Abstract
Integral membrane proteins are amphipathic molecules crucial for all cellular life. The structural study of these macromolecules starts with protein extraction from the native membranes, followed by purification and crystallisation. Detergents are essential tools for these processes, but detergent-solubilised membrane proteins often denature and aggregate, resulting in loss of both structure and function. In this study, a novel class of agents, designated mannitol-based amphiphiles (MNAs), were prepared and characterised for their ability to solubilise and stabilise membrane proteins. Some of MNAs conferred enhanced stability to four membrane proteins including a G protein-coupled receptor (GPCR), the β2 adrenergic receptor (β2AR), compared to both n-dodecyl-D-maltoside (DDM) and the other MNAs. These agents were also better than DDM for electron microscopy analysis of the β2AR. The ease of preparation together with the enhanced membrane protein stabilisation efficacy demonstrates the value of these agents for future membrane protein research.
Keywords: amphiphile design, electron microscopy, membrane proteins, novel detergents, protein stabilization
Membrane proteins are key players in a number of cellular processes, such as nutrient transport, signal transduction, cell adhesion and cell-to-cell communication. Due to their critical roles in biological systems, more than 50% of drug molecules target these macromolecules.[1] As a result, a clear understanding of both the structure and function of membrane proteins is of utmost importance for biological insight as well as for rational drug design. Despite their importance in biology and for drug discovery, only a few hundred membrane proteins have been structurally characterised, a tiny number compared with that of soluble proteins with known structure.[2] This discrepancy in the number of known structures between membrane and soluble proteins reflects the fact that handling these biomolecules is extremely difficult.[3] Their low natural abundance, instability in non-native environments and highly dynamic nature necessary for proper function all contribute to the comparatively slow progress.[4]
Conventional detergents, exemplified by n-octyl-D-glucoside (OG), lauryldimethylamine-N-oxide (LDAO) and n-dodecyl-D-maltoside (DDM), are widely used in membrane protein manipulation including protein extraction, purification and crystallisation.[5] Detergent micelles have a tendency to associate with membrane proteins, giving rise to protein-detergent complexes (PDCs).[6] However, many membrane proteins solubilised even in these popular conventional detergents undergo denaturation and aggregation, making structural study impossible. Furthermore, there are a vast number of membrane proteins that vary considerably in their propensity to aggregate and denature, characteristics dependent on their exact three-dimensional protein structures. Thus, currently available conventional detergents are limited in efficacy and scope, mainly as a result of narrow diversity in detergent structure.[7,8] Conventional detergents typically comprise of a single flexible alkyl chain attached to a large hydrophilic headgroup. Therefore, there is a great interest in developing a set of novel amphiphiles with distinct architecture that preserves the native structures of membrane proteins.[8]
Many novel agents have been developed over the past two decades, to address the narrow scope of utility of conventional detergents. These include small amphipathic agents such as tripod amphiphiles (TPAs),[9a,b] glyco-diosgenin (GDN),[9c] and hemifluorinated surfactants (HFSs),[9d] neopentyl glycol-derived triglucosides (NDTs),[9e] medium-sized peptide-based amphiphiles (e.g., lipopeptide detergents (LPDs),[10a] short peptides[10b] and β-peptides (BPs)[10c]) as well as large nanoassemblies such as amphipols (Apols),[11a,b] nanodiscs (NDs)[11c] and nanolipodisqs.[11d] Facial amphiphiles (FAs)[12a–c] glucose or maltose neopentyl glycols (GNGs or MNGs)[13a–e] are particularly attractive because these agents have been highly successful in membrane protein structural study. For instance, MNG-3 and GNG-3 have contributed to more than 20 new crystal structures of membrane proteins including several G protein-coupled receptors (GPCRs), clearly demonstrating the crucial role of novel amphiphiles in advancing membrane protein structural studies.[14a–j] In these studies, a novel class of agents, mannitol-based amphiphiles (MNAs) are designed, prepared, and evaluated in terms of their efficacy for membrane protein solubilisation and stabilisation (Figure 1). When four membrane proteins including a GPCR, the β2 adrenergic receptor (β2AR), were utilised, MNA-C13 and MNA-C14 were better than or at least comparable to a conventional detergent (DDM) and the other MNAs in maintaining structure and function.
Figure 1.
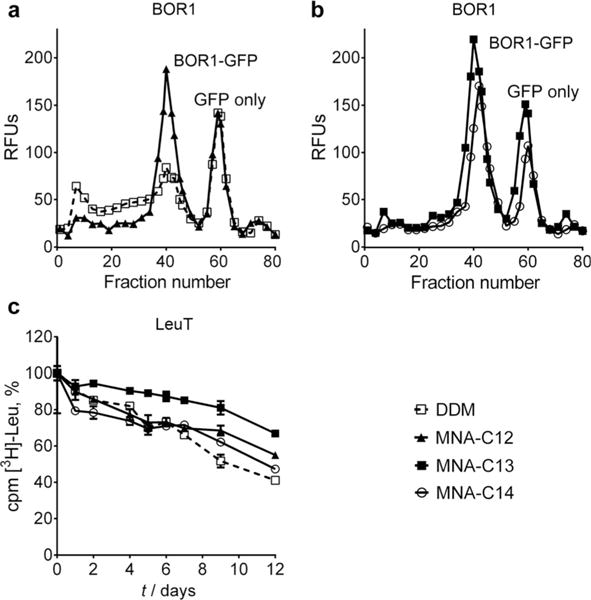
Protein stabilisation efficacy of representative MNAs (MNA-C12, MNA-C13 and MNA-C14) and DDM. a,b) FSEC of BOR1-GFP fusion protein solubilised by DDM and individual MNAs (MNA-C12, MNA-C13 and MNA-C14). The data were obtained after detergent-solubilised BOR1 was heated for 10 min at 40°C prior to loading onto the SEC column. The data shown is representative of two independent experiments. c) Long-term activity of LeuT solubilised in DDM or MNAs (MNA-C12, MNA-C13 and MNA-C14). LeuT stability was measured based on radiolabelled ligand ([3H]-Leu) binding via scintillation proximity assay (SPA) over the course of 12 days. The results are expressed as % activity relative to the activity on day 0 (mean SEM, n=2). All detergents were used at CMC +0.04 wt.% for comparison.
The newly designed agents have two flexible alkyl chains connected with four glucose groups by a mannitol linker (Scheme 1). Two alkyl chains were introduced to increase detergent affinity to the membrane protein surface, which could play a favourable role in membrane protein solubilisation and stabilisation. The alkyl chain length of MNAs varies from C8 to C14. This allowed us to screen for the optimal detergent hydrophile–lipophile balance (HLB). These agents were prepared in a straightforward synthetic protocol comprising four efficient steps with a high overall yield of ≈50%; alkylation, acetonide group removal, glycosylation, and benzoyl group de-protection. Note that the first and second steps could be combined without compromising synthetic yield, thus increasing synthetic convenience (see the Supporting Information for details). The critical micelle concentrations (CMCs) of the MNAs were estimated through a hydrophobic fluorescent dye, diphenylhexatriene (DPH),[15] and their micelle sizes were determined using dynamic light scattering (DLS). The summarised results are shown in Table 1. The CMCs of all the new agents are much lower (1–50 μM) than DDM (170 μM) and tend to decrease with increasing alkyl chain length. The exception is MNA-C8, which has a CMC value comparable to that of DDM. All MNAs form micelles comparable to or smaller than DDM and micelles formed by these agents increased with increasing alkyl chain length, giving hydrodynamic radii (Rh) of 2.3 and 3.3 nm for MNA-C8 and MNA-C14, respectively (Table 1). It is likely that MNA-C14 with the longest alkyl chain (C14) has the most cylindrical shape, and therefore forms the largest micelles. Similar to DDM, the DLS profiles of MNA micelles showed a single set of micelles (Figure S1, Supporting Information).
Scheme 1.
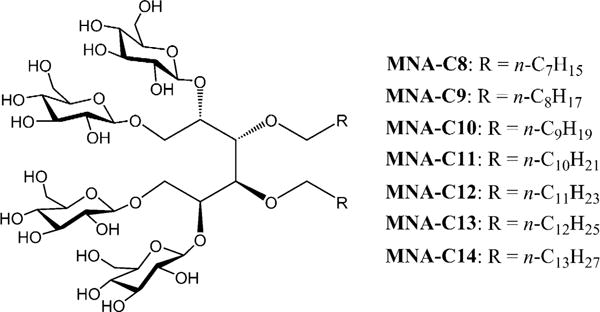
Chemical structures of the novel mannitol-based amphiphiles with different alkyl chain length (MNAs; MNA-C8-C14).
Table 1.
Molecular weights (MWs), critical micelle concentrations (CMCs) of MNAs and a conventional detergent (DDM) and hydrodynamic radii (Rh; n=4) of their micelles.
| Detergent | MW[a] | CMC [μM] | CMC [wt.%] | Rh [nm][b] |
|---|---|---|---|---|
| MNA-C8 | 1055.2 | ≈ 150 | ≈ 0.016 | 2.3 ± 0.13 |
| MNA-C9 | 1083.2 | ≈ 50 | ≈ 0.0054 | 2.5 ± 0.05 |
| MNA-C10 | 1111.3 | ≈ 15 | ≈ 0.0017 | 2.7 ± 0.04 |
| MNA-C11 | 1139.3 | ≈ 6 | ≈ 0.0007 | 2.9 ± 0.12 |
| MNA-C12 | 1167.4 | ≈ 4 | ≈ 0.0005 | 3.0 ± 0.01 |
| MNA-C13 | 1195.4 | ≈ 2 | ≈ 0.0002 | 3.3 ± 0.04 |
| MNA-C14 | 1233.5 | ≈ 1 | ≈ 0.0001 | 3.3 ± 0.08 |
| DDM | 510.1 | ≈ 170 | ≈ 0.0087 | 3.4 ± 0.03 |
Molecular weight of detergents.
Hydrodynamic radius of micelles determined at 1.0 wt.% by dynamic light scattering.
The newly synthesised MNAs were first evaluated using a boron transporter (BOR1) from Arabadopsis thaliana, expressed as a C-terminal GFP fusion protein in Saccharomyces cerevisiae.[16] Plant-based studies have shown that BOR1-GFP retains boron-transporting activity, demonstrating that this is a relevant form for both structural and functional analysis.[17] Detergent efficacy was compared by fluorescence size-exclusion chromatography (FSEC) after heat treatment.[18] Membranes (5.6 mg mL−1) containing BOR1-GFP fusion protein were treated with DDM or MNAs at 1.0 wt%. Following solubilisation and centrifugation to remove insoluble materials, the detergent-solubilised protein was heated at 40°C for 10 min. The sample was again briefly centrifuged and the soluble protein loaded onto the SEC column. The fluorescence of each elution fraction was measured. DDM-solubilised BOR1 underwent substantial protein aggregation, evidenced by the high intensity of the void peak (fraction numbers 0 to 20) and a concomitant reduction in the size of the main, monodispersed protein peak (≈fraction number 40) (Figure 1a). In addition, there were substantial losses during centrifugation due to heavy aggregation. For BOR1 solubilised in MNA-C8, MNA-C9, or MNA-C10, the fluorescent signal corresponding to monodispersed fusion protein was weaker than DDM (Figure S2, Supporting Information). These shorter alkyl chain MNAs extracted the protein from the membrane with solubilisation efficiencies of 40 ≈60% compared to 80% for DDM. Taken together, these findings indicate that MNA-C8, MNA-C9 and MNA-C10 are comparatively poor at both BOR1 solubilisation and stabilisation. In the case of MNA-C11, while this agent was inferior to DDM at extracting the fusion protein (solubilisation efficiency of ≈60 vs. ≈80% for DDM), its stabilisation efficacy was comparable to DDM (Figure S2, Supporting Information). Notably, MNA-C12, MNA-C13 and MNA-C14 with longer alkyl chains were markedly better at maintaining the protein in solution following heating compared to DDM. MNA-C13 was the best-performing amphiphile (Figure 1b). MNA-C12, MNA-C13 and MNA-C14 were also highly effective at extracting the protein from the membrane (solubilisation efficiencies of 70≈80%). This enhanced behaviour of these MNAs (MNA-C12, MNA-C13 and MNA-C14) compared to DDM is attributed, at least in part, to the presence of two alkyl chains which could strengthen interactions of these agents with BOR1. These MNAs were further compared with MNG-3, one of the most successful novel agents for membrane protein stabilisation and structural studies, using the same assay conditions. When the assay was performed at 40°C, the relative efficacy differences between MNAs and MNG-3 were small with MNA-C12 and MNA-C13 being slightly better at stabilising BOR1 than MNG-3 (Figure S3a, Supporting Information). When the assay temperature was increased to 45°C, the MNAs (MNA-C12, MNA-C13 and MNA-C14) maintained the protein in a monodisperse state; however, almost no monodispersed fusion protein was detectable in MNG-3. MNA-C14 showed the best performance, followed by MNA-C13 and MNA-C12 (Figure S3b, Supporting Information). These results indicate that the MNAs are substantially better at stabilising BOR1-GFP than both the standard detergent DDM and the significantly optimised MNG-3.
The superior stability of BOR1-GFP in MNA-C12, MNA-C13 and MNA-C14 encouraged us to further evaluate these agents for LeuT and LHI-RC complex. To test these agents with the bacterial leucine transporter (LeuT) from Aquifex aeolicus,[19,20] the transporter was initially solubilised by DDM at 1.0% and purified in 0.05% of the same detergent. DDM-purified transporter was then transferred into the individual solutions including DDM, MNA-C12, MNA-C13 or MNA-C14. The activity of LeuT was estimated by measuring the ability to bind [3H]-leucine via scintillation proximity assay (SPA) during a 12-day incubation period at room temperature.[21] When we used the detergents at CMC+0.04 wt. %, MNA-C13-solubilised LeuT showed enhanced ligand binding activity compared to DDM-solubilised protein (Figure 1c). Interestingly, MNA-C12 outperformed DDM only in the latter part of the incubation period (from day 7 to 12). In the case of MNA-C14, overall efficacy was similar to that of DDM in terms of retaining [3H]-leucine binding activity of the transporter. The difference between MNA-C13 and DDM is not great. However, taking into account the small standard errors of the data, the overall trend is that MNA-C13 is better than DDM in maintaining the ligand binding ability of this transporter. Following nine days of incubation, only half of the DDM-solubilised receptor was effective for [3H]-leucine binding while more than 80% LeuT protein maintained its binding ability in MNA-C13. When detergent concentration was increased to CMC+0.2 wt.%, the best stabilisation efficacy for this transporter was obtained by MNA-C12. In the case of MNA-C13 and MNA-C14, detergent efficacy was dependent on the incubation period, similar to that observed for MNA-C12 at CMC+0.04 wt% (Figure S4, Supporting Information). Overall, MNA-C12 and MNA-C13 performed better than DDM in terms of maintaining the [3H]-leucine binding activity of the transporter long-term.
The favourable behaviour of the MNAs (MNA-C12, MNA-C13 and MNA-C14) was also found in detergent evaluation with Rhodobacter capsulatus superassembly, comprising light-harvesting complex I and the reaction centre complex (LHI-RC).[22] LHI-RC complexes were first extracted with 1.0% DDM from the membrane and purified using 1×CMC DDM by Ni2+-NTA affinity chromatography. DDM-purified LHI-RC complex was diluted into detergent solutions containing individual detergents (MNA-C12, MNA-C13, MNA-C14, DDM and OG) to give the final detergent concentrations of CMC+0.04 wt% and CMC +0.2 wt%. The residual DDM concentration was thus far below its CMC (≈0.0087 wt%) after dilution. Long-term protein stability was measured over 15 days by monitoring absorbance at 875 nm (A875), mainly arising from the presence of multiple co-factors such as chlorophylls and carotenoids embedded in the native conformation of the complex. Upon protein denaturation, these cofactors readily dissociate from the complex, leading to a marked decrease in A875. As can be seen in Figure 2a, DDM was better than OG at stabilising the LHI-RC complex. DDM-solubilised LHI-RC complex showed a gradual loss in its integrity while OG-solubilised protein gave fast degradation at CMC+0.04 wt%. In contrast, all the three MNAs (MNA-C12, MNA-C13 and MNA-C14) clearly outperformed DDM, with the best performance by MNA-C13. The difference between MNAs and DDM in terms of detergent efficacy further increased when detergent concentration was increased to CMC + 0.2 wt%. This result indicates the favourable architecture of the MNAs compared to the two conventional detergents (OG and DDM) for stability of the complex.
Figure 2.
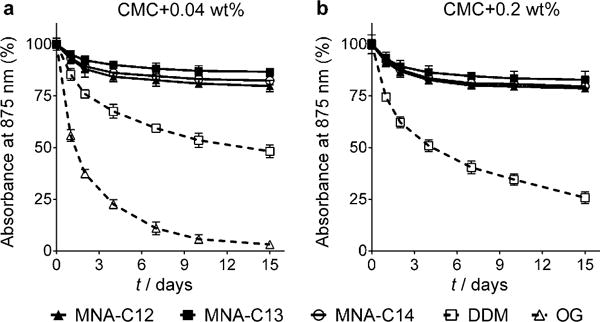
Long-terms stability of R. capsulatus superassembly solubilised in the MNAs (MNA-C12, MNA-C13 and MNA-C14) or two conventional detergents (DDM and OG). Individual detergents were evaluated at a) CMC+ 0.04 wt.% and b) CMC+0.2 wt. %. Protein stability was monitored over 15 days by measuring the absorbance value at 875 nm (A875) during incubation at 25°C. The results are expressed as % absorbance value relative to the value on day 0 (mean SEM, n=2).
The human β2 adrenergic receptor (β2AR) is an excellent target protein to explore the potential of the MNAs for structural studies of G-protein-coupled receptors (GPCRs).[23] For this purpose, we used bimane conjugated-β2AR in which the bimane moiety is covalently attached to cysteine 265 located at the cytoplasmic end of transmembrane helix 6 (TM6).[24] The subtle conformational change in β2AR induced upon ligand and Gs-protein binding was precisely monitored by measuring the fluorescence change of the bimane fluorophore.[24,25] Note that the monobromobimane (mBBr)-conjugated β2AR is functionally active.[24] For this assay, mBBr-β2AR was first solubilised and purified in 0.1% DDM. The DDM-purified mBBr-β2AR was diluted into individual detergent solutions to reach detergent concentrations of CMC+0.04 wt% or CMC+0.2 wt%. The bimane fluorescence spectra of these samples were measured in the absence or presence of a high affinity agonist, BI–167107. Among seven tested MNAs, use of MNA-C12, MNA-C13 and MNA-C14 resulted in the bimane fluorescence spectra of the receptor similar to DDM, indicative of the favourable behaviour of these MNAs (Figure S5, Supporting Information). This favourable performance of these MNAs is in agreement with the results obtained for the other membrane proteins (BOR1, LeuT and LHI-RC complex). We continued to investigate the effect of these MNAs on the conformational change of β2AR upon binding of another agonist and/or Gs-protein.[14a] The receptor requires Gs-protein coupling in addition to binding of a full agonist (e.g., BI) for full activation.[14a] A similar trend was also observed for each MNA-solubilised β2AR. In the presence of the full agonist, isoproterenol (ISO), each MNA-solubilised receptor showed bimane fluorescence spectra similar to the DDM-solubilised receptor, indicating that the receptor in these MNAs was partially activated (Figure 3a, b and Figure S6, Supporting Information). When ISO and Gs-protein were used in combination, mBBr-β2AR solubilised in MNA-C13 and MNA-C14 showed bimane fluorescence spectra which matched well with that seen for DDM-purified β2AR (Figure 3b). In the case of MNA-C12, the bimane fluorescence spectrum of the receptor differed slightly from that of the DDM-purified protein (Figure S6, Supporting Information). The spectral changes observed for the protein in the different detergents (reduction in fluorescence intensity and the shift in maximal emission wavelength) are associated with the transition from the inactive to active state (Figure 3b).[14a] This encouraging result prompted us to compare the effects of the MNAs (MNA-C12, MNA-C13 and MNA-C14) and DDM on receptor stabilisation at detergent concentration far below their respective CMCs. When detergent concentration was decreased to 1/50th of the individual CMCs by a dilution method, DDM-solubilised β2AR underwent an obvious conformational change as indicated by the large spectral change (Figure 3c). In contrast, little conformational change was observed for β2AR solubilised in MNA-C13 and MNA-C14. In the case of MNA-C12, there was a slight conformational change in the protein at this dilution (Figure S6b, Supporting Information). This result indicates that the new tested MNAs form stronger interactions with the hydrophobic protein surface compared to DDM and that they produce PDCs more resistant to reductions in detergent concentration to below the CMC. We further characterised β2AR by a ligand binding assay after detergent exchange. The receptor activity purified in DDM or MNAs (MNA-C12, MNA-C13 and MNA-C14) was measured using [3H]-dihydroalprenolol ([3H]-DHA) (Figure S7a, Supporting Information). The receptor purified in MNA-C13 and MNA-C14 showed ligand binding activity comparable to the DDM-purified receptor. MNA-C12-purified receptor exhibited reduced ligand binding activity compared to the DDM-purified protein. Finally, the β2AR purified in MNA-C13 and MNA-C14 was analysed by SEC in order to obtain information on the PDC size formed by these agents. As seen in Figure S7b, Supporting Information, these MNAs produced PDCs smaller than DDM. The favourable properties of MNA-C13 and MNA-C14 such as strong binding affinity, small PDCs and improved protein stabilisation suggest that they are suitable for electron microscopy (EM) analysis of membrane proteins. Indeed, electron micrographs of β2AR purified in either MNA-C13 or MNA-C14 were homogeneous and exhibit little background clutter compared to DDM-purified protein (Figure 4).
Figure 3.
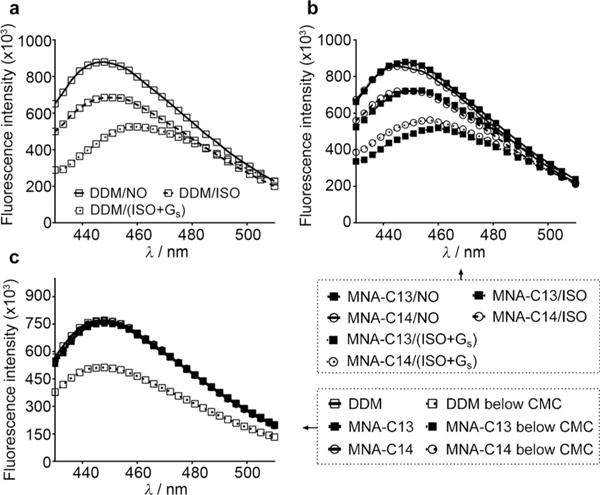
Fluorescence spectra of monobromobimane-labeled β2AR (mBBr-β2AR) solubilised in DDM or MNAs (MNA-C13 and MNA-C14). a,b) Fluorescence spectra of mBBr-β2AR in the absence (detergent/NO), the presence of a full agonist (isopreoterenol (ISO); detergent/ISO) or a combination of ISO and Gs-protein (detergent/(ISO+Gs)). c) Fluorescence spectral changes of mBBr-β2AR when detergent concentrations were changed from 20×CMC to (1/50)×CMC by 1000-fold dilution.
Figure 4.
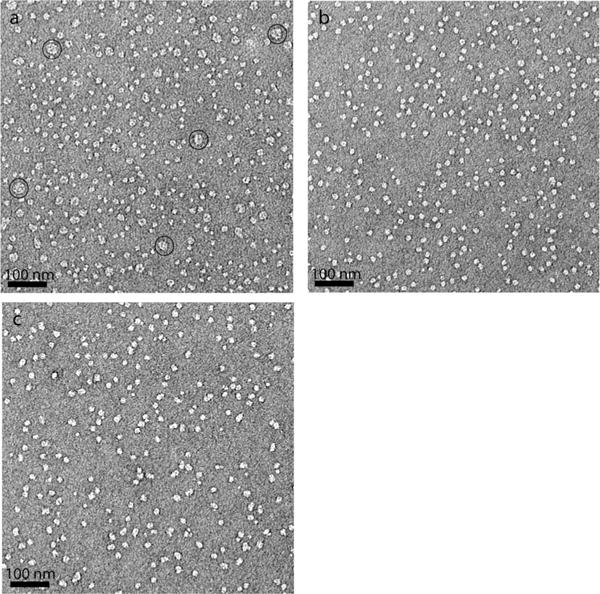
Negative stain electron micrographs for β2AR purified in DDM (a), MNA-C13 (b) and MNA-C14 (c). Individual samples (3 μL) were stained with 1.0 wt.% uranyl formate for the electron micrographs. A number of large particles present in (a) indicate occurrence of receptor aggregation in the DDM sample. Some large particles as representatives were indicated by open circles.
Membrane protein stability and PDC size are the two most important determinants in obtaining high-resolution crystal structures of membrane proteins.[7a,13a] Maintaining the integrity of a target membrane protein is key to the outcome of a biologically and pharmaceutically relevant protein structure. The formation of small PDCs is often essential to generation of well diffracting protein crystals through the production of membrane proteins with a large exposed hydrophilic surface area.[7b] The hydrophilic–hydrophilic interactions between membrane proteins facilitates crystal lattice formation. However, conventional detergents with good protein stabilisation efficacy (e.g., DDM) tend to form large PDCs while small PDC-forming detergents (e.g., OG and LDAO) often destabilize target membrane proteins.[7a] Most novel amphiphiles developed thus far often improve membrane protein stability, but are less suited to the formation of small PDCs. For instance, MNG-3 is generally superior to DDM for membrane protein stability, but PDCs formed by this agent tend to be larger than those formed by DDM.[13a] In contrast, the GNGs tend to form small PDCs, but their efficacy for membrane protein stabilisation is inferior to DDM.[13c] Thus, it is a challenging task to design novel agents with the ability to preserve the integrity of membrane proteins and to form small PDCs. Of the seven MNAs, three agents (MNA-C12, MNA-C13 and MNA-C14) were superior to DDM at stabilising most target membrane proteins. When BOR1-GFP was selected for efficacy comparison with MNG-3, these MNAs were even superior to MNG-3. MNA-C13 generally produced the most stable protein, followed by MNA-C12 or MNA-C14 depending on the target membrane protein. Note that DDM was the detergent selected for comparison as this has been shown to be the best of a number of classical detergents in stabilising our target membrane proteins including LeuT, β2AR and LHI-RC complex. In contrast, OG, LDAO and/or phosphocholines (e.g., Fos-choline-14) are not effective in maintaining the proteins in a stable state in solution.[9a,13c] When we measured PDC size, MNA-C13 and MNA-C14 produced smaller PDCs with β2AR than DDM. The small PDC-forming character of these MNAs is likely to be due to the presence of the glucoside headgroup instead of a maltoside group; glucoside detergents (e.g., OG) tend to form smaller PDCs than maltoside detergents (e.g., DDM).[7a] Note that most conventional or novel glucoside detergents (e.g., GNGs and TPAs) have failed to show enhanced protein stability relative to DDM for LeuT and β2AR.[13a] The current results indicate that both MNA-C13 and MNA-C14 have favourable properties distinct from conventional and most novel detergents.
Some MNA agents (MNA-C12, MNA-C13 and MNA-C14) were effective at solubilising membrane proteins from the membranes, as demonstrated by FSEC analysis of BOR1-GFP fusion protein. Since membrane proteins are produced, even recombinantly, at low levels and there are significant losses incurred during purification, membrane protein solubilisation efficiency is also an important detergent property for membrane protein research.[26] It is noteworthy that MNA-C13 or MNA-C14 maintained strong interactions with β2AR even when detergent concentrations were reduced to below their respective CMCs. This unique property could be useful when membrane protein structures were analysed by methods employing non-aqueous environments such as single particle techniques. In the current study, MNA-C13 and MNA-C14 were used to investigate the quality of β2AR particles generated by these agents. This analysis implied that these MNAs have significant potential for the structural analysis of a GPCR using this technique. This analytical tool is becoming increasingly relevant to the high-resolution determination of bio-macromolecular structures.[27a–c] Finally, MNA agents could be prepared by a short protocol comprising four synthetic steps. The accessibility of MNAs is essential for widespread use of these agents in the membrane protein community.
In summary, MNA-C13 and MNA-C14 showed a range of favourable properties such as ease of preparation, enhanced protein stabilisation, small PDC formation, efficient protein solubilisation, and the ability to form strong interactions with the protein surface. Combining such diverse properties into a simple molecular architecture is challenging; therefore, these MNAs could represent invaluable tools for membrane protein research.
Experimental Section
Details on the synthesis and characterisation of novel amphiphiles and membrane protein stability assays can be found in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grant number 2008–0061891 and 2013R1A2A2A03067623 to P.S.C., H.H., and H.E.B.). N.J.S. is in receipt of a BBSRC Doctoral Training Programme studentship awarded to B.B.
Footnotes
Supporting information and ORCIDs from the authors for this article are available on the WWW under http://dx.doi.org/10.1002/chem.201600533.
References
- 1.a) Overington JP, Al-Lazikani B, Hopkins AL. Nat Rev Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]; b) Lagerstrom MC, Schioth HB. Nat Rev Drug Discovery. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 2.http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html
- 3.a) White SH, Wimley WC. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; b) Bowie JU. Curr Opin Struct Biol. 2001;11:39–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 4.Lacapere JJ, Pebay-Peyroula E, Neumann JM, Etchebest C. Trends Biochem Sci. 2007;32:59–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 5.a) Newstead S, Ferrandon S, Iwata S. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Newstead S, Hobbs J, Jordan D, Carpenter EP, Iwata S. Mol Membr Biol. 2008;25:631–638. doi: 10.1080/09687680802526574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Garavito RM, Ferguson-Miller SJ. Biol Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]; b) Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Proc Natl Acad Sci USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Moller JV, le Maire M. J Biol Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]
- 7.a) Privé GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]; b) Zhang Q, Tao H, Hong WX. Methods. 2011;55:318–323. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadaf A, Cho KH, Byrne B, Chae PS. In: Amphipathic Agents for Membrane Protein Study in Methods in Enzymology. Shukla AK, editor. Vol. 557. Academic Press; Burlington: 2015. pp. 57–94. [DOI] [PubMed] [Google Scholar]
- 9.a) Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chae PS, Cho KH, Wander MJ, Bae HE, Gellman SH, Labile PD. Biochim Biophys Acta. 2014;1838:278–286. doi: 10.1016/j.bbamem.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Kruse AC, Nurva S, Gether U, Guan L, Loland CJ, Byrne B, Kobilka BK, Gellman SH. Chem Eur J. 2012;18:9485–9490. doi: 10.1002/chem.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Breyton C, Chabaud E, Chaudier Y, Pucci B, Popot JL. FEBS Lett. 2004;564:312–318. doi: 10.1016/S0014-5793(04)00227-3. [DOI] [PubMed] [Google Scholar]; e) Sadaf A, Mortensen JS, Capaldi S, Tikhonova E, Hariharan P, Ribeiro O, Loland CJ, Guan L, Byrne B, Chae PS. Chem Sci. 2016;7:1933–1939. doi: 10.1039/c5sc02900g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) McGregor CL, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Privé GG. Nat Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; b) Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Proc Natl Acad Sci USA. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tao H, Lee SC, Moeller A, Roy RS, Siu FY, Zimmermann J, Stevens RC, Potter CS, Carragher B, Zhang Q. Nat Methods. 2013;10:759–761. doi: 10.1038/nmeth.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Tribet C, Audebert R, Popot JL. Proc Natl Acad Sci USA. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Popot JL, Althoff T, Bagnard D, Baneres JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, Cremel G, Dahmane T, de la Maza LM, Ebel C, Gabel F, Giusti F, Gohon Y, Goormaghtigh E, Guittet E, Kleinschmidt JH, Kuhlbrandt W, Le Bon C, Martinez KL, Picard M, Pucci B, Sachs JN, Tribet C, van Heijenoort C, Wien F, Zito F, Zoonens M. Annu Rev Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]; c) Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]; d) Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A. Nano Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 12.a) Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. Angew Chem Int Ed. 2007;46:7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2007;119:7153–7155. [Google Scholar]; b) Lee SC, Bennett BC, Hong WX, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, Stout CD, Yeager MJ, Zhang Q. Proc Natl Acad Sci USA. 2013;110:E1203–1211. doi: 10.1073/pnas.1221442110. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chae PS, Gotfryd K, Pacyna J, Miercke LJW, Rasmussen SGF, Robbins RA, Rana RR, Loland CJ, Kobilka BK, Stroud R, Byrne B, Gether U, Gellman SH. J Am Chem Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Chae PS, Rana RR, Gotfryd K, Rasmussen SGF, Kruse AC, Cho KH, Apaldi S, Arlsson E, Kobilka BK, Loland CJ, Gether U, Banerjee S, Byrne B, Lee JK, Gellman SH. Chem Commun. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cho KH, Bae HE, Das M, Gellman SH, Chae PS. Chem Asian J. 2014;9:632–638. doi: 10.1002/asia.201301303. [DOI] [PubMed] [Google Scholar]; c) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot JL, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka BK, Gellman SH. Nat Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Cho KH, Byrne B, Chae PS. ChemBioChem. 2013;14:452–455. doi: 10.1002/cbic.201200759. [DOI] [PubMed] [Google Scholar]; e) Cho KH, Husri M, Amin A, Gotfryd K, Lee HJ, Go J, Kim JW, Loland CJ, Guan L, Byrne B, Chae PS. Analyst. 2015;140:3157–3163. doi: 10.1039/c5an00240k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Rasmussen SGF, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kellosalo J, Kajander T, Kogan K, Pokharel K, Goldman A. Science. 2012;337:473–476. doi: 10.1126/science.1222505. [DOI] [PubMed] [Google Scholar]; e) Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, Garcia KC, Kobilka BK. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Miller PS, Aricescu AR. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Karakas E, Furukawa H. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Science. 2014;344:304–307. doi: 10.1126/science.1248571. [DOI] [PubMed] [Google Scholar]; i) Dickson VK, Pedi L, Long SB. Nature. 2014;516:213–218. doi: 10.1038/nature13913. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Shukla AK, Westfield GH, Xiao K, Reis RI, Huang L-Y, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie AN, Dosey An M, Su M, Liang C-R, Gu L-L, Shan J-M, Chen X, Hanna R, Choi M, Yao XJ, Klink BU, Kahsai AW, Sidhu SS, Koide S, Penczek PA, Kossiakoff AA, Woods VL, Jr, Kobilka BK, Skiniotis G, Lefkowitz RJ. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chattopadhyay A, London E. Anal Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 16.Drew D, Newstead S, Sonoda Y, Kim H, von Heijne G, Iwata S. Nat Protoc. 2008;3:784–798. doi: 10.1038/nprot.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Kasai K, Takano J, Miwa K, Toyoda A, Fujiwara T. J Biol Chem. 2011;286:6175–6183. doi: 10.1074/jbc.M110.184929. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, Onouchi H, Naito S, Fujiwara T. Proc Natl Acad Sci USA. 2010;107:5220–5225. doi: 10.1073/pnas.0910744107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung J, Karachaliou M, Alves C, Diallinas G, Byrne B. Protein Expression Purif. 2010;72:139–146. doi: 10.1016/j.pep.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 20.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 21.Quick M, Javitch JA. Proc Natl Acad Sci USA. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laible PD, Kirmaier C, Udawatte CS, Hofman SJ, Holten D, Hanson DK. Biochemistry. 2003;42:1718–1730. doi: 10.1021/bi026959b. [DOI] [PubMed] [Google Scholar]
- 23.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Bobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 24.a) Yao X, Parnot C, Deupi X, Ratnala VRP, Swaminath G, Farrens D. Nat Chem Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]; b) Yao X, Ruiz GV, Whorton MR, Rasmussen SGF, DeVree BT, Deupi X, Sunahara RK, Kobilka BK. Proc Natl Acad Sci USA. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansoor SE, McHaourab HS, Farrens DL. Biochemistry. 2002;41:2475–2484. doi: 10.1021/bi011198i. [DOI] [PubMed] [Google Scholar]
- 26.Schlegel S, Lofblom J, Lee C, Hjelm A, Klepsch M, Strous M, Drew D, Slotboom DJ, de Gier JW. J Mol Biol. 2012;423:648–659. doi: 10.1016/j.jmb.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 27.a) Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kim J, Wu S, Tomasiak TM, Mergel C, Winter MB, Stiller SB, Robles-Colmanares Y, Stroud RM, Tampe R, Craik CS, Cheng Y. Nature. 2015;517:396–400. doi: 10.1038/nature13872. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JLS, Subramaniam S. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


