Abstract
Background
Sinonasal biofilms have been demonstrated in specimens collected from chronic rhinosinusitis (CRS) patients. Mounting evidence suggests that biofilms contribute to therapeutically recalcitrant CRS. Recently, the bitter taste receptor T2R38 has been implicated in the regulation of the sinonasal mucosal innate immune response. TAS2R38 gene polymorphisms affect receptor functionality and contribute to variations seen in sinonasal innate defense as well as taste perception reflected in gustatory sensitivity to the bitter compound phenylthiocarbamide (PTC). In a population of CRS patients with active infection or inflammation, we sought to determine if a correlation between T2R38 phenotype and in vitro biofilm formation existed.
Methods
Endoscopically guided sinonasal swabs were obtained prospectively from CRS (±polyp) patients with evidence of persistent inflammation or mucopurulence. In vitro biofilm formation was assessed with a modified Calgary Biofilm Detection Assay. Patients' phenotypic (functional) expression of the bitter taste receptor T2R38 was evaluated with a taste test including the compound PTC. Linear regression was used to determine the level of significance between mean in vitro biofilm formation levels and mean PTC taste test intensity ratings across CRS patients.
Results
Sinonasal swabs were obtained from 59 patients, with 42 of the 59 samples demonstrating in vitro biofilm formation. Analysis revealed an inverse linear association between in vitro biofilm formation and PTC taste intensity ratings (p = 0.019) for all patients. This association was exclusively driven by nonpolypoid CRS patients (p = 0.0026).
Conclusion
In vitro biofilm formation from sinonasal clinical isolates is inversely correlated with PTC taste sensitivity in nonpolypoid CRS patients.
Keywords: biofilm, Calgary biofilm detection, chronic rhinosinusitis, Pseudomonas aeruginosa, bitter taste receptor, T2R38 polymorphism, phenylthiocarbamide, nitric oxide, genetics, supertaster
Rhinosinusitis is a common disorder accounting for an estimated 13 million physician office visits in the United States each year.1 The aggregated cost of rhinosinusitis is approximately 8.6 billion dollars annually, and the disease affects an estimated 16% of the population in the United States.1, 2 In addition to the financial burden on the health system, quality of life and health utility studies have shown significant morbidity associated with rhinosinusitis, comparable to that of other chronic illnesses such as congestive heart failure and chronic obstructive pulmonary disease (COPD).3, 4 The loss of the sense of smell, which often accompanies chronic rhinosinusitis (CRS), further compromises quality of life.5
CRS has many known contributing factors including environmental exposures, impaired mucociliary clearance, and presumed underlying genetic predispositions.6–8 Recently, a novel arm of respiratory innate immunity mediated via taste receptors has been described.9–13 Our prior work demonstrated that the bitter taste receptor T2R38, a G-protein coupled receptor expressed by upper airway ciliated cells, is stimulated by gram-negative quorum sensing molecules and results in nitric oxide (NO) production.14 NO in the airway functions in innate immunity through two downstream effects: increased mucociliary clearance and direct bacterial killing.9–11, 13, 14 The T2R38 receptor has common polymorphisms at amino acid residues at 49, 262, and 296, which alter the activity of the receptor.15 A proline (P), alanine (A), and valine (V) at these positions respectively confer a functional receptor (in both the tongue and airway) whereas an alanine (A), valine (V) and isoleucine (I) at these positions confers a nonfunctional receptor.15 These substitutions tend to segregate together (PAV vs AVI) and are nearly a balanced polymorphism in the white population.16,17 Thus, the receptor is nonfunctional in roughly one-quarter of this population (AVI/AVI), somewhat functional in one-half of this population (PAV/AVI), and completely functional in the remaining one-quarter of this population (PAV/PAV).16 Phenylthiocarbamide (PTC) is a synthetic compound that activates and is specific for the T2R38 bitter taste receptor.18 In the tongue, activation of T2R38 elicits a bitter taste in individuals with at least 1 PAV allele, with the bitter intensity proportional to the amount PAV expressed19 and ultimately the TAS2R38 genotype,15 whereas in sinonasal cells, it stimulates NO production in a TAS2R38 genotype dependent manner.14 Thus, patient genotype, or at least an approximation of the degree of PAV T2R38 that is expressed in the tongue, can be quickly assessed by a taste test that elicits ratings of bitter taste intensity to PTC on a continuum (0 to 12),20 without collection of a biologic specimen nor the time and expense of molecular biology diagnostics. Thus, a taste test of T2R38 functionality may be developed into a “point of service” assay that could inform real time clinical decisions, rather than waiting for results of genetic testing, which may be less accurate at assessing gene expression.
Biofilms have been implicated in persistent bacterial infections and have been shown to be present in patients with therapeutically recalcitrant CRS.21–26 Bacterial biofilms are formed from layers of bacteria and their extruded exopolysaccharide matrix.27 Biofilms provide a chronic source of shedding bacteria, toxins, and antigens that stimulate the immune system. Biofilms in CRS have been associated with persistent infection and inflammation as well as worse postoperative outcomes,21, 24,26 and they may be chronically present in the nasal mucosa of some CRS patients, serving as a nidus for acute infections.28 At times, the immune response is insufficient in responding to the bacteria and bacterial products shed by biofilms, and an acute infection may result. Taste receptor–mediated immune function may decrease biofilm formation, as well as prevent planktonic bacterial shedding from a previously established biofilm.
To date, there has been no study investigating the interplay of T2R38 function and sinonasal biofilm formation. The goal of our investigation was 2-fold: (1) to determine whether CRS patients with the nonfunctional T2R38 form (AVI) are more likely to harbor bacteria within their sinonasal cavity with biofilm-forming capacity; and (2) whether taste testing for PTC could be used as a proxy for T2R38 functionality in the sinonasal cavity. We test our hypothesis by assessing in vitro biofilm formation from bacteria recovered from CRS patients and correlating biofilm predilection with PTC taste sensitivity and TAS2R38 genotype.
Patients and methods
Institutional Review Board approval was obtained to enroll adult patients who met objective and subjective guidelines for the diagnosis of CRS by the clinical practice guideline for adult sinusitis set forth by the Academy of Otolaryngology–Head and Neck Surgery.29 All patients were recruited from the Department of Otorhinolaryngology–Head and Neck Surgery at the University of Pennsylvania. Selection for the study was limited to immune competent CRS patients over age 18 years with endoscopic evidence of sinonasal inflammation (mucosal edema or polypoid degeneration) or overt mucopurulence. Exclusion criteria included patients with genetic disorders affecting mucociliary clearance, history of chemotherapy, immune deficiencies, or rhinologic granulomatous disease. Basic demographic data (age, gender, and race) were also collected based on self-reported identification in the patient chart. Information regarding medical history and use of nasal therapeutics was also collected. This included number of previous functional endoscopic sinus surgeries (# FESS), antibiotic use in the preceding month prior to presentation, topical and systemic steroid use, nasal irrigation use, hypertension (HTN), diabetes (DM), smoking status, 22-item Sino-Nasal Outcome Test (SNOT-22) score and allergic rhinitis. Smoking status was divided into smoke-naive (never smoked) and smoke-exposed groups (active or former smokers). A culture swab was obtained from the sinonasal location of purulence or inflammation from each patient at the time of the visit and used to carry out the in vitro biofilm assay. There was no standardization between groups or anatomic location of sampling.
Biofilm detection assay
Each nasal swab was evaluated with the modified Calgary Biofilm Detection Assay described by Goldstein-Daruech et al.30 and Moskowitz et al.31 Bacterial samples were individually placed in 100% Luria-Bertani (LB) broth (Fischer Scientific, Hanover Park, IL) and grown for 24 hours at 37°C in a shaker at 200 rpm to reach log phase growth. Optical density (OD595) of the bacterial inoculates was measured in a spectrophotometer and titrated to 0.1 OD595 in 100% LB broth. The resultant bacterial inoculate was further diluted to 1:100 in 100% LB broth. After standardization, each sample was transferred to a well of a flat-bottom 96-well microtiter plate (Costar-Corning, Sigma Aldrich Corp., St. Louis, MO) in quadruplicate. For each 96-well plate, positive and negative control bacterial strains (see the next paragraph) were also plated in quadruplicate. Bacterial biofilms were then allowed to form in an incubator for 24 hours at 37°C.
Pseudomonas aeruginosa (PAO1) served as the positive control for the experiment as a known biofilm-producing strain. Two negative controls were used: surface-attachment deficient strain 31 (sad-31), a pseudomonas mutant with defective type IV pili gene that cannot auto-aggregate,32 and sad-36, another mutant with defective flagella K gene that cannot produce functional flagella and attach to its surroundings.32 Each bacterial control strain was incubated, standardized, and diluted in the same manner as patient samples in other studies.24,30 Mutant strains were obtained from George O'Toole (Dartmouth College).
After incubation, the 96-well plates were decanted. Each well was washed (3×) with sterile phosphate buffered saline (50% PBS; pH 7.2) at room temperature. Each washing step was followed by flicking of the plate to completely empty the wells. Care was taken to prevent contamination of the surrounding environment and formation of aerosol. The wells were allowed to drain fully by inverting the plate until dry. Next, the cells were heat fixed by incubating at 60°C for 60 minutes. Each well was then filled with 10% crystal violet (Harleco, Gibbstown, NJ) and incubated for 30 minutes at room temperature. The plates were decanted and washed again with sterile water (3×); each time decanted by flicking the plate. The adherent material was resolubilized with 95% ethanol applied to each well at room temperature for 30 minutes. The ethanol elution was pipetted up and down thoroughly in order to dissolve the crystal violet completely. The resultant ethanol elution was measured at OD595 using the BioRad 680 plate reader (Hercules, CA). Each experiment was repeated at least twice.
PTC taste test
Subjects tasted and rated two, 5-mL samples of each of 6 solutions: distilled water (Wat), 0.35 M sucrose (Suc), 0.25 M sodium chloride (NaCl), 180 μM phenylthiocarbamide (PTC), 56 μM quinine-HCl (Qui), and 1.8 μM denatonium benzoate (DB).33 Concentrations were selected to be of moderate intensity and detectable to most people, as suggested by previous research or, for DB, by pilot testing.33, 34 The water used to prepare the taste solutions was obtained from a Millipore purification system (Billerica, MA). Taste compounds were obtained from Sigma Life Science (St. Louis, MO). Solutions were prepared in advance at the Monell Chemical Senses Center in Philadelphia, PA, dispensed into glass vials, tightly capped, packaged into small boxes for convenience, and taken to the testing location. The samples were presented twice in a fixed order: Wat, Qui, NaCl, PTC, Suc, DB, NaCl, DB, Wat, Qui, Suc, and PTC. Subjects were provided with a 25.3–fluid oz. bottle of 365™ Spring Water, commercially available from Whole Foods Market, a large plastic cup for expectoration, and a paper ballot on which they recorded their responses. Subjects were instructed to rinse their mouth once with water before and after tasting each solution and to rate the intensity of the stimulus on a validated, 13-point category scale with equidistant verbal descriptors: “Extremely Intense” (#12), “Very intense” (#9), “Moderately intense” (#6), “Slightly intense” (#3), and “No intensity at all” (#0). This scale has been used extensively in clinical studies at the Monell Chemical Senses Center.20 Subjects were also asked to identify the quality of each sample: salty, sour, bitter, sweet, or no flavor. The ratings of bitterness were averaged for vials containing PTC, yielding a measure of subjective taste intensity.
TAS2R38 genotype
In a subset of patients (36) participating in this study, sinonasal tissue or saliva was collected as part of a larger study and genomic DNA extracted and TAS2R38 genotype determined as described.14,35–37
Statistical analysis
Bacterial cultures from patients and positive and negative controls were tested in quadruplicate and concurrently in 96-well plates. The repeated OD595 readings from the sample wells were averaged. Patient derived bacteria were compared to positive (PAO1) and negative (sad-31, sad-36) controls that have been used in previous studies.24, 30 Samples demonstrating growth greater than PAO1 were designated as severe biofilm formers. Samples with biofilm growth between PAO1 and sad-31/sad-36 were designated as moderate biofilm formers, while samples with biofilm growth less than sad-31 identified as non-biofilm formers. Biofilm cultures identified as moderate or severe biofilm formers were categorized together as biofilm formers as described.24
All analyses were carried out with STATA 13 statistical software (StataCorp., College Station, TX). OD595 readings were determined to follow standard statistical distribution patterns and were characterized by a mean and standard deviation. The level of significance of the correlation between patient PTC mean phenotype (scale rating, 0 to 12) and mean biofilm forming potential (OD595) was calculated with linear regression. Pearson's chi square was used to analyze patient demographic and clinical data variability (asthma, nasal polyposis, diabetes, allergic rhinitis, tobacco exposure, previous FESS, oral antibiotics, systemic antibiotics, oral steroids, and nasal steroid use) to assess relationships to biofilm formation (non-formers and formers). History of medical therapy in the last month (antibiotic, oral steroid, and nasal steroid use was reduced to binary evaluation (yes or no). Student t test was used to determine the relationship between SNOT-22 scores and biofilm growth potential. For statistical analysis α was set at 5% and a p value < 0.05 was considered statistically significant.
Results
Patient demographics
This study is comprised of a total of 59 CRS patients who met study inclusion criteria. Thirty-two (54.2%) patients were male and 27 (45.8%) were female, with average ages of 52.2 (male) and 52.7 (female), p = 0.27 (see Table 1). Patient ethnicity breakdown was as follows: 93.2% white, 3.3% African American, and 1.7% Black Hispanic. Patient age, gender, and race showed no associations with biofilm formation (non-formers, formers) with the following respective p values: 0.59, 0.2, and 0.29 (see Table 1).
Table 1. Patient characteristics and biofilm formation.
| Characteristic | Non-biofilm former | Biofilm former | p |
|---|---|---|---|
| Patients (n) | 17 | 42 | |
| Age (years), mean | 50 | 53.4 | 0.59 |
| Age (years): mean 52.4; maximum 85; minimum 18 | |||
| Gender | |||
| Female | 10 | 17 | 0.20 |
| Male | 7 | 25 | |
| SNOT-22 score, mean | 36.9 | 30.1 | 0.26 |
| Race | 0.29 | ||
| African American | 0 | 2 | |
| Black Hispanic | 1 | 0 | |
| White | 16 | 39 | |
| East Indian | 0 | 1 | |
| Number of FESS | |||
| 0 | 5 | 7 | 0.53 |
| 1 | 6 | 13 | |
| 2 | 4 | 10 | |
| 3 | 1 | 7 | |
| 4 | 1 | 1 | |
| 5+ | 0 | 4 | |
| Steroid use | |||
| No | 11 | 22 | 0.39 |
| Yes | 6 | 20 | |
| Antibiotic use | |||
| No | 14 | 37 | 0.56 |
| Yes | 3 | 5 | |
| Nasal irrigation use | |||
| No | 5 | 15 | 0.64 |
| Yes | 12 | 27 | |
| HTN | |||
| No | 12 | 33 | 0.51 |
| Yes | 5 | 9 | |
| Asthma | |||
| No | 12 | 32 | 0.65 |
| Yes | 5 | 10 | |
| GERD | |||
| No | 12 | 31 | 0.80 |
| Yes | 5 | 11 | |
| DM | |||
| No | 16 | 40 | 0.86 |
| Yes | 1 | 2 | |
| Smoking status | |||
| Naive | 5 | 28 | 0.009 |
| Exposed | 12 | 14 | |
| Allergies | |||
| No | 5 | 20 | 0.20 |
| Yes | 12 | 22 | |
| Polyps | |||
| No | 9 | 32 | 0.079 |
| Yes | 8 | 10 |
DM = diabetes mellitus; FESS = functional endoscopic sinus surgery; GERD = gastroesophageal reflux disease; HTN = hypertension; SNOT-22 = 22-item Sino-Nasal Outcome Test.
Clinical data
Patients' self-reported histories of comorbidities demonstrated prevalence of asthma at 25.4%, allergic rhinitis at 57.6%, DM at 5.1%, HTN at 23.7%, gastroesophageal reflux disease (GERD) at 27.1%, and history of polyps at 30.1% in the group. Breakdown of these comorbidities in relationship to biofilm forming status (non-formers, formers) is shown in Table 1; no significant relationships were noted for the aforementioned comorbidities in relationship to biofilm deposition. CRS patients reported various medical and surgical interventions. Overall, 79.7% reported a history of at least 1 prior FESS procedure and 13.6% of patients reported antibiotic use within 1 month of presentation. Another 44.1% of patients used some form of oral or nasal steroids. Chi square analysis showed no relationship between biofilm forming status (non-formers, formers) on the following: number of FESS procedures (1 to 5+), p = 0.53; steroid use, p = 0.39 (see Table 1). There were 55.9% smoke-naive patients in the study; analysis of smoking history in relationship to biofilm formation demonstrated significance (p = 0.009), with more smoke-naive patients yielding culture swabs generating in vitro biofilms than smokers or smoke-exposed patients.
Antibiotic use did not reveal significance differences in biofilm forming status p = 0.56 (see Table 1).
Biofilm detection assay findings
The results of the biofilm formation assay for CRS patient swab samples in comparison to control bacterial strains are shown (Fig. 1). The assay determined control strain bacterial mass (OD595): PAO1 (3.26), sad-31 (0.34), and sad-36 (0.2). The biofilm assay showed that 42 of 59 (71.2%) CRS patients yielded cultures that were biofilm formers. The specific breakdown of severe biofilm formers with growth greater than PAO1 control (positive control) was 1 of 59 (1.7%) patient samples. The assay identified 41 of 59 (69.5%) patients as moderate biofilm formers with growth between PAO1 control and sad-31 (negative control). A further 7 of 59 (11.9%) patients were identified as negative biofilm formers with growth less than sad-31 and greater than sad-36. An additional 10 of 59 (16.9%) patients were also identified as negative biofilm formers with growth less than sad-36.
Figure 1.

In vitro biofilm assay from consecutive endoscopically guided sinonasal cultures. Patients evaluated in the outpatient clinic who were found to have sinonasal mucopurulence or mucosal inflammation were cultured in duplicate. While 1 sample was analyzed by the hospital microbiology laboratory for culture and sensitivity, the duplicate swab was processed for detection of biofilm-forming capacity. Concomitant positive and negative controls were performed with the pseudomonal species PAO-1 (wt), Sad-31, and Sad-36. Severe biofilm formation is categorized by OD595 greater than PAO-1 (wt), moderate OD595 less than wt but greater than Sad-31. OD595 = optical density measured at 595 nm; PAO1 = Pseudomonas aeruginosa; wt = wild-type.
Distribution of PTC taste sensitivity by TAS2R38 genotype
TAS2R38 genotype data was available for 36 of the 59 patients. As expected, PTC taste sensitivity significantly segregated by the genotype (p < 0.0001) (Fig. 2), as has been reported.38
Figure 2.

PTC taste sensitivity correlates with TAS2R38 genotype. ANOVA testing on taste test scores yielded significant variation in PTC taste sensitivity among genotype groups, F(3, 36) = 74.59, p < 0.0001. A post hoc Tukey test yielded that PAV/PAV group and the AVI/AVI group differed significantly (mean difference = 9.55; 95% CI, 7.35 to 11.74) at p < 0.001; the PAV/AVI group and PAV/PAV group differed significantly (mean difference = 3.68; 95% CI, 1.62 to 5.73) at p < 0.001; and the PAV/AVI group and AVI/AVI group differed significantly (mean difference = 5.87; 95% CI, 4.20 to 7.54) at p < 0.001. ANOVA = analysis of variance; AVI = alanine-valine-isoleucine; CI = confidence interval; PCT = phenylthiocarbamide; PAV = proline-alanine-valine.
Correlation of in vitro biofilm formation with PTC taste sensitivity
Linear regression revealed a significant correlation between patients' overall biofilm mass measured by OD595 and their PTC ratings, p = 0.019, adjusted r2 = 0.093 (Fig. 3A). We further divided our data based on the clinical evidence of nasal polyps at the time of swab collection. In nonpolyp patients (n = 41), a significant correlation was observed (p = 0.0026, r2 = 0.214) (Fig. 3B), which was not demonstrated in the polyp patients (n = 18) (p = 0.971, r2 = 0.0001) (Fig. 3C). When PTC taste sensitivity of nonpolyp patients with available genotypes was organized into equivalent tasting ranges (taste score 1 to 6, taste score 7 to 12), functionally distinguishing “non-tasters” (n = 13) and “tasters” (n = 12), there was a significant difference in observed overall biofilm mass (p = 0.012) (Fig. 4A), and this difference became even more pronounced when analyzing all 41 nonpolyp patients, including those without genotype data (p < 0.001). When the same group of nonpolyp patients was subdivided by genotype (n = 25), significance was not reached (Fig. 4B). NaCl and quinine, the other 2 tastants tested in this study, showed no correlation with overall biofilm mass, even when patients were subdivided based on endoscopic evidence of nasal polyps (Supporting Figs. 1 and 2).
Figure 3.

In vitro biofilm formation correlates to PTC taste sensitivity in CRS patients. (A) Biofilm formation was assessed (OD595) on 59 clinical isolates from CRS patients and compared to their PTC taste sensitivity. A significant correlation was observed (p = 0.019, r2 = 0.093). (B, C) Subjects were stratified based on the endoscopic presence of nasal polyps. In non-polyp patients (B) a significant correlation was observed (p = 0.0026, r2 = 0.214) while in the polyp patients (C) no significant correlation was observed (p = 0.971, r2 = 0.001). CRS = chronic rhinosinusitis; OD595 = optical density measured at 595 nm; PCT = phenylthiocarbamide.
Figure 4.
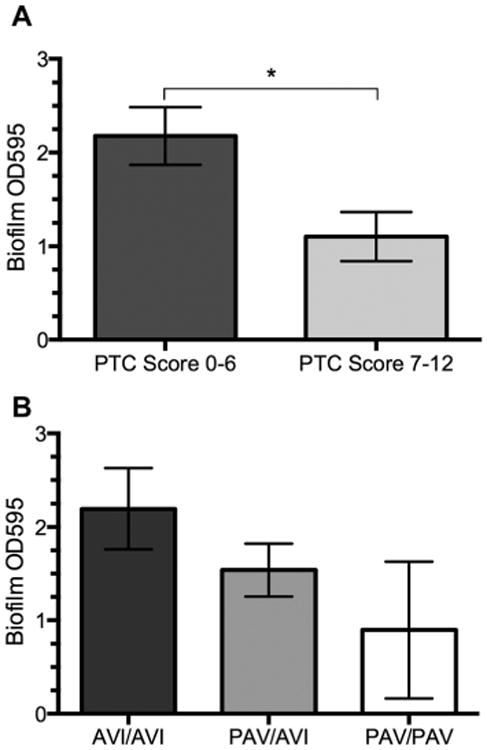
PTC taste score predicts in vitro biofilm formation in nonpolyp CRS patients. In vitro biofilm formation was assessed (OD595) and compared to TAS2R38 genotype and PTC score for nonpolyp CRS patients, with 25 of those patients having been genotyped. PTC score was broadly divided into 2 categories, score 0 to 6 (n = 13) and score 7 to 12 (n = 12). (A) Patients with a PTC score of 7 to 12 had a significantly lower level of in vitro biofilm formation (p = 0.012, 2-tailed t test). (B) Comparison of TAS2R38 genotype with in vitro biofilm formation did not reach significance (1-way ANOVA, p = 0.204). ANOVA = analysis of variance; CRS = chronic rhinosinusitis; OD595 = optical density measured at 595 nm; PCT = phenylthiocarbamide.
Discussion
The symptoms of CRS impact many individuals, resulting in physician office visits, surgeries, and repeated exposure to antibiotics. This disease is not caused by 1 initiating process, but rather is thought to be multifactorial.7, 39, 40 Patient innate immunity is partially determined by genetics. In this study, the interplay of host genetics, innate immunity, and bacterial biofilm formation potential are examined to better understand their relationships in CRS.
CRS has a tendency to run in families, leading to the idea that there is a strong genetic component.41–43 There is also a presumed microbial component of the disease.44–46 Investigation of the interaction between host genetics and the microbial contribution to CRS has become especially relevant in light of increasing bacterial resistance to antibiotics and the recalcitrant nature of CRS, which often results in multiple rounds of antibiotics without durable symptomatic relief. As the most common diagnosis to lead to the prescription of outpatient antibiotics in adults, rhinosinusitis accounts for approximately 15% to 21% of all antibiotic prescriptions.47 The interaction of host genetic makeup dictated by a Toll-like receptor polymorphism and infection susceptibility has been demonstrated in many areas of medicine, including predisposition to urinary tract infections, Helicobacter pylori infection, and Clostridium difficile infection.48–50 A bacterial sensing receptor that results in an immune response has been identified in each of these disease processes.48–50 The receptor's function is dictated by specific genetic polymorphisms in those sentinel receptors. TAS2R38 taste receptor gene polymorphisms resulting in decreased T2R38 function may make patients more susceptible to sinonasal infection through mechanisms much like those in the diseases mentioned earlier in this paragraph. In addition, an understanding of the genetics of patient defense mechanisms may eventually lead to the development of novel, individually targeted therapeutics and diagnostics, thus curtailing the rising trend of antibiotic resistant microbes.
The results of this study suggest several findings. First, CRS patients without nasal polyps who have increased T2R38 activity may have less recoverable bacteria capable of in vitro biofilm formation. This further corroborates the mounting evidence that the T2R38 receptor plays a sentinel role in innate defense mechanisms against microbes in the sinonasal cavity.9–12, 14, 51 Second, it continues to support the notion that CRS with and without polyps are 2 distinct disease entities with different environmental and host interactions. Third, we can use taste perception of PTC as a proxy for functionality of the T2R38 receptor in the sinonasal cavity. This last point is critical for future investigation. Although genetic testing has become cost affordable in many situations, for TAS2R38, it does not tell the whole picture as patients with the same genotype can have vastly different expression of the PAV allele and thus different T2R38 functionality, and a test identifying the level of expression/functionality is likely more informative than just identifying the genotype itself. In our study, the low number of PAV/PAV nonpolyp patients (n = 3) made PTC sensitivity a more powerful assessment of biofilm formation potential than genotype itself. This is additional evidence that a PTC taste test may be even more accurate than genotyping in assessing predilection to pathology.
We previously described in vitro studies showing that the T2R38 receptor, expressed on sinonasal ciliated cells, is activated by acyl-homoserine lactones (AHLs), gram-negative quorum-sensing molecules critical for the biofilm life cycle.14 Once activated by these AHLs, the T2R38 receptor generates a calcium-dependent increase in nitric oxide (NO) production, which subsequently increases mucociliary clearance (MCC) and diffuses into the overlying mucus layer where it also has bactericidal effects.14 This effect may contribute to the lower levels of biofilm-forming bacteria recovered from CRS patients who have increased T2R38 activity (high mean PTC taste test score). Our prior work has demonstrated sinonasal cell cultures with PAV/PAV genotypes have increased NO production and mucociliary clearance compared to cultures with PAV/AVI and AVI/AVI genotypes in response to AHLs. It therefore follows that the more PAV allele that is expressed, the more robust the “taster” phenotype.19 Likewise, as less PAV and more AVI is expressed, the “non-taster” phenotype becomes evident. Clinically this has been demonstrated in skewed population distribution data demonstrating that AVI/AVI patients make up a higher than expected portion of the population of CRS patients failing medical therapy and going on to sinus surgery, and conversely PAV/PAV patients make up a lower than expected portion of patients failing medical therapy and undergoing sinus surgery.36, 37
Although our data demonstrate a direct inverse relationship between mean PTC score and in vitro biofilm mass in samples derived from CRS patients without polyps, this may not be reflective of in vivo biofilms, and even if it was, biofilms are just 1 component of recalcitrant chronic rhinosinusitis that must be viewed in context of other genetic and environmental influences yet to be understood. Furthermore, prior data suggests that planktonic bacteria recovered from CRS patients are not necessarily reflective of the bacterial species residing within the biofilm in vivo.52 However, our results support that higher mean PTC taste test ratings, and therefore increased T2R38 activity, may be associated with the inhibition of in vivo biofilm formation, providing an extra layer of protection in innate immunologic defense. Our study suggests that patients with greater PTC taste sensitivity (and thus increased activity of T2R38) have decreased recovery of biofilm-forming bacteria from the sinonasal cavity. Because there may be incomplete penetrance at the TAS2R38 gene, the PAV/AVI genotype is not always predictive of activity at the T2R38 receptor and perception of bitterness.19 This may in fact increase the predictive value of a taste test, as the phenotype is a continuum based on PAV allele expression compared to genotype, which is a fixed ternary value.53
Tobacco smoke impacts sinonasal epithelial function,54–57 and several epidemiologic studies have demonstrated that smokers have an increased prevalence of CRS.58,59 A previous study reported increased biofilm mass in cultures from smokers when the cultures were repetitively exposed to exogenous tobacco smoke, compared to cultures taken from nonsmokers.30 The same study showed a reversal of this tobacco effect on in vitro biofilm mass in the absence of exogenous smoke.30 Further experiments investigating this phenomenon in Staphylococcus aureus demonstrated that the oxidative stress generated by tobacco smoke exposure induced the pilF attachment gene.60 In the current study, we evince a greater proportion of biofilm formers in the cultures from smoke-naive patients. However, we did not expose bacterial samples to smoke and thus the dearth of biofilm-forming cultures from patients with a smoking history may be reflective of the prior observation; ie, in the absence of tobacco smoke, microbes harvested from smokers do not form biofilms in vitro.30
There are several limitations to this study that require further evaluation. It must be stressed that this study evaluated in vitro biofilm growth. Although the assay used here is a semiquantitative measurement of biofilm formation, it has been validated in multiple studies.24, 30, 31 Other more direct biofilm growth assays available are confocal laser scanning microscopy or in situ hybridization.25,61, 62 These methods are limited in use due to their sampling bias and labor-intensive requirements. It must also be noted that the positive and negative biofilm controls were only of P. aeruginosa. It is possible that our in vitro growth conditions are optimized for P. aeruginosa biofilm formation and not for other common microbes; however, additional work in our laboratory has demonstrated that under these conditions we are able to form robust biofilms from, but not limited to, S. aureus, Streptococcus pneumoniae, Klebsiella pneumoniae, and Serratia marcescens.30
Conclusion
The interplay between genetic variability in host innate immune response and infection has recently been demonstrated in several disease processes.48–50 Bacterial biofilms in particular have been demonstrated in many chronic diseases; one of which is CRS. This research is the first to demonstrate a relationship between T2R38 bitter taste receptor activity (assessed by PTC taste sensitivity), a genetic trait, and in vitro biofilm formation in clinical isolates recovered from CRS patients. The differences in in vitro biofilm deposition may be explained by patients' genetic differential T2R38 receptor activity that activates the sinonasal innate immune response, with different degrees of defensive activity being permissive to different forms of microbes; ie, low T2R38 activity is more permissive to robust biofilm-forming microbes. Of even greater clinical significance, assessing the T2R38 phenotype (ie, the amount of PAV being expressed) determined by a PTC taste test appears to be more informative than genotyping T2R38, which does not quantify the degree of PAV expression. This finding supports a rationale for the development of therapeutics targeting the T2R mediated innate immune response pathway, as well as for potentially using the PTC taste test as a proxy for sinonasal innate immune function.
Supplementary Material
Acknowledgments
Funding sources for the study: RLG Foundation, Inc. (to N.A.C.); USPHS (R01DC013588 to N.A.C.).
Footnotes
Additional Supporting Information may be found in the online version of this article.
Potential conflict of interest: N.A.C. has a patent pending “Therapy and Diagnostics for Respiratory Infection.”
Presented at the Annual ARS Meeting, on September 25–26, 2015, in Dallas, TX.
References
- 1.Bhattacharyya N, Grebner J, Martinson NG. Recurrent acute rhinosinusitis: epidemiology and health care cost burden. Otolaryngol Head Neck Surg. 2012;146:307–312. doi: 10.1177/0194599811426089. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya N. Incremental healthcare utilization and expenditures for allergic rhinitis in the United States. Laryngoscope. 2011;121:1830–1833. doi: 10.1002/lary.22034. [DOI] [PubMed] [Google Scholar]
- 3.Khalid AN, Quraishi SA, Kennedy DW. Long-term quality of life measures after functional endoscopic sinus surgery. Am J Rhinol. 2004;18:131–136. [PubMed] [Google Scholar]
- 4.Luk LJ, Steele TO, Mace JC, Soler ZM, Rudmik L, Smith TL. Health utility outcomes in patients undergoing medical management for chronic rhinosinusitis: a prospective multiinstitutional study. Int Forum Allergy Rhinol. 2015;5:1018–1027. doi: 10.1002/alr.21588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simopoulos E, Katotomichelakis M, Gouveris H, Tripsianis G, Livaditis M, Danielides V. Olfaction-associated quality of life in chronic rhinosinusitis: adaptation and validation of an olfaction-specific questionnaire. Laryngoscope. 2012;122:1450–1454. doi: 10.1002/lary.23349. [DOI] [PubMed] [Google Scholar]
- 6.Hsu J, Peters AT. Pathophysiology of chronic rhinosinusitis with nasal polyp. Am J Rhinol Allergy. 2011;25:285–290. doi: 10.2500/ajra.2011.25.3680. [DOI] [PubMed] [Google Scholar]
- 7.Kern RC, Conley DB, Walsh W, et al. Perspectives on the etiology of chronic rhinosinusitis: an immune barrier hypothesis. Am J Rhinol. 2008;22:549–559. doi: 10.2500/ajr.2008.22.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antunes MB, Gudis DA, Cohen NA. Epithelium, cilia, and mucus: their importance in chronic rhinosinusitis. Immunol Allergy Clin North Am. 2009;29:631–643. doi: 10.1016/j.iac.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Lee RJ, Cohen NA. The emerging role of the bitter taste receptor T2R38 in upper respiratory infection and chronic rhinosinusitis. Am J Rhinol Allergy. 2013;27:283–286. doi: 10.2500/ajra.2013.27.3911. [DOI] [PubMed] [Google Scholar]
- 10.Lee RJ, Cohen NA. Taste receptors in innate immunity. Cell Mol Life Sci. 2015;72:217–236. doi: 10.1007/s00018-014-1736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee RJ, Cohen NA. Role of the bitter taste receptor T2R38 in upper respiratory infection and chronic rhinosinusitis. Curr Opin Allergy Clin Immunol. 2015;15:14–20. doi: 10.1097/ACI.0000000000000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee RJ, Cohen NA. Bitter and sweet taste receptors in the respiratory epithelium in health and disease. J Mol Med. 2014;92:1235–1244. doi: 10.1007/s00109-014-1222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee RJ, Cohen NA. Sinonasal solitary chemosensory cells “taste” the upper respiratory environment to regulate innate immunity. Am J Rhinol Allergy. 2014;28:366–373. doi: 10.2500/ajra.2014.28.4077. [DOI] [PubMed] [Google Scholar]
- 14.Lee RJ, Xiong G, Kofonow JM, et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 2012;122:4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bufe B, Breslin PA, Kuhn C, et al. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat. 2005;26:199–204. doi: 10.1002/humu.20203. [DOI] [PubMed] [Google Scholar]
- 17.Kim UK, Drayna D. Genetics of individual differences in bitter taste perception: lessons from the PTC gene. Clin Genet. 2005;67:275–280. doi: 10.1111/j.1399-0004.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 18.Wiener A, Shudler M, Levit A, Niv MY. BitterDB: a database of bitter compounds. Nucleic Acids Res. 2012;40:D413–D419. doi: 10.1093/nar/gkr755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipchock SV, Mennella JA, Spielman AI, Reed DR. Human bitter perception correlates with bitter receptor messenger RNA expression in taste cells. Am J Clin Nutr. 2013;98:1136–1143. doi: 10.3945/ajcn.113.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowart BJ. Relationships between taste and smell across the adult life span. Ann N Y Acad Sci. 1989;561:39–55. doi: 10.1111/j.1749-6632.1989.tb20968.x. [DOI] [PubMed] [Google Scholar]
- 21.Bendouah Z, Barbeau J, Hamad WA, Desrosiers M. Biofilm formation by Staphylococcus aureus and Pseudomonas aeruginosa is associated with an unfavorable evolution after surgery for chronic sinusitis and nasal polyposis. Otolaryngol Head Neck Surg. 2006;134:991–996. doi: 10.1016/j.otohns.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Cryer J, Schipor I, Perloff JR, Palmer JN. Evidence of bacterial biofilms in human chronic sinusitis. ORL J Otorhinolaryngol Relat Spec. 2004;66:155–158. doi: 10.1159/000079994. [DOI] [PubMed] [Google Scholar]
- 23.Perloff JR, Palmer JN. Evidence of bacterial biofilms on frontal recess stents in patients with chronic rhinosinusitis. Am J Rhinol. 2004;18:377–380. [PubMed] [Google Scholar]
- 24.Prince AA, Steiger JD, Khalid AN, et al. Prevalence of biofilm-forming bacteria in chronic rhinosinusitis. Am J Rhinol. 2008;22:239–245. doi: 10.2500/ajr.2008.22.3180. [DOI] [PubMed] [Google Scholar]
- 25.Psaltis AJ, Ha KR, Beule AG, Tan LW, Wormald PJ. Confocal scanning laser microscopy evidence of biofilms in patients with chronic rhinosinusitis. Laryngoscope. 2007;117:1302–1306. doi: 10.1097/MLG.0b013e31806009b0. [DOI] [PubMed] [Google Scholar]
- 26.Psaltis AJ, Weitzel EK, Ha KR, Wormald PJ. The effect of bacterial biofilms on post-sinus surgical outcomes. Am J Rhinol. 2008;22:1–6. doi: 10.2500/ajr.2008.22.3119. [DOI] [PubMed] [Google Scholar]
- 27.Richards JJ, Melander C. Controlling bacterial biofilms. Chembiochem. 2009;10:2287–2294. doi: 10.1002/cbic.200900317. [DOI] [PubMed] [Google Scholar]
- 28.Cohen M, Kofonow J, Nayak JV, et al. Biofilms in chronic rhinosinusitis: a review. Am J Rhinol Allergy. 2009;23:255–260. doi: 10.2500/ajra.2009.23.3319. [DOI] [PubMed] [Google Scholar]
- 29.Rosenfeld RM, Andes D, Bhattacharyya N, et al. Clinical practice guideline: adult sinusitis. Otolaryngol Head Neck Surg. 2007;137:S1–S31. doi: 10.1016/j.otohns.2007.06.726. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein-Daruech N, Cope EK, Zhao KQ, et al. Tobacco smoke mediated induction of sinonasal microbial biofilms. PLoS One. 2011;6:e15700. doi: 10.1371/journal.pone.0015700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moskowitz SM, Foster JM, Emerson J, Burns JL. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J Clin Microbiol. 2004;42:1915–1922. doi: 10.1128/JCM.42.5.1915-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 33.Knaapila A, Hwang LD, Lysenko A, et al. Analysis of heritability and candidate gene associations of chemosensory traits in human twins. Chem Senses. 2012;37:869–881. doi: 10.1093/chemse/bjs070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen JL, Reed DR, Wright MJ, Martin NG, Breslin PA. Heritability and genetic covariation of sensitivity to PROP, SOA, quinine HCl, and caffeine. Chem Senses. 2006;31:403–413. doi: 10.1093/chemse/bjj044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adappa ND, Farquhar D, Palmer JN, et al. TAS2R38 genotype predicts surgical outcome in nonpolypoid chronic rhinosinusitis. Int Forum Allergy Rhinol. 2016;6:25–33. doi: 10.1002/alr.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adappa ND, Howland TJ, Palmer JN, et al. Genetics of the taste receptor T2R38 correlates with chronic rhinosinusitis necessitating surgical intervention. Int Forum Allergy Rhinol. 2013;3:84–87. doi: 10.1002/alr.21140. [DOI] [PubMed] [Google Scholar]
- 37.Adappa ND, Zhang Z, Palmer JN, et al. The bitter taste receptor T2R38 is an independent risk factor for chronic rhinosinusitis requiring sinus surgery. Int Forum Allergy Rhinol. 2014;4:3–7. doi: 10.1002/alr.21253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 39.Fokkens WJ, Lund VJ, Mullol J, et al. EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists. Rhinology. 2012;50:1–12. doi: 10.4193/Rhino12.000. [DOI] [PubMed] [Google Scholar]
- 40.Peters AT, Spector S, Hsu J, et al. Diagnosis and management of rhinosinusitis: a practice parameter update. Ann Allergy Asthma Immunol. 2014;113:347–385. doi: 10.1016/j.anai.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 41.Greisner WA, 3rd, Settipane GA. Hereditary factor for nasal polyps. Allergy Asthma Proc. 1996;17:283–286. doi: 10.2500/108854196778662192. [DOI] [PubMed] [Google Scholar]
- 42.Lockey RF, Rucknagel DL, Vanselow NA. Familial occurrence of asthma, nasal polyps and aspirin intolerance. Ann Intern Med. 1973;78:57–63. doi: 10.7326/0003-4819-78-1-57. [DOI] [PubMed] [Google Scholar]
- 43.Cohen NA, Widelitz JS, Chiu AG, Palmer JN, Kennedy DW. Familial aggregation of sinonasal polyps correlates with severity of disease. Otolaryngol Head Neck Surg. 2006;134:601–604. doi: 10.1016/j.otohns.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 44.Abreu NA, Nagalingam NA, Song Y, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4:151ra24. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramakrishnan VR, Hauser LJ, Feazel LM, Ir D, Robertson CE, Frank DN. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J Allergy Clin Immunol. 2015;136:334–342.e1. doi: 10.1016/j.jaci.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Biswas K, Hoggard M, Jain R, Taylor MW, Douglas RG. The nasal microbiota in health and disease: variation within and between subjects. Front Microbiol. 2015;9:134. doi: 10.3389/fmicb.2018.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith SS, Evans CT, Tan BK, Chandra RK, Smith SB, Kern RC. National burden of antibiotic use for adult rhinosinusitis. J Allergy Clin Immunol. 2013;132:1230–1232. doi: 10.1016/j.jaci.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawn TR, Scholes D, Li SS, et al. Toll-like receptor polymorphisms and susceptibility to urinary tract infections in adult women. PLoS One. 2009;4:e5990. doi: 10.1371/journal.pone.0005990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li P, He CY, Xu Q, Sun LP, Ha MW, Yuan Y. Effect of the -2081G/A polymorphism of the TLR4 gene and its interaction with Helicobacter pylori infection on the risk of gastric cancer in Chinese individuals. Genet Test Mol Biomarkers. 2014;18:610–615. doi: 10.1089/gtmb.2014.0047. [DOI] [PubMed] [Google Scholar]
- 50.Hung YP, Lin HJ, Wu TC, et al. Risk factors of fecal toxigenic or non-toxigenic Clostridium difficile colonization: impact of Toll-like receptor polymorphisms and prior antibiotic exposure. PLoS One. 2013;8:e69577. doi: 10.1371/journal.pone.0069577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee RJ, Cohen NA. Bitter taste bodyguards. Sci Am. 2016;314:38–43. doi: 10.1038/scientificamerican0216-38. [DOI] [PubMed] [Google Scholar]
- 52.Sanderson AR, Leid JG, Hunsaker D. Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope. 2006;116:1121–1126. doi: 10.1097/01.mlg.0000221954.05467.54. [DOI] [PubMed] [Google Scholar]
- 53.Desai H, Smutzer G, Coldwell SE, Griffith JW. Validation of edible taste strips for identifying PROP taste recognition thresholds. Laryngoscope. 2011;121:1177–1183. doi: 10.1002/lary.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen NA, Zhang S, Sharp DB, et al. Cigarette smoke condensate inhibits transepithelial chloride transport and ciliary beat frequency. Laryngoscope. 2009;119:2269–2274. doi: 10.1002/lary.20223. [DOI] [PubMed] [Google Scholar]
- 55.Savitski AN, Mesaros C, Blair IA, Cohen NA, Kreindler JL. Secondhand smoke inhibits both Cl− and K+ conductances in normal human bronchial epithelial cells. Respir Res. 2009;10:120. doi: 10.1186/1465-9921-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virgin FW, Azbell C, Schuster D, et al. Exposure to cigarette smoke condensate reduces calcium activated chloride channel transport in primary sinonasal epithelial cultures. Laryngoscope. 2010;120:1465–1469. doi: 10.1002/lary.20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexander NS, Blount A, Zhang S, et al. Cystic fibrosis transmembrane conductance regulator modulation by the tobacco smoke toxin acrolein. Laryngoscope. 2012;122:1193–1197. doi: 10.1002/lary.23278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Dales R, Lin M. The epidemiology of chronic rhinosinusitis in Canadians. Laryngoscope. 2003;113:1199–1205. doi: 10.1097/00005537-200307000-00016. [DOI] [PubMed] [Google Scholar]
- 59.Lieu JE, Feinstein AR. Confirmations and surprises in the association of tobacco use with sinusitis. Arch Otolaryngol Head Neck Surg. 2000;126:940–946. doi: 10.1001/archotol.126.8.940. [DOI] [PubMed] [Google Scholar]
- 60.Antunes MB, Chi JJ, Liu Z, et al. Molecular basis of tobacco-induced bacterial biofilms: an in vitro study. Otolaryngol Head Neck Surg. 2012;147:876–884. doi: 10.1177/0194599812447263. [DOI] [PubMed] [Google Scholar]
- 61.Healy DY, Leid JG, Sanderson AR, Hunsaker DH. Biofilms with fungi in chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2008;138:641–647. doi: 10.1016/j.otohns.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Karygianni L, Hellwig E, Al-Ahmad A. Multiplex fluorescence in situ hybridization (M-FISH) and confocal laser scanning microscopy (CLSM) to analyze multispecies oral biofilms. Methods Mol Biol. 2014;1147:65–72. doi: 10.1007/978-1-4939-0467-9_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


