Abstract
Pathway refactoring serves as an invaluable synthetic biology tool for natural product discovery, characterization and engineering. However, the complicated and laborious molecular biology techniques largely hinder its application in natural product research, especially in a high-throughput manner. Here we report a plug-and-play pathway refactoring workflow for high-throughput, flexible pathway construction and in both Escherichia coli and Saccharomyces cerevisiae. Biosynthetic genes were firstly cloned into preassembled helper plasmids with promoters and terminators, resulting in a series of expression cassettes. These expression cassettes were further assembled using Golden Gate reaction to generate fully refactored pathways. The inclusion of spacer plasmids in this system would not only increase the flexibility for refactoring pathways with different number of genes, but also facilitate gene deletion and replacement. As a proof of concept, a total of 96 pathways for combinatorial carotenoid biosynthesis were built successfully and shown to be functional. This workflow should be generally applicable to different classes of natural products produced by various organisms.
Keywords: natural products, silent biosynthetic gene clusters, pathway refactoring, synthetic biology
Introduction
Natural products are an indispensable source for drug discovery (Newman and Cragg 2016). However, due to the ever-increasing rediscovery rate of natural products by traditional methods, an indispensable approach to expand the natural product library for screening new drug candidates is the combinatorial biosynthesis (Sun et al. 2015). Thanks to the great efforts on natural product research in the past decades, biosynthetic genes with various functions have been continuously characterized. Moreover, the advances in next generation sequencing and bioinformatics have enabled rapid prediction of numerous biosynthetic gene clusters (BGCs) with the potential to generate novel natural products (Gomez-Escribano et al. 2016; Weber and Kim 2016). Nevertheless, most studies in combinatorial biosynthesis are limited to only a small fraction of all possibilities, mainly due to the lack of a high-throughput pathway refactoring method (Cobb et al. 2014). Besides combinatorial biosynthesis, another intriguing part of natural product research is the investigation of biosynthetic mechanisms, which usually requires gene deletion in a BGC. However, gene deletion by constructing pathway variants through existing DNA assembly methods usually affects the context of other genes, resulting in repetitive cloning work. Therefore, it is highly desirable to develop a fast, high-throughput and flexible pathway refactoring method for natural product research.
Pathway refactoring can be technically accomplished through DNA assembly. Golden Gate reaction is a DNA assembly method based on the activity of Type IIs restriction enzymes, which cut outside their recognition sites and generate single-strand DNA overhangs (Engler et al. 2009). When designed appropriately, the overhangs can guide the corresponding DNA fragments to be ligated in a designated order by a DNA ligase. Previously, we optimized the condition of Golden Gate assembly with high and consistent fidelity (above 95%) and successfully applied it for high-throughput synthesis of transcription activator-like effectors (TALEs) (Liang et al. 2014). In this work, we extended this strategy to establish a scalable pathway refactoring workflow. Briefly, multiple helper plasmids are constructed with well characterized promoters and terminators between which biosynthetic genes can be inserted. The constructed cassettes in the helper plasmid can then be assembled in a defined order with the help of 4 bp overhangs. Spacer plasmids, which share the same 4 bp overhangs with their corresponding helper plasmids but only encode a 20 bp random DNA sequence are also constructed and help the refactoring workflow adapt to pathways with different number of genes. We employed this workflow to refactor the zeaxanthin biosynthetic pathway and achieved high fidelity. Because of the highly modularized design, the gene deletion and replacement can also be realized by appropriately using the spacer plasmids. As proof of concept, 96 functional pathways for combinatorial carotenoid biosynthesis were successfully built using this workflow.
Results and Discussion
Design of the plug-and-play pathway refactoring workflow
As shown in Figure 1, the workflow consists of two tiers of Golden Gate reaction, which are catalyzed by BbsI (1st tier) and BsaI (2nd tier) respectively. The biosynthetic gene can be either synthesized or PCR amplified with two BbsI cleavage sites at both ends, which help in generating the general overhangs AATG (at the side of start codon) and CGGT (at the side of stop codon) respectively. Other BbsI and BsaI cleavage sites in the biosynthetic gene need to be removed at the same time through silent mutations. The same BbsI cleavage sites and the corresponding overhangs also exist on the helper plasmid, flanking a counter-selection marker (ccdB). Those BbsI cleavage sites on the helper plasmid are also flanked by a promoter and a terminator (Figure 1A). In the 1st tier BbsI catalyzed Golden Gate reaction, the ccdB marker on the helper plasmid is replaced by the biosynthetic gene, resulting in an expression cassette. In the overhang AATG between the promoter and the biosynthetic gene, the first “A” comes from the last nucleotide of the promoter followed by the start codon “ATG” of the biosynthetic gene. This design allows the seamless connection of the promoter and the biosynthetic gene.
Figure 1.
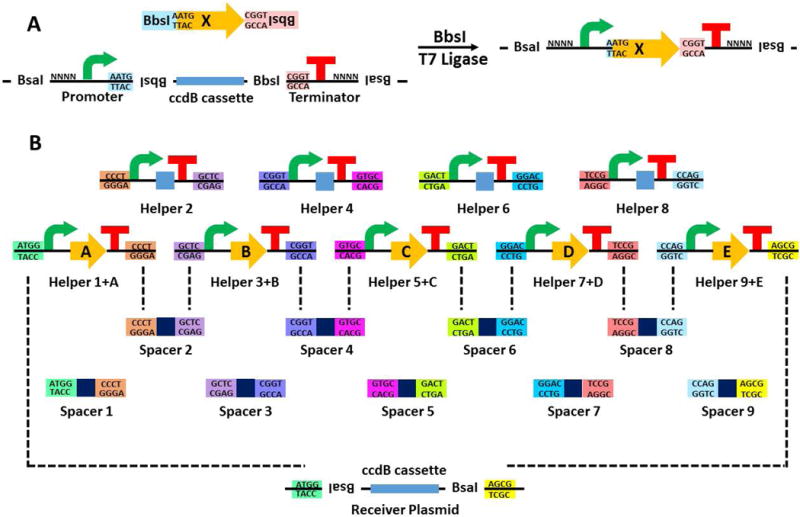
Scheme of the plug-and-play pathway refactoring workflow. (A) The 1st tier Golden Gate reaction. The gene is either synthesized or PCR amplified with BbsI cleavage sites at both ends and cloned into a helper plasmid through BbsI catalyzed Golden Gate reaction. (B) The 2nd tier Golden Gate reaction. Helper plasmids harboring the corresponding genes are mixed with the appropriate spacer plasmids and the receiver plasmid and assembled into the final construct through BsaI catalyzed Golden Gate reaction. All helper plasmids and spacer plasmids share the pUC19 backbone, while the receiver plasmid has either a pET28a backbone (for expression in E. coli) or pRS416 backbone with the ampicillin resistance gene replaced by the kanamycin resistance gene (for expression in S. cerevisiae).
After the 1st tier reaction, all the helper plasmids containing expression cassettes can be ligated together by their unique overhangs generated by BsaI in the 2nd tier reaction. However, as the number of biosynthetic genes varies from one BGC to another, gaps can appear once the number of biosynthetic genes is less than the number of helper plasmids (Figure 1B). Therefore, another set of plasmids called “spacer plasmids” are included in the system which have the same overhangs with their corresponding helper plasmids, but only a random 20bp DNA sequence in the middle. When the helper plasmid is not used in the 1st tier reaction, the corresponding spacer plasmid can be used to “fill the gap” in that position (Figure 1B). As a result, the 4 bp overhangs (ATGG and AGCG) flanking the ccdB marker on the receiver plasmid are not affected and the same receiver plasmid can be used for assembling pathways with various number of expression cassettes.
Refactoring of the zeaxanthin biosynthetic pathway using S. cerevisiae promoters and terminators
As proof of concept, we applied this workflow to refactor the zeaxanthin biosynthetic pathway in S. cerevisiae (Shao et al. 2009). Nine helper plasmids were built using the promoters and terminators from S. cerevisiae and the corresponding spacer plasmids were also built with the 20 bp random sequences designed by R2oDNA (Table S1) (Casini et al. 2014). We first checked the fidelity of the 1st tier reaction by blue-white screening. The lacZ cassette was cloned into one of the helper plasmids and the reaction mixture was transformed into NEB10-beta and spread on an LB+Amp plate with X-gal and IPTG. After overnight incubation, all the colonies on plate were blue, indicating the fidelity of the 1st tier reaction was 100% (Figure S1). By using the same reaction condition, all of the five genes from the zeaxanthin pathway were cloned into different S. cerevisiae helper plasmids respectively and 6 colonies were picked from each reaction. The plasmids isolated from these clones were digested by BsaI and all of them showed the correct digestion patterns (Figure 2A). Then the 5 plasmids with the corresponding expression cassettes were mixed with the other 4 spacer plasmids and the receiver plasmid for the 2nd tier Golden Gate reaction. After transformation into NEB10-beta and overnight incubation, constructs isolated from all 20 transformants showed the expected digestion patterns, indicating 100% assembly fidelity (Figure 2C).
Figure 2.
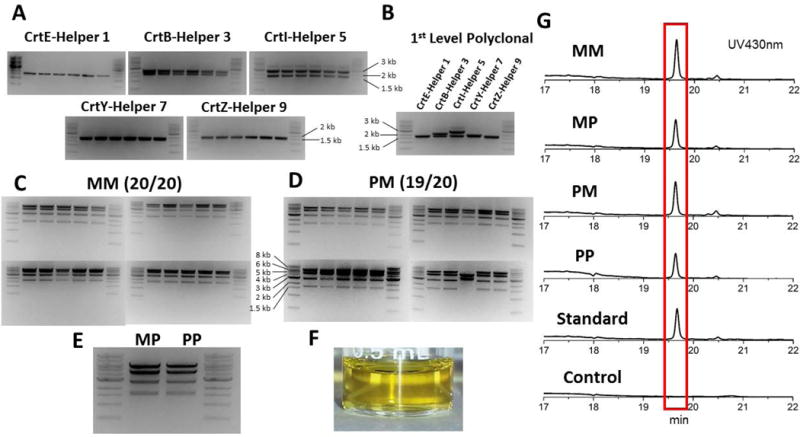
(A) The fidelity of the 1st tier reaction (monoclonal) checked by restriction digestion (expected band sizes: CrtE-Helper 1: 1.7 kb + 1.7 kb; CrtB-Helper 3: 2.2 kb + 1.7 kb; CrtI-Helper 5: 2.6 kb + 1.7 kb; CrtY-Helper 7: 1.9 kb+ 1.7 kb; CrtZ-Helper 9: 1.7 kb + 1.6 kb). (B) The fidelity of the 1st tier reaction (polyclonal) checked by restriction digestion. All the plasmids obtained from the 1st tier reactions were double-checked by Sanger sequencing. (C-E) The fidelity of the 2nd tier reaction (scenarios I-IV) checked by restriction digestion (expected band sizes: 5.7 kb + 4.3 kb + 2.7 kb + 1.6 kb). (F) The crude extract of zeaxanthin producing CEN.PK2-1C. (G) HPLC profile of zeaxanthin producing CEN.PK2-1C strains.
Since both the 1st and 2nd tier Golden Gate reactions showed very high fidelity, we hypothesized that polyclonal plasmids could also be used for cloning and expression, which would save the time for colony growth on the plate and the labor for colony picking. As a polyclonal plasmid can be obtained from either the 1st or 2nd tier reaction, there are four scenarios to obtain the final construct (I: monoclonal-monoclonal; II: monoclonal-polyclonal; III: polyclonal-monoclonal; IV: polyclonal-polyclonal). Polyclonal plasmids from the 1st tier reaction were digested and showed no significant difference from their monoclonal counterparts (Figure 2B), and these plasmids were used for 2nd tier reaction for scenarios III and IV. In scenario III, 20 colonies from the 2nd tier reaction were picked for plasmid isolation and digestion, 19 out of 20 showed a correct pattern (Figure 2D). In scenarios II and IV, polyclonal plasmids from the 2nd tier reaction were also checked by restriction digestion and no significant difference from the monoclonal one can be observed (Figure 2E). The final constructs from scenario I to scenario IV were transformed into S. cerevisiae CEN.PK2-1C for expression. Cells were extracted by acetone and analyzed by HPLC. All samples showed peaks at 430 nm with the same retention time as the zeaxanthin standard, which confirmed the production of zeaxanthin (Figure 2F–G). By using polyclonal plasmids, the workflow can be finished in two days, which is suitable for quick check of the pathway’s function. However, we recommend using monoclonal plasmids for quantitative analysis of the pathway’s function, which only requires some additional waiting time (1 or 2 overnight) for colony growth.
Construction of various zeaxanthin pathways with gene deletions using spacer plasmids
Because of its high modularity, this workflow also enables researchers to delete genes in the final construct for biosynthetic mechanistic studies with minimal cloning effort. Because every expression cassette should have the same overhangs that are generated by BsaI with the corresponding spacer plasmid, inclusion/deletion of a gene can be readily accomplished by using either the expression cassette or the corresponding spacer plasmid in the 2nd tier reaction and no repetitive cloning effort in the 1st tier is needed. As proof of concept, the same expression cassettes for the construction of the zeaxanthin pathway were used to build pathways producing phytoene, lycopene and β-carotene, which are intermediates in zeaxanthin biosynthesis (Schmidt-Dannert et al. 2000). The final constructs were again transformed into CEN.PK2-1C and extracted by acetone. The expected colors associated with these products were observed for all the three samples (Figure 3C). The samples were further analyzed with HPLC and LC/MS and the production of phytoene (1), lycopene (2) and β-carotene (3) was verified (Figure 3 and Figure S2).
Figure 3.
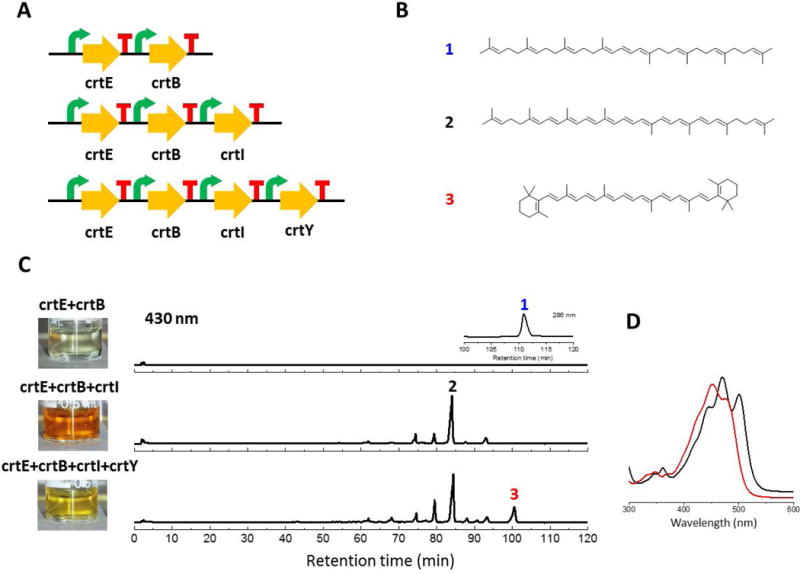
Analysis of the zeaxanthin producing pathway variants with gene deletion expressed in S. cerevisiae. (A) Pathways refactored for analysis. (B) Structures of carotenoids expected to be produced. (C) HPLC-UV traces. (D) UV absorption of the corresponding peaks. MS data is shown in Figure S2.
Construction of pathways for combinatorial carotenoid biosynthesis in E. coli
Because the overhangs generated by BsaI are independent from the sequence of the biosynthetic gene, every gene in the final construct can be easily replaced by another gene sharing the same helper plasmid. As a demonstration, two C30 carotenoid pathways with only one gene difference were selected (Umeno et al. 2005) (Figure 4A). CrtM and CrtN produce the intermediate diapolycopene shared by those two pathways and then CrtA or CrtOx add a ketone group (and a hydroxyl group) at different positions (Lee et al. 2010; Mijts et al. 2005). As all the four enzymes showed little activity in S. cerevisiae, another set of helper plasmids with T7 promoters and terminators (four in total) was built for the expression in E. coli. The final constructs were transformed into BL21(DE3) for expression and the yellow, orange and red colors were observed in the corresponding crude extracts (Figure 4C). The crude extracts were again analyzed by the HPLC and LC/MS. The diapolycopene (4) was observed from CrtM+CrtN at 430 nm. The expected product compound 5 and 6 were verified from CrtM+CrtN+CrtA by LC/MS while compounds 7, 8 and 9 were verified from CrtM+CrtN+CrtOx by LC/MS as well (Figure 4 and Figure S2).
Figure 4.
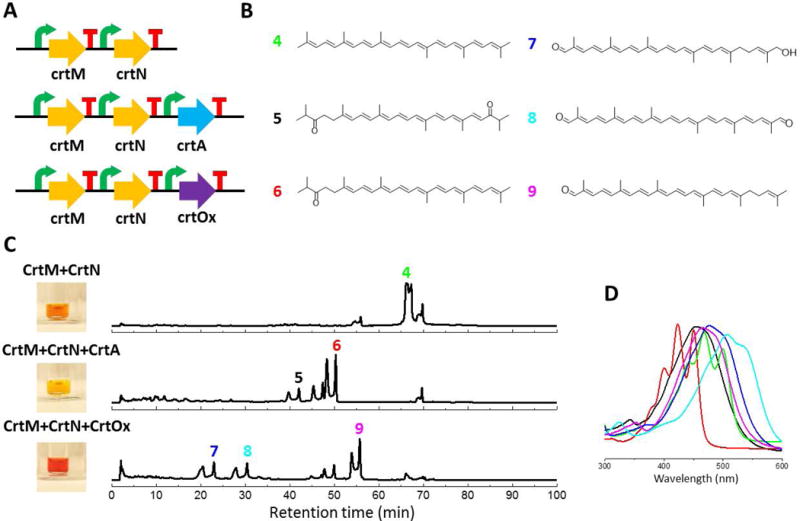
Analysis of the C30 carotenoid producing pathways expressed in E. coli. (A) Pathways refactored for analysis. (B) Structures of carotenoids expected to be produced. (C) HPLC-UV traces. (D) UV absorption of the corresponding peaks. MS data is shown in Figure S2.
Finally, to investigate whether the plug-and-play workflow has the potential to be scaled up and applied in combinatorial biosynthesis, a total of 96 pathways were created for the biosynthesis of carotenoids. Carotenoid biosynthesis can be split into three steps: 1) backbone synthesis, 2) desaturation, and 3) cyclization or other modification (Schmidt-Dannert et al. 2006; Umeno et al. 2005). Taking the biosynthesis of β-carotene as an example, the CrtE and CrtB first catalyze the formation of the C40 backbone phytoene, then the double bonds are introduced by CrtI, and finally the two ring structures are formed by CrtY. Accordingly, we collected 15 carotenoid biosynthetic genes and mutants and divided them into 3 groups. The backbone synthases group (group 1) includes four subgroups which can produce carotenoid backbones ranging from C30 to C50 (Umeno and Arnold 2003; Umeno and Arnold 2004). The desaturases group (group 2) contains wild type C40 desaturase CrtI and its two mutants and wild type C30 desaturase CrtN (Schmidt-Dannert et al. 2000). The cyclases and others group (group 3) consists of six enzymes which can catalyze different cyclization and oxidation reactions (Cunningham and Gantt 2001; Schmidt-Dannert et al. 2000) (Figure 5A). As the longest pathway consists of four genes (two from group 1, one from group 2 and one from group 3), the 4 helper plasmids that we built for the expression in E. coli should be sufficient for constructing all of the 96 combinations. Genes from group 1 were assigned to either helper plasmid 1 or helper plasmid 2, while genes from group 2 and 3 were assigned to helper plasmids 3 and 4 respectively. Successful assembly of the 96 constructs were confirmed using restriction digestion, and the plasmids were transformed into BL21(DE3) for carotenoid production. Carotenoids with different colors were generated after overnight induction by IPTG (Figure 5B).
Figure 5.
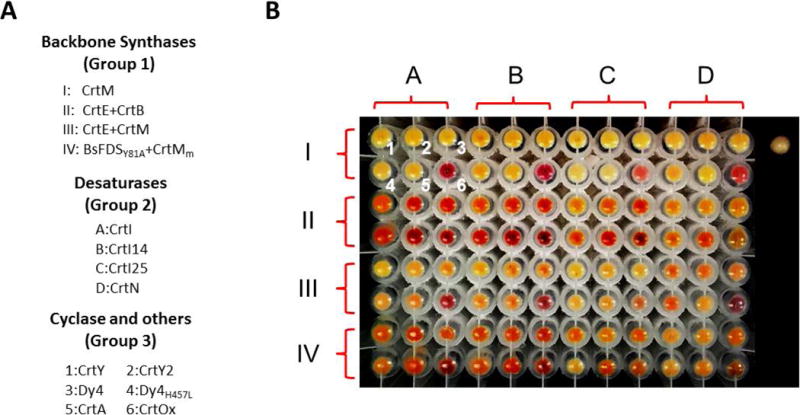
(A) The genes included in the 96 combinatorial pathways for carotenoid biosynthesis. Negative control (BL21(DE3) with empty pET28a plasmid) at the right side. (B) Cell pellets of BL21(DE3) after the induction by IPTG.
Conclusion
In this work we have developed a pathway refactoring workflow based on hierarchical Golden Gate assembly. Two sets of helper plasmids and one set of spacer plasmids with standardized 4 bp overhangs were constructed, which enabled the addition, deletion and exchange of expression cassettes in the final construct with minimal cloning effort. By using the optimized condition of Golden Gate assembly, high-throughput pathway refactoring was accomplished and demonstrated by the successful construction of 96 combinatorial pathways for carotenoid biosynthesis.
Prior to this work, a few Golden Gate reaction based multigene assembly systems have been developed, such as the GoldenBraid and the modular cloning (MoClo) system (Sarrion-Perdigones et al. 2011; Weber et al. 2011). Among all these systems, the most representative one is the MoClo and currently there are multiple part libraries available to enable its application in various hosts, such as E. coli and S. cerevisiae (Iverson et al. 2016; Lee et al. 2015). By its delicate design, MoClo enables researchers to assemble multigene constructs with high flexibility, as all of the parts, such as promoters and terminators, are modularized and free to be changed. However, because natural product research is typically focused on discovery of new natural products and study of biosynthetic mechanisms, it is unnecessary to find out the best combination of promoters for the BGC and in most cases strong promoters are arbitrarily chosen. Therefore, in our workflow, all the elements required for gene expression are included in the helper plasmids. In addition, MoClo includes many end-linkers and destination vectors to help its users add or delete genes continuously from both ends in their constructs, but it is unable to delete the gene in the middle without including the selectable marker. However, in the study of biosynthetic mechanisms, researchers usually do not know the gene functions in the BGC. As a result, genes at any positions of their constructs need to be deleted. Thus, our workflow includes the spacer plasmids, which makes gene deletion more flexible. Overall, because our workflow has fewer working plasmids and a simpler and more flexible design than MoFlo, it is uniquely positioned for high throughput natural product research.
In addition to the applications for biosynthetic mechanism investigation and combinatorial biosynthesis as demonstrated in this work, another potential application of this workflow would be the activation of silent BGCs (Luo et al. 2013), which is currently underway in our laboratory. In summary, this pathway refactoring workflow can serve as a powerful tool for the natural product research in the future.
Materials and Methods
Strains, media and cell cultivation
All chemicals were purchased from Sigma-Aldrich or ThermoFisher Scientific. E. coli NEB® 10-beta (New England Biolabs, Ipswich, MA) was used for DNA manipulation. Plasmids containing ccdB marker are propagated in E. coli DB3.1 (gyrA462 endA1 Δ(sr1-recA) mcrB mrr hsdS20 glnV44 (=supE44) ara14 galK2 lacY1 proA2 rpsL20 xyl5 leuB6 mtl1). S. cerevisiae strain CEN.PK2-1C (MATa ura3–52 trp1–289 leu2–3,112 his3Δ1 MAL2–8C SUC2) (EUROSCARF) and E. coli BL21(DE3) (Cell Media Facility, University of Illinois at Urbana–Champaign, Urbana, IL) were used for carotenoid production. E. coli was cultured in Lysogeny broth (LB) (ThermoFisher Scientific, Waltham, MA) containing appropriate antibiotics at 37 °C and 250 rpm unless specified otherwise. S. cerevisiae was cultured in synthetic complete medium (SC-URA) consisting of 0.17% Difco yeast nitrogen base without amino acids, 0.5% ammonium sulfate, and 0.083% amino acid drop out mixture without uracil (MP Biomedicals, Solon, OH) supplemented with 2% glucose was used for DNA homologous recombination and zeaxanthin fermentation. The plasmids containing S. cerevisiae promoters and terminators and the genes (crtE, crtB, crtI, crtY, crtZ) were described elsewhere (Shao et al., 2009). The plasmids containing other carotenoid biosynthetic genes were gifts from Frances Arnold, California Institute of Technology and Claudia Schmidt-Dannert, University of Minnesota.
1st Tier Golden Gate reaction
100 ng helper plasmid was mixed with an insert in 1:3 molar ratio. The reaction condition was as follows: a 20 μL reaction system contained 2 μL 10× Tango buffer (ThermoFisher Scientific, Waltham, MA), 1 μL 10 mM DTT, 1 μL 10 mM ATP, 1 μL FastDigest BbsI (ThermoFisher Scientific, Waltham, MA), 0.5 μL T7 ligase (Enzymatics, Beverley, MA), and ddH2O to 20 μL. The following temperature cycles were set up: 6 cycles of 5 min at 37 °C and 5 min at 21 °C. Then the reaction mix was treated by Plasmid-Safe-ATP-Dependent DNase (Epicentre, Madison, WI) by adding 0.25 μL enzyme and 1 μL 25 mM ATP and then incubating at 37 °C for 30 min. 10 μL reaction mix was then transformed into 100 μL NEB® 10-beta competent cell by standard heat-shock transformation. After recovery in 1 mL LB at 37 °C for 1 h, 5 μL was inoculated into 5 mL LB with 100 μg/mL ampicillin for polyclonal plasmid isolation or spread on LB+Amp plate and incubate overnight at 37 °C. Single colony was inoculated into 5 mL LB liquid culture with 100 μg/mL ampicillin next day for monoclonal plasmid isolation. Plasmids were isolated using QIAGEN Plasmid Mini Kit (QIAGEN, Germantown, MD). The maps and sequences of the helper plasmids used in this work are available in Figures S3–S4.
2nd Tier Golden Gate reaction
100 ng receiver plasmid (~0.035 nmol) was mixed with the same molar amount of helper plasmids with corresponding inserts cloned in and the appropriate spacer plasmids. The reaction condition was as follows: a 20 μL reaction system contained 2 μL 10× T4 DNA Ligase Reaction Buffer (New England Biolabs, Ipswich, MA), 0.8 μL BsaI (New England Biolabs, Ipswich, MA), 0.2 μL T4 DNA Ligase (M0202T, New England Biolabs, Ipswich, MA), and ddH2O to 20 μL. The following temperature cycles were set up: 5 min at 37 °C, 30 cycles of 5 min at 37 °C and 10 min at 16 °C, 30 min at 16 °C, 45 min at 37 °C and 5 min at 80 °C. The reaction was then treated by Plasmid-Safe-ATP-Dependent DNase (Epicentre, Madison, WI) and transformed into 100 μL NEB® 10-beta competent cell, which was similar to the 1st tier Golden Gate reaction. Monoclonal and polyclonal plasmids were isolated by the same method that described in the 1st tier Golden Gate reaction. Plasmids were isolated using QIAGEN Plasmid Mini Kit (QIAGEN, Germantown, MD). The maps and sequences of the spacer and receiver plasmids used in this work are available in Figures S5–S7.
Design of the spacer plasmid inserts
All insert sequences were designed using R2oDNA (http://www.r2odna.com/) with the following criteria: GC content of 60%; no start codons, common multiple cloning sites (MCS), or assembly restriction sites; no bacterial promoter-like sequences; and no similar sequences within E. coli and S. cerevisiae genome. Detailed sequences of 18 unique inserts utilized in this study are listed in Table S1.
Functional analysis of the assembled pathways
For carotenoid pathways expressed in S. cerevisiae, cell pellets were harvested from either 50 mL (for analysis of zeaxanthin) or 500 mL (for analysis of phytoene, lycopene and β-carotene) Sc-Ura liquid media after fermentation at 30 °C and 250 rpm for 72 h. The extraction was performed as previously described (Shao et al., 2009). Briefly the cell pellet was resuspended by 50 mL acetone and passed through French press at 10,000 psi three times.
Pathways for carotenoid biosynthesis in E. coli was induced by 0.1 mM IPTG when the OD600 reached 0.6–0.8 and the cell pellets containing the CrtM+CrtN, CrtM+CrtN+CrtA, or CrtM+CrtN+CrtOx pathways were harvested from 500 mL LB liquid media after fermentation at 30 °C and 250 rpm for 24 h. The cell pellet was resuspended in 50 mL acetone and vortexed for 1 min.
After extraction from either S. cerevisiae or E. coli, the supernatant was collected after centrifugation and evaporated to dryness. After resuspension in 200 μL THF, 20 μL was analyzed on an Agilent 1100 series HPLC with a Photodiode Array (PDA) detector. H2O + 0.1% formic acid (solvent A) and acetonitrile + 0.1% formic acid (solvent B) were used as mobile phase and different reverse-phase columns were used for different samples. The zeaxanthin containing crude extract was loaded onto Kinetex® 2.6 μm EVO C18 100Å 150×2.1 mm column (Phenomenex, Torrance, CA) and eluted at 0.2 mL/min with the following gradient: 50% B for 5 min, to 100% B in 10 min, hold at 100% B for 10 min and return to 50% B over 10 min. The other samples were loaded onto Kinetex® 5 μm EVO C18 100Å 250×4.6 mm column (Phenomenex, Torrance, CA) and eluted at 1 mL/min with the following gradient: 70% B for 15 min, to 100% B in 60 min, hold at 100% B for 30 min and return to 70% over 15 min. The same condition was used for the analysis of carotenoids by LC/MS (Agilent). Mass spectra were acquired in ultra scan mode using atmospheric pressure chemical ionization with positive polarity. The MS system (Agilent 1100 LC/MSD Trap XCT Plus) was operated using a drying temperature of 350 °C, a nebulizer pressure of 35 psi, a drying gas flow of 8.5 L/min, and a capillary voltage of 4500 V.
The 96 combinatorial pathways for carotenoid biosynthesis in E. coli were similarly induced by 0.1 mM IPTG and fermented at 30 °C and 250 rpm for 24 h. Then 1 mL culture was taken from 5 mL culture for each pathway and placed into the corresponding position in a 96 deep well plate. Cell pellets were harvested by centrifugation.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM077596). We thank Ran Chao and Dr. Tong Si for valuable discussion. We also thank Dr. Frances H. Arnold and Dr. Claudia Schmidt-Dannert for providing plasmids. All Zhao Group members are appreciated for critical comments.
Footnotes
Competing Interest Statement
The authors declare that they have no competing financial interests.
Supplementary Data
Supplementary Data are available online.
References
- Casini A, Christodoulou G, Freemont PS, Baldwin GS, Ellis T, MacDonald JT. R2oDNA designer: computational design of biologically neutral synthetic DNA sequences. ACS Synth Biol. 2014;3(8):525–8. doi: 10.1021/sb4001323. [DOI] [PubMed] [Google Scholar]
- Cobb RE, Ning JC, Zhao H. DNA assembly techniques for next-generation combinatorial biosynthesis of natural products. J Ind Microbiol Biotechnol. 2014;41(2):469–77. doi: 10.1007/s10295-013-1358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FX, Jr, Gantt E. One ring or two? Determination of ring number in carotenoids by lycopene epsilon-cyclases. Proc Natl Acad Sci U S A. 2001;98(5):2905–10. doi: 10.1073/pnas.051618398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden Gate shuffling: a one-pot DNA shuffling method based on Type IIs restriction enzymes. Plos One. 2009;4(5):e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Alt S, Bibb MJ. Next generation sequencing of actinobacteria for the discovery of novel natural products. Mar Drugs. 2016;14(4):78. doi: 10.3390/md14040078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson SV, Haddock TL, Beal J, Densmore DM. CIDAR MoClo: improved MoClo assembly standard and new E. coli part library enable rapid combinatorial design for synthetic and traditional biology. ACS Synth Biol. 2016;5(1):99–103. doi: 10.1021/acssynbio.5b00124. [DOI] [PubMed] [Google Scholar]
- Lee ME, DeLoache WC, Cervantes B, Dueber JE. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth Biol. 2015;4(9):975–86. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- Lee PC, Holtzapple E, Schmidt-Dannert C. Novel activity of Rhodobacter sphaeroides spheroidene monooxygenase CrtA expressed in Escherichia coli. Appl Environ Microbiol. 2010;76(21):7328–31. doi: 10.1128/AEM.01606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Chao R, Abil Z, Bao Z, Zhao H. FairyTALE: a high-throughput TAL effector synthesis platform. ACS Synth Biol. 2014;3(2):67–73. doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijts BN, Lee PC, Schmidt-Dannert C. Identification of a carotenoid oxygenase synthesizing acyclic xanthophylls: combinatorial biosynthesis and directed evolution. Chem Biol. 2005;12(4):453–60. doi: 10.1016/j.chembiol.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juarez P, Fernandez-del-Carmen A, Granell A, Orzaez D. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. Plos One. 2011;6(7):e21622. doi: 10.1371/journal.pone.0021622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Dannert C, Lee PC, Mijts BN. Creating carotenoid diversity in E. coli cells using combinatorial and directed evolution strategies. Phytochem Rev. 2006;5(1):67–74. [Google Scholar]
- Schmidt-Dannert C, Umeno D, Arnold FH. Molecular breeding of carotenoid biosynthetic pathways. Nat Biotechnol. 2000;18(7):750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37(2):e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Liu Z, Zhao H, Ang EL. Recent advances in combinatorial biosynthesis for drug discovery. Drug Des Devel Ther. 2015;9:823–33. doi: 10.2147/DDDT.S63023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeno D, Arnold FH. A C35 carotenoid biosynthetic pathway. Appl Environ Microbiol. 2003;69(6):3573–9. doi: 10.1128/AEM.69.6.3573-3579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeno D, Arnold FH. Evolution of a pathway to novel long-chain carotenoids. J Bacteriol. 2004;186(5):1531–6. doi: 10.1128/JB.186.5.1531-1536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeno D, Tobias AV, Arnold FH. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol Mol Biol Rev. 2005;69(1):51–78. doi: 10.1128/MMBR.69.1.51-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. Plos One. 2011;6(2):e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Kim HU. The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth Syst Biotechnol. 2016;1(2):69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


