated the anti-proliferative effects of EL on NSCLC cells. The selected cell lines harbor different somatic mutations that influence their growth properties and ultimately their response to cancer therapies (Table 1). All the cell lines in this study carry wild-type EGFR. A549 and H441 cells are KRAS mutant, while H520 cells are wild-type KRAS. Further, A549 cells express wild-type p53, while H441 and H520 cells exhibit mutant p53. We found EL to have similar growth-inhibitory effects in all three lung cancer cell lines, which suggests that the growth-inhibitory properties of EL for these cells are independent of their KRAS and p53 gene status. In contrast to the significant growth-inhibitory effects of EL observed in lung cancer cells, no obvious growth-inhibitory effects of EL were detected in normal lung fibroblast cells.
Table 1.
Information on Non-Small Cell Lung Cancer (NSCLC) Cell Lines
| Cell lines | Histologic subtype | Genetic background | ||
|---|---|---|---|---|
| TP53 | KRAS | EGFR | ||
| A549 | Adenocarcinoma | WT | MT | WT |
| H441 | Adenocarcinoma | MT | MT | WT |
| H520 | Squamous cell carcinoma | MT | WT | WT |
WT - Wild type
MT- Mutant type
Interestingly, our findings suggest that EL (≥50 μM, 48h) mediates suppression of NSCLC cell growth. Similarly, previous studies have also shown that high concentrations of EL are needed to inhibit in vitro growth of breast, colon, and prostate cancers. Xiong et al. (2015) demonstrated that EL with an IC50 of 261.9 ± 10.5 μM inhibited the growth of MDA-MB-231 breast cancer cells after 48 h of treatment [21]. Similarly, Danbara et al. (2005) and Chen et al. (2007) reported that treatment of COLO 201 colon cancer cells and LNCaP prostate cancer cells with 118.4 and 75 μM EL, respectively, for 72 h significantly inhibited their proliferation by 50% [20, 30]. Furthermore, our results suggest that the in vitro inhibition of lung cancer cell growth is not due to induction of cell death. This result is in contrast to in vitro studies of colon and prostate cancers that have shown that EL treatment results in a concentration- and time-dependent increase in apoptosis. Danbara et al. (2005) demonstrated that EL treatment (118.4 μM, 72 h) in COLO 201 colon cancer cells caused an increase in the percentage of cells in the sub-G1 phase of the cell cycle which is indicative of apoptosis [20]. They further demonstrated that EL increased the expression of the cleaved form of caspase-3 protein, a marker of apoptosis, and simultaneously decreased the expression of Bcl-2 and PCNA proteins, markers of cell proliferation. Similarly, Chen et al. (2007) reported that exposure of LNCaP prostate cancer cells to increasing concentrations of EL (0 –100 μM, 72 h) led to a gradual increase in sub-G0/G1 DNA content (apoptotic cells) from 2.6% to 56.3% [30]. Further, they showed that treatment of LNCaP cells with EL resulted in an increase in expression of cleaved caspase-3 and PARP proteins that initiate apoptosis. Although we observed slightly more dead cells in H520 cell line with 50 and 100 μM EL treatment (48 h, but not 72 h), the number of dead cells did not make up for the difference in cell count observed between the control and EL treatment, suggesting a cytostatic rather than cytotoxic effect of EL. It is possible that the cytotoxic effects of EL are dependent upon cancer cell type.
We did observe that EL treatment arrested lung cancer cells in the G1-phase of the cell cycle. Our results are in partial agreement with the study by McCann et al. (2013) who observed G1 and S phase arrest along with an increase in percentage of cells in the sub-G0/G1 phase in several prostate cancer cell lines after treatment with EL (20 μM) for 48 h [31]. However, differing results have been reported with regards to the stage-specific cell cycle arrest caused by EL. Studies by Xiong et al. (2015) and McCann et al. (2008) have shown that the treatment of MDA-MB-231 breast cancer cells with increasing concentrations of EL (50, 100, and 200 μM) and LNCaP prostate cancer cells with 60 μM EL for 48 and 72 h, respectively, induced accumulation of cells in S-phase [21, 32]. These findings may suggest that cell cycle arrest caused by EL is cell-type specific and concentration- and time-dependent.
Progression of cells through the G1, S, G2, and M cell cycle phases is positively regulated by a family of proteins called cyclins and cyclin dependent kinases (CDK), and negatively regulated by CDK inhibitors (CDKI). Cyclin D1 interacts with CDK4 and CDK6 to drive progression of cells through G1 [23, 24]. Therefore, it is not surprising that we noticed down-regulation of cyclin D1 and CDK4 mRNA, which would explain arrest of lung cancer cells in the G1-phase. Further, the association of cyclin E with CDK2 is active at the G1-S transition and guides entry of cells into the S phase. We found that EL-treatment down-regulated CDK2 mRNA expression. On the other hand, CDKIs such as p21WAF1/CIP1 and p27KIP1 stop cell cycle progression to S-phase [23, 24]. They inhibit kinase activity of cyclins-CDKs complexes by binding to CDKs, and prevent their association with cyclins. We found that EL-treatment resulted in elevated levels of p21WAF1/CIP1 mRNA in lung cancer cell lines. In addition, cdc25A phosphatases, members of the tyrosine phosphatase family, catalyze dephosphorylation and activation of CDKs which play a role in G1-S phase transition [24]. We found EL reduced cdc25A mRNA expression in lung cancer cell lines. Previous research by Xiong et al. (2015) found that EL treatment (0–200 μM, 48 h) in MBA-MD-231 breast cancer cell line significantly decreased mRNA levels of cyclin E1, cyclin A2, cyclin B1, and cyclin B2 which play a role in S and G2/M phases in the cell cycle [21]. They observed no significant change in mRNA levels of cyclin D1, CDK2, and CDK4.
The tumor suppressor protein pRb plays a critical role in the G0/G1 and G1-S phase transitions. In quiescent cells, pRb exists in an activated/hypo-phosphorylated state, and sequesters members of the E2F gene family of transcription factors, which suppress cell-cycle progression [23]. Phosphorylation of pRb leads to the disruption of the pRb-E2F transcription complex, and the release of active E2F, which promotes cell-cycle progression [23]. We observed that EL-treatment caused a significant reduction in phosphorylated p-pRb protein in H520 and H441 cell lines. Phosphorylation of pRb is normally initiated by the cyclin D1-CDK4/6 complex. As expected, we found EL treatment resulted in reduced expression of CDK4 and CDK6 protein in lung cancer cell lines. At the time of G1-S phase transition cyclin E is actively synthesized and accumulated to activate CDK2, leading to further pRb phosphorylation resulting in E2F activation and subsequent entry of the cells into S-phase [23]. We observed that EL-treatment led to a significant reduction in cyclin E protein expression in lung cancer cell lines, as was CDK2 at certain time points.
The findings from this study suggest that exposure to high concentrations of EL (≥ 50 μM) is required to inhibit in vitro growth of lung cancer cells. Achieving such a high plasma concentration of EL from flaxseed consumption may prove difficult given that 25–50 g of flaxseed per day, which is equivalent to consumption of 500 mg/day, results in no more than 2 μM EL in the plasma [33, 34]. Consumption of purified SDG may be more effective at achieving high plasma EL concentrations than consumption of flaxseed. Setchell et al. (2013) showed that increasing SDG dose (25–173 mg/day) concomitantly increased plasma EL levels [35]. This suggests that consumption of purified SDG may be needed to achieve EL concentrations capable of lung cancer cell growth inhibition. It is not clear what the tolerable dose of SDG is for human consumption or what plasma EL levels are achievable. Further, it is likely that a clinical application of EL would involve combination with other anti-cancer agents. For example, EL combined with tamoxifen led to enhanced inhibition of breast cancer cell adhesion and invasion [36].
CONCLUSIONS
In summary, EL inhibits cell proliferation by inducing G1-phase cell cycle arrest. EL treatment results in down-regulation of cell cycle promoting cyclins and CDKs and up-regulation of their inhibitors, leading to decreased phosphorylation of pRb protein and thereby, decreased expression of genes that promote G1 to S-phase transition (Fig. 5). Development of cancer therapies that are effective against KRAS and p53 mutant cancers have proven clinically challenging. Therefore, identification of new non-toxic natural chemotherapeutic agents that target downstream effectors of mutant-KRAS and p53 signaling appears to be a novel treatment strategy for NSCLC.
Figure 5.
Proposed molecular mechanism of EL-induced G1-phase cell cycle arrest. EL suppresses expression of cyclin D1-CDK4/6 and cyclinE-CDK2 proteins by up-regulating expression of the cell cycle inhibitor p21WAF1/CIP1. EL-mediated down-regulation of cyclin-CDKs complexes suppresses phosphorylation of pRb. Hypo-phosphorylated pRb binds to transcription factor E2F, and represses its transcriptional activity and the activation of genes required for the G1-S phase transition, such as cdc25A. This leads to G1-phase cell cycle arrest and growth inhibition.
Acknowledgments
We would also like to thank Dr. Devendra Chikara for editing the paper.
FUNDING
This work was supported by grants from National Science Foundation (No. HRD-0811239) and National Institute of Health (No. P30 GM103332-01) from the National Center for Research Resources.
References
- 1.Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011;32:605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun S, Schiller JH, Spinola M, Minna JD. New molecularly targeted therapies for lung cancer. J Clin Invest. 2007;117:2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Khor TO, Shu L, Su ZY, Fuentes F, et al. Plants vs. cancer: a review on natural phytochemicals in preventing and treating cancers and their druggability. Anticancer Agents Med Chem. 2012;12:1281–1305. doi: 10.2174/187152012803833026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toure A, Xu XM. Flaxseed Lignans: Source, Biosynthesis, Metabolism, Antioxidant Activity, Bio-Active Components, and Health Benefits. Compr Rev in Food Sci and Food Saf. 2010;9:261–269. doi: 10.1111/j.1541-4337.2009.00105.x. [DOI] [PubMed] [Google Scholar]
- 5.Lowcock EC, Cotterchio M, Boucher BA. Consumption of flaxseed, a rich source of lignans, is associated with reduced breast cancer risk. Cancer Causes Control. 2013;24:813–816. doi: 10.1007/s10552-013-0155-7. [DOI] [PubMed] [Google Scholar]
- 6.Demark-Wahnefried W, Polascik TJ, George SL, Switzer BR, Madden JF, Hars V, Albala DM, et al. Flaxseed Supplementation (Not Dietary Fat Restriction) Reduces Prostate Cancer Proliferation Rates on Men Presurgery. Cancer Epidemiol Biomarkers & Prev. 2008;17:3577–3587. doi: 10.1158/1055-9965.EPI-08-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demark-Wahnefried W, Robertson CN, Walther PJ, Polascik TJ, Paulson DF, et al. Pilot study to explore effects of low-fat, flaxseed-supplemented diet on proliferation of benign prostatic epithelium and prostate-specific antigen. Urology. 2004;63:900–904. doi: 10.1016/j.urology.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Thompson LU, Chen JM, Li T, Strasser-Weippl K, Goss PE. Dietary flaxseed alters tumor biological markers in postmenopausal breast cancer. Clin Cancer Res. 2005;11:3828–3835. doi: 10.1158/1078-0432.CCR-04-2326. [DOI] [PubMed] [Google Scholar]
- 9.Serraino M, Thompson LU. The Effect of Flaxseed Supplementation on the Initiation and Promotional Stages of Mammary Tumorigenesis. Nutr Cancer. 1992;17:153–159. doi: 10.1080/01635589209514182. [DOI] [PubMed] [Google Scholar]
- 10.Chen JM, Tan KP, Ward WE, Thompson LU. Exposure to flaxseed or its purified lignan during suckling inhibits chemically induced rat mammary tumorigenesis. Exp Biol Med. 2003;228:951–958. doi: 10.1177/153537020322800811. [DOI] [PubMed] [Google Scholar]
- 11.Rickard SE, Yuan YV, Chen J, Thompson LU. Effect of flaxseed and its lignan precursor on MNU-induced mammary tumorigenesis. Faseb J. 1998;12:A829–A829. [Google Scholar]
- 12.Jenab M, Thompson LU. The influence of flaxseed and lignans on colon carcinogenesis and beta-glucuronidase activity. Carcinogenesis. 1996;17:1343–1348. doi: 10.1093/carcin/17.6.1343. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez-Salazar M, Guevara-Gonzalez R, Cruz-Hernandez A, Guevara-Olvera L, Bello-Perez LA, et al. Flaxseed (Linum usitatissimum L.) and Its Total Non-digestible Fraction Influence the Expression of Genes Involved in Azoxymethane-induced Colon Cancer in Rats. Plant Foods for Hum Nutr. 2013;68:259–267. doi: 10.1007/s11130-013-0372-y. [DOI] [PubMed] [Google Scholar]
- 14.Ansenberger K, Richards C, Zhuge Y, Barua A, Bahr JM, et al. Decreased severity of ovarian cancer and increased survival in hens fed a flaxseed-enriched diet for 1 year. Gynecol Oncol. 2010;117:341–347. doi: 10.1016/j.ygyno.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X, Gingrich JR, Bao WJ, Li J, Haroon ZA, et al. Effect of flaxseed supplementation on prostatic carcinoma in transgenic mice. Urology. 2002;60:919–924. doi: 10.1016/s0090-4295(02)01863-0. [DOI] [PubMed] [Google Scholar]
- 16.Thompson LU, Seidl MM, Rickard SE, Orcheson LJ, Fong HHS. Antitumorigenic effect of a mammalian lignan precursor from flaxseed. Nutr Cancer. 1996;26:159–165. doi: 10.1080/01635589609514472. [DOI] [PubMed] [Google Scholar]
- 17.Thompson LU, Rickard SE, Orcheson LJ, Seidl MM. Flaxseed and its lignan and oil components reduce mammary tumor growth at a late stage of carcinogenesis. Carcinogenesis. 1996;17:1373–1376. doi: 10.1093/carcin/17.6.1373. [DOI] [PubMed] [Google Scholar]
- 18.Serraino M, Thompson LU. The Effect of Flaxseed Supplementation on Early Risk Markers for Mammary Carcinogenesis. Cancer Lett. 1991;60:135–142. doi: 10.1016/0304-3835(91)90220-c. [DOI] [PubMed] [Google Scholar]
- 19.Chen LH, Fang J, Li H, Demark-Wahnefried W, Lin X. Enterolactone induces apoptosis in human prostate carcinoma LNCaP cells via a mitochondrial-mediated, caspase-dependent pathway. Mol Cancer Ther. 2007;6:2581–2590. doi: 10.1158/1535-7163.MCT-07-0220. [DOI] [PubMed] [Google Scholar]
- 20.Danbara N, Yuri T, Tsujita-Kyutoku M, Tsukamoto R, Uehara N, et al. Enterolactone induces apoptosis and inhibits growth of Colo 201 human colon cancer cells both in vitro and in vivo. Anticancer Res. 2005;25:2269–2276. [PubMed] [Google Scholar]
- 21.Xiong XY, Hu XJ, Li Y, Liu CM. Inhibitory Effects of Enterolactone on Growth and Metastasis in Human Breast Cancer. Nutr Cancer. 2015;67:1324–1332. doi: 10.1080/01635581.2015.1082113. [DOI] [PubMed] [Google Scholar]
- 22.Gerard C, Goldbeter A. The balance between cell cycle arrest and cell proliferation: control by the extracellular matrix and by contact inhibition. Interface Focus. 2014;4 doi: 10.1098/rsfs.2013.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 24.Dubravka D, Scott DW. Regulation of the G1 phase of the mammalian cell cycle. Cell Research. 2000;10:1–16. doi: 10.1038/sj.cr.7290031. [DOI] [PubMed] [Google Scholar]
- 25.McCann MJ, Gill CI, Linton T, Berrar D, McGlynn H, et al. Enterolactone restricts the proliferation of the LNCaP human prostate cancer cell line in vitro. Mol Nutr Food Res. 2008;52:567–580. doi: 10.1002/mnfr.200700052. [DOI] [PubMed] [Google Scholar]
- 26.Dukes F, Kanterakis S, Lee J, Pietrofesa R, Andersen ES, et al. Gene expression profiling of flaxseed in mouse lung tissues-modulation of toxicologically relevant genes. BMC Complement Altern Med. 2012;12:47–57. doi: 10.1186/1472-6882-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinniry P, Amrani Y, Vachani A, Solomides CC, Arguiri E, et al. Dietary flaxseed supplementation ameliorates inflammation and oxidative tissue damage in experimental models of acute lung injury in mice. J Nutr. 2006;136:1545–1551. doi: 10.1093/jn/136.6.1545. [DOI] [PubMed] [Google Scholar]
- 28.Lee JC, Bhora F, Sun J, Cheng G, Arguiri E, et al. Dietary flaxseed enhances antioxidant defenses and is protective in a mouse model of lung ischemia-reperfusion injury. Am J Physiol Lung Cell Mol Physiol. 2008;294:L255–L265. doi: 10.1152/ajplung.00138.2007. [DOI] [PubMed] [Google Scholar]
- 29.Lee JC, Krochak R, Blouin A, Kanterakis S, Chatterjee S, et al. Dietary flaxseed prevents radiation-induced oxidative lung damage, inflammation and fibrosis in a mouse model of thoracic radiation injury. Cancer Biol Ther. 2009;8:47–53. doi: 10.4161/cbt.8.1.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen LH, Fang J, Li HX, Demark-Wahnefried W, Lin X. Enterolactone induces apoptosis in human prostate carcinoma LNCaP cells via a mitochondrial-mediated, caspase-dependent pathway. Mol Cancer Ther. 2007;6:2581–2590. doi: 10.1158/1535-7163.MCT-07-0220. [DOI] [PubMed] [Google Scholar]
- 31.McCann MJ, Rowland IR, Roy NC. Anti-proliferative effects of physiological concentrations of enterolactone in models of prostate tumourigenesis. Mol Nutr Food Res. 2013;57:212–224. doi: 10.1002/mnfr.201200362. [DOI] [PubMed] [Google Scholar]
- 32.McCann MJ, Gill CIR, Linton T, Berrar D, McGlynn H, et al. Enterolactone restricts the proliferation of the LNCaP human prostate cancer cell line in vitro. Mol Nutr Food Res. 2008;52:567–580. doi: 10.1002/mnfr.200700052. [DOI] [PubMed] [Google Scholar]
- 33.Saarinen NM, Smeds AI, Penalvo JL, Nurmi T, Adlercreutz H, et al. Flaxseed ingestion alters ratio of enterolactone enantiomers in human serum. J Nutr Metab. 2010 doi: 10.1155/2010/403076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallund J, Ravn-Haren G, Bugel S, Tholstrup T, Tetens I. A lignan complex isolated from flaxseed does not affect plasma lipid concentrations or antioxidant capacity in healthy postmenopausal women. J Nutr. 2006;136:112–116. doi: 10.1093/jn/136.1.112. [DOI] [PubMed] [Google Scholar]
- 35.Setchell KD, Brown NM, Zimmer-Nechemias L, Wolfe B, Jha P, et al. Metabolism of secoisolariciresinol-diglycoside the dietary precursor to the intestinally derived lignan enterolactone in humans. Food Funct. 2014;5:491–501. doi: 10.1039/c3fo60402k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Thompson LU. Lignan and tamoxifen, alone or in combination, reduce breast cancer cell adhesion, invasion and migration in vitro. Breast Cancer Res Treat. 2003;80:163–179. doi: 10.1023/A:1024513815374. [DOI] [PubMed] [Google Scholar]